Abstract
Secreted proteins such as growth factors, cytokines and chemokines play important roles in tumor development. Through expression microarray and bioinformatic analysis, we discovered a novel secreted protein, neuroblastoma derived secretory protein (NDSP). The NDSP gene is found on chromosome 1q25.2 and encodes a 167 amino acid protein with a putative signal peptide. Using real-time PCR and immunoblotting we find that NDSP is specifically overexpressed in neuroblastoma at much higher levels than other adult and pediatric malignancies and normal tissues. NDSP is an 18 kDa protein that can be secreted by NDSP-transfected HEK-293T cells, as well as, neuroblastoma cell lines endogenously expressing NDSP. Inhibiting NDSP expression in neuroblastoma cell lines with retrovirally transduced NDSP small hairpin interfering RNA (shRNA), sh-NDSP, results in decreased cellular proliferation and colony formation. We also find inhibited ERK1/2 phosphorylation in the sh-NDSP cell line. Treating the parental cell line with MEK1/2 inhibitors which diminish ERK1/2 phosphorylation results in decreased cell proliferation. Culturing these transduced cells with recombinant NDSP, reintroducing NDSP overexpression in the knockdown cell line, or inducing Ras oncogene overexpression for constitutive ERK1/2 activation results in a reversal of the growth inhibited phenotype and proliferation rates similar to the control cells. In addition, reintroduction of NDSP overexpression in the sh-NDSP cell line results in ERK1/2 phosphorylation similar to control. We conclude that NDSP is specifically overexpressed in neuroblastoma and actively secreted from tumor cells. Furthermore, NDSP serves as a growth factor for neuroblastoma tumor cells through activation of the ERK-mediated proliferation pathway.
Keywords: NDSP, secreted protein, neuroblastoma, extracellular signal regulated kinase
Introduction
Biomarker discovery has become a critical aspect of translational oncology research and has been successful in identifying useful markers for diagnosis and monitoring of various cancers. Additionally, markers unique to tumor cells will also serve as effective targets for therapies. Within the last 30 years, multiple useful clinical markers have been identified such as prostate specific antigen (PSA) for prostate cancer (1, 2), ERBB2 overexpression for breast cancer (3), BCR-ABL fusion gene expression for chronic myelocytic leukemia, and MYCN amplification for neuroblastoma (4).
Neuroblastoma is the most common extracranial solid tumor in children (5). The most valuable predictors of prognosis within neuroblastoma are age, stage, histopathology, DNA ploidy number, and MYCN amplification. The Children's Oncology Group (COG) risk stratification system utilizes all of these factors to assign patients to risk-specific treatment protocols (6). Patients in the highest risk category have a particularly poor outcome; unfortunately this group constitutes approximately half of the children with neuroblastoma. Multiple serum and urine markers have been studied in neuroblastoma including serum ferritin, tyrosine hydroxylase, neuron specific enolase, lactate dehydrogenase (LDH), and urine catecholamines (7). The majority of these markers is nonspecific and can be found in other oncologic and non-oncologic disease states, such as prematurity, sepsis, and stress (8, 9).
As understanding of the molecular cell biology of neuroblastoma increases, more specific markers of disease will be discovered. With knowledge of tumor specific markers, it is possible to design less toxic targeted therapies. Some of the biologic therapies that are currently in pre-clinical and clinical trials for neuroblastoma are small-molecule inhibitors designed to inhibit histone deacetylase (suberoylanilide hydroxamic acid, SAHA, and valproate) and the receptor tyrosine kinases (CEP-701), anti-GD2 monoclonal antibody therapy, 13 Cis-retinoic acid, and fenretinide (10-15). With such a large percentage of the patients having a poor prognosis, it is imperative to find more tumor-specific markers for neuroblastoma that may serve as effective therapeutic targets in conjunction with other molecular therapies already in trials.
In the following report, we describe the cloning and characterization of a novel protein in neuroblastoma that we named neuroblastoma derived secretory protein (NDSP). Through extensive expression microarray and bioinformatics analysis as well as expression validation experiments, we find that NDSP is specifically overexpressed in neuroblastoma compared to low level expression in a number of normal tissues and other adult and pediatric malignancies. In addition, we demonstrate that NDSP is secreted from neuroblastoma cell lines and promotes neuroblastoma cell proliferation through an ERK-mediated pathway.
Materials and Methods
Cell culture
Human embryonic kidney 293T (HEK293T) cells were obtained from Edge Biosystems (Gaithersberg, MD, USA). These cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum and 100 units/ml streptomycin/penicillin (Gibco, Grand Island, NY, USA). SK-N-MC, SK-N-AS, SK-N-SH, SH-SY5Y, IMR-32, LAN-1, SMS-KCN (ATCC, Manassas, VA, USA), NB-16 (neuroblastoma, donated by A. Davidoff) were cultured in modified Eagle's medium or DMEM or MEM supplemented with 10% fetal bovine serum and 100 units/ml streptomycin/penicillin (Gibco). Additionally, NCI-H-1299 (non-small cell lung cancer, ATCC), MD-MB-231 (breast cancer, ATCC), SHEP (neuroblastoma, donated by J. Shohet), NB19, PCL-5134, PCL-3014 (neuroblastoma, donated by A. Davidoff), and JF (neuroblastoma, donated by M. Brenner) were used for RNA extraction only. All cells were incubated at 37°C in a humidified atmosphere of 5% CO2.
Human Tumor Tissues
All research on patient tumor tissue was performed under protocols approved by the Baylor College of Medicine Institutional Review Board. Parent/legal guardian/patient consent was obtained prior to tumor removal. Tumor tissue was categorized according to stage determined by the International Neuroblastoma Staging System (7).
Reagents
Antibodies used for Western blots include: anti-V5 antibody (Invitrogen, Carlsbad, CA), anti-NDSP antibodies (anti-NDSP-Ab1 and -Ab2, generated by Genemed Synthesis Inc., South San Francisco, CA, USA); anti-β-actin, anti-phospho-ERK1/2, anti-ERK1/2 (Cell Signaling, Danvers, MA), mouse monoclonal antibody (Sigma, St. Louis, MO, USA); and horseradish peroxidase (HRP) conjugated, goat anti-rabbit secondary antibody (Jackson ImmunoResearch Laboratories, West Grove, PA, USA). The anti-NDSP antibodies were tested and validated using immunoblotting and immunostaining (Supp. Fig. 1). Recombinant nerve growth factor-β (NGF- β) (Sigma) was used to stimulate neuroblastoma cells as indicated. The MEK1/2 inhibitors U0126 and PD98059 were purchased from EMD Biosciences/Calbiochem and dissolved in DMSO.
Bioinformatics
The following online databases were used to characterize NDSP: BLASTn, BLASTp, reverse position specific-BLAST, HomoloGene, Serial Analysis of Gene Expression (SAGE), Cancer Genome Anatomy Project (CGAP) (http://www.ncbi.nlm.nih.gov); Scanps 2.3 (http://www.ebi.ac.uk/scanps/); and iPSORT (http://www.hypothesiscreator.net/iPSORT/index.html). Using the IMAGE Consortium ID# 1641875 for NDSP, a normal tissue and pediatric malignancy cDNA microarray databases were queried for NDSP mRNA expression. These databases were constructed by Dr. Javed Khan and colleagues at the Oncogenomics Section of the Pediatric Oncology Branch, National Cancer Institute, National Institutes of Health, Bethesda, Maryland (http://home.ccr.cancer.gov/oncology/oncogenomics/). The methods for creating the normal tissue database and pediatric malignancy database is presented in references by Son et al (16) and Whiteford et al (17).
