Abstract
Riboswitches are regulatory elements commonly found in the 5'-untranslated regions of bacterial mRNAs that bind cellular metabolites to direct expression at either the transcriptional or translational level. The effectors of these RNAs are chemically diverse, including nucleobases and nucleosides, amino acids, cofactors, and second messenger molecules. Over the last few years, a number of structures have revealed the architectural means by which RNA creates binding pockets of high affinity and specificity for these compounds. For most effectors, there is a single class of associated riboswitches. However, eight individual classes of S-adenosylmethionine (SAM) and/or S-adenosylhomocysteine (SAH) responsive riboswitches that control various aspects of sulfur metabolism have been validated, revealing a diverse set of solutions to the recognition of these ubiquitous metabolites. This review focuses upon the structures of RNAs that bind SAM and SAH and how they discriminate between these compounds.
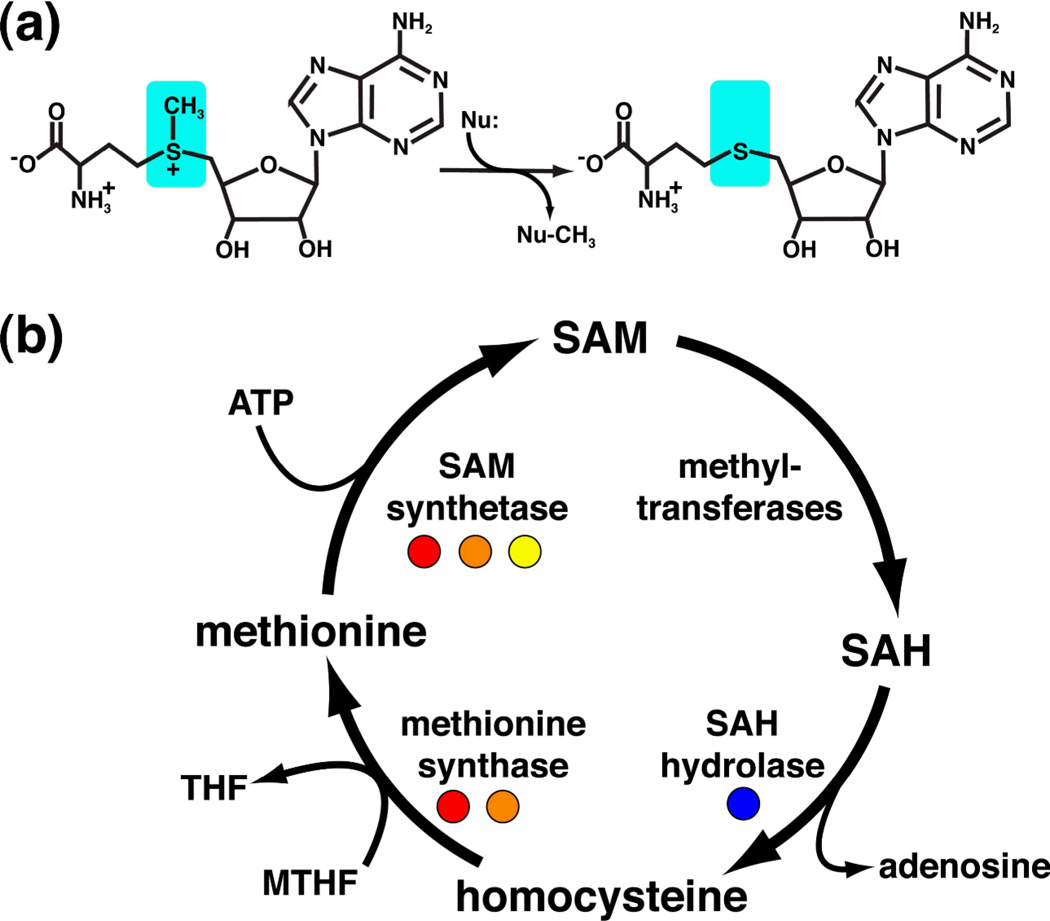
S-adenosylmethionine (SAM) is second only to ATP in terms of its use as an enzyme substrate1. Most commonly, SAM is employed as a methyl group donor (Figure 1a), but also participates in a wide variety of chemical reactions in which it is a source of methylene, ribosyl, amino, and aminoalkyl groups and free radicals2,3. Furthermore, SAM is strongly coupled to methionine through its synthesis and recycling (Figure 1b) and is thus tied to most metabolic processes involving sulfur in the cell4. As a consequence of its central biological importance, intracellular methionine and SAM concentrations are tightly regulated.
Figure 1.
(a) Chemical structures of S-adenosyl-L-methionine (reactant) and S-adenosyl-L-homocysteine (product). SAM is the primary donor of methyl groups in biology via a nucleophilic attack (Nu) on its activated methyl group (cyan), such that SAH is the leaving group. (b) The SAM regeneration cycle. Enzymes regulated by SAM and SAH riboswitches are highlighted with colored circles (red, SAM-I; orange, SAM-II; yellow, SAM-III; blue, SAH).
In E. coli, SAM is a corepressor of the metJ protein that regulates transcription of a number of genes involved in methionine biosynthesis (reviewed by Weissbach and Brot5). However, in other bacteria the regulatory mechanisms associated with SAM and methionine were unknown at this time. In 1998, Grundy and Henkin characterized a sequence they called the "S-box" that appears upstream of a number of genes involved in sulfate assimilation into cysteine and methionine biosynthesis6. This pioneering study determined that this element acts at the level of RNA which folds into a cloverleaf-like structure important for function. These results clearly indicated that RNA structure in the 5' leader of mRNA plays an important role in the regulation of gene expression. However, no associated protein could be identified that could serve as an intermediary between intracellular methionine concentrations and the regulatory activity of the RNA.
At the same time, other regulatory elements were being identified that had similar properties7,8. For example, Gelfand and coworkers described a sequence commonly found upstream of riboflavin biosynthetic genes in a broad spectrum of bacterial species that also appeared to be a regulatory element7. The authors, noting the puzzling lack of an identifiable protein associated this sequence, conjectured that "an intriguing possibility is that in this case the direct binding of the reaction product to the RNA structure is involved"7. Within a few years, the Breaker group discovered an element in the E. coli cobB mRNA capable of directly binding adenosylcobalamin that directed a downstream secondary structural switch dictating the expression fate of the mRNA9. This type of regulatory element became known as a riboswitch. Not shortly afterward, three groups independently demonstrated that the S-box (also called SAM-I, for reasons that will become clear) bound SAM to direct expression10–12. Since then, at least twenty classes of riboswitches have been validated that interact with a diverse set of cellular metabolites, including amino acids, nucleobases and nucleosides, and protein cofactors (reviewed by Roth and Breaker13).
Most bacterial riboswitches appear to have a common organization allowing them to regulate gene expression at either the transcription or translational levels14. On the 5'-side of bacterial riboswitches is the aptamer domain whose function is to directly and specifically bind the effector compound. The secondary and tertiary structure of the aptamer domain of each class is specific to the metabolite bound by the mRNA (reviewed by Montange and Batey15). The structure of SAM-binding aptamer domains is the principal focus of this review, with an emphasis on the architecture of the RNA and how it allows for a high degree of discrimination against the product form of methylation by SAM, S-adenosylhomocysteine (SAH) (Figure 1a).
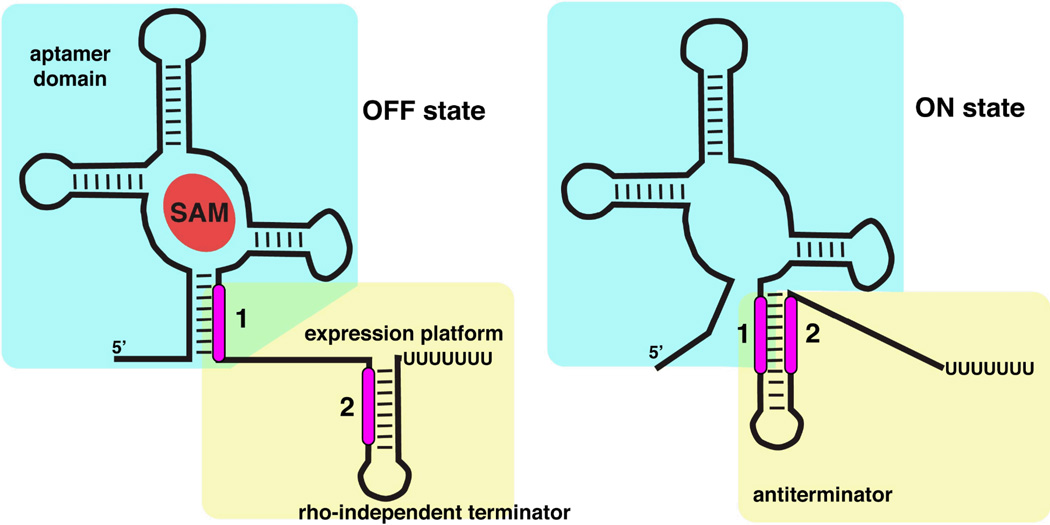
Regulation by SAM-binding riboswitches occurs by a number of mechanisms. Immediately downstream of the aptamer domain in most riboswitches is the expression platform that contains a mutually exclusive secondary structural switch14 (Figure 2). In the case of transcription, this switch adopts the form of an antiterminator/terminator pair that directs transcription to proceed or abort, respectively. Each of these signals is a stem-loop structure; the terminator is a rho-independent termination signal composed of a stem-loop followed by four to six uridine residues that causes polymerase to disengage16,17. The hallmark of the mutually exclusive switch is that the two helices share a sequence (marked "2" in Figure 2) such that formation of one stem-loop precludes formation of the other. For translational regulators, the secondary structural switch involves exposing or occluding sequence elements necessary for loading the ribosome onto the mRNA. More recently, several other modes of SAM-binding riboswitch mediated regulation have been discovered. In Clostridium acetobutylicum, a SAM-I riboswitch regulates the expression of the ubiG locus by directing the expression of an antisense RNA that interferes with transcription18. Another example is found in Listeria monocytognes, in which a SAM-I riboswitch transcript anneals to the mRNA encoding the master regulator of virulence, PrfA, and thereby regulates expression through an antisense mechanism19.
