Abstract
Telomeres protect chromosome termini to maintain genomic stability and regulate cellular lifespan. Maintenance of telomere length is required for neoplastic cells after the acquisition of mutations that deregulate cell cycle control and increase cellular proliferation, and can occur through expression of the enzyme telomerase or in a telomerase-independent manner termed alternative lengthening of telomeres (ALT). The precise mechanisms that govern the activation of ALT or telomerase in tumor cells are unknown, although cellular origin may favor one or the other mechanisms. ALT pathways are incompletely understood to date; however, recent publications have increasingly broadened our understanding of how ALT is activated, how it proceeds, and how it influences tumor growth. Specific mutational events influence ALT activation, as mutations in genes that suppress recombination and/or alterations in the regulation of telomerase expression are associated with ALT. Once engaged, ALT uses DNA repair proteins to maintain telomeres in the absence of telomerase; experiments that manipulate the expression of specific proteins in cells using ALT are illuminating some of its mechanisms. Furthermore, ALT may influence tumor growth, as experimental and clinical data suggest that telomerase expression may favor tumor progression. This review summarizes recent findings in mammalian cells and models, as well as clinical data, that identify the genetic mutations permissive to ALT, the DNA repair proteins involved in ALT mechanisms and the importance of telomere maintenance mechanisms for tumor progression. A comprehensive understanding of the mechanisms that permit tumor cell immortalization will be important for identifying novel therapeutic targets in cancer.
Keywords: alternative lengthening of telomeres, BLM, DNA repair, immortalization, telomerase, telomere, telomere maintenance, tumor, WRN
1. Introduction
Telomeres are DNA-protein structures that protect chromosome ends and are composed of (TTAGGG)n sequence repeats in vertebrates [1]. Telomeres protect DNA by concealing the chromosome end in a looped structure resembling a replication D-loop. The 3’ single-stranded telomere overhang folds back and invades a double-stranded telomere region to create a T-loop [2]. This structure is maintained by a number of telomere-binding proteins, including telomere repeat binding factor 1 (TRF1), telomere repeat binding factor 2 (TRF2), and protection of telomeres 1 (POT1). These proteins, along with adrenocortical dysplasia protein homolog known as TPP1, TRF1-interacting nuclear protein 2 (TIN2), and TRF2-interacting telomeric protein 1 (RAP1), form the shelterin complex [3]. Shelterin functions as a telomere cap that distinguishes the chromosome end from a DNA break, protecting chromosomal integrity.
Telomeres prevent the eventual loss of coding DNA due to the end replication problem, a replicative limitation that yields progressive telomere shortening with each round of cell division [4]. Progressive telomere shortening accompanies organismal aging [4–6], while rapidly dividing cells actively maintain their telomeres. For example, the high proliferation rate of neoplastic cells necessitates the maintenance of telomere length to facilitate immortalization and tumorigenesis. Telomere maintenance is thus not only critical for genomic stability, but also represents a key step in tumor cell immortalization.
In human cells, two telomere maintenance mechanisms are known: expression of telomerase or activation of telomerase-independent pathways termed alternative lengthening of telomeres (ALT). Telomerase is an enzyme expressed during development that catalyzes the addition of telomere repeats to chromosome ends. The enzyme is primarily composed of a reverse transcriptase catalytic subunit (TERT) and an RNA template (TERC). Post-neonatal human somatic cells repress telomerase expression [7, 8], although telomerase continues to be expressed in proliferative cells such as germ cells and stem cells. Most neoplastic cells de-repress telomerase expression to support immortalization and tumor formation [7, 9], although some neoplastic cells use telomere recombination or ALT [10] for the addition of telomere repeats without telomerase activity. High levels of telomere recombination characterize ALT cells [11]; both inter-telomeric [12] and intra-telomeric [13] recombination have been observed in immortalized human ALT cell lines, demonstrating that telomeres can use unique templates in their maintenance. ALT cells contain both linear and circular extrachromosomal telomeric repeats (ECTR) [14] that may represent other telomeric templates for recombination.
ALT cells are characterized by the ability to maintain telomeres in the absence of telomerase and by highly heterogeneous [10, 15, 16] telomere lengths [17, 18]. The majority of ALT cells contain ALT-associated promyelocytic (PML) bodies (APBs) that differ from PML bodies in other cell types by the inclusion of telomeric DNA and proteins [19]. ALT cells often display spontaneous DNA damage at the telomere, localized to foci called telomere-dysfunction-induced foci (TIF), and sometimes over-express mitochondrial regulators due to prevailing mitochondrial dysfunction [20]. Although a definitive enzymatic assay for ALT recombination has not been developed, ALT is characterized by heterogeneous telomere lengths, APBs, telomere recombination and/or ECTR.
