SUMMARY
SIX1 interacts with EYA to form a bipartite transcription factor essential for development. Loss of function of this complex causes branchio-oto-renal syndrome (BOR), while re-expression of SIX1 or EYA promotes metastasis. Here we describe the 2.0 Å structure of SIX1 bound to EYA2, which suggests a novel DNA binding mechanism for SIX1 and provides a rationale for the effect of BOR syndrome mutations. The structure also reveals that SIX1 uses predominantly a single helix to interact with EYA. Substitution of a single amino acid in this helix is sufficient to disrupt the SIX1–EYA interaction, SIX1-mediated epithelial-mesenchymal transition and metastasis in mouse models. Given that SIX1 and EYA are co-overexpressed in many tumor types, our data indicate that targeting the SIX1–EYA complex may be a potent approach to inhibit tumor progression in multiple cancer types.
SIX1 belongs to the mammalian Six family of homeobox genes which are homologues of the Drosophila sine oculis (so) gene. These genes encode transcription factors that play vital roles in proliferation and survival of progenitor cells during organogenesis1. The Six family consists of 6 members (SIX1-6) that share a homologous Six-type DNA binding homeodomain (HD) and a conserved but novel Six-domain (SD), which is involved in protein-protein interactions2.
Because SIX1 is a transcription factor that lacks intrinsic activation domains, it requires the EYA family of co-factors to mediate transcriptional activation. The four EYA proteins (EYA1-4) are mammalian homologues of the Drosophilaeyes absent (eya) gene, and are characterized by a highly conserved ~270 amino acid C-terminal Eya domain (ED)3, which interacts with the SD of SIX12. The ED contains signature motifs of the haloacid dehalogenase (HAD) hydrolases, and has Mg2+-dependent protein tyrosine phosphatase activity4–6. The crystal structure of the EYA2ED revealed that it is comprised of the catalytic sub-domain and a helix bundle motif (HBM) that is hypothesized to play a role in protein-protein interactions7. Like SIX1, EYA proteins are critical for the development of multiple organs in part by promoting proliferation and survival of progenitor cell populations4. Loss of function of SIX1 and EYA is implicated in multiple developmental disorders8,9. Mutations in either SIX1 or EYA1 cause branchio-oto-renal (BOR) syndrome, an autosomal dominant developmental disorder characterized by hearing loss, branchial fistulae and renal anomalies8. Mutations in EYA4 are also the cause of sensorineural hearing loss within the DFNA10 locus10–12. Additionally, mutations in EYA4 have been shown to cause cardiomyopathy12 and SIX1 and EYA have recently been implicated in cardiac hypertrophy13.
SIX1 is down-regulated after organ development is complete; thus its expression is low or undetectable in most normal adult tissues14. However, SIX1 is re-expressed in a number of cancers and its overexpression strongly correlates with disease progression in many tumor types15–21. Our laboratory has shown that SIX1 overexpression in the mouse mammary gland leads to highly aggressive mammary tumors that display oncogenic EMT and stem cell phenotypes22. Additionally, we have shown that SIX1 can induce EMT and cancer stem cell (CSC) phenotypes as well as metastasis through upregulation of the TGF-β signaling pathway16,23. Finally, we have recently demonstrated that SIX1 affects metastasis via additional mechanisms including upregulation of VEGF-C and induction of lymphangiogenesis24. These observations suggest that SIX1 is a global regulator of tumor progression and that disruption of SIX1 function would be therapeutically relevant in many different cancers. Indeed, knockdown of SIX1 in breast24 and hepatocellular carcinoma25, as well as in rhabdomyosarcoma15, leads to a dramatic decrease in tumor size and metastasis in animal models. Since it is traditionally difficult to target transcription factor–DNA interactions26 we set out to investigate if inhibiting the transcriptional complex formed by SIX1 and its EYA co-activator would serve as a viable approach to inhibit SIX1-mediated tumor progression.
Multiple studies imply that SIX1 and EYA act together in cancer. Overexpression of both SIX1 and EYA is observed in Wilms’ Tumor27, acute leukemia28 and malignant peripheral nerve sheath tumors29. SIX1 and EYA2 have also both been independently implicated in ovarian cancer21,30. In breast tumors, high levels of SIX1 and EYA2 together (but neither gene alone) significantly correlate with reduced time to relapse and metastasis, and with decreased survival31. Furthermore, SIX1 and EYA have independently been shown to contribute to metastasis in breast cancer cells16,32, and EYA2 is required for many of the SIX1 induced pro-metastatic phenotypes in breast cancer cell lines31. However, their coordinated action in cancer has never been shown in vivo, and it remains controversial whether the direct physical interaction between SIX1 and EYA is required since EYA’s phosphatase activity is critical for breast cancer-associated metastasis and this function of EYA may be independent of SIX132. In spite of the clear importance of the SIX1–EYA complex in development and disease, the atomic structure of SIX1, the molecular details of the SIX1–EYA interaction, as well as the functional significance of their direct physical interaction in cancer have remained unexplored. To fill these significant voids, we performed structural and functional analyses of the human SIX1–EYA2 complex.
RESULTS
The SIX1–EYA2ED structure
To gain insight into the molecular details of the SIX1–EYA interaction and the atomic structure of SIX1, we determined the crystal structure of the human SIX1–EYA2ED complex. We employed the maltose binding protein (MBP) fusion/surface entropy reduction technique that uses MBPs with surface mutations to aid crystallization33. We fused MBP_B (E172A N173A mutant, which will be referred to as MBP, for simplicity)to the amino-terminus of a SIX1 construct that includes both the SD and HD (residues 1-189) but excludes the predicted unstructured carboxyl-terminus (Fig. 1a). We confirmed that the MBP–SIX1 protein was functionally active in a DNA binding assay (Supplementary Fig. 1).
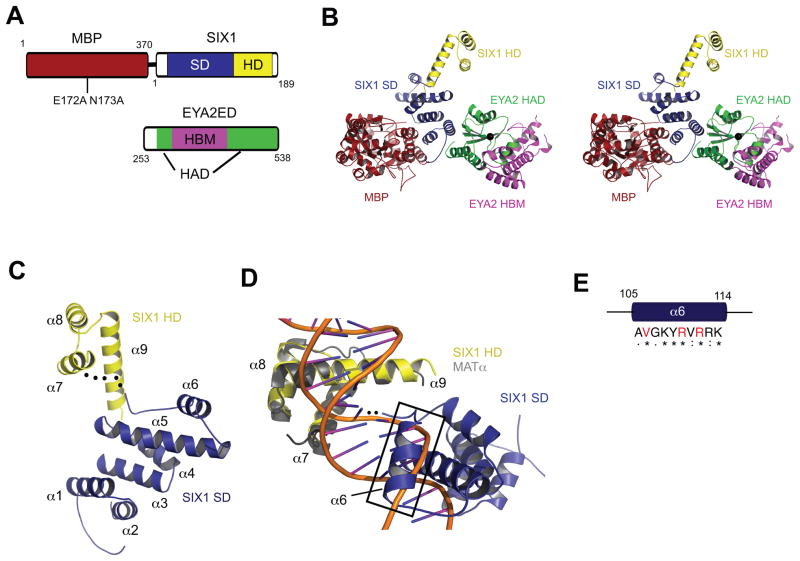
Figure 1. Structure of the MBP-SIX1–EYA2ED Complex Supports a Novel DNA binding Mechanism for SIX1.
(A) Domain organization of MBP, SIX1 and EYA2 used for crystallization. Numbers correspond to amino acid positions.
(B) Stereo view of the overall structure of the MBP–SIX1–EYA2ED complex. MBP is shown in red, SIX1SD in blue, SIX1HD in yellow, EYA2HAD in green and EYA2HBM in magenta. Mg2+ ion at the tyrosine phosphatase active site is shown as a black sphere.
(C) Cartoon representation of SIX1 (colored as in B), with HD and SD; the flexible linker between the SD and HD is shown as a black dotted line.
(D) Cartoon representation of SIX1 (colored as in B) superimposed on the MATα–DNA complex (MATα colored gray, DNA backbone colored orange) showing the close proximity of α6 of SIX1(black box)to the DNA major groove.
(E) Conservation of amino acids in the α6 helix of SIX1 among Six family members. Strictly conserved residues are indicated with “*”. Residues with strongly similar properties are indicated with “:”. Residues with weakly similar properties are indicated by “.”. Residues mutated in BOR are indicated in red.