Figure 2.
Alternative secondary structures of a riboswitch that regulates transcription. The "off" state (left) is characterized by SAM bound to the aptamer domain (cyan box) which incorporates a sequence that is shared with the downstream expression platform (marked as "1"). This forces the expression platform (yellow box) to form a terminator stem-loop. In the "on" state (right) the lack of SAM binding allows the expression platform to form an antiterminator element using the sequence shared between the two domains. Note the mutual exclusivity of formation of the antiterminator and terminator through differential partitioning of sequences "1" and "2".
Underscoring the importance of regulation of intracellular methionine and SAM concentrations by riboswitches in bacteria, six classes of SAM-binding riboswitches have been identified (called SAM-I through -V, and -I/IV)20–23. These regulatory motifs are broadly distributed across bacterial phylogeny, with the SAM-I motif being the most prevalent24. In addition, there is an SAH-binding riboswitch found in proteobacteria that upregulates S-adenosylhomocysteine recycling enzymes25 and one that appears to recognize SAM and SAH with equal affinity20. To illuminate how these riboswitches bind their effector with high affinity and specificity, atomic resolution structures of three classes of these elements have been solved26–28.
The structure of the SAM-I riboswitch
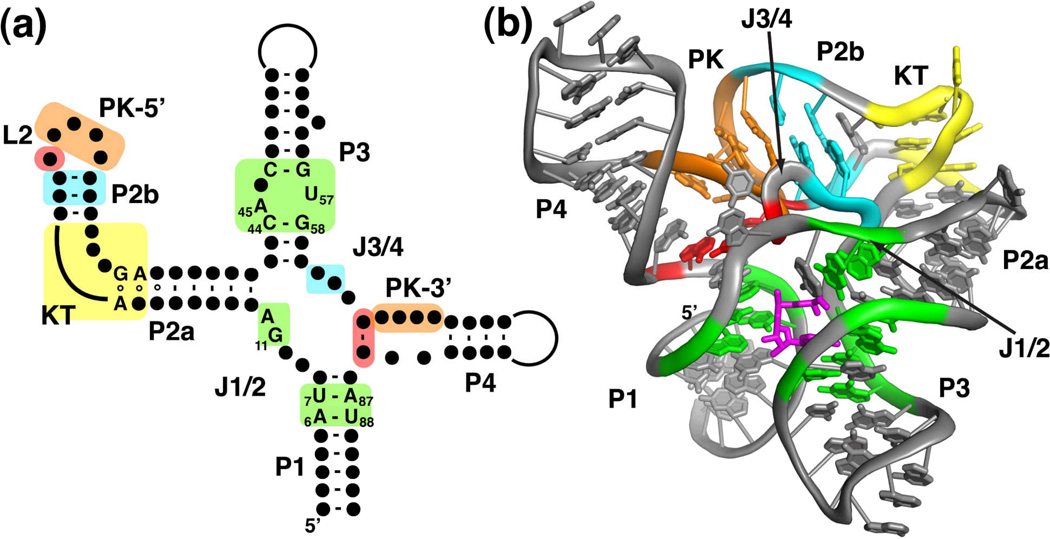
The conserved consensus sequence of the SAM-I riboswitch aptamer folds into a secondary structure in which most of the phylogenetically conserved nucleotides cluster around a four-way junction (Figure 3a)6,29. Embedded within this secondary structure are several motifs commonly found in biological RNAs. P2 contains a widely distributed motif called a kink-turn (reviewed by Tiedge30), defined by two non-canonical G-A base pairs adjacent to a three-nucleotide bulge31,32. Introduction of mutations into this motif severely compromises both SAM binding33 and regulatory function32. Another feature is a pseudoknot motif (the structure of pseudoknots is reviewed by Staple and Butcher34) formed through Watson-Crick base pairing between L2 and J3/46,35. Similar to the kink-turn motif, disruption of the pseudoknot results in loss of SAM binding and regulatory function35. Further conserved nucleotides are interspersed around the four-way junction and its associated helices, with most invariant nucleotides found in P1, J1/2, and P314 (nucleotides whose identity is >97% conserved in alignments of over 2900 SAM-I sequences29,36 are highlighted in Figure 3a).
Figure 3.
Structure of the SAM-I riboswitch. (a) Phylogenetically conserved primary and secondary structure of the class I SAM riboswitch. Nucleotides that are >97% conserved are specified by their identity; conserved secondary structure is shown as dots. Colored boxes refer to elements of conserved tertiary architecture: a kink-turn motif (yellow), a pseudoknot (orange), adenosine minor triples (cyan), and a base triple (red) as well as structure directly involved in SAM binding (green). Note that the all of the conserved residues are either in the critical G-A pairs that define the kink-turn motif or in the SAM binding pocket. The numbering of key nucleotides is consistent with that in Montange and Batey26. (b) Cartoon representation of the X-ray crystal structure of the SAM-I riboswitch aptamer domain bound to SAM. This panel and other molecular representations of the SAM-I riboswitch were made using the coordinates in PDB accession number 3GX5. The color scheme is the same as in section (a), emphasizing the placement of the conserved tertiary architecture and the SAM binding pocket. SAM is shown as magenta sticks.
The three-dimensional structure of the aptamer domain of the SAM-I riboswitch that regulates the Thermoanaerobacter tengcongensis metF2-metH (methylenetetrahydrofolate reductase and cobalamin-independent methionine synthase) operon revealed a compact fold26. While the sequence of this RNA was heavily engineered to promote crystallization37, including truncation of helices, addition of GNRA type tetraloops and internal point mutations, all of the sequence elements >80% conserved in phylogeny are present26,38. Thus, this structure represents the conserved core of all SAM-I riboswitches and very likely all RNAs within the SAM family of riboswitches (vide infra).
The architecture of the SAM-I riboswitch aptamer domain is defined by the organization of the principal helices into two coaxial stacks: P1/P4 and P2/P3--a common organizing principle in large RNAs39 (Figure 3b). The two stacks are tied together through a set of tertiary interactions involving the two long joining regions, J1/2 and J3/4. The kink-turn motif (yellow, Figure 3b) in the middle of P2 positions the terminal loop (L2) towards P4 by introducing a ~120° bend between helices P2a and 2b. As predicted by biochemical and genetic experiments6,40, four bases in L2 pair with nucleotides in J3/4 to form a pseudoknot that lies between the two coaxial stacks (orange, Figure 3b). Conserved adenosine residues in J3/4 support the pseudoknot (PK) by forming critical two base triples with conserved G-C pairs in P2b adjacent to L2 (cyan, Figure 3). These adenosine minor triples are different from the commonly observed Type I/II triples41,42 in that they use the Watson-Crick face of the adenosine rather than its sugar edge. This region of the aptamer is also further defined by a base triple between nucleotides in J3/4, L2, and J4/1 (red, Figure 3). The role of the second joining region, J1/2, is to form a set of triple interactions with P3 that primarily serve to define part of the ligand-binding pocket. A recent study of the folding of the SAM-I aptamer showed that P4, PK, and P2a/b form a subdomain that folds independently and stably in the absence of SAM43.
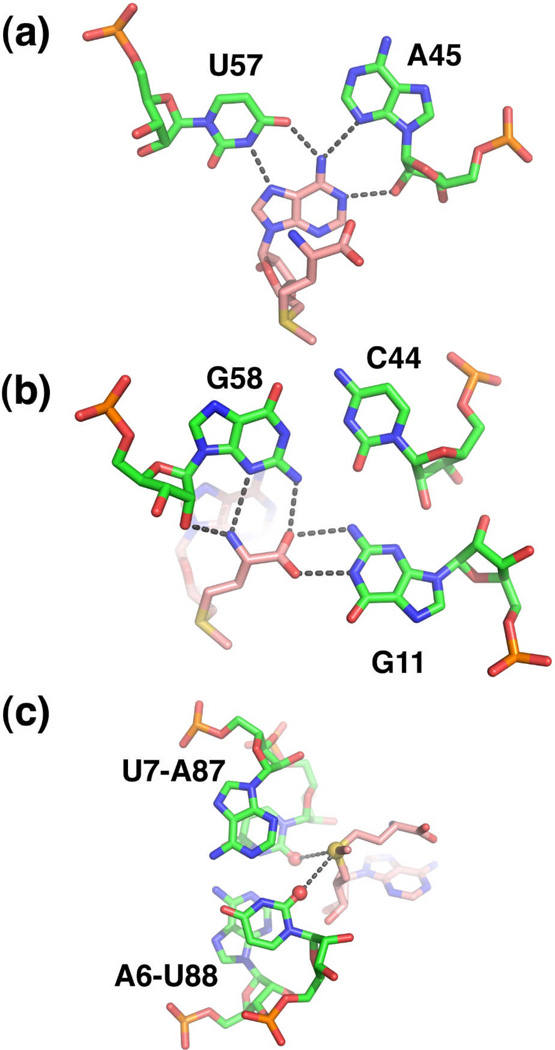
Within the core of the RNA is a bipartite SAM-binding pocket consisting of elements of P1 and P326. This mode of recognition is made possible in part by SAM adopting a cis-configuration in which the methionine moiety stacks upon the adenine ring. One "face" of SAM presents the adenine and methionine moiety to the P3 helix. The adenine moiety participates in a base triple with A45 and U57 (numbering scheme of important nucleotides is given in Figure 3a) and the main chain atoms of methionine interact with the G11•(C44–G58) triple (Figure 4a, b). The other face of SAM, comprising the ribose sugar and the positively charged sulfonium ion, is recognized by the minor groove of the P1 helix. The sulfonium group is adjacent to two universally conserved base pairs in the P1 helix, A6-U88 and U7-A87 such that the sulfur atom is positioned ~4 Å from the carbonyl oxygens of the uracils (Figure 4c). The O2 and O4 carbonyl oxygens (as well as the oxygen of the 2'-hydroxyl group) each carry approximately one half negative charge44, suggesting that the sulfonium ion of SAM interacts with the RNA through an electrostatic interaction.
