Abstract
Replicative cellular senescence is an important tumor suppression mechanism and also contributes to aging. Progression of both cancer and aging include significant epigenetic components, but the chromatin changes that take place during cellular senescence are not known. We used formaldehyde assisted isolation of regulatory elements (FAIRE) to map genome-wide chromatin conformations. In contrast to growing cells, whose genomes are rich with features of both open and closed chromatin, FAIRE profiles of senescent cells are significantly smoothened. This is due to FAIRE signal loss in promoters and enhancers of active genes, and FAIRE signal gain in heterochromatic gene poor regions. Chromatin of major retrotransposon classes, Alu, SVA and L1, becomes relatively more open in senescent cells, affecting most strongly the evolutionarily recent elements, and leads to an increase in their transcription and ultimately transposition. Constitutive heterochromatin in centromeric and peri-centromeric regions also becomes relatively more open, and the transcription of satellite sequences increases. The peripheral heterochromatic compartment (PHC) becomes less prominent, and centromere structure becomes notably enlarged. These epigenetic changes progress slowly after the onset of senescence, with some, such as mobilization of retrotransposable elements, becoming prominent only at late times. Many of these changes have also been noted in cancer cells.
Introduction
Replicative cellular senescence was first described as an irreversible growth arrest triggered by the accumulation of cell divisions (Hayflick & Moorhead, 1961). Subsequently it has emerged as a potent tumor suppression mechanism, and recent evidence points to important connections with aging (Collado et al., 2007; Baker et al., 2011). Progression of both cancer and aging includes a significant epigenetic component, such as changes in DNA methylation and chromatin remodeling (Decottignies & d’Adda di Fagagna, 2011).
Euchromatin is permissive for transcription, whereas heterochromatin, by virtue of its higher compaction, exerts an inhibitory effect. Constitutive heterochromatin, located at repetitive elements such as centromeres, telomeres and retrotransposons, is present in all somatic cells in a largely fixed manner. Facultative heterochromatin is typically established during cellular differentiation and can be reversible. Formation of a distinctive type of heterochromatin known as senescence-associated heterochromatin foci (SAHF) is an important component of cellular senescence (Narita et al., 2003; Zhang et al., 2005). On the other hand, DNA methylation of constitutive heterochromatin (Wilson & Jones, 1983; Zhang et al., 2008) decreases in senescence, and is indicative of reduced heterochromatinization. While these disparate observations point to widespread chromatin changes, their genome-wide distribution is not known.
FAIRE is a simple but effective method for genome-wide mapping of open chromatin regions (Giresi & Lieb, 2009). It exploits the observation that when formaldehyde crosslinked chromatin is extracted with phenol-chloroform, the majority of genomic DNA partitions into the organic phase, by virtue of being extensively crosslinked with histones. Only regions that are relatively free of nucleosomes appear in the aqueous phase. FAIRE-enriched peaks are highly correlated with transcriptional start sites (TSS) and DNase hypersensitive sites (Giresi et al., 2007), and global FAIRE enrichment correlates negatively with nucleosome occupancy (Hogan et al., 2006).
Results
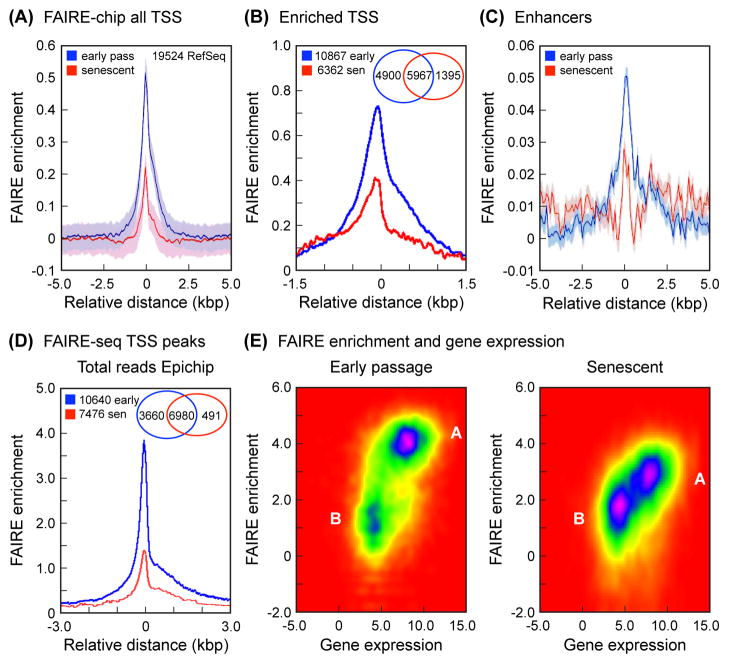
We performed FAIRE on early passage and senescent (Fig. S1) normal human diploid fibroblasts (HDF) and interrogated the genome using Affymetrix Human Tiling arrays (FAIRE-chip). We first computed average FAIRE enrichments of all RefSeq genes in a TSS-centered window (Fig. 1A). As expected, we found a strong peak located directly over the TSS. Interestingly, senescent cells displayed lower enrichment than early passage cells. To examine in more detail how FAIRE enrichment changes during senescence, we performed k-means clustering of the interpolated FAIRE signals of all the TSS in our dataset (Fig. 1B). The FAIRE-enriched cluster contained notably more genes in early passage cells compared to senescent cells (56% and 33% of total genes, respectively). In addition, reinforcing the analysis of all TSS, the magnitude of the signal in FAIRE-enriched genes was decreased in senescent cells. We also examined FAIRE signal over predicted enhancers (Heintzman et al., 2009) and found that it decreases in senescent cells (Fig. 1C). Finally, we obtained gene expression data from corresponding early passage and senescent HDF using Affymetrix U133 microarrays, and intersected these data with the FAIRE clustering analysis. As expected, we found a positive correlation between FAIRE enrichment and gene expression (Fig. S2).
Fig. 1.
