Abstract
Modern yeast living in fleshy fruits rapidly convert sugars into bulk ethanol through pyruvate. Pyruvate loses carbon dioxide to produce acetaldehyde, which is reduced by alcohol dehydrogenase 1 (Adh1) to ethanol, which accumulates. Yeast later consumes the accumulated ethanol, exploiting Adh2, an Adh1 homolog differing by 24 (of 348) amino acids. As many microorganisms cannot grow in ethanol, accumulated ethanol may help yeast defend resources in the fruit1. We report here the resurrection of the last common ancestor2 of Adh1 and Adh2, called AdhA. The kinetic behavior of AdhA suggests that the ancestor was optimized to make (not consume) ethanol. This is consistent with the hypothesis that before the Adh1-Adh2 duplication, yeast did not accumulate ethanol for later consumption but rather used AdhA to recycle NADH generated in the glycolytic pathway. Silent nucleotide dating suggests that the Adh1-Adh2 duplication occurred near the time of duplication of several other proteins involved in the accumulation of ethanol, possibly in the Cretaceous age when fleshy fruits arose. These results help to connect the chemical behavior of these enzymes through systems analysis to a time of global ecosystem change, a small but useful step towards a planetary systems biology.
Generating ethanol from glucose in the presence of dioxygen, only to then reoxidize the ethanol, is energetically expensive (Fig. 1). For each molecule of ethanol converted to acetyl-coenzyme A, a molecule of ATP is used. This ATP would not be ‘wasted’ if pyruvate made initially from glucose were delivered directly to the citric acid cycle. This implies that yeast has a reason, transcending simple energetic efficiency, for rapidly converting available sugar in fruit to bulk ethanol in the presence of dioxygen.
Figure 1.
The pathway by which yeast make, accumulate and then consume ethanol. Enzymes in red are associated with gene duplications that, according to the transition redundant exchange clock18, arose nearly contemporaneously. The make-accumulate-consume pathway is boxed. The shunting of the carbon atoms from pyruvate into (and then out of, blue arrows) ethanol is energy-expensive, consuming a molecule of ATP (green) for every molecule of ethanol generated. This ATP is not consumed if pyruvate is oxidatively decarboxylated directly to acetyl-coenzyme A to enter the citric acid cycle directly (dashed arrow to the right). If dioxygen is available, the recycling of NADH does not need the acetaldehyde-to-ethanol reduction.
One hypothesis to explain this ‘inefficiency’ holds that yeast, which are relatively resistant to ethanol toxicity, may accumulate ethanol to defend resources in the fruit from competing microorganisms3. Although the ecology of wine yeast is certainly more complex than this simple hypothesis implies4, fleshy fruits offer a large reservoir of carbohydrate, and this resource must have value to competing organisms as well as to yeast. For example, humans have exploited the preservative value of ethanol since prehistory5.
Both the timing of Adh expression in Saccharomyces cerevisiae and the properties of the expressed proteins are consistent with this hypothesis. The yeast genome encodes two alcohol dehydrogenases (Adhs) that interconvert ethanol and acetaldehyde (Fig. 1)6. The first (Adh1) is expressed at high levels constitutively. Its kinetic properties optimize it as a catalyst to make ethanol from acetaldehyde7,8. The Michaelis constant (KM) for ethanol for Adh1 is high (17,000–20,000 µM), consistent with ethanol being a product of the reaction. After the sugar concentration drops, the second dehydrogenase (Adh2) is derepressed. This paralog oxidizes ethanol to acetaldehyde with kinetic parameters suited for this role. The KM for ethanol for Adh2 is low (600–800 µM), consistent with ethanol being its substrate.
Adh1 and Adh2 are homologs8 differing by 24 of 348 amino acids. Their common ancestor, called AdhA, had an unknown role. If AdhA existed in a yeast that made, but did not accumulate, ethanol, its physiological role would presumably have been the same as that of lactate dehydrogenase in mammals during anaerobic glycolysis: to recycle NADH generated by the oxidation of glyceraldehyde-3-phosphate (Fig. 1)9. Lactate in human muscle is removed by the bloodstream; ethanol would be lost by the yeast to the environment. If so, AdhA should be optimized for ethanol synthesis, as is modern Adh1. The kinetic behaviors of AdhA should resemble those of modern Adh1 more than those of modern Adh2, with a high KM for ethanol.
