Abstract
C. elegans molts at the end of each of its four larval stages but this cycle ceases at the reproductive adult stage. We have identified a regulator of molting, pqn-47. Null mutations in pqn-47 cause a developmental arrest at the first larval molt, showing that this gene activity is required to transit the molt. Mutants with weak alleles of pqn-47 complete the larval molts but fail to exit the molting cycle at the adult stage. These phenotypes suggest that pqn-47 executes key aspects of the molting program including the cessation of molting cycles. The pqn-47 gene encodes a protein that is highly conserved in animal phylogeny but probably misannotated in genome sequences due to much less significant homology to a yeast transcription factor. A PQN-47∷GFP fusion gene is expressed in many neurons, vulval precursor cells, the distal tip cell (DTC), intestine, and the lateral hypodermal seam cells but not in the main body hypodermal syncytium (hyp7) that underlies, synthesizes, and releases most of the collagenous cuticle. A functional PQN-47∷GFP fusion protein localizes to the cytoplasm rather than the nucleus at all developmental stages, including the periods preceding and during ecdysis when genetic analysis suggests that pqn-47 functions. The cytoplasmic localization of PQN-47∷GFP partially overlaps with the endoplasmic reticulum, suggesting that PQN-47 is involved in the extensive secretion of cuticle components or hormones that occurs during molts. The mammalian and insect homologues of pqn-47 may serve similar roles in regulated secretion.
Keywords: molt (moult), C11orf9 MRF pqn-47, poly glutamine protein, endocrine secretion
Introduction
C. elegans undergoes four larval stage molts between hatching and adulthood, releasing the collagenous cuticle of the previous larval stage from its epidermis and pharynx, and exposing the newly synthesized slightly larger cuticle of the next developmental stage. Nematode cuticle is tough, flexible, and mainly composed of proteins (over 80% is collagen). The major component of arthropod cuticles, the polysaccharide chitin, is only found in the nematode egg shell, buccal capsule, and the pharynx (Zhang et al., 2005). Intestinal cell endoreduplications and pulses of protein synthesis are also coordinated with each C. elegans molt (Kipreos, 2005).
Molting is not strictly a hypodermal phenomenon; rather it requires precise coordination of events throughout the animal, including developmental synchronization between tissues and the coordination of special behaviors, all acting in concert with the degradation and synthesis of the exoskeleton. Hyp7 is a large post-mitotic syncytial hypodermal cell that covers the body of the worm and synthesizes, secretes, and underlies the collagenous cuticle (Page and Johnstone, 2007). During the process of molting, epithelial cells like Hyp7 synthesize and secrete a new collagenous cuticle that underlies the old. Connections to the old cuticle are released prior to ecdysis, while the worm lies still in a sleep-like state called lethargus (Raizen et al., 2008; Van Buskirk and Sternberg, 2007). The animal eventually resumes movement and escapes by breaking through the anterior tip of the preexisting cuticle (ecdysis). The proliferative seam cells divide in a stem cell-like lineage prior to each larval molt, where the daughter seam cell endoreduplicates its DNA (Hedgecock and White, 1985) and fuses with the syncytial hyp7 cell (Podbilewicz and White, 1994). The lateral cuticle is secreted by seam cells, has distinct ridges called alae that are each distinctive signatures at the L1, dauer, and adult stages. Although the seam cells divide again before the final molt into adulthood, they thereafter terminally differentiate and fuse to their neighboring seam cells, forming two bilateral syncytia with 16 nuclei each (Sulston and Horvitz, 1977). During the L4 to adult molt, fusion of the somatic cells of the germ line with the epidermis occurs to allow the laying of eggs; this coincides with reproductive maturity, and then animals exit the molting cycle.
The number or timing of molts may be modulated by timing pathways autonomous to epithelial cells as well as by probable neuroendocrine cues that might be related to the better characterized ecdysone and juvenile hormones of insects (Ambros and Horvitz, 1984; Ruaud et al., 2011; Tennessen et al., 2010). In insects, cascades of hormones orchestrate molting cycles; neurosecretory cells in the brain release a peptide hormone that modulates the release of the steroid hormone ecdysone from glandular organs, which in turn triggers molting. Epithelial-derived endocrine tissues release Juvenile Hormone, a sesquiterpenoid, which determines if a molt will be larval to larval or larval to adult (metamorphic). The titer of ecdysone regulates the secretion of Eclosion Hormones, which stimulate the behaviors required for the animal to escape its old cuticle, or ecdyse. The titer of the steroid hormone ecdysone serves as the main point where physiological and environmental cues that influence the period between molts are integrated (Gilbert et al., 2002). Ecdysone receptors are widely expressed, and specificity of responses to ecdysone may be achieved through interaction with other temporally and spatially regulated nuclear hormone receptors (Kozlova and Thummel, 2002). Ecdysone receptor-mediated transcriptional activation initiates cascades of gene expression that can be visualized by endoreduplicated salivary puffs in Drosophila and which ultimately trigger molting and metamorphosis. Although orthologous ecdysone receptors have recently been identified in parasitic nematode genomes (Graham et al., 2010; Tzertzinis et al., 2010), none has been identified in free-living nematodes like C. elegans. However, C. elegans does have many nuclear hormone receptors including daf-12 (Ruaud and Bessereau, 2006), nhr-23/CHR3 (Brooks et al., 2003; Hayes et al., 2006) and nhr-25 (Hada et al., 2010) that have roles in molting, some of which are similar to those required for molting in insects (Gissendanner et al., 2004; Gissendanner and Sluder, 2000; Kostrouchova et al., 1998; Kostrouchova et al., 2001). Even though non-parasitic nematodes have not been observed to respond to exogenous ecdysone, it is known that cholesterol is required for molting in C. elegans (Entchev and Kurzchalia, 2005; Martin et al., 2010), suggesting that steroid hormones could be ligands for receptors involved in regulating nematode molting. Different species of nematodes undergo molts at different intervals and in response to different environmental triggers (Lee, 2002), further supporting the likelihood of nematode molting specific hormones. Moreover, C. elegans have glandular cells and neuroendocrine cells that may respond to and release systemic signals, though none have been definitively linked to the regulation of molting cycles.
The C. elegans heterochronic gene regulatory network is coupled to the molting cycle. Exit from the molting cycle, accompanied by terminal differentiation of the epidermis, normally coincides with reproductive maturity. In heterochronic mutants, exit from the molting cycle may occur precociously or be delayed or prevented (Ambros and Horvitz, 1984; Moss, 2007; Pasquinelli and Ruvkun, 2002; Rougvie, 2005). In the precocious lin-14, lin-28, hbl-1, and lin-41 mutants, epithelial cells cease proliferation early and fewer molts occur. Conversely, in the retarded lin-4, let-7, and lin-29 mutants, epithelial tissues fail to differentiate and animals undergo supernumerary molts. In most retarded heterochronic mutants, the earlier larval stage division and fusion pattern of seam cells is repeated prior to the L4/L5 molt, delaying the final differentiation program of the seam cells and the production of alae until after the 5th molt (Ambros and Horvitz, 1984; Bettinger et al., 1996; Frasch, 2008; Moss, 2007; Resnick et al., 2010). Although the heterochronic pathway converges on repression of NHR-23 and NHR-25 in adults (Hayes et al., 2006), the underlying mechanism is not well understood. Also, the extent to which some heterochronic phenotypes arise from loss of systemic control of molting cycles, rather than from cell-autonomous defects within the hypodermis, is not yet known.
Key components of endocrine and possibly neuroendocrine pathways regulating the molting cycle may have evaded detection in a previous high-throughput, RNAi-based screen for animals unable to fully remove the larval cuticles (Frand et al., 2005). This screen identified many genes that are cyclically expressed in the hypodermis and regulated by NHR-23. We reasoned that genes that act upstream in the molting pathway could be identified using forward genetic screens for mutants that molt at inappropriate times. We therefore conducted a screen for mutants that continue molting after reproductive maturity. Here, we show that pqn-47, which emerged from this screen, is essential for both the cessation of molting in adults and the completion of larval molts. The glutamine and asparagine rich PQN-47 protein is expressed in neurons, other secretory cells and seam cells, and is highly conserved across animal phylogeny. We propose that PQN-47 in many animal species may function in the regulated secretion of endocrine signals and tissue remodeling enzymes during major developmental transitions.
Results
Isolation of pqn-47 in a genetic screen for mutations that cause supernumerary molts
The gene mlt-10 was identified in a genetic screen for molting defective mutants. The mlt-10 promoter is activated early in the molting program and tracks with the transcription of genes involved in the synthesis of new cuticles (Frand et al., 2005; Meli et al., 2010). The expression of a mlt-10p∷gfp-pest fusion gene bearing a PEST destabilization sequence is undetectable in the adult stage after the cessation of the molting cycle. Use of a PEST-destabilized GFP allowed the dynamics of the molting cycle to be visualized, because the GFP produced each molt is degraded before the onset of the next molt. To identify genes required for the cessation of molting, we screened mutagenized animals for adult-stage reanimation of the mlt-10p∷gfp-pest fusion gene (Fig. 1A), for example, as a supernumerary molt was initiated (Frand et al., 2005). Transgenic but otherwise wild type animals express GFP during the L4-to-adult molt, but not thereafter (Fig. 1B). From a screen of approximately 112,000 EMS-mutagenized genomes, we identified 91 candidate mutants that expressed the mlt-10 reporter as young or gravid adults, and we established lines from the fertile candidates. In subsequent generations, adult-stage expression of GFP was observed in 70 of the corresponding lines. The partial shedding of cuticles was also observed in 31of these strains. Based on preliminary complementation analysis and mapping data, we estimate that this screen uncovered a minimum of 6 distinct loci (to be described elsewhere). This report focuses on the mg412 mutant, which had a highly penetrant supernumerary molt out of an adult cuticle (described below), segregated as a single locus through backcrosses, and which we establish below as an allele of pqn-47/F59B10.1. Further characterization of additional alleles (described below) of pqn-47 shows that this gene is required for the completion of larval molts, as well as the cessation of molting cycles in reproductively mature animals.
Figure 1. Phenotypes of hypomorph pqn-47(mg412).