FAIRE analysis of TSS and enhancers. (A) FAIRE-chip: average FAIRE signal at TSS decreases in senescent cells. Enrichment (Y-axis) was calculated as the averaged log2 (FAIRE/input) signal for each probe within a 10 kbp window (X-axis) centered on all the selected TSS (corrected for background, which was calculated by interpolating over 530,000 random 5 kbp regions across the genome). The 19,524 RefSeq genes were selected from the 26,083 genes annotated in the hg18 build of the human genome because they contained unique TSS. The interpolated values are shown as a solid line (blue, early passage; red, senescent), and the shaded regions represent the standard deviation envelopes above and below the mean. (B) FAIRE-chip: signal at TSS partitions into 2 clusters and decreases in the enriched cluster in senescent cells. The analysis was performed using 2 clusters and a window of 3 kbp (see Experimental Procedures). Shown here is the FAIRE-enriched cluster, which contains 10,867 genes in early passage cells (blue) and 6,362 genes in senescent cells (red). Interpolated values are shown as in (A). The Venn diagram shows the distribution of genes in the FAIRE enriched cluster: 5,967 genes are enriched in both early passage and senescent cells, 4,900 genes are uniquely enriched in early passage cells, and 1,395 genes are uniquely enriched in senescent cells. (C) FAIRE-chip: enrichment at enhancers decreases in senescent cells. Enrichment was calculated as in (A) using a database of predicted enhancers (Heintzman et al., 2009) (see Experimental Procedures). (D) FAIRE-seq: average FAIRE signal at TSS decreases in senescent cells. All uniquely mapping reads were processed with EpiChIP software (Hebenstreit et al., 2011), using a window of 6 kbp centered on all the TSS in the human genome hg18 or hg19 annotation files. The FDR cutoffs for both signal and noise were set at 0.05. This analysis identified significant TSS peaks in 10,640 genes in early passage cells, and 7,476 genes in senescent cells. The Venn diagram shows the distribution of genes: 6,980 genes have peaks in both early passage and senescent cells, 3,660 genes have peaks only in early passage cells, and 491 genes have peaks only in senescent cells. (E) FAIRE-seq: positive correlation of FAIRE enrichment at TSS with gene expression in early passage (left) and senescent (right) cells. The FAIRE enrichment dataset was intersected with the Affymetrix gene expression dataset and visualized using EpiChIP software. FAIRE enrichment (Y-axis) is shown as reads per gene. Gene expression (X-axis) is shown as PLIER scores. Note that early passage cells contain two well demarcated clusters of genes, one (A) with higher FAIRE and gene expression signals, and one (B) with lower values for both. Cluster A is notably larger. In senescent cells, cluster A loses FAIRE enrichment, and cluster B becomes more prominent.
Since the resolution and dynamic range of tiling arrays are limited, we examined FAIRE-extracted DNA by high-throughput sequencing (FAIRE-seq). These experiments broadly confirmed all the findings from the tiling arrays, in that both the number of FAIRE-enriched genes as well as the relative magnitude of the signal were significantly decreased in senescent cells (Fig. 1D), and that FAIRE signal correlated positively with gene expression (Fig. 1E). We also independently verified the FAIRE enrichment over TSS and enhancers in the FAIRE-seq datasets (Fig. S3), and used a bootstrap randomization method to confirm statistical significance.
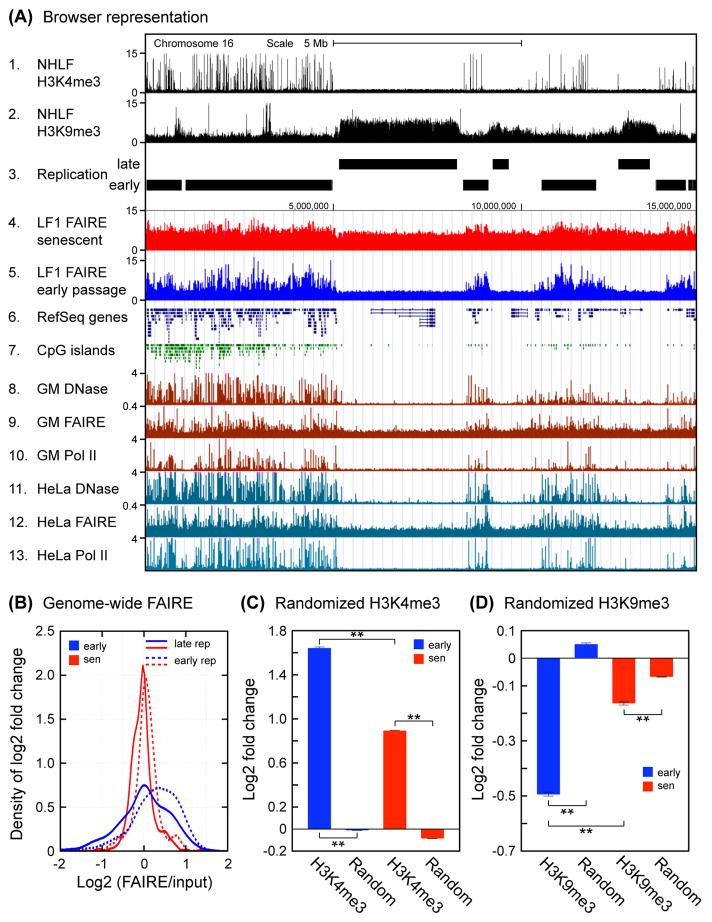
A whole chromosome view of the FAIRE-chip data showed that while early passage cells displayed distinct banding of high and low FAIRE enrichment, these patterns were noticeably smoothened in senescent cells (Fig. S4). This was reminiscent of the genome-wide redistribution of SIRT1 protein in response to DNA damage (Oberdoerffer et al., 2008). When we examined the chromosomal distribution of the FAIRE signal in the FAIRE-seq data we again noted a pronounced smoothening in senescent cells. A genome browser view showed that in early passage cells the FAIRE signal was higher in gene-rich regions and showed numerous peaks, while it was lower and more uniform over gene-poor regions (Fig. 2A, track 5). In senescent cells, the signal over gene-rich regions notably decreased, while the signal over gene-poor regions increased (Fig. 2A, track 4). These changes were specific to senescent cells, as the FAIRE profile in quiescent cells was very similar to early passage cells (Fig. S5).
Fig. 2.