RNA extraction, RT-PCR, and quantitative PCR
Total RNA was extracted using the Trizol reagent (Invitrogen). In addition, pooled RNA from normal tissues was purchased commercially (Clontech, Mountain View, CA). Reverse transcription was performed with random hexamer and Super-script II reverse transcriptase (RT) (Invitrogen). PCR was carried out with the following primers: NDSP- forward: 5′-CACCATGACAGCGGGAACGGTTG-3′ and reverse: 5′-CACGTCTGTACTAATAGCCC-3′; and GAPDH- forward: 5′-AAGGTGAAGGTCGGAGTCAA-3′ and reverse: 5′-TGAGTACGTCGTGGAGTCCA-3′. Standard PCR methods were followed for cDNA amplification (18). The Quantitect SYBR Green PCR kit (Qiagen, Valencia, CA, USA) was used for quantitative PCR as previously performed (19). For all SYBR Green PCR, CT values were normalized to either β-actin or GAPDH. All real time quantitative RT-PCR results represent the mean and standard deviations of two to three replicate experiments.
Mammalian expression constructs and cloning
Full length cDNA clone containing the NDSP gene was obtained from ATCC using IMAGE Consortium ID# 1641875. NDSP was localized from the cDNA clone using PCR amplification (forward: 5′-CACCATGACAGCGGGAACGGTTG-3′ and reverse: 5′-CACGTCTGTACTAATAGCCC-3′) and cloned into the pcDNA3.1D/V5-His-TOPO vector (Invitrogen) creating a V5-tagged NDSP fusion protein. A different reverse primer, 5′-TTACACGTCTGTACTAATAGCCC-3′, was used to incorporate the native “TAA” stop codon in order to create a full length NDSP open reading frame without a tag. These primers were also used to clone the full length NDSP open reading frame into the pBabe vector for retroviral transduction and overexpression (pBabe-NDSP-imm). A similar reverse primer containing the sequence for a 6 copy histidine tag followed by the TAA stop codon was used to purify recombinant NDSP with a C-terminus histidine tag (NDSP-6xHis).
NDSP small-hairpin interfering RNA (shRNA) expression was achieved by cloning the following ligated primers into the pSuper-PURO and pSuper-NEO retrovirus expression vectors per manufacturer's protocol (Oligoengine, Seattle, WA): sh-NDSP sense: 5′-GATCCCCATGGATGAAAGACCACAATAATTCAAG AGATTATTGTGGTCTTTCATCCATTTTTTGGAAA-3′ and antisense: 5′-AGCTTTTCCAAAAA ATGGATGAAAGACCACAATAATCTCTTGAATTATTGTGGTCTTTCATCCATGGG-3′. The following nontargeting shRNA primer was used as a control for the shRNA experiments: sh-Control sense: 5′-GATCCCCCGTCTTTTCGGACTTAGAGAGTTCAAGAGA CTCTCTAAGTCCGAAAAGACGTTTTTGGAAA-3′ and antisense: 5′-AGCTTTTCCAAAAACGTCTTTTCGGACTTAGAGAGT CTCTTGAACTCTCTAAGTCCGAAAAGACGGGG-3′. The sh-NDSP target sequence was chosen from a 3′ untranslated region of the mRNA sequence; therefore, the pBabe-NDSP-immune (pBabe-NDSP-imm) transduction results in NDSP transcription which is immune to the sh-NDSP inhibition effect due to the lack of this untranslated region in the NDSP-imm construct. The pBabe-RasV12 plasmid was generously donated by Dr. Scott W. Lowe. The pBabe-vector is the pBabe construct ligated to itself without an insertion sequence.
Transient transfection and retroviral transduction
For transient transfections, 1 to 2μg of NDSP expression plasmids were transfected using LipoFectamine 2000 (Invitrogen). For retroviral transduction, either the pSuper shRNA contructs (shControl and shNDSP-1) or pBabe contructs (pBabe and NDSP-imm) were co-transfected with gag-polymerase (Peg-Pam3) and viral envelope (RDF) plasmids (generously contributed by Dr. Gianpietro Dotti) into a packaging cell line (HEK-293T). A total of 10μg of DNA was transfected into the packaging cell line using FuGene 6 per manufacturer's protocol (Roche Diagnostics, Nutley, NJ). Viral supernatant was collected at 48 and 72 hours and flash frozen. Subconfluent SK-N-AS cells were grown in viral supernatant with 4μg/ml of polybrene (Sigma) for 48 hours followed by a 24 hour period in regular culture media. Cells were then selected with puromycin or neomycin (Sigma) for 48 hours. All assays were carried out using freshly transduced and selected cells. Student's T-test was used to determine statistical significance with a p-value < 0.05 considered statistically significant.
Western blotting
For Western blot, cells were lysed with protein lysis buffer (20mM Na Phosphate, pH 7.4, 150mM NaCl, 5mM EDTA, and 1% Triton-X) and 1mM dithiothreitol (DTT), 10μg/mL aprotinin, 10μg/mL leupeptin, and 1mM phenylmethylsulfonyl fluoride (PMSF). Tumor lysate was made by cutting a piece of frozen tissue and lysing in protein lysis buffer as above. Culture supernatant was concentrated using a 10,000 MWCO spin column (Millipore, Billerica, MA, USA) centrifuged at 15,000 × g for 20 minutes at 4°C. Media not used for cell culture was used as a control. Lysate or supernatant were then mixed with reducing (β-mercaptoethanol) or non-reducing 2× Laemelli sample buffer (BioRad), boiled at 100°C, and resolved by sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Western blots were visualized with enhanced chemiluminescence substrate (Amersham, Piscataway, NJ, USA). Densitometry was used to quantify NDSP band intensity versus β-Actin and reported as a ratio of NDSP/β-Actin.
Proliferation and soft agar assays
SK-N-AS and SH-SY5Y cells were plated in 96-well plates at a concentration of 1 × 103 cells/well. Using the Cell Counting Kit-8 (CCK-8, Dojindo Molecular Technologies, Gaithersburg, MD), cells were stained on indicated days in replicates of 3 or 6. After a 2 hour incubation period, optical density was measured at 450nm using a standard plate reader. Student's T-test was used to determine statistical significance with a p-value < 0.05 considered statistically significant.
For the soft agar assays, a layer of 0.5% agarose/DMEM media was plated and allowed to solidify. SK-N-AS and SH-SY5Y cells were suspended at a concentration of 1 × 104 cells in 0.3% agarose/DMEM media and plated on top of the 0.5% layer. 500μl of DMEM media was added on top of the agarose to prevent drying of the soft agar. Wells were photographed after 14 days of growth and colonies (>30 cells) were counted. All experiments were carried out in triplicate with the mean and standard deviation reported. Student's T-test was used to determine statistical significance with a p-value < 0.05 considered statistically significant.
Purification of Recombinant NDSP
Subconfluent HEK-293T cells were transfected with the NDSP-6xHis construct and cultured for 48 hours. Fifty milliliters of supernatant was collected at that time and new media was added to each plate. An additional 50ml harvest was completed after 24 hours. The 100ml of cultured supernatant containing NDSP-6xHis was incubated with the HIS-Select affinity gel (Sigma) and recombinant protein was eluted from the affinity gel according to manufacturer's protocol. For the proliferation assays, indicated concentrations of NDSP-6xHis protein was added to each well 24 hours after plating 1 × 103 cells/well in a 96 well plate in a triplicate fashion. Student's T-test was used to determine statistical significance with a p-value < 0.05 considered statistically significant.
MEK1/2 inhibitor assays
SK-N-AS cells were seeded in a 6 well plates at 3 × 105 cells and incubated for 24 hours, various concentrations of U0126 (5μM, 10μM), PD98059(10μM, 20μM) were added to the cell growth medium and incubation was continued for an additional 24 hours. Dimethyl sulfoxide only was administered as the control treatment. Cells were harvested and submitted to immunoblotting for phospho-ERK1/2 and total ERK1/2. The growth inhibitory effects of MEK1/2 inhibitors U0126 and PD98059 was tested on cultured NB cells in vitro. SK-N-AS cells were plated in 96-well flat-bottom plates at 1 × 104 cells/well. Twenty-four hours later, U0126 (10μM) or PD98059 (60μM) were added to the cell growth medium and incubation was continued for an additional 3 days. Cytotoxic activity was determined by the CCK-8 assay. *p<0.001 compared to the DMSO only treatment group.
