Abstract
Acs2p is one of two acetyl-coenzyme A synthetases in Saccharomyces cerevisiae. We have prepared and characterized a monoclonal antibody specific for Acs2p and find that Acs2p is localized primarily to the nucleus, including the nucleolus, with a minor amount in the cytosol. We find that Acs2p is required for replicative longevity: an acs2Δ strain has a reduced replicative life span compared to wild-type and acs1Δ strains. Furthermore, replicatively aged acs2Δ cells contain elevated levels of extrachromosomal rDNA circles, and silencing at the rDNA locus is impaired in an acs2Δ strain. These findings indicate that Acs2p-mediated synthesis of acetyl-CoA in the nucleus functions to promote rDNA silencing and replicative longevity in yeast.
Keywords: Acetyl-coenzyme A, Aging, Nucleus, Saccharomyces cerevisiae
Introduction
Acetyl-coenyzme A (acetyl-CoA) synthetase (ACS) enzymes play a central role in cellular metabolism by regulating acetate utilization. Two types of ACS enzymes are known: AMP forming and ADP forming. AMP-forming enzymes have a wide distribution in nature and are present in eubacteria, archaea, and eukaryotes, including fungi, plants, and animals [1]. All catalyze the reaction: acetate + CoA + ATP → acetyl-CoA + AMP + PPi. Acetyl-CoA formed in this manner is used for biosynthesis and energy production. For example, this reaction is required for yeast to utilize ethanol as the sole carbon source for growth, which is an important aspect of the metabolic strategy yeast employ to protect sugar resources in their environment [2].
Eukaryotes usually have two AMP-forming ACS isoforms that are specifically localized in the cytosol and the mitochondrion. Cytosolic ACS produces acetyl-CoA that is utilized for biosynthetic processes, such as fatty acid synthesis in the endoplasmic reticulum. Mitochondrial ACS synthesizes acetyl-CoA for delivery to the tricarboxylic acid and glyoxylate cycles. In yeast, ACS isozymes encoded by ACS1 and ACS2 are known as the “aerobic” (gluconeogenic) and “anerobic” (glycolytic) isoforms, respectively [3]. Consistent with this, ACS1 expression is subject to glucose repression, making Acs2p the primary enzyme that is active during growth in glucose-containing media [3]. Until recently, Acs1p and Acs2p were thought to function in mitochondria and the cytosol, respectively, based on cell fractionation studies [4-6]. However, recent studies suggest that the majority of Acs2p is localized to the nucleus [7, 8], where it has been implicated in synthesis of acetyl-CoA for use in histone acetylation and silencing [8], which are known to play an important role in determining yeast replicative life span (RLS) [9].
We have investigated the localization of Acs2p and its role in RLS. A novel monoclonal antibody specific for Acs2p demonstrates nuclear localization of Acs2p using cell fractionation and immunofluorescence localization approaches. The RLS of an acs2Δ null strain is significantly shorter than that of wild-type (WT) and acs1Δ strains. Consistent with this, levels of extrachromosomal rDNA circles (ERCs) are elevated in seven generation old acs2Δ cells, and silencing at the rDNA locus is impaired in an acs2Δ strain. We also investigated modification of Acs2p by acetylation at lysine-637 based on previous studies [10-12], but found no evidence for this modification. These findings broaden our understanding of yeast Acs2p function to include roles in silencing at the rDNA locus and in promoting replicative longevity in yeast.
Materials and methods
Yeast strains
T2-3D (HO/HO ACS1/ACS1 ACS2/ACS2) is a homozygous diploid strain [13], from which strains GG621 (HO/HO acs1∷APT1/acs1∷APT1 ACS2/ACS2) and GG625 (HO/HO ACS1/ACS1 acs2∷Tn5ble/acs2∷Tn5ble) were derived [14]. The following strains were obtained from EUROSCARF: BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) hoΔ∷KanMX4, BY4741 acs1Δ∷KanMX4, BY4741 sir2Δ∷KanMX4, and BY4743 (MATa/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 lys2Δ0/LYS2 MET15/met15Δ0 ura3Δ0/ura3Δ0) ACS2/acs2Δ∷KanMX4. Yeast were grown in YPD-, YPE-, or YPAG-media containing 2% glucose, 2% ethanol, or 1% potassium acetate plus 1% glycerol, respectively [15]. The aminoglycoside antibiotic G418 sulfate (Geneticin) was used at a final active concentration of 250 μg/ml.
Strains YJPA56, YJPA57, YJPA58, and YJPA59 were prepared by transforming BY4741 hoΔ (WT), BY4741 acs1Δ, BY4741 sir2Δ, and BY4743 acs2Δ/ACS2, respectively, with plasmid pJPA118 that was linearized with SfiI. pJPA118 contains a complete 9.1 kb rDNA repeat (with XmaI endpoints) and was constructed by inserting an XhoI-BamHI fragment from pJPA105 [16] into pRS306 [17] digested with XhoI and BamHI. YJPA59 was sporulated and dissected on YPAG agar. Heterogeneity in haploid colony size was observed following dissection of YJPA59 tetrads, which may reflect the presence of uncharacterized mutations in the ACS2/acs2Δ strain. One of the large Ura+ G418 resistant acs2Δ haploid colonies was isolated as YJPA59 and used for the pin stamp experiment in Fig. 5.
Monoclonal antibody production and library screening
Monoclonal antibody (mAb) 40E10 was generated against a nucleolus-enriched fraction prepared as described [18]. Mouse immunization, hybridoma cell culture, and antibody production were done using standard methods as described [19, 20] in conjunction with the Hybridoma Core Laboratory at the University of Florida Interdisciplinary Center for Biotechnology Research (UF ICBR). A yeast genomic library (Clontech) in the expression vector λgt11 was screened using standard techniques with mAb 40E10 ascites fluid diluted 1/1000 as described [19, 20]. λ DNAs from positive clones were amplified by PCR from purified plaques and sequenced using primers flanking the EcoRI site as described [19]. Escherichia coli strain Y1089 was lysogenized with a positive clone and used to prepare a protein lysate for SDS-PAGE following induction of protein expression with 1 mM isopropyl thiogalactopyranoside.
