Abstract
Objective
Deficiency or reduced expression of signal transduction and activation of RNA family protein Quaking (Qki) is associated with developmental defects in neural and vascular tissues and the development of debilitating human diseases including colorectal cancer (CRC). However, the mechanisms underlying the aberrant downregulation or deficiency of Qki were uncertain.
Design
Expression of miR-574-5p, Qki5/6/7/7b splicing variants, β-catenin and p27Kip1 was determined in mouse and human CRC cells and tissues to investigate the post-transcriptional regulation of Qki isoforms by miR-574-5p and its impact on β-catenin/p27Kip1 signalling, cell cycle progression, proliferation, migration, invasion and tumour growth.
Results
In the CRC tissues of C57BL/6-Apcmin/+ mice, miR-574-5p was found to be significantly upregulated and negatively correlated with the expression of Qki but positively correlated with the expression of β-catenin. In mouse and human CRC cells, miR-574-5p was shown to regulate Qki isoforms (Qki6/7 in particular) post-transcriptionally and caused altered expression in β-catenin and p27Kip1, increased proliferation, migration and invasion and decreased differentiation and cell cycle exit. Furthermore, in clinical CRC tissues, miR-574-5p was shown to be greatly upregulated and inversely correlated with the expression of Qkis. Finally, inhibition of miR-574-5p was shown to suppress the growth of tumours in the nude mice.
Conclusions
Together, these novel findings suggest that miR-574-5p is a potent ribo-regulator for Qkis and that aberrant miR-574-5p upregulation can be oncogenic.
Keywords: miR-574-5p, quaking, colorectal cancer, β-catenin, p27Kip1, abdominal surgery, hepatic encephalopathy, gut inflammation, colorectal cancer, cell biology, colorectal cancer genes, gene mutation
Significance of this study.
What is already known on this subject?
Downregulation of QKI5 and QKI6 was shown to be associated with the development of colorectal cancer (CRC).
QKI5/6 might contribute to the development of CRC by controlling β-catenin and p27Kip1 signalling.
DNA methylation in the promoter region of Qki was suggested to contribute in part to Qki's aberrant down regulation in CRC.
What are the new findings?
miR-574-5p regulates Qki6/7 negatively and post-transcriptionally.
QKI7 is the most predominant isoform in human CRC cells. QKI5/6/7 proteins all regulate β-catenin post-transcriptionally.
miR-574-5p expression correlates inversely with that of Qkis in CRC cells and tissues.
Aberrant upregulation of miR-574-5p can be oncogenic and its inhibition is beneficial.
How might it impact on clinical practice in the foreseeable future?
The discovery of this miR-574-5p-QKI-β-catenin/p27Kip1 axis of signal transduction will enable better understanding of the pathogenic mechanisms in human diseases associated with the abnormal expression of miR-574-5p, QKI and QKI-regulated genes.
miR-574-5p and Qkis might serve as diagnostic or therapeutic targets for CRC.
Introduction
Quaking (QKI) is a RNA binding protein belonging to the signal transduction and activation of RNA (STAR) protein family that has been implicated in the regulation of cellular processes such as cell cycle and differentiation,1 apoptosis,2 cell fate decision and development3–5 and angiogenesis.6 7 Aberrant expression of the STAR proteins has been associated with a number of developmental defects and human diseases.8 9 Studies have shown that Src associated in mitosis, of 68 kDa (Sam68), a prototype of the STAR proteins, is a multifunctional player in human cancers.10–14 Genetic deletion in the Qki locus, on the other hand, was found to be associated with human glioblastoma or astrocytic glioma15–17 mental retardation and autism.18 Haploinsufficiency of Qki has also been linked with the 6q terminal deletion syndrome.19 Furthermore, reduced Qki expression has been implicated in the pathogenesis of human disorders such as schizophrenia20 21 and major depressive disorders.22 Yang et al found that QKI5 and QKI6 were greatly reduced or even absent in colorectal cancer (CRC) cells and tissues and that downregulation of QKI5/6 was associated with abnormal regulation of β-catenin and p27Kip1 signalling.23 More recently, QKI expression was found to be significantly reduced in several types of tumours, including testis, lung, breast, bladder, cervix, ovary and colon cancer, which was shown to be associated with an abnormal decrease in the histone variant macroH2A1.1.24
Despite the potentially dire biological consequences of dysregulation in the expression of Qki, the exact genetic and epigenetic mechanisms governing Qki expression are poorly understood.8 25 Klempan et al studied the effects of genetic and epigenetic variation on the promoter function and expression of Qki in the human brains of the suicide victims with major depressive disorders.22 While significant reductions in Qki expression were found in the cortical regions, the hippocampus and the amygdala of suicide victims, analysis of variations in and methylation state of the promoter sequence did not identify differences at genetic and epigenetic levels between depressed suicide victims and controls. In a later study with 288 individuals diagnosed with schizophrenia and 288 controls, no association was found between seven genetic markers (including single-nucleotide polymorphism and haplotype) around the promoter region of the Qki gene and schizophrenia.26 In the study conducted by Yang et al,23 while characterisations of the Qki promoter region in a few CRC cell lines suggested enhanced promoter DNA methylation, increased methylation was found in only 4 out of 10 CRC tissue samples tested. Moreover, methylation was often detected in both CRC tissues and the neighbouring normal tissues from the same patient. Furthermore, DNA methylation is unlikely to be the major mechanism responsible for the downregulation of Qki as it cannot adequately explain the temporal, spatial and isoform-specific regulation of Qki in many cases of cell development and diseases. Collectively these observations suggest that genetic variations and epigenetic modifications in the Qki promoter region might not be the major determinants responsible for reduced Qki expression that is associated with human diseases such as schizophrenia, depression and cancers.
MicroRNAs (miRNAs) are non-coding small RNAs that serve as important regulators of gene expression by targeting the 3′ untranslated region (UTR) of messenger RNA (mRNA) and inducing mRNA degradation and suppression of translation.27 28 The 3′UTRs of Qki mRNAs are evolutionally conserved and thus may be important to Qki's post-transcriptional regulation.29–31 In this current study, we found that miR-574-5p negatively regulates Qki6/7/7b through interactions with miRNA recognition elements (MREs) on the corresponding 3′UTRs. In mouse and human CRC tissues, very significant upregulation of miR-574-5p is correlated with substantial reductions in Qki6/7/7b, which might contribute significantly to the aberrant regulation of β-catenin and p27Kip1 and to the pathogenesis or progression of CRC. These results unravel a novel and important mechanism for the regulation of Qkis and the development of CRC.
Materials and methods
For details of the following, please see the supplemental materials and methods section in online supplementary information: plasmid construction, cell culture, transient transfections, western blot, immunohistochemistry, immunofluorescence staining, in situ hybridisation analyses; cell cycle, proliferation, migration, colony formation and invasion assays; luciferase reporter assays, intestinal alkaline phosphatase (IAP) enzyme assays and real-time quantitative RT-PCR (qPCR) analyses of mRNAs and miRNAs.
