Abstract
The development of mucosal-associated invariant T (MAIT) cells is dependent upon the class Ib molecule MHC-related protein 1 (MR1), commensal bacteria, and a thymus. Furthermore, recent studies have implicated MR1 presentation to MAIT cells in bacteria recognition, although the mechanism remains undefined. Surprisingly, however, surface expression of MR1 has been difficult to detect serologically, despite ubiquitous detection of MR1 transcripts and intracellular protein. In this article, we define a unique mAb capable of stabilizing endogenous mouse MR1 at the cell surface, resulting in enhanced mouse MAIT cell activation. Our results demonstrated that under basal conditions, endogenous MR1 transiently visits the cell surface, thus reconciling the aforementioned serologic and functional studies. Furthermore, using this approach, double-positive thymocytes, macrophages, and dendritic cells were identified as potential APCs for MAIT cell development and activation. Based on this pattern of MR1 expression, it is intriguing to speculate that constitutive expression of MR1 may be detrimental for maintenance of immune homeostasis in the gut and/or detection of pathogenic bacteria in mucosal tissues.
Major histocompatibility complex-related protein 1 (MR1) is a class Ib molecule encoded by a single functional, monomorphic Mr1 gene (1–3). The Mr1 gene is not Mhc linked, is highly conserved, and seems to be unique to mammals (2, 4). As striking evidence for interspecies conservation, the predicted amino acid sequences of mouse MR1 (mMR1) and human MR1 are 89/90% identical in their α1/α2 domains (2, 5). By contrast, mouse and human MHC-linked class Ia and Ib molecules are 69/70% and 51/41% identical, respectively (6). The high level of polymorphism of classical MHC molecules allows them to present diverse peptides to T cells during the adaptive immune response to pathogens. By contrast, the remarkable conservation of MR1 suggests that it evolved under strong negative selection, possibly imposed by immune responses to pathogens (7). MR1 message and protein are ubiquitously expressed in different tissues (1, 2). Enigmatically, endogenous MR1 has yet to be detected on the plasma membrane of cells from murine or human origins using available mAbs (8–10). However, surface expression of MR1 can be achieved using transfection or transduction to overexpress an MR1-encoding cDNA in mouse or human cell lines (11–13). The failure to detect endogenous MR1 at the cell surface could reflect limited ligand supply as is the case with H2-M3 (14). Alternatively, this failure might be attributable to the lack of suitable mAbs to MR1.
MR1 expression is required for the in vivo development of a novel population of T cells with an invariant TCRα-chain (one Vα-Jα combination) (12). These invariant T cells express Vα19-Jα33 in mice and cattle and the homologous Vα7.2-Jα33 in humans (15). The development of mouse Vα19i T cells is dependent on β2-microglobulin (β2m) but not class Ia molecules or TAP (12). Based on quantitative PCR analysis, these invariant T cells were found to preferentially reside in the lamina propria of the intestine and the lung in mice and humans. Thus, they were assigned the acronym MAIT (mucosal-associated invariant T). MAIT cell transcripts were not detected in mice completely devoid of B cells, nor were they detected in germ-free mice (12). More recent studies showed that B cells are required for the homeostatic expansion of mature MAIT cells in the gut (16). Alternatively, MAIT cell ontogeny is not dependent upon B cells, but it does require MR1 expression on hematopoietic cells and a thymus (16). These latter observations raise the interesting possibility that non-B cells may present an endogenous ligand during MAIT cell thymic development. Upon TCR stimulation, transgenic Vα19i MAIT cells were found to rapidly secrete several cytokines, including IFN-γ, IL-4, IL-5, and IL-10 (17). Thus, MAIT cells, like invariant NKT (iNKT) cells, secrete Th1 and Th2 cytokines, are restricted by a monomorphic class Ib molecule, and express an invariant TCRα and limited TCRβs. Based on their common features, it was proposed that MAIT cells may function as innate T cells, similar to iNKT cells, but servicing different immune compartments (i.e., MAIT cells regulate the mucosal compartment, whereas iNKT cells regulate peripheral lymphoid tissues) (8, 18).
As evidence for physiological relevance, two recent reports demonstrated that MAIT cells have antimicrobial activity. More specifically, the study by Gold et al. (19) showed that human MAIT cells are activated in an MR1-restricted manner by Mycobacterium tuberculosis-infected cells and certain, but not all, other bacterial strains. In a second study, Le Bourhis et al. (20) showed that mouse MAIT cells expressing transgenic TCRs can be activated by cells infected with a variety of bacterial strains and yeast. Furthermore, both studies showed that MAIT cells accumulate in the lungs of individuals with ongoing M. tuberculosis infections (19, 20). Notably, however, surface induction of endogenous MR1 expression was not convincingly shown by either study.
In the current study, we demonstrate that endogenous MR1 transiently visits the cell surface in uninfected cells. The implications of this finding on the Ag-presentation function of MR1 to MAIT cells are discussed.
Materials and Methods
Mice and cell lines
TCRα−/− and TCRβ−/− mice (C57BL/6 background) were gifts from P. Allen and C. Hsieh, respectively (Washington University). All mouse studies were approved by the Animal Studies Committee of Washington University. Embryonic fibroblast cell lines WT3 (derived from C57BL/6 mice) and FMR1−/− (derived from MR1 knockout [KO] mice) were described previously (21, 22). Mouse pre-B leukemia A70.2, myeloma P3X63Ag8.653 (P3X), and B1 B lymphoma line CH27 were gifts from B. Sleckman, M. Diamond, and P. Allen, respectively (Washington University). Mouse B cell lymphoma A20 was purchased from American Type Culture Collection (TIB-208). WT3 and CH27 cells overexpressing MR1 were described previously (11, 13, 22). MAIT cell hybridomas 6C2 and 8D12 were gifts from O. Lantz (Département de Biologie des Tumeurs, Institut Curie, Paris, France) and have been readily used for monitoring MR1 expression (13).