Gel electrophoresis and blotting
Protein extracts from whole cells and subcellular fractions were prepared as described [18] and separated by SDS-PAGE using Criterion gels (Bio-Rad Laboratories). Western blots were prepared by semidry transfer to nitrocellulose membranes [21] and probed with mAb 40E10 ascites fluid diluted 1/10,000 followed by ECL detection [22].
DNA was extracted from yeast cells using a glass beads/phenol method, digested with PstI (New England Biolabs), analyzed in 1% agarose gels, and capillary transferred to positively charged nylon membrane under alkaline conditions as described [16]. 32P-labeled probe was generated by random-primed labeling (New England Biolabs) using a 9.1 kb rDNA repeat as template [16]. Southern data were acquired with a Typhoon 9400 PhosphorImager using ImageQuant software (Amersham Biosciences).
Immunofluorescence
Immunofluorescence localization was done as described [19, 20] using cell culture supernatant diluted 1/5 and ascites fluid diluted 1/500. Affinity purified polyclonal antibody against Nop2p (APpAb3) [23] was diluted 1/40. Secondary Cy2-conjugated antirabbit and Cy3-conjugated antimouse antibodies (Jackson ImmunoResearch Laboratories) were diluted 1/200. DAPI (4′,6-diamidino-2-phenylindole) was present in mounting medium at a final concentration of 0.1 μg/ml. mAb 32D6 against Nsp1p and related nucleoporins was diluted 1/1000. Digital images were acquired with a Zeiss Axiophot microscope equipped with a CoolSnap HQ CCD camera (Photometrics) and IP Lab software (Scanalytics). Optical sections were acquired using a Leica DMR microscope and computationally processed to remove out-of-focal-plane light.
Immunoprecipitation
Cells in IP buffer (50 mM Tris–HCl, pH 8, 150 mM NaCl, 1 mM EDTA, 0.5% Nonidet-P40) plus 5 mM sodium butyrate, 10 μM trichostatin A, 5 mM nicotinamide, 1 mM DTT, and protease inhibitors [18] were vortexed with 0.5 mm acid-washed glass beads for 30 min at 4°C. Lysates were clarified by centrifugation for 5 min at 13,200 rpm (16,100gmax) and added to 20 μl of Protein G-Sepharose beads (Pharmacia) with bound mAb 40E10. After an overnight binding step, beads were washed five times with IP buffer, once with IP buffer lacking NP-40, resuspended in SDS sample buffer, and boiled 5 min. Proteins were separated in a 10.5–14% Criterion gel (Bio-Rad Laboratories), and coomassie blue stained Acs2p bands were excised, reduced with DTT, alkylated with iodoacetamide, and digested in-gel to yield peptides that were analyzed by the mass spectrometry (MS) core facility at the UF ICBR.
Replicative aging methods
Replicative life span determinations were done by micro-dissection using a Zeiss Axiolab microscope as described [16]. Replicatively aged yeast cells were obtained following biotinylation, overnight growth in rich medium, and sorting with streptavidin magnetic beads as described [16].
Results
Monoclonal antibody 40E10 is specific for Acs2p
During the preparation of monoclonal antibodies to a nucleolus-enriched fraction, we identified an antibody, 40E10, that exhibited strong reactivity toward a nuclear antigen in the primary round of screening using immunofluorescence (Fig. 1a). mAb 40E10 recognizes a single protein with an apparent molecular mass of ~75 kDa in whole cell lysates and in fractions highly enriched in nuclei or nucleoli (Fig. 1b). The nuclear and nucleolar fractions used in this analysis have been extensively characterized [18, 19, 24]. Nucleoli are prepared by digestion of nuclei with DNase I in buffer containing a low Mg++ concentration, followed by treatment with heparin, which reduces the amounts of DNA and histones in the nucleolus-enriched fraction [18]. This suggests that the association of Acs2p with the nucleolus may not be entirely dependent on the presence of chromatin in the nucleolus.
Fig. 1. Characterization of mAb 40E10.
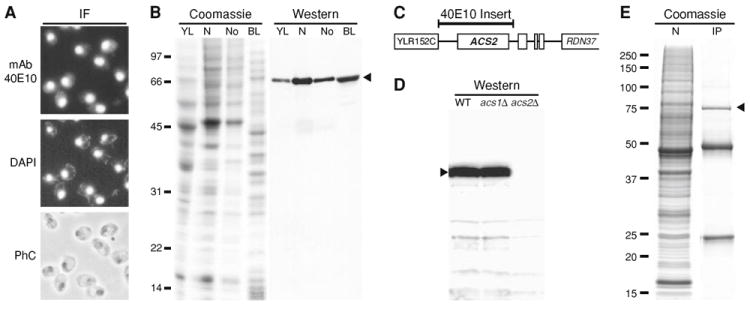
a Immunofluorescence (IF) localization with hybridoma supernatant. Staining with mAb 40E10 or DAPI or visualization by phase contrast (PhC) is shown. b Cell fractionation analysis. A whole cell yeast lysate (YL), isolated nuclei (N), isolated nucleoli (No), or a bacterial lysate (BL) from a λ lysogen were subject to SDS-PAGE, followed by coomassie staining or western blotting. Approximately 200 μg of total protein was loaded in each lane. c Summary of results from screening a λgt11 expression library with mAb 40E10. The position of the ~3.1 kb insert in a positive library clone is indicated by the bar over a region of chromosome XII near the rDNA repeat (RDN37) (not to scale). d Western blotting with mAb 40E10 of whole cell extracts from strains T2-3D (WT), GG621 (acs1Δ), and GG625 (acs2Δ). e Immunoprecipitation (IP) with mAb 40E10 from a yeast nuclear fraction (N) was followed by SDS-PAGE and coomassie staining. Lane N represents ~5% of the total used for lane IP. Immunoglobulin heavy and light chains migrate at ~50 and ~25 kDa, respectively. Positions of molecular weight markers are shown (in kDa). Arrowheads point to bands of ~75 kDa
In order to characterize the 40E10 antigen, we screened an expression library containing yeast genomic DNA inserts in λgt11. Three positive clones were identified and fell into two overlapping classes based on restriction analysis. DNA sequencing showed that the largest positive λ clone contained a ~3.1 kb region of chromosome XII near the rDNA repeat (approximate coordinates 444700 to 447800) (Fig. 1c). This region contains the entire ACS2 open reading frame on the Crick strand (i.e., the library clone did not encode a β-galactosidase fusion protein). Expression of full-length Acs2p was confirmed by analyzing a protein lysate from an E. coli lysogen carrying the clone with the ~3.1 kb insert. The ~75 kDa protein expressed by the λ lysogen co-migrated with protein detected in yeast (Fig. 1b). Acs2p has a predicted molecular weight of 75,491 Da, which is in excellent agreement with the apparent size of ~75 kDa on SDS gels.