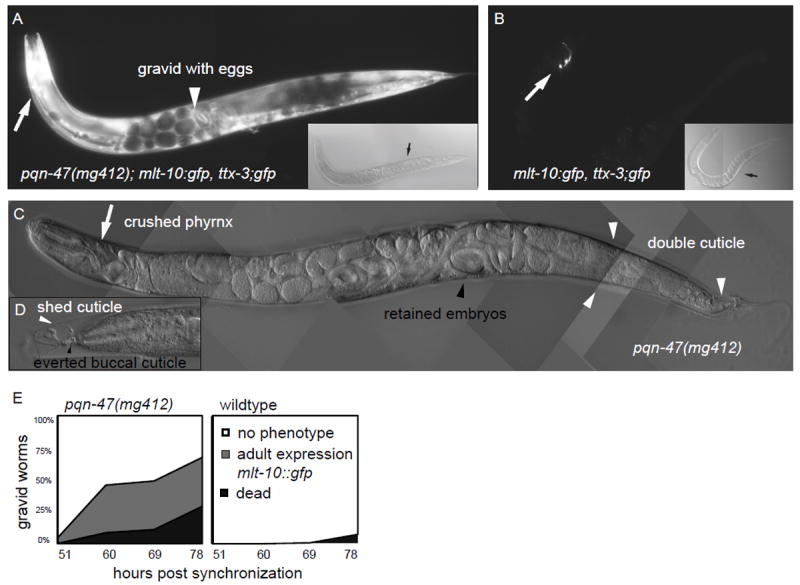
pqn-47(mg412) gravid adults (A) re-animate the molting reporter gene mlt-10p∷gfp-pest, whereas age matched wildtype gravid adults (B) do not. Wildtype only expresses the co-injection marker ttx-3p∷gfp (white arrows). GFP images in A and B were taken at the same exposure, and in both cases are the same focal plane on the same worm as the DIC image shown as an inset. Note the eggs within the gravid mg412 adults (black arrow) and the accumulating older unhatched worms (white arrow head) that have already turned on mlt-10 expression on their own, which in contrast only happens after they are laid in wildtype. (C) An older worm after the supernumerary molt has begun. Often the pharynx is bent (white arrow) due to futile attempts to release the everted buccal cuticle by pulling back against it. Gravid adults release cuticle from their entire body (see white arrows for outer cuticle along posterior body and tail). (D) An even older worm that successfully detached from the tip of the old cuticle (white arrow head), but whose buccal cavity has everted and has damaged pharyngal tissue (black arrow head). (E) Synchronized hatchlings of the indicated genotypes (mg412 n=368, wild type n=136) were raised at 25° C and scored for animation of the mlt-10 molting reporter and death due to bagging, supernumerary molt, and bursting. mg412 gravid adults first turn on the molting reporter, and then after 5-10 hours initiate a supernumerary molt that ultimately results in death. In wildtype, the reporter does not come back on, and the worms survive for weeks if not allowed to starve out the plate.
pqn-47(mg412) mutants exhibit inappropriate reanimation of the mlt-10p∷gfp-pest reporter in gravid adults (Fig 1AB) prior to the execution of a supernumerary molt that culminates in an aberrant ecdysis (Fig. 1A-E). Such an adult stage molt could be due to an inappropriate induction of a larval molting cycle or a more global transformation of cell fates to an earlier larval stage, such as has been observed in the retarded class of heterochronic mutants. For example, a reiteration of larval stage fates has been observed in the retarded heterochronic mutants lin-29, let-7(mg279), and mir-48;mir-84 double mutant animals. These mutants also express mlt-10p∷gfp-pest during the supernumerary adult molt. The lin-29 mutants and, to a lesser degree, mir-48;mir-84 double mutants successfully complete the extra molt (Abbott et al., 2005; Hayes et al., 2006; Papp et al., 1991; Reinhart et al., 2000) in contrast to pqn-47(mg412) mutants.
Although pqn-47(mg412) animals initiate an extra molt as gravid adults, the mutants are unable to completely shed the preexisting cuticle and perish. The cuticle along the head and body of the animal is released (Fig. 1CD). Although animals were able to evert the buccal cuticle from the buccal cavity (the oral cavity lined with cuticle), they were often unable to release it, as indicated by a bundle of tissue and/or cuticle connecting the tip of the head to the partially shed cuticle (Fig. S1A). Aside from the initiation of an incomplete supernumerary molt in pqn-47(mg412) mutants, earlier developmental events appear normal. Populations synchronized at the first larval stage enter the L1 molt, execute the L4 to adult molt and begin egg laying at approximately the same time as wildtype animals (data not shown). pqn-47(mg412) mutants do not display significant embryonic or larval lethality compared to wild type animals, even though the null phenotype of pqn-47 is penetrant larval lethality (see below).
To identify the gene affected by the mg412 lesion, the inappropriate expression of mlt-10p∷gfp-pest in adults characteristic of the mg412 mutant was used to map the corresponding mutation using a standard SNP mapping protocol to an gene rich interval in the middle of chromosome II. RNAi of genes in the vicinity identified several genes whose inactivation caused either adult-stage expression of mlt-10p∷gfp-pest, bursting, or caused larval molting defects (data not shown). Sequencing of these open reading frames identified a missense mutation in pqn-47/F59B10.1 in mg412 mutants. The mg412 mutation is an A-G transition in the DNA sequence that encodes the fourth exon, and specifies the substitution of threonine 364, which is a conserved residue and a possible phosphorylation site, with alanine (Table S1). Multiple independent lines transformed with genomic PCR fragments of wild type pqn-47 show significant rescue of inappropriate mlt-10p∷gfp-pest expression as well as the supernumerary molt phenotype of mg412 (Fig. S1B). Thus, mg412 is a mutation in pqn-47.
Analysis of multiple alleles uncovers an essential role for pqn-47 in larval molting cycles
Identification and characterization of additional pqn-47 alleles have revealed that mg412 is a weak pqn-47 allele, and that pqn-47 gene function is required for the earlier larval molts in addition to the cessation of molting in adults. We characterized this defect in detail using a null allele. A deletion allele pqn-47(tm2707) which deletes 186 amino acids, encoded by exon four, and adds 4 new amino acids in frame downstream of the deletion had been isolated by the Mitani lab and the National BioResource Project of Japan (Table S1). This pqn-47(tm2707) deletion allele was known to be lethal but had not been characterized further. We observed that homozygous pqn-47(tm2707) progeny derived from a balanced heterozygous strain showed a fully penetrant early larval arrest at the L1 stage, accompanied by a molting-defective phenotype (Fig. 2A). pqn-47(tm2707) homozygous animals enter into the L1 stage lethargus, a brief period of quiescence that accompanies molting. Apolysis then occurs around the head (arrowheads Fig. 2A), body and tail region, but the mutants are unable to fully escape the L1 cuticle. Arrested animals fail to completely release everted pharyngeal cuticle from the buccal cavity. Animals continue to struggle to escape from the ensheathment and can be seen straining against the shed cuticle, but they are unable to eat and grow and therefore eventually perish. These pharyngeal phenotypes are similar to those seen in the mg412 adults that perish attempting a supernumerary molt.
Figure 2. Strong loss of function mutant pqn-47(tm2707) terminally arrests during the L1 molt.
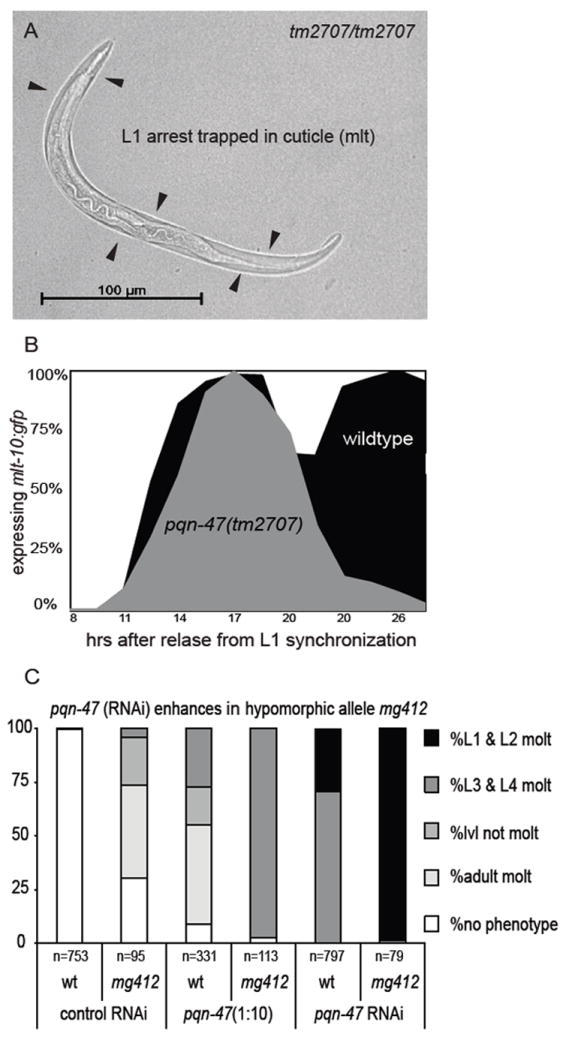
(A) at 48 hours, a tm2707 homozygote arrested in the L1 molt (by absence of rescuing transgene). (B) tm2707 homozygous animals recover from L1 starvation and enter into the first larval molt with apparently normal kinetics as reflected by their expression of the molting reporter mlt-10p∷gfp-pest. However, unlike wildtype animals that quickly finish the molt and turn mlt-10p∷gfp-pest off as L2s, only to then repeat the molting cycle, tm2707 homozygotes initiate but are unable to complete the L1 molt, remain trapped in the cuticle and turn off mlt-10p∷gfp-pest. Time course at 25°C post synchronization, wild type n>190 and tm2707 n>80 (C) pqn-47 (RNAi) recapitulates the phenotypes of strong and weak loss of function alleles of pqn-47 as well as enhancing the hypomorphic allele mg412. Synchronized L1s were plated on to RNAi plates and grown at 25°C for 66 hrs. Gravid adults expressing mlt-10p∷gfp-pest or animals in a supernumerary molt were scored as adult molt.
pqn-47(tm2707) hatchlings arrest growth in the absence of food and restart growth upon re-feeding with approximately normal kinetics as reflected in the dynamics of mlt-10p∷gfp-pest expression at the end of the L1 stage (Fig. 2B). Not only do pqn-47(tm2707) homozygotes activate mlt-10p∷gfp-pest in the hypodermis during the first larval molt, the mutants also progress through the molting cycle to repress the transgene within a few hours of the aberrant ecdysis, as wild type animals do after completion of the molt. Thus the molting program is initiated normally in pqn-47(tm2707) null mutants and even begins to resolve a cycle, but a late stage event, possibly the proteolytic release of cuticle from the buccal cavity or the behavioral routines of ecdysis, fails. The pqn-47(tm2707) homozygous animals accumulate in the molt such that at 24 hours post release from synchronization most are arrested and trapped in cuticle as L1s.
pqn-47 mutants’ phenotypes indicate that pqn-47 has roles throughout development and is required for all ecdyses as well as the cessation of molting in adults, and are also recapitulated by RNAi knockdown experiments. mg412 is a reduction of function allele. The mg412/+ and tm2707/+ heterozygotes have no molting phenotypes, suggesting recessive inheritance. tm2707 is a stronger loss-of-function allele than mg412 (Fig. 2C). The mg412/tm2707 trans-heterozygote reaches adulthood and exhibits the adult-stage phenotype characteristic of mg412 homozygotes (Fig. S1D). The mg412/tm2707 trans-heterozygotes do not arrest as larvae, indicating that even a single copy of pqn-47(mg412) provides enough function to support the larval molting cycle. Interestingly the adult re-animation of mlt-10p∷gfp-pest and supernumerary molt occur earlier in the mg412/tm2707 trans-heterozygote than in pqn-47(mg412) homozygous animals, indicating that levels of pqn-47 influence the timing of molts. Furthermore, RNAi of pqn-47 in mg412 animals shifts their phenotype from an extra adult molt to failed larval ecdsyes (Fig. 2C). Stronger RNAi knockdown using concentrated bacterial cultures (see Material and Methods) caused further enhancement as indicated by earlier molt phenotypes, and weaker RNAi treatment (using diluted RNAi cultures) cause weaker phenotypes as indicated by later larval or adult molt phenotypes. These variable strength RNAi experiments support the idea that levels of pqn-47 determine how early in development the pqn-47 phenotype is expressed (Fig. 2C and data not shown). Similarly, the L1 molt arrest phenotype of tm2707 is rescued by an extra chromosomal array of pqn-47(mg412) (data not shown).