Production of recombinant chimeric mMR1/Ld molecule
Recombinant MR1a/Ld was produced as a soluble protein in insect cells using the pFastbac Dual baculoviral transfer vector (Invitrogen, Carlsbad, CA). Some modifications were made: the p10 promoter was replaced with the pBasic early promoter, and the eGFP gene was cloned downstream of it to allow for visual identification of recombinant baculovirus (23). The foot and mouth disease virus 2A peptide linkage was used to enable coexpression of the MR1a/Ld chimera and mouse β2m (24). Briefly, a single cistron with the following elements, starting from the N terminus, was cloned downstream of the pH promoter: honey bee melittin signal peptide for enhanced secretion (25), residues 1–179 of the mature portion of mMR1a (α1 + α2 domains), residues 184–284 of the mature portion of H-2Ld (α3 domain), thrombin recognition site/hexahistidine tag/foot and mouth disease virus 2A peptide (ASSVPRGSHHHHHHSGSVNFDLLKLAGDVESNPGP), and the entire coding sequence of mouse β2mb (signal peptide + mature peptide). Recombinant baculovirus was generated in Sf9 cells, according to the instructions from the Bac-to-Bac Baculovirus Expression System manual (Invitrogen). P3 or P4 baculoviral supernatants were used to infect Hi5 insect cells in serum-free Ex-Cell 405 medium (JRH Biosciences, Lenexa, KS) supplemented with antibiotics. The Hi5 cells were infected at 1 × 106/ml in 1-l bottles (final volume of 200 ml/bottle) at 27°C on an orbital shaker. Seventy-two hours postinfection, the Hi5 cells were spun down (>3000 × g for 20 min), and the supernatant containing the secreted MR1a/Ld/b2mb complex was filtered, concentrated, and buffer exchanged using the Ultralab Systems with Centramate Cassette (Pall Life Sciences, Port Washington, NY). The recombinant protein was in a final volume of 500 ml Ni-binding buffer (50 mM sodium monobasic phosphate monohydrate, 50 mM sodium citrate, 300 mM sodium chloride [pH 8]). The His-tagged MR1a/Ld/b2mb was affinity purified on a Ni-NTA Superflow resin (Qiagen, Valencia, CA)-packed column. The His-tagged protein eluted in 250 mM imidazole was further purified by size-exclusion chromatography using the AKTA FPLC System coupled to a Superdex 200 column equilibrated in 20 mM HEPES, 150 mM sodium chloride (pH 7.2) (GE Life Sciences, Piscataway, NJ). Fractions containing the purified complex, as revealed by staining of SDS-PAGE gels, were pooled and concentrated.
Abs
Anti-MR1 mAbs 12.2 and 26.5 were reported previously (13). To generate new mAbs to MR1, MR1 KO mice (12) were immunized with soluble, recombinant protein of the chimeric mMR1/Ld molecule (mMR1 [α1-α2 domain]/Ld [α3-cytoplasmic tail]). Hybridomas were obtained by fusing immunized splenocytes with myeloma P3X cells. Culture supernatants were screened by flow cytometry using mMR1/Ld-transduced WT3 cells (WT3-mMR1/Ld) and FMR1−/−. The clones that recognized WT3-mMR1/Ld, but not FMR1−/−, were further subcloned and screened. Two new mAbs, 8F2.F9 and 8H9.D11, are discussed in this article. Mouse IgG1 isotype control (15H6) was purchased from Southern Biotech.
Flow cytometry
A FACSCalibur (BD Biosciences) and Cell Quest and FlowJo software were used for flow cytometry analysis, as previously reported (22). Cells were treated with purified rat anti-mouse FcγRII/III Ab (mouse BD Fc Block) before staining with anti-MR1 mAbs. PE-conjugated goat anti-mouse Ig and rat anti-mouse IgG1 Abs (BD Pharmingen) were used to visualize the primary Ab staining. Intracellular staining was performed after cells were fixed with 2% paraformaldehyde and permeabilized with 0.5% saponin.
MAIT cell-activation assay
A total of 2 × 104 APCs and 105 MAIT cell hybridomas was cocultured in a 96-well flat-bottom plate overnight, as previously described (7, 13, 22). Purified or culture supernatant of anti-mMR1 mAbs was added into co-culture to block or enhance MAIT cell activation. IL-2 secreted by activated MAIT cell hybridomas was measured by ELISA (BioLegend).
Results
Select B cell lines express endogenous MR1 at the cell surface
There has been a major inconsistency in MR1 expression between biochemical/serological findings and functional studies (2, 11–13, 22). More specifically, biochemical studies demonstrated that MR1 is detected almost exclusively in Endo H-sensitive forms, suggesting endoplasmic reticulum (ER) residence (11), and prior flow cytometric studies failed to detect endogenous MR1 at the cell surface using previously described mAbs (9–11, 13, 19, 20). By contrast, the in vivo dependency on MR1 of MAIT cell development and peripheral expansion was highly suggestive that MR1 was expressed at the cell surface (12, 16, 19, 20), and the in vitro MAIT cell hybridoma activation by MR1-overexpressing cell lines was shown to occur at the surface, based on Ab blocking (13). Two previously reported mAbs detecting mMR1, mAbs 12.2 and 26.5 (13), are of the IgG2a isotype and result in strong FcR-mediated staining, as determined by staining MR1-deficient cells (W.-J. Chua and N. Meyers, unpublished observations). To solve this problem, we generated new mAbs to MR1 that do not have background staining on MR1-deficient cells and, thus, are suitable for flow cytometry analysis. These new mAbs were obtained by immunizing MR1 KO mice with soluble, recombinant mMR1 secreted by insect cells. Hybridoma cells secreting MR1-specific mAbs were then selected by flow cytometry based on reactivity with cell lines overexpressing mMR1 and not with MR1-deficient cells. In the current study, we focus on two new mAbs of the IgG1 isotype: 8F2.F9 and 8H9.D11. mAb 8F2.F9 has cross-reactivity against overexpressed MR1 from four species tested, mouse, rat, bovine, and human, whereas mAb 8H9.D11 recognizes only mMR1 (Supplemental Table I).