The majority of human tumors express telomerase. However, the likelihood of one or the other telomere maintenance mechanism varies with specific tumor types—cancers that are mesenchymal in origin more frequently activate ALT, while carcinomas, epithelial in origin, more frequently express telomerase [21, 22]. Osteosarcomas have the highest incidence of ALT, with 59% of cases exhibiting ALT characteristics. Many, but not all, soft tissue sarcomas display ALT characteristics (27% overall), along with gastric carcinoma (38%), low-grade (grade 1–3) astrocytoma (37%), and diffuse malignant pleural mesothelioma (17%). The variation of ALT frequency associated with tumor type suggests that cell-specific differences favor one telomere maintenance mechanism over another. For example, mesenchymal stem cells express little to no detectable telomerase [23] and may predispose cells from this lineage to use ALT due to a continued chromatin-mediated repression of telomerase expression [24]. Although cell type-specific differences may exist, the question of how cells activate one mechanism over the other remains unresolved.
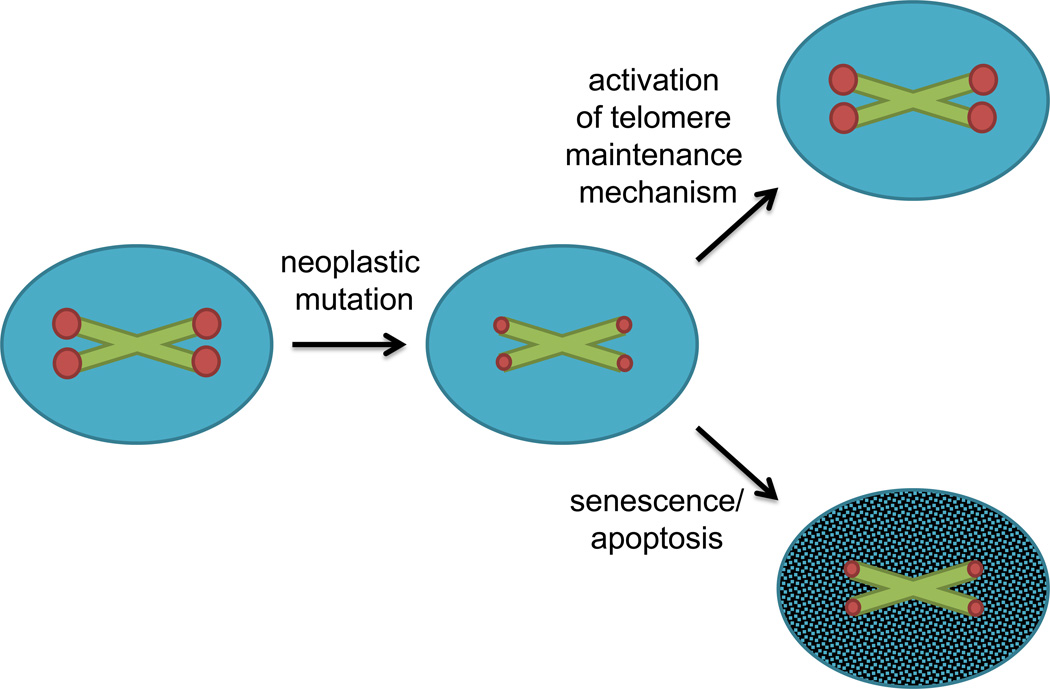
Experimental evidence from cell culture and mouse models demonstrates that following the primary genomic alterations that confer an increased proliferative capacity and a high tolerance for subsequent genomic alterations, neoplastic cells activate a telomere maintenance mechanism. Primary human adrenocortical cells transformed with H-RAS and SV40 form tumors following implantation in immunodeficient mice, although subsequent activation of telomerase is required to prevent crisis or the cellular senescence induced by telomere shortening [25]. In a hepatocellular carcinoma mouse model, genomic instability in the absence of telomerase expression favors tumor initiation, although these tumors display a limited ability for progression [26]. Telomere dysfunction in Terc−/−, p53+/− or Terc−/−, p53−/− cells is associated with a loss of cell cycle checkpoint control in mouse embryonic fibroblasts (MEFs) [27] and the promotion of epithelial tumors in mice [28]. These tumors exhibit greater genomic instability than tumors arising without telomere dysfunction [29], suggesting that telomere dysfunction may permit tumor initiation and promote the acquisition of additional mutations that drive tumor progression. Furthermore, histologically dysplastic but non-invasive regions of human skin, breast and colon cancers demonstrate colocalization of DNA damage markers with telomeres, although telomere dysfunction is absent in the adjacent malignant or invasive tumor [30]. These data support a model in which early mutations in neoplastic cells enhance proliferative capacity, genomic instability and telomere shortening, creating a bottleneck for the activation of a telomere maintenance mechanism to promote tumor cell immortalization (Figure 1). Activation of telomere maintenance represents a key target for limiting tumorigenesis.