Animal experiments
All experimental procedures involving animals were performed in accordance with animal protocols approved by the Institutional Animal Use and Care Committee of Xiamen University. C57BL/6-Apcmin/+ mice and controls were generous gifts from Dr Boan Li, Xiamen University. Five-week-old male athymic BALB/c nude mice were obtained from the SLAC Laboratory Animals Co Ltd (Shanghai, China).
For xenograft tumour growth, 5×106 SW480 cells pre-infected with control lentiviruses or lentiviruses carrying shRNA for miR-574-5p were injected subcutaneously into the dorsal flanks of mice. Beginning from the 10th day after the injection, the size of the tumour was measured every 3 days by a Vernier calliper along two perpendicular axes. The volume of the tumour was calculated with the formula: volume (mm3) = length (mm) × width (mm)2×0.52. Twenty-eight days after the injection, the mice were sacrificed and the tumours were dissected for further analyses.
For bioluminescence imaging of tumour growth in live animals, see supplemental materials and methods section in online supplementary information.
Clinical samples
All clinical samples were collected with the informed consent of the patients and study protocols that were in accordance with the ethical guidelines of the Declaration of Helsinki (1975) and were approved by the Institutional Medical Ethics Committee of Xiamen University. CRC pathological diagnosis was verified by at least two pathologists. Sixty human CRC specimens and paired adjacent epithelial tissues and serum samples from seven patients with CRC and seven normal controls were obtained from the First Affiliated Hospital of Xiamen University from 25 January 2010 to December 2011. Tumour and non-cancerous tissues were confirmed histologically by haematoxylin and eosin (H&E) staining.
Bioinformatics, data acquisition, image processing and statistical analyses
See supplemental materials and methods section in online supplementary information.
Results
Negative correlation between miR-574-5p and Qki expression and positive correlation between miR-574-5p and β-catenin in the CRC tissues of C57BL/6-Apcmin/+ mice
Our bioinformatics analyses suggested the presence of multiple MREs on the 3′UTR of mouse or human Qki5/6/7/7b mRNAs, including those for miR-574-5p, miR-17 and miR-20a (figure S1, online supplementary information). We therefore performed qPCR and western blotting to analyse the expression levels of Qki mRNAs and proteins and miRNAs in the CRC tissues from C57BL/6-Apcmin/+ (Apc-mutant) mice and in colorectal tissues from normal control mice. Compared with the expression levels in the normal tissues, total Qki mRNA/protein (pan-Qki/pan-QKI) and Qki5/6/7 mRNA/protein isoforms were all greatly downregulated while β-catenin was greatly upregulated in the CRC tissues of the Apc-mutant mice (figure 1A–C). Analyses of miRNA expression indicated that miR-574-5p, miR-717 and miR-20a were significantly upregulated in mouse CRC tissues but miR-200b, miR-466g and miR-17 were not (figure 1D). Among the three significantly upregulated miRNAs, miR-574-5p had the highest fold change (∼3.7-fold, p<0.001). Interestingly, MREs for miR-574-5p were found on the 3′UTRs of mouse Qki6/7 but not on that of Qki5 mRNA (online figure S1), suggesting that miR-574-5p might be involved in the negative regulation of Qki6/7 in mouse colorectal tissues. Remarkably, the expression levels of two putative intronic miR-574-5p-hosting genes, nervous system overexpressed protein-20 (Noxp20,32 and miRBase) and procollagen type III, α-1 (Col3a1)33 and the level of a transcription factor SRY-like HMG box-2 (Sox2) were also significantly upregulated in mouse CRC tissues (figure 1C). This result suggests that miR-574-5p might be co-regulated with Noxp20 or Col3a1 in the mouse tissues.
Figure 1.
miR-574-5p expression correlates negatively with the expression of Qki but correlates positively with β-catenin in C57BL/6-Apcmin/+ mice (16-week old males, n=3). Data are represented as mean ± SEM. *p<0.05; ***p<0.001. (A) Total QKI and QKI5/6/7 protein expression levels were greatly downregulated in mouse colorectal cancer (CRC) tissues. (B) Total Qki and Qki5/6/7 messenger RNA (mRNA) expression levels were significantly downregulated in mouse CRC tissues. (C) The expression levels of β-catenin, Noxp20, Col3a1 and Sox2 mRNAs were greatly increased in mouse CRC tissues. Col3a1, procollagen type III, α-1; Noxp20, nervous system overexpressed protein-20; Sox2, SRY-like HMG box-2. (D) The expression levels of miR-574-5p, miR-717 and miR-20a were significantly upregulated in mouse CRC tissues, whereas that of miR-200b, miR-466g and miR-17 were not. See also online figure S1. miRNA, microRNA; NS, not significant.
miR-574-5p represses the expression of Qki6/7/7b through a post-transcriptional mechanism
To demonstrate the silencing effects of miR-574-5p on Qkis, we performed transient transfection studies in cultured CRC cells (figure 2). Our qPCR analyses indicated that, compared with the control group, transfection of CT26 cells with a plasmid overexpressing miR-574 precursor (pmiR-574) or with chemically synthesised mimics for miR-574-5p (see supplemental materials and methods section in online supplementary information) led to a 49–59% reduction in total Qki mRNA expression (p<0.001, figure 2A). Conversely, inhibition of miR-574-5p with an anti-sense inhibitor (see supplemental materials and methods section in online supplementary information) resulted in a 92% increase in Qki mRNA expression (p<0.001). Similar effects were found for Qki5/6/7 mRNA isoforms. Parallelling the increase in Qki mRNAs following the inhibition of miR-574-5p, total QKI and QKI5/6/7 proteins were all significantly increased as indicated by western blots (p<0.001, figure 2B). Moreover, similar effects were observed for human HCT116, SW480 and SW620 cells (figure S2, online supplementary information).
Figure 2.
miR-574-5p represses Qki6/7/7b. CT26 cells were transfected with a microRNA (miRNA)-overexpressing plasmid or a miRNA mimics or a miRNA inhibitor, in the absence or presence of a luciferase reporter together with pSV40-β-galactosidase (4:3:1). Cells were harvested for quantitative PCR (qPCR), western blotting and luciferase activity assays 24 h after the transfections. For qPCR and the luciferase reporter assays, n=3–4. Statistical comparisons were made between a control (pFlag-CMV, mimics control, or inhibitor control)-transfected cells and a particular treatment. Data are represented as mean ± SEM. **p<0.01; ***p<0.001. (A) Overexpression of miR-574-5p or transfection with its mimics caused significant downregulation of total Qki messenger RNA (mRNA) and Qki5/6/7 mRNA isoforms, whereas inhibition of miR-574-5p significantly increased the level of total Qki and Qki5/6/7 mRNA isoforms as determined by qPCR. (B) Inhibition of miR-574-5p significantly increased the levels of total QKI and QKI5/6/7 protein as determined by western blots. (C) miR-574-5p regulates Qki6/7/7b (but not Qki5) by interacting with the 3′ untranslated region of Qki6/7/7b. β-galactosidase activity was determined and used for the calibration of luciferase reporter activity. NS, not significant. See also online figure S2. RLU, relative light units.