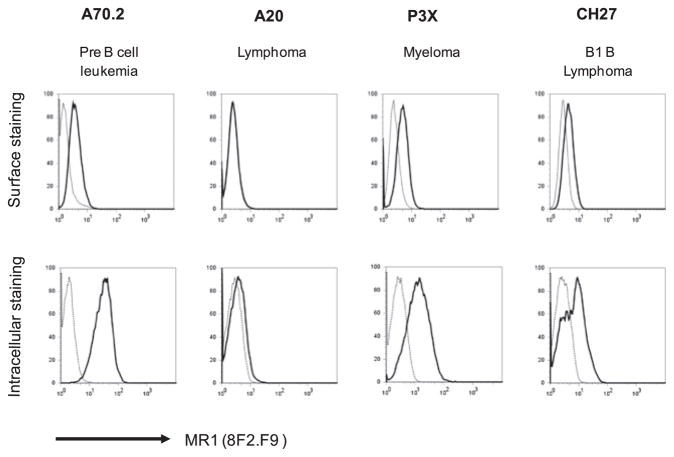
With these newly generated mAbs, we first characterized endogenous MR1 expression on various B cell lines, because B cells are required for homotypic expansion of MR1-restricted MAIT cells in the peripheral mucosal tissues (16). We monitored surface and intracellular (total) expression of endogenous MR1 by flow cytometry analysis. The staining results showed that among different B cell lines tested, A70.2 (a pre-B cell line), P3X (a myeloma line), and CH27 (a B1 B cell line) express high levels of intracellular (total) MR1 (Fig. 1). Consistent with the intracellular expression, A70.2, P3X, and CH27 cells express surface MR1, albeit at low levels. We believe that this is the first time that endogenous MR1 has been detected serologically at the cell surface. By contrast, A20 (a B lymphoma line) had little, if any, total MR1 protein and no detectable surface MR1 (Fig. 1). These combined findings are consistent with the model that under basal conditions, high levels of intracellular MR1 are required to attain surface MR1 expression. Such a notion is consistent with detection of surface MR1 on overexpressing cell lines, because, even in these cells, most overexpressed MR1 remains in the ER (11).
FIGURE 1.
B cells express a low level of endogenous MR1 at the cell surface. Different B cell lineages were stained by anti-MR1 mAb 8F2.F9 and analyzed by flow cytometry. Surface staining (upper panels) of endogenous MR1 is low, whereas intracellular (total) staining (lower panels) of endogenous MR1 is abundant in pre B, myeloma, and B1 B cell lines. By contrast, the A20 cell line does not express endogenous MR1. In the graphs (representing one of five independent experiments), the solid line indicates MR1 staining and the dotted line indicates the background of PE–anti-mouse IgG1 secondary Ab.
Detection of a unique mAb that enhances, rather than blocks, MAIT cell activation
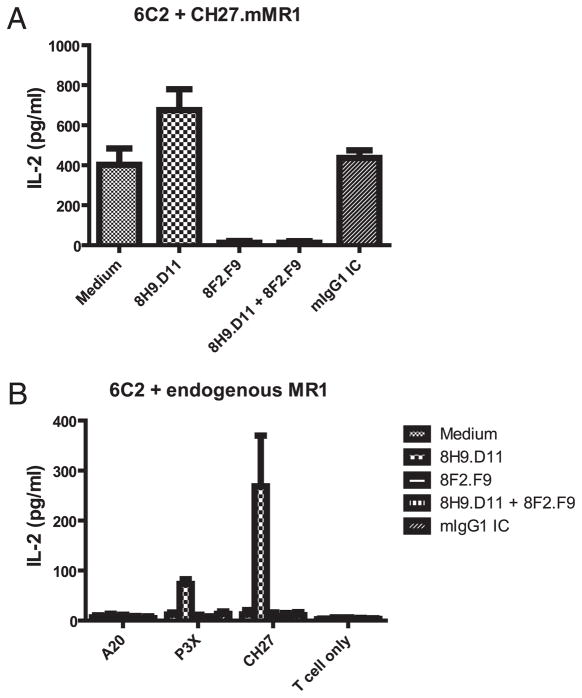
Published studies implicating MR1 in MAIT cell activation showed that the previously described mAbs 12.2 and 26.5 blocked MAIT cell hybridoma activation by cell lines overexpressing MR1 (13, 22). Therefore, it was of interest to determine whether the new mAbs 8F2.F9 and 8H9.D11 could also block MAIT cell activation. As shown in Fig. 2A, mAb 8F2.F9 blocked activation of the MAIT cell hybridoma 6C2 by CH27 cells overexpressing mMR1 (CH27.mMR1). By contrast, mAb 8H9.D11 did not block activation of the 6C2 hybridoma, but it seemed to modestly enhance 6C2 activation. To extend these findings, we also tested a second MAIT cell hybridoma, 8D12. New MR1 mAb 8F2.F9 blocked the modest 8D12 activation, and mAb 8H9.D11 resulted in a dramatic enhancement of MAIT cell activation by overexpressed mMR1 (Supplemental Fig. 1A). Interestingly, adding both anti-MR1 mAbs resulted in blocking and not enhancement. These findings suggested that binding of mAb 8H9.D11 does not effectively compete with the TCR of MAIT cells engaging MR1 but that it somehow enhances MR1-specific recognition by MAIT cells.
FIGURE 2.
MAIT cell hybridoma activation is blocked by mAb 8F2.F9 but enhanced by mAb 8H9.D11 in the coculture with B cells. The MAIT cell hybridoma 6C2 was cocultured with B cell lines in the absence or presence of anti-MR1 mAbs 8F2.F9 and 8H9.D11. The IL-2 production of MAIT cell hybridomas in the coculture with CH27.mMR1 (A) or various B cell lines (B) is shown for three independent experiments. Experiments were performed in triplicate for each group. Error bars represent SD.
To determine whether endogenous MR1 detected on the cell surface is functional, we subjected different B cell lines to MAIT cell activation in the presence or absence of mAb 8H9.D11. Despite the low level of surface MR1 expression, B cell lines P3X and CH27 were unable to activate the MAIT cell hybridoma 6C2 (Fig. 2B). However, in the presence of mAb 8H9.D11, P3X and CH27 cells displayed substantial levels of MAIT cell activation (Fig. 2B). Furthermore, 8H9.D11-induced MAIT cell activation was blocked by mAb 8F2.F9, demonstrating that the MAIT cells were being specifically activated by endogenous MR1 (Fig. 2B). Consistent with the failure to detect surface MR1 on A20 cells, incubation with mAb 8H9.D11 failed to induce MAIT cell activation by A20 cells. A similar result was obtained with the MAIT cell hybridoma 8D12 (Supplemental Fig. 1B). The combined findings suggested that select B cell lines express low levels of endogenous MR1 that is not sufficient in quality and/or quantity to elicit activation of the MAIT cell hybridomas. The observation that a <10-kDa eluate from recombinant MR1 (putative MR1 ligand) did not enhance MAIT cell activation by endogenous MR1 on B cells (7) argues that sufficiency in quality and quantity may be required for endogenous MR1 to activate MAIT cells. However, mAb 8H9.D11 has the unique ability to induce and/or stabilize MR1 molecules that are capable of MAIT hybridoma cell activation.