Acs1p and Acs2p in S. cerevisiae are 56% identical (82% similar) in a FASTA alignment over the entire length of Acs2p (683 amino acids) (data not shown). Given this similarity, we determined if 40E10 specifically recognized Acs2p by carrying out immunoblotting, immunoprecipitation, and immunofluorescence experiments using acs1Δ and acs2Δ deletion strains. Immunoblotting shows that 40E10 recognizes a ~75 kDa band in whole cell extracts from WT and acs1Δ strains, but not from an acs2Δ strain (Fig. 1d). A band of 75 kDa was also immunoprecipitated by 40E10 from extracts prepared from isolated nuclei (Fig. 1e) and whole cells (data not shown). The 75 kDa protein was identified by MS as Acs2p, with over 50% coverage of predicted tryptic peptides (data not shown). No Acs1p was detected in the MS analyses (data not shown). Thus, mAb 40E10 is specific for Acs2p and shows no detectable cross reactivity with Acs1p.
Acs2p is localized primarily to the nucleus
Preliminary immunofluorescence data indicated that mAb 40E10 reacted with a nuclear antigen (Fig. 1a), but did not shed light on the intranuclear localization of Acs2p. Thus, we studied the intracellular localization of Acs2p in greater detail. Immunofluorescence showed that 40E10 detects a predominantly nuclear signal in WT and acs1Δ strains, but not in the acs2Δ strain (Fig. 2a, e, i). Long exposures of acs2Δ cells shown in Fig. 2i showed only background levels of staining comparable to control samples not incubated with primary antibody (data not shown). Furthermore, in some cells, nuclei are oriented in such a manner that the nucleoplasm (stained with DAPI) can be seen on one side of the nucleus, and the nucleolus (stained with anti-Nop2p antibody) can be seen on the other side. In these cells, the nucleolus appears crescent shaped or semicircular. In order to illustrate this relationship, pairs of arrowheads are used in Fig. 2 to denote side-by-side positioning of nucleoplasm and nucleolus. Comparison of the 40E10 staining pattern to positions of the nucleoplasm and nucleoli reveals that Acs2p is localized in both the nucleoplasm and nucleolus (Fig. 2a, e, m).
Fig. 2. Intracellular localization of Acs2p.
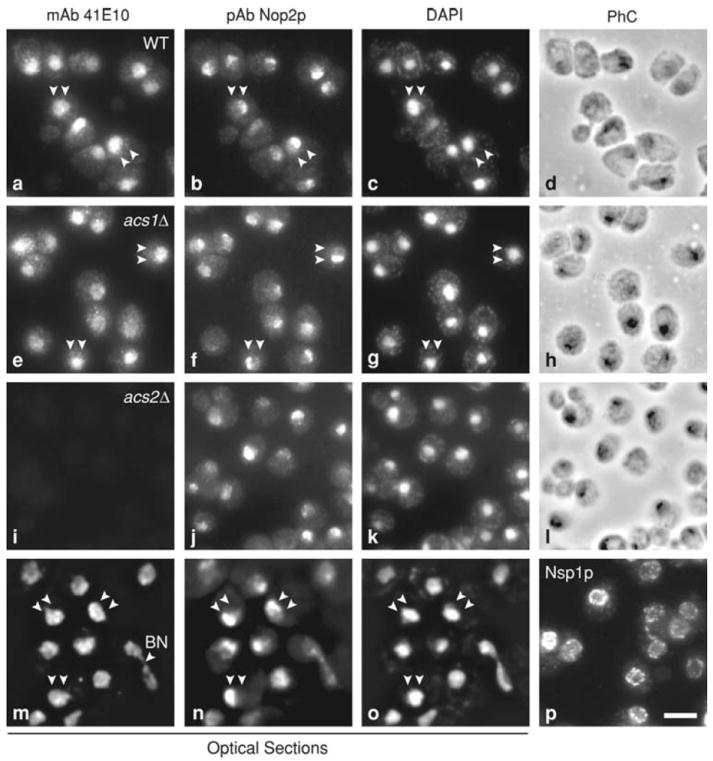
MAb 40E10 was used for immunofluorescence localization of Acs2p in wild-type (WT), acs1Δ, and acs2Δ strains (a, e, i, m). Cells were double immunostained with affinity purified polyclonal antibody (pAb) against the nucleolar protein Nop2p (b, f, j, n) and counter stained with the DNA-binding dye DAPI (c, g, k, o). Panels d, h, and l show phase contrast images (PhC). The same fields of cells are shown in a–d, e–h, i–l, and m–o. Panels m–o are images of 0.2 μm optical sections through WT cells processed with “deconvolution” software to minimize out-of-focus light. Nuclear pore complexes were revealed with mAb 32D6 (p). Bar = 5 μm. All panels are shown at the same final magnification
In order to better resolve the intranuclear localization of Acs2p, we examined optical sections collected with a “deconvolution” microscope. Optical sections through the middle of the nucleus reveal that Acs2p is distributed in a somewhat heterogeneous pattern in the nuclear interior in both the nucleoplasm and nucleolus, as expected (Fig. 2m). Importantly, there is no evidence of colocalization of Acs2p with mitochondria, which contain nucleoids that are stained with DAPI (see punctate cytoplasmic staining in Fig. 2c, g, k, o).