To determine whether miR-574-5p regulates Qkis by targeting Qki-3′UTR, we constructed mouse Qki-3′UTR-luciferase reporter plasmids (see supplemental materials and methods section in online supplementary information) and used them for CRC cell transfections. Plasmid pmQki7-3′UTR-luc carries the 3′UTR for Qki7 mRNA that contains a putative miR-574-5p MRE, which is also shared by Qki6-3′UTR and Qki7b-3′UTR (figure S1, online supplementary information). In contrast, plasmid pmQki5-3′UTR-luc carries the Qki5-3′UTR that does not contain any MRE for miR-574-5p. Consistent with the bioinformatics prediction, however, increased miR-574-5p or inhibition of miR-574-5p did not lead to significant changes in Qki5-3′UTR activity, suggesting that miR-574-5p does not regulate Qki5 mRNA through 3′UTR-mediated mechanisms (figure 2C). As anticipated, however, increased miR-574-5p led to significant suppression of Qki7-3′UTR activity in CT26 cells. Conversely, inhibition of miR-574-5p led to significant activation of Qki7-3′UTR (figure 2C), suggesting that Qki6/7/7b mRNAs are all under the post-transcriptional control of miR-574-5p. Mutational analyses of the miR-574-5p MRE on Qki7-3′UTR (pMT-miR-574-5p-MRE-luc (mutated MRE) versus pWT-miR-574-5p-MRE-luc (wildtype MRE)) further confirmed the specific interaction between miR-574-5p and Qki6/7/7b-3′UTR (figure 2C).
QKI5/6/7 represses β-catenin expression and activity in CRC cells
β-catenin is one of the predicted targets of QKI proteins,34 with two putative QREs on its mRNA (figure S3, online supplementary information). However, a previous study suggested that QKI5/6 suppressed the expression of β-catenin protein but not that of mRNA.23 We therefore wanted to further determine how QKI5/6/7 would affect the expression of β-catenin mRNA and protein and the β-catenin/Wnt signalling activity in CRC cells. To this end, we first transiently transfected CT26 cells with three plasmids overexpressing mouse Qki5/6/7 respectively (see supplemental materials and methods section in online supplementary information). Overexpression of each QKI caused significant downregulation of β-catenin mRNA and protein expression (figure 3A,C). To determine whether QKIs regulate β-catenin through post-transcriptional silencing, we constructed a mouse β-catenin-3′UTR-luciferase reporter (pmCtnnb1-3′UTR-luc) that contains two putative QREs (figure S3 and supplemental materials and methods section in online supplementary information). Co-transfection studies indicated that overexpression of QKI5/6/7 all dose dependently reduced the reporter activity (figure 3B), suggesting that these QKIs are all capable of silencing β-catenin expression post-transcriptionally. Consistent with QKI-mediated downregulation of β-catenin, the TOP/FOP-Flash analyses indicated that these three QKIs could all suppress the activity of TOP-Flash reporter but not the FOP-Flash reporter (figure 3D), suggesting that β-catenin/Wnt signalling is regulated in part by the STAR family proteins QKI5/6/7.
Figure 3.
QKI5, QKI6 and QKI7 negatively regulate β-catenin expression and activity. For western blots, CT26 cells were transfected with pFlag-CMV or QKI overexpressing plasmids (pFlag-mQki5 or pFlag-mQki6 or pFlag-mQki7). For luciferase reporter assays, cells were co-transfected with a Qki-overexpressing plasmid (pFlag-mQki5 or pFlag-mQki6 or pFlag-mQki7), a luciferase reporter plasmid (pmCtnnb1-3′UTR-luc or TOP-Flash or FOP-Flash), Wnt1, together with pSV40-β-galactosidase (3:2:2:1). Cells were harvested for analyses 24 h after the transfections. α-Tubulin expression and β-galactosidase activity were determined and used for the calibration of western blots and luciferase reporter assay respectively. For the luciferase reporter assays, n=3. Data are represented as mean ± SEM. *p<0.05; **p<0.01; ***p<0.001. (A) QKI5/6/7 repress the expression of β-catenin messenger (mRNA) in CT26 cells dose dependently. (B) QKI5/6/7 dose-dependently repress the expression of β-catenin by interacting with β-catenin-3′ untranslated region. (C) QKI5/6/7 repress β-catenin protein expression dose dependently. Plasmid-derived proteins and endogenous proteins appeared to be resolved as two separate bands. (D) QKI5/6/7 suppress β-catenin/Wnt signalling dose dependently. See also online figure S3. RLU, relative light units.
miR-574-5p positively regulates β-catenin but negatively regulates p27Kip1
To determine how miR-574-5p might affect β-catenin and p27Kip1, two putative targets for QKIs, we performed qPCR and western blot analyses with CRC cells. In CT26 and SW480 cells, overexpression of miR-574-5p caused significant upregulation of mRNA and protein levels in β-catenin, whereas inhibition of miR-574-5p caused significant downregulation (figure 4A–C). In contrast, overexpression of miR-574-5p caused downregulation of p27Kip1 and inhibition of miR-574-5p caused upregulation of p27Kip1 mRNA and protein (figure 4D–F). These results suggest that miR-574-5p positively regulates β-catenin but negatively regulates p27Kip1, both largely through downregulating the QKIs.
Figure 4.
miR-574-5p positively regulates β-catenin but negatively regulates p27Kip1. SW480 and CT26 cells were transfected with a miRNA-overexpressing plasmid or a microRNA (miRNA) inhibitor. Cells were harvested for quantitative PCR and western blot assays 24 h after the transfections (n=3). Statistical comparisons were made between a control ((pFlag-CMV or inhibitor control)-transfected cells) and a particular treatment. Data are represented as mean ± SEM. *p<0.05; **p<0.01; ***p<0.001. (A, B) Overexpression of miR-574-5p significantly increased messenger RNA (mRNA) or protein expression of β-catenin, whereas inhibition of miR-574-5p significantly reduced the levels of β-catenin mRNA or protein in SW480 and CT26 cells. (C) Western blots for β-catenin in SW480 and CT26 cells. (D, E) Overexpression of miR-574-5p significantly decreased mRNA or protein expression of p27Kip1 in SW480 and CT26 cells, whereas inhibition of miR-574-5p significantly increased the levels of p27Kip1 mRNA or protein in SW480 and CT26 cells. (F) Western blots for p27Kip1 in SW480 and CT26 cells.