Figure 4.
Recognition of SAM (pink carbon backbone) by the SAM-I subfamily (green carbon backbone). (a) Interaction of the adenine moiety with nucleotides in P3 (A45 and U57) to form a base triple. (b) The main chain atoms of methionine form hydrogen-bonding interactions with the P3 helix (G58) and J1/2 (G11). (c) Electrostatic interaction between the sulfonium ion of SAM (yellow sphere) and two carbonyl oxygens (red spheres) in the P1 helix.
A key aspect of SAM-dependent genetic regulation by RNA and proteins is the ability to discriminate against SAH. These two compounds differ only around the sulfur atom; SAM contains the activated methyl group that imparts a positive charge on the sulfur (Figure 1a). The B. subtilis yitJ SAM-I riboswitch is capable of >100-fold discrimination between SAM and SAH, on par with proteins12; the T. tengcongensis sequence displays over 500-fold selectivity for SAM38. Two lines of evidence suggest that discrimination is mediated by an electrostatic interaction between the RNA and ligand. The Breaker group investigated the binding of a series of thioether-modified SAM analogs to the B. subtilis yitJ SAM-I45. Strikingly, the tightest binding of these analogs was one in which the sulfonium ion was replaced with a positively charged quaternary amine, which binds with same affinity as SAM. On the other hand, an analog with a neutral tertiary amine bound with a 40-fold lower affinity than SAM, indicating the importance of an electrostatic component to ligand binding45. A separate study looked at discrimination from the perspective of the conserved A6-U88/U7-A87 pairs in the P1 helix (Figure 4c)38. Mutagenesis of these A–U pairs by transversion or replacement with a G-C pair showed that altered placement of carbonyl oxygens weakened the affinity of the RNA for SAM while not affecting its affinity for SAH. The most severe of the mutations (C6-G88/G7-C87) resulted in an RNA that binds SAM with only ~3-fold selectivity over SAH, demonstrating that these two universally conserved A–U pairs are directly responsible for discrimination38.
Variations of the SAM-I riboswitch: the SAM superfamily
In a search for novel classes of riboswitches, a distinct set of elements were uncovered that display clear similarities to the SAM-I riboswitch22. This class, the SAM-IV riboswitch, is primarily found in Actinomycetales (Mycobacterium tuberculosis, for example) upstream of genes involved in sulfur metabolism22. SAM-IV can be folded into a secondary structure similar to that of SAM-I, with the notable exception that SAM-IV lacks the P4 helix of SAM-I (Figures 3, 5). In addition, there is significant similarity between the ligand binding cores of the SAM-I and -IV riboswitches in that key nucleotides involved in SAM binding are conserved in both classes. This suggests that they recognize SAM in an almost identical fashion. The only difference is in the sulfonium ion binding site within the P1 helix. The more important pair for SAM binding and discrimination in the SAM-I riboswitch (U7-A87) is preserved, however the A6–U88 pair is a G-C pair in SAM-IV (Figure 5)22. Nonetheless, analysis of binding of SAM and SAH to the Streptomyces coelicolor SAM-IV riboswitch revealed a comparable level of affinity and specificity for SAM as SAM-I riboswitches22.
Figure 5.
Secondary structures of the SAM superfamily of riboswitches: (a) SAM-I, (b) SAM-IV, and (c) SAM I/IV. While many of the critical nucleotide identities are retained in all three classes (red), the pattern of pseudoknot formation around the core (blue and green) is different in each.
The significant differences between the two classes are found in the peripheral tertiary architecture of the aptamer domain22. First, the conserved kink-turn found in the P2 helix of SAM-I riboswitches is replaced with an asymmetric internal loop that likely also introduces a bend in the helix (P2, Figure 5b). However, it is not known whether the two turns are structurally similar. Furthermore, while L2 and J3/4 form a pseudoknot interaction in SAM-IV, it is predicted to contain six Watson-Crick base pairs (versus four in SAM-I). Second, the terminal loop of P3 is involved in a second predicted pseudoknot interaction with the 3'-end of the aptamer. Finally, the P4 helix, which is universally present in SAM-I riboswitches, is absent in the SAM-IV class. Instead, a new helix (P4', Figure 5b) is found downstream of the P1 helix. Together, these differences lead to an altered organization of helices that scaffold the binding pocket. More recently, a third class of the SAM riboswitches has been found, "SAM-I/IV", that also bears strong similarities to the SAM-I class (Figure 5c)20. This motif preserves the identities of nucleotides directly in contact SAM in the SAM-I class but differs from SAM-I in that it lacks an internal loop in P2 and the associated pseudoknot with J3/4. Thus, the conserved core of the SAM-I riboswitch can be supported by different peripheral elements without impairing SAM binding activity or regulatory function.
Given the similarity of these RNAs, it has been suggested that they might be evolutionarily related. In one proposed model, the widespread distribution of the SAM-I riboswitch as opposed to SAM-IV or SAM-I/IV hints at a common ancestor that would have been similar to SAM-I20. Divergence occurred through loss of peripheral tertiary interactions and acquisition of new ones. Thus, this model proposes that these three classes (or more accurately, families) are members of an evolutionarily related superfamily of SAM-riboswitches. A related model has also been proposed in which modern SAM-I riboswitches emerged from an ancient SAM synthetase and/or SAM-dependent methyltransferase ribozymes from an RNA world46. This primordial RNA is speculated to have had a minimal SAM binding core lacking supporting peripheral tertiary interactions, which were added later as the RNA diversified into the modern riboswitches.
Structure of the unbound SAM-I riboswitch
Over the last six years, crystal structures of over ten distinct classes of riboswitches have been solved in complex with their effector ligand. While these structures reveal atomic-level details of recognition, by themselves they do not yield clear insights into the structure of the free aptamer and the presumed conformational changes that accompany binding. The first example of a structure of an unliganded riboswitch was the lysine riboswitch, revealing that the RNA's structure on both a global and local level is nearly identical with the lysine-bound form47,48. This finding, validated using solution methods, suggested that the unliganded aptamer can adopt conformations extremely similar to the bound state49–51. On the opposite side of the spectrum are RNAs like the preQ149–51 and TPP riboswitches52–54, which are proposed to undergo large conformational changes upon productive interaction with their cognate ligand. Thus, a somewhat murky picture of this aspect of riboswitch structure and its relevance to functional switching has emerged. Nonetheless, it is critically important to understand the nature of the structure of unbound riboswitch aptamers as conformational changes between the unbound and bound states are suggested to be a critical component of communication with the secondary structural switch55.
Similar to the lysine riboswitch, the crystal structure of the unliganded SAM-I riboswitch revealed a global and local architecture nearly identical to the bound form43. However, a formal possibility is that the crystal lattice actively trapped the RNA in a bound-like conformation. Thus, the relevance of the structure to understanding the nature of the free state remained in question. To address this issue, the conformation of the RNA was investigated using small angle X-ray scattering (SAXS), a solution technique that yields information about the size and shape of the scattering macromolecule56,57. Analysis of the SAM-I riboswitch RNA demonstrated that the RNA undergoes a large magnesium-dependent compaction, followed by a smaller SAM-dependent compaction, with the dimensions of the SAM-bound form in solution consistent with the crystal structure58.