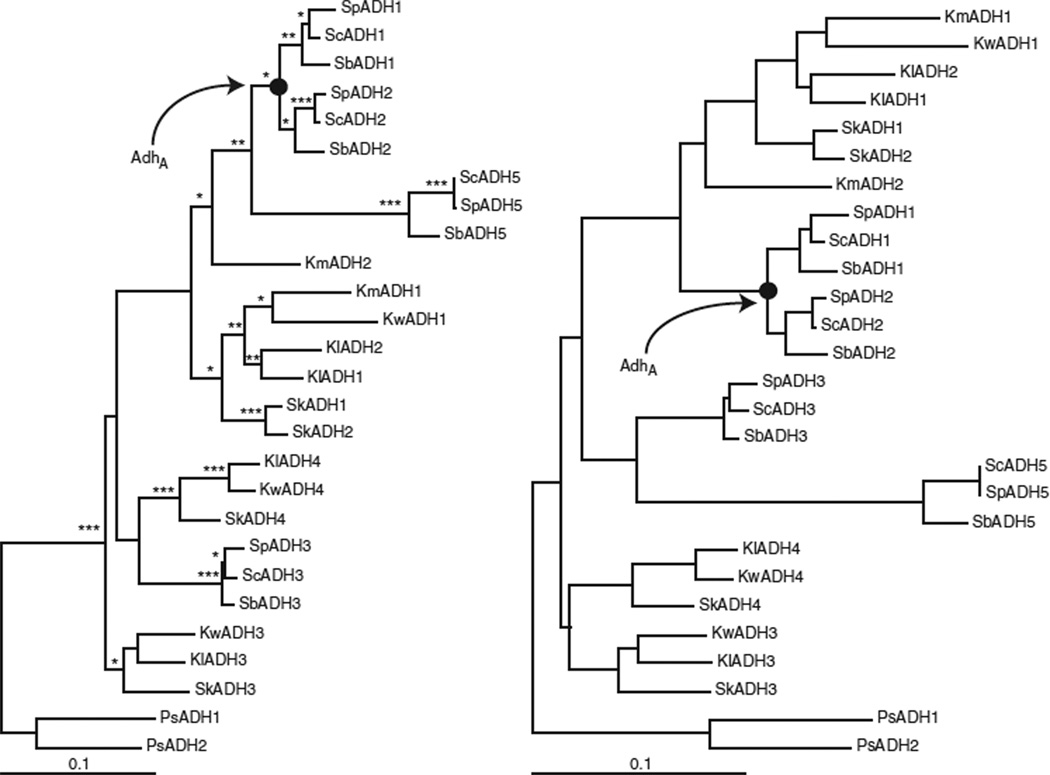
We tested this hypothesis using a paleobiochemical experiment2. We cloned and sequenced 15 additional homologs of Adh from yeasts related to S. cerevisiae (Supplementary Table 1 online). We constructed a maximum likelihood evolutionary tree using PAUP*4.0 (Fig. 2)10 to combine these with sequences already in the database. We then reconstructed maximum likelihood sequences for AdhA using both codon and amino acid models in PAML11 (Table 1). When the posterior probability that a particular amino acid occupied a particular site was > 80%, we assigned that amino acid at that site in AdhA.
Figure 2.
Maximum likelihood trees interrelating sequences determined in this work with sequences in the publicly available database. Shown are the two trees with the best (and nearly equal) ML scores using the following parameters estimated from the data: substitutions A↔C, A↔T, C↔G and G↔T = 1.00; A↔G = 2.92; and C↔T = 5.89; empirical base frequencies; and proportion of invariable sites and the shape parameter of the gamma distribution set to 0.33 and 1.31, respectively. The scale bar represents the number of substitutions per codon per unit evolutionary time. Single, double and triple asterisks represent bootstrap values greater than 50%, 70% and 90%, respectively.
Table 1.