Effects of miR-574-5p on CRC cell proliferation, differentiation, invasion, migration and colony formation
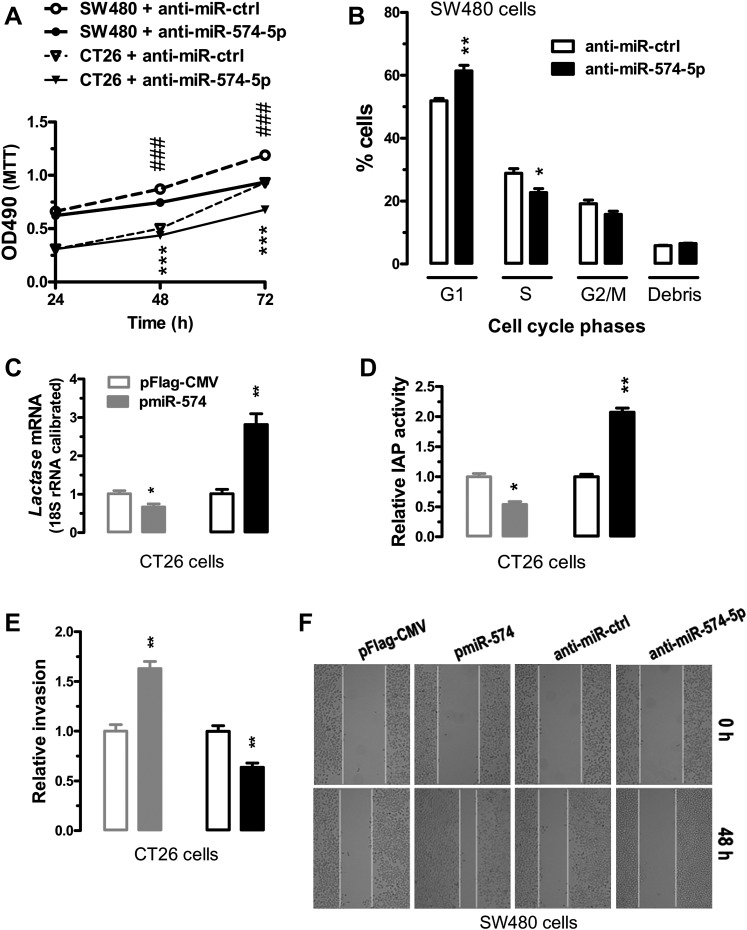
To evaluate the impact of miR-574-mediated regulation of QKIs, β-catenin and p27Kip1, we inhibited or overexpressed miR-574-5p in CT26 cells or SW480 cells. In human and mouse cells, inhibition of miR-574-5p greatly reduced cell proliferation as determined by the MTT assays (figure 5A). In SW480 cells, the G1 cells were significantly increased while the S cells were significantly decreased 72 h after miR-574-5p inhibition, suggesting G1/S arrest (figure 5B). We analysed the expression of Lactase mRNA and the enzyme activity of intestinal alkaline phosphatase (IAP), two important markers of intestinal differentiation. In CT26 cells, overexpression of miR-574-5p significantly reduced the expression of Lactase mRNA and the activity of IAP, whereas inhibition of miR-574-5p increased both (figure 5C,D). Meanwhile, overexpression of miR-574-5p significantly increased invasion likelihood in CT26 cells, whereas inhibition of miR-574-5p decreased invasion likelihood (figure 5E). Furthermore, overexpression of miR574-5p in SW480 cells greatly enhanced cell migration, whereas inhibition of miR-574-5p suppressed the migration (figure 5F). Additionally, specific inhibition of miR-574-5p also greatly decreased the ability of colony formation in SW480 cells (figure 6B). Together these data suggest that when overexpressed, miR-574-5p might be oncogenic.
Figure 5.
Effects of miR-574-5p on cell cycle, proliferation, differentiation, invasion and migration. SW480 or CT26 cells were transfected with a microRNA (miRNA)-overexpressing plasmid or a miRNA inhibitor (see supplemental materials and methods section in online supplementary information). Cells were harvested for analyses 24 h after the transfections (n=3). Statistical comparisons were made between a control ((pFlag-CMV or inhibitor control)-transfected cells) and a particular treatment. Data are represented as mean ± SEM. *p<0.05; **p<0.01; *** or ###, p<0.001. (A) Inhibition of miR-574-5p significantly reduced the rate of proliferation in CT26 and SW480 cells. ### represents comparisons among SW480 cells whereas *** represents comparisons among CT26 cells. (B) Inhibition of miR-574-5p caused G1/S arrest in SW480 cells. (C) Overexpression of miR-574-5p significantly decreased messenger RNA (mRNA) of Lactase, whereas inhibition of miR-574-5p significantly increased the mRNA level of Lactase in CT26 cells. (D) Overexpression of miR-574-5p significantly decreased the activity of intestinal alkaline phosphatase (IAP), whereas inhibition of miR-574-5p significantly increased IAP activity in CT26 cells. (E) Overexpression of miR-574-5p significantly enhanced cell invasion, whereas inhibition of miR-574-5p significantly suppressed invasion in CT26 cells as determined by the Matrigel invasion assays. (F) Overexpression of miR-574-5p significantly enhanced migration in SW480 cells, whereas inhibition of miR-574-5p suppressed cell migration as determined by wound-healing assays. Results are typical for three independent experiments. Similar results were obtained for CT26 cells but not included. Original magnification, ×100.
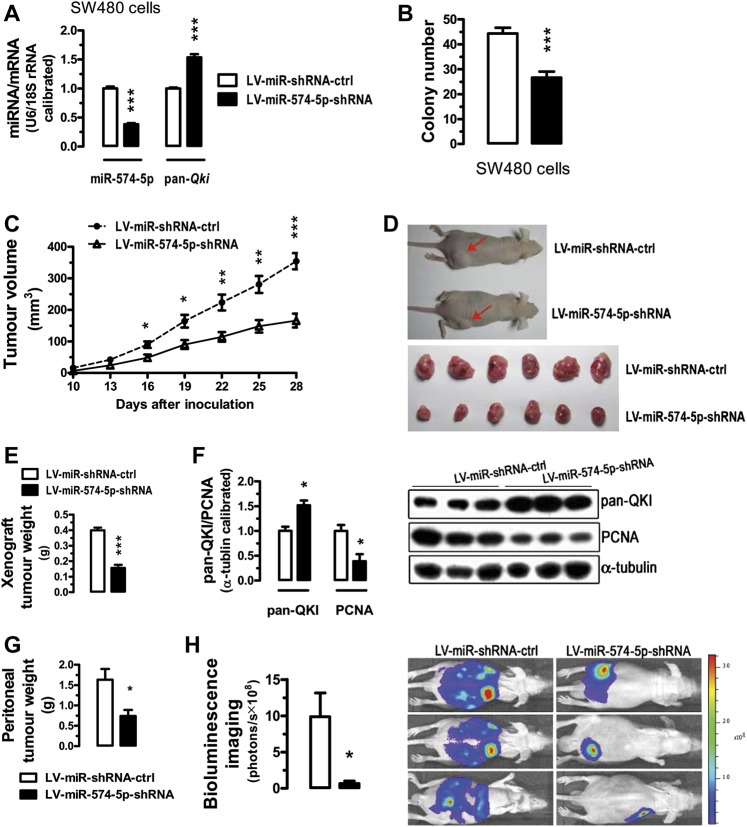
Figure 6.