Results
Identification and bioinformatic analysis of NDSP
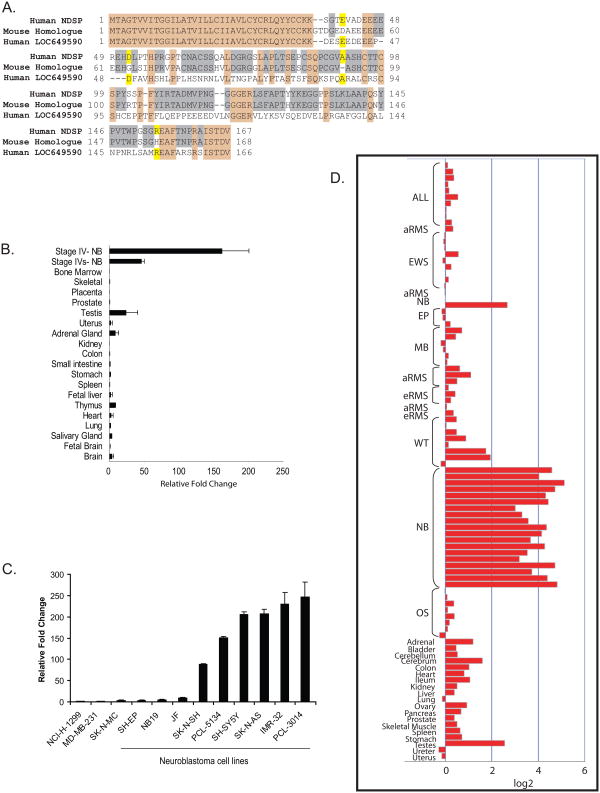
We had previously conducted a cDNA microarray experiment comparing an International Neuroblastoma Staging System (INSS) Stage IV, COG Risk Group-high risk tumor sample to a Stage IVS, COG Risk Group-low risk tumor sample (data not shown). This experiment was simply a comparison a disseminated neuroblastoma with a good prognosis versus a disseminated neuroblastoma with a poor prognosis. No control was used for this experiment. The results of this microarray study revealed an expressed sequenced tag (EST) that showed a six-fold increase in expression in the high risk sample and a negative one fold expression in the low risk sample. This EST corresponds to a 504 base pair open reading frame (ORF) and 167 amino acid sequence (Supp. Fig. 2). The locus of the candidate gene, NDSP, is chromosome 1q25.2. A well conserved mouse homologue was identified. The encoded mouse protein, A230106N23, shares over 80% protein homology with human NDSP (Fig. 1A). An additional human gene was identified encoding a protein, LOC649590, with 43% sequence identity to NDSP. Among these three sequences, the first 37 amino acids are identical including a 30 amino acid signal peptide and a C-C motif.
Figure 1.
Bioinformatic analysis of NDSP and validation of expression in neuroblastoma. A. NDSP protein sequence aligned to the mouse homologue (A230106N23) and additional human protein (LOC649590). Tan shading denotes sequence identity among all sequences; grey shading denotes sequence identity between human NDSP and the mouse homologue; and yellow shading denotes sequence identity between human NDSP and human LOC649590 protein. B. SYBR Green quantitative PCR was used to compare NDSP mRNA expression in pooled normal tissue cDNA samples to expression in two neuroblastoma tissue samples. ΔΔCT's were calculated relative to NDSP expression in the small intestine. C. SYBR Green quantitative PCR was used to analyze NDSP mRNA levels in 9 neuroblastoma cell lines compared to the other cancer cell lines that had no detectable NDSP transcript by conventional RT-PCR. D. The IMAGE clone number for NDSP (1641875) was used to query a normal tissue and pediatric malignancy microarray array database (16,17). Acute lymphocytic leukemia (ALL), alveolar (aRMS) and embryonal (eRMS) rhabdomyosarcoma, Ewing's sarcoma (EW), ependymoma (EP), medulloblastoma (MB), Wilms' tumor (WT), neuroblastoma (NB), and osteosarcoma (OS).
Validation of NDSP expression
We tested NDSP mRNA expression in 18 normal adult and fetal tissue RNA samples. Compared to the two neuroblastoma tumor samples used in the original microarray experiment, NDSP transcript levels in the normal tissues are relatively low (Fig. 1B). Of note, NDSP is detected in adult brain, thymus, adrenal gland, uterus, and testes. In addition, northern blot for NDSP mRNA expression was also performed and no detectable signal is seen in a panel of normal tissue RNA (data not shown).
Next, we tested NDSP mRNA expression in a panel of 45 cell lines representing 13 cancer types (Supplemental Figure 4). NDSP is expressed in 5 of 13 cancers including retinoblastoma, melanoma, small cell lung cancer, Ewing's sarcoma and neuroblastoma. Real time PCR was used to test NDSP expression in 9 neuroblastoma cell lines. Six out of the nine cell lines express NDSP at high levels compared to the negative controls, non-small cell lung cancer, breast cancer, and Ewing's sarcoma (Fig. 1C).
A normal human tissue and pediatric malignancy microarray database (16,17) was queried for NDSP mRNA expression (Figure 1D). NDSP has a baseline level of expression in normal human tissues such as adrenal gland, cerebrum, colon, ileum, and ovary. The highest expression in normal tissue is in testes. These findings are similar to the real time PCR data presented above. On average, the neuroblastoma tissue samples have NDSP expression levels that are 2 to 4 fold higher than the normal tissues and the majority of the other cancers.
NDSP protein is overexpressed in neuroblastoma
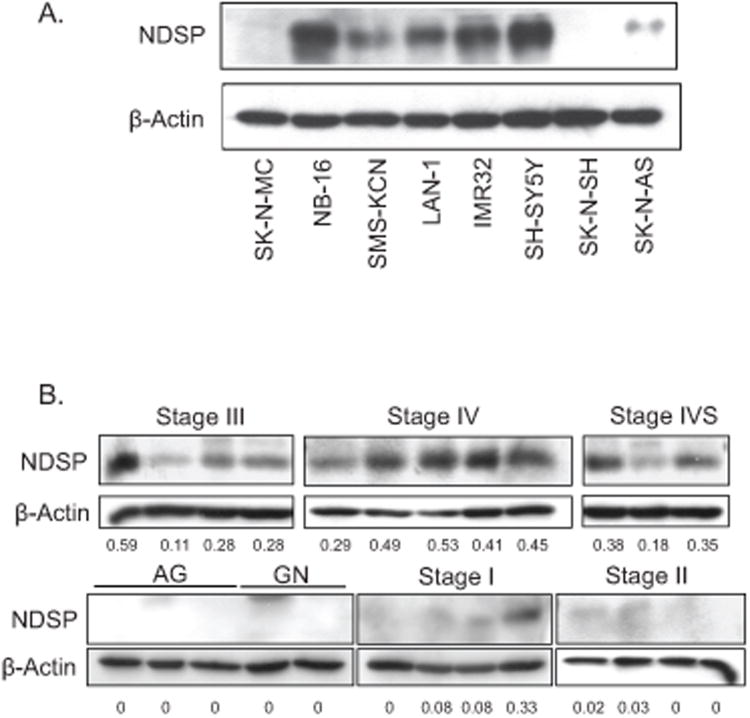
In light of the mRNA data showing the specificity of NDSP overexpression in neuroblastoma, we tested endogenous NDSP protein levels in neuroblastoma cell lines. Using a rabbit-polyclonal antibody against NDSP (anti-NDSP-Ab1), we find varying NDSP protein expression in 6 out of the 7 neuroblastoma cell lines tested (Fig. 2A). NDSP expression in neuroblastoma tumor tissue samples was then analyzed in a similar fashion. Tumor stage was determined by individual patient characteristics and prognositic factors according to the International Neuroblastoma Staging System (7). The normal adrenal gland (AG), ganglioneuroma (GN), and Stage I and II (localized disease) samples have very little NDSP expression; however, the Stage III (advanced locoregional disease), Stage IV (metastatic, disseminated disease, poor prognosis), and Stage IVS (metastatic, disseminated disease that spontaneously regresses by 1 year of age) samples all have much higher expression (Fig. 2B).
Figure 2.