The intranuclear staining pattern appears relatively uniform in some cells, but granular or punctate in other cells (Fig. 2a, e, m). This granular or punctate staining pattern is different from the punctate staining pattern associated with nuclear pore complexes, which are distributed at the nuclear periphery (Fig. 2p). Although the nuclear distribution pattern of Acs2p is different than the distribution of the pore complexes, we cannot rule out the possibility that some Acs2p is localized near or at the nuclear envelope. In some cells, Acs2p is partly localized in the cytosol, where a punctate or granular staining pattern is seen (Fig. 2a, e). The staining pattern of Acs2p does not change significantly over the course of the cell cycle. Unbudded cells in G1, small budded cells in S phase, and large budded cells in G2-M show similar staining patterns (Fig. 2). During G2-M, Acs2p colocalizes with the elongate nucleus that extends through the bud neck (Fig. 2m, BN, bud neck). Finally, the distribution of Acs2p appears the same in WT and acs1Δ cells (Fig. 2a, e), indicating that Acs1p is not required for nuclear localization of Acs2p.
ACS2 is required for a normal replicative life span
Nuclear localization of Acs2p lead us to consider the effects of nuclear acetyl-CoA synthesis on RLS. Acetyl-CoA is utilized by histone acetyltransferases during chromatin modification, which is known to regulate silencing and influence RLS [8, 9]. In order to analyze RLS, it was necessary to use an agar medium containing ethanol as the carbon source rather than glucose. acs2Δ strains are inviable on glucose media because ACS1 is glucose repressed and yeast require at least one functional acetyl-CoA synthetase (i.e., a double acs1Δ, acs2Δ mutant is inviable) [14]. Thus, the RLS of WT and mutant strains were determined using standard techniques on rich ethanol (YPE) medium.
The WT strain T2-3D exhibited a long RLS (Fig. 3a). The mean RLS for T2-3D was ~55 generations, making this strain one of the longest lived strains reported in the literature. In contrast, the acs2Δ strain GG625 had a comparatively short life span, with a mean RLS of ~17 generations (Fig. 3a). The acs1Δ strain GG621 had a mean RLS of ~47 generations on YPE medium, which was similar to strain T2-3D (Fig. 3a), but distinguishable from it by the Wilcoxon signed pair rank test (P < 0.05).
Fig. 3. Life span analysis.
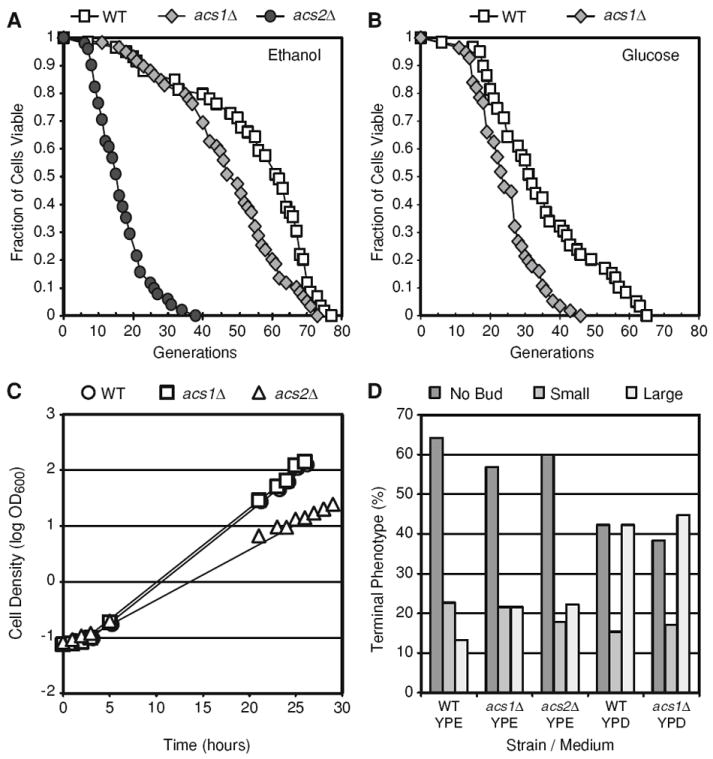
a and b Numbers of daughter cells (generations) produced per mother cell are plotted as a function of fraction of viable mother cells. The number of mother cells (n) analyzed on YPE medium equals 59, 59, and 51 for wild-type (WT), acs1Δ, and acs2Δ strains, respectively. The number of mother cells (n) analyzed on YPD medium equals 59 and 56 for WT and acs1Δ strains, respectively. All curves in each panel are distinguishable by Wilcoxon two-sample paired signed rank tests (P < 0.05). c Growth on YPE liquid medium. Log phase growth trend lines are shown (R2 > 0.99). d Analysis of cell cycle stage (“terminal phenotype”) of senescent cells. Mother cells (n ≥ 45 for each strain/medium) at the end of the life span were scored for bud presence and size as described [16, 25]
Given the long RLS of the WT and acs1Δ strains on medium-containing ethanol, we examined RLS on YPD medium containing 2% glucose, which is widely used for yeast RLS assays. The life spans of WT and acs1Δ strains were reduced by approximately 25–33% on YPD medium compared to YPE medium (Fig. 3a, b). The average RLS for T2-3D on YPD was ~35 generations, and the average RLS of the acs1Δ strain GG621 on YPD was ~23 generations. Thus, both WT and acs1Δ strains exhibited a shorter RLS on glucose-containing medium compared to ethanol-containing medium. This may be a result of a higher respiratory rate and/or upregulation of stress-response pathways on ethanol-containing medium.
One explanation for the short RLS of the acs2Δ mutant is that the absence of ACS2 leads to a physiological deficit that shortens life span. In order to evaluate this, we examined the growth rate and cell cycle phase at senescence (i.e., the “terminal cell phenotype”) of the acs2Δ strain. In YPE medium, the growth rate of the acs2Δ strain was 54% slower than the WT and acs1Δ strains (Fig. 3c). Specifically, the T2-3D (WT) and GG621 (acs1Δ) strains had log phase doubling times of 2.2 h, whereas the GG625 (acs2Δ) strain had a doubling time of 3.4 h. However, the acs2Δ strain achieved the same cell density at saturation in YPE medium as the WT and acs1Δ strains (OD600 = 1.7–1.8). In order to examine the cell cycle phase at senescence, the bud size was determined for each senescent cell in the RLS experiments (Fig. 3a, b). Bud size is a reliable predictor of cell cycle stage (i.e., no bud = G1, small bud = S, large bud = G2/M). Typically, a preponderance of senescent yeast is unbudded cells in G1 [25]. Consistent with this, we found that the majority of WT, acs1Δ, and acs2Δ senescent cells were unbudded on YPE medium (Fig. 3d). On YPD, WT and acs1Δ strains stopped dividing with roughly equal numbers of unbudded and large-budded cells (Fig. 3d). These data indicate that the acs2Δ mutant exhibits reduced fitness during log phase growth, but that growth to saturation and cell cycle progression prior to senescence in the acs2Δ strain is comparable to WT and acs1Δ strains.