Inhibition of miR-574-5p suppresses tumour growth in the nude mice. For a xenograft tumour model, SW480 cells were pre-infected with LV-miR-shRNA-ctrl or LV-miR-574-5p-shRNA (see supplemental materials and methods section in online supplementary information). Subsequently, the LV-infected cells were harvested and suspended in phosphate-buffered saline and then inoculated subcutaneously (∼5×106 cells/mouse) into the right side of the posterior flank of 5-week-old nude mice. For bioluminescence imaging of tumour growth in live animals, CT26 cells stably overexrpressing luciferase or its controls were injected into the nude mice intraperitoneally (5×105 cells/mouse). Starting from the third day after CT26 cell inoculation, each mouse was injected with 2×107 transducing units of control lentiviruses or lentiviruses carrying short hairpin RNA (shRNA) for miR-574-5p once a week for 2 weeks. Peritoneal tumour growth was monitored by bioluminescence imaging (described in supplemental materials and methods section in online supplementary information). Data are represented as mean ± SEM. *p<0.05; **p<0.01; ***p<0.001. (A) Downregulation of miR-574-5p and upregulation of total Qki messenger RNA (mRNA) 96 h after lentiviral infection (n=3). (B) Lentivirus-miR-shRNA-mediated inhibition of miR-574-5p significantly decreased colony formation in SW480 cells. SW480 cells were transfected with LV-miR-shRNA-ctrl or LV-miR-574-5p-shRNA. Ninety-six hours after the transfection, 50 transfected cells were seeded on six-well plates and maintained in DMEM containing 10% FBS for 2 weeks before colony counting (n=3). (C) Time-dependent growth of xenograft tumour tissues in the nude mice (n=6). (D, E) Tumour tissues dissected 28 days after the inoculation and the comparison of the tumour weights (n=6). (F) Altered expression of total QKI and proliferating cell nuclear antigen in the dissected tumour tissues (n=3). (G) Weights of visible and dissected peritoneal tumours following gene therapy with lentiviruses carrying shRNA for miR-574-5p or its controls for 24 days (n=3). (H) Intraperitoneal tumour growth as demonstrated by bioluminescence imaging in the live nude mice inoculated with CT26 cells stably overexpressing a luciferase reporter gene 24 days after treatment with lentiviruses carrying shRNA for miR-574-5p or its control (n=3). miRNA, microRNA. DMEM, Dulbecco's Modified Eagle Medium; FBS, fetal bovine serum; PCNA, proliferating cell nuclear antigen.
To verify that miR-574-5p-mediated repression of QKIs is important for the oncogenic function of miR-574-5p, we co-expressed one of the three mouse QKIs (QKI5/6/7) with miR-574-5p in CT26 cells (online figure S4). In comparison with cells overexpressing miR-574-5p only, cells co-overexpressing miR-574-5p and QKI5/6/7 in general all appeared to lead to increased mRNA expression of p27Kip1, reduced mRNA expression of β-catenin, reduced ability in cell invasion and slightly reduced cell proliferation, although the re-introduction of QKI5 only caused relatively minor and in some cases not significant effects. These results confirm that the miR-574-5p-mediated repression of Qkis contributes significantly to its oncogenic function.
Inverse correlation between the expression of miR-574-5p and the expression of total Qki mRNA/protein in CRC clinical samples
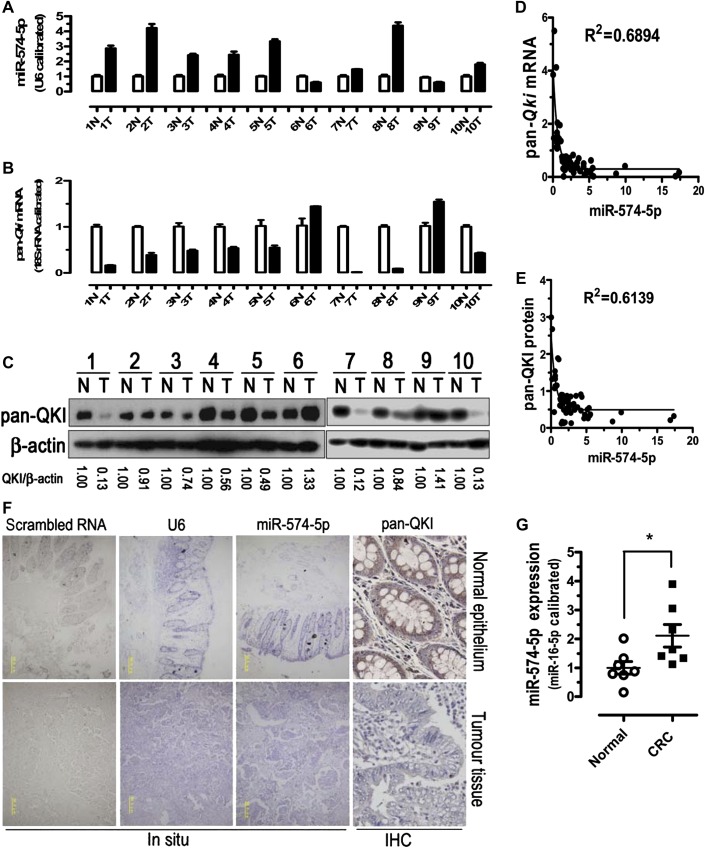
To explore the relationship between the expression of miR-574-5p and the expression of total Qki mRNA/protein, we performed qPCR and western blots with 60 pairs of clinical samples. In about 76.7% of human CRC tissues (46 out of 60), miR-574-5p was found to be significantly upregulated in comparison with the controls, while total Qki mRNA and protein expression levels were greatly reduced (figure 7A–E). These results were further confirmed by immunohistochemistry analyses of QKI protein and in situ hybridisation analyses of miR-574-5p (figure 7F). Largely consistent with the downregulation in total Qki, Qki5/6/7/7b mRNA and protein isoforms were all significantly downregulated in most CRC tissues (online figure S5). Pearson's correlation analyses demonstrated significant inverse correlations between the expression of miR-574-5p and the expression of total Qki mRNA (Pearson r=−0.4082, p<0.05) or QKI protein (Pearson r=−0.4406, p<0.05) in the 60 pairs of CRC samples tested (figure 7D,E). No apparent association, however, was found between miR-574-5p expression and patient gender, patient age, tumour location, TNM staging or lymph node status (online table S1). As a matter of fact, the highest levels (5.737±2.546-fold of the normal level) of miR-574-5p expression were detected for TNM stage I cancers that decreased subsequently but maintained at moderate levels (2.4–2.6-fold) for TNM stages II, III and IV cancers. A similar trend was observed for lymph node status.