Figure 1.
Neoplastic mutations increase proliferation and accelerate telomere shortening. Once telomeres shorten critically, a cell activates a telomere maintenance mechanism (telomerase or ALT) or it will undergo cell cycle arrest, senescence and/or apoptosis. The green X’s represent chromosomes and red dots represent telomeres at each end; blue circles represent proliferating cells, while the shaded blue circle represents a cell that has exited the cell cycle. The smaller red dots represent shortened telomeres.
2. How is ALT activated?
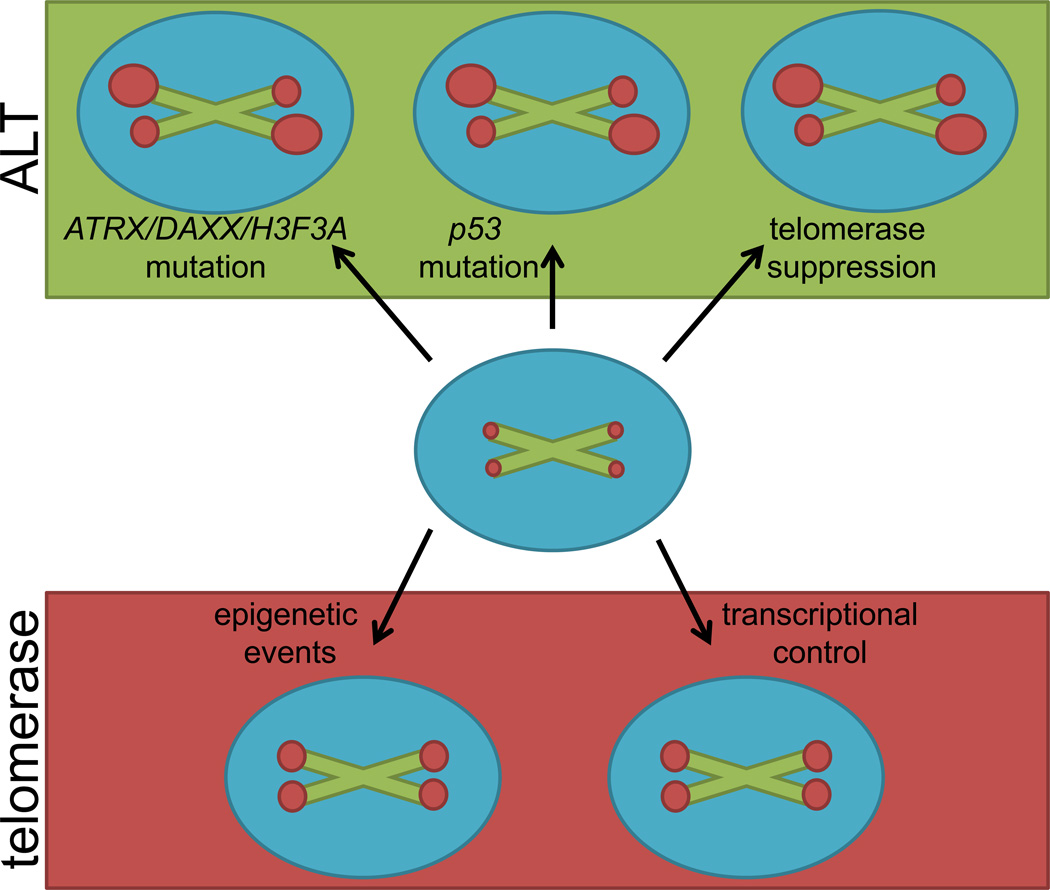
Specific genetic alterations in tumors can be associated with a higher likelihood for telomerase- or ALT-associated mechanisms of telomere maintenance (Figure 2). The regulation of genes encoding telomerase components has been extensively studied and is governed by complex interactions of transcription factors, signaling pathways [31] and epigenetic regulation [32]. These data suggest that mutations affecting telomerase regulation may facilitate ALT activation. Additionally, mutations associated with ALT activation are found in genes that suppress recombination, also facilitating recombination-mediated telomere maintenance. These genes include p53, ATRX, DAXX and H3F3A, the latter three encoding proteins that affect chromatin remodeling at the telomere.