NDSP protein level in neuroblastoma. A. Cell lysates for seven different neuroblastoma cell lines and a Ewing's sarcoma cell line (SK-N-MC) were resolved by SDS-PAGE and immunoblotted with anti-NDSP-Ab1. Additionally, blots were stained for β-actin. B. Tissue lysate was prepared from banked neuroblastoma tumor samples. Lysates were resolved by SDS-PAGE, and immunoblotted with anti-NDSP-Ab1 and anti-β-actin antibody. Densitometry was used to quantify both the NDSP and β-actin bands and present an NDSP/β-actin ratio (underneath each lane). Adrenal gland (AG), ganglioneuroma (GN), local disease or isolated tumor (Stage I), locoregional disease (Stage II), advanced locoregional disease (Stage III), disseminated, metastatic disease, poor prognosis (Stage IV), disseminated, metastatic disease that spontaneously regresses by 1 year of age, good prognosis (Stage IVS). C. Immunohistochemistry was performed on frozen tissue sections with anti-NDSP-Ab2 and HRP-conjugated, biotinylated anti-rabbit secondary antibody. Slides were viewed under 40× magnification. A tumor from a patient with stage I, low risk, localized disease and another from a child with stage IV, high risk, disseminated disease are presented.
NDSP is a secreted protein
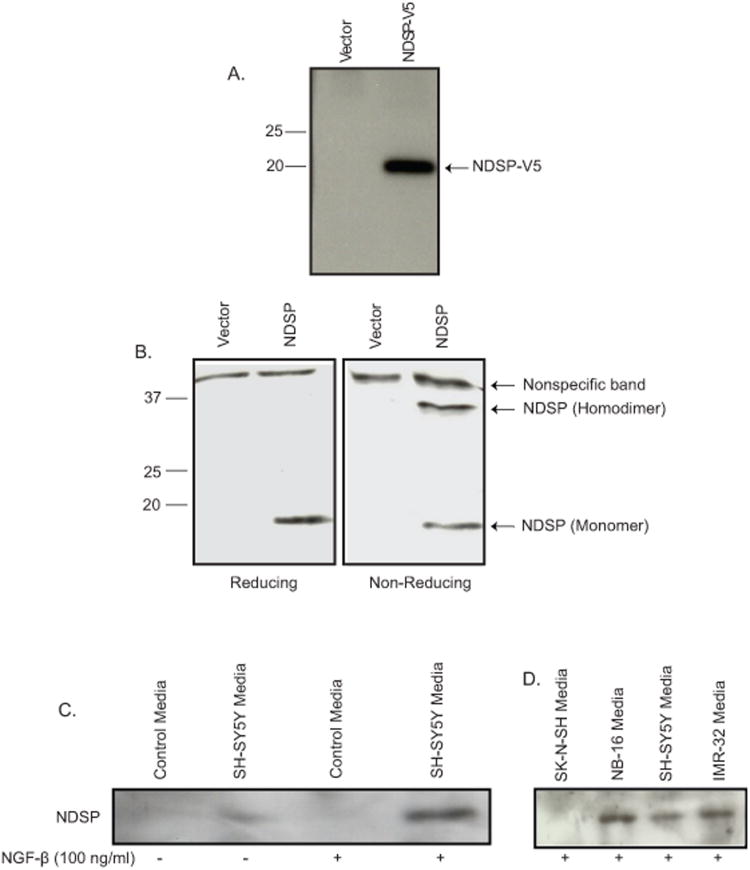
Sequence analysis of the NDSP protein revealed that the first 30 amino acids are consistent with a signal peptide with a putative cleavage site between leucine 30 and glutamine 31. This sequence is also highly conserved when compared to the NDSP mouse homologue and the related human protein LOC649590 (Figure 1A). To test the functionality of this signal peptide, a fusion protein was created by cloning the NDSP ORF into a mammalian expression vector with a C-terminus V5-tag (NDSP-V5). Supernatant from transfected HEK-293T cells reveal a distinct 20 kDa band (Figure 3A). We then collected the supernatant from non-tagged NDSP transfected HEK-293T cells and analyzed the supernatant by immunoblotting. Under reducing conditions, secreted NDSP appears as an 18 kDa monomer; however, under non-reducing conditions, secreted NDSP appears as both a 36 kDa homodimer and an 18 kDa monomer (Figure 3B).
Figure 3.
Characterizing NDSP as a secreted protein. A. HEK-293T cells were transfected with control (vector) or NDSP-V5 expression plasmids and supernatant was collected 48 hours after transfection. Supernatants were resolved by SDS-PAGE and immunoblotted with anti-V5 monoclonal antibody. B. Supernatants from HEK-293T cells transfected with control or NDSP expression plasmids were resolved by SDS-PAGE under both reducing (sample buffer with β-mercaptoethanol) and non-reducing (sample buffer without β-mercaptoethanol) conditions. Immunoblotting was performed with anti-NDSP-Ab1. C. Supernatant from SH-SY5Y cells without and with 100ng/ml NGF-β treatment was resolved on SDS-PAGE and immunoblotted with anti-NDSP-Ab1. Control media was simply culture media incubated without cells. D. Additional cell lines were treated with 100ng/ml NGF-β and supernatants were resolved on SDS-PAGE and immunoblotted with anti-NDSP-Ab1.
In order to test whether NDSP is secreted from neuroblastoma cell lines, we attempted to augment NDSP synthesis by exposing neuroblastoma cell lines to an array of cytokines and growth factors to simulate the tumor microenvironment in vivo. Quantitative PCR reveals that NGF-β increased NDSP mRNA levels 3-4 fold above control (Supp. Fig. 3). Immunoblotting culture supernatant from SH-SY5Y cells reveals a faint band without NGF-β; whereas, 100ng/ml NGF-β treatment for 48 hours results in a significant increase in secreted NDSP (Fig. 3C). In addition, NDSP is detected in the culture supernatant of the NB16 and IMR32 neuroblastoma cell lines treated with NGF-β (Fig. 3D). The negative control SK-N-SH which has no detectable protein level of NDSP, shown in above experiments, cannot be stimulated to secrete NDSP into the culture media.
NDSP promotes neuroblastoma cell proliferation and anchorage-independent growth
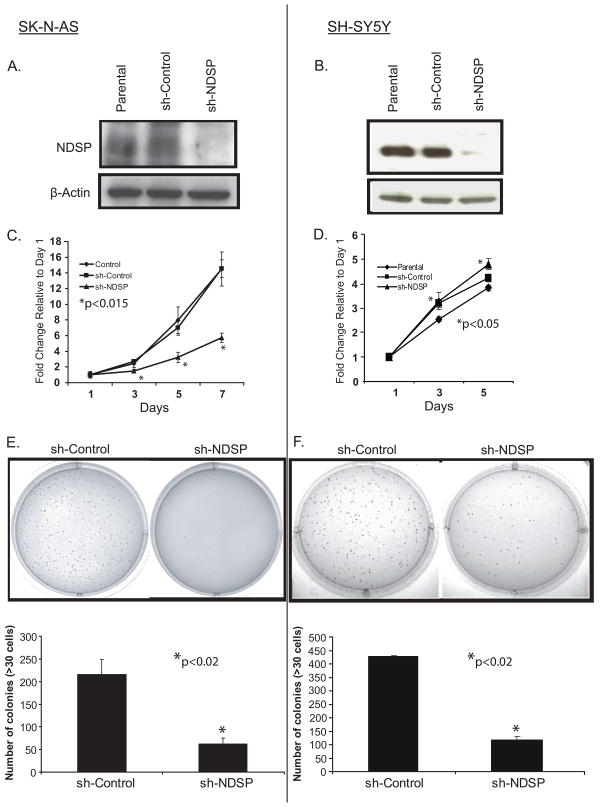
To determine the function of NDSP in neuroblastoma, the SK-N-AS and SH-SY5Y neuroblastoma cell lines were transduced with retroviral vector expressing a small hairpin RNA against NDSP (sh-NDSP) to suppress the expression of NDSP in these cell lines. In this experiment, a non-targeting sequence vector was used to transduce these cell lines creating a control (sh-Control). After selection, cells were assayed for NDSP expression. Compared to the parental control cell line, the sh-Control cells do not display NDSP knockdown; however, the sh-NDSP cells show a significant decrease in NDSP protein levels (Fig. 4A & 4B).