Large scale genome-wide distribution of FAIRE signals. (A) A genome browser view is shown for a representative 15 Mb region of the left arm of chromosome 16. FAIRE signal from early passage and senescent LF1 cells is shown in tracks 4 and 5; other tracks present selected data from public databases. Note that FAIRE enrichment in senescent cells becomes more uniform and increases in heterochromatic late-replicating regions. H3K4me3 and H3K9me3 data for normal human lung fibroblasts (tracks 1, 2) were reported by (Ernst et al., 2011) and obtained from ENCODE. The early and late replicating tracks (tracks 3, 4) were generated from data reported by (Hansen et al., 2010). The DNase hypersensitivity, FAIRE, and RNA Pol II ChIP-seq data for the GM12878 lymphoblastoid and HeLa cell lines (tracks 8–13) were taken from the ENCODE Open Chromatin tracks, release 3 (Mar 2010). RefSeq genes and CpG islands (tracks 6,7) are from the UCSC genome browser. (B) Genome-wide FAIRE enrichment profile of early and late replicating regions in early passage and senescent cells. Genomic feature coverage was computed for reads mapping to unique locations. FAIRE-seq and input sample read counts were normalized to total unique mapping reads and used to calculate log2 fold changes for senescent and early passage cells. Kernel smoothing density estimation (Bowman & Azzalini, 1997) was applied to the distributions of log2 fold changes. These changes were significant using the bootstrap randomization method (below). (C) Genome-wide FAIRE enrichment of regions marked by H3K4me3 modification. Analysis was performed as in (B) using coordinates for H3K4me3 modification and represented in bar graph format. Statistical validation was performed using a bootstrap randomization of the coordinate files. Log2 (FAIRE/input) values of the original and randomized datasets were computed, and a two-tailed t-test was applied assuming unequal variances. **The results were significant in all cases (p < 0.01). (D) Genome-wide FAIRE enrichment of regions marked by H3K9me3 modification. Analysis was performed as described in (C).
It is well documented in public databases that gene-rich regions are demarcated with numerous features of open chromatin, such as activating histone marks (H3K4me3), DNaseI-hypersensitive sites, or RNA polymerase II localization (Fig. 2A). In contrast, gene-poor regions are enriched for features of heterochromatin, such as repressive histone marks (H3K9me3), and are known to replicate late (Hansen et al., 2010). These features are constant in very different cell lines (HDF, HeLa, lymphoblastoid, etc.) and likely represent basic architectural features of the human genome (Fig. 2A).
Changes in the distribution of FAIRE signal were confirmed in a genome-wide analysis. When analyzed in the context of early and late replicating regions, the FAIRE signal distributions were relatively broad in early passage cells, indicative of considerable local variation, and early replicating regions were enriched for FAIRE signal (Fig. 2B). In senescent cells the distributions became much sharper and moved closer together. To specifically address changes taking place in the context of euchromatin and heterochromatin, FAIRE signal was additionally analyzed in regions demarcated by H3K4me3 and H3K9me3 modifications. FAIRE signal was positively enriched in H3K4me3 regions (Fig. 2C), and this enrichment decreased in senescent cells, suggesting that these regions were becoming relatively more closed. In contrast, FAIRE signal was negatively enriched in H3K9me3 regions (Fig. 2D), but here relative enrichment increased in senescent cells, suggesting that these regions were becoming more open. Statistical validation using bootstrap randomization showed that all changes were highly significant. It should also be mentioned that because of the manner in which FAIRE-chip and FAIRE-seq data are normalized, the resultant comparisons reflect the relative distribution of open and closed chromatin across a single epigenome (and hence indirectly between proliferating and senescent cells), rather than a direct comparison of openness between proliferating and senescent cells.
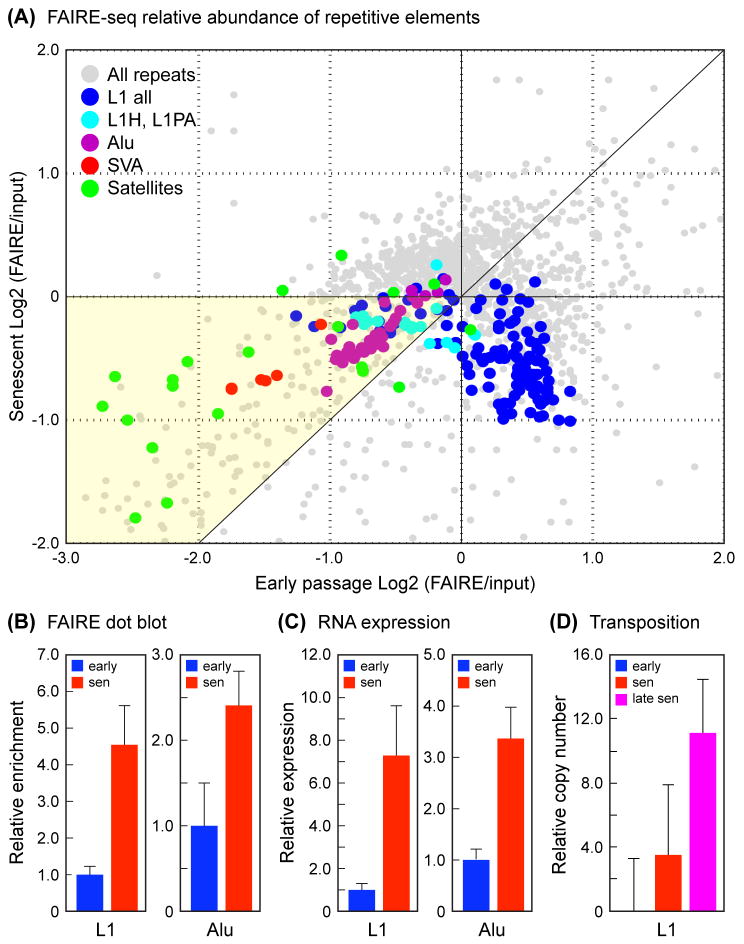
To gain more insight into the consequences of the changes in heterochromatic gene-poor regions, we extended our analysis to repetitive sequences, which are typically heavily heterochromatinized and comprise over 50% of our genomes. We applied recently developed software (Day et al., 2010) for assessing repeat sequence coverage from high throughput sequencing data. We first focused our analysis on SINE and LINE retrotransposons, and specifically the Alu and L1 families, which together comprise some 30% of the human genome, and include elements that can actively transpose (Batzer & Deininger, 2002; Deininger & Batzer, 2002). Random transposition is deleterious, and heterochromatinization of retrotransposons is a key mechanism for limiting their spread. As expected, all subfamilies of Alu elements were under-represented in FAIRE-enriched DNA relative to input, but became more abundant in senescent cells (Fig. 3A, Fig. S6). Alu elements continue to transpose and evolve in the human germline genome, and the most evolutionarily recent subfamilies showed the highest degree of depletion in FAIRE as well as the largest increase in senescence. Another class of recent and active retrotransposons, the SVA elements, showed the same pattern of changes.
Fig. 3.