We determined the structure of the MBP–SIX1 in complex with EYA2ED, and the final model was refined to 2.0 Å (Table 1). The MBP–SIX1–EYA2ED crystal contains one copy of each protein in the asymmetric unit (Fig. 1b). Three regions (40-45, 124-133 and 186-189) in SIX1 and two regions (253-267 and 356-371) in EYA2ED are disordered and not visible in the electron density map.
Table 1.
Data collection, phasing and refinement statistics
| Native | SeMet | |
|---|---|---|
| Data collection | ||
| Space group | P21212 | P21212 |
| Cell dimensions | ||
| a, b, c (Å) | 123.2, 150.2, 53.9 | 123.2, 150.2, 53.9 |
| α, β, γ (°) | 90 | 90 |
|
Peak
|
||
| Wavelength | 0.97912 | 0.97912 |
| Resolution (Å) | 36.0-2.0 | 47.6-2.0 |
| Rmerge (%) | 3.1 (55) | 5 (45) |
| I/σI | 66.0 (3.4) | 55.2 (4.1) |
| Completeness (%) | 100 | 100 |
| Redundancy | 9.4 (8.3) | 6.6 (6.7) |
| Refinement | ||
| Resolution (Å) | 36.0-2.0 | |
| No. reflections | 68,728 | |
| Rwork/Rfree | 18.4/22.4 | |
| No. atoms | ||
| Protein | 6,295 | |
| Ligand/ion | 24 | |
| Water | 812 | |
| B-factors | ||
| Protein | 30.0 | |
| Ligand/ion | 20.8 | |
| Water | 36.5 | |
| r.m.s deviations | ||
| Bond lengths (Å) | 0.007 | |
| Bond angles (°) | 1.045 | |
One crystal was used for each data set. Values in parentheses are for highest-resolution shell.
The structure of SIX1 is organized into two distinct domains (SD and HD) connected by a flexible linker (Fig. 1c). The novel SD consists of six alpha-helices, α1-α6 (Fig. 1c), and does not show structural homology with any other proteins in a DALI server search34. The HD of SIX1 contains 3alpha helices (α7-α9), forming the signature HD fold35. The N-terminal arm, as expected, is flexible in the absence of DNA and is therefore missing from the electron density (dotted lines in Fig. 1c).
A putative DNA binding mechanism for SIX1
Canonical HD proteins interact with DNA through a mechanism where the HD recognition helix (α3 in canonical HDs or α9 in SIX1) binds the major groove and the N-terminal arm contacts the minor groove35. The N-terminal arm typically contains basic residues at positions 2 and 3 that form hydrogen bonds with the DNA backbone, and an arginine at position 5 that makes extensive contacts with the nucleotide base and sugar. However, the N-terminal arm of SIX1 lacks basic amino acids at these three positions, with two of these residues replaced with acidic residues. This suggests that the N-terminal arm of SIX1 does not bind the minor groove in the canonical manner, if it binds this groove at all. Indeed, deletion of the N-terminal arm of SIX6 only slightly decreases its ability to bind DNA36, whereas in typical HD containing proteins, deletion of the N-terminal arm dramatically decreases binding affinity (~100 fold)36–38. Recent analyses have shown that regions outside of the HD are necessary to recognize DNA sequences bound by SIX1 within the cell39 and regions upstream of the N-terminal arm enhance DNA binding in SIX440 and SIX541. Finally, the HDs of SIX2 and SIX6 alone are unable to bind the MEF3 consensus sequence36. These observation sled to the hypothesis that the DNA binding activity of the Six family of homeoproteins extends beyond the HD.
Several lines of evidence from our structural and biochemical analyses support this hypothesis. First, when the SIX1 structure is superimposed on MATα–DNA (PDB ID 1K61, chain A), a known homeodomain–DNA complex structure, α6 of the SD (the first helix upstream of the HD) could easily interact with the DNA major groove with a minor positional adjustment (which is feasible since the SD and HD are connected by a flexible linker) (Fig. 1c,d). Second, this 10-residue helix (sequence AVGKYRVRRK) contains five positively charged residues and amino acids in this helix are highly conserved in the Six family (Fig. 1e and Supplementary Fig. 2). Third, three SIX1 residues that are mutated in BOR syndrome (Val106, Arg110, Arg112) that we previously demonstrated to abrogate DNA binding42, reside within this helix (Fig. 1e and Supplementary Fig. 2). Taken together, these observations suggest a novel DNA binding mechanism for Six family proteins that involves both the HD and SD. Formal proof of this hypothesis awaits future analyses of the structure of SIX1 bound to DNA.
Molecular details of the SIX1–EYA interface
The structure of the SIX1–EYA2ED complex unexpectedly reveals that SIX1 interacts with the HAD catalytic domain and not the HDM of EYA2ED as was previously proposed (Fig. 1b)7, with a combined buried surface area of 1600 Å2 (~800 Å2 on the SIX1 interface and ~800 Å2 on EYA). Strikingly, the interaction between SIX1 and EYA is mediated predominantly by a single SIX1 helix (α1) that fits into a binding groove on EYA2ED through a combination of hydrophobic interactions, hydrogen bonds, and salt bridges (Fig. 2a–c). This interaction mode demonstrates an interesting and unexpected resemblance to two other protein-protein interactions (p53–MDM2 and BH3–BCL-XL)43,44, both of which use an amphipathic α-helix to bind in a hydrophobic groove and both of which have been successfully targeted by small molecules as potential cancer therapeutics45,46. The binding groove in EYA is formed with the β1 and β4 strands constituting the floor and the β5-turn-α12 structure constituting the binding groovewall (Fig. 2a). The binding groove is largely hydrophobic and lined with Val496, Val498, Pro516, Phe517, Trp518 and Leu538 (Fig. 2b, orange surface). Cys16 and Val20 from SIX1 α1 protrude into the hydrophobic groove (Fig. 2b). There are several hydrogen bonds between SIX1 α1 and EYA2ED including between the sulfhydryl group of the centrally located cysteine, SIX1 Cys16, and the carbonyl oxygen of EYA2ED Phe517, while the hydrophobic binding groove is flanked by two salt bridges (Fig. 2c). The N-terminal tail immediately upstream of α1 and the α2 helix of SIX1 forms additional van der Waals interactions with a small surface patch on EYA2ED (Fig. 2b, gray surface). Multiple sequence alignments of Six and Eya family members from diverse eukaryotic organisms show a high degree of sequence conservation in their interacting regions, highlighting the importance of these residues for proper function of the complex (Supplementary Fig. 2,3).
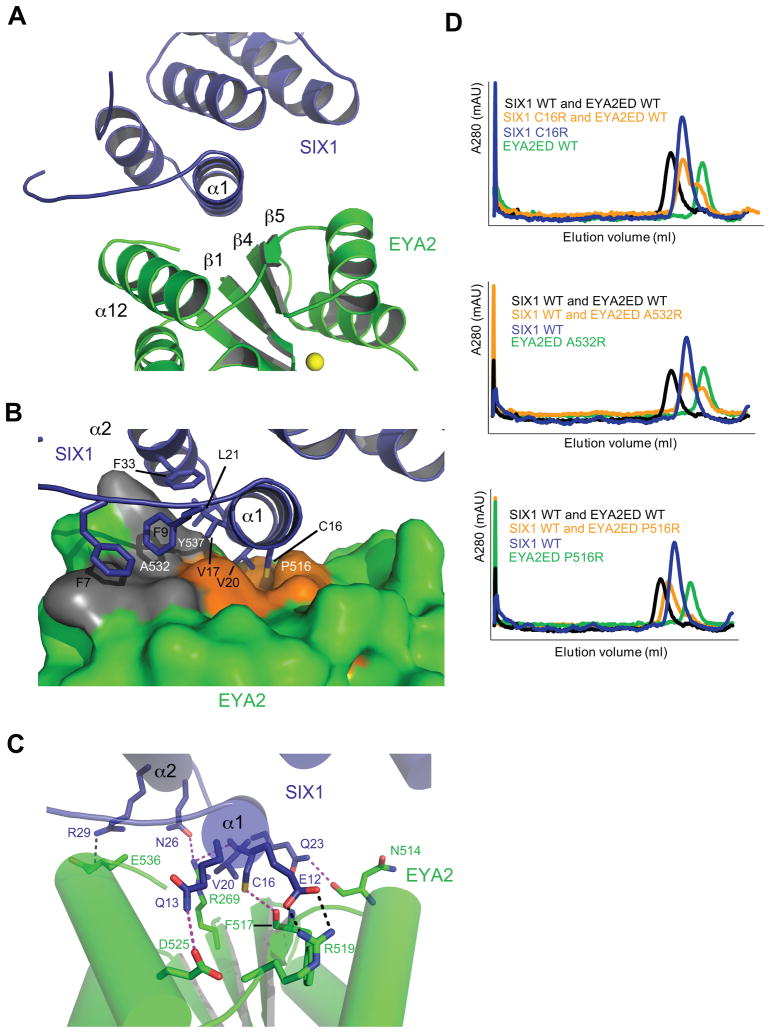
Figure 2. Interacting Surfaces of Human SIX1 and EYA2ED.