Blocking and enhancing mAbs detect different epitopes on MR1
To probe the mechanism by which mAb 8H9.D11 enhances MAIT cell activation, we characterized the epitope on MR1 that this mAb detects. First, we performed a competitive-binding study to see whether mAb 8H9.D11 detects a spatially distinct epitope of MR1 from the blocking mAb 8F2.F9. To this end, binding of mAb 8F2.F9 directly conjugated with Alexa Fluor 647 (8F2.F9-AF647) to CH27.mMR1 cells was tested in the presence of unlabeled mAb 8F2.F9 or 8H9.D11. As expected, unlabeled mAb 8F2.F9 blocked staining of labeled 8F2.F9 to CH27.mMR1 in a dose-dependent manner (Fig. 3). However, unlabeled mAb 8H9.D11 did not compete for staining of 8F2.F9-AF647 to CH27.mMR1, demonstrating that these two anti-MR1 mAbs detect spatially distinct MR1 epitopes. Interestingly, unlabeled anti-MR1 mAb 26.5 also failed to block binding of 8F2.F9-AF647 to CH27.MR1 cells. Because mAb 26.5 and 8F2.F9 block MAIT cell activation, these data indicated that they are blocking TCR engagement by binding spatially distinct MR1 epitopes. These combined findings demonstrated that mAb 8H9.D11 binds a unique site on MR1 to enhance MAIT cell activation and that this site is spatially distinct on MR1 from the interface engaged by the TCR on MAIT cells. Indeed, this conclusion explains why the blocking mAbs act dominantly to the enhancing mAb.
FIGURE 3.
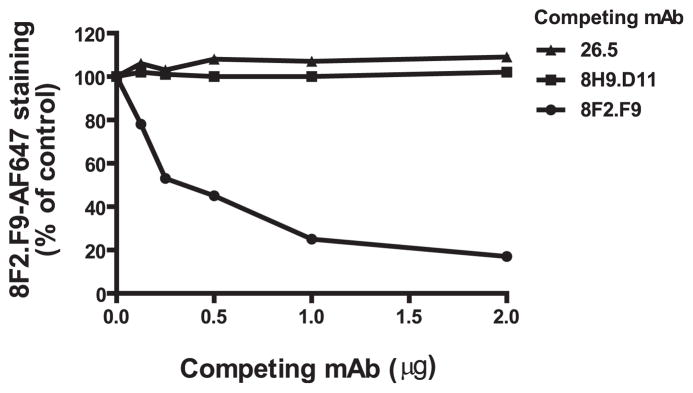
Blocking and enhancing mAbs to MR1 do not compete for binding to MR1. CH27.mMR1 were stained with 8F2.F9-AF647 in the absence or presence of various unlabeled competing mAbs and analyzed by flow cytometry. 8F2.F9-AF647 staining (MFI) at different concentrations of competing mAbs is shown as the percentage of staining without a competing mAb group. Data represent the results from one of three independent experiments.
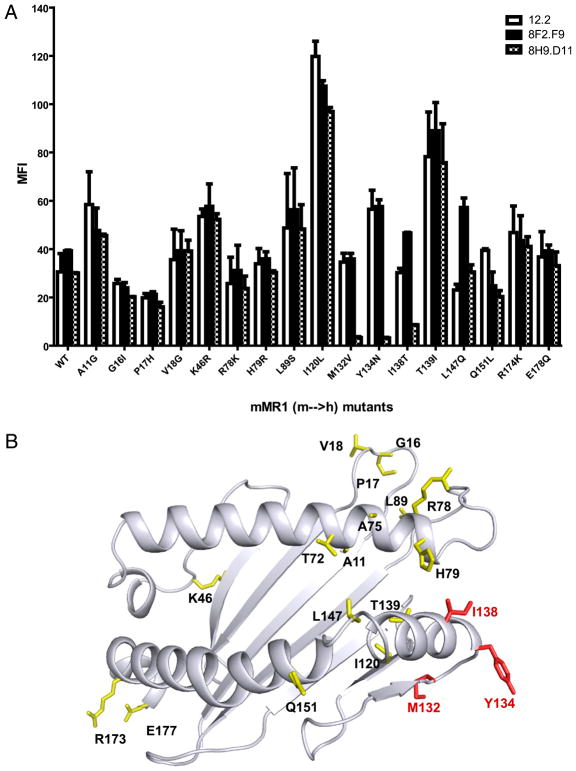
We used a panel of cells expressing MR1 mutations with single amino acid substitutions to further map the precise MR1 epitope detected by mAb 8H9.D11. We chose mutations of mMR1, with substitution of human MR1 residues in the α1 and α2 domains (7), because mAb 8H9.D11 recognizes mMR1 but not human MR1 (Supplemental Table I). The epitope mapping showed that the blocking and enhancing mAbs recognize different epitopes (Fig. 4A). More specifically, three mutations (M132V, Y134N, and I138T) specifically abolished mAb 8H9.D11 recognition of MR1 (Fig. 4A). Given the high probability that MR1 will have an MHC fold, these three residues likely reside on a loop connecting helical residues with the β-sheet structure (Fig. 4B) (26). By contrast, the blocking mAbs (12.2 and 26.5) detect MR1 epitopes defined by helical residues pointed toward the predicted TCR contact interface (13).
FIGURE 4.
Epitope mapping revealed that the enhancing mAb recognizes a putative hinge region in the α2 domain of MR1. A, Anti-MR1 mAbs 8F2.F9 and 8H9.D11 were tested for recognition of WT3 cells expressing wild-type mMR1 or mutants with single amino acid substitutions (mouse to human residues in α1 and α2 domains of mMR1). Anti-MR1 mAb 12.2 was used as a staining control for each mMR1 construct. The staining of anti-MR1 mAbs was visualized by PE–anti-mouse Ig secondary Ab followed by flow cytometry analysis. The plot shows the mean MFI (± SD) from three independent experiments. B, An mMR1-threading model was described previously (7). The mouse residues that were mutated to human residues in α1 and α2 domains of MR1 are labeled. The epitope residues of the enhancing mAb are labeled in red.