Kinetic properties of Adh1, Adh2 and candidate ancestral AdhA
| KM (µM) | ||||
|---|---|---|---|---|
| Samplea | Ethanol | NAD+ | Acetaldehyde | NADH |
| Adh1 | 20,060 | 218 | 1,492 | 164 |
| MKD | 17,280 | 511 | 1,019 | 144 |
| MKN | 13,750 | 814 | 1,067 | 1,106 |
| MRD | 11,590 | 734 | 1,265 | 287 |
| MRN | 10,960 | 554 | 1,163 | 894 |
| MTD | 10,740 | 467 | 959 | 190 |
| MTN | NA | NA | NA | NA |
| RKD | 8,497 | 449 | 1,066 | 142 |
| RKN | 7,238 | 407 | 1,085 | 735 |
| RRD | 7,784 | 400 | 1,074 | 203 |
| RRN | 8,403 | 172 | 1,156 | 1,142 |
| RTD | 6,639 | 254 | 1,083 | 316 |
| RTN | 7,757 | 564 | 1,158 | 477 |
| Adh1b | 24,000 | 240 | 3,400 | 140 |
| Adh1c | 17,000 | 170 | 1,100 | 110 |
| Adh2b | 2,700 | 140 | 45 | 28 |
| Adh2c | 810 | 110 | 90 | 50 |
| Adh3c | 12,000 | 240 | 440 | 70 |
| Adh1c (S. pombe) | 14,000 | 160 | 1,600 | 100 |
| Adh1 (M270L)c | 19,000 | 630 | 1,000 | 80 |
| KlP20369d | 27,000 | 2,800 | 1,200 | 110 |
| KlX64397d | 23,000 | 2,200 | 1,700 | 180 |
| KlX62766d | 2,570 | 310 | 100 | 20 |
| KlX62767d | 1,560 | 200 | 3,100 | 30 |
The three letters designate the amino acids at positions 168, 211 and 236 (e.g., MKD = Met168 Lys211 Asp236). The remaining residues were the same as in Adh1, except for the following changes (using sequence numbering of Adh1 from S. cerevisiae): Asn15 Pro30 Thr58 Ala74 Glu147 Leu213 Ile232 Cys259 Val265 Leu270 Ser277 Asn324.
From ref. 8.
From ref. 16.
From ref. 30.
Kl, Kluyveromyces lactis.
When the posterior probability was < 80% or the most probabilistic ancestral states estimated using the codon and amino acid models were not in agreement, we considered the site to be ambiguous and constructed alternative ancestral sequences. For example, the posterior probabilities for two amino acids (methionine and arginine) were nearly equal at site 168 in AdhA, three amino acids (lysine, arginine and threonine) were plausibly present at site 211, and two (aspartic acid and asparagine) were plausible for site 236. To handle these ambiguities, we generated twelve (all 2 × 2 × 3 combinations) candidate AdhA proteins by constructing genes that encoded them, using these genes to transform a strain of S. cerevisiae12 from which both Adh1 and Adh2 had been deleted, and then expressing them from the Adh1 promoter. After showing that the ancestral sequences could rescue the double deletion phenotype (Supplementary Fig. 1 online), we isolated the candidate ancestral proteins, purified them to homogeneity on a Cibracon-blue agarose column13 and then analyzed their kinetic behaviors14 (Table 1).
To assess the quality of the data, we calculated Haldane values (Keq = VfKiqKp/VrKiaKb)15, where Vf and Vr are forward and reverse maximal velocities; Kia and Kiq are disassociation constants for NAD+ and NADH, respectively; and Kb and Kp are Michaelis constants for ethanol and acetaldehyde, respectively) from the experimental data. These reproduced the literature equilibrium constant for the reaction to within a factor of two. One variant, called MTN, had very low catalytic activity in both directions. We inferred that this particular candidate ancestor was not present in the ancient yeast.
Notably, the kinetic properties of the remaining ancestral AdhA candidates resembled those of Adh1 more than those of Adh2 (Table 1). From this, we inferred that the ancestral yeast did not have an Adh specialized for the consumption of ethanol, similar to modern Adh2, but rather had an Adh specialized for making ethanol, similar to modern Adh1. This suggests that before the Adh1-Adh2 duplication, the ancestral yeast did not consume ethanol. This implies that the ancestral yeast also did not accumulate ethanol under aerobic conditions for future consumption and that the make-accumulate-consume strategy emerged after Adh1 and Adh2 diverged. These interpretations are robust with respect to the ambiguities in the reconstructions.
For modern Adh1, reported KM values range from 17,000 to 24,000 µM for ethanol (our value is 20,000 µM), from 170 to 240 µM for NAD+ (ours is 220 µM), from 1,100 to 3,400 µM for acetaldehyde (ours is 1,500 µM) and from 110 to 140 µM for NADH (ours is 164 µM)14. These comparisons, together with the Haldane analysis, provide a view of the experimental error in the kinetic parameters reported here. Our interpretations are based on differences well outside of this error.
When paralogs are generated by duplication, the duplicate acquiring the new functional role is believed to evolve more rapidly than the one retaining the primitive role16. If this were generally true, one might identify the functionally innovative duplicate by a bioinformatics analysis. This may be true for many genes, but chemical principles do not obligate this outcome, and it is not manifest with these Adh paralogs. Here, the rate of evolution is not markedly faster in the lineage leading to Adh2 (having the derived function) than in the lineage leading to Adh1 (having the primitive function). Thus, a paleobiochemistry experiment was necessary to assign the primitive behavior.