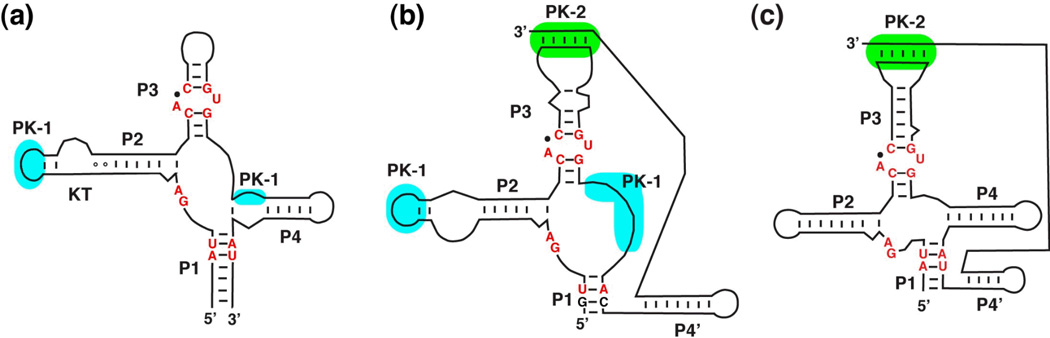
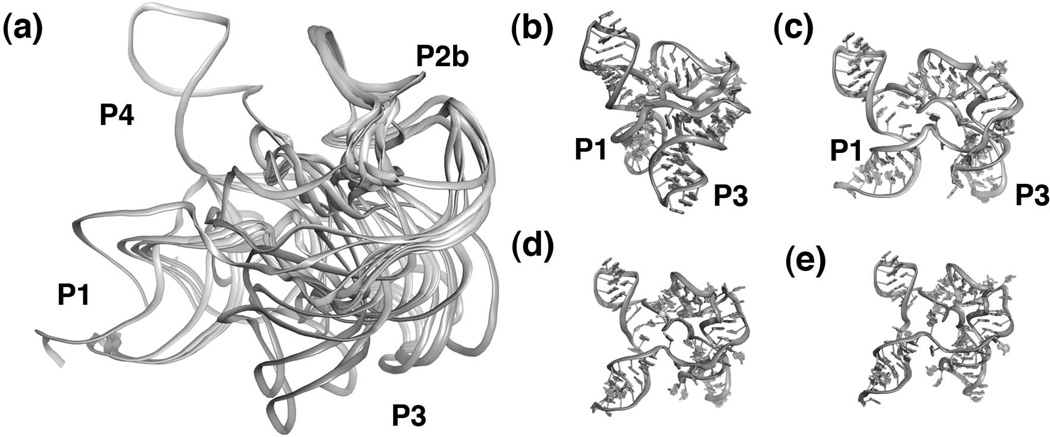
Rather than being a single, discrete structure, the unbound form of the SAM-I aptamer likely samples a multitude of conformations in solution. To understand the nature of these conformations, the scattering data for the unbound RNA was analyzed using an "ensemble optimization method"59,60. This technique yields a ensemble of structural models that best describes the experimental scattering curve. The resulting ensemble of models for the SAM-I riboswitch (Figure 6a) includes both "open" conformations in which P1 and P3 are apart (Figure 6c–e) and "closed" conformations in which P1 and P3 exhibit a near-bound conformation (Figure 6b)43. These data support the idea that riboswitches in solution sample bound-like configurations in the absence of their effector. This view is supported by extensive chemical probing of SAM-I RNA as a function of magnesium, SAM, and temperature43 using SHAPE chemistry61,62.
Figure 6.
Structure of the free SAM-I riboswitch. (a) Superposition of 13 models that best describes experimental SAXS data. Note that while the P4, pseudoknot, P2b, and the kink turn superimpose quite well, the P1 and P3 helices are relatively disordered. Individual models extracted from the ensemble including a bound-like state (b) as well as three distinct "open" states (c–e) in which the P1 and P3 helices are much further apart.
The illusion of large scale ligand-induced folding is given by the observation that the majority of the ensemble adopts "open" conformations and only moderately samples "closed" conformations. Yet, these near-bound conformations are likely the active form of the aptamer. When unbound SAM-I crystals were soaked with SAM, the resulting structure clearly revealed electron density corresponding to the ligand, indicating that the closed form, with its buried pocket, is a binding active state43. Since the global closed conformation of SAM-I is enforced by the crystal lattice, SAM can only access its binding pocket through local conformational flexibility in the RNA. Therefore, we currently favor a model in which ligands do not promote large scale conformational changes in the RNA, but rather associate by conformational selection of near-bound states. Similar conclusions were reached in studies of the guanine riboswitch63,64. Further in support of this view, NMR studies of the HIV transactivation region (TAR) binding small molecules65,66 and an artificial neomycin riboswitch67 have also shown that near-bound conformers of the RNA are the binding-productive states. The "conformational selection" model of binding is currently emerging as a important paradigm for understanding molecular recognition by proteins as well68.
The SAM-II riboswitch
The SAM-II riboswitch is primarily found in α-proteobacteria that lack the SAM-I element69--the two are never found in the same genome24. This regulatory element is distinctly different from the SAM superfamily in that it is a much smaller motif centered around a classic "H-type" pseudoknot (Figure 7a)69. Within the pseudoknot are a number of nearly invariant residues in loop regions between the two principal helices. Despite being half the size of the SAM-I motif, the SAM-II aptamer binds SAM with nearly the same affinity and level of discrimination against SAH69.
Figure 7.
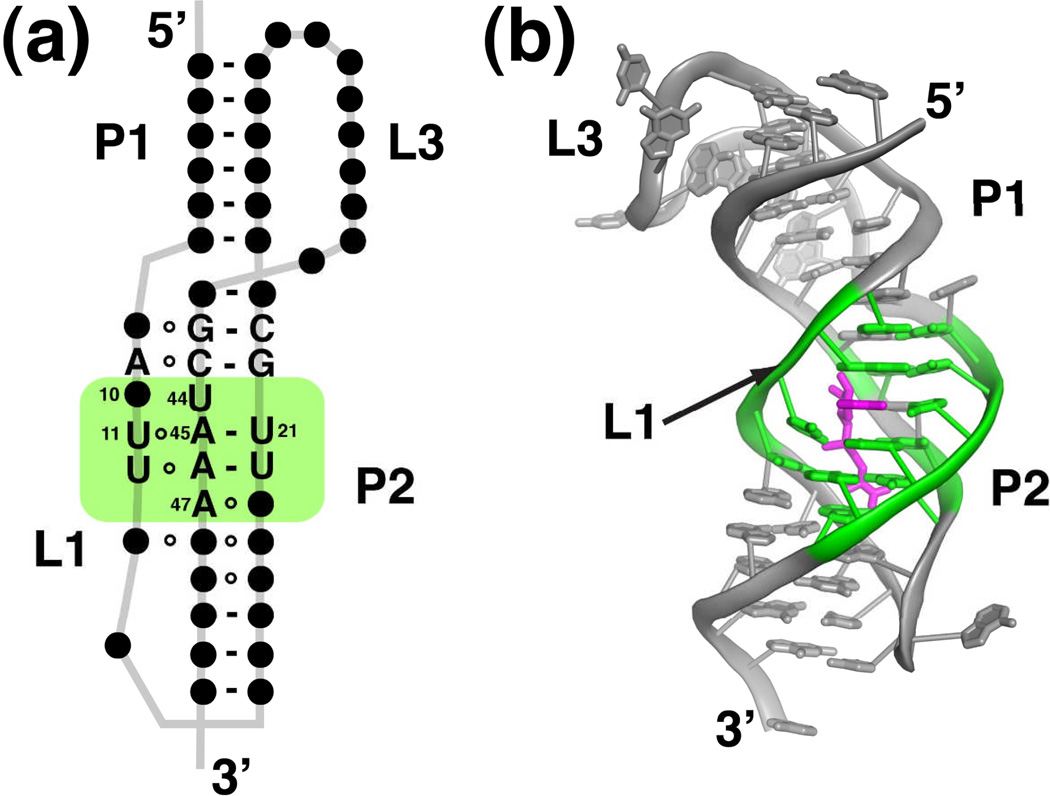
Structure of the SAM-II riboswitch. (a) Phylogenetically conserved primary and secondary structure of the class II SAM riboswitch. Nucleotides that are >97% conserved are specified by their identity; conserved secondary structure is shown as dots. The green box represents the elements of structure that are directly in contact with SAM. The numbering of key nucleotides is consistent with that in Gilbert et al.27 (b) Cartoon representation of the X-ray crystal structure of the SAM-II riboswitch bound to SAM. This panel and other molecular representations of the SAM-II riboswitch were made using the coordinates in PDB accession number 2QWY. Nucleotides highlighted in green comprise the SAM binding pocket, and SAM is shown as magenta sticks.
The crystal structure of a SAM-II riboswitch that regulates the homoserine acetyltransferase (metX) gene of a sequence within the Sargasso Sea metagenome revealed a distinctly different means of binding SAM27. As predicted, the RNA adopts a pseudoknot structure in which two helices (P1 and P2) are coaxially stacked upon one another. The central region of the P2 helix was not predicted from covariation analysis of a sequence alignment69 because most of the nucleotides in this region are invariant27. The helices form a number of triple base pairing interactions with two loop strands, L1 and L3 that cross the major groove of P2 and the minor groove of P1, respectively. This overall fold is not unusual, as it was previously observed in other RNAs such as the human telomerase pseudoknot70, several in vitro selected aptamers71,72, and more recently, the preQ1 riboswitch49–51.
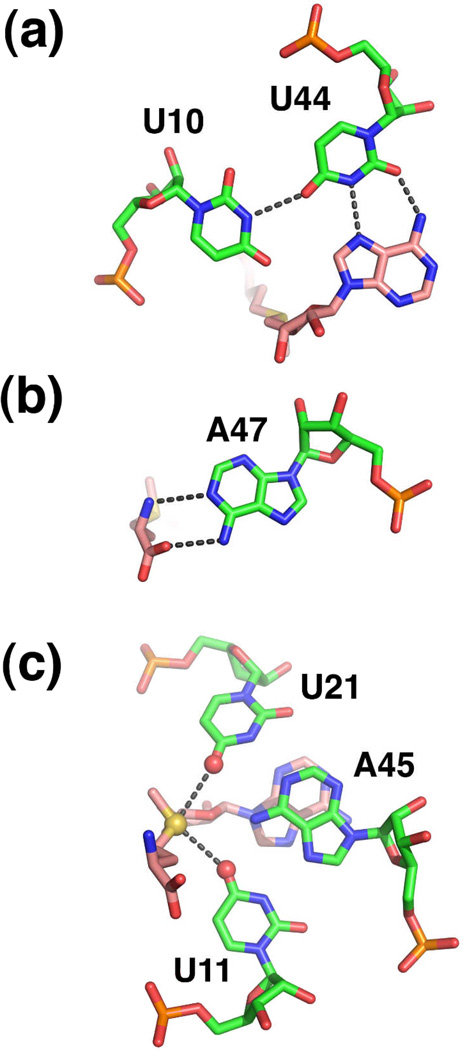
The SAM-binding pocket is embedded within the nearly perfect triplex formed by L1 and P227. Several facets of SAM recognition differ between SAM-I and -II; in SAM-II the ligand is positioned in the major groove and is in an extended (trans) configuration (Figure 7b). Despite these differences, at the functional group level there are similarities between the two complexes. Like SAM-I, the adenine moiety participates in the formation of a base triple (Figure 8a). This interaction likely acts as the keystone that stabilizes both P2 and the major groove triplex. The sulfonium ion of SAM is positioned adjacent to two carbonyl oxygens supplied by two uridines of the adjacent Hoogsteen U11•A45-U21 triple (Figure 8b). This electrostatic interaction is likely to be the primary means of discrimination between SAM and SAH. Finally the main chain atoms of the methionine moiety participate in hydrogen bonding interactions with the Watson-Crick face of a highly conserved adenosine residue (A47) (Figure 8c). In the particular riboswitch used for X-ray crystallography, the Shine-Dalgarno sequence is embedded within the SAM-binding pocket as part of the 3' strand of P227. Presumably, this riboswitch controls translation by preventing the ribosome access to the mRNA upon SAM binding.