Figure 2.
Critically short telomeres necessitate the activation of a telomere maintenance mechanism for immortalization. Activation of ALT (top, green) is estimated to occur in about 10% of human tumors. ALT may be initiated by mutations in ATRX, DAXX, H3F3A, p53 and/or through telomerase suppression; it results in heterogeneous telomere lengths. Telomerase expression (bottom, red) is present in about 90% of human tumors. De-repression of telomerase may be preceded by changes in epigenetic events and/or in transcriptional controls; it results in homogenous telomere lengths. The green X’s represent chromosomes with red dots representing telomeres at each end. The varying sizes of the dots represent heterogeneous or homogenous telomere lengths.
2.1 p53 mutation
Alteration or loss of function of the tumor suppressor p53 is a common event in many tumors and tumor-derived cell lines regardless of the telomere maintenance mechanism employed. In vitro and in vivo studies suggest, however, that impaired p53 may be associated with ALT activation. p53 is a DNA binding and signaling protein that alters transcription of target genes, controls cell cycle progression and facilitates DNA repair [33]. p53 mutation or an otherwise impaired p53 pathway is present in more than 95% of immortalized ALT cell lines [21]. Mechanistically, p53 mutations alter homologous recombination processes and could be permissive for ALT through a loss of recombination suppression [34]. In vitro reconstitution of expression of a transactivation-incompetent p53, unable to activate the transcription of its target genes but retaining the ability to suppress homologous recombination, inhibits DNA synthesis and cell proliferation in immortalized human p53-deficient ALT cell lines [35], suggesting that p53 mutations could permit ALT. Furthermore, hotspot mutations in codons 281, 273, 248, 175 or 143 of p53 are found in human tumors and functionally elevate recombination frequencies up to 26-fold in vitro [36].
Studies also suggest that p53 mutation in the context of telomere dysfunction allows cells to bypass checkpoint controls that would otherwise limit the proliferation of cells with shortened telomeres in vivo. Heterozygosity for p53 mutation in Tert−/− mouse models of hepatocellular carcinoma permits the formation of tumors in late generation mice, while wildtype p53 limits tumor progression [37]. Similar results were obtained using a K-RasG12D, Terc−/− lung cancer mouse model in which p53 heterozygosity is associated with tumor progression and metastasis, characteristics not associated with and presumably inhibited by wildtype p53 [38]. These mouse models suggest p53 mutation disrupts cell cycle checkpoint control and DNA damage signaling, and, in the absence of telomerase, permits tumor progression, likely through activation of ALT.
Clinical data demonstrate that p53 mutations may be permissive for ALT in human tumors as well. GBM patients with ALT-positive tumors have a longer median survival than GBM patients with telomerase-positive tumors [39]. Mutations of p53 are associated with ALT in human gliomas of the brain and spinal cord [40], a tumor type with a 26% incidence of ALT. The mutational status of p53 has been correlated with telomere maintenance mechanism in 110 patients with GBM: mutant p53 was identified in 14 of 18 ALT-positive tumors, 7 of 33 telomerase-positive tumors and 7 of 59 tumors without a confirmed telomere maintenance mechanism [40]. These p53-mutant tumors were sequenced and found to carry mutations in codons 273 and 248 [40]—two of the hotspot mutations that increase recombination frequencies in vitro [36]—suggesting that these mutations may permit ALT activation in human tumors. No differences in median survival between ALT-positive/mutated p53 and ALT-positive/wildtype p53 were observed, suggesting that the survival advantage in GBM is an ALT-associated effect. It should again be noted that p53 mutations are present in a high percentage of telomerase-positive tumors, with an incidence of 38%–50% in ovarian, esophageal, colorectal, larynx and lung tumors [41], a reminder that p53 has numerous tumor suppressor functions outside of those affecting telomere maintenance mechanisms.