(A) A close up view of the interface in the SIX1–EYA2ED complex (colored as in Figure 1). The location of the tyrosine phosphatase active site is indicated by the Mg2+ ion (yellow sphere).
(B) Surface representation of the EYA2ED hydrophobic binding pocket and cartoon representation of SIX1. SIX1 residues that extend into the hydrophobic groove (Cys16 and Val20) or cover the surface patch (Phe7, Phe9, Val17, Leu21 and Phe33) are shown as sticks. SIX1 residues are labeled black and EYA residues are labeled white.
(C) Residues involved in potential salt bridges(black dashed lines)or hydrogen bonds(magenta dashed lines)are shown in stick representation.
(D) Analytical gel filtration profiles of SIX1 and EYA2ED, wild type or mutant proteins. The profile of SIX1–WT EYA2ED WT was overlaid to show normal complex formation.
To confirm the binding interface, we performed structure-based mutagenesis of SIX1 and EYA2ED. We have previously reported that a naturally occurring SIX1 BOR mutant, V17E, disrupts the formation of the SIX1–EYA complex42. In the crystal structure, Val17 resides in the middle of the SIX1 α1 helix and extends into the binding interface making van der Waals contacts with Tyr537 in EYA (Fig. 2b), providing a clear explanation of how this mutation disrupts the ability of SIX1 to bind EYA. In the current study, we have mutated the centrally located cysteine residue (C16R) in SIX1 and two EYA2ED residues (P516R and A532R) that reside at the interface (Fig. 2b). We found that SIX1 C16R and EYA2ED A532R completely abolish complex formation, while EYA2ED P516R severely compromises SIX1–EYA2ED binding based on size exclusion chromatography analyses (Fig. 2d). These observations validate the SIX1–EYA binding interface and demonstrate that single point mutations can significantly reduce SIX1–EYA binding.
Structural basis of BOR mutations
EYA1 is the most commonly mutated gene in BOR syndrome and there are at least 14 reported missense mutations within the ED of EYA18,47–49. While our structure was determined with the ED of EYA2, EYA1ED and EYA2ED share over 90% sequence similarity (Supplementary Fig. 3,4). Importantly, mammalian EYA1 and EYA2 can both complement Drosophila eya mutations with comparable efficiency50,51 and EYA1 and EYA2 have been shown to be functionally redundant during myogenesis52. Furthermore, of the 14 BOR mutations found in EYA1ED, 12 residues are identical between EYA1 and EYA2 (Supplementary Fig. 3,4). These data suggest that our SIX1–EYA2ED structure can be used as a framework to predict the molecular mechanisms of the EYA1 BOR mutants, serving as valuable models for directing future functional studies.
We first evaluated the impact of each missense mutation on protein structure and stability using the Site Directed Mutator (SDM) program, a program that was validated using 855 mutations from 17 different proteins53. SDM predicts that 6 of the 14 missense mutations destabilize the EYA2ED structure (Table 2). The remaining mutations (we will refer these as non-destabilizing mutants) that are solvent exposed may affect protein function by disrupting substrate binding, catalysis, or binding to SIX1 or other co-factors. One of these mutations, E309V, was previously predicted to be on the SIX1 binding surface7. However, our structure demonstrates that this residue is in fact distant from the actual SIX1–EYA interface (Fig. 3a). Instead, this amino acid resides on the same face as the active site pocket (Fig. 3a), leaving open the possibility that it is involved in substrate binding. This residue is conserved in all human and mouse Eya family members as well as Drosophilaeyes absent, consistent with its potential importance for EYA function (Supplementary Fig. 3). Another non-destabilizing mutant, N433P resides in the middle of α8 of the catalytic domain. While not predicted by SDM to destabilize the overall protein structure, it likely alters the secondary structure of α8, which resides behind the active site and its disruption could affect phosphatase activity. Supporting this hypothesis, EYA proteins containing this mutation are able to bind SIX1 in cells54,55, but have severely compromised phosphatase activity55,56. Two other non-destabilizing mutants, D375G and R386Q, both mapped to α5 near a flexible loop (residues 356-371), may represent residues involved in interactions with a yet to be identified co-factor. Indeed, Asp375 and a positively charged amino acid at position 386 are conserved across all Eya family members (Supplementary Fig. 3).
Table 2.
Prediction of structural effect of EYA1 and SIX1 BOR mutations
| EYA1 residuea | EYA1 residueb | EYA2 residuec | Location | Solvent accessibility | SDM ΔΔG | SDM prediction |
|---|---|---|---|---|---|---|
| I293N | I326N | V272N | β1 | buried | −4.96 | destabilizing |
| E330V | E363V | E309V | α3 | partially accessible | 0.49 | non-destabilizing |
| G393S | G426S | G372S | loop | accessible | 2.60 | stabilizing |
| D396G | D429G | D375G | α5 | accessible | −1.20 | non-destabilizing |
| R407Q | R440Q | R386Q | α5 | partially accessible | −0.72 | non-destabilizing |
| S454P | S487P | N433P | α8 | accessible | 0.61 | non-destabilizing |
| R459P | R492P | R438P | loop | partially accessible | −4.66 | destabilizing |
| L472R | L505R | L451R | α9 | buried | −3.25 | destabilizing |
| V479F | V512F | V458F | α9 | buried | −0.39 | non-destabilizing |
| L480P | L513P | L459P | α9 | buried | −4.85 | destabilizing |
| Y494C | Y527C | Y473C | β3 | buried | −0.82 | non-destabilizing |
| V517E | V550E | V496E | β4 | partially accessible | −1.55 | non-destabilizing |
| M536T | M569T | M515T | loop | buried | −1.86 | non-destabilizing |
| L550P | L583P | L529P | α12 | buried | −4.85 | destabilizing |
|
| ||||||
| SIX1 residue | Location | Solvent accessibility | SDM ΔΔG | SDM prediction | ||
|
| ||||||
| V17E | α1 | partially accessible | −0.75 | non-destabilizing | ||
| H73P | loop | partially accessible | −2.50 | destabilizing | ||
| V106G | α6 | accessible | −2.10 | destabilizing | ||
| R110W | α6 | accessible | 1.29 | non-destabilizing | ||
| R110Q | α6 | accessible | 0.54 | non-destabilizing | ||
| R112C | α6 | partially accessible | 0.67 | non-destabilizing | ||
Previous BOR literature have mainly used residues numbered according to either the 559 amino acid encoding EYA1 construct NM_172060.2a or the longest EYA1 construct, 592 amino acids NM_000503.3b. Residues are listed for both numbering systems for clarity in this table.
The corresponding EYA1 BOR mutations in the EYA2 sequence NM_005244.4
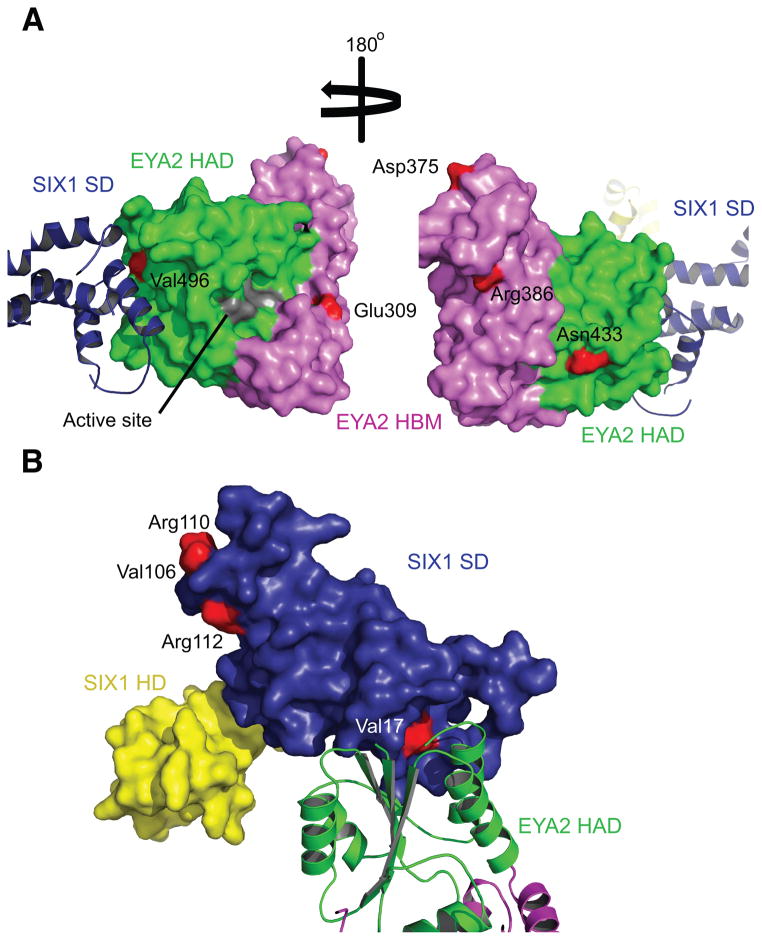
Figure 3. Mapping BOR Mutations on EYA2ED and SIX1.