Figure 7.
Expression of miR-574-5p and total Qki messenger RNA (mRNA)/protein levels in control and colorectal cancer (CRC) samples. Arabic numbers indicate individual sample pairs. (A) miR-574-5p expression was significantly upregulated in human CRC tissues. (B, C) Total Qki mRNA and protein were significantly downregulated in human CRC tissues. N, normal adjacent epithelial tissue; T, tumour tissue. (D, E) Inverse correlation between the expression of miR-574-5p and the expression of Qki mRNA/protein in 60 pairs of clinical samples. Nonlinear regression (exponential) was indicated. (F) Typical expression patterns of pan-QKI protein in human CRC tissues and normal adjacent epithelial tissues as determined by immunohistochemistry (IHC, original magnification, ×1000) and miR-574-5p expression patterns in human CRC tissues and normal adjacent epithelial tissues as determined by in situ hybridisation (original magnification, ×100; yellow bars represent 50.0 μm). See also online figure S5. (G) miR-574-5p expression was significantly upregulated in serum samples from patients with CRC. Normal, normal controls, n=7; CRC, patients with CRC, n=7.
Remarkably, serum miR-574-5p was also found to be significantly elevated in patients with CRC with respect to the controls (2.11±0.39 for patients with CRC vs 1.00±0.22 for controls, p<0.05, figure 7G).
Qki isoform subcellular localisation and relative expression levels in CRC cells and tissues
We performed western blot and immunofluorescence analyses to determine the subcellular localisation of QKI isoforms in the CRC cells. Largely consistent with previous studies in neural cells35 and HeLa cells,2 36 QKI5 was found to be present in the nucleus and cytoplasm of CT26 and SW620 cells, whereas QKI6/7 proteins were exclusively localised in the cytoplasm (online figure S6A,B). To determine the relative expression levels of Qki mRNA isoforms, we performed qPCR analyses for mouse CT26, human HCT116, SW480 and SW620 cells (online figure S6C), CRC tissues and normal adjacent epithelial tissues from 10 patients with CRC (online figure S6D). In cultured CT26 cells, Qki6 mRNA was 210-fold higher than that of Qki5 mRNA while Qki7 was 308 times higher. In three lines of cultured human CRC cells and most of the human tissues (CRC and normal colorectal tissues), Qki7 mRNA was consistently more than 100-fold higher than that of any other isoforms (including Qki5/6/7b), with only a few exceptions. These results highlight that Qki6 and Qki7 are the predominant isoforms in mouse CRC cells but in human CRC cells and tissues, Qki7 is the most abundant splicing variant.
To evaluate the effects of increased miR-574-5p on the expression and subcellular distribution of β-catenin, we performed both the western blots and immunofluorescent staining in CT26 cells overexpressing miR-574-5p (online figure S7). Our western blots indicated that following miR-574-5p overexpression, β-catenin protein was significantly increased in the nuclear and membrane fractions while not much increase was found in the cytoplasm. Immunofluorescent staining also confirmed significant increases in β-catenin in the nucleus.
Inhibition of miR-574-5p suppresses tumour growth in the nude mice
To further investigate the role of miR-574-5p in tumour growth, we assessed the effects of inhibition of miR-574-5p on the growth of xenograft tumours in nude mice. Mice were injected with SW480 cells that were pre-infected with control lentiviruses or lentiviruses carrying shRNA for miR-574-5p. Lentivirus-mediated knockdown of miR-574-5p caused significant upregulation in total Qki (figure 6A). Probably as a consequence, the ability to form colonies was significantly reduced in miR-574-5p knockdown cells (figure 6B). In nude mice, tumour growth with miR-574-5p-knockdown cells was much slower than that with the control SW480 cells (figure 6C). When dissected at the end of the study (day 28), the average tumour weight of the miR-574-5p knockdown group was only 39% of that of the control group (p<0.001) (figure 6D,E). In the miR-574-5p knockdown tumour tissues, total QKI protein was greatly increased whereas the proliferating cell nuclear antigen expression was significantly decreased (figure 6F). In a more systemic in vivo miR-574-5p inhibition experiment, CT26 cells stably overexpressing luciferase were injected into the nude mice intraperitoneally. The mice were then treated with control lentiviruses or lentiviruses carrying shRNA for miR-574-5p. As shown in figure 6G,H, miR-574-5p inhibition greatly reduced peritoneal tumour growth in the nude mice as monitored by bioluminescent imaging on the 24th day. Together these results suggest that in vivo inhibition of miR-574-5p suppresses CRC tumour growth and this effect is largely due to the increased expression in QKI proteins.
Discussion
QKI is emerging as a major regulator for RNA metabolism and cell signalling.37 38 Dysregulation of QKIs has been associated with a number of important human diseases such as schizophrenia, ataxia and cancers.8 24 However, the regulatory mechanisms underlying the dysregulation of QKIs were elusive. We showed in this current study that miR-574-5p represses the expression of Qki6/7/7b mRNAs by interacting with the MREs on their 3′UTRs. As predicted by bioinformatics analyses, however, miR-574-5p did not regulate the expression of Qki5 through this miR-574-5p-3′UTR interaction. Nevertheless, Qki5 expression very often was downregulated when miR-574-5p was overexpressed but upregulated when miR-574-5p was inhibited. This suggests that other indirect and unknown mechanisms might be involved in the regulation of Qki5 that require further investigation. Importantly, we demonstrated in human CRC cells that Qki7 mRNA expression is usually more than 100-fold higher than any other isoform, with only rare exceptions (figure S6D). This is in contrast to Qki6 being the most abundant isoforms in the brain39 and probably has important physiological implications. Surprisingly, we did not find increased apoptosis in Qki7-overexpressing CT26 cells or human cells with abundant QKI7 protein, contrary to previous reported QKI7-induced apoptosis in fibroblast cells and oligodendrocytes.2 40 QKI5 and QKI6 were previously shown to exert a suppressive effect on β-catenin/Wnt signalling by reducing the level of β-catenin protein and affecting its subcellular redistribution, but they did not appear to affect mRNA expression of β-catenin. 23 By contrast, we showed in our current study that QKI5/6/7 proteins are all capable of repressing mRNA and protein expression of β-catenin and suppressing β-catenin/Wnt signalling in CRC cells. The reason for the discrepancy in β-catenin mRNA expression between the previous study and our current study is not clear but might be related to the semi-quantitative real-time PCR used in the previous study23 compared with the qPCR used in our current study. In light of the relative abundance of QKI7 and its ability to regulate β-catenin/p27Kip1 expression and their signalling activity, however, our novel results suggest a prominent role for QKI7 in QKI-mediated signal transduction and activation of RNA and in colorectal cell differentiation and the development of CRC which to date had been missed from the research.