Elevated levels of extrachromosomal rDNA circles (ERCs) in acs2Δ cells
The short RLS of the acs2Δ strain suggests that levels of ERCs may be elevated during replicative aging in acs2Δ cells compared to the more long-lived WT and acs1Δ strains. This expectation was confirmed by Southern blotting (Fig. 4). For this analysis, WT, acs1Δ, and acs2Δ strains were grown on YPE medium, and cells with average ages of 8.6, 7.8, and 7.4 generations, respectively, were isolated using a standard sorting method [16]. Young cells were isolated in parallel and had average ages of ~2 generations. ERC monomers and multimers were evident in ~7-generation-old acs2Δ cells, but not in ~8–9-generation-old WT and acs1Δ cells (Fig. 4, arrowheads), which is consistent with the short life span of the acs2Δ strain. The absence of ERCs in 8–9-generation-old WT and acs1Δ cells was unexpected, given that WT W303 strains contain detectable levels of ERCs at this age [16]. However, the T2-3D strain background is more long-lived than W303 (or BY) strains when grown on rich medium containing ethanol as a carbon source, and the absence of ERCs in WT and acs1Δ strains is consistent with this. As expected, ERCs were not detected in young cells from the different strains (Fig. 4).
Fig. 4. Southern blot analysis of DNA from young and replicatively aged cells.
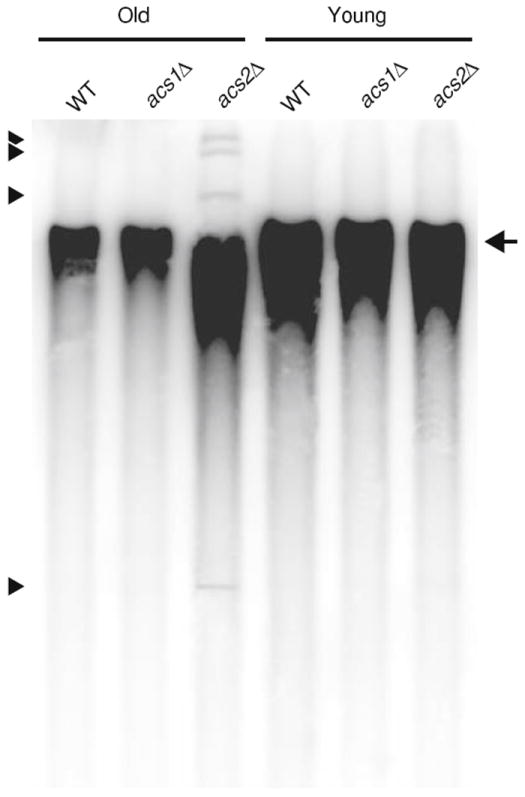
Yeast cells were isolated by biotinylation and magnetic sorting [16]. Bud scars were stained with calcofluor and counted to determine average replicative age. The average age of sorted cells from wild-type (WT), acs1Δ, and acs2Δ strains was 8.6, 7.8, and 7.4 generations, respectively. DNA was extracted using standard methods and analyzed by Southern blotting using an rDNA probe. Individual chromosomes are not resolved in this agarose gel and migrate as a single broad band (arrow). Monomeric extrachromosomal rDNA circles (ERCs) migrate faster than the chromosomal band, whereas multimeric ERCs migrate slower (arrowheads denote ERC bands)
Silencing of the rDNA locus is reduced in the absence of ACS2
The short RLS and presence of elevated levels of ERCs during replicative aging suggested that silencing at the rDNA locus was compromised in the acs2Δ mutant. In order to test this directly, reporter strains were generated by integrating plasmid pJPA118 containing the URA3 gene into the rDNA locus of WT, acs1Δ, acs2Δ, and sir2Δ strains (see Materials and methods). WT and acs1Δ strains containing rDNA∷pJPA118(URA3) grew poorly in the absence of uracil due to silencing of the integrated copy of URA3 (Fig. 5, −Uracil). For the same reason, WT and acs1Δ rDNA∷pJPA118(URA3) strains grew well on medium containing the drug 5-fluoro-orotic acid (Fig. 5, +FOA). In contrast, the rDNA∷pJPA118(URA3) acs2Δ strain grew well in the absence of uracil, but poorly in the presence of FOA (Fig. 5). sir2Δ strains are well known to exhibit reduced silencing at the rDNA locus, increased ERC production, and reduced RLS [26]. Because of this, a sir2Δ rDNA∷pJPA118(URA3) strain was used as a basis of comparison and found to grow in the absence of uracil, but fail to grow in the presence of FOA, as expected (Fig. 5). These data indicate that deletion of ACS2 impairs silencing at the rDNA locus and to an extent that is roughly equivalent to the impairment observed in a sir2Δ strain.
Fig. 5. Analysis of rDNA silencing.