Figure 8.
Recognition of SAM by the SAM-II riboswitch. (a) Interaction of the adenine moiety as part of a base triple in the center of the P2 helix. (b) Recognition of the methionine main chain atoms by the Watson-Crick face of a conserved adenine residue. (c) Electrostatic interaction between the sulfonium ion (yellow sphere) with carbonyl oxygens (red spheres) of a Hoogsteen base triple in P2.
Like the SAM-I riboswitch, SAM-II appears to have multiple, bioinformatically distinct families23. Applying a new computational approach to finding riboswitches in intergenic regions, the Breaker group discovered a number of novel RNA motifs in the genome of the marine α-proteobacterium Cadidatus Pelagibacter ubique73. One of these was associated with methionine biosynthesis genes and found to bind SAM with ~150 µM affinity23. While its affinity is substantially lower than other SAM-binding riboswitches, this RNA still retains selectivity over SAM analogs23. Like SAM-II, this "SAM-V" motif can be folded into an H-type pseudoknot with a majority of the conserved nucleotides clustered in P2 and L1. The positioning and identity of these nucleotides suggests a SAM-binding pocket that is similar to SAM-II, but this remains to be confirmed through a structural analysis23. Thus, SAM-II can be considered a superfamily consisting of the SAM-II and SAM-V families.
The SAM-III (SAMMK) riboswitch
A third distinct family of SAM-binding riboswitches is found in lactic acid bacteria (Enterococcus, Streptococcus and Lactococcus species)74. The consensus sequence for this motif clusters around a three-way junction and an adjacent bulge motif (Figure 9a), indicating that it uses a different means of recognizing SAM than the SAM or SAM-II superfamilies. This riboswitch, SAM-III/SAMMK, regulates translation of mRNAs encoding SAM synthetase (metK) by incorporating the Shine-Dalgarno sequence into the aptamer domain75. In the presence of SAM, the riboswitch undergoes a structural rearrangement that promotes pairing of an anti-Shine-Dalgarno sequence with the Shine-Dalgarno, thereby blocking access of the 30S subunit to the mRNA.
Figure 9.
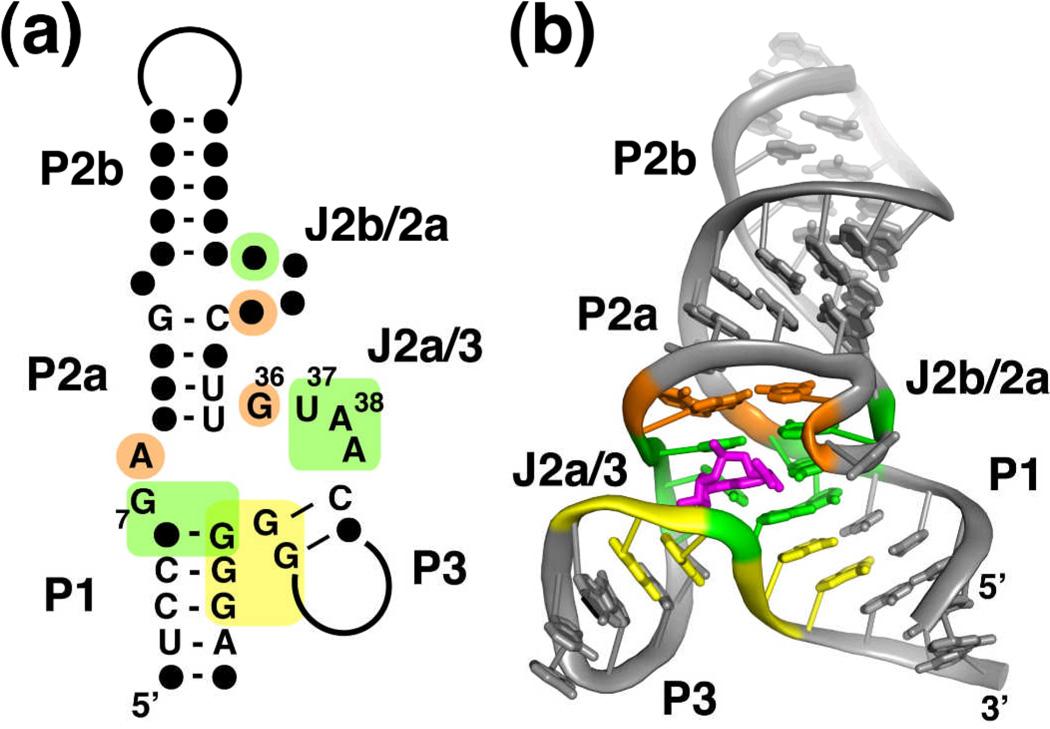
Structure of the SAM-III riboswitch. (a) Phylogenetically conserved primary and secondary structure of the class III SAM riboswitch. Nucleotides that are >97% conserved are specified by their identity; conserved secondary structure is shown as dots. The colored boxes represent the nucleotides in direct contact with SAM (green), a tertiary interaction (orange box), and the Shine-Dalgarno sequence (yellow). The numbering of key nucleotides is consistent with Lu et al28. (b) Cartoon representation of the X-ray crystal structure of the SAM-III riboswitch bound to SAM. This panel and other molecular representations of the SAM-III riboswitch were made using the coordinates in PDB accession number 3E5C. The coloring scheme is the same as in part (a), and SAM is shown as magenta sticks.
The three-dimensional structure of the SAM-III riboswitch in complex with SAM revealed that the ligand is specifically recognized by a set of conserved nucleotides within a three-way junction28. Globally, the RNA folds such that two helices (P1 and P2a/b) coaxially stack upon one another with the third (P3) nearly perpendicular to the others (Figure 9b). This helical organization is typical of "Family A" three-way junctions, which are defined as junctions whose length of J3/1 is greater than that of J2/376. The structure of the junction is supported by interactions between invariant nucleotides in J2a/2b that pair with nucleotides in J2/328. Together, ten invariant nucleotides in the core of the junction create the binding pocket for SAM28,74.
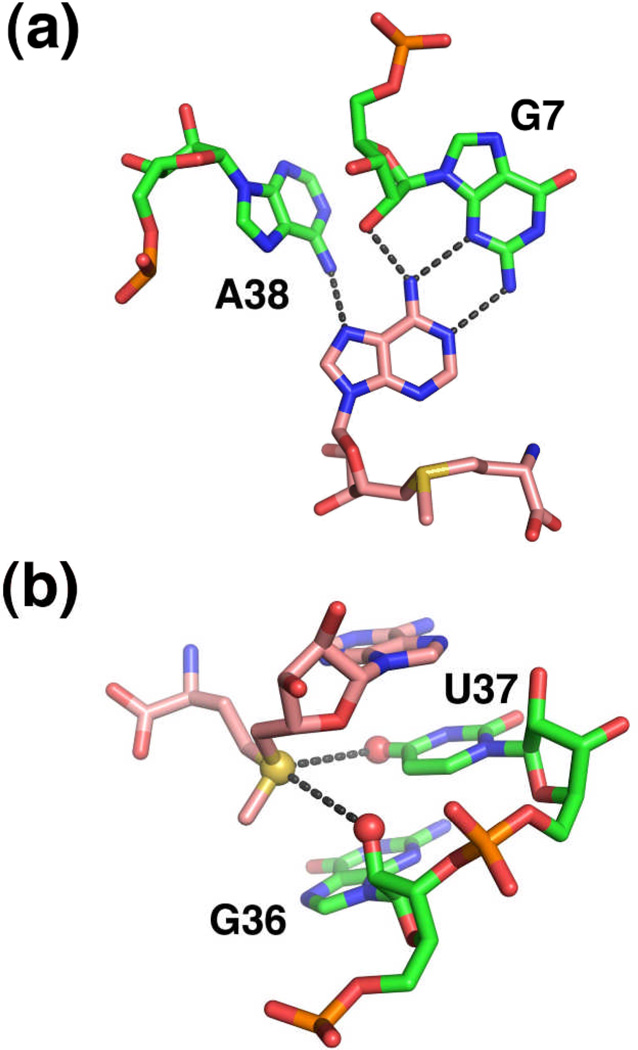
The ligand is recognized through a pattern of local interactions that bear a resemblance to those observed in SAM-I and SAM-II28. The adenosyl moiety is recognized as part of a base triple through formation of a sheared G•A pair with G7 and a direct hydrogen bond with A38 (Figure 10a). To achieve this pairing arrangement, the adenosyl moiety of SAM adopts an unfavorable syn conformation. The ribose sugar of SAM directly interacts with the RNA through hydrogen bonding between the 2'-hydroxyl and N7 of G47 and its 3'-hydroxyl with a non-bridging phosphate oxygen of G4728. Strikingly, the methionine moiety is not recognized by the RNA, as indicated by both a lack of electron density beyond the sulfonium moiety and binding assays using SAM analogs that lack this group28. This is in distinct contrast to the previously described riboswitches.