2.2 ATRX, DAXX and H3F3A mutations
ATRX and DAXX form a heterodimeric chromatin-remodeling complex that modulates chromatin changes, including telomeric chromatin, during S-phase [42]. ATRX encodes the α-thalassemia mental retardation X-linked protein, a member of the Switch 2, sucrose non-fermenting 2 (SWI2/SNF2) family of helicases/ATPases that exhibits chromatin-remodeling activity. DAXX encodes the death-associated protein 6, another component of ATRX heterochromatin-positive regions of repetitive G-rich regions in the genome, such as telomeres [43]. The ATRX-DAXX complex is required for chromatin deposition of histone H3.3, a histone variant associated with transcriptionally active open chromatin, transcription factor binding sites and telomeres [44–47]. Mutations in H3F3A, encoding H3.3, may affect telomere dynamics by altering histone incorporation throughout the genome with subsequent changes in chromatin remodeling and gene expression. Loss of ATRX or DAXX would limit H3.3 incorporation into telomeric chromatin, in turn disrupting telomeric heterochromatin and facilitating telomere recombination.
ALT-associated mutations in ATRX and/or DAXX have been observed in vivo, with or without accompanying p53 mutations. An analysis of 41 human pancreatic neuroendocrine tumors (PanNETs) identified a significant correlation of ATRX or DAXX mutations with the ALT phenotype [48]. ALT-positive PanNETs negative for point mutations, insertions or deletions of ATRX or DAXX did not display nuclear expression of ATRX or DAXX. These observations emphasize a relationship between ATRX/DAXX mutation and ALT, although the mechanism by which ATRX/DAXX loss contributes to the selection or maintenance of ALT remains unclear. Analysis of other CNS tumor types shows that 3 of 21 pediatric GBM, 8 of 112 adult GBM, 1 of 13 oligodendrogliomas and 1 of 65 medulloblastomas—all cancer types with a prevalence of ALT—carry ATRX mutations. The ALT phenotype precisely correlated with ATRX mutation in these cases [48], although the sample size is limited.
ATRX and DAXX mutations are also present in pancreatic tumors from individuals with multiple endocrine neoplasia type 1 (MEN1) syndrome. These individuals harbor a germline mutation in MEN1, encoding a component of the histone methyltransferase complex protein menin, and are predisposed to PanNETs [49]. Loss of ATRX and/or DAXX expression in 3 of 50 MEN1 PanNETs correlated precisely with the ALT phenotype [50], although again the sample size is limited. Analyses of 22 human immortalized ALT cell lines from a variety of tumor types demonstrate that 16 of 22 have undetectable ATRX expression, with 6 cell lines harboring ATRX deletion. DAXX expression is low or undetectable in 4 of the 22 ALT cell lines, although no mutations or deletions were identified in DAXX [51].
Mutations of H3F3A occur in pediatric GBM tumors, often in combination with mutations of ATRX, DAXX and p53. Whole-exome sequencing of 48 pediatric GBMs identified 15 tumors with mutations in H3F3A that encode amino acid substitutions (K27M, G34R/G34V) within the histone tail; 21 tumors were characterized with at least one mutation in ATRX, DAXX or H3F3A. Heterozygous mutations in H3F3A affect protein regions at or extremely close to positions in the amino-terminal tail of H3.3, a post-translationally modified region tied to transcriptional repression or activation. p53 mutation was identified in 18 of the 21 tumors with ATRX, DAXX or H3F3A mutations [52], suggesting a potential synergy between these mutations; again, the numbers are small.
2.3 Telomerase regulation
ALT-permissive mutations include those that affect telomerase regulation. Recent work suggests that ALT arises when telomerase expression is inhibited and that ALT cells actively repress telomerase expression through complex signaling networks [20]. Gene expression profiling of various human tumor cell lines and human liposarcomas has associated a signature pattern of 297 genes that distinguishes telomerase-positive and ALT-positive cells [53]. Interaction models using gene signatures of ALT-positive cells predict TERT repression; correspondingly, TERT expression is low and the TERT repressor E2F1 is upregulated in ALT-positive cells in comparison to telomerase-positive cells [53].
Further network modeling studies of gene signatures have identified several signaling networks that impact telomere maintenance mechanism in cell lines including c-MYC, HNF4-α, p53, SP1 and STAT3 [54]. c-MYC activates telomerase by transcriptionally inducing TERT expression [55]. Telomerase-positive cells exhibit increased binding of c-MYC to the TERT promoter in comparison to ALT-positive cells. Conversely, the c-MYC competitive inhibitor TCEAL7 is upregulated in ALT cell lines and tumors [54], suggesting that a balance of these factors contributes to the determination of telomerase expression versus ALT activation.
2.4 Further considerations
Numerous studies have evaluated telomere maintenance mechanisms in different human tumor types [21, 22]. While the majority of these tumors are distinctly classified as telomerase-positive or ALT-positive, almost all studies have identified a small subset of tumors with characteristics of both or neither. These tumors exhibit telomerase activity and APBs, or telomere lengths of indeterminate size. It was unknown whether these characteristics represented distinct populations of tumors cells or cells that used both telomere maintenance mechanisms.