In human and mouse, the miR-574 gene is intronic. Initially, miR-574-5p was thought to be hosted by the first intron of the gene encoding Noxp20 32 on human chromosome 4 or mouse chromosome 5. A recent study, however, also suggested that miR-574-5p might be encoded by the first intron of Col3a1. Interestingly, alterations in the expression of miR-574-5p have been found to be associated with a variety of diseases, including many types of cancers (online table S2). Ranade et al found that miR-574-5p is one of a few miRNAs that were associated with chemoresistance and decreased survival in patients with small cell lung cancer.41 In serum samples of patients with early stage non-small cell lung cancer, they also found that miR-1254 and miR-574-5p were significantly elevated in the patients with cancer compared with controls,42 which suggests miR-574-5p might be used as a biomarker for non-small cell lung cancer. Very recently, Meyers-Needham et al also showed that miR-574-5p mediated the silencing of tumour suppressor ceramide synthase-1 (CerS1) in head and neck squamous cell carcinoma by selectively targeting one of its alternatively spliced mRNA variants, CerS1-2.43 Moreover, inhibition of miR-574-5p was shown to reconstitute CerS1-2 expression and C18-cermide generation in multiple lines of cancer cells, which subsequently led to suppressed proliferation and anchorage-independent growth of cancer cells. Together these results strongly suggest that miR-574-5p is oncogenic and its inhibition might be beneficial.
We demonstrated in our current investigation that miR-574-5p was significantly elevated in colorectal tissues and serum samples from patients with CRC, especially for cancers at TNM stage I. Consistent with the lack of association between miR-92a and TNM stage in CRC,44 the abnormal upregulation of miR-574-5p in the CRC tissues of our current study was not found to be associated with TNM stage or lymph node status. One possible explanation for the lack of association between miR-574-5p and TNM stage or lymph node status is that miR-574-5p probably promotes CRC tumourigenesis or progression primarily at the early stages. Moreover, it is probably not necessary for miR-574-5p to be continuously increased with CRC progression because a moderate twofold to threefold increase appeared to be sufficient to knockdown Qki significantly (figure 7D,E). Indeed, QKI protein expression was maintained at low levels for cancers at TNM stages II, III and IV. We further demonstrated that aberrantly upregulated miR-574-5p probably contributes to the development of CRC in part by selectively targeting tumour suppressors Qki6/7/7b post transcriptionally, leading to the dys-regulation of β-catenin/Wnt signalling. Whether miR-574-5p also regulates macroH2A1.1 and CerS1-2 in the CRC cells to contribute to CRC development or not, however, remains to be determined. Despite this, findings from our current study have important implications for the elucidation of pathogenic mechanisms and for clinical practice. Theoretically, the identification of this novel regulatory mechanism of Qki isoforms mediated by miR-574-5p sheds lights on the elucidation of pathogenic mechanisms underlying the development of important human diseases associated with the aberrant expression of miR-574-5p, QKIs and more than 1400 putative QKI-regulated genes (online figure S8). From a clinical perspective, our demonstration that serum miR-574-5p is significantly elevated in most patients with CRC suggests that miR-574-5p might serve as a novel diagnostic biomarker for early detection of CRC. As importantly, our demonstration that inhibition of miR-574-5p ameliorated CRC development or progression suggests that miR-574-5p might be an important therapeutic target for the prevention or treatment of CRC and other miR-574-5p-associated cancers.
Supplementary Material
Supplementary Material
Acknowledgments
The authors thank Dr Qiao Wu for the TOP/FOP-flash luciferase reporters, Dr Boan Li for the Apc-mutant mice and Dr Bixing Zhao and Mr Lianwei Yang at the National Institute of Diagnostic and Vaccine Development (Xiamen University) for technical assistance. We also thank Professor Longping Wen for critical reading of the manuscript.
Footnotes
Contributors: SJ, GY, JZ, LW, TW, ZW, TZ and GW performed the experiments, collected and analysed the data; YL and ZG designed the experiments, analysed the data and revised the paper; JC designed and supervised the project, analysed the data and revised the paper; JYY conceived, designed and supervised the project, analysed the data, wrote and revised the paper.
Funding: This work was supported in part by grants from the National Science Foundation of China (30970649), the 973 Program of China (2009CB941601), the Fujian Provincial Department of Science and Technology (2010L0002), and the 111 Project of Education of China (B06016).
Competing interests: None.
Ethics approval: Institutional Medical Ethics Committee of Xiamen University.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1. Biedermann B, Hotz HR, Ciosk R. The Quaking family of RNA-binding proteins: coordinators of the cell cycle and differentiation. Cell Cycle 2010;9:1929–33 [DOI] [PubMed] [Google Scholar]
- 2. Pilotte J, Larocque D, Richard S. Nuclear translocation controlled by alternatively spliced isoforms inactivates the QUAKING apoptotic inducer. Genes Dev 2001;15:845–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Larocque D, Galarneau A, Liu HN, et al. Protection of p27(Kip1) mRNA by quaking RNA binding proteins promotes oligodendrocyte differentiation. Nat Neurosci 2005;8:27–33 [DOI] [PubMed] [Google Scholar]
- 4. Li Z, Takakura N, Oike Y, et al. Defective smooth muscle development in qkI-deficient mice. Dev Growth Differ 2003;45:449–62 [DOI] [PubMed] [Google Scholar]
- 5. Lobbardi R, Lambert G, Zhao J, et al. Fine-tuning of Hh signaling by the RNA-binding protein Quaking to control muscle development. Development 2011;138:1783–94 [DOI] [PubMed] [Google Scholar]
- 6. van MA, Grundmann S, Goumans MJ, et al. MicroRNA-214 inhibits angiogenesis by targeting Quaking and reducing angiogenic growth factor release. Cardiovasc Res. Published Online First: 6 February 2012. doi:10.1093/cvr/cvs003 [DOI] [PubMed] [Google Scholar]
- 7. Noveroske JK, Lai L, Gaussin V, et al. Quaking is essential for blood vessel development. Genesis 2002;32:218–30 [DOI] [PubMed] [Google Scholar]
- 8. Chenard CA, Richard S. New implications for the QUAKING RNA binding protein in human disease. J Neurosci Res 2008;86:233–42 [DOI] [PubMed] [Google Scholar]
- 9. Richard S. Reaching for the stars: linking RNA binding proteins to diseases. Adv Exp Med Biol 2010;693:142–57 [PubMed] [Google Scholar]
- 10. Bielli P, Busa R, Paronetto MP, et al. The RNA-binding protein Sam68 is a multifunctional player in human cancer. Endocr Relat Cancer 2011;18:R91–102 [DOI] [PubMed] [Google Scholar]