Figure 10.
Recognition of SAM by the SAM-III riboswitch. (a) Participation of the adenine residue of SAM in a base triple within the center of the three-way junction. Note that the nucleotide numbering in PDB 3E5C is inconsistent with the numbering used in the figures found in Lu et al.28 The numbering in the text and figures is consistent with PDB ID 3E5C. (b) Recognition of the sulfonium ion (yellow sphere) by a carbonyl oxygen (U37) and a 2'-hydroxyl group (G36) (red spheres). Note that this riboswitch does not appear to specifically interact with the methionine moiety.
Recognition of the positively-charged sulfonium ion is achieved through electrostatic interactions with the adjacent O4 carbonyl of U37 and the 2'-hydroxyl group of G36 (Figure 10b)28. Like other SAM riboswitches, this placement of the positively charged sulfonium ion yields a discrimination mechanism between SAM and SAH based upon formation of favorable electrostatics. Backsoaking of the SAM-RNA crystals with saturating concentrations of SAH resulted in a structure that shows that while the adenosyl moiety occupies an identical position, the glycosidic bond rotates such that SAH adopts the more favorable anti configuration. This places the neutral sulfoether group in a distinctly different position than the sulfonium ion. Thus, both the ribose sugar and sulfur make an alternative set of interactions with the RNA that are presumably significantly weaker than those observed in the SAM-RNA complex, which augments discrimination between these two compounds28.
The SAH and SAM/SAH riboswitches
As illustrated above, there are multiple solutions to the problem of recognizing SAM with sufficient affinity to drive a regulatory response and to discriminate against its product form, SAH. The opposite is also a biologically important problem, as SAH levels in the cell need to be strictly regulated since it acts as a potent competitive inhibitor of many SAM-utilizing enzymes and at high levels is toxic77. The above structures do not provide insight into how RNA can achieve selectivity towards SAH or non-selectivity. In the case of SAM-I, mutations within the sulfonium ion binding site in P1 lead to a significant loss in selectivity, but not completely38. Fortunately, recent discoveries of new classes of riboswitches will lead to further insights into mechanisms of selectivity.
Upstream of the SAM recycling enzymes SAH hydrolase (ahcY) (Figure 1b) and 5-methyl-THF:L-homocysteine methyltransferase (metH) in a number of proteobacteria, the Breaker group identified an SAH-binding riboswitch21. The observed consensus sequence comprises three or four helices (one helix is present in only 67% of sequences) organized into an unusual "LL" type pseudoknot motif (as defined by Han and Byun78). Like other riboswitches, conserved nucleotides cluster in the joining regions between helical elements. Using standard approaches such as in-line probing and equilibrium dialysis, it was shown that several representatives of this motif bind SAH with high affinity (~20 nM) and selectivity over SAM (>1000-fold)21. Furthermore, analysis of binding of a variety of SAM and SAH analogs reveals that the RNA effectively recognizes almost every feature of the ligand.
A recent crystal structure of this RNA complexed with SAH reveals that the RNA achieves selectivity using a steric mechanism rather than through electrostatics79. Modeling of SAM in this site indicates that its methyl group would encounter severe steric hindrance with several nucleotides that define the binding pocket. This is a common means of discrimination in biology, and it is not surprising that the SAH riboswitch employs it. For example, in the theophylline aptamer, selectivity between theophylline and caffeine is the result of a single methyl group on the N7 position of caffeine that prevents it from forming a productive interaction80,81. More recently, a survey that identified 104 new candidate structured RNAs in bacteria, archaea, and various metagenomes revealed a new motif that cannot distinguish between SAM and SAH20. This RNA, observed exclusively in Rhodobacterales is generally found upstream of SAM synthetase (metK), contains a consensus sequence that can be folded into an H-type pseudoknot like SAM-II. In this case, however, in-line probing and equilibrium dialysis revealed that this riboswitch aptamer, called SAM/SAH, binds SAM and SAH with equal affinity (~10 µM)20. An explanation is that, like the SAM-III riboswitch, the RNA ignores a significant amount of the ligand--in this case, the full methionine moiety. While methionine is not a competitor of SAM binding, adenosine was not tested20, leaving this a possibility. As a testament to how easy it is to generate adenine-binding motifs, an attempt to select aptamers that bind SAM yielded RNAs that recognized only the adenosyl moiety using a motif similar to that of the ATP aptamer82.
Conclusions
Since their discovery, one of the most active areas of research into riboswitches has been the determination of structures in complex with effector. The aptamers that recognize SAM and SAH are particularly interesting case studies in understanding the mechanisms of small molecule recognition by RNA because of the diverse motifs identified that recognize these compounds. Within the SAM family, the peripheral architecture supporting a common SAM binding site diverges substantially. This is similar to what is observed in other biological RNAs, such as group I splicing introns which contain a common catalytic core surrounded by varying peripheral elements that facilitate folding and/or catalysis83. Despite clear differences in the global architecture of different classes of SAM riboswitches, at the functional group level how they recognize the principal moieties in SAM is very similar. This is exemplified by recognition of the sulfonium cation by electrostatic interactions with carbonyls in all three RNAs, which is also found in SAM-binding proteins38.
These structures point to a common mechanism of communication between the aptamer and the expression platform. In each case, the effector binding site incorporates a sequence element shared with the downstream expression platform. Ligand binding serves to stably partition this sequence into the aptamer, thereby dictating the downstream secondary structure that determines the expression fate of the mRNA. SAM-II and -III directly achieve this by sequestering the Shine-Dalgarno sequence within the SAM binding pocket. SAM-I achieves the same end through stable incorporation of the 3'-side of the P1 helix (Figure 2) that enforces formation of a rho-independent transcriptional terminator.
Several aspects of SAM-dependent regulation remain unresolved. A recent study by Henkin and coworkers demonstrated that the eleven different SAM-I riboswitches in B. subtilis have different regulatory characteristics in vivo, despite all bearing highly conserved aptamer domains84. Importantly, it was observed that the various riboswitches are "tuned" to meet specific regulatory needs of the genes that they control. SAM-I riboswitches controlling methionine transport appeared to be less stringent in their response then their counterparts that control biosynthetic processes84. The molecular basis for genetic tuning remains unknown, but is likely to be an important aspect of riboswitch function, as it has been observed for purine riboswitches as well85.
Henkin and coworkers also point out that the affinity of the riboswitch for SAM does not necessarily translate into the concentration of effector needed for regulatory control. These data, along with a study of the FMN riboswitch86, reveals that there is a complex relationship between transcription, RNA folding, effector binding affinity and rates, and structural switching that has yet to be fully understood for any riboswitch. To fully define their mechanisms of regulatory function, riboswitches must be studied in the context of transcription, and in the case of translational regulators, cotranslation. The structures described in this review represent only a single snapshot in the life of a riboswitch--that is, events that occur when RNA polymerase has just finished transcribing the aptamer. What happens beyond in the time domain is poorly understood and where the future challenges lay.
Acknowledgements
I would like to thank all of the members of my laboratory who have performed the research on SAM-binding riboswitches that serves as the backbone of this review. This work has been supported by a grant to the author from the National Institutes of Health (GM083895).