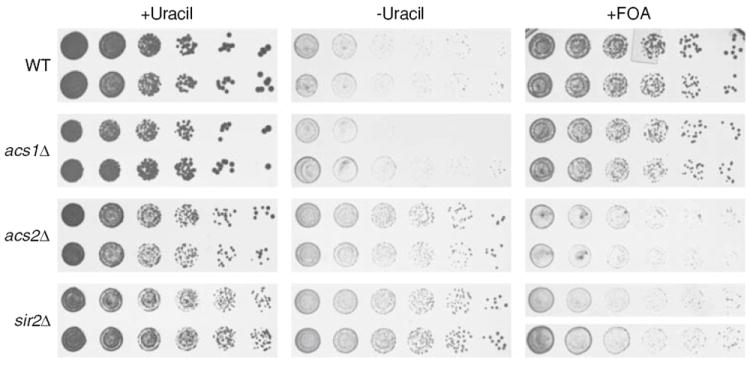
Wild-type (WT), acs1Δ, acs2Δ, and sir2Δ strains containing the URA3 gene integrated at the rDNA locus were grown in YPAG-rich medium containing acetate and glycerol as a carbon source. Fivefold serial dilutions were pin-stamped onto SAG minimal synthetic medium containing acetate, glycerol, G418, and either essential nutrients (+Uracil), essential nutrients lacking uracil (−Uracil) or essential nutrients plus 1 mg/ml 5-fluoro-orotic acid (+FOA). Grown results are shown for two independent transformants of each strain following incubation for 5–7 days at 30°C
No evidence for acetylation of Lysine-637 in Acs2p
The acetyl-coA synthetase 2 enzymes in S. enterica and mammalian mitochondria are post-translationally regulated by acetylation of a conserved lysine residue, and enzyme activation requires deacetylation by sirtuin family deacetylases. Lysine-637 (K637) in yeast Acs2p lies in a ~25 amino acid stretch that is conserved from bacteria to S. cerevisiae and aligns with the acetylated lysine (K609) in S. enterica Acs [27]. We investigated acetylation at K637 using mAb 40E10 to immunoprecipitate Asc2p with high specificity and yield (see Fig. 1e). Immunoprecipitations were done using single and multiple sirtuin gene knockout strains [28, 29] grown under a range of conditions to maximize Acs2p lysine acetylation (e.g., different carbon sources, with and without nicotinamide). Microgram amounts of Acs2p were immunoblotted with an anti-acetylated lysine antibody (Cell Signaling Technology mAb Ac-K-103), but only negative results were obtained (data not shown; nanogram quantities of acetylated BSA produced strong positive control signals). Acs2p was also analyzed by MS. Digestion with Asp-N, Lys-C, or trypsin yielded peptides that corresponded to 21–54% coverage of Acs2p, unequivocally identifying the immunoprecipitated protein (data not shown). However, only Asp-N permitted good detection of the region of interest around K637. Multiple forms of the Asp-N-generated peptide DLPRTRSGK637IMRRVLRKVASNEAEQLG were observed, including peptides due to cleavage on the N-terminal side of E650 and E652. In all cases, only the unacetylated form of this peptide was identified with confidence, including verification by manual interpretation of MS/MS spectra (Supplementary Figure and data not shown).
Discussion
Acetyl-CoA functions as an acetyl donor in numerous biosynthetic and regulatory pathways in cells. Among these is post-translational modification of proteins by acetylation, which has been implicated in regulating gene activity and aging [30]. We have prepared and characterized a monoclonal antibody against Acs2p and used it to investigate Acs2p localization and function. Cell fractionation and immunofluorescence studies indicate that Acs2p is localized primarily in the nucleus, including the nucleolus, with a minor amount in the cytosol. The nuclear distribution of Acs2p agrees with results from genome-wide protein localization studies and GFP-fusion protein studies that conclude that the majority of Acs2p is present in the nucleus [8, 31, 32]. Nuclear localization may explain one or more of the interactions between Acs2p and other nuclear proteins that have been reported in the literature [33-36]. Acs2p does not contain a canonical nuclear localization sequence (NLS). Thus, either Acs2p contains a non-canonical NLS that mediates direct transport to the nucleus or Acs2p is transported to the nucleus indirectly via an interaction with an NLS-containing nuclear protein. In addition, while its steady-state distribution is predominately nuclear, Acs2p may dynamically shuttle between the nucleus and cytoplasm.
The nuclear localization of Acs2p indicates that the nucleus is a site of active acetyl-CoA synthesis in yeast. This raises the interesting question: why is acetyl-CoA synthesized in the nucleus? Recent studies have shown that acetyl-CoA synthesized in the nucleus is required for histone acetylation and global transcriptional control [8, 37]. In addition, acetyl-CoA synthesized in the nucleus may be used for biosynthetic purposes. A number of enzymes involved in lipid synthesis have been localized to the nucleus, including the nuclear interior as well as the nuclear envelope [32]. For example, we have previously shown that the homocitrate synthase isozymes Lys20p and Lys21p are localized to the nucleus in yeast [19]. Lys20p and Lys21p carry out the first step in lysine biosynthesis, which requires acetyl-CoA. And, acetyl-CoA carboxylase (Acc1p) catalyzes the initial, acetyl-CoA consuming, rate-limiting step in fatty acid synthesis and is associated with the nuclear envelope and endoplasmic reticulum [38].
Our studies show that Acs2p is required for yeast replicative longevity. We initially hypothesized that the absence of Acs2p would extend RLS based on our understanding of how SIR2 regulates life span. Sir2p is an NAD+-dependent deacetylase that promotes longevity by maintaining silencing of the rDNA repeat cluster [39]. We hypothesized that deletion of ACS2 would reduce acetyl-CoA levels in the nucleus, which would promote silencing and extend RLS. However, contrary to this hypothesis, we found that an acs2Δ mutant had a short RLS. An acs1Δ mutant was also more short-lived than the WT strain, but not to the same extent as the acs2Δ mutant. Thus, extension of RLS was not observed in either mutant. Consistent with its effect on RLS, only the acs2Δ mutation lead to increased levels of ERCs in aging cells and impaired silencing at the rDNA locus. Given the intracellular localization of Acs2p, we conclude that nuclear acetyl-CoA synthetase activity is required for rDNA silencing and replicative longevity in yeast.
We note that the WT, acs1Δ, and acs2Δ strains used in our life span studies were in a diploid prototrophic genetic background. The lack of auxotrophies in these strains may promote longevity to some extent compared to more commonly used strains (e.g., W303, BY) carrying auxotrophic markers. However, diploidy in these strains probably does not contribute to longevity given that diploids have been found to be no more long lived than haploids [40].