Figure 4.
Inhibiting NDSP expression results in a proliferation defect. SK-N-AS and SH-SY5Y cell lines were transduced with the sh-Control and sh-NDSP constructs. After transduction for 48 hours, cells were selected with puromycin and grown to confluence. A and B, NDSP protein levels were analyzed in the parental, sh-Control, and sh-NDSP cell lines by immunoblotting for NDSP with the anti-NDSP-Ab1. Immunoblotting for β-actin served as a loading control. C and D, The SK-N-AS and SH-SY5Y parental, sh-Control, and sh-NDSP cell lines were plated in 96-well plates at 1 × 103 cells/well. The CCK-8 assay was used to quantify cellular proliferation relative to Day 1 absorbance measured at 450nm. These experiments were performed with six replicates and reported as the mean with standard deviations. Student's T-test was used to determine statistical significance with a p-value < 0.05 considered statistically significant. *Indicates statistical significance comparing sh-NDSP to parental and sh-Control (C) and sh-NDSP to parental (D). E and F, SK-N-AS and SH-SY5Y sh-Control and sh-NDSP stable transduced cell lines were plated in 0.3% agarose/DMEM media on top of a 0.5% agarose/DMEM layer. After 14 days of growth, colonies were stained with MTT and colonies >30 cells were counted. These experiments were performed in triplicate and reported as the mean with standard deviations. Student's T-test was used to determine statistical significance with a p-value < 0.05 considered statistically significant. *Indicates statistical significance comparing sh-NDSP to sh-Control.
In a proliferation assay, the SK-N-AS sh-NDSP cell line grows significantly (p<0.015) slower compared to both the parental and sh-control cell lines over a 7 day period (Fig. 4C). The same phenomenon is seen with anchorage-independent growth in soft agar in which SK-N-AS sh-NDSP colony formation is significantly (p<0.02) less compared to the sh-Control cells (Fig. 4E). These two assays reveal inherent differences between the neuroblastoma cell lines. The proliferation assay with SH-SY5Y sh-NDSP cells does not display an inhibited growth pattern as seen with SK-N-AS (Fig. 4D). There is no significant difference in growth between the sh-Control and parental cells; however, the sh-NDSP cells grew significantly (p<0.05) faster than the parental cells but there is no difference when compared to the sh-Control cells. When assayed in soft agar, the SH-SY5Y sh-NDSP cells grow significantly (p<0.02) fewer colonies than the sh-Control cells showing that NDSP knockdown in both cell lines consistently affects anchorage-independent growth (Fig. 4F).
NDSP overexpression or recombinant NDSP can reverse shRNA effects
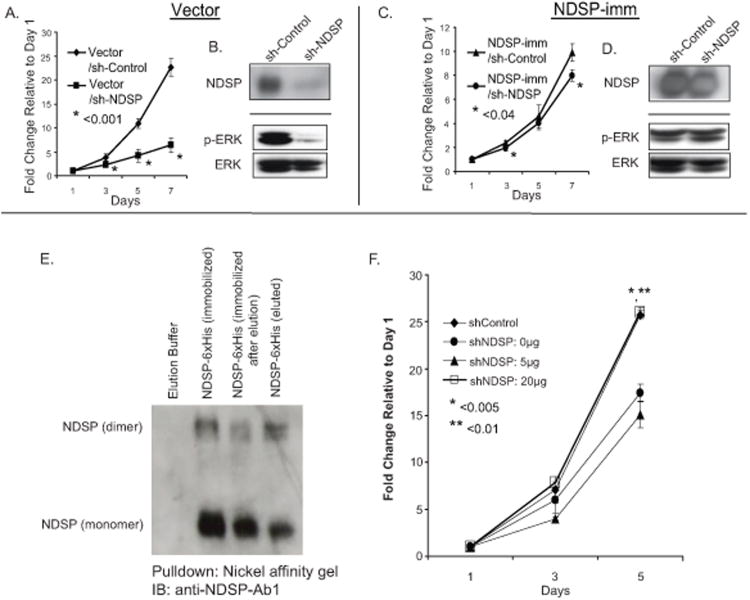
In order to further confirm that inhibition of NDSP expression is responsible for decreased proliferation, we tested whether this phenotype could be reversed by re-introducing NDSP to the sh-NDSP cell line. We accomplished this in two ways. First we transduced the SK-N-AS cell line with either a retroviral pBabe-vector control (vector) or pBabe-NDSP-shRNA-immune expression vectors (NDSP-imm). The NDSP-imm vector forces overexpression of the full length NDSP without the 3′ untranslated region which contains the target sequence for the sh-NDSP. Therefore NDSP expression from this expression vector is “immune” to the knockdown effect of sh-NDSP (Fig. 5B & 5D). Using these cells in a proliferation assay, we find that overexpression of NDSP-imm reverses the proliferation defect seen in the vector control group (Fig. 5A & 5C).
Figure 5.
Proliferation defect in sh-NDSP cells is reversible with overexpression of NDSP or treatment with recombinant NDSP. SK-N-AS cells were transduced with control (vector) or the NDSP shRNA-immune vector (NDSP-imm). After selection these separate cell lines were transduced with sh-Control or sh-NDSP vectors. A and C, These four cell lines, vector/sh-Control, vector/sh-NDSP, NDSP-imm/sh-Control; NDSP-imm/sh-NDSP were selected and subsequently used in the CCK-8 cell proliferation assays. These experiments were performed in triplicate and reported as the mean with standard deviations. Student's T-test was used to determine statistical significance with a p-value < 0.05 considered statistically significant. *Indicates statistical significance comparing the indicated sh-NDSP cell line (vector vs. NDSP-imm) to sh-Control. B and D, During the exponential growth phase, the cell lines were harvested, lysed and immunoblotting was performed with anti-NDSP-Ab1, anti-phospho-ERK1/2-Ab, and anti-ERK1/2-Ab. E, Recombinant NDSP-6xHis was purified by transfecting HEK293T cells with the NDSP-6xHis expression vector and collecting the supernatant. The supernatant was then incubated with nickel affinity gel and the NDSP-6xHis protein was immobilized. The protein was then eluted and immunoblotting (IB) with anti-NDSP-Ab1 was performed with the various phases of the purification process. F, SK-N-AS sh-Control and sh-NDSP cell lines were plated in 96-well plates at 1 × 103 cells/well. Recombinant NDSP-6xHis was added to the media of the sh-NDSP cells in varying concentrations and the CCK-8 assay was used to quantify cellular proliferation relative to Day 1 absorbance measured at 450nm. These experiments were performed in triplicate and reported as the mean with standard deviations. Student's T-test was used to determine statistical significance with a p-value < 0.05 considered statistically significant. *Indicates statistical significance comparing sh-Control (
 ) to sh-NDSP: 0ug (
) to sh-NDSP: 0ug (
 ), **Indicates statistical significance comparing sh-NDSP: 20ug (
), **Indicates statistical significance comparing sh-NDSP: 20ug (
 ) to sh-NDSP: 0ug (
) to sh-NDSP: 0ug (
 ).
).
We were also able to purify recombinant NDSP by overexpressing a six histidine-tagged NDSP construct in HEK293T cells. The supernatant was collected after 72 hours of culture and NDSP-6xHis was immobilized with nickel affinity gel and eluted (Fig. 5E). A proliferation assay was performed with sh-Control and sh-NDSP SK-N-AS cells where varying concentrations of recombinant NDSP were added to the sh-NDSP cells. The proliferation defect seen in the sh-NDSP cells is effectively reversed with the addition of 20μg recombinant NDSP-6xHis displaying a similar fold change in growth compared to the sh-Control at 5 days (Fig. 5F).