Our recent work demonstrates that some human sarcomas contain two types of tumor cells—those that express telomerase and those that use ALT [56]. Tumors were simultaneously analyzed for expression of the telomerase subunit TERT and APBs through colocalization of TRF2 and PML using the Nuance multispectral imaging system. Distinct cells expressing TERT and distinct cells with APBs were observed within the same tumor in 14/27 osteosarcomas, 3/4 osteoclastomas and 9/26 chondrosarcomas. Five fresh frozen osteosarcomas were evaluated using the telomere repeat amplification protocol (TRAP) assay to measure telomerase activity and telomere-specific Southern blotting to characterize telomere characteristics; three of these tumors were characterized by mixed phenotypes [56]. These results demonstrate that some tumors can be mosaic for cells using different telomere maintenance mechanisms. These data re-emphasize some intriguing questions: 1) are there some tumors that do not arise clonally; 2) can each tumor cell activate ALT or telomerase independently of its lineage; 3) can tumor cells actively switch between ALT mechanisms and telomerase; and 4) if so, how does this occur?
3. What proteins are required to maintain telomeres by ALT?
Once activated, ALT pathways require functions of many DNA repair proteins to maintain telomeres by recombination. These DNA repair proteins have other normal cellular functions—in replication, repair, recombination, chromatin remodeling, resolution of DNA intermediates and DNA processing—that are also useful in coordinating telomere recombination and maintaining telomere lengths. Some of these proteins are essential for telomere lengthening as their loss results in telomere shortening in ALT cells; others contribute to the maintenance of telomeres but are not required as their loss induces characteristics of telomere dysfunction (Table 1).
Table 1.
Many DNA repair proteins are involved in ALT. Loss of these proteins in ALT cells produces the telomere phenotypes listed. (APB=ALT-associated PML body; ss=single-stranded; ECTR=extrachromosomal telomeric repeats)
| Protein(s) | ALT cell line tested | Result |
|---|---|---|
| MRN complex (MRE11, RAD50, NBS1) | Saos-2 [61]; IIICF/c [61, 62]; GM847, Saos-2, WI-38 VA-13/2RA [63] | Loss of ALT characteristics and/or telomere shortening [61, 62, 63] |
| SMC5/6 complex (SMC5, SMC6, NSE1-NSE6) | U-2 OS, SUSM1 | Loss of ALT characteristics and/or telomere shortening; senescence in SUSM1 [64] |
| BLM | Saos-2, WI-38 VA-13/2RA | Loss of ALT characteristics and telomere shortening; growth arrest [65] |
| Topoisomerase IIIα | MRC5-V1, U-2 OS | Inhibited BLM and TRF2 stability, telomere dysfunction [66] |
| WRN | Saos-2, WI-38 VA-13/2RA, U-2 OS | Telomere shortening & loss of APBs in WI-38 VA-13/2RA; loss of APBs in U-2 OS; no effects in Saos-2 [67] |
| FEN1 | U-2 OS | Telomere dysfunction [69] |
| MUS81 | GM847, Saos-2, U-2 OS | Growth arrest, increased signal-free telomere ends, inhibition of telomere recombination [70] |
| RPA | U-2 OS, GM847 | Growth arrest, accumulation of ss telomere ends [71] |
| FANCD2 | GM847, U-2 OS | Telomere loss, inhibition of telomere recombination [72] |
| FANCA | GM847, U-2 OS | Telomere loss, inhibition of telomere recombination [72] |
| Ku70/80 heterodimer | CCL75.1 | Growth arrest, reduced ECTR [73] |
| XRCC3 | GM847, WI-38 VA-13/2RA | Reduced ECTR [63] |
| RAD51D | WI-38 VA-13/2RA | Telomere dysfunction, apoptosis [74] |