References
- 1.Cantoni GL. Biological methylation: selected aspects. Annu Rev Biochem. 1975;44:435–451. doi: 10.1146/annurev.bi.44.070175.002251. [DOI] [PubMed] [Google Scholar]
- 2.Fontecave M, Atta M, Mulliez E. S-adenosylmethionine: nothing goes to waste. Trends Biochem Sci. 2004;29:243–249. doi: 10.1016/j.tibs.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 3.Wang SC, Frey PA. S-adenosylmethionine as an oxidant: the radical SAM superfamily. Trends Biochem Sci. 2007;32:101–110. doi: 10.1016/j.tibs.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 4.Sekowska A, Kung HF, Danchin A. Sulfur metabolism in Escherichia coli and related bacteria: facts and fiction. J Mol Microbiol Biotechnol. 2000;2:145–177. [PubMed] [Google Scholar]
- 5.Weissbach H, Brot N. Regulation of methionine synthesis in Escherichia coli. Mol Microbiol. 1991;5:1593–1597. doi: 10.1111/j.1365-2958.1991.tb01905.x. [DOI] [PubMed] [Google Scholar]
- 6.Grundy FJ, Henkin TM. The S box regulon: a new global transcription termination control system for methionine and cysteine biosynthesis genes in gram-positive bacteria. Mol Microbiol. 1998;30:737–749. doi: 10.1046/j.1365-2958.1998.01105.x. [DOI] [PubMed] [Google Scholar]
- 7.Gelfand MS, Mironov AA, Jomantas J, Kozlov YI, Perumov DA. A conserved RNA structure element involved in the regulation of bacterial riboflavin synthesis genes. Trends Genet. 1999;15:439–442. doi: 10.1016/s0168-9525(99)01856-9. [DOI] [PubMed] [Google Scholar]
- 8.Miranda-Rios J, Navarro M, Soberon M. A conserved RNA structure (thi box) is involved in regulation of thiamin biosynthetic gene expression in bacteria. Proc Natl Acad Sci U S A. 2001;98:9736–9741. doi: 10.1073/pnas.161168098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nahvi A, Sudarsan N, Ebert MS, Zou X, Brown KL, Breaker RR. Genetic control by a metabolite binding mRNA. Chem Biol. 2002;9:1043. doi: 10.1016/s1074-5521(02)00224-7. [DOI] [PubMed] [Google Scholar]
- 10.Epshtein V, Mironov AS, Nudler E. The riboswitch-mediated control of sulfur metabolism in bacteria. Proc Natl Acad Sci U S A. 2003;100:5052–5056. doi: 10.1073/pnas.0531307100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McDaniel BA, Grundy FJ, Artsimovitch I, Henkin TM. Transcription termination control of the S box system: direct measurement of S-adenosylmethionine by the leader RNA. Proc Natl Acad Sci U S A. 2003;100:3083–3088. doi: 10.1073/pnas.0630422100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Winkler WC, Nahvi A, Sudarsan N, Barrick JE, Breaker RR. An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nat Struct Biol. 2003;10:701–707. doi: 10.1038/nsb967. [DOI] [PubMed] [Google Scholar]
- 13.Roth A, Breaker RR. The structural and functional diversity of metabolite-binding riboswitches. Annu Rev Biochem. 2009;78:305–334. doi: 10.1146/annurev.biochem.78.070507.135656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Winkler WC, Breaker RR. Genetic control by metabolite-binding riboswitches. Chembiochem. 2003;4:1024–1032. doi: 10.1002/cbic.200300685. [DOI] [PubMed] [Google Scholar]
- 15.Montange RK, Batey RT. Riboswitches: emerging themes in RNA structure and function. Annu Rev Biophys. 2008;37:117–133. doi: 10.1146/annurev.biophys.37.032807.130000. [DOI] [PubMed] [Google Scholar]
- 16.Epshtein V, Cardinale CJ, Ruckenstein AE, Borukhov S, Nudler E. An allosteric path to transcription termination. Mol Cell. 2007;28:991–1001. doi: 10.1016/j.molcel.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 17.Gusarov I, Nudler E. The mechanism of intrinsic transcription termination. Mol Cell. 1999;3:495–504. doi: 10.1016/s1097-2765(00)80477-3. [DOI] [PubMed] [Google Scholar]
- 18.Andre G, Even S, Putzer H, Burguiere P, Croux C, Danchin A, Martin-Verstraete I, Soutourina O. S-box and T-box riboswitches and antisense RNA control a sulfur metabolic operon of Clostridium acetobutylicum. Nucleic Acids Res. 2008;36:5955–5969. doi: 10.1093/nar/gkn601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loh E, Dussurget O, Gripenland J, Vaitkevicius K, Tiensuu T, Mandin P, Repoila F, Buchrieser C, Cossart P, Johansson J. A trans-acting riboswitch controls expression of the virulence regulator PrfA in Listeria monocytogenes. Cell. 2009;139:770–779. doi: 10.1016/j.cell.2009.08.046. [DOI] [PubMed] [Google Scholar]
- 20.Weinberg Z, Wang JX, Bogue J, Yang J, Corbino K, Moy RH, Breaker RR. Comparative genomics reveals 104 candidate structured RNAs from bacteria, archaea, and their metagenomes. Genome Biol. 2010;11:R31. doi: 10.1186/gb-2010-11-3-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang JX, Breaker RR. Riboswitches that sense S-adenosylmethionine and S-adenosylhomocysteine. Biochem Cell Biol. 2008;86:157–168. doi: 10.1139/O08-008. [DOI] [PubMed] [Google Scholar]
- 22.Weinberg Z, Regulski EE, Hammond MC, Barrick JE, Yao Z, Ruzzo WL, Breaker RR. The aptamer core of SAM-IV riboswitches mimics the ligand-binding site of SAM-I riboswitches. RNA. 2008;14:822–828. doi: 10.1261/rna.988608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poiata E, Meyer MM, Ames TD, Breaker RR. A variant riboswitch aptamer class for S-adenosylmethionine common in marine bacteria. RNA. 2009;15:2046–2056. doi: 10.1261/rna.1824209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barrick JE, Breaker RR. The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome Biol. 2007;8:R239. doi: 10.1186/gb-2007-8-11-r239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang JX, Lee ER, Morales DR, Lim J, Breaker RR. Riboswitches that sense S-adenosylhomocysteine and activate genes involved in coenzyme recycling. Mol Cell. 2008;29:691–702. doi: 10.1016/j.molcel.2008.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Montange RK, Batey RT. Structure of the S-adenosylmethionine riboswitch regulatory mRNA element. Nature. 2006;441:1172–1175. doi: 10.1038/nature04819. [DOI] [PubMed] [Google Scholar]
- 27.Gilbert SD, Rambo RP, Van Tyne D, Batey RT. Structure of the SAM-II riboswitch bound to S-adenosylmethionine. Nat Struct Mol Biol. 2008;15:177–182. doi: 10.1038/nsmb.1371. [DOI] [PubMed] [Google Scholar]
- 28.Lu C, Smith AM, Fuchs RT, Ding F, Rajashankar K, Henkin TM, Ke A. Crystal structures of the SAM-III/S(MK) riboswitch reveal the SAM-dependent translation inhibition mechanism. Nat Struct Mol Biol. 2008;15:1076–1083. doi: 10.1038/nsmb.1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Griffiths-Jones S, Moxon S, Marshall M, Khanna A, Eddy SR, Bateman A. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 2005;33:D121–D124. doi: 10.1093/nar/gki081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tiedge H. K-turn motifs in spatial RNA coding. RNA Biol. 2006;3:133–139. doi: 10.4161/rna.3.4.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klein DJ, Schmeing TM, Moore PB, Steitz TA. The kink-turn: a new RNA secondary structure motif. EMBO J. 2001;20:4214–4221. doi: 10.1093/emboj/20.15.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winkler WC, Grundy FJ, Murphy BA, Henkin TM. The GA motif: an RNA element common to bacterial antitermination systems, rRNA, and eukaryotic RNAs. RNA. 2001;7:1165–1172. doi: 10.1017/s1355838201002370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heppell B, Lafontaine DA. Folding of the SAM aptamer is determined by the formation of a K-turn-dependent pseudoknot. Biochemistry. 2008;47:1490–1499. doi: 10.1021/bi701164y. [DOI] [PubMed] [Google Scholar]
- 34.Staple DW, Butcher SE. Pseudoknots: RNA structures with diverse functions. PLoS Biol. 2005;3:e213. doi: 10.1371/journal.pbio.0030213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McDaniel BA, Grundy FJ, Kurlekar VP, Tomsic J, Henkin TM. Identification of a mutation in the Bacillus subtilis S-adenosylmethionine synthetase gene that results in derepression of S-box gene expression. J Bacteriol. 2006;188:3674–3681. doi: 10.1128/JB.188.10.3674-3681.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Griffiths-Jones S, Bateman A, Marshall M, Khanna A, Eddy SR. Rfam: an RNA family database. Nucleic Acids Res. 2003;31:439–441. doi: 10.1093/nar/gkg006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gilbert SD, Montange RK, Stoddard CD, Batey RT. Structural studies of the purine and SAM binding riboswitches. Cold Spring Harb Symp Quant Biol. 2006;71:259–268. doi: 10.1101/sqb.2006.71.015. [DOI] [PubMed] [Google Scholar]
- 38.Montange RK, Mondragon E, van Tyne D, Garst AD, Ceres P, Batey RT. Discrimination between closely related cellular metabolites by the SAM-I riboswitch. J Mol Biol. 2010;396:761–772. doi: 10.1016/j.jmb.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holbrook SR. Structural principles from large RNAs. Annu Rev Biophys. 2008;37:445–464. doi: 10.1146/annurev.biophys.36.040306.132755. [DOI] [PubMed] [Google Scholar]
- 40.McDaniel BA, Grundy FJ, Henkin TM. A tertiary structural element in S box leader RNAs is required for S-adenosylmethionine-directed transcription termination. Mol Microbiol. 2005;57:1008–1021. doi: 10.1111/j.1365-2958.2005.04740.x. [DOI] [PubMed] [Google Scholar]
- 41.Doherty EA, Batey RT, Masquida B, Doudna JA. A universal mode of helix packing in RNA. Nat Struct Biol. 2001;8:339–343. doi: 10.1038/86221. [DOI] [PubMed] [Google Scholar]
- 42.Nissen P, Ippolito JA, Ban N, Moore PB, Steitz TA. RNA tertiary interactions in the large ribosomal subunit: the A-minor motif. Proc Natl Acad Sci U S A. 2001;98:4899–4903. doi: 10.1073/pnas.081082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stoddard CD, Montange RK, Hennelly SP, Rambo RP, Sanbonmatsu KY, Batey RT. Free state conformational sampling of the SAM-I riboswitch aptamer domain. Structure. 2010;18:787–797. doi: 10.1016/j.str.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saenger W. Principles of Nucleic Acid Structure. New York: Springer-Verlag; 1984. [Google Scholar]