A survey of previously mapped lethal mutations residing in the same genetic region as pqn-47 identified additional alleles, including let-25(mn25) (Herman, 1978). We found a substitution mutation in the pqn-47 open reading frame of let-25(mn25) mutants, which specifies the substitution of the conserved neutral residue glycine 471 with a positively charged arginine, likely disrupting folding within this essential region (Fig. 3 & Table S1). Homozygous let-25(mn25) animals exhibit a fully penetrant L1 arrest and molting defective phenotype identical to tm2707. A second pqn-47(ok3445) knockout allele, which creates a deletion and early stop, also causes a fully penetrant L1 arrest and molting defective phenotype (Fig. 3 & Table S1). The fact that three independent mutations in pqn-47 cause arrest at the L1 to L2 molt strongly argues that the phenotype is due to pqn-47 lesions and not linked mutations. Moreover, the L1 lethality and molting defects of pqn-47(tm2707) were rescued with a simple extra-chromosomal array of wild-type pqn-47 (Fig. S1C and Table S3A).
Figure 3. pqn-47 mutations and alignment with animal orthologues and spurious yeast homologues.
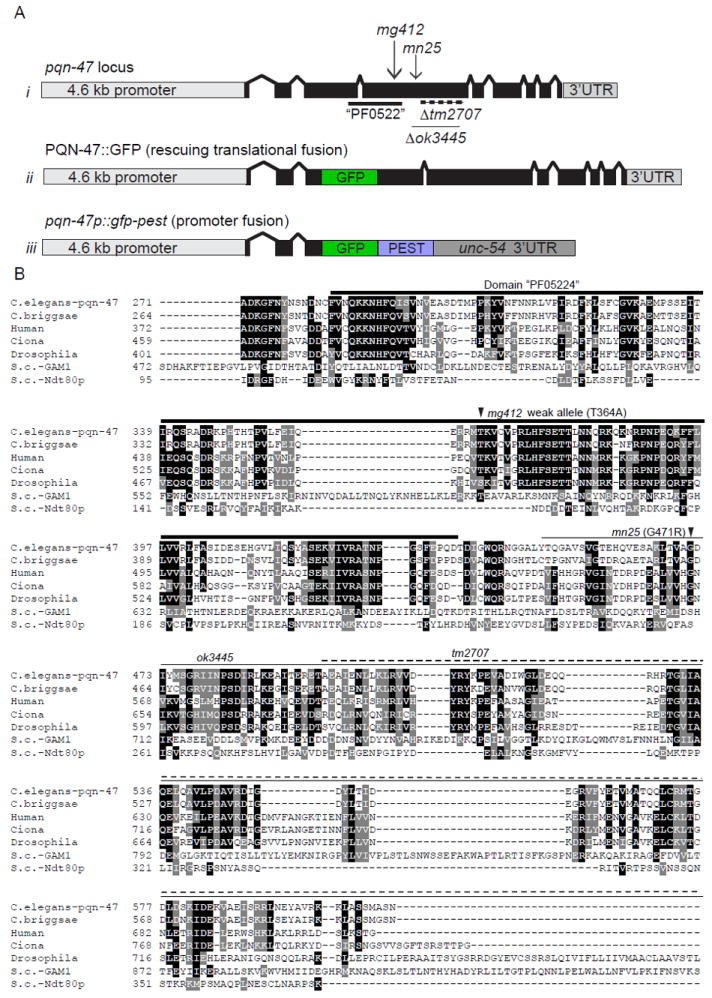
(A) schematic of i) the pqn-47 locus with location of alleles (described in Table. S1) ii) PQN-47∷GFP, a rescuing translational fusion with GFP inserted into a nonconserved region of the third exon iii) a pested promoter pqn-47 fusion. (B) The PQN-47 protein is highly conserved in animals, especially around the central domain PF05224 (thick line) in which all of our mutations are located, but not significantly in fungi. Identical residues shaded black and similar with gray. Arrow heads indicate locations of point mutations mg412 and mn25, and deletions are indicated by a thin line (ok3445) or dashed line (tm2707). GFP was inserted in frame in the coding sequence in an area of low conservation to generate the translational protein fusion used in this study, as C terminal fusions were not able to rescue the mutant phenotypes (data not shown). Full protein sequence shown in figure S2. Protein alignment by CLUSTAL W (2), then BOXSHADE 3.21
PQN-47 is highly conserved but has no domains of known function
PQN-47, prion-like-(Q/N-rich)-domain-bearing protein, is named for the over-represented number of glutamine and asparagine residues. The pqn-47 gene encodes a 931 amino acid protein with strong conservation over a large percent of the protein between animal species as diverse as nematoda, insects, mammals, and the tunicate Ciona, but poorly annotated domains and function. The homologues detected in these species are likely orthologues, in that they are the top homologue detected in each species and when that top homologue is compared to C. elegans, it detects pqn-47 as its top hit---that is they are reciprocal top BLAST hits. The probability of these BLAST scores occurring by chance were 2.0e-85 between human and C. elegans, and the conserved region covered 72% of the C. elegans protein and is similarly strong for mouse (8.0e-85, over 72% of the protein) and Drosophila (6.0e-77, over 53% of the protein) and all the other animal species tested, as shown in the alignment (Fig. 3, Table S2).
Probable PQN-47 orthologues are identified even in the single-celled animal choanoflagellates, such as Monosiga (8.0e-45 over 28% of the C. elegans protein that likely represents a truncated open reading frame in Monosiga), that do not have nervous systems. The only animal, in 26 non-nematode genomes queried, where a PQN-47 orthologue was not found was in the mosquito species Aedes aegypti and Anopheles gambiae genomes, although it is formally possible but unlikely that both of these putatively complete genome sequences are incomplete at this same locus. The amoeboid Dictyostelium is the only protist, of 10 queried, with a potential orthologue. Each of the pqn-47 alleles described above introduces amino acid changes or deletions within the conserved region, supporting the hypothesis presented by conservation data that this region is essential for PQN-47 function (Fig.3).
PQN-47 has a well-conserved C. elegans paralogue F21A10.2 (Table S2), also found in some animals, though presumably with a distinct and non-redundant function, as mutations in pqn-47 have penetrant C. elegans phenotypes. RNAi against the C. elegans paralogue F21A10.2 did not cause molting defects or other developmental abnormalities.
Neither C. elegans PQN-47 nor its orthologues or paralogues have conserved signals for specific cellular localization, including NLS, or signal peptides, beside 1-2 potential transmembrane domains (See Table S4 for a list of in silico analysis of pqn-47 and orthologues and paralogues).
The 3’ UTR of pqn-47 has many predicted miRNA binding sites including let-7 family member mir-84/48, and has been identified in vitro as a target for let-7 (Andachi, 2008). Similarly the human orthologues have been identified as targets of multiple miRNAs that are regulated during the differentiation of oligodendrites (Letzen et al., 2010).
pqn-47 is expressed in diverse somatic tissues throughout development
A GFP fusion to the full length and functional pqn-47 gene was constructed to reveal its cellular pattern of expression during development of the animal, its subcellular localization within those cells that express the gene, and any dynamic regulation of cellular or subcellular localization during the molting cycle. GFP was fused in frame to an internal coding region that encodes a poorly conserved portion of the protein (Fig. 3A). The entire genomic pqn-47 locus, including the 5’ flanking region up to the next gene, all exons and introns, and a complete 3’UTR including potential let-7 sites are included. This transgene fully rescues the lethal pqn-47(tm2707) allele such that this allele can now be made homozygous from a balancer chromosome heterozygote, as long as the transgene is also segregated to this homozygote (Table S3A). An identical transgene bearing this pqn-47 wild type region without the GFP can also rescue the homozygous pqn-47(tm2707) allele. An alternative construct bearing a C-terminal GFP fusion was not competent to rescue, presumably because it disrupts an essential conserved region of the protein. At high gene dosage, the rescuing PQN-47∷GFP fusion transgene causes over-expression phenotypes (described below).
The PQN-47∷GFP data presented is from a tm2707 homozygous strain that has functional pqn-47 only from the GFP translational fusion gene. We also used a promoter fusion construct, bearing the promoter region of pqn-47 fused to GFP-PEST to visualize the dynamic expression of the gene. Promoter and translational reporters show pqn-47 expression in numerous somatic cells, including cells uniquely poised to mediate or transmit signal(s) involved in the regulation of molting, some of which have been implicated in molting. For example, many cells expressing PQN-47 have significant exposure to the pseudocoelom, and as such are candidates to transmit or detect endocrine signals; the H-shaped excretory cell and its ducts, which form extensive gap junctions with the hypodermis and lie against the pseudocoelom along the entire body of the worm (Nelson and Riddle, 1984), the head mesodermal cell (hmc) lies in the pseudocoelom up against the (excretory) gland cell and forms gap junctions with them and muscle, and the VPI cells at the juncture of the pharynx and intestine are bathed by the pseudocoelom, as well as the intestine itself. Head and nerve ring neurons, pharyngeal cells, ventral nerve cord cells, vulval precursor cells, seam (though interestingly not hyp7), as well as cells in the tail show the strongest pqn-47 expression (Fig. 4). Muscle, intestine, the distal tip cells of the gonad (Fig. 4D), the spermatheca, and a large neuron that may be CAN that is essential for survival but of unknown function near the vulva (also bathed in pseudocoelom fluid, and next to the seam and canal cells), as well as a subset of the ciliated neurons of the head (amphid neurons ASI, ADL, ASK, or AWB) and tail including phasmid cilia PHA and PHB (Fig. 4C), also express pqn-47. We could not detect expression in the pharyngeal glands as reported for a different promoter pqn-47 fusion construct made as part of a high-throughput analysis of gene expression, although other tissues did show similar patterns (Hunt-Newbury et al., 2007).
Figure 4. PQN-47∷GFP is expressed in many tissues.