Analysis of repetitive elements in early passage and senescent cells. (A) Relative abundance of Alu, L1, SVA and satellite elements in FAIRE-seq datasets. The representation of RepeatMasker annotated repetitive elements was computed for FAIRE-seq datasets using the software pipeline of (Day et al., 2010). Read counts were normalized to the total number of mapping reads, and fold changes were calculated as the Log2 (FAIRE/input). The graph shows each repetitive element subfamily as a point by plotting early passage changes along the X axis versus senescent changes along the Y axis. The relative enrichments of the four quadrants are (clockwise from top left): 1) depleted in early FAIRE and enriched in senescent FAIRE (X<0, Y>0), top left; 2) enriched in both early and senescent FAIRE (X>0, Y>0), top right; 3) enriched in early FAIRE and depleted in senescent FAIRE (X>0, Y<0), bottom right; and 4) depleted in both early and senescent FAIRE (X<0, Y<0), bottom left. The great majority of elements are found in quadrants 3 and 4. Quadrant 3 contains mostly ancient L1 elements, and quadrant 4 contains the majority of Alu, SVA and satellite elements, and the evolutionarily recent L1 elements. The diagonal in quadrant 4 demarcates the regions of relative enrichment in early versus senescent FAIRE, the yellow portion being the region where derepression occurs in senescent cells, and where the majority of potentially active Alu, SVA and L1 elements as well as the satellite elements are located. Alternative representations as bar graphs are shown in Fig. S6-S7. (B) Alu and L1 sequences are enriched in FAIRE DNA extracted from senescent cells. DNA preparations (FAIRE and input) were spotted onto nylon membranes and probed with 32P-labeled Alu and L1 probes. FAIRE signals were subtracted for background and normalized to the amount of DNA spotted. Data are shown normalized to input, and expressed as fold-change relative to early passage cells. The means and standard deviations of 3 independent experiments are shown (p < 0.01). (C) Expression of Alu and L1 RNAs is increased in senescent cells. Total RNA was prepared from early passage and senescent cells, exhaustively depleted of DNA, reverse transcribed, and quantified by qPCR. Data are shown normalized to GAPDH, and expressed as fold-change relative to early passage cells. The means and standard deviations of 3 independent experiments are shown (p < 0.01). (D) Copy number of L1 elements is increased in genomic DNA of senescent cells. Total DNA was prepared from early passage, senescent, and late senescent (12–14 weeks) cells, and L1 copy number was quantified using multiplex qPCR as described (Coufal et al., 2009). Data shown were normalized to 5S rDNA (normalization to L1 5′ UTR produced the same results), and are expressed as % increase relative to early passage cells (shown as 0%). The means and standard deviations of 3 independent experiments are shown. The 3–4% increase in senescent cells was reproducible in repeated experiments but not statistically significant; the 11% increase in late senescent cells was significant at p < 0.01. Based on in silico PCR simulations with the L1 ORF2 primers used, an increase of 11% corresponds to approximately 150 new insertions.
L1 elements are a large and diverse class, but most of its members have been extensively truncated in the human genome. Only the most recent primate (L1P) and human (L1H) subfamilies contain full-length elements that are believed to be capable of autonomous transposition. A large proportion (~75%) of L1 subfamilies were enriched in FAIRE relative to input in early passage cells and became depleted in senescence, the same pattern that was noted for the majority of RefSeq genes (Fig. 3A, Fig. S7). The remaining 25% of L1 subfamilies showed the same behavior as the Alu and SVA elements, namely, depleted in FAIRE relative to input and more abundant in senescence. Interestingly, this group was comprised almost entirely of primate and human subfamilies (Fig. 3A, Fig. S8), with the most recent members (such as L1HS, L1PA2, L1PA3, L1PA4) being among the most extreme in this trend. Thus, for all the major classes of retrotransposons (Alu, SVA, L1), the most recent subfamilies showed evidence of the strongest heterochromatinization in normal cells (which is reasonable given that they pose the greatest risk of transposition), and importantly, the most profound relative opening in senescent cells.
To further validate these bioinformatic inferences, we biochemically probed FAIRE-extracted DNA for representation of repetitive sequences using a dot-blotting strategy, and found that both Alu and L1 sequences were relatively enriched in senescent cells (Fig. 3B). We then examined the expression of Alu and L1 RNA by qRT-PCR, and found that it was correspondingly increased in senescent cells (Fig. 3C). Finally, we investigated the copy number of L1 elements, and found a statistically significant increase of 11% in late senescent cells (Fig. 3D). These PCR experiments employed primers designed to consensus sequences but are biased to interrogate recent primate and human subfamilies. (Coufal et al., 2009).
Given the apparent loosening of chromatin in late-replicating heterochromatic regions, we wished to investigate how this may affect the retrotransposons located there. We designed primer pairs specific to individual elements of the recent AluYb9, L1PA3 and L1PA4 subfamilies that are located in these regions, and performed qPCR on FAIRE samples from early passage and senescent cells. The design of these primers was based on the presence of single nucleotide polymorphisms found in individual elements, and all primers pairs were empirically verified to amplify their targets with single-copy kinetics. Six to seven individual elements of each subfamily on several different chromosomes were investigated, and in all cases we observed a marked increase of signal in FAIRE DNA extracted from senescent cells (Fig. S10, 11). Finally, the same primer pairs were used in qRT-PCR experiments, where in most cases we observed notable increases in the RNA expressed from these elements (Fig. S13, 14). Overall, the magnitude of the changes observed for the individual elements, both at the DNA and RNA levels, exceeded the changes seen with consensus primers or bioinformatic analysis of FAIRE-seq data, suggesting that retrotransposons located in heterochromatic late-replicating regions may be particularly prone to derepression and activation.
Centromeres are the most prominent sites of constitutive heterochromatin in the genome. Centromeres and pericentromeres are comprised mostly of satellite repeat sequences, which play important roles in their heterochromatinization and interphase nuclear location (Pezer et al., 2012). In the FAIRE-seq datasets satellite sequences were depleted in FAIRE and became prominently enriched in senescent cells (Fig. 3A, Fig. S9). Satellites are known to be transcribed in many organisms (Pezer et al., 2012), which is believed to play an important role in their heterochromatinization (Volpe et al., 2002). We next designed PCR primers specific for individual elements of the prominent human pericentromeric satellite II (hSATII), using the strategies described above. We found a significant enrichment on several chromosomes in FAIRE DNA extracted from senescent cells (Fig. S12). Finally, we used the same primers in qRT-PCR assays, and found remarkable increases in hSATII RNA (in some cases 100 to 1000-fold) in senescent cells (Fig. S15).