NDSP induced cell proliferation is mediated by ERK1/2 activation
In addition to testing proliferation, we assayed the SK-N-AS vector/sh-control, vector/sh-NDSP cells, NDSP-imm/sh-Control, and NDSP-imm/sh-NDSP cell lines during the exponential growth phase for ERK1/2 phosphorylation. We find constitutively activated ERK1/2 in the SK-N-AS vector/sh-control cells and a significant inhibition of ERK1/2 phosphorylation in the vector/sh-NDSP cells (Fig. 5B). Inhibited ERK1/2 phosphorylation is effectively reversed by overexpressing the NDSP-imm construct in the setting of NDSP knockdown correlating with a similar proliferation rate to the NDSP-imm/sh-Control cells (Fig. 5D).
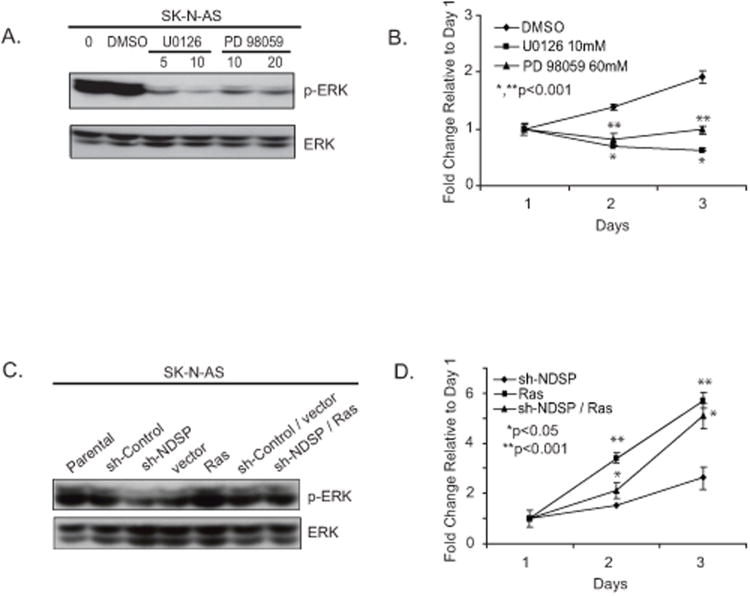
In order to test the importance of ERK1/2 activation on proliferation, we used MEK1/2 small-molecule inhibitors on the parental SK-N-AS cell line. ERK1/2 is constitutively active in SK-N-AS cells (Fig. 6A). Both U0126 and PD98059 MEK1/2 inhibitors were used in a proliferation assay with SK-N-AS cells. Figure 6B shows a significant decrease in proliferation of the SK-N-AS cell line with MEK1/2 inhibitor treatment. This correlates with the Western data showing significant decrease in ERK1/2 phosphorylation with MEK1/2 inhibitor treatement (Fig 6A).
Figure 6.
MEK1/2 inhibitors cause decreased ERK activation in SK-N-AS cells resulting in a prolifereation defect. A, The MEK1/2 inhibitor assays were performed by placing SK-N-AS cells in culture at 3 × 105 cells/well and incubating for 24 hours. Various concentrations of U0126 (5μM, 10μM), PD98059(10μM, 20μM) were added to the cell growth medium and incubation was continued for an additional 24 hours. Dimethyl sulfoxide only was administered as the control treatment. Cells were harvested and immunoblotted for phospho-ERK1/2 and total ERK1/2. B, To test proliferation, SK-N-AS cells were plated in 96-well flat-bottom plates at 1 × 104 cells/well. Twenty-four hours later, various concentrations of U0126 (10μM) or PD98059 (60μM) were added to the cell growth medium and incubation was continued for an additional 3 days. Cytotoxic activity was determined by the CCK-8 assay. These experiments were performed in triplicate and reported as the mean with standard deviations. Student's T-test was used to determine statistical significance with a p-value < 0.05 considered statistically significant. *Indicates statistical significance comparing the U0126 group to the DMSO control. **Indicates statistical significance comparing the PD98059 group to the DMSO control C, In order to test the effects of constitutive ERK1/2 activation, SK-N-AS parental cells were cultured at 3× 105 cells/well and were transiently transfected with sh-control, sh-NDSP, pBabe-vector control (vector), pBabe-RasV12 (Ras), sh-Control/vector, and sh-NDSP/Ras. Forty-eight hours after transfection, the cells were collected and immunoblotted for phospho-ERK1/2 and total ERK1/2. D, The cells were also re-seeded into 96-well plates at 1 × 104 cells/well and allowed to grow for an additional 4 days to compare growth among the experimental groups. These experiments were performed in triplicate and reported as the mean with standard deviations. Student's T-test was used to determine statistical significance with a p-value < 0.05 considered statistically significant. *Indicates statistical significance comparing the sh-NDSP/Ras group to the sh-NDSP group. **Indicates statistical significance between the Ras group and the sh-NDSP group.
Having established that NDSP knockdown results in decreased ERK1/2 activation and decreased proliferation, we tested the effect of constitutive ERK1/2 activation in these cells and if the proliferation defect was reversible. For constitutive ERK1/2 activation, we used a Ras oncogene overexpression plasmid, pBabe-RasV12. This experiment was performed with transient transfection of parental SK-N-AS cells with sh-Control, sh-NDSP, pBabe-vector (vector), and pBabe-RasV12 (Ras) expression plasmids. Cells were assayed for Western blotting and proliferation. Diminished ERK1/2 phosphorylation induced by NDSP knockdown is reversed by overexpression of Ras (Fig. 6C). This results in increased proliferation of the sh-NDSP/Ras contransfected cell line compared to the sh-NDSP transfected cell line (Fig 6D).
Discussion
As various regulators of transformation, proliferation, survival, resistance, and metastasis are defined, a multi-modal approach targeting multiple pathways will enable more efficient and effective eradication of cancer. Determining the functions of the proteins within a particular pathway also helps define the most effective therapeutic approach, whether it is in the form of a small-molecule inhibitor, inhibitory peptide, monoclonal antibody, or systemic shRNA delivery. The focus of this report is to establish NDSP as a neuroblastoma-specific protein and begin to define the function of NDSP in neuroblastoma cell proliferation and the molecular pathways involved.
The NDSP gene was initially identified from an expressed sequence tag (EST) that was overexpressed in a metastatic neuroblastoma tissue sample based on expression microarray analysis. The EST corresponded to an open reading frame encoding the NDSP protein. Our methods for testing NDSP expression in neuroblastoma began with an extensive bioinformatic analysis of NDSP mRNA expression using multiple online databases. The NCBI databases, SAGE and CGAP, were used to perform initial expression analysis of NDSP; however, these databases are lacking in data from pediatric cancers. Son et al. began the process of strengthening these databases by compiling microarray data on multiple normal human tissues that can be searched with a cDNA sequence for the gene of interest (16). This group has furthered these efforts by creating a pediatric malignancy database with the same search capabilities (17). A query of the pediatric malignancy microarray database revealed very specific NDSP overexpression in neuroblastoma samples and low expression in normal tissue and other cancer types.
Our own mRNA analysis of cancer cell lines and normal tissue cDNA confirm the bioinformatic findings. The analysis of normal tissues revealed some expression in brain, adrenal gland, and ovaries/testes. In a series of cancer cell lines, we see NDSP expression in melanoma, small cell lung cancer, retinoblastoma, Ewing's sarcoma, and neuroblastoma. This limited expression pattern shows specificity to cancers originating from neural crest-derived cells. The results of these initial studies illustrate the utility of cDNA microarray screens and online bioinformatics in identifying markers and molecular targets in cancer.