The mAb that enhances MAIT cell activation also increases MR1 surface expression
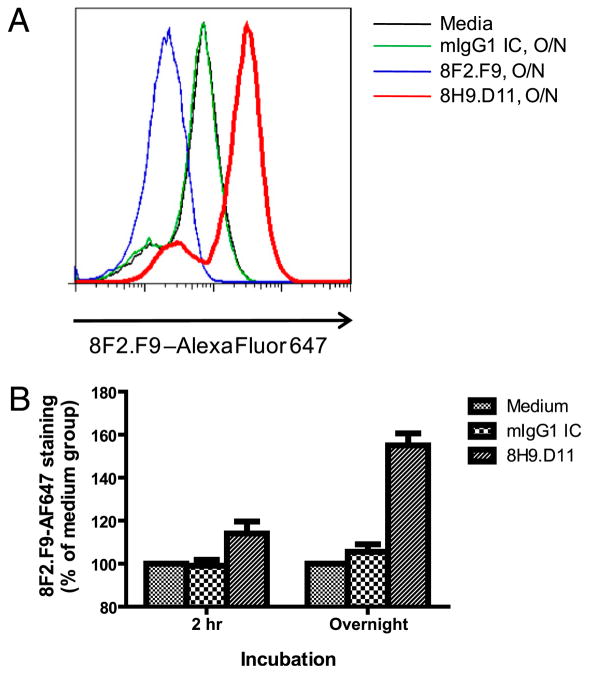
We used flow cytometry analysis of CH27.mMR1 to determine whether mAb 8H9.D11 traps mMR1 at the cell surface. We reasoned that the increased amount of surface MR1 trapped by mAb 8H9.D11 could be monitored by the staining of mAb 8F2.F9, because these two mAbs do not compete for binding to MR1. In this analysis, we incubated CH27.mMR1 with unlabeled mAb 8H9. D11 for 2 h or overnight before staining the cells with 8F2.F9-AF647. Interestingly, incubation with mAb 8H9.D11 overnight dramatically increased the staining signal of CH27.mMR1 (Fig. 5A). It should be noted that the only FcR for IgG identified on B cells is FcγRIIB1, which is the nonendocytic isoform of FcγRIIB (27, 28). Moreover, the Fab fragments of mAb 8H9.D11 also enhanced MAIT cell activation by endogenous MR1 (data not shown). Therefore, the ability of mAb 8H9.D11 to trap MR1 and to enhance MAIT cell activation is very likely a cell-surface phenomenon. Furthermore, the results also showed that the 2-h incubation with mAb 8H9.D11 marginally increased the surface staining of mMR1 (Fig. 5B). It suggested that only low levels of MR1 exit the ER to transit to the plasma membrane and require a longer period of time to accumulate when trapped by mAb 8H9. D11 on the cell surface.
FIGURE 5.
The enhancing mAb traps MR1 at the cell surface. CH27. mMR1 were incubated with medium alone, mouse IgG1 isotype control, 8F2.F9, or 8H9.D11 for 2 h or overnight before staining with 8F2.F9-AF647 and analysis by flow cytometry. A, Staining of CH27.mMR1 from the different overnight incubations from one of three experiments. B, 8F2. F9-AF647 staining of CH27.mMR1 from the 2-h or overnight incubations is shown as the percentage of the staining of the medium control in A. The plot shows the mean percentage (± SD) from three independent experiments.
If ligand occupancy is the defining factor for stable expression of MR1 at the cell surface, it is attractive to speculate that the enhancing Ab is preserving MR1 H chains assembled with endogenous ligand. There is a precedent for such a mechanism based on studies of class Ia molecules. Ortiz-Navarrete and Hammerling (29) found that some mAbs to H-2b molecules trap and increase Kb and Db molecules on the cell surface of RMA-S cells, presumably by preserving ligand occupancy. The unique characteristic of mAb 8H9.D11 is that it does not interfere with TCR engagement, whereas most conformation mAbs to class Ia molecules also block T cell activation. In fact, the 8H9.D11 epitope corresponds exactly with the putative tapasin-binding site in the class Ia structure (26). The residues of the 8H9D.11 epitope are located where the α2 helical region forms a loop that transitions to β-sheet conformation (Fig. 4B). This region has been proposed to be a hinge region whose conformation is altered when high-affinity ligands bind to the class Ia molecules. Based on this model, it is possible that mAb 8H9.D11 binding to the hinge region induces a more favorable conformation for endogenous ligand (which may be low affinity) binding to MR1 H chains. Therefore, mAb 8H9.D11 can stabilize the MR1 complexes on the cell surface without directly contacting ligand. In any case, the availability of a mAb that stabilizes the assembled form of MR1 could be a tremendous resource for ligand identification and structural resolution.
Use of the enhancing mAb to identify physiological APCs for MAIT cells
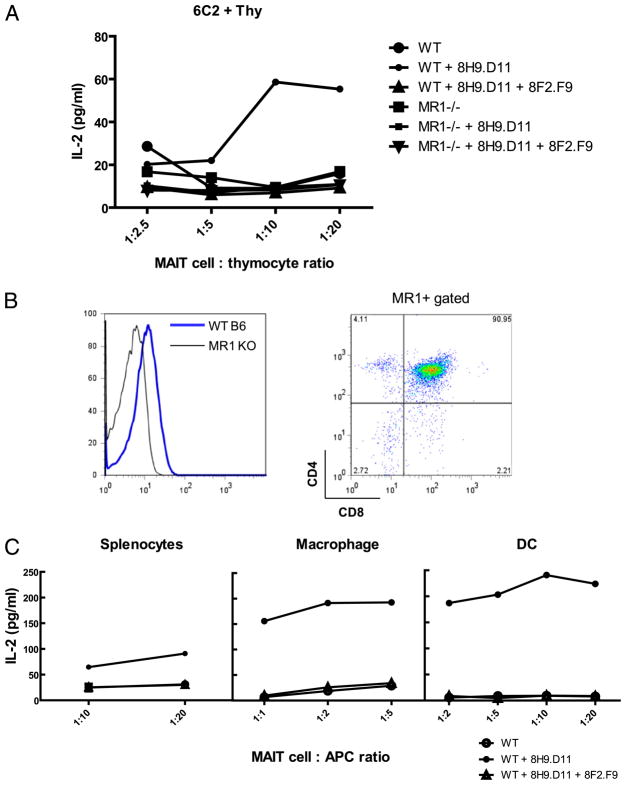
The ability to stabilize endogenous MR1 should also be a great help in defining physiologic MR1-expressing APCs that activate MAIT cells in vivo. Unlike conventional T cells, MAIT cell development does not require MR1 expression on thymic epithelial cells (12). Martin et al. (16) also showed that MAIT cells were not selected by B cells. Interestingly, iNKT cells are selected by CD1d-expressing cortical double-positive (DP) thymocytes (30, 31). Thymocytes constitutively express CD1d on the cell surface and readily activate NKT cell hybridomas (30, 31). To test whether thymocytes can also activate MAIT cells, we used a similar system to coculture thymocytes and MAIT cell hybridomas. The results showed that thymocytes alone were unable to activate MAIT cell hybridomas (Fig. 6A), consistent with the flow cytometry analysis showing that there is no detectable endogenous MR1 on the cell surface of thymocytes (data not shown). However, thymocytes treated with mAb 8H9.D11, which traps endogenous MR1, activated MAIT cell hybridomas (Fig. 6A, Supplemental Fig. 2). Interestingly, DP thymocytes revealed high levels of intracellular MR1 (Fig. 6B), and thymocytes from TCRα-deficient mice (that are arrested at the DP stage) robustly stimulated MAIT cell hybridomas in the presence of the trapping mAb (Supplemental Table II). Thus, DP thymocytes may be important APCs for MAIT cell development, establishing another parallel with iNKT cells. To further characterize physiological APCs for MAIT cells in the periphery, we used the same approach to coculture the MAIT cell hybridoma 6C2 with splenocytes, macrophages, or dendritic cells (DCs). The results showed that although APCs alone did not activate 6C2, they readily activated 6C2 in an MR1-specific manner in the presence of mAb 8H9.D11 (Fig. 6C). These combined results suggested that mAb 8H9.D11 has the unique ability to trap MR1 molecules transiently expressed on APCs to activate MAIT cells. Therefore, DP thymocytes may be physiological APCs selecting MAIT cells in their thymic development, whereas B cells, macrophages, and DCs may be responsible for the expansion and/or activation of MAIT cells in the periphery.