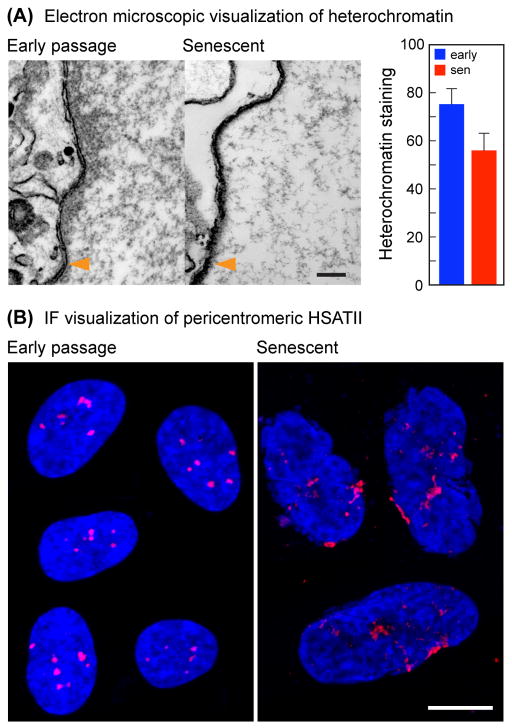
A considerable fraction of heterochromatin, including some late-replicating regions and centromeres, are localized close to the nuclear envelope in the peripheral heterochromatic compartment (PHC) (Carone & Lawrence, 2012). The PHC was first described on the basis of its dark staining in electron microscopic (EM) studies. Using EM we found significantly less staining of the PHC in senescent cell nuclei, indicative of a decrease of heterochromatin (Fig. 4A). We therefore examined centromeric structure by FISH using a hSATII probe. We found compact centromeric signals in early passage cells, which became distinctly enlarged in senescent cells (Fig. 4B). By this assay centromeres and pericentromeres thus appear to adopt a relatively more open conformation.
Fig. 4.
EM and centromere FISH analysis of senescent cell nuclei. (A) The peripheral heterochromatic compartment decreases in senescent cells. Representative electron micrographs (left panels) show the distribution of heterochromatin (dark staining) on the inside (right side) of the nuclear membrane (indicated with orange arrow heads). Scale bar = 200 nm. The graph on the right shows the quantification of the heterochromatic staining within a 200 nm region adjacent to the nuclear membrane (p < 0.01). (B) Centromeric regions become disorganized and enlarged in senescent cells. FISH was performed with a probe to hSATII. Each panel is a composite of several representative nuclei from images of early passage (left) and senescent (right) cells. Scale bar = 10 μm.
Discussion
Our studies reveal that the fundamental architecture of the genome undergoes profound alterations during replicative cellular senescence. One consequence is an overall closing of chromatin in euchromatic gene-rich regions, as evidenced by decreased FAIRE enrichment and associated dampening of gene activity, although some specific genes oppose this trend and become expressed at higher levels in senescent cells. Another, somewhat paradoxical trend, is an overall relative opening of chromatin in mostly heterochromatic gene-poor regions, as evidenced by increased FAIRE enrichment. The latter is associated with increased transcription of transposable elements, culminating in active transposition, as evidenced by an increase in copy number of L1 elements.
Telomere dysfunction drives a state of persistent DNA damage, which chronically activates the ATM and p53 pathways, leading to the cell cycle arrest that is the hallmark of senescence. Many forms of oncogene-induced senescence have also been associated with the induction of the DNA damage response (Rodier & Campisi, 2011). Transcription of retrotransposons and satellite elements can be induced by stresses such as DNA damage, heat shock, and even ethanol consumption (Li et al., 1999; Hagan et al., 2003; Jolly et al., 2004; Valgardsdottir et al., 2005). Induction of Alu transcription has been noted in replicative senescence (Wang et al., 2011), as has been demethylation (Suzuki et al., 2002) and transcription (Enukashvily et al., 2007) of human satellite sequences, and L1 activation has been documented in aging brain tissue (Coufal et al., 2009), Active transposases introduce numerous genomic double strand breaks even in the absence of transposition (Wallace et al., 2008), suggesting the possibility of self-reinforcing rounds of damage and degeneration. Thus, DNA damage caused by telomere dysfunction could induce the transcription of retrotranposable elements, which in turn could cause further damage by producing active transposases and even new insertional events.
Retrotransposons are mostly located in highly repressed heterochromatic regions of the genome, and are expressed at very low levels in normal unstressed cells. Their activation is opposed at multiple levels, including DNA methylation and miRNA targeting of transcripts (Belancio et al., 2010; Shalgi et al., 2010). It has been known for some time that total genomic DNA methylation decreases during aging, both in vivo in tissues as well as in cell culture during replicative senescence, and that this occurs largely in repetitive DNA sequences (Sedivy et al., 2008). One reason is that the expression of the DNA methyltransferase DNMT1 decreases as cells approach senescence (Young et al., 2003). The causes of this decrease are not well understood, and the consequent loss of DNA methylation could lead to a relaxation of constitutive heterochromatin, which could then either promote transposable element expression, or cooperate with other inducing events, such as DNA damage, to allow such expression to occur. It has also been reported that lamin B1 expression declines in cellular senescence (Freund et al., 2012). This could lead to structural perturbations of heterochromatin, especially in the late-replicating regions and centromeres that are located in the PHC, and cooperate with changes in DNA methylation.
Using L1 retrotransposon reporters it was found that active transposition was inhibited in quiescent and senescent cells, and was associated with a decrease in L1 transcription (Shi et al., 2007). It was suggested that this constitutes a defense mechanism to inhibit retrotransposition in nondividing cells to protect somatic tissue. It is possible that this mechanism involves the targeting of transcripts by retrotransposon-specific miRNAs, which have been shown to be very abundant (Yuan et al., 2011). Recently it was reported that Dicer was downregulated with age in mouse adipose tissue, resulting in global decreases in many miRNA species (Mori et al., 2012), and that DNA damage or oxidative stress downregulated Dicer in cell culture. We think it is therefore possible that the controls that oppose retrotransposition, most importantly, heterochromatinization and the targeting of their transcripts by miRNAs, are gradually subverted and relaxed as cells persist in a senescent state, and hence allow transposition to occur at late times.
The sequence of these events remains to be worked out in detail, and some of the effects that we have documented may well be adaptive and/or compensatory, for example, increased RNA transcription of satellites could be an attempt to reinforce heterochromatinization of these regions. Efforts to maintain control of retrotransposable elements could spill over to the rest of the genome, and could lead to the repression of gene activity in gene-rich regions that we have observed.
In our studies many of the effects we observed, including chromatin changes and expression of transposable elements, were evident in cells that had been senescent for a considerable period of time (6–8 weeks after cessation of proliferation). For example, cultures that had just reached the proliferative barrier (or had passed it for a short time such as 2–3 weeks) showed minimal, if any changes in FAIRE or retrotransposon expression. Retrotransposition, as evidenced by an increase in copy number, was the latest event (reaching significance only at 3–4 months after entry into senescence). This is likely the result of the redundant mechanisms that repress retrotransposons, all (or most) of which need to be defeated for active transposition to occur. Collectively, we view these as degenerative changes that affect genome integrity, which are likely to be the consequence, rather than the cause of cellular senescence.