NDSP protein expression correlated well with the PCR data in the neuroblastoma cell lines tested. Some divergence was seen in the SK-N-AS cell line where mRNA expression was high and the protein expression in lysate was lower than the other cell lines tested. This may be explained by differences in NDSP protein metabolism (synthesis/breakdown) or more efficient secretion of NDSP in the SK-N-AS cell line as compared to SH-SY5Y or IMR32. NDSP inhibition by sh-RNA was able to cause phenotypic changes in the SK-N-AS and SH-SY5Y cell lines indicating that NDSP plays an important role regardless of the absolute protein level in cell lysates. Our analysis of neuroblastoma tissues revealed that the neuroblastic tumors had much higher expression than that seen in either ganglioneuroma or normal adrenal gland samples. High NDSP levels correlated with the advanced locoregional disease and disseminated, metastatic disease groups compared to the localized Stage I and II tumors. At the mRNA level, we found that 96% of neuroblastoma tissue samples tested had overexpression of NDSP mRNA relative to controls and that higher mRNA expression correlated with the high risk, poor prognosis group (19). We did not find an obvious correlation with MYCN-amplification or other prognostic factors within our sampling of patients. We also performed real time PCR expression analysis of a MYCN-inducible cell line and could not detect a higher NDSP mRNA in the induced cells further supporting no direct correlation between the two proteins (data not shown). In order to prove that NDSP is a useful biomarker for neuroblastoma and to investigate its utility as a predictor of prognosis, a prospective clinical trial assessing NDSP expression levels at diagnosis, post-treatment, and recurrence must be performed.
The next part of this report demonstrates that NDSP is a secreted protein which appears to be involved in neuroblastoma cell proliferation. The NDSP protein sequence exhibits a highly conserved signal peptide seen in both a mouse homologue and a related human protein with 100% sequence identity. NDSP is secreted as a homodimer as well as a monomer. Downstream from the signal peptide, there is a C-C motif at position 34 and 35 and six additional cysteine groups suggesting that NDSP may be a part of the C-C family of chemokines (20). This C-C motif is also well conserved in both the related human protein and the mouse homologues.
We did find that a known ligand, NGF-β, can stimulate secretion of NDSP from multiple neuroblastoma cell lines. Tyrosine kinase A (Trk A) receptor expression and its primary ligand, NGF-β, are associated with a well differentiated phenotype and localized, more benign neuroblastoma (21, 22). Trk B expression and its primary ligand, brain derived growth factor (BDNF) are associated with a more malignant phenotype and poor prognosis (23-25). However, the TrkA receptor was originally identified as a proto-oncogene and found to be oncogenic in prostate cancer, papillary thyroid carcinoma, and sarcomas (26). There is recent evidence to suggest that aberrant signaling through TrkA isoforms results in a more malignant and metastatic form of neuroblastoma (27-29). We speculate that NDSP may be a transcriptional target gene regulated by the more aberrant form of NGF-TrkA signaling.
Because NDSP is actively secreted from neuroblastoma cells, we hypothesize that this protein may serve as an extracellular ligand with growth factor properties. Insulin-like growth factor (IGF) and BDNF are two growth factors associated with the malignant phenotype in neuroblastoma. IGF is a ligand for the IGF receptor (IGFR) that acts through an Akt-mediated pathway to increase proliferation, migration, and invasiveness of neuroblastoma (30-33). The BDNF and Trk B ligand/receptor complex also leads to activated survival pathways through Akt resulting in increased chemoresistance and cellular invasion of neuroblastoma cell lines (34). Anchorage independent growth is dependent on both the PI3K/Akt and ERK1/2 mediated survival pathways that inhibit anoikis (25, 35). We found that NDSP inhibition with shRNA results in decreased colony formation in both SK-N-AS and SH-SY5Y cell lines. We also found that NDSP shRNA causes inhibited ERK1/2 phosphorylation during SK-N-AS proliferation which is reversible with overexpression of NDSP in this setting. It seems that SK-N-AS has constitutive ERK1/2 activation; therefore, the proliferation effects are seen with NDSP inhibition in both 2-dimensional (growth in a culture dish) and 3-dimensional growth (growth in soft agar) assays. The defect in proliferation was also reproducible when commercially available MEK1/2 inhibitors were added to parental SK-N-AS cells in culture further emphasizing the importance of ERK1/2 activation in proliferation. However, we found that SH-SY5Y has very little ERK1/2 phosphorylation in normal 2-dimensional proliferation possibly explaining why the effect on growth was not seen in this setting (data not shown). We hypothesize that NDSP induces ERK1/2 phosphorylation during anchorage independent growth as a mechanism for inhibiting anoikis; therefore, inhibiting NDSP expression in SH-SY5Y during a soft agar assay results in decreased colony formation. These findings together suggest that the endogenous NDSP expression seen in neuroblastoma may contribute to increased tumor growth and progression through an ERK-mediated proliferation pathway.
We believe that one of the most important findings in this study is the reversibility of the NDSP shRNA effects with NDSP overexpression, the addition of recombinant NDSP, and induction of constitutive ERK1/2 activation. These findings show that the shRNA is specific for NDSP and not a result of an “off target” effect. Additionally this provides evidence that there is a membrane bound receptor for NDSP expressed in neuroblastoma that may trigger an ERK-mediated proliferation pathway downstream. Identification of this receptor would be advantageous in designing effective treatments to inhibit NDSP signaling.
In conclusion, our studies have effectively shown that NDSP is overexpressed with high specificity in neuroblastoma, is involved in tumor cell proliferation, and may serve as a useful therapeutic target for this disease. Our laboratory is currently trying to further define the molecular mechanisms responsible for the pro-proliferative phenotype, and identify the NDSP specific membrane-bound receptor. The results of these experiments will help design effective peptide and small molecule inhibitors engineered to block NDSP signaling in hopes of decreasing the malignant progression of neuroblastoma tumors.
Supplementary Material
Acknowledgments
We would like to thank Dr. Jason M. Shohet and the members of the Neuroblastoma Research Program at the Texas Children's Cancer Center for helpful discussions and critical review of this paper. We would also like to thank Kristen Kaiser for her assistance in preparing this manuscript for submission.
Grant support: Children's Oncology Group Translational Research Award (to J.G. N.), National Cancer Institute grant 1R21CA106513-01A2 (J. Yang), Baylor College of Medicine Cancer Center Pilot Project grant (J. Yang), American Cancer Society grant RSG-06-070-01-TBE (J. Yang), Hope Street Kids Foundation (J. Yang), Bear Necessities Pediatric Cancer Foundation (J. Yang), Society of University Surgeons Ethicon Resident Scholarship (S.A.Vasudevan), and the NIH/NCI-NRSA training grant 1F32CA113059-01A1 (S.A.Vasudevan).
Abbreviations list
- NDSP
Neuroblastoma Derived Secretory Protein
- NGF
nerve growth factor
- shRNA
small hairpin interfering RNA
- ERK
extracellular signal regulated kinase
Footnotes
Disclosure of Potential Conflicts of Interest: No potential conflicts of interest were disclosed.