Domains colored as in Figure 1. Residues mutated in BOR are shown in red on the surface diagrams. (A) Mapping of BOR mutations onto the surface of EYA2ED (colored as in Figure 1). The phosphatase active site shown in gray on the surface diagram. (B) Mapping of BOR mutations onto the surface of SIX1 (colored as in Figure 1).
Three BOR mutations (V496E, delV499 and L529P) map to secondary structures that make up the SIX1 binding groove in EYA (Fig. 3a and Supplementary Fig. 3). EYA2ED Val496 resides at the beginning of β4, which forms part of the floor of the hydrophobic binding groove, and makes van der Waals contacts with SIX1 Gln23 and Gly24. This mutation to a charged amino acid with an elongated side chain will likely result in electrostatic and steric clashes with SIX1. EYA2ED Val499 is not solvent accessible and is located in the center of β4 and deletion of this residue would be predicted to alter the structure of this beta strand. EYA2ED Leu529 resides in the middle of α12 and a leucine to proline change likely disrupts the secondary structure of this helix, and indeed SDM predicts that aproline in this buried position would destabilize the protein (Table 2).
Missense mutations in SIX1 have also been identified in BOR patients9. Our previous functional analyses on 6 of these mutations demonstrate that 5 of them disrupt DNA binding42. All 5 mutations map either to the HD itself (Y129C and delE133) or to α6 in the SD (V106G, R110W, R112C) (Fig. 3b and Supplementary Fig. 2). As stated previously, the single SIX1 BOR mutation that we found to inhibit binding to EYA, V17E, resides on the SIX1 helix (α1) that interacts with EYA (Fig. 2b,3b). Finally, SDM predicts that one of the SIX1 mutations that we were unable to express efficiently, H73P, would deleteriously affect protein stability (Table 2).
SIX1–EYA binding is required for metastatic phenotypes
In contrast to the loss of function mutations observed in BOR, overexpression of either SIX1 or EYA has independently been associated with the development and progression of numerous cancers. For example, SIX1 and EYA have been shown to induce pro-EMT characteristics and metastasis in multiple breast cancer studies16,31,32. However, the relevance of the direct physical interaction between SIX1 and EYA in metastasis has remained unclear, particularly since EYA has intrinsic phosphatase activity that may act independently of its co-activator function with SIX1 in promoting metastasis32. To address this question, we capitalized on the information obtained from the structure and biochemical analyses of the SIX1–EYA complex and generated stably transfected MCF7 cells overexpressing either wild type SIX1 (WT), SIX1 V17E (the mutation that abrogates SIX1 binding to EYA but still allows for DNA binding42), SIX1 delE133 (a mutation which abrogates DNA binding but not EYA binding42), and chloramphenicol acetyltransferase as a control (Ctrl). Of note, the SIX1 V17E mutant is likely to inhibit binding to all EYAs, since SIX1 can bind to all members of the Eya family11,42,57 and residues involved in SIX1 binding are well conserved (identical or similar) in all Eya family members(Supplementary Fig. 3). To control for insertion site effects, 3 stable clonal isolates (unpooled)for each construct were selected with similar SIX1 expression levels (Supplementary Fig. 5)and are used for all subsequent experiments.
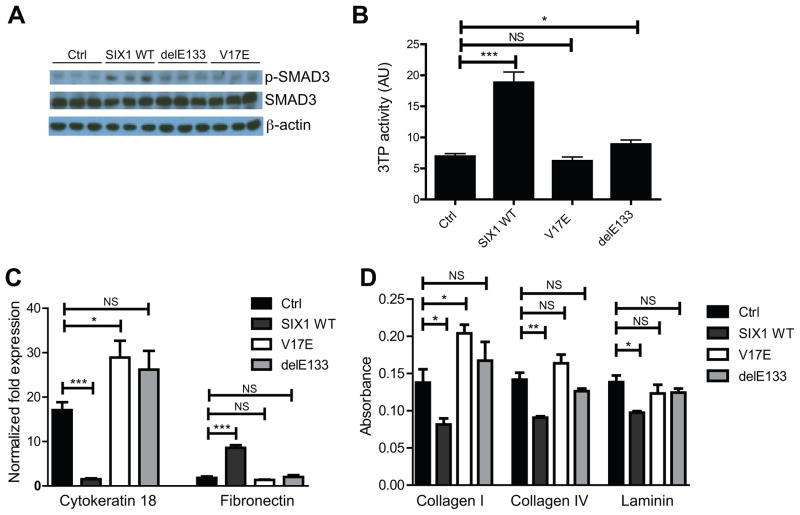
We first evaluated if the direct SIX1–EYA interaction is essential for SIX1-mediated TGF-β signaling, a pathway that is required for the EMT and metastatic phenotypes induced by SIX1 in MCF7 breast cancer cells16. While SIX1 WT is able to induce phospho-SMAD3 protein levels (an effector of TGF-β signaling) and TGF-β signaling (as measured by the 3TP-Lux luciferase reporter which contains TGF-β/Smad-responsive elements from the plasminogen-activator inhibitor-1 and collagenase I genes58), both the SIX1–EYA binding mutant (V17E) as well as the DNA binding mutant (delE133) are unable to activate this signaling pathway (Fig. 4a,b). These data demonstrate that the direct interaction of SIX1 with EYA (and as expected, of SIX1 with DNA) is required to activate TGF-β signaling.
Figure 4. Disruption of the SIX1–EYA Transcriptional Complex Inhibits SIX1-mediated TGF-β Signaling and Characteristics of EMT.
(A) MCF7-V17E and delE133 cells fail to induce p-SMAD3 protein to the same levels as SIX1 WT, as demonstrated by Western blot performed on whole cell lysates. β–actin was used as a loading control.
(B) MCF7-V17E and delE133 cells fail to induce TGF-β responsive 3TP-luciferase reporter activity to levels observed in SIX1 WT cells. Values represent biological replicates with SEM error bars (n=3) and are normalized to renilla luciferase levels.
(C) MCF7-V17E and delE133 cells fail to reduce cytokeratin 18 levels and induce fibronectin levels, as is observed in MCF7-SIX1 WT cells. Expression of cytokeratin 18 and fibronectin were determined using qRT-PCR.
(D) Expression of SIX1 V17E and delE133 in MCF7 cells fails to decrease the adhesion of the cells to the matrix proteins Collagen I, Collagen IV and Laminin, whereas expression of SIX1 WT leads to decreased adhesion to these matrices. Relative adherence measured by crystal violet staining. P-values represent statistical analysis using a paired t-test.
We next examined whether the SIX1 and EYA interaction is required to induce EMT-like characteristics, by analyzing the aforementioned cell lines for expression of the epithelial marker cytokeratin 18 and the mesenchymal marker fibronectin. Consistent with previous observations16, overexpression of SIX1 WT leads to decreased expression of cytokeratin 18 and concomitant increased expression of fibronectin (Fig 4c). However, in the MCF7-V17E and MCF7-delE133 cells, expression of cytokeratin 18 remained high and expression of fibronectin remained low, similar to what is observed in MCF7-Ctrl cells. Similarly, expression of SIX1 V17E and delE133 in MCF7 cells fails to significantly decrease cell-matrix adhesion to collagen I, collagen IV, and laminin (Fig. 4d). In contrast, expression of SIX1 WT significantly decreases adhesion to all three matrices, another characteristic of EMT. In all cases, the SIX1 V17E and delE133 mutants behave similarly to the MCF7-Ctrl cells, strongly suggesting that a direct interaction between SIX1 and EYA is required to mediate TGF-β signaling, ultimately enabling pro-EMT characteristics in cultured cells.