The Haldane ratio relates various kinetic parameters (kcat, KM, Kdiss) that can change as a result of changing the amino acid sequence to the overall equilibrium constant, which the enzyme (being a catalyst) cannot change. Thus, if a lower KM for ethanol is selected, other terms in the Haldane equation must change to keep the ratio the same. This is observed in data for the ancestral proteins prepared here and the natural enzymes.
The assignment of a primitive function to AdhA raises a broader historical question: did the Adh1-Adh2 duplication, and the accumulate-consume strategy that it presumably enabled, become fixed in response to a particular selective pressure? Connecting molecular change to organismic fitness is always difficult17 but is necessary if reductionist biology is to move through systems biology to a planetary biology that determines why as well as how changes occurred18. Hypothetically, the emergence of a make-accumulate-consume strategy may have been driven by the domestication of yeast by humans selecting for yeast that accumulate ethanol. Alternatively, the strategy might have been driven by the emergence of fleshy fruits that offered a resource worth defending using ethanol accumulation. We might distinguish between the two by estimating the date of the Adh1-Adh2 duplication. Even with large errors in the estimate, a distinction should be possible, as human domestication occurred in the past million years, whereas fleshy fruits arose in the Cretaceous age, after the first angiosperms appeared in the fossil record 125 million years ago19 but before the extinction of the dinosaurs 65 million years ago20.
The topology of the evolutionary tree (Fig. 2) suggests that the Adh1-Adh2 duplication occurred before the divergence of the sensu strictu species of Saccharomyces21 but after the divergence of Saccharomyces and Kluyveromyces. The date of divergence of Saccharomyces and Kluyveromyces is unknown but is estimated to have occurred 80 ± 15 million years ago22. This date is consistent with a transition redundant exchange clock23, which exploits the fractional identity (f2) of silent sites in conserved twofold redundant codon systems to estimate the time since the divergence of two genes (Supplementary Note online). Between pairs of presumed orthologs from Saccharomyces and Kluyveromyces, f2 is typically 0.82, not much lower than the f2 value separating Adh1 and Adh2 (0.85)18 but much lower than paralog pairs within the Saccharomyces genome that seem to have arisen by more recent duplication (~0.98)24.
Notably, Adh1 and Adh2 are not the only pair of paralogs with an f2 value between 0.80 and 0.86 (ref. 18). We analyzed ~350 pairs of paralogs in the yeast genome that shared at least 100 silent sites and diverged by less than 120 point-accepted replacements per 100 sites, and we identified 15 pairs with f2 values between 0.80 and 0.86. These represent eight duplications that occurred near the time of the Adh1-Adh2 duplication, if f2 values are assumed to support a clock.
These duplications are not randomly distributed in the yeast genome. Rather, six of the eight duplications involve proteins that participate in the conversion of glucose to ethanol (Table 2). Furthermore, the enzymes arising from the duplicates are those that seem, from expression analysis, to control flux from hexose to ethanol1,25. These include proteins that import glucose, pyruvate decarboxylases that generate acetaldehyde from pyruvate, the transporter that imports thiamine for these decarboxylases and the Adhs (Fig. 1). If the f2 clock (within its expected variance) is assumed to date paralogs in yeast, this cluster suggests that several proteins other than Adh duplicated as part of the emergence of the new make-accumulate-consume strategy, near the time that fleshy fruit arose.
Table 2.