Exactly how acetyl-CoA synthetase activity in the nucleus is functionally linked to rDNA silencing and replicative longevity remains an interesting question. Acetyl-CoA synthesized in the nucleus is used by histone acetyl-transferases to regulate gene expression [8], but reduced histone acetylation is predicted to extend RLS. This suggests that acetylation of other nuclear proteins involved in the regulation of silencing may be the mechanism by which Acs2p promotes longevity. The relatively small effect of the acs1Δ mutation on RLS may be related to retrograde signaling. Activation of the retrograde pathway is associated with replicative longevity in yeast [41], and ACS1 is known to be upregulated in respiration deficient yeast (ρ0) in a retrograde pathway dependent manner [42]. Thus, elimination of ACS1 may reduce the contribution of retrograde signaling to replicatively longevity. It is also possible that acetyl-CoA is utilized for biosynthetic steps that play a role in silencing and longevity. Consistent with this, an enzyme required for ceramide biosynthesis, Lag1p, has been shown to be required for replicative longevity in yeast [43]. It will be interesting to see how future studies of acetyl-CoA utilization in the nucleus resolve the paradoxical effect of the acs2Δ mutation on RLS.
Supplementary Material
Acknowledgments
Strains T2-3D, GG621, and GG625 were kindly provided by Raymond Brandt and H. Yde Steensma, Leiden University. Proteolytic fragmentation and mass spectrometry were done by Scott H. McClung and Stanley M. Stevens at the University of Florida Interdisciplinary Center for Biotechnology Research (UF ICBR). This study was supported by the Ellison Medical Foundation grant AG-NO-0014 to AAF and the NIH grant AG023719 to JPA.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s11010-009-0209-z) contains supplementary material, which is available to authorized users.
References
- 1.Starai VJ, Escalante-Semerena JC. Acetyl-coenzyme A synthetase (AMP forming) Cell Mol Life Sci. 2004;61:2020–2030. doi: 10.1007/s00018-004-3448-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thomson JM, Gaucher EA, Burgan MF, De Kee DW, Li T, Aris JP, Benner SA. Resurrecting ancestral alcohol dehydrogenases from yeast. Nat Genet. 2005;37:630–635. doi: 10.1038/ng1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van den Berg MA, de Jong-Gubbels P, Kortland CJ, van Dijken JP, Pronk JT, Steensma HY. The two acetyl-coenzyme A synthetases of Saccharomyces cerevisiae differ with respect to kinetic properties and transcriptional regulation. J Biol Chem. 1996;271:28953–28959. doi: 10.1074/jbc.271.46.28953. [DOI] [PubMed] [Google Scholar]
- 4.Klein HP, Jahnke L. Cellular localization of acetyl-coenzyme A synthetase in yeast. J Bacteriol. 1968;96:1632–1639. doi: 10.1128/jb.96.5.1632-1639.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klein HP, Jahnke L. Variations in the localization of acetyl-coenzyme A synthetase in aerobic yeast cells. J Bacteriol. 1971;106:596–602. doi: 10.1128/jb.106.2.596-602.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klein HP, Jahnke L. Effects of aeration on formation and localization of the acetyl coenzyme A synthetases of Saccharomyces cerevisiae. J Bacteriol. 1979;137:179–184. doi: 10.1128/jb.137.1.179-184.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O’Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 8.Takahashi H, McCaffery JM, Irizarry RA, Boeke JD. Nucleocytosolic acetyl-coenzyme a synthetase is required for histone acetylation and global transcription. Mol Cell. 2006;23:207–217. doi: 10.1016/j.molcel.2006.05.040. [DOI] [PubMed] [Google Scholar]
- 9.Sinclair D, Mills K, Guarente L. Aging in Saccharomyces cerevisiae. Annu Rev Microbiol. 1998;52:533–560. doi: 10.1146/annurev.micro.52.1.533. [DOI] [PubMed] [Google Scholar]
- 10.Hallows WC, Lee S, Denu JM. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc Natl Acad Sci USA. 2006;103:10230–10235. doi: 10.1073/pnas.0604392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwer B, Bunkenborg J, Verdin RO, Andersen JS, Verdin E. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc Natl Acad Sci USA. 2006;103:10224–10229. doi: 10.1073/pnas.0603968103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Starai VJ, Celic I, Cole RN, Boeke JD, Escalante-Semerena JC. Sir2-dependent activation of acetyl-CoA synthetase by deacetylation of active lysine. Science. 2002;298:2390–2392. doi: 10.1126/science.1077650. [DOI] [PubMed] [Google Scholar]
- 13.Wenzel TJ, van den Berg MA, Visser W, van den Berg JA, Steensma HY. Characterization of Saccharomyces cerevisiae mutants lacking the E1 alpha subunit of the pyruvate dehydrogenase complex. Eur J Biochem. 1992;209:697–705. doi: 10.1111/j.1432-1033.1992.tb17338.x. [DOI] [PubMed] [Google Scholar]
- 14.van den Berg MA, Steensma HY. ACS2, a Saccharomyces cerevisiae gene encoding acetyl-coenzyme A synthetase, essential for growth on glucose. Eur J Biochem. 1995;231:704–713. doi: 10.1111/j.1432-1033.1995.tb20751.x. [DOI] [PubMed] [Google Scholar]
- 15.Sherman F. Getting started with yeast. Methods Enzymol. 2002;350:3–41. doi: 10.1016/s0076-6879(02)50954-x. [DOI] [PubMed] [Google Scholar]
- 16.Falcon AA, Aris JP. Plasmid accumulation reduces life span in Saccharomyces cerevisiae. J Biol Chem. 2003;278:41607–41617. doi: 10.1074/jbc.M307025200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dove JE, Brockenbrough JS, Aris JP. Isolation of nuclei and nucleoli from the yeast Saccharomyces cerevisiae. In: Berrios M, editor. Nuclear structure and function. Academic Press; London: 1998. pp. 33–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen SP, Brockenbrough JS, Dove JE, Aris JP. Homocitrate synthase is located in the nucleus in the yeast Saccharomyces cerevisiae. J Biol Chem. 1997;272:10839–10846. doi: 10.1074/jbc.272.16.10839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu P, Brockenbrough JS, Metcalfe AC, Chen S, Aris JP. Nop5p is a small nucleolar ribonucleoprotein component required for pre-18 S rRNA processing in yeast. J Biol Chem. 1998;273:16453–16463. doi: 10.1074/jbc.273.26.16453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current protocols in molecular biology. Greene Publishing and Wiley-Interscience; New York: 2009. p. 4800. [Google Scholar]