SIX1–EYA binding is required for SIX1-mediated metastasis
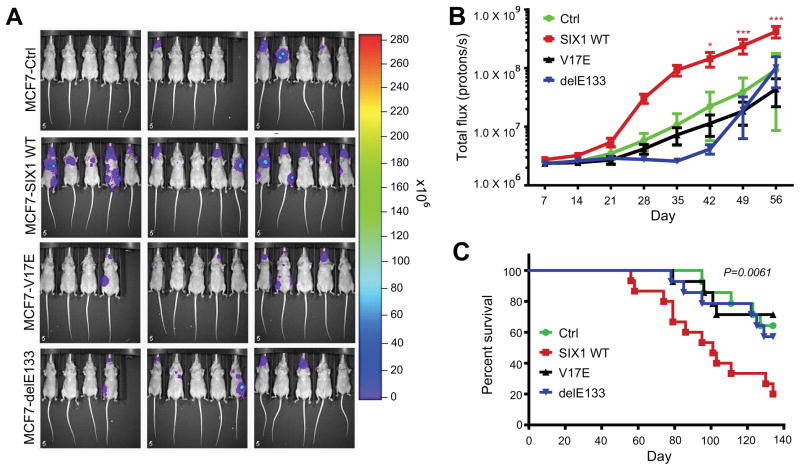
We have previously demonstrated that the involvement of SIX1 in later stages of metastasis is dependent on the induction of TGF-β signaling16. Additionally, there is increasing evidence for EMT as an important mechanism of the metastatic process59. In the current work, we show that the SIX1–EYA interaction (and as expected, the SIX1–DNA interaction) is required for enhanced TGF-β signaling and pro-EMT characteristics (Fig. 4). To determine if this interaction is also necessary for enhanced late stage metastasis induced by SIX1, we performed experimental metastasis assays using MCF7-Ctrl, -SIX1 WT, -V17E, and -delE133 cells tagged with firefly luciferase to visualize in vivo metastatic spread. Three clonal isolates of each line were injected into the arterial bloodstream of nude mice. In agreement with previous work16, the MCF7-SIX1 WT cells metastasized significantly more than MCF7-Ctrl cells as measured by total body bioluminescence (Fig. 5a,b). Additionally, mice injected with MCF7-SIX1 WT cells had an overall shortened survival compared to mice injected with MCF7-Ctrl cells (Fig. 5c). Importantly, the SIX1 V17E and SIX1 delE133 mutants did not enhance metastasis above that observed with MCF7-Ctrl cells (Fig. 5a,b) or alter overall survival (Fig. 5c). These data demonstrate that disruption of the SIX1–EYA interaction potently inhibits the ability of SIX1 to induce metastasis.
Figure 5. Disruption of the SIX1–EYA Transcriptional Complex Inhibits SIX1-mediated Metastasis.
(A) Bioluminescent imaging of nude mice at day 56 post injection of MCF7-Ctrl, MCF7-SIX1 WT, MCF7 SIX1 V17E or MCF7 SIX1 delE133 cells into the left ventricle.
(B) Quantification of the total body luminescence signal (photons/sec) at all time points recorded. Injection of MCF7 cells expressing the SIX1 V17E mutant, as well as the delE133 DNA binding mutant, did not enhance metastasis above that observed with MCF7-Ctrl cells.
(C) Kaplan-Meier curves representing overall survival of the injected mice. Mice injected with MCF7-V17E or MCF7-delE133 cells did not display a decrease in overall survival compared with MCF7-Ctrl cells. In contrast, survival of mice injected with MCF7-SIX1 WT cells was significantly worse. Statistical analysis was performed using the log-rank test. (*p<0.05, **p<0.01, *** p<0.001)(n=15).
DISCUSSION
Although it has been traditionally difficult to target transcription factors in the clinic, inhibiting the protein-protein interaction in these transcriptional complexes has been increasingly realized as a strategy that holds great promise26. Our crystal structure reveals that SIX1 uses predominantly a single amphipathic α-helix to bind a hydrophobic cleft in EYA, a binding mechanism that is highly similar to two other protein-protein interactions that have been successfully targeted in cancer with small molecules. These small molecules include nutlin, which is an efficient p53–MDM2 interaction inhibitor45, and navitoclax, which inhibits the interactions between members of the B-cell lymphoma 2 (Bcl-2) family of proteins46. Here we demonstrate that a single amino acid mutation on the amphipathic α-helix of SIX1 that can disrupt the SIX1–EYA interaction, potently inhibits SIX1-mediated TGF-β signaling, EMT and metastasis. The combination of our structure and functional data strongly suggest that disruption of the SIX1–EYA transcriptional complex using small molecules may be a feasible approach in developing novel anti-cancer agents.
Such anti-cancer therapeutics could potentially be effective in breast cancer, Wilms’ Tumor, acute leukemia, malignant peripheral nerve sheath tumors and ovarian cancer, where SIX1 and EYA are both over-expressed16,21,24,27,29–32. Further analysis of SIX1–EYA expression in the Oncomine database demonstrates that SIX1 and EYA are both significantly overexpressed in numerous additional tumor types (Supplementary Table 1), suggesting that inhibition of the SIX1–EYA transcriptional complex may be a broad anti-cancer therapeutic approach. Importantly, SIX1–EYA overexpression correlates with decreased overall survival in brain, breast, head-neck, lung, and pancreatic carcinomas as well as lymphoma (Supplementary Table 1). High SIX1–EYA expression also correlates with recurrence in glioblastoma, breast, and ovarian carcinomas, with metastasis in breast, prostate and head-neck carcinomas as well as melanoma, with higher grade malignancies in bladder, breast, colon, hepatocellular, and endometrial carcinomas, and with more advanced stage in breast, cervical, colon, head and neck, lung and ovarian carcinomas (Supplementary Table 1). Since the SIX1–EYA transcriptional complex is highly expressed during development, downregulated in the adult, and re-expressed in many tumor types, inhibiting the SIX1–EYA interaction may be a unique approach to inhibit tumor progression, while conferring limited side effects, in the many tumor types that overexpress this complex.
ONLINE METHODS
Protein Expression and Purification
Human SIX1 (residues 1-189) was subcloned into the pMALX_B plasmid33 using the NotI and XbaI restriction sites and transformed into E. coli strain XA-90. Selenomethionine (Se-Met) incorporatation was carried out by growing cells in M9 minimal medium containing 0.04mg/ml of each of the 19 amino acids excluding methionine at 37°C. When the cells reached an A600 = 0.4–0.6, 100mg each of threonine, lysine-HCL, phenylalanine, cysteine, 50mg each of leucine, isoleucine, valine, tryptophan and 120mg of DL-seleno-methionine were added. The temperature was shifted to 25°C and grown for an additional 30 minutes before 0.2mM IPTG was added to induce protein expression overnight. Cells were harvested by centrifugation and lysed by sonication in 50mM HEPES pH 8.0, 250mM MgCl2, 0.5mM TCEP (Bond-Breaker, Thermo Scientific) containing protease inhibitors phenylmethylsulfonyl fluoride, pepstatin, and leupeptin. Native protein expression was carried out as described42.
After centrifugation, SIX1-containing supernatant was treated with 0.4% polyethyleneimine to precipitate out contaminating DNA, which was pelleted by centrifugation. The resulting supernatant was loaded onto an amylose resin column, washed and eluted with 40mM maltose. The protein was further purified using a Superdex 200 column (GE Healthcare) in 50mM HEPES pH 8.0, 250mM MgCl2, 0.5mM TCEP, 5mM maltose. The purified protein was adjusted to 5mM TCEP.
Se-Met EYA2ED was purified in a protocol similar to SIX1 except the cell pellet supernatant was passed over glutathione-Sepharose 4B resin (GE Healthcare), and the EYA2ED protein was eluted after cleavage with PreScission protease. The protein was purified further using the Superdex 200 column (GE Healthcare) in 50mM HEPES pH 8.0, 250mM NaCl and 0.5mM TCEP. The purified protein was adjusted to 5mM TCEP.
The SIX1–EYA2ED complex was formed by mixing the purified MBPB-SIX1 and EYA2ED in a 1:1 molar ratio. The complex was purified using a Superdex 200 column in 20mM HEPES pH 8.0, 200mM MgCl2, 0.5mM TCEP and 5mM maltose. The peak fractions were adjusted to a final 5mM TCEP concentration.