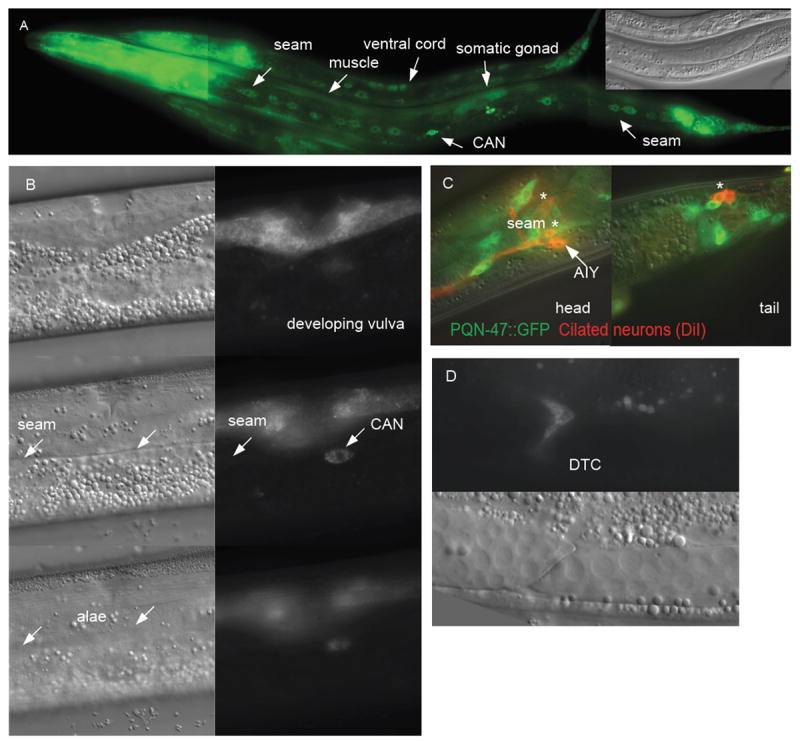
(A) L1 and L2 animals in a long exposure image to show PQN-47∷GFP expression (translational fusion) outside the pharynx and tail. Inset Nomarski image shows gonad for staging (B) 3 serial focal planes showing that once seam cells have made alae (left lower NOM image), they no longer express PQN-47∷GFP (see focal plane matched GFP image on lower right). Middle left nomarski (NOM) shows seam cells in focus, though no GFP expression (focal plane matched GFP). PQN-47∷GFP is however expressed in developing (L4) vulval cells (top row of images). (C) PQN-47∷GFP is expressed in senosry neurons in head and tail. The head cells were identified by position as chemoattractive ASI, AKS and/or chemorepulsive neuron ADL or sheath cell AWB and the tail neurons are chemorepulsive neurons PHA and PHB. (D) migrating distal tip cells.
Expression becomes detectable around the comma stage of embryogenesis and persists through adulthood (Fig. S2). Expression in vulval precursor cells is strong and can first be seen in L3. PQN-47∷GFP is expressed in seam cells, peaking at L2 and ceasing after the seam cells differentiate in late L4, concurrent with the appearance of alae (Fig. 4B). Unlike the molting pathway genes, mlt-10, mlt-8, and nas-37, which are expressed in the hypodermis and whose expression fluctuates with the molting cycle, pqn-47 is not expressed in hyp7, and its overall expression level does not fluctuate with the molting cycle based on the promoter reporter using destabilized GFP or the functional PQN-47∷GFP fusion protein (Fig. S3). This suggests that PQN-47 activity may be regulated by mechanisms other than protein abundance. The intestine shows variably undetectable to low pqn-47 expression (always less than in the neurons) and gets dimmer as development progresses, especially after L3. The two bulbs of the pharynx, specifically pharyngeal muscle cells pm3-8 (not pm6), are variably bright. Overall expression levels are lower in adults than younger animals, with only some expression in head and tail neurons remaining (Fig. S3). pqn-47 expression is very low in starved hatchlings and becomes stronger as animals resume growth on food (Fig. S4). pqn-47 expression (from either promoter or translational fusion transgenes) is not reanimated, as mlt-10p∷gfp-pest is, prior to a supernumerary molt caused by RNAi inactivation of lin-29, or mab-10/R166.1 and lin-66 (SR and GR unpublished observations) suggesting that these genes act downstream of pqn-47 or in parallel pathways.
PQN-47 is a cytoplasmic protein
The functional PQN-47∷GFP protein fusion is localized to cytoplasmic and perinuclear dots (Fig. 5). The perinuclear dots of PQN-47∷GFP localization suggest endoplasmic reticulum or golgi localization. We used double labeling experiments with a CHERRY fused to a KDEL motif that retains the RFP protein in the ER as a navigation aid for the localization of PQN-47. PQN-47∷GFP partially overlaps with this particular marker in neurons (Fig. 5), consistent with an ER or golgi location, although the small cytoplasmic volume makes this cell type non-ideal for co-localization studies. In seam cells, the ER is a larger, more sheet like structure, and PQN-47 is in puncta (Fig. 5). However, we occasionally observed PQN-47 in a more sheet-like pattern (see muscle cell in Fig. 5B). We also noticed that PQN-47∷GFP expression is restricted to the basal cytoplasm in the seam cells.
Figure 5. PQN-47∷GFP is in cytoplasmic puncta.
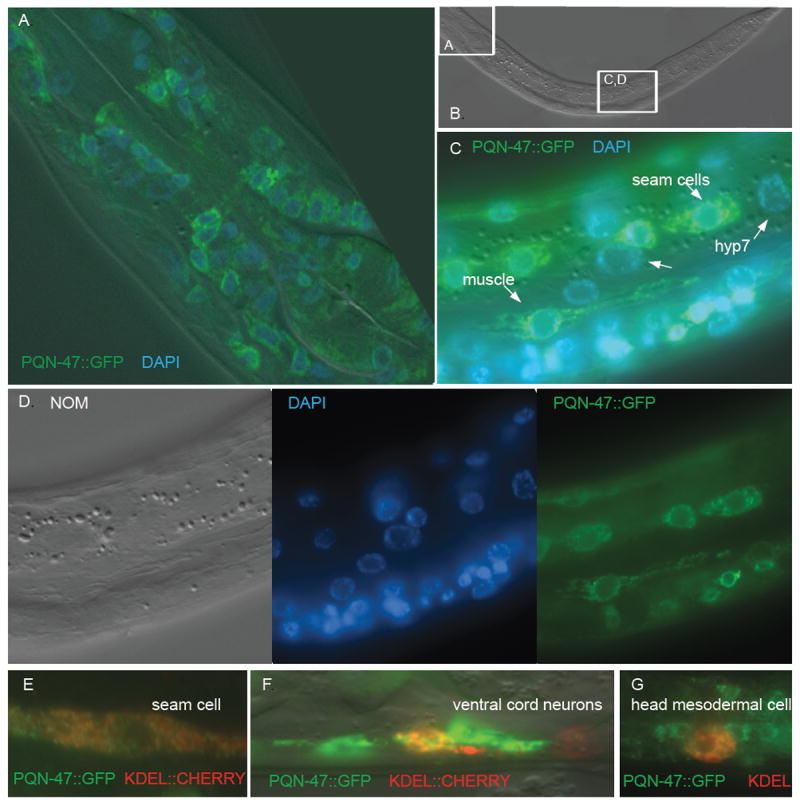
Same worm shown in panels A through D. PQN-47∷GFP is excluded from the nucleus, as indicated by the lack of overlap with DAPI staining. (A) nerve ring neurons and pharyngeal muscle cells (with Apitome sectioning). (B) Nomarski. (C) seam and body wall muscle cells show a punctate non-nuclear pattern. (D) For clarity, separate channels shown of image in C. Partial ER co- localization (punc-129∷ssCHERRY-KDEL), (E) in seam cell, (F) ventral cord neurons, and (G) the head mesodermal cell.
Our cytoplasmic localization of a functional PQN-47∷GFP fusion gene does not agree with the reported nuclear localization of the mouse pqn-47 orthologue myelin gene regulatory factor, or MRF (Emery et al., 2009). This mouse tissue culture analysis of a myc tagged MRF that has not been shown to be functional, for example by rescuing a mutation, was consistent with an initial genome misannotation of the PQN-47/mammalian MRF gene family as homologous to a yeast transcription factor (see Discussion), but is inconsistent with our immunofluorescence analysis of the C. elegans orthologue which suggests a cytoplasmic function. Because the GFP fusion protein that we used for sub-cellular localization is able to rescue the mutant, we think it likely reflects the localization of the endogenous protein, however we cannot rule out the possibility that a small sub-fraction of it is in fact nuclear and is actually responsible for the rescuing activity. Given the mouse finding of the mammalian orthologue MRF in the nucleus, we looked very carefully for nuclear localization of PQN-47∷GFP in particular cells or at particular times during the molting cycle. However, PQN-47∷GFP appeared to be excluded from the nucleus in all cell types and all developmental stages. In neurons where the cytoplasm is small and compact, as well as in cells with larger volumes of cytoplasm, for example seam and muscle cells, PQN-47∷GFP was not found in the nucleus. Although PQN-47 does not have a nuclear localization signal (NLS) nor a nuclear export signal (NES) (Table S4), the protein could in principle be shuttled in and out of the nucleus via interaction with another protein that does. Reasoning that PQN-47 might therefore spend a small fraction of time in the nucleus, we tried to trap the protein in the nucleus by treatment with Leptomycin B, a drug that specifically inhibits the nuclear export of proteins from the nucleus by CRM1/xpo-1 (Kudo et al., 1998; Ossareh-Nazari et al., 1997). Treatment conditions that were sufficient to drive the FoxO factor DAF-16 into the nucleus did not cause any accumulation of PQN-47∷GFP in the nucleus of any cells examined, including head neurons, seam or muscle cells, or intestinal cells (Fig. 6).
Figure 6. Leptomycin B, an inhibitor of export from the nucleus, does not drive PQN-47∷GFP into the nucleus.