It should also be noted that many of the changes we documented are also prevalent in cancer cells, such as demethylation of retrotransposons and satellites and high levels of their transcription (Belancio et al., 2010; Hansen et al., 2011; Berman et al., 2012). Although long suspected, recent genomic data show that the well-known plasticity and rearrangements of cancer genomes are associated with the transposition of retrotransposons (Lee et al., 2012). We find it intriguing that some these processes are already activated in senescent cells.
Experimental procedures
Cell culture
The human diploid fibroblast (HDF) cell strain derived from embryonic fetal lung, designated LF1 (Brown et al., 1997), was grown at 37°C in an atmosphere of 5% CO2 and 2.5% O2 in F-10 nutrient medium (Thermo Scientific) supplemented with 15% fetal bovine serum, penicillin, streptomycin and L-glutamine (Herbig et al., 2004). Cells were serially passaged by trypsinization at 1:4 dilution when reaching 80–90% confluence. To obtain senescent cells, cultures were propagated to replicative exhaustion (passage 50–52). Cultures were monitored by microscopic observation every 2–3 days to assess cell division. When replication ceased, the cultures were incubated further for at least 6–8 weeks prior to harvesting, unless otherwise indicated. Senescence was verified by staining for senescence-associated β-galactosidase (SA-β-Gal) (Itahana et al., 2007) (Fig. S1).
FAIRE
For the preparation of FAIRE DNA, cells were grown in 10 or 15 cm culture dishes. The procedure of Giresi and Lieb (Giresi & Lieb, 2009) was followed with minor modifications. For a detailed protocol, see Supplemental Information.
Tiling microarrays
1 μg of FAIRE extracted (or input) DNA was amplified with Klenow (3′ exo-minus) enzyme using a random oligonucleotide priming kit (NEB) in a 50 μl reaction volume containing dNTPs at 100 μM final concentration for 2 hr at 37ΰC. When necessary, multiple reactions were pooled to obtain enough material for the microarrays. The Affymetrix GeneChip Human Tiling 2.0R Array Set was processed by the Brown University Genomics Core Facility. This set covers the human genome on 7 arrays at an average resolution of 35 bp (45 million total 25-mer oligonucleotides tiled with a gap of approximately 10 bp between probes). Data were analyzed using the Tiling Array Software (TAS) from Affymetrix. Briefly, after removing probes mapping to multiple locations, all arrays (early passage FAIRE, early passage input, senescent FAIRE, senescent input) were quantile normalized together to equalize the signal levels across all arrays.
Expression microarrays
Whole cell total RNA was harvested in three independent experiments from early passage and senescent cells using Trizol reagent (Invitrogen). Affymetrix Human Genome U133 Plus 2.0 Arrays were processed by the Brown Genomics Core. The data from all expression arrays were quantile normalized and processed using full model GCRMA software with mismatch probes (Wu & Irizarry, 2005). The probesets were annotated using all of the mappings in hgu133plus2 RefSeq from Bioconductor. For each gene, the overall expression was calculated as the median value of all the probesets that mapped to that gene. Fold changes and p-values were calculated using Limma (Smyth, 2005).
FAIRE-seq
DNA prepared by the FAIRE method was sequenced on an Illumina GAIIx instrument, and later on an Illumina HighSeq 2000 instrument by the Brown Genomics Core. For a detailed protocol, see Supplemental Information.
Computation of FAIRE enrichment in genomic features
The FAIRE and input samples were first aligned to the unmasked reference human genome (build hg18 or hg19) using the Bowtie short read aligner (version 0.12.7) (Langmead et al., 2009), requiring reads to map uniquely to the genome. Late and early replicating coordinate BED files were generated using published datasets of replication timing in human cells (Hansen et al., 2010). The H3K9me3 and H3K4me3 genomic feature BED files were generated from broadPeak files describing regions of ChIP-seq enrichment for normal human lung fibroblasts (NHLF) (Ernst et al., 2011) which were obtained from ENCODE. For a detailed protocol, see Supplemental Information.
Computation of repetitive element enrichment
The software pipeline described by Day et al. (Day et al., 2010) was used to examine differences in repetitive element enrichment in FAIRE-seq datasets. For a detailed protocol, see Supplemental Information.
Dot blotting and hybridization
For a detailed protocol, see Supplemental Information.
Quantitative real-time PCR
qPCR of both DNA and RNA was performed using the SYBR Green system (Applied Biosystems) on the ABI 7900 Fast Sequence Detection instrument, according to the manufacturer’s specifications, with the exception of the multiplex assay to detect retrotransposition events, which was performed using the Taqman Gene Expression system (Applied Biosystems), as described by Coufal et al. (Coufal et al., 2009). Primer design is described below, and all primers are listed in Table S1. For qPCR of DNA, purified genomic or FAIRE DNA was used with the indicated primers. For qPCR of RNA, total RNA was harvested from cells using Trizol reagent (Invitrogen) according to the manufacturer’s instructions. 1 μg of total RNA was transcribed into cDNA in 50 μl reactions using the Taqman kit (Applied Biosystems), according to the manufacturer’s protocol. 1 μl of this reaction was used in subsequent qPCR reactions. GAPDH was used as the normalization control. For the measurement repetitive DNA transcription, total RNA was exhaustively digested with RNase free DNase, and further cleaned up on RNeasy columns, prior to the synthesis of cDNA. Effectiveness of the DNase digestion was assessed using controls that omitted reverse transcriptase.
Design of PCR primers
See Table S1 for a listing of all primers. For detailed methods, see Supplemental Information.
Chromatin immunoprecipitation
All procedures followed the protocols in the Magna ChIP kit (Millipore). For a detailed protocol, see Supplemental Information.
Electron microscopy
Cells were grown in 10 cm dishes as indicated. For a detailed protocol, see Supplemental Information.
Fluorescence in situ hybridization
Cells were grown on coverslips, fixed with 4% paraformaldehyde for 20 min at room temperature, washed 3x in phosphate buffered saline (PBS), and stored in 70% ethanol at 4°C until used. For a detailed protocol, see Supplemental Information.