FIGURE 6.
Thymocytes, macrophages, and DCs express endogenous MR1 and activate MAIT cell hybridomas in the presence of the enhancing mAb. Thymocytes from wild-type (WT) and MR1 KO mice were used as APCs in the coculture with the MAIT cell hybridoma 6C2 in the absence or presence of anti-MR1 mAb 8H9.D11. A, The IL-2 production of 6C2 was measured by ELISA. The plot represents data from one of three independent experiments that were performed using one mouse for each genotype and set up in triplicate. B, Thymocytes were stained for surface expression of CD4 and CD8, followed by intracellular staining of mAb 8F2.F9 for MR1. In the graph (left panel), the blue line indicates the staining in WT thymocytes; the black line indicates the background in MR1 KO thymocytes. Right panel, MR1+ WT thymocytes were mostly CD4+CD8+. FACS analysis represents staining results of one mouse for each genotype in one representative experiment. C, Total cells (RBCs lysed), macrophages, and DCs from the spleen of WT mice were used as APCs to activate 6C2 in the presence of mAb 8H9.D11 using the same method described in A.
Discussion
We showed in this study using mAb trapping that low levels of MR1 exit the ER and transit to the plasma membrane. Furthermore, these mAb-trapped surface MR1 proteins activated mouse MAIT cell hybridomas. Under basal conditions, surface MR1 was not detected, except at very low levels on certain B cell lines that were incapable of activation of MAIT hybridoma cells without mAb trapping. Based on these findings, we speculate that bacterial infection might alter the intracellular trafficking and/or ligand binding of endogenous MR1, allowing it to accumulate at the cell surface at sufficient levels to activate MAIT cells. Although the MR1 ligands have not been identified, there is accumulating evidence suggesting that MR1 has an Ag-presentation function (6). This evidence was based initially on a limited number of mouse MAIT cell hybridomas that can be activated by MR1-over-expressing cell lines (13). Although the chemical nature of the ligand bound by MR1 has not been identified, a <10-kDa eluate from recombinant MR1 can activate MAIT cells in an MR1-dependent manner (7). Furthermore, extensive mutagenesis analyses suggested that MR1 binds a putative ligand and engages the TCR in a manner similar to classical MHC class I molecules (13). In addition, all other αβ T cells studied thus far detect ligand bound to MHC or MHC-like structures.
The in vivo development of MR1-dependent MAIT cells was shown to be TAP independent, and the in vitro activation of the above-mentioned mouse MAIT hybridoma cells by MR1-overexpressing cell lines was TAP independent (12, 22). Similarly, the activation of human MAIT cells by bacterial-infected cells was TAP independent (19). Interestingly, mMR1 molecules were detected in association with Ii, and this association promoted endosomal trafficking and MAIT hybridoma cell activation (22). The endosomal trafficking is of interest in light of the fact that certain bacteria reside in endocytic compartments. Despite this consensus agreement of TAP independence, the source and the chemical nature of a putative MR1 ligand remain undefined. Regarding MR1 ligand source, the ability of subpopulations of mouse and human MAIT cells to be activated by MR1-overexpressing cell lines suggests that MR1 may present a self (nonmicrobial)-ligand to MAIT cells (7). Alternatively, human MAIT cells with antibacterial activity were activated by an M. tuberculosis cell wall fraction (19). Mouse MAIT cells with antibacterial activity were reported to detect a conserved exogenous Ag after coculture with infected cells (20). The chemical nature of the MR1 ligand also remains unclear. The self ligand(s) capable of activating certain mouse MAIT cell hybridomas were not sensitive to proteasome and protease (7, 22), whereas activation of antibacterial human MAIT cells by the M. tuberculosis cell wall fraction was reported to be protease sensitive (19). Thus, MR1 activation of MAIT cells may be similar to CD1d activation of NKT cells, in that both present a variety of self and microbial-derived ligands (32). However, also like NKT cells, MAIT cells probably do little ligand discrimination based on their conserved TCR expression and the highly cross-reactive nature of microbial-infected cells (19, 20, 22, 33, 34).
Prior to this study, expression of endogenous MR1 was not detected at the cell surface. Indeed, we have now repeated this finding with an extensive panel of anti-MR1–reactive mAbs and different tumor and primary cells. Furthermore, little, if any, surface MR1 is detected serologically on bacterial-infected cells, suggesting that only a low level of MR1 loaded with appropriate Ags is required for MAIT cell activation. Based on these findings, it is attractive to speculate that constitutive MR1 expression at the cell surface under nonstress conditions could be detrimental. Such a model would be similar to how MHC class I chain-related A/B has been proposed to regulate immune responses (35). Perhaps the simplest way to induce stable expression of MR1 would be to improve the supply or quality of ligand, similar to the case with H2-M3 (14). Although plasma membrane recycling may be important for MR1 acquiring an antigenic ligand, it should be noted that the cytoplasmic tails of MR1 do not contain known sorting tyrosine or dileucine motifs.