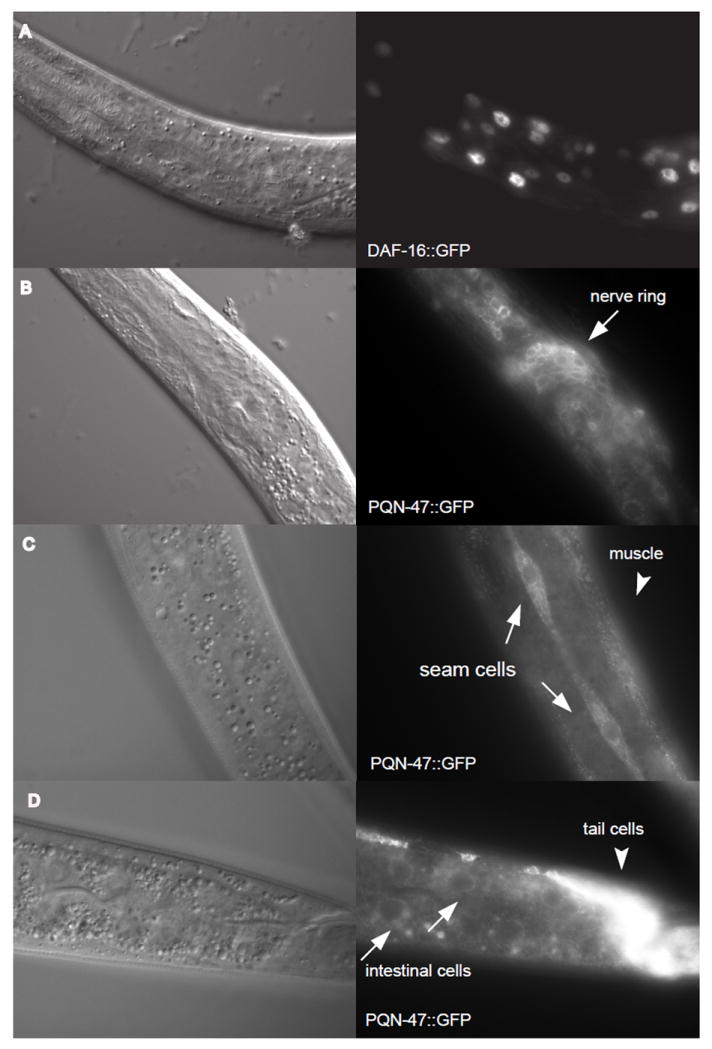
Nomarski images taken at the same focal plane as the florescent images to the right. (A) After 24 hours of LMB treatment 20 out of 20 worms observed had DAF-16∷GFP in the nucleus (representative image is shown), whereas DAF-16∷GFP was cytoplasmic in 20 out of 20 animals treated with methanol control (counted on a dissecting scope). DAF-16∷GFP begins to accumulate in the nucleus upon mounting on a slide and imaging, so over time the even the methanol control treated worms turns positive, as the stress of paralysis and imaging drives it into the nucleus (no image shown). Three hours of LMP treatment is sufficient to drive DAF-16∷GFP into the nucleus of hyp7 and intestinal cells in 9 out of 10 worms counted. Synchronized L1s are small but superficially healthy worms after 24 hours on LMP treatment compared to the methanol controls (personal observation). (B) PQN-47∷GFP was not seen in the nucleus at 24 hours of LMB treatment, nor under any conditions or cells examined. (C) Even in the larger seam cells, there is little PQN-47∷GFP in the nucleus, even after 3 hrs of LMB treatment, when 12 out of 13 animals have entirely nuclear localization of DAF-16∷GFP. (D) Although there is only faint cytoplasmic PQN-47 expression in the intestine, it remains cytosolic after 3 hrs of LMB treatment.
However, analysis of the mouse oligodendrocyte MRF gene knockout phenotype as defective in myelin secretion can be interpreted in light of our hypothesis that pqn-47 functions in the highly secretory demanding processes of molting (see Discussion).
Interactions with other molting (mlt) genes
We screened mlt genes, including those identified in a genome wide RNAi screen for mlt genes (Frand et al., 2005), for suppression of pqn-47 null and “weak” adult phenotypes. Reduction of the nhr-23 molting regulatory gene function by RNAi at larval stage 4 produced a significant reduction in the percentage of pqn-47(mg412) animals inappropriately expressing mlt-10p∷gfp-pest as adults (84% to 47%, n>60) and their subsequent death due to the supernumerary molt (82% to 10%) (Fig. S5). We did not find any molting gene inactivations that suppressed the tm2707 arrest and allowed animals to complete the L1 molt when fed in the same generation.
We determined if RNAi inactivation of mlt genes influence the expression of pqn-47 promoter and translational GFP reporters. Synchronized L1s were observed over the course of their development into adults. Inactivation of rpl-23, rps-22, rps-23, which encode ribosomal proteins, and rab-1 and kin-2 reduced expression of the promoter reporter. Inactivation of pas-6, pbs-5, and rpn-7, which encode proteasome subunits, increased the expression level of promoter pqn-47 fusion genes. Inactivation of alg-1, vha-15, F43G9.12, and smgl-1 increased expression of only the translational pqn-47 reporter, while inactivation of pqn-47 reduced it (data not shown).
pqn-47(tm2707) arrests as an L1 larvae in the molt with the mlt-10p∷gfp-pest reporter on, but the reporter is quickly down-regulated in arrested animals, suggesting that either pqn-47 acts downstream of mlt-10 or that the two genes act in parallel pathways, or that maternal pqn-47 is sufficient until after the induction of mlt-10. Because the hypomorphic allele mg412 causes reanimation of the mlt-10p∷gfp-pest reporter in adults, we reasoned that pqn-47 is likely not downstream of mlt-10. Therefore, we screened the subset of mlt genes that arrest with the mlt-10p∷gfp-pest reporter off, and therefore are upstream of mlt-10. RNAi inactivations of xrn-1,nhr-23, mlt-8, or nas-37 do not cause down-regulation of our pqn-47 promoter or translational reporters (data not shown).
Interactions with the heterochronic genes
The supernumerary adult molt of pqn-47(mg412) animals has elements of heterochrony, in that it is a reiteration of a fate normally associated with earlier larval stages, the molt. However, pqn-47(mg412) mutant animals are distinct in that they attempt to shed a properly specified adult cuticle rather than a cuticle that has features of the larval cuticle. pqn-47(mg412) mutants secrete adult cuticle alae as L4s (seen on the detached cuticle during an adult molt in Fig. S1A, also see Table S5A). Whereas let-7 and lin-29 animals have increased numbers of seam cells relative to wild type animals (resulting from an extra round of division), pqn-47(mg412) mutants do not (Table S4B). This suggests that division of seam cells, which occurs after the previous molt, may not be absolutely required for the initiation of a subsequent molt, and thus is separable from the molting cycle. Finally, proper temporal regulation of col-19, an adult-specific collagen, confirms that an adult cuticle has been synthesized by the hypodermis in pqn-47(mg412) adult animals (Table S1C). In contrast to lin-29 and other retarded heterochronic mutants that do not express col-19∷GFP as adults (Abrahante et al., 1998). The heterochronic mir-48;mir-84 double mutant also does not express the col-19∷GFP transgene appropriately in hyp7 cells as adults (Abbott et al., 2005), although they do make alae at the appropriate time. Therefore, the adult molt observed in the pqn-47(mg412) mutant appears not to be a reiteration of an earlier larval molt as are those of let-7 and lin-29, but a genuine adult molt. Thus pqn-47(mg412) defines a mutant with defects in regulation of molting, rather than a heterochronic mutant.
However, RNAi knockdown of precocious heterochronic genes lin-41, lin-28, and lin-14 completely suppress the hypomorphic pqn-47(mg412) retarded adult expression of mlt-10 and subsequent supernumerary molt (Fig. S5B and C), suggesting that pqn-47 can only specify the supernumerary adult stage molt if the animal has developed though a normal pattern of earlier larval molts.
The genetic interactions of pqn-47 with other stage-specific phenotypes were also characterized. We found that the cuticle fluid filled blister (bli) phenotypes of bli-1, a cuticle collagen that is required for proper strut formation, and bli-5, a protease inhibitor required for cuticle integrity, were suppressed by the weak loss of function allele pqn-47(mg412) (Fig. S5D). The process of making a new cuticle and molting precludes the formation of these blisters since larvae and mutants that continue to molt, for example, let-7(mg279), do not form them. Alternately, the absence of these blisters in pqn-47(mg412) adults may indicate that the cuticle does retain some larval characteristics. Sensitivity to bli RNAi clones may represent an easy and sensitive assay for adult molting.
Somatic cell proliferation ends at the adult stage of wild type animals, and the DNA replication inhibitor FUDR can be used to stop DNA replication. However, the adult expression of mlt-10p∷gfp-pest and the supernumerary molt of pqn-47(mg412) are completely suppressed by FUDR (Fig. S6), suggesting that this molt requires either cell divisions or DNA endoreduplication to be triggered. pqn-47(mg412) young adults have a normal number of seam cells and have alae, reflecting that the seam cells do not divide and fuse again after their last normal division between the L3 to L4 molt. Therefore a preceding seam cell division is not required to enter into a molt; rather, a DNA replication event is required for the supernumerary molt of pqn-47(mg412). It is possible that intestinal endoreduplications which occur before each molt, leading to a 32× DNA content in adult intestinal nuclei, contribute to keeping time of the age of the animal or may be required for the completion of a molt. If these endoreduplications are inhibited by FUDR, the threshold of DNA content in the intestine may not be sufficient to trigger a molt. The suppression of the supernumerary molt in the pqn-47(mg412) mutant by the precocious heterochronic mutants may be an indirect effect of the intestinal endoreduplication defects of these mutants. For example, lin-14 mutants do not have the L1 intestinal duplications (Hong et al., 2000).
Over expression of PQN-47 causes precocious phenotypes
Higher than endogenous levels of PQN-47 is likely toxic, as pqn-47 transgene injected animals produced mostly dead eggs and only a few viable F1s. We were only able to obtain transmitting lines when injecting a small dose (0.1ng/ul) of transgenic DNA into mutant animals. Over-expression of PQN-47 causes precocious development of adult alae. In wild type animals, cuticle alae are first discernable under the soon-to-be-shed larval stage four cuticle as the new adult cuticle is formed by the hypodermis. Wild type late L3 animals synthesize their L4 cuticle, which is devoid of alae. However, precocious heterochronic mutants such as lin-41 synthesize an L4 cuticle at the late L3 stage with discernable “adult” alae, usually with gaps. Strains over-expressing PQN-47 form alae precociously on the L4 cuticle, as evident from when the alae can first be discerned (Table S2B) and by their appearance on discarded L4 cuticles. Since heterochronic mutants also influence the developmental timing of gonadal migration (Tennessen et al., 2006) and vulval morphogenesis (Johnson et al., 2009) with respect to each other and the molting cycle, we examined both in order to control for stage in the determination of when alae develop. Over-expression of PQN-47 causes precocious alae in animals staged either by extent of gonad migration or vulval development. It is interesting to note that we did not observe gapped alae, but rather saw complete alae, weak alae, or none at all, perhaps suggesting that it is a non cell autonomous switch toggled by PQN-47 levels.
pqn-47(tm2707)IsPQN-47∷GFP strains burst, after the L4-A molt, through a vulva that has already started laying eggs. An independent laboratory also observed that a pqn-47 construct (with a basically identical promoter to our rescuing and expression reporters but lacking the endogenous 3’UTR) exhibits 1% bursting (Andachi, 2008). We see higher levels of bursting (25%, see Table S2C) with our constructs that do contain the endogenous 3’UTR, suggesting that let-7 binding to the over-expressed pqn-47 3’UTR might deplete let-7 enough to cause bursting.
PQN-47 over-expression lines also yield animals that lay older embryos than do wild type animals. About 25% of F1 progeny from over-expressing mothers hatch at 0-4 hours post egg lay whereas wild type mothers lay eggs that hatch 9 hours later (data not shown). The simplest interpretation is that the mother retains the developing embryos longer prior to laying than wild type mothers do. F1s survive synchronization by starvation and resume growth on food normally (Fig. 2C), so this method was used rather than using egg lays for any studies needing comparison of developmental timing.
Discussion
We have established that the conserved PQN-47 protein mediates an essential step in the molting program of C. elegans. The null phenotype of pqn-47 is a penetrant arrest at the first larval molt. The arrested animals begin their molting cycle normally, activating the cyclically expressed molting gene mlt-10p∷gfp-pest normally, and activating release of the L1 cuticle. But the release of this cuticle fails and the animal arrests at the larval stage one to larval stage two molt. Smaller decreases in pqn-47 gene activity, defined by the pqn-47(mg412) point mutant and recapitulated by pqn-47 inactivation by RNAi, allows successful larval molts but a failure to cease the molting cycle in adults. It is possible that an aberrant last molt in these weak mutants fails to signal an end to molting.