Duplication in the Saccharomyces cerevisiae genome where 0.80 < f2 < 0.86
| SGD name | gi number | Trivial name | Annotation and comments |
|---|---|---|---|
| Inosine-5′-monophosphate dehydrogenase family (3 paralogs, 3 pairs, 2 duplications)a | |||
| f2 = 0.803b | Pair associated with Wolfe duplication blocks 1 and 44 | ||
| YAR073W | gi|456156 | IMD1 | Nonfunctional homolog, near telomere, not expressed |
| YLR432W | gi|665971 | IMD3 | Inosine-5′-monophosphate dehydrogenase |
| f2 = 0.825b | |||
| YLR432W | gi|665971 | IMD3 | Inosine-5′-monophosphate dehydrogenase |
| YHR216W | gi|458916 | IMD2 | Inosine-5′-monophosphate dehydrogenase |
| Subfamily pair: YHR216W and YAR073W | f2 = 0.93 (proposed recent duplication creating a pseudogene) | ||
| Sugar transporter family A (4 paralogs, 4 pairs, 3 duplications)c | |||
| f2 = 0.805b | Pair not associated with any duplication block | ||
| YJR158W | gi|1015917 | HXT16 | Sugar transporter repressed by high glucose levels |
| YNR072W | gi|1302608 | HXT17 | Sugar transporter repressed by high glucose levels |
| f2 = 0.806b | Pair not associated with any duplication block | ||
| YDL245C | gi|1431418 | HXT15 | Sugar transporter induced by low glucose, repressed by high glucose |
| YNR072W | gi|1302608 | HXT17 | Sugar transporter repressed by high glucose levels |
| f2 = 0.809b | Pair not associated with any duplication block | ||
| YJR158W | gi|1015917 | HXT16 | Sugar transporter repressed by high glucose levels |
| YEL069C | gi|603249 | HXT13 | Sugar transporter induced by low glucose, repressed by high glucose |
| f2 = 0.810b | Pair not associated with any duplication block | ||
| YEL069C | gi|603249 | HXT13 | Sugar transporter induced by low glucose, repressed by high glucose |
| YDL245C | gi|1431418 | HXT15 | Sugar transporter |
| Subfamily pair: YEL069C and YNR072W | f2 = 0.932 (proposed recent duplication) | ||
| Subfamily pair: YJR158W and YDL245C | f2 = 1.000 (proposed very recent duplication) | ||
| Chaperone family A (2 paralogs, 1 pair, 1 duplication)a | |||
| f2 = 0.81 | Pair associated with Wolfe duplication block 48 | ||
| YMR186W | gi|854456 | HSC82 | Cytoplasmic chaperone induced 2–3 fold by heat shock |
| YPL240C | gi|1370495 | HSP82 | Cytoplasmic chaperone, pheromone signaling, Hsf1p regulation |
| Phosphatase/thiamine transport family A (2 paralogs, 1 pair, 1 duplication)c | |||
| f2 = 0.818 | Pair not associated with any duplication block | ||
| YBR092C | gi|536363 | PHO3 | Acid phosphatase implicated in thiamine transport |
| YBR093C | gi|536365 | PHO5 | Acid phosphatase, one of three repressible phosphatases |
| Pyruvate decarboxylase family A (2 paralogs, 1 pair, 1 duplication)c | |||
| f2 = 0.835 | Pair not associated with any duplication block | ||
| YlR044C | gi|1360375 | PDC1 | Pyruvate decarboxylase, major isoform |
| YlR134W | gi|1360549 | PDC5 | Pyruvate decarboxylase, minor isoform |
| By ortholog analysis, Saccharomyces bayanus (gi|515236) diverged from S. cerevisiae after the f2 = 0.835 duplication; Kluyveromyces diverged before. | |||
| Glyceraldehyde-3-phosphate dehydrogenase family (3 paralogs, 3 pairs, 2 duplications)c | |||
| f2 = 0.845b | Pair not associated with any duplication block | ||
| YJL052W | gi|1008189 | TDH1 | Glyceraldehyde-3-phosphate dehydrogenase |
| YGR192C | gi|1323341 | TDH3 | Glyceraldehyde-3-phosphate dehydrogenase |
| f2 = 0.845b | Pair not associated with any duplication block | ||
| YJL052W | gi|1008189 | TDH1 | Glyceraldehyde-3-phosphate dehydrogenase |
| YJR009C | gi|1015636 | TDH2 | Glyceraldehyde-3-phosphate dehydrogenase |
| Subfamily pair: YJR009C and YGR192C | f2 = 0.991 (proposed very recent duplication) | ||
| Alcohol dehydrogenase family (2 paralogs, 1 pair, 1 duplication)c | |||
| f2 = 0.848 | Pair not associated with any duplication block | ||
| YMR303C | gi|798945 | Adh2 | Alcohol dehydrogenase, glucose-repressible |
| YOL086C | gi|1419926 | Adh1 | Alcohol dehydrogenase, constitutive |
| Spermine transporter family (2 paralogs, 1 pair, 1 duplication)a | |||
| f2 = 0.86 | Pair associated with Wolfe duplication block 34 | ||
| YGR138C | gi|1323230 | TPO2 | Spermine transporter activity |
| YPR156C | gi|849164 | TPO3 | Spermine transporter activity |
| Sugar transporter family B (3 paralogs, 3 pairs, 2 duplications)c | |||
| f2 = 0.847b | Pair not associated with any duplication block | ||
| YDR343C | gi|1230670 | HXT6 | Sugar transporter, high-affinity high basal levels |
| YDR345C | gi|1230672 | HXT3 | Sugar transporter, low-affinity glucose transporter |
| f2 = 0.854b | Pair not associated with any duplication block | ||
| YDR342C | gi|1230669 | HXT7 | Sugar transporter, high-affinity, high basal levels |
| YDR345C | gi|1230672 | HXT3 | Sugar transporter, low affinity |
| Subfamily pair: YDR342C and YDR343C | f2 = 0.994 (proposed very recent duplication) | ||
Not associated with fermentation. These are associated with duplication blocks in the yeast genome16, where the high value of f2 (typically equilibrated in block paralog pairs) may reflect either variance or selective pressure to conserve silent sites in individual codons.