Supplementary Material
Acknowledgments
P.D. Adams and his lab (Beatson Institute) are gratefully acknowledged for a productive ongoing collaboration and communication of unpublished data and reagents. This work was supported by NIH/NIA grant R37 AG016694 to J.M.S. M.D.C. was supported in part by Dottorato di Ricerca in “Biotecnologie, Farmacologia e Tossicologia”, PFDR in “Biotecnologie Cellulari e Molecolari” fellowship of Bologna University. S.W.C. was supported in part by NIH/NIGMS Institutional Research Training Grant T32 GM007601. J.A.K. was supported in part by NIH/NCRR grant P20 RR015578-10S1 and a Mentored Research Scientist Development Award from the NIH/NIA K01 AG039410. N.N. was supported by a Mentored Quantitative Research Development Award from the NIH/NIA K25 AG028753 and K25 AG028753-03S1. The Genomics Core Facility in the Laboratories for Molecular Medicine at Brown University was supported in part by the COBRE award from the NIH/NIGMS P30 GM0103410. J.M.S. is a Senior Scholar of the Ellison Medical Foundation and a recipient of the Glenn Award for Research on the Biological Mechanisms of Aging from the Glenn Medical Foundation.
References
- Baker DJ, Wijshake T, Tchkonia T, LeBrasseur NK, Childs BG, van de Sluis B, Kirkland JL, van Deursen JM. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature. 2011;479:232–236. doi: 10.1038/nature10600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batzer MA, Deininger PL. Alu repeats and human genomic diversity. Nat Rev Genet. 2002;3:370–379. doi: 10.1038/nrg798. [DOI] [PubMed] [Google Scholar]
- Belancio VP, Roy-Engel AM, Deininger PL. All y’all need to know ‘bout retroelements in cancer. Semin Cancer Biol. 2010;20:200–210. doi: 10.1016/j.semcancer.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman BP, Weisenberger DJ, Aman JF, Hinoue T, Ramjan Z, Liu Y, Noushmehr H, Lange CP, van Dijk CM, Tollenaar RA, Van Den Berg D, Laird PW. Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina-associated domains. Nat Genet. 2012;44:40–46. doi: 10.1038/ng.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman AW, Azzalini A. Applied smoothing techniques for data analysis. New York: Oxford University Press; 1997. [Google Scholar]
- Brown JP, Wei W, Sedivy JM. Bypass of senescence after disruption of p21CIP1/WAF1 gene in normal diploid human fibroblasts. Science. 1997;277:831–834. doi: 10.1126/science.277.5327.831. [DOI] [PubMed] [Google Scholar]
- Carone DM, Lawrence JB. Heterochromatin instability in cancer: From the Barr body to satellites and the nuclear periphery. Semin Cancer Biol. 2012 doi: 10.1016/j.semcancer.2012.06.008. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado M, Blasco MA, Serrano M. Cellular senescence in cancer and aging. Cell. 2007;130:223–233. doi: 10.1016/j.cell.2007.07.003. [DOI] [PubMed] [Google Scholar]
- Coufal NG, Garcia-Perez JL, Peng GE, Yeo GW, Mu Y, Lovci MT, Morell M, O’Shea KS, Moran JV, Gage FH. L1 retrotransposition in human neural progenitor cells. Nature. 2009;460:1127–1131. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day DS, Luquette LJ, Park PJ, Kharchenko PV. Estimating enrichment of repetitive elements from high-throughput sequence data. Genome Biol. 2010;11:R69. doi: 10.1186/gb-2010-11-6-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decottignies A, d’Adda di Fagagna F. Epigenetic alterations associated with cellular senescence: a barrier against tumorigenesis or a red carpet for cancer? Semin Cancer Biol. 2011;21:360–366. doi: 10.1016/j.semcancer.2011.09.003. [DOI] [PubMed] [Google Scholar]
- Deininger PL, Batzer MA. Mammalian retroelements. Genome Res. 2002;12:1455–1465. doi: 10.1101/gr.282402. [DOI] [PubMed] [Google Scholar]
- Enukashvily NI, Donev R, Waisertreiger IS, Podgornaya OI. Human chromosome 1 satellite 3 DNA is decondensed, demethylated and transcribed in senescent cells and in A431 epithelial carcinoma cells. Cytogenet Genome Res. 2007;118:42–54. doi: 10.1159/000106440. [DOI] [PubMed] [Google Scholar]
- Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, Ku M, Durham T, Kellis M, Bernstein BE. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund A, Laberge RM, Demaria M, Campisi J. Lamin B1 loss is a senescence-associated biomarker. Mol Biol Cell. 2012;23:2066–2075. doi: 10.1091/mbc.E11-10-0884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giresi PG, Kim J, McDaniell RM, Iyer VR, Lieb JD. FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res. 2007;17:877–885. doi: 10.1101/gr.5533506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giresi PG, Lieb JD. Isolation of active regulatory elements from eukaryotic chromatin using FAIRE (Formaldehyde Assisted Isolation of Regulatory Elements) Methods. 2009;48:233–239. doi: 10.1016/j.ymeth.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan CR, Sheffield RF, Rudin CM. Human Alu element retrotransposition induced by genotoxic stress. Nat Genet. 2003;35:219–220. doi: 10.1038/ng1259. [DOI] [PubMed] [Google Scholar]
- Hansen KD, Timp W, Bravo HC, Sabunciyan S, Langmead B, McDonald OG, Wen B, Wu H, Liu Y, Diep D, Briem E, Zhang K, Irizarry RA, Feinberg AP. Increased methylation variation in epigenetic domains across cancer types. Nat Genet. 2011;43:768–775. doi: 10.1038/ng.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen RS, Thomas S, Sandstrom R, Canfield TK, Thurman RE, Weaver M, Dorschner MO, Gartler SM, Stamatoyannopoulos JA. Sequencing newly replicated DNA reveals widespread plasticity in human replication timing. Proc Natl Acad Sci USA. 2010;107:139–144. doi: 10.1073/pnas.0912402107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayflick L, Moorhead PS. The serial cultivation of human diploid cell strains. Exp Cell Res. 1961;25:585–621. doi: 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- Hebenstreit D, Gu M, Haider S, Turner DJ, Lio P, Teichmann SA. EpiChIP: gene-by-gene quantification of epigenetic modification levels. Nucleic Acids Res. 2011;39:e27. doi: 10.1093/nar/gkq1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, Ching KA, Antosiewicz-Bourget JE, Liu H, Zhang X, Green RD, Lobanenkov VV, Stewart R, Thomson JA, Crawford GE, Kellis M, Ren B. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbig U, Jobling WA, Chen BP, Chen DJ, Sedivy JM. Telomere shortening triggers senescence of human cells through a pathway involving ATM, p53, and p21(CIP1), but not p16(INK4a) Mol Cell. 2004;14:501–513. doi: 10.1016/s1097-2765(04)00256-4. [DOI] [PubMed] [Google Scholar]