A functional PQN-47∷GFP fusion protein is localized to puncta in the cytoplasm of many neurons, gland like cells, and other cells that might regulate the molting cycle of C. elegans. As in other molting organisms, there is evidence for a neurosecretory component to the regulation of the molting cycle (Frand et al., 2005). pqn-47 in neurons could contribute to that function. Given the strong expression of pqn-47 and its human orthologues in neurons, we expect that it is required in them. However we failed to see convincing rescue of pqn-47(tm2707) under a limited set of tissue specific promoters, including the pan-neruonal unc-119 promoter (data not shown). Perhaps pqn-47 function is required in tissues or particular cells not covered by the promoters we surveyed. However, we saw penetrant L1 arrest only in two generation feeding of pqn-47 (RNAi) in enhanced RNAi (eri) strains, suggesting perhaps that neuronal function is required for the earliest larval arrest, or that low levels of PQN-47 provide enough function (Fig. S7). The PQN-47 cytoplasmic puncta partially colocalize with endoplasmic reticulum and could represent sites of secretory activity, perhaps part of the propagation or response to a molting signal. As in other molting organisms, there is evidence for an endocrine component to the regulation of the molting cycle (Frand et al., 2005). pqn-47 in neurons could contribute to that function. The expression of pqn-47 is not cyclical with the molting cycle, nor did we observe its localization within any of the cells that express it change with the molting cycle, suggesting that it may not be a regulated component of the molting cycle but instead necessary for the secretion or response to other regulated components.
About 15% of over 90 pqn genes have been reclassified as activated in blocked unfolded protein response (abu) and are now implicated in ER function (Urano et al., 2002). Non-canonical UPR protein and pqn-47 expression is repressed by OCTR-1 (a putative octopamine G-protein coupled catecholamine receptor) activity in sensory neurons (Sun et al., 2011). Therefore pqn-47 is either regulated in distal tissues via an endocrine signals, or since pqn-47 is expressed in the same sensory neurons as octr-1, its expression could be massively induced in the sensory neurons, which would have been diluted to four fold when pooled with whole animals RNA for analysis.
The genome sequence annotation of the protein orthologue family of which PQN-47 is a member is misleading. PQN-47 has strong conservation in animals, including humans, annotated as C11orf9, but this protein family was initially assigned a functional annotation based on much weaker homology to the NDT80/PhoG DNA binding domain of yeast, and the domain has been annotated as PF05224 NDT80/PhoG. Reciprocal BLASTp and PSI searches performed manually do not identify NDT80 or PhoG as having any similarity to PQN-47, giving a score that lies below standard cut-off thresholds (Table S2). Alignment over the PF05224 region (Fig. 3B) shows weak conservation between the yeast genes and PQN-47 with an insignificant E value of .86 between yeast and C. elegans, in contrast to the almost 100 logs more significant sequence similarity between PQN-47 and the human or other animal family members (complete alignment in Fig. S2). More recently, the mammalian orthologue of pqn-47, C11orf9/Gm98, has been identified via knockout mutations directed to oligodendrocytes, myelinated glial cells that support neurons by insulating them, as a key factor in oligodendrocyte differentiation, and is now called myelin gene regulatory factor (MRF)(Emery et al., 2009). This gene is clearly an orthologue where the yeast genes are not.
A high throughput study using commercial antibodies to the human MRF protein shows cytoplasmic localization by antibody staining in normal tissue, with occasional nuclear localization mainly seen in cell lines. Furthermore expression and cytoplasmic immunostaining is especially strong in adrenal cortex and macrophages, both of which are secretory tissues (Berglund et al., 2008) (see http://www.proteinatlas.org/ENSG00000124920/normal/adrenal+gland). While a tissue culture experiment with tagged Myelin Regulating Factor/GM98/C11orf9, the mouse orthologue of PQN-47, showed nuclear localization (Emery et al., 2009), our cytological characterization of a functional PQN-47∷GFP protein fusion, and the very marginal homology that PQN-47 bears to the yeast NTD80 transcription factor, suggest that it resides mainly in the cytoplasm. The formal possibility remains that only a small fraction of (undetectable) PQN-47∷GFP in the nucleus is functional and responsible for the complete rescue of the mutant and that the overwhelming preponderance of PQN-47∷GFP we observe in the cytoplasm corresponds to nonfunctional or inactive proteins. However, when over- expressed polyglutamine proteins form aggregates, they are usually non-functional (Michelitsch and Weissman, 2000). Future experiments with antibody staining of endogenous protein, forced localization of PQN-47 into the nucleus with an engineered NLS, or detection of DNA binding might establish if nuclear rather than cytoplasmic localization is required for function, and could help resolve this apparent inconsistency with the localization of a construct containing the mouse homolog MRF (not shown to be rescuing). Based on the significant conservation between the animal orthologues, we expect PQN-47/MRF to have similar function.
One attractive model for pqn-47 function in the endoplasmic reticulum is that each C. elegans molt is a time of maximal secretion of collagen from the pharynx and hypodermis, but also of proteases for the release of the previous collagenous cuticle. Similarly, the mouse functions most implicated in the function of the pqn-47 orthologue MRF is secretion of myelin basic protein and various co-regulated secretions from oligodendrocytes (Emery et al., 2009). Myelin regulatory factor emerged from a differential expression screen of oligodendrocytes and a knockout mutation in developing oligodendrocytes that causes a dramatic decrease in the production of myelin sheath components, which are secreted proteins (Emery et al., 2009). PQN-47, by analogy, may mediate features of secretion that are peculiar to the molts, for example the massive export of collagens or collagenases. The arrest point at the time of proteolytic release of the cuticle suggests a defect in the secretion of proteases involved in release. However, an earlier misspecification of a molting program could trigger the later arrest during the aberrant molting cycle. Alternatively, PQN-47 may be required for the efficient secretion of endocrine signals either from neuroendocrine cells, or from hypodermal tissues, like the seam cells, that are remodeled during the molt to signal back to neurosecretory cells that a successful molting cycle has been executed.
Although molting is restricted to the Ecdysozoan animals, pqn-47 is distributed beyond the molting clade of animals, ranging from mammals to cnidarians to choanoflagellates. It is not conserved in fungi or most protists. The PQN-47 human orthologue, C11orf9, is up-regulated in malignant but not benign models of cancer and is significantly up-regulated in invasive and or metastatic cancers relative to healthy tissues. It is expressed widely, including in brain, kidney, pancreas, uterus, testis, fetal heart and liver, respiratory, and other tissues (Berglund et al., 2008; Kiemer et al., 2001; Stohr et al., 2000). A role in secretion of, for example, niche signal factors in these tumors and tissues could be a common theme.
pqn-47 could be involved in the propagation or response to a molting signal. As in other molting organisms, there is evidence for a neurosecretory component to the regulation of the molting cycle (Frand et al., 2005). pqn-47 in neurons could contribute to that function. About 15% of over 90 pqn genes have been reclassified as activated in blocked unfolded protein response (abu) and are now implicated in ER function (Urano et al., 2002). Non-canonical UPR protein and pqn-47 expression is repressed by OCTR-1 (a putative octopamine G-protein coupled catecholamine receptor) activity in sensory neurons (Sun et al., 2011). Therefore pqn-47 is either regulated in distal tissues via endocrine signals, or since pqn-47 is expressed in the same sensory neurons as octr-1, its expression could be massively induced in the sensory neurons.
Overexpression of PQN-47 is sufficient to cause misspecification of the cuticular identity. This is evidence that not only is PQN-47 required for molting, but its activity may be regulated during the molt to specify temporal cell fates. In this regard, PQN-47 appears to tie the molting cycle to the heterochronic gene pathway. Interestingly, precocious heterochronic mutants that mis-specify adult fates at earlier larval stages also suppress the molting phenotype of the pqn-47(mg412) weak allele, also suggesting a close tie between the molting cycle and specification of temporal fates. And weak alleles of the let-7 miRNA gene also cause supernumerary molts, like the weak pqn-47 allele. These miRNAs may regulate the activity of pqn-47 at later larval and adult molts. Although some links between hormones and heterochronic genes have been established, for example ecdysone regulates let-7 microRNA levels in Drosophila (Caygill and Johnston, 2008; Garbuzov and Tatar, 2010; Sempere et al., 2003), there is much more to learn about how they are integrated (Thummel, 2001).
Materials & Methods
Worm husbandry
Worms were grown on OP50 unless for RNAi, and at 15-25°C degrees as noted, using standard protocols.
Generation of transgenics
pqn-47 rescuing arrays used simple (bluescript vector DNA), rather than complex (genomic) DNA as filler. Injections, corroboration with multiple independent lines, integration by UV irradiation and subsequent backcrossing were done with standard protocols.
Rescue with the genomic locus of F59B10.1 PCR product from amplification of wildtype genomic DNA and oligos promoterpqn-47-7124low” attgctaaagccatcagaggg” and after3’utrpqn- 47low” aaacatcacaggtacaatcgg” was injected into mg412 or tm2707/mnCi animals at 0.1 ng/ul, with 85ng/ul bluescript vector DNA as carrier, and 10ng/ul myo-2p∷ gfp as a co-injection marker. In the case of tm2707, heterozygotes (balanced with a wild type copy on the balancer mnCi chromosome) gave multiple independent transmitting lines. Two days post egg lays, F1 worms trapped in the L1 molt were picked and examined under a UV light and scored for the co-injection marker, myo-3p∷gfp and compared to animals that had not arrested at the L1 molt. L1 molt animals usually did not have the array, where as animals developing normally always did, reflecting that a functional copy of pqn-47 is necessary and sufficient for viability in animals homozygous for tm2707. Animals that did have the co-injection marker but that arrested nonetheless are presumed to be mosaic for pqn-47 in tissues that require its activity. Different independent lines’ arrays may be less stably inherited, explaining why they have different rates of rescue, however, multiple lines show significant rescue. C terminal PQN-47∷CHERRY and GFP translational fusions were unable to rescue mg412 (and not used for any studies) therefore GFP was engineered into a non-conserved internal region of exon 3 instead, which did rescue. Multiple independent lines injected with 1ng/ul of PQN-47∷GFP showed significant rescue, and again the balancer was often spontaneously lost allowing for growth of animals only in the presence of the array (Table S3A). The PQN-47∷GFP construct used in all studies described has GFP inserted into an area of low conservation in exon 3 using a PCR based 3 step sewing strategy (see Figure 3A for cartoon, and Fig. S3 for exact insertion point between AA 171/172). The constructs contain all endogenous genomic features; the full promoter to upstream gene F59B10.2, introns, and full endogenous 3’UTR.