These pairs represent a family generated with a single duplication with f2 value between 0.80 and 0.86 and subsequent duplication(s) in the derived lineages. Trees are shown in Supplementary Figure 2 online. Paralog pairs were considered only if they have with at least 100 aligned silent sites and are not separated by more than 120 point-accepted mutations per 100 aligned amino acid sites.
Associated with the pathway to make, accumulate and then consume ethanol. Genes involved in the fermentation pathway that are not rate-limiting1,25 generally do not have duplicates in the yeast genome (e.g., hexokinase, glucose-6-phosphate isomerase, phosphofructokinase, aldolase, triose phosphate isomerase and phosphoglycerate kinase are all present in one isoform). Enolase has two paralogs (ENO1 and ENO2), where f2 = 0.946. These are distantly related to a homolog known as ERR1, with the silent sites equilibrated. Phosphoglycerate mutase has three paralogs, GM1, GM2 and GM3, with silent sites that are essentially equilibrated (Supplementary Note online).
The six duplications proposed to be part of the emergence of the make-accumulate-consume strategy (with f2 values between 0.80 and 0.86) are not associated with one of the documented blocks of genes duplicated in ancient fungi, possibly as part of a whole-genome duplication16,26. But two duplications in genes that are not associated with fermentation that have f2 values between 0.80 and 0.86 are part of a duplication block (Table 2 and Supplementary Note online). The silent sites for most gene pairs associated with blocks are nearly equilibrated (with the prominent exception of ribosomal proteins), suggesting that most blocks arose by duplications more ancient than those with f2 values between 0.80 and 0.86. Therefore, the hypothesis that a set of six time-correlated duplication (Fig. 1 and Table 2) generated the make-accumulate-consume strategy in yeast near the time when fermentable fruit emerged is not inconsistent with the whole-genome duplication hypothesis.
The ecology of fermenting fruit is complex. In rotting fruits, S. cerevisiae becomes dominant after fermentation begins, whereas osmotic stress and pH, as well as ethanol, seem to inhibit the growth of competing organisms1. Nevertheless, the emergence of bulk ethanol may not be unrelated to other changes in the ecosystem at the end of the Cretaceous age, which include the extinction of the dinosaurs and the emergence of mammals and fruit flies27,28. Thus, this paleogenetics experiment is a small but necessary step in connecting the chemical behavior of individual enzymes operating as part of a multienzyme system, through metabolism and physiology, to the ecosystem and the fitness of organisms in it. We expect that this combination of chemistry, enzymology and cell biology with genomics, geology, paleontology and planetary science will be a key activity in addressing the ‘why’ questions in contemporary biology.
METHODS
Yeast strains and genomic DNA preparation
We purchased strains from ATCC. We used isogenic S. cerevisiae strains BY4742 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) and BY4741 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0) for Adh1 and Adh2 deletions12. We used the double deletion strain YMT-1D (MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 adh1::URA3 adh2::URA3) to express AdhA. We purchased strains for cloning new ADH genes from ATCC.
ADH gene cloning12
We designed degenerate primers from an alignment of existing S. cerevisiaeand Kluyveromyces lactis Adh proteins. We obtained partial ADH gene sequences using degenerate primers NSDP-1-FOR (positions 127–151 of S. cerevisiae ADH1) and NSDP-2-REV (positions 357–381 of S. cerevisiae ADH1) and amplification parameters of 96 °C for 2.5 min; followed by 30 cycles of 94 °C for 30 s, 39–50 °C for 30 s and 72 °C for 1 min; followed by a final extension step of 72 °C for 5 min. We pooled PCR products and shotgun-cloned them into TOPO PCR4-TA cloning vector (Invitrogen). We sequenced random clones and used them to generate a consensus sequence for each allele. We then used the consensus sequences to generate gene-specific primers. We used a combination of partial inverse PCR and linker-mediated PCR to clone the remaining flanking DNA.