Electrophoretic Mobility Shift Assay
The DNA binding assay was essentially performed as described42, but with biotinylated oligonucleotides. Binding reactions were carried out with 15 fmole oligo and 1.6 pmole MBP-SIX1-189 or MBP. DNA was blotted onto a Biodyne B nylon membrane (Thermo Scientific) and detected with the Chemiluminescent Nucleic Acid Detection Module (Thermo Scientific).
Crystallization and Data Collection
All crystallization trials were carried out at 18°C using the hanging drop vapor diffusion method, with each drop consisting of 1μl complex and 1μl well solution. The final crystallization condition is 50mM MES pH 5.1, 10mM MgCl2, 13.75% PEG 8000. Crystals were fully grown after 1 week. The crystals were cryoprotected by serial transfer through increasing concentrations of PEG 8000 and glycerol to a final concentration of 15%/15%, and flash cooled in liquid nitrogen.
Diffraction data were collected at the Advance Photon Source, SBC beam line 19ID. Single wavelength anomalous dispersion data (SAD) was collected using the selenomethionine substituted complex at the Se peak wavelength 0.97912 Å. Data set for the native crystal was also collected at the 0.97912 Å wavelength.
Structure Determination and Refinement
All data were processed and scaled with HKL300060. The structure was determined by SAD phasing using the anomalous signal from Se atoms andHKL300060 with SHELX61, MLPHARE62 and DM63 using the peak data to 2.0 Å. The initial model was built usingarp/Warp64. This model was refined against data from the native crystals. Refinement and model building were carried out iteratively using Phenix65 and Coot66 until it converged to the Rwork factor of 18.4, and the Rfree of 22.4. The stereochemistry of the structure was checked with MOPROBITY67. 98.3% of the residues were in favored regions with 0% in disallowed regions in the Ramachandran plot. The structure has been deposited in the Protein Data Bank (PDB 4EGC). All structural figures were produced using PyMol (http://www.pymol.org).
Analyses of Complex Formation
Complex formation was analyzed by analytical size exclusion chromatography as previously described42, but using the MBP-SIX1 fusion protein.
Prediction of Structural Effects for Missense Mutations
The effect of missense mutations on structural stability was evaluated with the SDM server. Predicted stability score is analogous to the free energy difference between the wild-type and mutant protein. SDM utilizes a statistical approach by fitting variables such as substitution frequencies, distance potentials and residue environments to a potential energy function and this method has been shown to have an accuracy of 74% in predicting the sign of stability change and a linear correlation coefficient of 0.60 between predicted and observed ΔΔG values53. Positive values indicate an increase in the thermodynamic protein stability, while negative values indicate a decrease in stability. A cut-off of 2.0 kcal mol−1 in pseudo ΔΔG value indicates a significant effect on protein stability53.
Cell Culture
MCF7-Ctrl (chloramiphenicol transferase), MCF7-SIX1 WT and MCF7-SIX1 mutants (delE133, V17E) cell lines were newly generated the same way as previously described14, (the MCF7-Ctrl and MCF7-SIX1 WT cells were re-made at the same time as the MCF7-SIX1 mutants to negate differences due to passage time). Stable transfectants were selected using G418 (Invitrogen). For in vivo metastasis experiments, Bosc cells were co-transfected with the SFG-nes-GTL luciferase/GFP plasmid and pcL-Ampho retroviral packaging plasmid using Effectene (Qiagen). MCF7 clones were then transduced using subsequent viral particles. To reduce variation in expression levels between clones, cells were selected for similar GFP expression by FACS, expanded and re-sorted. LUC expression levels were then compared by plating the cells at ~75% confluency and imaged using IVIS.
3TP Luciferase Assay
Luciferase assays were performed as described16. Briefly, MCF7-CAT, -SIX1 WT, -delE133, and -V17E clones were co-transfected, in triplicate, with both 3TP and renilla luciferase constructs. Cells were lysed using a passive lysis buffer (Promega) at 48 hours. Lysates were analyzed using the Dual Luciferase Kit (Promega) on a Turner Biosystems Modulus.
Cell Adhesion Assay
Cell adhesion assays were performed as described16. Briefly, collagen I, collagen IV, or laminin coated 96 well plates (BD Biosciences) were blocked with 1% BSA for 1 hour. 105 Cells in 100 μl serum free DMEM were then added to each well in replicates of 6 per cell line, and incubated at 37 °C for 1 hour. The plates were then washed in cold PBS 3 times before being fixed with 100 μl ice-cold methanol for 10 minutes. The cells were then stained using 0.05% crystal violet for 40 minutes and washed 3 times with dH2O. The dye was then solubilized in 10% glacial acetic acid and absorbance was determined at 570nm using a Turner Biosystems Modulus.
In vivo Metastasis Assay
The intracardiac metastasis assay was carried out as previously described16. Briefly, 7-week-old female nude mice were implanted with an estrogen pellet. 105 Cells in 100 μl PBS were injected into the left ventricle of 5 mice per each clonal isolate (3 clonal isolates per Ctrl, WT, V17E and delE133), totaling 15 mice per group. Mice were injected with D-luciferin 10 minutes prior to intracardiac injections and imaged using the IVIS200 (Caliper LS) imaging system within 5 minutes post injection. Tumor size was monitored weekly by IVIS imaging, and mice were euthanized when moribund. Living image software was used for quantification of the experiment. The mice were housed at the Center for Comparative Medicine at the UCD Anschutz Medical Campus and treated in accordance with the humane care and use of laboratory animals guide. Work was approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Colorado Anschutz Medical Campus.
Western Blot Analysis
Whole cell lysates were prepared using RIPA buffer. Antibodies used include β-actinat 1:2000 (Sigma, AC-74), SMAD3 at 1:1000 (Invitrogen, 51-1500) and p-SMAD3 at 1:500 (Cell Signaling, C25A9). For analysis of SIX1 expression, SIX1, CAT, and mutant SIX1 over-expressing MCF7 clones were treated with 25 μM of the proteosome inhibitor MG-132 for 12 hours. Blots were then performed with nuclear extracts (Pierce Biotechnology Inc.) and probed using primary antibodies against SIX1 at 1:1000 and HDAC at 1:1000 (Santa Cruz). The SIX1 antibody was made as previously described68.
qPCR
qPCR was performed on a BIORAD CFX96. Expression levels of fibronectin, cytokeratin-18 and GAPDH were detected using SsoFast Evagreen (Biorad). SIX1 and GAPDH Amplicons were detected using Taqman primer/probe assays Hs00195590.m1 and Hs99999905.m1, respectively, per manufacturers instruction. Fibronectin 5′-GAGTGTGTGTGTCTTGGTAATGG-3′ (sense) and 5′-CCACGTTTCTCCGACCAC-3′ (anti-sense); Cytokeratin-18 5′-ATCTTGGTGATGCCTTGGAC-3′ (sense) and5′-CCTGCTTCTGCTGGCTTAAT-3′ (anti-sense); GAPDH 5′-CAACTACATGGTTTACATGTTC -3′ (sense) and 5′-GCCAGTGGACTCCACGAC -3′ (anti-sense). Targets were normalized to GAPDH.
Examination of Public Microarray Datasets and Patient Outcome Analysis
Publically available gene expression arrays were analyzed for SIX1 and EYA expression in various cancers using the Oncomine database69. Detailed descriptions of data normalization and statistical analysis can be found at www.oncomine.com. Briefly, normalization of data for fold-change analysis was completed by z-score normalization. Pre-processed microarray data was first log2 transformed, and then median centered to 0. The fold change statistic was calculated on a linear scale, and p-value was calculated using a student’s t-test for each gene and condition.
Supplementary Material
Acknowledgments
The authors would like to thank Drs. Mair Churchill, Douglas Micalizzi and Wayne Lilyestrom for critical reading of the manuscript, as well as members of the Ford and Zhao laboratories for their help and suggestions. We would like to thank the University of Colorado AMC Biomolecular X-ray Crystallography Core; the Biostatistics and Bioinformatics Shared Resource of The University of Colorado Comprehensive Cancer Center (P30CA046934) for access to databases through Oncomine; the Advanced Light Source beam line 4.2.2 at Berkeley and JayNix for help with earlier data collection. This work was supported by grants from the National Cancer Institute (2R01-CA095277 and R01CA157790) to H.L.F. and from the Department of Defense Synergistic IDEA award (W81XWH-09-1-0253), Breast Cancer Research Foundation-American Association for Cancer Research, State of Colorado (2009 and 2011) and the National Institute of Health (R03DA030559 and R03DA033174) to H.L.F. and R.Z. A.N.P. was supported by a Pediatric Hematology/Oncology Postdoctoral Fellowship (2T32082086-11A1). Data in this paper were collected at Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source operated by UChicago Argonne, LLC, for the U.S. Department of Energy, Office of Biological and Environmental Research under contract DE-AC02-06CH11357.