In summary, MR1 activation of MAIT cells seems to require only a low level of surface expression, even after bacterial infection. This property has made it very difficult to study several questions about MR1 and MAIT cell biology, including “What are the physiologic signals that induce surface MR1 expression?”, Which APCs in mucosal tissues activate MAIT cells?”, and “What are the antigenic MR1 ligands required for MAIT cell activation and are they of self and/or bacterial origin?” The ability of the unique anti-MR1 mAb to stabilize surface MR1 reported in this article should greatly aid in the resolution of these outstanding questions of the physiological role of MAIT cells in microbial infection.
Supplementary Material
Acknowledgments
We thank Drs. Barry Sleckman and Paul Allen for providing B cell lines, Dr. Olivier Lantz for providing the two MAIT cell hybridomas, Dr. Susan Gilfillan for advice and discussion, Dr. Xiaoli Wang for discussion and technical support, and Dr. Janet Connolly for reviewing the manuscript.
This work was supported by grants from the National Institutes of Health (AI0465530), the Midwest Regional Center of Excellence for Biodefense and Emerging Infectious Disease Research, and McDonnell International Scholars Academy (Washington University).
Abbreviations used in this article
- CH27.mMR1
CH27 cells overexpressing mouse MHC-related protein 1
- DC
dendritic cell
- DP
double positive
- ER
endoplasmic reticulum
- 8F2.F9-AF647
mAb 8F2.F9 directly conjugated with Alexa Fluor 647
- iNKT
invariant NKT
- KO
knockout
- β2m
β2-microglobulin
- MAIT
mucosal-associated invariant T
- mMR1
mouse MHC-related protein 1
- MR1
MHC-related protein 1
Footnotes
The online version of this article contains supplemental material.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Hashimoto K, Hirai M, Kurosawa Y. A gene outside the human MHC related to classical HLA class I genes. Science. 1995;269:693–695. doi: 10.1126/science.7624800. [DOI] [PubMed] [Google Scholar]
- 2.Riegert P, Wanner V, Bahram S. Genomics, isoforms, expression, and phylogeny of the MHC class I-related MR1 gene. J Immunol. 1998;161:4066–4077. [PubMed] [Google Scholar]
- 3.Parra-Cuadrado JF, Navarro P, Mirones I, Setién F, Oteo M, Martínez-Naves E. A study on the polymorphism of human MHC class I-related MR1 gene and identification of an MR1-like pseudogene. Tissue Antigens. 2000;56:170–172. doi: 10.1034/j.1399-0039.2000.560211.x. [DOI] [PubMed] [Google Scholar]
- 4.Yamaguchi H, Hirai M, Kurosawa Y, Hashimoto K. A highly conserved major histocompatibility complex class I-related gene in mammals. Biochem Biophys Res Commun. 1997;238:697–702. doi: 10.1006/bbrc.1997.7379. [DOI] [PubMed] [Google Scholar]
- 5.Yamaguchi H, Kurosawa Y, Hashimoto K. Expanded genomic organization of conserved mammalian MHC class I-related genes, human MR1 and its murine ortholog. Biochem Biophys Res Commun. 1998;250:558–564. doi: 10.1006/bbrc.1998.9353. [DOI] [PubMed] [Google Scholar]
- 6.Hansen TH, Huang S, Arnold PL, Fremont DH. Patterns of nonclassical MHC antigen presentation. Nat Immunol. 2007;8:563–568. doi: 10.1038/ni1475. [DOI] [PubMed] [Google Scholar]
- 7.Huang S, Martin E, Kim S, Yu L, Soudais C, Fremont DH, Lantz O, Hansen TH. MR1 antigen presentation to mucosal-associated invariant T cells was highly conserved in evolution. Proc Natl Acad Sci USA. 2009;106:8290–8295. doi: 10.1073/pnas.0903196106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Treiner E, Lantz O. CD1d- and MR1-restricted invariant T cells: of mice and men. Curr Opin Immunol. 2006;18:519–526. doi: 10.1016/j.coi.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 9.Gozalbo-López B, Gómez del Moral M, Campos-Martín Y, Setién F, Martín P, Bellas C, Regueiro JR, Martínez-Naves E. The MHC-related protein 1 (MR1) is expressed by a subpopulation of CD38+, IgA+ cells in the human intestinal mucosa. Histol Histopathol. 2009;24:1439–1449. doi: 10.14670/HH-24.1439. [DOI] [PubMed] [Google Scholar]
- 10.Goldfinch N, Reinink P, Connelley T, Koets A, Morrison I, Van Rhijn I. Conservation of mucosal associated invariant T (MAIT) cells and the MR1 restriction element in ruminants, and abundance of MAIT cells in spleen. Vet Res. 2010;41:62. doi: 10.1051/vetres/2010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miley MJ, Truscott SM, Yu YY, Gilfillan S, Fremont DH, Hansen TH, Lybarger L. Biochemical features of the MHC-related protein 1 consistent with an immunological function. J Immunol. 2003;170:6090–6098. doi: 10.4049/jimmunol.170.12.6090. [DOI] [PubMed] [Google Scholar]
- 12.Treiner E, Duban L, Bahram S, Radosavljevic M, Wanner V, Tilloy F, Affaticati P, Gilfillan S, Lantz O. Selection of evolutionarily conserved mucosal-associated invariant T cells by MR1. Nature. 2003;422:164–169. doi: 10.1038/nature01433. [DOI] [PubMed] [Google Scholar]
- 13.Huang S, Gilfillan S, Cella M, Miley MJ, Lantz O, Lybarger L, Fremont DH, Hansen TH. Evidence for MR1 antigen presentation to mucosal-associated invariant T cells. J Biol Chem. 2005;280:21183–21193. doi: 10.1074/jbc.M501087200. [DOI] [PubMed] [Google Scholar]