First, the 5’ fragment contains the promoter and first two and half exons from PCR of genomic DNA using oligos 1stpartexon3pqn-47low “agcagttcccggttggctc” and promoterpqn-47-7124low “attgctaaagccatcagaggg.” The 3’ Fragment is the C terminal part of pqn-47 (2nd half of exon 3 thru the 3’UTR) amplified from genomic DNA using oligos: 2ndpartexon3pqn-47up “gcagtcaatcaacctacaaaca” and after3’utrpqn-47low” aaacatcacaggtacaatcgg.” GFP was amplified from pD95_79 with oligos: gfptopqnlow “tgtttgtaggttgat tgactgctttgtatagttcatccatgcc” and gfptopqnup “gagccaaccgggaactgctatgagtaaaggagaaga acttttc.” The GFP and 3’ fragments were sewn together with oligos: after3’utrpqn-47low” aaacatcacaggtacaatcgg” and gfptopqnlow “tgtttgtaggttgattgactgctttgtatagttcatccatgcc” creating fragment GFP-3’. The 5’ fragment and the GFP-3’ fragment were subsequently sewn together using Oligos: promoterpqn-47-7124low “attgctaaagccatcagaggg” and after3’utrpqn-47low “aaacatcacaggtacaatcgg.” This final PCR product PQN-47∷GFP (5’-GFP-3’) was injected into tm2707/mnCi animals at 1ng/ul, with 40ng/ul bluescript carrier DNA, and 35ng/ul ttx-3p∷dsred as a co-injection marker. Promoter pqn-47 fusions to GFP were made with the same 4.6kb 5’ region as the rescuing translational GFP reporter described above. Oligos promoterpqn-47-7124low “attgctaaagccatcagaggg” and Endexon 2pqn-47 back “tcttctcc tttactcatctgaaaaatgccattttggttttgtc” were used to amplify genomic DNA. The gfp∷pest fragment was made by PCR using Pqn-47 exon2fusegfpup “caaaatggcatttttcagatgagtaaaggaga agaacttttcac” and CAW31 on pARF207 (Frand et al., 2005). The promoter and 1st two exon fragment and the gfp-pest fragments were sewn together using oligos promoterpqn-47-7124low and CAW32. This pqn-47p∷gfp-pest was injected at 2ng/ul, with 40ng/ul bluescript carrier DNA, and 33ng/ul ttx-3p∷dsred as a co-injection marker, and 4ng/ul of pha-1(+) into a pha-1 mutant strain (for selection of the array at 25°) to generate mgEx773 in GR1681. punc-129∷ssCHERRY-KDEL is Kaplan lab plasmid KP#1532 (McEwen and Kaplan, 2008).
Phenotypic characterization
For lethal alleles, homozygous animals were identified by the absence of the co-injection marker contained in the rescuing array. Adult phenotypes, unless otherwise specified, were scored by number of gravid animals expressing mlt-10p∷gfp-pest plus the number of gravid adults trapped in cuticle (supernumerary molt) plus dead gravid animals. Alae: animals were immobilized and staged based on gonad and vulval morphology and scored for alae.
RNAi
Unless otherwise noted, populations of L1 larvae were synchronized and plated on bacterial lawns expressing double stranded RNAi to the gene of interest according to standard protocols. Vector refers to the empty RNAi vector-containing strain, L4440, which in all cases was used as the control in RNAi experiments. Dilution of RNAi cultures: diluted with vector cultures such that 1:10 dilutions had one part (by volume of stationary phase culture) bacterial culture harboring vector targeting gene of interest and 9 parts of bacterial culture with control vector L4440. Conversely, control and experimental bacterial cultures were concentrated to 1/10th volume by centrifugation in experiments using concentrated RNAi. Animals were grown at 25°C unless otherwise noted. L4 feeding RNAi experiments used synchronized L1 animals grown up on OP50 and transferred onto RNAi plates as L4s. L4 animals were put on to RNAi plates and allowed to lay eggs for 24 hours, removed, and their F1 progeny were assayed for two-generation RNAi experiments.
Drug treatments
FUDR, final 100ng/ml (or water control) on plates, was allowed to soak in around OP50 lawns and dry prior to adding L4 stage young adults. Leptomycin B (LMB) (Sigma) or the same volume of solvent (70% methanol control) was added on top of the OP50 lawns on 35mm plates, such that after drying and complete equilibration throughout the plates, LMB was at 50ng/ml final concentration. Modified from (Bussing et al., 2010; Segal et al., 2001). Worms were kept on plates for 3-24 hours, observed on a dissecting scope or freeze cracked, methanol fixed, and washed in PBS with DAPI.
Microscopy
Animals were mounted on agarose pads and immobilized with BDM or Na Azide. Photos were taken on a Zeiss Axioplan or Zeiss Apitome. Ciliated neurons identified by DiI filling. (Perkins et al., 1986; Starich et al., 1995). The dye enters 6 amphid (ASI, ADL, ASK, AWB, ASH and ASJ) and the two phasmid (PHA and PHB) neurons (Herman, 1984). Live worms were imaged with a 10X objective under dissection stereomicroscopes.
Supplementary Material
Highlights.
pqn-47 is required in C. elegans to transit each molt
Null pqn-47 mutants fail to complete the first molt while a weak loss of function mutant molts an extra time as an adult
pqn-47 is highly conserved in animals, but not in fungi or plants, and does not have domains of known function
pqn-47 is expressed in neurons, gland-like cells, and seam cells, but not the much larger hyp7 hypodermal cells which underlie the cuticle
A PQN-47 reporter that fully rescues the mutants is in cytoplasmic puncta
Acknowledgments
We thank the Caenorhabditis elegans Genetic center, (CGC) and the knockout consortia of David Baillie, Don Moerman, and Bob Barstead, as well as the National Bioresource Project of Shohei Mitani for knockout strains. We thank Gabe Hayes, Justine Melo, Yuval Tabach, and John K. Kim, Mark R. Brown, and the entire Ruvkun lab for strains, reagents and useful conversations. We also thank Jason McEwen for the KDEL plasmid, the Kaplan lab, and Annie L. Conery and Jonah Larkins-Ford for sorting the worms for SF3. We thank Gabe Hayes and Justine Melo for helpful comments on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Sascha Russel, Email: russel@molbio.mgh.harvard.edu.
Alison R. Frand, Email: AFrand@mednet.ucla.edu.
Gary Ruvkun, Email: ruvkun@molbio.mgh.harvard.edu.
Bibliography
- Abbott AL, Alvarez-Saavedra E, Miska EA, Lau NC, Bartel DP, Horvitz HR, Ambros V. The let-7 MicroRNA Family Members mir-48, mir-84, and mir-241 Function Together to Regulate Developmental Timing in Caenorhabditis elegans. Dev Cell. 2005;9:403–414. doi: 10.1016/j.devcel.2005.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrahante JE, Miller EA, Rougvie AE. Identification of Heterochronic Mutants in Caenorhabditis elegans: Temporal Misexpression of a Collagen∷Green Fluorescent Protein Fusion Gene. Genetics. 1998;149:1335–1351. doi: 10.1093/genetics/149.3.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V, Horvitz HR. Heterochronic mutants of the nematode Caenorhabditis elegans. Science. 1984;226:409–416. doi: 10.1126/science.6494891. [DOI] [PubMed] [Google Scholar]
- Andachi Y. A novel biochemical method to identify target genes of individual microRNAs: Identification of a new Caenorhabditis elegans let-7 target. RNA. 2008;14:2440–2451. doi: 10.1261/rna.1139508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglund L, Björling E, Oksvold P, Fagerberg L, Asplund A, Al-Khalili Szigyarto C, Persson A, Ottosson J, Wernérus H, Nilsson P, Lundberg E, Sivertsson Å, Navani S, Wester K, Kampf C, Hober S, Pontén F, Uhlén M. A Genecentric Human Protein Atlas for Expression Profiles Based on Antibodies. Molecular & Cellular Proteomics. 2008;7:2019–2027. doi: 10.1074/mcp.R800013-MCP200. [DOI] [PubMed] [Google Scholar]
- Bettinger JC, Lee K, Rougvie AE. Stage-specific accumulation of the terminal differentiation factor LIN-29 during Caenorhabditis elegans development. Development. 1996;122:2517–2527. doi: 10.1242/dev.122.8.2517. [DOI] [PubMed] [Google Scholar]
- Brooks DR, Appleford PJ, Murray L, Isaac RE. An essential role in molting and morphogenesis of Caenorhabditis elegans for ACN-1, a novel member of the angiotensin-converting enzyme family that lacks a metallopeptidase active site. J Biol Chem. 2003;278:52340–52346. doi: 10.1074/jbc.M308858200. [DOI] [PubMed] [Google Scholar]
- Bussing I, Yang J-S, Lai EC, Groszhans H. The nuclear export receptor XPO-1 supports primary miRNA processing in C. elegans and Drosophila. EMBO J. 2010;29:1830–1839. doi: 10.1038/emboj.2010.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caygill EE, Johnston LA. Temporal Regulation of Metamorphic Processes in Drosophila by the let-7 and miR-125 Heterochronic MicroRNAs. Current biology : CB. 2008;18:943–950. doi: 10.1016/j.cub.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery B, Agalliu D, Cahoy JD, Watkins TA, Dugas JC, Mulinyawe SB, Ibrahim A, Ligon KL, Rowitch DH, Barres BA. Myelin gene regulatory factor is a critical transcriptional regulator required for CNS myelination. Cell. 2009;138:172–185. doi: 10.1016/j.cell.2009.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entchev EV, Kurzchalia TV. Requirement of sterols in the life cycle of the nematode Caenorhabditis elegans. Semin Cell Dev Biol. 2005;16:175–182. doi: 10.1016/j.semcdb.2005.01.004. [DOI] [PubMed] [Google Scholar]
- Frand AR, Russel S, Ruvkun G. Functional Genomic Analysis of C. elegans Molting. PLoS Biol. 2005;3:e312. doi: 10.1371/journal.pbio.0030312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasch M. A matter of timing: microRNA-controlled temporal identities in worms and flies. Genes & Development. 2008;22:1572–1576. doi: 10.1101/gad.1690608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbuzov A, Tatar M. Hormonal regulation of Drosophila microRNA let-7 and miR-125 that target innate immunity. Fly (Austin) 2010;4:306–311. doi: 10.4161/fly.4.4.13008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gissendanner CR, Crossgrove K, Kraus KA, Maina CV, Sluder AE. Expression and function of conserved nuclear receptor genes in Caenorhabditis elegans. Dev Biol. 2004;266:399–416. doi: 10.1016/j.ydbio.2003.10.014. [DOI] [PubMed] [Google Scholar]
- Gissendanner CR, Sluder AE. nhr-25, the Caenorhabditis elegans ortholog of ftz-f1, is required for epidermal and somatic gonad development. Dev Biol. 2000;221:259–272. doi: 10.1006/dbio.2000.9679. [DOI] [PubMed] [Google Scholar]
- Graham LD, Kotze AC, Fernley RT, Hill RJ. An ortholog of the ecdysone receptor protein (EcR) from the parasitic nematode Haemonchus contortus. Molecular and Biochemical Parasitology. 2010;171:104–107. doi: 10.1016/j.molbiopara.2010.03.003. [DOI] [PubMed] [Google Scholar]