Footnotes
Accession codes. Protein Data Bank: Structural data and coordinates for MBP-SIX1 in complex with EYA2ED have been deposited with accession number 4EGC.
AUTHOR CONTRIBUTIONS
A.N.P performed the structure and biochemical experiments. J.H.C performed the cell culture and animal experiments. A.L.S. performed the bioinformatic analyses within the manuscript. X.S.C and R.Z. designed and supervised the structure and biochemical experiments. H.L.F. designed and supervised the cell culture and animal experiments. A.N.P., R.Z. and H.L.F. designed and conceived the overall experimental design throughout the manuscript, and together wrote the manuscript. All authors participated in the preparation of the manuscript.
References
- 1.Christensen KL, Patrick AN, McCoy EL, Ford HL. The six family of homeobox genes in development and cancer. Adv Cancer Res. 2008;101:93–126. doi: 10.1016/S0065-230X(08)00405-3. [DOI] [PubMed] [Google Scholar]
- 2.Kawakami K, Sato S, Ozaki H, Ikeda K. Six family genes--structure and function as transcription factors and their roles in development. Bioessays. 2000;22:616–626. doi: 10.1002/1521-1878(200007)22:7<616::AID-BIES4>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 3.Rebay I, Silver SJ, Tootle TL. New vision from Eyes absent: transcription factors as enzymes. Trends Genet. 2005;21:163–171. doi: 10.1016/j.tig.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 4.Li X, et al. Eya protein phosphatase activity regulates Six1-Dach-Eya transcriptional effects in mammalian organogenesis. Nature. 2003;426:247–254. doi: 10.1038/nature02083. [DOI] [PubMed] [Google Scholar]
- 5.Tootle TL, et al. The transcription factor Eyes absent is a protein tyrosine phosphatase. Nature. 2003;426:299–302. doi: 10.1038/nature02097. [DOI] [PubMed] [Google Scholar]
- 6.Rayapureddi JP, et al. Eyes absent represents a class of protein tyrosine phosphatases. Nature. 2003;426:295–298. doi: 10.1038/nature02093. [DOI] [PubMed] [Google Scholar]
- 7.Jung SK, et al. Crystal structure of ED-Eya2: insight into dual roles as a protein tyrosine phosphatase and a transcription factor. FASEB J. 2010;24:560–569. doi: 10.1096/fj.09-143891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abdelhak S, et al. A human homologue of the Drosophila eyes absent gene underlies branchio-oto-renal (BOR) syndrome and identifies a novel gene family. Nat Genet. 1997;15:157–164. doi: 10.1038/ng0297-157. [DOI] [PubMed] [Google Scholar]
- 9.Ruf RG, et al. SIX1 mutations cause branchio-oto-renal syndrome by disruption of EYA1-SIX1-DNA complexes. Proc Natl Acad Sci USA. 2004;101:8090–8095. doi: 10.1073/pnas.0308475101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wayne S, et al. Mutations in the transcriptional activator EYA4 cause late-onset deafness at the DFNA10 locus. Hum Mol Genet. 2001;10:195–200. doi: 10.1093/hmg/10.3.195. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y, Knosp BM, Maconochie M, Friedman RA, Smith RJH. A comparative study of Eya1 and Eya4 protein function and its implication in branchio-oto-renal syndrome and DFNA10. J Assoc Res Otolaryngol. 2004;5:295–304. doi: 10.1007/s10162-004-4044-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schönberger J, et al. Mutation in the transcriptional coactivator EYA4 causes dilated cardiomyopathy and sensorineural hearing loss. Nat Genet. 2005;37:418–422. doi: 10.1038/ng1527. [DOI] [PubMed] [Google Scholar]
- 13.Delgado-Olguín P, et al. Epigenetic repression of cardiac progenitor gene expression by Ezh2 is required for postnatal cardiac homeostasis. Nat Genet. 2012 doi: 10.1038/ng.1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ford HL, Kabingu EN, Bump EA, Mutter GL, Pardee AB. Abrogation of the G2 cell cycle checkpoint associated with overexpression of HSIX1: a possible mechanism of breast carcinogenesis. Proc Natl Acad Sci USA. 1998;95:12608–12613. doi: 10.1073/pnas.95.21.12608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu Y, et al. Expression profiling identifies the cytoskeletal organizer ezrin and the developmental homeoprotein Six-1 as key metastatic regulators. Nat Med. 2004;10:175–181. doi: 10.1038/nm966. [DOI] [PubMed] [Google Scholar]
- 16.Micalizzi DS, et al. The Six1 homeoprotein induces human mammary carcinoma cells to undergo epithelial-mesenchymal transition and metastasis in mice through increasing TGF-beta signaling. J Clin Invest. 2009;119:2678–2690. doi: 10.1172/JCI37815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mimae T, et al. Upregulation of Notch2 and Six1 Is Associated with Progression of Early-Stage Lung Adenocarcinoma and a More Aggressive Phenotype at Advanced Stages. Clin Cancer Res. 2012 doi: 10.1158/1078-0432.CCR-11-1946. [DOI] [PubMed] [Google Scholar]
- 18.Zheng XH, et al. Expression and clinical implications of homeobox gene Six1 in cervical cancer cell lines and cervical epithelial tissues. Int J Gynecol Cancer. 2010;20:1587–1592. [PubMed] [Google Scholar]
- 19.Ng KT, et al. Clinicopathological significance of homeoprotein Six1 in hepatocellular carcinoma. Br J Cancer. 2006;95:1050–1055. doi: 10.1038/sj.bjc.6603399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ono H, et al. SIX1 promotes epithelial-mesenchymal transition in colorectal cancer through ZEB1 activation. Oncogene. 2012 doi: 10.1038/onc.2011.646. [DOI] [PubMed] [Google Scholar]
- 21.Behbakht K, et al. Six1 overexpression in ovarian carcinoma causes resistance to TRAIL-mediated apoptosis and is associated with poor survival. Cancer Res. 2007;67:3036–3042. doi: 10.1158/0008-5472.CAN-06-3755. [DOI] [PubMed] [Google Scholar]
- 22.McCoy EL, et al. Six1 expands the mouse mammary epithelialstem/progenitor cell pool and induces mammary tumors that undergo epithelial-mesenchymal transition. J Clin Invest. 2009;119:2663–2677. doi: 10.1172/JCI37691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iwanaga R, et al. Expression of Six1 in luminal breast cancers predicts poor prognosis and promotes increases in tumor initiating cells by activation of extracellular signal-regulated kinase and transforming growth factor-beta signaling pathways. Breast Cancer Res. 2012;14:R100. doi: 10.1186/bcr3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang CA, et al. SIX1 induces lymphangiogenesis and metastasis via upregulation of VEGF-C in mouse models of breast cancer. J Clin Invest. 2012;122:1895–1906. doi: 10.1172/JCI59858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ng KTP, et al. Suppression of tumorigenesis and metastasis of hepatocellular carcinoma by shRNA interference targeting on homeoprotein Six1. Int J Cancer. 2010;127:859–872. doi: 10.1002/ijc.25105. [DOI] [PubMed] [Google Scholar]
- 26.Darnell JE. Transcription factors as targets for cancer therapy. Nat Rev Cancer. 2002;2:740–749. doi: 10.1038/nrc906. [DOI] [PubMed] [Google Scholar]
- 27.Li CM, et al. Gene expression in Wilms’ tumor mimics the earliest committed stage in the metanephric mesenchymal-epithelial transition. Am J Pathol. 2002;160:2181–2190. doi: 10.1016/S0002-9440(10)61166-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang QF, et al. MLL fusion proteins preferentially regulate a subset of wild-type MLL target genes in the leukemic genome. Blood. 2011;117:6895–6905. doi: 10.1182/blood-2010-12-324699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller SJ, et al. Inhibition of Eyes Absent Homolog 4 expression induces malignant peripheral nerve sheath tumor necrosis. Oncogene. 2010;29:368–379. doi: 10.1038/onc.2009.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang L, et al. Transcriptional coactivator Drosophila eyes absent homologue 2 is up-regulated in epithelial ovarian cancer and promotes tumor growth. Cancer Res. 2005;65:925–932. [PubMed] [Google Scholar]