Depletion of the MRE11-RAD50-NBS1 (MRN) complex, the structural maintenance of chromosomes 5/6 (SMC5/6) complex, or the BLM helicase—all proteins or complexes involved in homologous recombination—results in telomere shortening, loss of APBs or alterations of other characteristics of ALT cell lines. The MRN complex functions in the detection, signaling and resolution of double-strand breaks throughout the genome. Suppression of the MRN complex in IIICF/c and Saos-2 immortalized human ALT cell lines by over-expression of SP100 results in shortened telomeres and loss of APBs [57]. Direct inhibition of only one component of the MRN complex, NBS1, by RNA-interference produces similar results [58] and also impairs the formation of ECTR in GM847, Saos-2 and WI-38 VA-13/2RA ALT cells [59]. The SMC5/6 complex promotes homologous recombination-mediated repair of DNA double strand breaks. The complex is required for localization of telomeric DNA to APBs in U-2 OS cells; RNA interference-mediated depletion results in shortened telomeres and cellular senescence in SUSM1 ALT cells [60]. BLM is a RecQ-like helicase that unwinds non-telomeric and telomeric DNA/DNA duplexes in vitro and DNA/RNA duplexes mimicking nucleolar substrates, and is required for the suppression of inter- and intra-chromosomal recombination in somatic cells. RNA interference-mediated inhibition of BLM induces telomere shortening, growth arrest and loss of APBs in Saos-2 and WI-38 VA-13/2RA ALT cells, but is dispensable for telomere maintenance in cells using telomerase [61]. In addition, immunoprecipitation and mass spectrometry of telomeric BLM complexes identified interactions with telomerase-associated protein 1 (TEP1), heat shock protein 90 (HSP90) and topoisomerase IIα in ALT cells but non telomerase-positive cells. Immunofluorescence demonstrates co-localization of these proteins with BLM and APBs; helicase assays demonstrate they modulate BLM helicase activity using telomeric substrates in vitro [61]. Depletion of topoisomerase IIIα by RNA interference reduces BLM and TRF2 protein levels, subsequently producing telomere dysfunction in MRC5-V1 and U-2 OS ALT cells [62]. In summary, these results demonstrate that the MRN complex, the SMC5/6 complex, and the BLM helicase directly participate in telomere recombination and are essential for ALT to maintain telomeres in the absence of telomerase.
The BLM helicase may be required to unwind telomeric DNA, allowing the MRN and SMC5/6 complexes access to telomeric DNA to promote homologous recombination. Normal somatic cells from persons with Bloom’s syndrome and lacking the BLM helicase are characterized by increased telomeric associations between homologous chromosome arms, suggesting an ability of telomeres to associate but not separate once strand invasion has occurred. Structural and functional similarities between BLM and the related RecQ-like WRN helicase do not predict the requirement for WRN in ALT processes. Additionally, the WRN helicase includes a domain with exonuclease function. WRN is a helicase that, like BLM, functions in numerous aspects of DNA repair and replication. In contrast, RNA interference-mediated depletion of WRN inhibits ALT in some, but not all, immortalized human ALT cell lines [63]. Long-term WRN knockdown in WI-38 VA-13/2RA and U-2 OS cells significantly reduces telomere length and inhibits APB formation. In Saos-2 cells, depletion of WRN does not inhibit cell growth, telomere lengthening or APB formation [63]. The results from Saos-2 are consistent with the observation of an ALT phenotype in an immortalized cell line from a person with Werner’s syndrome (WRN−/−) [64]. Data from these experiments suggest that WRN is required for telomere maintenance in some, but not all, ALT cell lines, and suggest a role for WRN in APB formation in some cells.
Other DNA repair proteins associated with ALT may not function directly in telomere recombination to maintain telomere lengths, as their loss does not result in telomere shortening. These proteins may function in other aspects of telomere maintenance such as cell cycle control, telomere damage signaling or structural integrity. These include flap endonuclease 1 (FEN1), MUS81, replication protein A (RPA), Fanconi anemia complementation group D protein 2 (FANCD2) and group A (FANCA), Ku70/80, X-ray repair cross-complementing protein 3 (XRCC3) and RAD51D. Results from deletion or knock-down experiments are summarized in Table 1 and remind us of the complexity of the processes that maintain the integrity of ALT telomeres independently of telomere elongation.