- Hogan GJ, Lee CK, Lieb JD. Cell cycle-specified fluctuation of nucleosome occupancy at gene promoters. PLoS Genet. 2006;2:e158. doi: 10.1371/journal.pgen.0020158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itahana K, Campisi J, Dimri GP. Methods to detect biomarkers of cellular senescence: the senescence-associated beta-galactosidase assay. Methods Mol Biol. 2007;371:21–31. doi: 10.1007/978-1-59745-361-5_3. [DOI] [PubMed] [Google Scholar]
- Jolly C, Metz A, Govin J, Vigneron M, Turner BM, Khochbin S, Vourc’h C. Stress- induced transcription of satellite III repeats. J Cell Biol. 2004;164:25–33. doi: 10.1083/jcb.200306104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E, Iskow R, Yang L, Gokcumen O, Haseley P, Luquette LJ, 3rd, Lohr JG, Harris CC, Ding L, Wilson RK, Wheeler DA, Gibbs RA, Kucherlapati R, Lee C, Kharchenko PV, Park PJ. Landscape of Somatic Retrotransposition in Human Cancers. Science. 2012;337:967–971. doi: 10.1126/science.1222077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Spearow J, Rubin CM, Schmid CW. Physiological stresses increase mouse short interspersed element (SINE) RNA expression in vivo. Gene. 1999;239:367–372. doi: 10.1016/s0378-1119(99)00384-4. [DOI] [PubMed] [Google Scholar]
- Mori MA, Raghavan P, Thomou T, Boucher J, Robida-Stubbs S, Macotela Y, Russell SJ, Kirkland JL, Blackwell TK, Kahn CR. Role of microRNA processing in adipose tissue in stress defense and longevity. Cell Metab. 2012;16:336–347. doi: 10.1016/j.cmet.2012.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita M, Nunez S, Heard E, Narita M, Lin AW, Hearn SA, Spector DL, Hannon GJ, Lowe SW. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003;113:703–716. doi: 10.1016/s0092-8674(03)00401-x. [DOI] [PubMed] [Google Scholar]
- Oberdoerffer P, Michan S, McVay M, Mostoslavsky R, Vann J, Park SK, Hartlerode A, Stegmuller J, Hafner A, Loerch P, Wright SM, Mills KD, Bonni A, Yankner BA, Scully R, Prolla TA, Alt FW, Sinclair DA. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell. 2008;135:907–918. doi: 10.1016/j.cell.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezer Z, Brajkovic J, Feliciello I, Ugarkovic D. Satellite DNA-mediated effects on genome regulation. In: Garrido-Ramos M, editor. Repetitive DNA. Basel: S. Karger AG; 2012. pp. 153–169. [DOI] [PubMed] [Google Scholar]
- Rodier F, Campisi J. Four faces of cellular senescence. J Cell Biol. 2011;192:547–556. doi: 10.1083/jcb.201009094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedivy JM, Banumathy G, Adams PD. Aging by epigenetics--a consequence of chromatin damage? Exp Cell Res. 2008;314:1909–1917. doi: 10.1016/j.yexcr.2008.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalgi R, Pilpel Y, Oren M. Repression of transposable-elements - a microRNA anti- cancer defense mechanism? Trends Genet. 2010;26:253–259. doi: 10.1016/j.tig.2010.03.006. [DOI] [PubMed] [Google Scholar]
- Shi X, Seluanov A, Gorbunova V. Cell divisions are required for L1 retrotransposition. Mol Cell Biol. 2007;27:1264–1270. doi: 10.1128/MCB.01888-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. Limma: Linear Models for Microarray Data. New York: Springer; 2005. [Google Scholar]
- Suzuki T, Fujii M, Ayusawa D. Demethylation of classical satellite 2 and 3 DNA with chromosomal instability in senescent human fibroblasts. Exp Gerontol. 2002;37:1005–1014. doi: 10.1016/s0531-5565(02)00061-x. [DOI] [PubMed] [Google Scholar]
- Valgardsdottir R, Chiodi I, Giordano M, Cobianchi F, Riva S, Biamonti G. Structural and functional characterization of noncoding repetitive RNAs transcribed in stressed human cells. Mol Biol Cell. 2005;16:2597–2604. doi: 10.1091/mbc.E04-12-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- Wallace NA, Belancio VP, Deininger PL. L1 mobile element expression causes multiple types of toxicity. Gene. 2008;419:75–81. doi: 10.1016/j.gene.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Geesman GJ, Hostikka SL, Atallah M, Blackwell B, Lee E, Cook PJ, Pasaniuc B, Shariat G, Halperin E, Dobke M, Rosenfeld MG, Jordan IK, Lunyak VV. Inhibition of activated pericentromeric SINE/Alu repeat transcription in senescent human adult stem cells reinstates self-renewal. Cell Cycle. 2011;10:3016–3030. doi: 10.4161/cc.10.17.17543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson VL, Jones PA. DNA methylation decreases in aging but not in immortal cells. Science. 1983;220:1055–1057. doi: 10.1126/science.6844925. [DOI] [PubMed] [Google Scholar]
- Wu Z, Irizarry RA. Stochastic models inspired by hybridization theory for short oligonucleotide arrays. J Comput Biol. 2005;12:882–893. doi: 10.1089/cmb.2005.12.882. [DOI] [PubMed] [Google Scholar]
- Young JI, Sedivy JM, Smith JR. Telomerase expression in normal human fibroblasts stabilizes DNA 5-methylcytosine transferase I. J Biol Chem. 2003;278:19904–19908. doi: 10.1074/jbc.M301685200. [DOI] [PubMed] [Google Scholar]
- Yuan Z, Sun X, Liu H, Xie J. MicroRNA genes derived from repetitive elements and expanded by segmental duplication events in mammalian genomes. PLoS One. 2011;6:e17666. doi: 10.1371/journal.pone.0017666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Poustovoitov MV, Ye X, Santos HA, Chen W, Daganzo SM, Erzberger JP, Serebriiskii IG, Canutescu AA, Dunbrack RL, Pehrson JR, Berger JM, Kaufman PD, Adams PD. Formation of MacroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA. Dev Cell. 2005;8:19–30. doi: 10.1016/j.devcel.2004.10.019. [DOI] [PubMed] [Google Scholar]
- Zhang W, Ji W, Yang J, Yang L, Chen W, Zhuang Z. Comparison of global DNA methylation profiles in replicative versus premature senescence. Life Sci. 2008;83:475–480. doi: 10.1016/j.lfs.2008.07.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.