- 31.Farabaugh SM, Micalizzi DS, Jedlicka P, Zhao R, Ford HL. Eya2 is required to mediate the pro-metastatic functions of Six1 via the induction of TGF-β signaling, epithelial-mesenchymal transition, and cancer stem cell properties. Oncogene. 2011 doi: 10.1038/onc.2011.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pandey RN, et al. The Eyes Absent phosphatase-transactivator proteins promote proliferation, transformation, migration, and invasion of tumor cells. Oncogene. 2010;29:3715–3722. doi: 10.1038/onc.2010.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moon AF, Mueller GA, Zhong X, Pedersen LC. A synergistic approach to protein crystallization: combination of a fixed-arm carrier with surface entropy reduction. Protein Sci. 2010;19:901–913. doi: 10.1002/pro.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holm L, Sander C. Protein structure comparison by alignment of distance matrices. J Mol Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 35.Andes SE, Sauer RT. Specificity of minor-groove and major-groove interactions in a homeodomain-DNA complex. Biochemistry. 1995;34:14601–14608. doi: 10.1021/bi00044a040. [DOI] [PubMed] [Google Scholar]
- 36.Hu S, Mamedova A, Hegde RS. DNA-binding and regulation mechanisms of the SIX family of retinal determination proteins. Biochemistry. 2008;47:3586–3594. doi: 10.1021/bi702186s. [DOI] [PubMed] [Google Scholar]
- 37.Percival-Smith A, Müller M, Affolter M, Gehring WJ. The interaction with DNA of wild-type and mutant fushi tarazu homeodomains. EMBO J. 1990;9:3967–3974. doi: 10.1002/j.1460-2075.1990.tb07617.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shang Z, et al. Design of a ‘minimAl’ homeodomain: the N-terminal arm modulates DNA binding affinity and stabilizes homeodomain structure. Proc Natl Acad Sci USA. 1994;91:8373–8377. doi: 10.1073/pnas.91.18.8373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Y, et al. Discovery, optimization and validation of an optimal DNA-binding sequence for the Six1 homeodomain transcription factor. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kawakami K, Ohto H, Ikeda K, Roeder RG. Structure, function and expression of a murine homeobox protein AREC3, a homologue of Drosophila sine oculis gene product, and implication in development. Nucleic Acids Res. 1996;24:303–310. doi: 10.1093/nar/24.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harris SE, Winchester CL, Johnson KJ. Functional analysis of the homeodomain protein SIX5. Nucleic Acids Res. 2000;28:1871–1878. doi: 10.1093/nar/28.9.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patrick AN, Schiemann BJ, Yang K, Zhao R, Ford HL. Biochemical and functional characterization of six SIX1 Branchio-oto-renal syndrome mutations. J Biol Chem. 2009;284:20781–20790. doi: 10.1074/jbc.M109.016832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kussie PH, et al. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. 1996;274:948–853. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 44.Sattler M, et al. Structure of Bcl-xL-Bak peptide complex: recognition between regulators of apoptosis. Science. 1997;275:983–986. doi: 10.1126/science.275.5302.983. [DOI] [PubMed] [Google Scholar]
- 45.Vassilev LT, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 46.Tse C, et al. ABT-263: a potent and orally bioavailable Bcl-2 family inhibitor. Cancer Res. 2008;68:3421–3428. doi: 10.1158/0008-5472.CAN-07-5836. [DOI] [PubMed] [Google Scholar]
- 47.Azuma N, Hirakiyama A, Inoue T, Asaka A, Yamada M. Mutations of a human homologue of the Drosophila eyes absent gene (EYA1) detected in patients with congenital cataracts and ocular anterior segment anomalies. Hum Mol Genet. 2000;9:363–366. doi: 10.1093/hmg/9.3.363. [DOI] [PubMed] [Google Scholar]
- 48.Orten DJ, et al. Branchio-oto-renal syndrome (BOR): novel mutations in the EYA1 gene, and a review of the mutational genetics of BOR. Hum Mutat. 2008;29:537–544. doi: 10.1002/humu.20691. [DOI] [PubMed] [Google Scholar]
- 49.Krug P, et al. Mutation screening of the EYA1, SIX1, and SIX5 genes in a large cohort of patients harboring branchio-oto-renal syndrome calls into question the pathogenic role of SIX5 mutations. Hum Mutat. 2011;32:183–190. doi: 10.1002/humu.21402. [DOI] [PubMed] [Google Scholar]
- 50.Bonini NM, Bui QT, Gray-Board GL, Warrick JM. The Drosophila eyes absent gene directs ectopic eye formation in a pathway conserved between flies and vertebrates. Development. 1997;124:4819–4826. doi: 10.1242/dev.124.23.4819. [DOI] [PubMed] [Google Scholar]
- 51.Bui QT, Zimmerman JE, Liu H, Bonini NM. Molecular analysis of Drosophila eyes absent mutants reveals features of the conserved Eya domain. Genetics. 2000;155:709–720. doi: 10.1093/genetics/155.2.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grifone R, et al. Eya1 and Eya2 proteins are required for hypaxial somitic myogenesis in the mouse embryo. Dev Biol. 2007;302:602–616. doi: 10.1016/j.ydbio.2006.08.059. [DOI] [PubMed] [Google Scholar]
- 53.Worth CL, Preissner R, Blundell TL. SDM--a server for predicting effects of mutations on protein stability and malfunction. Nucleic Acids Res. 2011;39:W215–22. doi: 10.1093/nar/gkr363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Buller C, Xu X, Marquis V, Schwanke R, Xu PX. Molecular effects of Eya1 domain mutations causing organ defects in BOR syndrome. Hum Mol Genet. 2001;10:2775–2781. doi: 10.1093/hmg/10.24.2775. [DOI] [PubMed] [Google Scholar]
- 55.Mutsuddi M, et al. Using Drosophila to decipher how mutations associated with human branchio-oto-renal syndrome and optical defects compromise the protein tyrosine phosphatase and transcriptional functions of eyes absent. Genetics. 2005;170:687–695. doi: 10.1534/genetics.104.039156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rayapureddi JP, Hegde RS. Branchio-oto-renal syndrome associated mutations in Eyes Absent 1 result in loss of phosphatase activity. FEBS Lett. 2006;580:3853–3859. doi: 10.1016/j.febslet.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 57.Ikeda K, Watanabe Y, Ohto H, Kawakami K. Molecular interaction and synergistic activation of a promoter by Six, Eya, and Dach proteins mediated through CREB binding protein. Mol Cell Biol. 2002;22:6759–6766. doi: 10.1128/MCB.22.19.6759-6766.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wrana JL, et al. TGF beta signals through a heteromeric protein kinase receptor complex. Cell. 1992;71:1003–1014. doi: 10.1016/0092-8674(92)90395-s. [DOI] [PubMed] [Google Scholar]
- 59.Drasin DJ, Robin TP, Ford HL. Breast cancer epithelial-to-mesenchymal transition: examining the functional consequences of plasticity. Breast Cancer Res. 2011;13:226. doi: 10.1186/bcr3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution--from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr. 2006;62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 61.Sheldrick GM. Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Crystallogr D Biol Crystallogr. 2010;66:479–485. doi: 10.1107/S0907444909038360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Otwinowski Z. Maximum likelihood refinement of heavy atom parameters. In: Evans P, Leslie A, editors. Isomorphous Replacement and Anomalous Scattering. SERC Daresbury Laboratory; Daresbury, UK: 1991. pp. 80–85. [Google Scholar]
- 63.Cowtan K. dm′: An automated procedure for phase improvement by density modification. Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography. 1994;31:34–38. [Google Scholar]
- 64.Cohen SX, et al. Towards complete validated models in the next generation of ARP/wARP. Acta Crystallogr D Biol Crystallogr. 2004;60:2222–2229. doi: 10.1107/S0907444904027556. [DOI] [PubMed] [Google Scholar]
- 65.Adams PD, et al. The Phenix software for automated determination of macromolecular structures. Methods. 2011;55:94–106. doi: 10.1016/j.ymeth.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ford HL, et al. Cell cycle-regulated phosphorylation of the human SIX1 homeodomain protein. J Biol Chem. 2000;275:22245–22254. doi: 10.1074/jbc.M002446200. [DOI] [PubMed] [Google Scholar]
- 69.Rhodes DR, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.