- 45.Lim J, Winkler WC, Nakamura S, Scott V, Breaker RR. Molecular-recognition characteristics of SAM-binding riboswitches. Angew Chem Int Ed Engl. 2006;45:964–968. doi: 10.1002/anie.200503198. [DOI] [PubMed] [Google Scholar]
- 46.Breaker RR. Riboswitches and the RNA World. In: Gesteland RF, Cech TR, Atkins JF, editors. The RNA World. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2006. pp. 89–107. [Google Scholar]
- 47.Garst AD, Heroux A, Rambo RP, Batey RT. Crystal structure of the lysine riboswitch regulatory mRNA element. J Biol Chem. 2008;283:22347–22351. doi: 10.1074/jbc.C800120200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Serganov A, Huang L, Patel DJ. Structural insights into amino acid binding and gene control by a lysine riboswitch. Nature. 2008;455:1263–1267. doi: 10.1038/nature07326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kang M, Peterson R, Feigon J. Structural Insights into riboswitch control of the biosynthesis of queuosine, a modified nucleotide found in the anticodon of tRNA. Mol Cell. 2009;33:784–790. doi: 10.1016/j.molcel.2009.02.019. [DOI] [PubMed] [Google Scholar]
- 50.Klein DJ, Edwards TE, Ferre-D'Amare AR. Cocrystal structure of a class I preQ1 riboswitch reveals a pseudoknot recognizing an essential hypermodified nucleobase. Nat Struct Mol Biol. 2009;16:343–344. doi: 10.1038/nsmb.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Spitale RC, Torelli AT, Krucinska J, Bandarian V, Wedekind JE. The structural basis for recognition of the PreQ0 metabolite by an unusually small riboswitch aptamer domain. J Biol Chem. 2009;284:11012–11016. doi: 10.1074/jbc.C900024200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Serganov A, Polonskaia A, Phan AT, Breaker RR, Patel DJ. Structural basis for gene regulation by a thiamine pyrophosphate-sensing riboswitch. Nature. 2006;441:1167–1171. doi: 10.1038/nature04740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thore S, Leibundgut M, Ban N. Structure of the eukaryotic thiamine pyrophosphate riboswitch with its regulatory ligand. Science. 2006;312:1208–1211. doi: 10.1126/science.1128451. [DOI] [PubMed] [Google Scholar]
- 54.Ali M, Lipfert J, Seifert S, Herschlag D, Doniach S. The ligand-free state of the TPP riboswitch: a partially folded RNA structure. J Mol Biol. 2010;396:153–165. doi: 10.1016/j.jmb.2009.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Batey RT, Gilbert SD, Montange RK. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature. 2004;432:411–415. doi: 10.1038/nature03037. [DOI] [PubMed] [Google Scholar]
- 56.Lipfert J, Doniach S. Small-angle X-ray scattering from RNA, proteins, and protein complexes. Annu Rev Biophys Biomol Struct. 2007;36:307–327. doi: 10.1146/annurev.biophys.36.040306.132655. [DOI] [PubMed] [Google Scholar]
- 57.Rambo RP, Tainer JA. Bridging the solution divide: comprehensive structural analyses of dynamic RNA, DNA, and protein assemblies by small-angle X-ray scattering. Curr Opin Struct Biol. 2010;20:128–137. doi: 10.1016/j.sbi.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rambo RP, Tainer JA. Improving small-angle X-ray scattering data for structural analyses of the RNA world. RNA. 2010;16:638–646. doi: 10.1261/rna.1946310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bernado P, Mylonas E, Petoukhov MV, Blackledge M, Svergun DI. Structural characterization of flexible proteins using small-angle X-ray scattering. J Am Chem Soc. 2007;129:5656–5664. doi: 10.1021/ja069124n. [DOI] [PubMed] [Google Scholar]
- 60.Putnam CD, Hammel M, Hura GL, Tainer JA. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q Rev Biophys. 2007;40:191–285. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- 61.Wilkinson KA, Merino EJ, Weeks KM. Selective 2'-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat Protoc. 2006;1:1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- 62.Wilkinson KA, Merino EJ, Weeks KM. RNA SHAPE chemistry reveals nonhierarchical interactions dominate equilibrium structural transitions in tRNA(Asp) transcripts. J Am Chem Soc. 2005;127:4659–4667. doi: 10.1021/ja0436749. [DOI] [PubMed] [Google Scholar]
- 63.Ottink OM, Rampersad SM, Tessari M, Zaman GJ, Heus HA, Wijmenga SS. Ligand-induced folding of the guanine-sensing riboswitch is controlled by a combined predetermined induced fit mechanism. RNA. 2007;13:2202–2212. doi: 10.1261/rna.635307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stoddard CD, Gilbert SD, Batey RT. Ligand-dependent folding of the three-way junction in the purine riboswitch. RNA. 2008;14:675–684. doi: 10.1261/rna.736908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stelzer AC, Kratz JD, Zhang Q, Al-Hashimi HM. RNA Dynamics by Design: Biasing Ensembles Towards the Ligand-Bound State. Angew Chem Int Ed Engl. 2010;49:5731–5733. doi: 10.1002/anie.201000814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang Q, Stelzer AC, Fisher CK, Al-Hashimi HM. Visualizing spatially correlated dynamics that directs RNA conformational transitions. Nature. 2007;450:1263–1267. doi: 10.1038/nature06389. [DOI] [PubMed] [Google Scholar]
- 67.Duchardt-Ferner E, Weigand JE, Ohlenschlager O, Schmidtke SR, Suess B, Wohnert J. Highly Modular Structure and Ligand Binding by Conformational Capture in a Minimalistic Riboswitch. Angew Chem Int Ed Engl. 2010;49:6216–6219. doi: 10.1002/anie.201001339. [DOI] [PubMed] [Google Scholar]
- 68.Boehr DD, Nussinov R, Wright PE. The role of dynamic conformational ensembles in biomolecular recognition. Nat Chem Biol. 2009;5:789–796. doi: 10.1038/nchembio.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Corbino KA, Barrick JE, Lim J, Welz R, Tucker BJ, Puskarz I, Mandal M, Rudnick ND, Breaker RR. Evidence for a second class of S-adenosylmethionine riboswitches and other regulatory RNA motifs in alpha-proteobacteria. Genome Biol. 2005;6:R70. doi: 10.1186/gb-2005-6-8-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Theimer CA, Blois CA, Feigon J. Structure of the human telomerase RNA pseudoknot reveals conserved tertiary interactions essential for function. Mol Cell. 2005;17:671–682. doi: 10.1016/j.molcel.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 71.Nix J, Sussman D, Wilson C. The 1.3 A crystal structure of a biotin-binding pseudoknot and the basis for RNA molecular recognition. J Mol Biol. 2000;296:1235–1244. doi: 10.1006/jmbi.2000.3539. [DOI] [PubMed] [Google Scholar]
- 72.Sussman D, Nix JC, Wilson C. The structural basis for molecular recognition by the vitamin B 12 RNA aptamer. Nat Struct Biol. 2000;7:53–57. doi: 10.1038/71253. [DOI] [PubMed] [Google Scholar]
- 73.Meyer MM, Ames TD, Smith DP, Weinberg Z, Schwalbach MS, Giovannoni SJ, Breaker RR. Identification of candidate structured RNAs in the marine organism 'Candidatus Pelagibacter ubique'. BMC Genomics. 2009;10:268. doi: 10.1186/1471-2164-10-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fuchs RT, Grundy FJ, Henkin TM. The S(MK) box is a new SAM-binding RNA for translational regulation of SAM synthetase. Nat Struct Mol Biol. 2006;13:226–233. doi: 10.1038/nsmb1059. [DOI] [PubMed] [Google Scholar]
- 75.Fuchs RT, Grundy FJ, Henkin TM. S-adenosylmethionine directly inhibits binding of 30S ribosomal subunits to the SMK box translational riboswitch RNA. Proc Natl Acad Sci U S A. 2007;104:4876–4880. doi: 10.1073/pnas.0609956104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lescoute A, Westhof E. Topology of three-way junctions in folded RNAs. RNA. 2006;12:83–93. doi: 10.1261/rna.2208106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ueland PM. Pharmacological and biochemical aspects of S-adenosylhomocysteine and S-adenosylhomocysteine hydrolase. Pharmacol Rev. 1982;34:223–253. [PubMed] [Google Scholar]
- 78.Han K, Byun Y. PSEUDOVIEWER2: Visualization of RNA pseudoknots of any type. Nucleic Acids Res. 2003;31:3432–3440. doi: 10.1093/nar/gkg539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Edwards AL, Reyes FE, Heroux A, Batey RT. Structural basis for recognition of S-adenosylhomocysteine by riboswitches. RNA. 2010;16:2144–2155. doi: 10.1261/rna.2341610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zimmermann GR, Jenison RD, Wick CL, Simorre JP, Pardi A. Interlocking structural motifs mediate molecular discrimination by a theophylline-binding RNA. Nat Struct Biol. 1997;4:644–649. doi: 10.1038/nsb0897-644. [DOI] [PubMed] [Google Scholar]
- 81.Jenison RD, Gill SC, Pardi A, Polisky B. High-resolution molecular discrimination by RNA. Science. 1994;263:1425–1429. doi: 10.1126/science.7510417. [DOI] [PubMed] [Google Scholar]
- 82.Burke DH, Gold L. RNA aptamers to the adenosine moiety of S-adenosyl methionine: structural inferences from variations on a theme and the reproducibility of SELEX. Nucleic Acids Res. 1997;25:2020–2024. doi: 10.1093/nar/25.10.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vicens Q, Cech TR. Atomic level architecture of group I introns revealed. Trends Biochem Sci. 2006;31:41–51. doi: 10.1016/j.tibs.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 84.Tomsic J, McDaniel BA, Grundy FJ, Henkin TM. Natural variability in S-adenosylmethionine (SAM)-dependent riboswitches: S-box elements in bacillus subtilis exhibit differential sensitivity to SAM In vivo and in vitro. J Bacteriol. 2008;190:823–833. doi: 10.1128/JB.01034-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mulhbacher J, Lafontaine DA. Ligand recognition determinants of guanine riboswitches. Nucleic Acids Res. 2007;35:5568–5580. doi: 10.1093/nar/gkm572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wickiser JK, Winkler WC, Breaker RR, Crothers DM. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol Cell. 2005;18:49–60. doi: 10.1016/j.molcel.2005.02.032. [DOI] [PubMed] [Google Scholar]