Ancestral ADH genes
We generated ancestral ADH genes by site-directed mutagenesis of S. cerevisiae ADH1 cloned into the pRS415 vector12. We constructed the expression plasmid using primers ADH1/−1605-For (corresponding to 1,605 nucleotides upstream of the ADH1 start site) and ADH1/+378-Rev (corresponding to 378 nucleotides downstream of the ADH1 stop codon). Amplification parameters were 96 °C for 2.5 min; followed by 20 cycles of 94 °C for 30 s, 58 °C for 45 s and 72 °C for 3.5 min; followed by a final extension step of 72 °C for 5 min. We digested the amplified product with XbaI and XhoI and cloned it into pRS415. We used the resulting construct pRS415-ADH1 as a template for creating the AdhA constructs using site-directed mutagenesis (Stratagene).
Expression strain construction12
We constructed single deletion strains using PCR. We used isogenic S. cerevisiaestrains BY4741 (MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) and BY4742 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0; ATCC) to create deletions of the two primary ADH alleles ADH2 and ADH1, respectively. We designed primers to amplify URA3 with 50 bp of homology to the 5′ and 3′ untranslated regions of ADH1 or ADH2. We used the URA3-containing vector pRS316 as a template and cycled 30 times at 94 °C for 30 s, 55 °C for 45 s and 72 °C for 1 min. We disrupted alleles as described29. We created the double deletion strain (YMT-1D) by mating the single deletion strains followed by tetrad dissection.
Protein purification12,13
We transformed the double deletion strain YMT-1D with either pRS415-ADH1 or pRS415-AdhA. We grew cells in 2% glucose yeast minimal medium to midlog phase, with A600 = 0.6. We applied extracts to a Cibracon blue–coupled agarose column and eluted protein with a gradient of 0–200 µM NADH. We pooled fractions and applied them to a Superdex 200 gel-filtration column to remove excess NAD+(H) and collect tetrameric Adh.
Kinetic measurements12,14
We measured kinetics with NAD+, ethanol, NADH and acetaldehyde by holding the concentration of one constant and varying the concentrations of the others. All reactions were done in potassium phosphate buffer (83 mM, pH 7.3) with KCl (40 mM) at 25 °C. Ethanol reactions contained semicarbazide (10 mM) to avoid product inhibition by acetaldehyde. We monitored reactions by measure UV absorbance at 340 nm on a Cary Varian spectrophotometer.
GenBank accession numbers
Adh homologs from yeasts related to S. cerevisiae, AY216989 through AY217003.
Supplementary Material
ACKNOWLEDGMENTS
We thank A. Falcon and T. Barnash for their assistance. Funding was provided by the US National Research Council, the US National Aeronautics and Space Administration’s Astrobiology Institute (E.A.G.) and the US National Aeronautics and Space Administration Exobiology program (S.A.B.).
Footnotes
Note: Supplementary information is available on the Nature Genetics website.
COMPETING INTERESTS STATEMENT
The authors declare that they have no competing financial interests.
References
- 1.Pretorius IS. Tailoring wine yeasts for the new millennium: novel approaches to the ancient art of winemaking. Yeast. 2000;16:675–729. doi: 10.1002/1097-0061(20000615)16:8<675::AID-YEA585>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 2.Thornton JW. Resurrecting ancient genes. Experimental analysis of extinct molecules. Nat. Rev. Genet. 2004;5:366–376. doi: 10.1038/nrg1324. [DOI] [PubMed] [Google Scholar]
- 3.Boulton B, Singleton VL, Bisson LF, Kunkee RE. Principles and Practices of Winemaking. New York: Chapman and Hall; 1996. Yeast and biochemistry of ethanol fermentation; pp. 139–172. [Google Scholar]
- 4.Fleet GH, Heard GM. Wine Microbiology and Biotechnology. Chur, Switzerland: Harwood Academic Publishers; 1993. Yeast growth during fermentation; pp. 27–54. [Google Scholar]
- 5.McGovern PE, et al. Fermented beverages of pre- and proto-historic China. Proc. Natl. Acad. Sci. USA. 2004;101:17593–17598. doi: 10.1073/pnas.0407921102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wills C. Production of yeast alcohol dehydrogenase isoenzymes by selection. Nature. 1976;261:26. doi: 10.1038/261026a0. [DOI] [PubMed] [Google Scholar]