- 11. Rajan P, Gaughan L, Dalgliesh C, et al. Regulation of gene expression by the RNA-binding protein Sam68 in cancer. Biochem Soc Trans 2008;36:505–7 [DOI] [PubMed] [Google Scholar]
- 12. Lukong KE, Richard S. Targeting the RNA-binding protein Sam68 as a treatment for cancer? Future Oncol 2007;3:539–44 [DOI] [PubMed] [Google Scholar]
- 13. Chen ZY, Cai L, Zhu J, et al. Fyn requires HnRNPA2B1 and Sam68 to synergistically regulate apoptosis in pancreatic cancer. Carcinogenesis 2011;32:1419–26 [DOI] [PubMed] [Google Scholar]
- 14. Song L, Wang L, Li Y, et al. Sam68 up-regulation correlates with, and its down-regulation inhibits, proliferation and tumourigenicity of breast cancer cells. J Pathol 2010;222:227–37 [DOI] [PubMed] [Google Scholar]
- 15. Li ZZ, Kondo T, Murata T, et al. Expression of Hqk encoding a KH RNA binding protein is altered in human glioma. Jpn J Cancer Res 2002;93:167–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yin D, Ogawa S, Kawamata N, et al. High-resolution genomic copy number profiling of glioblastoma multiforme by single nucleotide polymorphism DNA microarray. Mol Cancer Res 2009;7:665–77 [DOI] [PubMed] [Google Scholar]
- 17. Ichimura K, Mungall AJ, Fiegler H, et al. Small regions of overlapping deletions on 6q26 in human astrocytic tumours identified using chromosome 6 tile path array-CGH. Oncogene 2006;25:1261–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sukumar S, Wang S, Hoang K, et al. Subtle overlapping deletions in the terminal region of chromosome 6q24.2-q26: three cases studied using FISH. Am J Med Genet 1999;87:17–22 [PubMed] [Google Scholar]
- 19. Backx L, Fryns JP, Marcelis C, et al. Haploinsufficiency of the gene Quaking (QKI) is associated with the 6q terminal deletion syndrome. Am J Med Genet A 2010;152A:319–26 [DOI] [PubMed] [Google Scholar]
- 20. Aberg K, Saetre P, Lindholm E, et al. Human QKI, a new candidate gene for schizophrenia involved in myelination. Am J Med Genet B Neuropsychiatr Genet 2006;141B:84–90 [DOI] [PubMed] [Google Scholar]
- 21. Aberg K, Saetre P, Jareborg N, et al. Human QKI, a potential regulator of mRNA expression of human oligodendrocyte-related genes involved in schizophrenia. Proc Natl Acad Sci U S A 2006;103:7482–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Klempan TA, Ernst C, Deleva V, et al. Characterization of QKI gene expression, genetics, and epigenetics in suicide victims with major depressive disorder. Biol Psychiatry 2009;66:824–31 [DOI] [PubMed] [Google Scholar]
- 23. Yang G, Fu H, Zhang J, et al. RNA-binding protein quaking, a critical regulator of colon epithelial differentiation and a suppressor of colon cancer. Gastroenterology 2010;138:231–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Novikov L, Park JW, Chen H, et al. QKI-mediated alternative splicing of the histone variant MacroH2A1 regulates cancer cell proliferation. Mol Cell Biol 2011;31:4244–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bockbrader K, Feng Y. Essential function, sophisticated regulation and pathological impact of the selective RNA-binding protein QKI in CNS myelin development. Future Neurol 2008;3:655–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huang K, Tang W, Xu Z, et al. No association found between the promoter variations of QKI and schizophrenia in the Chinese population. Prog Neuropsychopharmacol Biol Psychiatry 2009;33:33–6 [DOI] [PubMed] [Google Scholar]
- 27. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281–97 [DOI] [PubMed] [Google Scholar]
- 28. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009;136:215–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Artzt K, Wu JI. STAR trek: an introduction to STAR family proteins and review of quaking (QKI). Adv Exp Med Biol 2010;693:1–24 [PubMed] [Google Scholar]
- 30. Zorn AM, Grow M, Patterson KD, et al. Remarkable sequence conservation of transcripts encoding amphibian and mammalian homologues of quaking, a KH domain RNA-binding protein. Gene 1997;188:199–206 [DOI] [PubMed] [Google Scholar]
- 31. Kondo T, Furuta T, Mitsunaga K, et al. Genomic organization and expression analysis of the mouse qkI locus. Mamm Genome 1999;10:662–9 [DOI] [PubMed] [Google Scholar]
- 32. Boucquey M, De PE, Locker M, et al. Noxp20 and Noxp70, two new markers of early neuronal differentiation, detected in teratocarcinoma-derived neuroectodermic precursor cells. J Neurochem 2006;99:657–69 [DOI] [PubMed] [Google Scholar]
- 33. Sterling KM. The procollagen type III, alpha 1 (COL3A1) gene first intron expresses poly-A+ RNA corresponding to multiple ESTs and putative miRNAs. J Cell Biochem 2011;112:541–7 [DOI] [PubMed] [Google Scholar]
- 34. Galarneau A, Richard S. Target RNA motif and target mRNAs of the Quaking STAR protein. Nat Struct Mol Biol 2005;12:691–8 [DOI] [PubMed] [Google Scholar]
- 35. Hardy RJ, Loushin CL, Friedrich VL, Jr, et al. Neural cell type-specific expression of QKI proteins is altered in quaking viable mutant mice. J Neurosci 1996;16:7941–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wu J, Zhou L, Tonissen K, et al. The quaking I-5 protein (QKI-5) has a novel nuclear localization signal and shuttles between the nucleus and the cytoplasm. J Biol Chem 1999;274:29202–10 [DOI] [PubMed] [Google Scholar]
- 37. Feng Y, Bankston A. The star family member QKI and cell signaling. Adv Exp Med Biol 2010;693:25–36 [PubMed] [Google Scholar]
- 38. Vernet C, Artzt K. STAR, a gene family involved in signal transduction and activation of RNA. Trends Genet 1997;13:479–84 [DOI] [PubMed] [Google Scholar]
- 39. Zhao L, Tian D, Xia M, et al. Rescuing qkV dysmyelination by a single isoform of the selective RNA-binding protein QKI. J Neurosci 2006;26:11278–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chen T, Richard S. Structure–function analysis of Qk1: a lethal point mutation in mouse quaking prevents homodimerization. Mol Cell Biol 1998;18:4863–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ranade AR, Cherba D, Sridhar S, et al. MicroRNA 92a-2*: a biomarker predictive for chemoresistance and prognostic for survival in patients with small cell lung cancer. J Thorac Oncol 2010;5:1273–8 [DOI] [PubMed] [Google Scholar]
- 42. Foss KM, Sima C, Ugolini D, et al. miR-1254 and miR-574-5p: serum-based microRNA biomarkers for early-stage non-small cell lung cancer. J Thorac Oncol 2011;6:482–8 [DOI] [PubMed] [Google Scholar]
- 43. Meyers-Needham M, Ponnusamy S, Gencer S, et al. Concerted functions of HDAC1 and microRNA-574-5p repress alternatively spliced ceramide synthase 1 expression in human cancer cells. EMBO Mol Med 2012;4:78–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Huang Z, Huang D, Ni S, et al. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer 2010;127:118–26 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.