- 14.Chiu NM, Chun T, Fay M, Mandal M, Wang CR. The majority of H2-M3 is retained intracellularly in a peptide-receptive state and traffics to the cell surface in the presence of N-formylated peptides. J Exp Med. 1999;190:423–434. doi: 10.1084/jem.190.3.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tilloy F, Treiner E, Park SH, Garcia C, Lemonnier F, de la Salle H, Bendelac A, Bonneville M, Lantz O. An invariant T cell receptor alpha chain defines a novel TAP-independent major histocompatibility complex class Ib-restricted alpha/beta T cell subpopulation in mammals. J Exp Med. 1999;189:1907–1921. doi: 10.1084/jem.189.12.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin E, Treiner E, Duban L, Guerri L, Laude H, Toly C, Premel V, Devys A, Moura IC, Tilloy F, et al. Stepwise development of MAIT cells in mouse and human. PLoS Biol. 2009;7:e54. doi: 10.1371/journal.pbio.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawachi I, Maldonado J, Strader C, Gilfillan S. MR1-restricted V alpha 19i mucosal-associated invariant T cells are innate T cells in the gut lamina propria that provide a rapid and diverse cytokine response. J Immunol. 2006;176:1618–1627. doi: 10.4049/jimmunol.176.3.1618. [DOI] [PubMed] [Google Scholar]
- 18.Treiner E, Duban L, Moura IC, Hansen T, Gilfillan S, Lantz O. Mucosal-associated invariant T (MAIT) cells: an evolutionarily conserved T cell subset. Microbes Infect. 2005;7:552–559. doi: 10.1016/j.micinf.2004.12.013. [DOI] [PubMed] [Google Scholar]
- 19.Gold MC, Cerri S, Smyk-Pearson S, Cansler ME, Vogt TM, Delepine J, Winata E, Swarbrick GM, Chua WJ, Yu YY, et al. Human mucosal associated invariant T cells detect bacterially infected cells. PLoS Biol. 2010;8:e1000407. doi: 10.1371/journal.pbio.1000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Le Bourhis L, Martin E, Péguillet I, Guihot A, Froux N, Coré M, Lévy E, Dusseaux M, Meyssonnier V, Premel V, et al. Antimicrobial activity of mucosal-associated invariant T cells. [Published erratum appears in 2010 Nat. Immunol. 11: 969.] Nat Immunol. 2010;11:701–708. doi: 10.1038/ni.1890. [DOI] [PubMed] [Google Scholar]
- 21.Pretell J, Greenfield RS, Tevethia SS. Biology of simian virus 40 (SV40) transplantation antigen (TrAg). V In vitro demonstration of SV40 TrAg in SV40 infected nonpermissive mouse cells by the lymphocyte mediated cytotoxicity assay. Virology. 1979;97:32–41. doi: 10.1016/0042-6822(79)90370-2. [DOI] [PubMed] [Google Scholar]
- 22.Huang S, Gilfillan S, Kim S, Thompson B, Wang X, Sant AJ, Fremont DH, Lantz O, Hansen TH. MR1 uses an endocytic pathway to activate mucosal-associated invariant T cells. J Exp Med. 2008;205:1201–1211. doi: 10.1084/jem.20072579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Philipps B, Forstner M, Mayr LM. Baculovirus expression system for magnetic sorting of infected cells and enhanced titer determination. Biotechniques. 2004;36:80–83. doi: 10.2144/04361ST03. [DOI] [PubMed] [Google Scholar]
- 24.Kokuho T, Watanabe S, Yokomizo Y, Inumaru S. Production of biologically active, heterodimeric porcine interleukin-12 using a monocistronic baculoviral expression system. Vet Immunol Immunopathol. 1999;72:289–302. doi: 10.1016/s0165-2427(99)00141-5. [DOI] [PubMed] [Google Scholar]
- 25.Tessier DC, Thomas DY, Khouri HE, Laliberté F, Vernet T. Enhanced secretion from insect cells of a foreign protein fused to the honeybee melittin signal peptide. Gene. 1991;98:177–183. doi: 10.1016/0378-1119(91)90171-7. [DOI] [PubMed] [Google Scholar]
- 26.Hansen TH, Lybarger L, Yu L, Mitaksov V, Fremont DH. Recognition of open conformers of classical MHC by chaperones and monoclonal antibodies. Immunol Rev. 2005;207:100–111. doi: 10.1111/j.0105-2896.2005.00315.x. [DOI] [PubMed] [Google Scholar]
- 27.Miettinen HM, Rose JK, Mellman I. Fc receptor isoforms exhibit distinct abilities for coated pit localization as a result of cytoplasmic domain heterogeneity. Cell. 1989;58:317–327. doi: 10.1016/0092-8674(89)90846-5. [DOI] [PubMed] [Google Scholar]
- 28.Miettinen HM, Matter K, Hunziker W, Rose JK, Mellman I. Fc receptor endocytosis is controlled by a cytoplasmic domain determinant that actively prevents coated pit localization. J Cell Biol. 1992;116:875–888. doi: 10.1083/jcb.116.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ortiz-Navarrete V, Hämmerling GJ. Surface appearance and instability of empty H-2 class I molecules under physiological conditions. Proc Natl Acad Sci USA. 1991;88:3594–3597. doi: 10.1073/pnas.88.9.3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bendelac A. Positive selection of mouse NK1+ T cells by CD1-expressing cortical thymocytes. J Exp Med. 1995;182:2091–2096. doi: 10.1084/jem.182.6.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wei DG, Lee H, Park SH, Beaudoin L, Teyton L, Lehuen A, Bendelac A. Expansion and long-range differentiation of the NKT cell lineage in mice expressing CD1d exclusively on cortical thymocytes. J Exp Med. 2005;202:239–248. doi: 10.1084/jem.20050413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mattner J, Debord KL, Ismail N, Goff RD, Cantu C, III, Zhou D, Saint-Mezard P, Wang V, Gao Y, Yin N, et al. Exogenous and endogenous glycolipid antigens activate NKT cells during microbial infections. Nature. 2005;434:525–529. doi: 10.1038/nature03408. [DOI] [PubMed] [Google Scholar]
- 33.Mallevaey T, Scott-Browne JP, Matsuda JL, Young MH, Pellicci DG, Patel O, Thakur M, Kjer-Nielsen L, Richardson SK, Cerundolo V, et al. T cell receptor CDR2 beta and CDR3 beta loops collaborate functionally to shape the iNKT cell repertoire. Immunity. 2009;31:60–71. doi: 10.1016/j.immuni.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pellicci DG, Patel O, Kjer-Nielsen L, Pang SS, Sullivan LC, Kyparissoudis K, Brooks AG, Reid HH, Gras S, Lucet IS, et al. Differential recognition of CD1d-alpha-galactosyl ceramide by the V beta 8.2 and V beta 7 semi-invariant NKT T cell receptors. Immunity. 2009;31:47–59. doi: 10.1016/j.immuni.2009.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Groh V, Bahram S, Bauer S, Herman A, Beauchamp M, Spies T. Cell stress-regulated human major histocompatibility complex class I gene expressed in gastrointestinal epithelium. Proc Natl Acad Sci USA. 1996;93:12445–12450. doi: 10.1073/pnas.93.22.12445. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.