- 7.Fersht AR. Enzyme Structure and Mechanism. San Francisco: W.H. Freeman; 1977. [Google Scholar]
- 8.Ellington AD, Benner SA. Free energy differences between enzyme bound states. J. Theor. Biol. 1987;127:491–506. doi: 10.1016/s0022-5193(87)80145-5. [DOI] [PubMed] [Google Scholar]
- 9.Stryer L. Biochemistry. 3rd edn. New York: Freeman; 1988. [Google Scholar]
- 10.Swofford DL. PAUP* Phylogenetic Analysis Using Parsimony Version 4. Sunderland, Massachusetts: Sinauer Associates; 1998. [Google Scholar]
- 11.Yang ZH. PAML. A program package for phylogenetic analysis by maximum likelihood. CABIOS. 1997;15:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- 12.Thomson JM. Ph.D. dissertation. Gainesville: University of Florida; 2002. Interpretive Proteomics: Experimental Paleogenetics as a Tool to Analyze Function and Discover Pathways in Yeast. [Google Scholar]
- 13.Weinhold EG, Glasfeld A, Ellington AD, Benner SA. Structural determinants of the stereospecificity of alcohol dehydrogenase from yeast. Proc. Natl. Acad. Sci. USA. 1991;88:8420–8424. doi: 10.1073/pnas.88.19.8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ganzhorn AJ, Green DW, Hershey AD, Gould RM, Plapp BV. Kinetic characterization of yeast alcohol dehydrogenases. Amino acid residue 294 and substrate specificity. J. Biol. Chem. 1987;262:3754–3761. [PubMed] [Google Scholar]
- 15.Segel IH. Enzyme Kinetics. New York: Wiley; 1975. [Google Scholar]
- 16.Kellis M, Birren BW, Lander ES. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature. 2004;428:617–624. doi: 10.1038/nature02424. [DOI] [PubMed] [Google Scholar]
- 17.Kreitman M, Akashi H. Molecular evidence for natural selection. Ann. Rev. Ecol. Systematics. 1995;26:403–422. [Google Scholar]
- 18.Benner SA, Caraco MD, Thomson JM, Gaucher EA. Planetary biology. Paleontological, geological, and molecular histories of life. Science. 2002;293:864–868. doi: 10.1126/science.1069863. [DOI] [PubMed] [Google Scholar]
- 19.Sun G, et al. Archaefructaceae, a new basal angiosperm family. Science. 2002;296:899–904. doi: 10.1126/science.1069439. [DOI] [PubMed] [Google Scholar]
- 20.Collinson ME, Hooker JJ. Fossil evidence of interactions between plants and plant-eating mammals. Philos. Trans. R. Soc. London Ser. B. 1991;333:197–208. doi: 10.1098/rstb.1991.0068. [DOI] [PubMed] [Google Scholar]
- 21.Fernandez-Espinar MT, Barrio E, Querol A. Analysis of the genetic variability in the species of the Saccharomyces sensu stricto complex. Yeast. 2003;20:1213–1226. doi: 10.1002/yea.1034. [DOI] [PubMed] [Google Scholar]
- 22.Berbee ML, Taylor JW. Dating the evolutionary radiations of the true fungi. Can. J. Bot. 1993;71:1114–1127. [Google Scholar]
- 23.Benner SA. Interpretive proteomics. Finding biological meaning in genome and proteome databases. Adv. Enzyme Regul. 2003;43:271–359. doi: 10.1016/s0065-2571(02)00024-9. [DOI] [PubMed] [Google Scholar]
- 24.Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 25.Schaaff I, Heinisch J, Zimmerman FK. Overproduction of glycolytic enzymes in yeast. Yeast. 1989;5:285–290. doi: 10.1002/yea.320050408. [DOI] [PubMed] [Google Scholar]
- 26.Wolfe KH, Shields DC. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 2001;387:708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- 27.Ashburner M. Speculations on the subject of alcohol dehydrogenase and its properties in Drosophila and other flies. Bioessays. 1998;20:949–954. doi: 10.1002/(SICI)1521-1878(199811)20:11<949::AID-BIES10>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 28.Barrett PM, Willis KJ. Did dinosaurs invent flowers? Dinosaur-angiosperm coevolution revisited. Biological Rev. 2001;76:411–447. doi: 10.1017/s1464793101005735. [DOI] [PubMed] [Google Scholar]
- 29.Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bozzi A, Saliola M, Falcone C, Bossa F, Martini F. Structural and biochemical studies of alcohol dehydrogenase isozymes from Kluyveromyces lactis. Biochim. Biophys. Acta. 1997;1339:133–142. doi: 10.1016/s0167-4838(96)00225-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.