- Hada K, Asahina M, Hasegawa H, Kanaho Y, Slack FJ, Niwa R. The nuclear receptor gene nhr-25 plays multiple roles in the Caenorhabditis elegans heterochronic gene network to control the larva-to-adult transition. Dev Biol. 2010;344:1100–1109. doi: 10.1016/j.ydbio.2010.05.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes GD, Frand AR, Ruvkun G. The mir-84 and let-7 paralogous microRNA genes of Caenorhabditis elegans direct the cessation of molting via the conserved nuclear hormone receptors NHR-23 and NHR-25. Development. 2006;133:4631–4641. doi: 10.1242/dev.02655. [DOI] [PubMed] [Google Scholar]
- Hedgecock EM, White JG. Polyploid tissues in the nematode Caenorhabditis elegans. Dev Biol. 1985;107:128–133. doi: 10.1016/0012-1606(85)90381-1. [DOI] [PubMed] [Google Scholar]
- Herman RK. Crossover suppressors and balanced recessive lethals in Caenorhabditis elegans. Genetics. 1978;88:49–65. doi: 10.1093/genetics/88.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y, Lee RC, Ambros V. Structure and Function Analysis of LIN-14, a Temporal Regulator of Postembryonic Developmental Events in Caenorhabditis elegans. Mol Cell Biol. 2000;20:2285–2295. doi: 10.1128/mcb.20.6.2285-2295.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt-Newbury R, Viveiros R, Johnsen R, Mah A, Anastas D, Fang L, Halfnight E, Lee D, Lin J, Lorch A, McKay S, Okada HM, Pan J, Schulz AK, Tu D, Wong K, Zhao Z, Alexeyenko A, Burglin T, Sonnhammer E, Schnabel R, Jones SJ, Marra MA, Baillie DL, Moerman DG. High-Throughput In Vivo Analysis of Gene Expression in Caenorhabditis elegans. PLoS Biol. 2007;5:e237. doi: 10.1371/journal.pbio.0050237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RW, Liu LY, Hanna-Rose W, Chamberlin HM. The Caenorhabditis elegans heterochronic gene lin-14 coordinates temporal progression and maturation in the egg-laying system. Developmental Dynamics. 2009;238:394–404. doi: 10.1002/dvdy.21837. [DOI] [PubMed] [Google Scholar]
- Kiemer AK, Takeuchi K, Quinlan MP. Identification of genes involved in epithelial-mesenchymal transition and tumor progression. Oncogene. 2001;20:6679–6688. doi: 10.1038/sj.onc.1204872. [DOI] [PubMed] [Google Scholar]
- Kipreos ET. C. elegans cell cycles: invariance and stem cell divisions. Nat Rev Mol Cell Biol. 2005;6:766–776. doi: 10.1038/nrm1738. [DOI] [PubMed] [Google Scholar]
- Kostrouchova M, Krause M, Kostrouch Z, Rall JE. CHR3: a Caenorhabditis elegans orphan nuclear hormone receptor required for proper epidermal development and molting. Development. 1998;125:1617–1626. doi: 10.1242/dev.125.9.1617. [DOI] [PubMed] [Google Scholar]
- Kostrouchova M, Krause M, Kostrouch Z, Rall JE. Nuclear hormone receptor CHR3 is a critical regulator of all four larval molts of the nematode Caenorhabditis elegans. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:7360–7365. doi: 10.1073/pnas.131171898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlova T, Thummel CS. Spatial patterns of ecdysteroid receptor activation during the onset of Drosophila metamorphosis. Development. 2002;129:1739–1750. doi: 10.1242/dev.129.7.1739. [DOI] [PubMed] [Google Scholar]
- Kudo N, Wolff B, Sekimoto T, Schreiner EP, Yoneda Y, Yanagida M, Horinouchi S, Yoshida M. Leptomycin B Inhibition of Signal-Mediated Nuclear Export by Direct Binding to CRM1. Experimental Cell Research. 1998;242:540–547. doi: 10.1006/excr.1998.4136. [DOI] [PubMed] [Google Scholar]
- Lee DL. The biology of nematodes. Taylor & Francis; London: 2002. [Google Scholar]
- Letzen BS, Liu C, Thakor NV, Gearhart JD, All AH, Kerr CL. MicroRNA Expression Profiling of Oligodendrocyte Differentiation from Human Embryonic Stem Cells. PLoS ONE. 2010;5:e10480. doi: 10.1371/journal.pone.0010480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin R, Entchev EV, Kurzchalia TV, Knolker HJ. Steroid hormones controlling the life cycle of the nematode Caenorhabditis elegans: stereoselective synthesis and biology. Org Biomol Chem. 2010;8:739–750. doi: 10.1039/b918488k. [DOI] [PubMed] [Google Scholar]
- McEwen JM, Kaplan JM. UNC-18 Promotes Both the Anterograde Trafficking and Synaptic Function of Syntaxin. Mol Biol Cell. 2008;19:3836–3846. doi: 10.1091/mbc.E08-02-0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meli VS, Osuna B, Ruvkun G, Frand AR. MLT-10 defines a family of DUF644 and proline-rich repeat proteins involved in the molting cycle of Caenorhabditis elegans. Mol Biol Cell. 2010;21:1648–1661. doi: 10.1091/mbc.E08-07-0708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelitsch MD, Weissman JS. A census of glutamine/asparagine-rich regions: Implications for their conserved function and the prediction of novel prions. Proceedings of the National Academy of Sciences. 2000;97:11910–11915. doi: 10.1073/pnas.97.22.11910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss EG. Heterochronic genes and the nature of developmental time. Curr Biol. 2007;17:R425–434. doi: 10.1016/j.cub.2007.03.043. [DOI] [PubMed] [Google Scholar]
- Nelson FK, Riddle DL. Functional study of the Caenorhabditis elegans secretory-excretory system using laser microsurgery. J Exp Zool. 1984;231:45–56. doi: 10.1002/jez.1402310107. [DOI] [PubMed] [Google Scholar]
- Ossareh-Nazari B, Bachelerie Fo, Dargemont C. Evidence for a Role of CRM1 in Signal-Mediated Nuclear Protein Export. Science. 1997;278:141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- Page AP, Johnstone IL. The cuticle. In: Johnstone IL, editor. WormBook. The C elegans Research Community; 2007. [Google Scholar]
- Papp A, Rougvie AE, Ambros V. Molecular cloning of lin-29, a heterochronic gene required for the differentiation of hypodermal cells and the cessation of molting in C.elegans. Nucleic Acids Res. 1991;19:623–630. doi: 10.1093/nar/19.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli AE, Ruvkun G. Control and developmental timing by microRNAs and their targets. Annual Review of Cell and Developmental Biology. 2002;18:495–513. doi: 10.1146/annurev.cellbio.18.012502.105832. [DOI] [PubMed] [Google Scholar]
- Podbilewicz B, White JG. Cell Fusions in the Developing Epithelia of C. elegans. Developmental Biology. 1994;161:408–424. doi: 10.1006/dbio.1994.1041. [DOI] [PubMed] [Google Scholar]
- Raizen DM, Zimmerman JE, Maycock MH, Ta UD, You Y-j, Sundaram MV, Pack AI. Lethargus is a Caenorhabditis elegans sleep-like state. Nature. 2008;451:569–572. doi: 10.1038/nature06535. [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Resnick TD, McCulloch KA, Rougvie AE. miRNAs give worms the time of their lives: small RNAs and temporal control in Caenorhabditis elegans. Dev Dyn. 2010;239:1477–1489. doi: 10.1002/dvdy.22260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougvie AE. Intrinsic and extrinsic regulators of developmental timing: from miRNAs to nutritional cues. Development. 2005;132:3787–3798. doi: 10.1242/dev.01972. [DOI] [PubMed] [Google Scholar]
- Ruaud A-F, Katic I, Bessereau J-L. Insulin/Insulin-Like Growth Factor Signaling Controls Non-Dauer Developmental Speed in the Nematode Caenorhabditis elegans. Genetics. 2011;187:337–343. doi: 10.1534/genetics.110.123323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruaud AF, Bessereau JL. Activation of nicotinic receptors uncouples a developmental timer from the molting timer in C. elegans. Development. 2006;133:2211–2222. doi: 10.1242/dev.02392. [DOI] [PubMed] [Google Scholar]
- Segal SP, Graves LE, Verheyden J, Goodwin EB. RNA-Regulated TRA-1 Nuclear Export Controls Sexual Fate. Developmental Cell. 2001;1:539–551. doi: 10.1016/s1534-5807(01)00068-5. [DOI] [PubMed] [Google Scholar]
- Sempere LF, Sokol NS, Dubrovsky EB, Berger EM, Ambros V. Temporal regulation of microRNA expression in Drosophila melanogaster mediated by hormonal signals and Broad-Complex gene activity. Developmental Biology. 2003;259:9–18. doi: 10.1016/s0012-1606(03)00208-2. [DOI] [PubMed] [Google Scholar]
- Stohr H, Marquardt A, White K, Weber BH. cDNA cloning and genomic structure of a novel gene (C11orf9) localized to chromosome 11q12-->q13.1 which encodes a highly conserved, potential membrane-associated protein. Cytogenet Cell Genet. 2000;88:211–216. doi: 10.1159/000015552. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- Sun J, Singh V, Kajino-Sakamoto R, Aballay A. Neuronal GPCR Controls Innate Immunity by Regulating Noncanonical Unfolded Protein Response Genes. Science. 2011 doi: 10.1126/science.1203411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennessen JM, Gardner HF, Volk ML, Rougvie AE. Novel heterochronic functions of the Caenorhabditis elegans period-related protein LIN-42. Developmental Biology. 2006;289:30–43. doi: 10.1016/j.ydbio.2005.09.044. [DOI] [PubMed] [Google Scholar]
- Tennessen JM, Opperman KJ, Rougvie AE. The C. elegans developmental timing protein LIN-42 regulates diapause in response to environmental cues. Development. 2010;137:3501–3511. doi: 10.1242/dev.048850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thummel CS. Molecular mechanisms of developmental timing in C. elegans and Drosophila. Dev Cell. 2001;1:453–465. doi: 10.1016/s1534-5807(01)00060-0. [DOI] [PubMed] [Google Scholar]
- Tzertzinis G, Egaña AL, Palli SR, Robinson-Rechavi M, Gissendanner CR, Liu C, Unnasch TR, Maina CV. Molecular Evidence for a Functional Ecdysone Signaling System in Brugia malayi. PLoS Negl Trop Dis. 2010;4:e625. doi: 10.1371/journal.pntd.0000625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano F, Calfon M, Yoneda T, Yun C, Kiraly M, Clark SG, Ron D. A survival pathway for Caenorhabditis elegans with a blocked unfolded protein response. The Journal of Cell Biology. 2002;158:639–646. doi: 10.1083/jcb.200203086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Buskirk C, Sternberg PW. Epidermal growth factor signaling induces behavioral quiescence in Caenorhabditis elegans. Nat Neurosci. 2007;10:1300–1307. doi: 10.1038/nn1981. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Foster JM, Nelson LS, Ma D, Carlow CKS. The chitin synthase genes chs-1 and chs-2 are essential for C. elegans development and responsible for chitin deposition in the eggshell and pharynx, respectively. Developmental Biology. 2005;285:330–339. doi: 10.1016/j.ydbio.2005.06.037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