References
- 1.Kuriyama M, Wang MC, Lee CI, et al. Use of human prostate-specific antigen in monitoring prostate cancer. Cancer Res. 1981;41:3874–6. [PubMed] [Google Scholar]
- 2.Kuriyama M, Wang MC, Lee CL, et al. Multiple marker evaluation in human prostate cancer with the use of tissue-specific antigens. J Natl Cancer Inst. 1982;68:99–105. [PubMed] [Google Scholar]
- 3.Paik S, Hazan R, Fisher ER, et al. Pathologic findings from the National Surgical Adjuvant Breast and Bowel Project: prognostic significance of erbB-2 protein overexpression in primary breast cancer. J Clin Oncol. 1990;8:103–12. doi: 10.1200/JCO.1990.8.1.103. [DOI] [PubMed] [Google Scholar]
- 4.Brodeur GM, Seeger RC, Schwab M, et al. Amplification of N-myc in untreated human neuroblastomas correlates with advanced disease stage. Science. 1984;224:1121–4. doi: 10.1126/science.6719137. [DOI] [PubMed] [Google Scholar]
- 5.Brodeur GM. Neuroblastoma: biological insights into a clinical enigma. Nat Rev Cancer. 2003;3:203–16. doi: 10.1038/nrc1014. [DOI] [PubMed] [Google Scholar]
- 6.London WB, Castleberry RP, Matthay KK, et al. Evidence for an age cutoff greater than 365 days for neuroblastoma risk group stratification in the Children's Oncology Group. J Clin Oncol. 2005;23:6459–65. doi: 10.1200/JCO.2005.05.571. [DOI] [PubMed] [Google Scholar]
- 7.Matthay KK. Neuroblastoma: a clinical challenge and biologic puzzle. CA Cancer J Clin. 1995;45:179–92. doi: 10.3322/canjclin.45.3.179. [DOI] [PubMed] [Google Scholar]
- 8.Bell S, Parker L, Craft AW, et al. False positive results in a neuroblastoma screening programme. Med Pediatr Oncol. 1994;22:181–6. doi: 10.1002/mpo.2950220306. [DOI] [PubMed] [Google Scholar]
- 9.Mathieu P, Favrot M, Frappaz D, et al. Neuroblastoma in children: clinical and biological aspects. An experience of screening in France. Ann Biol Clin (Paris) 1993;51:665–88. [PubMed] [Google Scholar]
- 10.Butler LM, Zhou X, Xu WS, et al. The histone deacetylase inhibitor SAHA arrests cancer cell growth, up-regulates thioredoxin-binding protein-2, and down-regulates thioredoxin. Proc Natl Acad Sci USA. 2002;99:11700–5. doi: 10.1073/pnas.182372299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hrzenjak A, Moinfar F, Kremser ML, et al. Valproate inhibition of histone deacetylase 2 affects differentiation and decreases proliferation of endometrial stromal sarcoma cells. Mol Cancer Ther. 2006;5:2203–10. doi: 10.1158/1535-7163.MCT-05-0480. [DOI] [PubMed] [Google Scholar]
- 12.Marshall JL, Kindler H, Deeken J, et al. Phase I trial of orally administered CEP-701, a novel neurotrophin receptor-linked tyrosine kinase inhibitor. Invest New Drugs. 2005;23:31–7. doi: 10.1023/B:DRUG.0000047103.64335.b0. [DOI] [PubMed] [Google Scholar]
- 13.Raffaghello L, Marimpietri D, Pagnan G, et al. Anti-GD2 monoclonal antibody immunotherapy: a promising strategy in the prevention of neuroblastoma relapse. Cancer Lett. 2003;197:205–9. doi: 10.1016/s0304-3835(03)00100-9. [DOI] [PubMed] [Google Scholar]
- 14.Matthay KK, Villablanca JG, Seeger RC, et al. Treatment of high-risk neuroblastoma with intensive chemotherapy, radiotherapy, autologous bone marrow transplantation, and 13-cis-retinoic acid Children's Cancer Group. N Engl J Med. 1999;341:1165–73. doi: 10.1056/NEJM199910143411601. [DOI] [PubMed] [Google Scholar]
- 15.Ribatti D, Raffaghello L, Marimpietri D, et al. Fenretinide as an anti-angiogenic agent in neuroblastoma. Cancer Lett. 2003;197:181–4. doi: 10.1016/s0304-3835(03)00105-8. [DOI] [PubMed] [Google Scholar]
- 16.Son CG, Bilke S, Davis S, et al. Database of mRNA gene expression profiles of multiple human organs. Genome Res. 2005;15:443–50. doi: 10.1101/gr.3124505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whiteford CC, Bilke S, Greer BT, et al. Credentialing preclinical pediatric xenograft models using gene expression and tissue microarray analysis. Cancer Res. 2007;67:32–40. doi: 10.1158/0008-5472.CAN-06-0610. [DOI] [PubMed] [Google Scholar]
- 18.Vasudevan SA, Skoko J, Wang K, et al. MKP-8, a novel MAPK phosphatase that inhibits p38 kinase. Biochem Biophys Res Commun. 2005;330:511–8. doi: 10.1016/j.bbrc.2005.03.028. [DOI] [PubMed] [Google Scholar]
- 19.Vasudevan SA, Russell HV, Okcu MF, et al. Neuroblastoma-derived secretory protein messenger RNA levels correlate with high-risk neuroblastoma. J Pediatr Surg. 2007;42:148–52. doi: 10.1016/j.jpedsurg.2006.09.064. [DOI] [PubMed] [Google Scholar]
- 20.Schall TJ, Mak JY, DiGregorio D, et al. Receptor/ligand interactions in the C-C chemokine family. Adv Exp Med Biol. 1993;351:29–37. doi: 10.1007/978-1-4615-2952-1_4. [DOI] [PubMed] [Google Scholar]
- 21.Baker DL, Reddy UR, Pleasure D, et al. Analysis of nerve growth factor receptor expression in human neuroblastoma and neuroepithelioma cell lines. Cancer Res. 1989;49:4142–6. [PubMed] [Google Scholar]
- 22.Nakagawara A, rima-Nakagawara M, Scavarda NJ, et al. Association between high levels of expression of the TRK gene and favorable outcome in human neuroblastoma. N Engl J Med. 1993;328:847–54. doi: 10.1056/NEJM199303253281205. [DOI] [PubMed] [Google Scholar]
- 23.Brodeur GM, Nakagawara A, Yamashiro DJ, et al. Expression of TrkA, TrkB and TrkC in human neuroblastomas. J Neurooncol. 1997;31:49–55. doi: 10.1023/a:1005729329526. [DOI] [PubMed] [Google Scholar]
- 24.Nakagawara A, Azar CG, Scavarda NJ, et al. Expression and function of TRK-B and BDNF in human neuroblastomas. Mol Cell Biol. 1994;14:759–67. doi: 10.1128/mcb.14.1.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Douma S, Van LT, Zevenhoven J, et al. Suppression of anoikis and induction of metastasis by the neurotrophic receptor TrkB. Nature. 2004;430:1034–9. doi: 10.1038/nature02765. [DOI] [PubMed] [Google Scholar]
- 26.Geiger TR, Peeper DS. The neurotrophic receptor TrkB in anoikis resistance and metastasis: a perspective. Cancer Res. 2005;65:7033–6. doi: 10.1158/0008-5472.CAN-05-0709. [DOI] [PubMed] [Google Scholar]
- 27.Pierotti MA, Greco A. Oncogenic rearrangements of the NTRK1/NGF receptor. Cancer Lett. 2006;232:90–8. doi: 10.1016/j.canlet.2005.07.043. [DOI] [PubMed] [Google Scholar]
- 28.Tacconelli A, Farina AR, Cappabianca L, et al. TrkA alternative splicing: a regulated tumor-promoting switch in human neuroblastoma. Cancer Cell. 2004;6:347–60. doi: 10.1016/j.ccr.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 29.Tacconelli A, Farina AR, Cappabianca L, et al. TrkAIII. A novel hypoxia-regulated alternative TrkA splice variant of potential physiological and pathological importance. Cell Cycle. 2005;4:8–9. doi: 10.4161/cc.4.1.1349. [DOI] [PubMed] [Google Scholar]
- 30.Pollak MN, Schernhammer ES, Hankinson SE. Insulin-like growth factors and neoplasia. Nat Rev Cancer. 2004;4:505–18. doi: 10.1038/nrc1387. [DOI] [PubMed] [Google Scholar]
- 31.Meyer GE, Shelden E, Kim B, et al. Insulin-like growth factor I stimulates motility in human neuroblastoma cells. Oncogene. 2001;20:7542–50. doi: 10.1038/sj.onc.1204927. [DOI] [PubMed] [Google Scholar]
- 32.Meyer A, van Golen CM, Kim B, et al. Integrin expression regulates neuroblastoma attachment and migration. Neoplasia. 2004;6:332–42. doi: 10.1593/neo.03445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Noujaim D, van Golen CM, van Golen KL, et al. N-Myc and Bcl-2 coexpression induces MMP-2 secretion and activation in human neuroblastoma cells. Oncogene. 2002;21:4549–57. doi: 10.1038/sj.onc.1205552. [DOI] [PubMed] [Google Scholar]
- 34.Matsumoto K, Wada RK, Yamashiro JM, et al. Expression of brain-derived neurotrophic factor and p145TrkB affects survival, differentiation, and invasiveness of human neuroblastoma cells. Cancer Res. 1995;55:1798–806. [PubMed] [Google Scholar]
- 35.Reginato MJ, Mills KR, Paulus JK, et al. Integrins and EGFR coordinately regulate the pro-apoptotic protein Bim to prevent anoikis. Nat Cell Biol. 2003;5:733–40. doi: 10.1038/ncb1026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.