- 22.Yakunin AF, Hallenbeck PC. A luminol/iodophenol chemiluminescent detection system for western immunoblots. Anal Biochem. 1998;258:146–149. doi: 10.1006/abio.1998.2571. [DOI] [PubMed] [Google Scholar]
- 23.de Beus E, Brockenbrough JS, Hong B, Aris JP. Yeast NOP2 encodes an essential nucleolar protein with homology to a human proliferation marker. J Cell Biol. 1994;127:1799–1813. doi: 10.1083/jcb.127.6.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aris JP, Blobel G. Isolation of yeast nuclei. Methods Enzymol. 1991;194:735–749. doi: 10.1016/0076-6879(91)94056-i. [DOI] [PubMed] [Google Scholar]
- 25.McVey M, Kaeberlein M, Tissenbaum HA, Guarente L. The short life span of Saccharomyces cerevisiae sgs1 and srs2 mutants is a composite of normal aging processes and mitotic arrest due to defective recombination. Genetics. 2001;157:1531–1542. doi: 10.1093/genetics/157.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sinclair DA, Guarente L. Extrachromosomal rDNA circles—a cause of aging in yeast. Cell. 1997;91:1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- 27.Starai VJ, Takahashi H, Boeke JD, Escalante-Semerena JC. A link between transcription and intermediary metabolism: a role for Sir2 in the control of acetyl-coenzyme A synthetase. Curr Opin Microbiol. 2004;7:115–119. doi: 10.1016/j.mib.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 28.Starai VJ, Takahashi H, Boeke JD, Escalante-Semerena JC. Short-chain fatty acid activation by acyl-coenzyme A synthetases requires SIR2 protein function in Salmonella enterica and Saccharomyces cerevisiae. Genetics. 2003;163:545–555. doi: 10.1093/genetics/163.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lamming DW, Latorre-Esteves M, Medvedik O, Wong SN, Tsang FA, Wang C, Lin SJ, Sinclair DA. HST2 mediates SIR2-independent life-span extension by calorie restriction. Science. 2005;309:1861–1864. doi: 10.1126/science.1113611. [DOI] [PubMed] [Google Scholar]
- 30.North BJ, Sinclair DA. Sirtuins: a conserved key unlocking AceCS activity. Trends Biochem Sci. 2007;32:1–4. doi: 10.1016/j.tibs.2006.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Habeler G, Natter K, Thallinger GG, Crawford ME, Kohlwein SD, Trajanoski Z. YPL.db: the yeast protein localization database. Nucleic Acids Res. 2002;30:80–83. doi: 10.1093/nar/30.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Natter K, Leitner P, Faschinger A, Wolinski H, McCraith S, Fields S, Kohlwein SD. The spatial organization of lipid synthesis in the yeast Saccharomyces cerevisiae derived from large scale green fluorescent protein tagging and high resolution microscopy. Mol Cell Proteomics. 2005;4:662–672. doi: 10.1074/mcp.M400123-MCP200. [DOI] [PubMed] [Google Scholar]
- 33.Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, Yang L, Wolting C, Donaldson I, Schandorff S, Shewnarane J, Vo M, Taggart J, Goudreault M, Muskat B, Alfarano C, Dewar D, Lin Z, Michalickova K, Willems AR, Sassi H, Nielsen PA, Rasmussen KJ, Andersen JR, Johansen LE, Hansen LH, Jespersen H, Podtelejnikov A, Nielsen E, Crawford J, Poulsen V, Sorensen BD, Matthiesen J, Hendrickson RC, Gleeson F, Pawson T, Moran MF, Durocher D, Mann M, Hogue CW, Figeys D, Tyers M. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 34.Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci USA. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oender K, Loeffler M, Doppler E, Eder M, Lach S, Heinrich F, Karl T, Moesl R, Hundsberger H, Klade T, Eckl P, Dickinson JR, Breitenbach M, Koller L. Translational regulator RpL10p/Grc5p interacts physically and functionally with Sed1p, a dynamic component of the yeast cell surface. Yeast. 2003;20:281–294. doi: 10.1002/yea.963. [DOI] [PubMed] [Google Scholar]
- 36.Fromont-Racine M, Rain JC, Legrain P. Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat Genet. 1997;16:277–282. doi: 10.1038/ng0797-277. [DOI] [PubMed] [Google Scholar]
- 37.Carrozza MJ, Utley RT, Workman JL, Cote J. The diverse functions of histone acetyltransferase complexes. Trends Genet. 2003;19:321–329. doi: 10.1016/S0168-9525(03)00115-X. [DOI] [PubMed] [Google Scholar]
- 38.Ivessa AS, Schneiter R, Kohlwein SD. Yeast acetyl-CoA carboxylase is associated with the cytoplasmic surface of the endoplasmic reticulum. Eur J Cell Biol. 1997;74:399–406. [PubMed] [Google Scholar]
- 39.Blander G, Guarente L. The Sir2 family of protein deacetylases. Annu Rev Biochem. 2004;73:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- 40.Muller I. Parental age and the life-span of zygotes of Saccharomyces cerevisiae. Antonie Van Leeuwenhoek. 1985;51:1–10. doi: 10.1007/BF00444223. [DOI] [PubMed] [Google Scholar]
- 41.Jazwinski SM. Rtg2 protein: at the nexus of yeast longevity and aging. FEMS Yeast Res. 2005;5:1253–1259. doi: 10.1016/j.femsyr.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 42.Epstein CB, Waddle JA, Hale WIV, Dave V, Thornton J, Macatee TL, Garner HR, Butow RA. Genome-wide responses to mitochondrial dysfunction. Mol Biol Cell. 2001;12:297–308. doi: 10.1091/mbc.12.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang JC, Kirchman PA, Allen M, Jazwinski SM. Suppressor analysis points to the subtle role of the LAG1 ceramide synthase gene in determining yeast longevity. Exp Gerontol. 2004;39:999–1009. doi: 10.1016/j.exger.2004.03.026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