Some proteins listed in Table 1 may contribute to cell cycle control and/or ameliorate inherent telomeric dysfunction as their loss induces growth arrest of ALT cells. MUS81 is a DNA endonuclease that processes replication intermediates, suggesting normal functions in maintaining DNA replication fidelity. MUS81 loss induces growth arrest, increases signal-free telomere ends and inhibits telomere sister chromatid exchange in GM847, Saos-2 and U-2 OS ALT cells, although telomere shortening is not observed after more than three weeks of MUS81 reduction [65]. The telomeric functions of MUS81 are specific to ALT cells as MUS81 reduction does not affect telomerase-positive cells, suggesting this protein may also process telomere recombination intermediates. RPA binds to single stranded DNA to stabilize intermediate structures in DNA replication, repair and recombination. RPA knockdown induces growth arrest and accumulation of single-stranded telomeric DNA in U-2 OS and GM847 ALT cells [66], suggesting functions in telomere end processing. Knockdown of the Ku70/80 heterodimer, which binds to broken DNA ends to facilitate non-homologous end joining, impairs the growth of CCL75.1 ALT cells and reduces ECTRs, although telomere shortening does not occur [67]. Likewise, XRCC3, a component of the homologous recombination machinery that resolves Holliday junctions, is required for the maintenance of ECTRs in GM847 and WI-38 VA-13/2RA ALT cells [59], as its knock-down reduces ECTRs in these cell lines. ECTRs may reflect aberrant recombination at telomere ends, suggesting roles for Ku70/80 and XRCC3 in suppressing inappropriate recombination. MUS81, RPA, Ku70/80 and XRCC3 are not required for telomere length maintenance, but are clearly implicated in maintaining telomere dynamics to preserve the fidelity or structure of ALT telomeres, perhaps with consequent impact on cell growth.
Some proteins listed in Table 1 may maintain ALT telomere structure or coordinate other ALT processes as their loss increases telomeric dysfunction in ALT cells. RNA interference-mediated reduction of FEN1, a protein normally involved in processing DNA replication intermediates, in U-2 OS ALT cells increases the telomeric DNA damage response [68]. This study suggests that FEN1 contributes to processing telomeric DNA replication intermediates to promote faithful telomere replication. The FANC proteins, identified by the study of the various complementation groups of the autosomal recessive inherited disorder Fanconi anemia, are important modulators of base excision DNA repair and are required for genomic stability. FANCD2 functions in the pairing of homologous chromosomes during meiosis, double strand break repair and cell cycle checkpoint activation; FANCA may function in cell cycle checkpoint and resolution of interstrand crosslinks. Knockdown of FANCD2 or FANCA induces telomere loss and inhibits telomere sister chromatid exchange in GM847 and U-2 OS ALT cells [69]. Knockdown of RAD51D, another homologous recombination protein required to resolve Holliday junctions, in WI-38 VA-13/2RA ALT cells induces telomere dysfunction and apoptosis [70].
Experiments combining methionine restriction and RNA interference technologies show that PML, TRF1, TRF2, TIN2, RAP1 and the MRN complex are required for APB formation in IIICF/c ALT cells [71]. TRF2 is also required for ALT in U-2 OS cells, as loss induces telomere shortening and activates senescence [72]. The telomere-protective shelterin complex includes TRF1, TRF2, TIN2 and RAP1 and is required for telomere maintenance in all cells rather than specifically in ALT-positive cells. Other proteins are implicated in ALT through their co-localization with telomeric DNA in ALT cells; these proteins are discussed in Nabetani and Ishikawa [73] and will not be categorized here. Further studies will determine the functions of these proteins and how they may contribute to ALT mechanisms.
4. How does ALT impact tumor progression?
Activation of either telomere maintenance mechanism is required for tumor cell immortalization, although ALT and telomerase may be unequal in facilitating tumor progression. The ability of ALT to enhance tumor progression may be less than that of telomerase. Oncogenic H-RAS expression in immortalized human GM847 ALT fibroblasts does not confer transformation, as these cells cannot form tumors following injection into immunocompromised mice; however, expression of telomerase in these cells permits tumor formation [9]. Telomerase activity increases with tumor progression in multiple mouse models [74, 75], suggesting again that telomere maintenance mechanism may impact tumor growth and metastasis in vivo. Telomerase is required for tumor progression in some mouse models. Late generation mTerc−/−, Ink4a−/− mice display a reduced susceptibility to tumor formation; tumor cells from these mice display impaired growth and reduced colony formation in focus-forming assays that measure oncogenic potential. All in vitro and in vivo phenotypes can be reversed by mTerc expression [76]. Late generation mTerc−/−, Ink4a/Arf−/− mouse embryonic fibroblasts using ALT can form tumors following injection into immunocompromised mice, although these tumors lack metastatic potential. Reconstitution of mTerc promotes the formation of metastatic lesions [77].
Other mouse models demonstrate that faithful expression and activity of telomerase are required for progression of epithelial tumors. Prostate tumors in mTert−/− and mTert+/− mice with conditional prostate-specific Pten/p53 knockouts show that telomere dysfunction constrains the ability of tumors to progress beyond initiation: only mTert+/− tumors develop into invasive adenocarcinomas. Expression of an inducible mTert knock-in allele in mTert−/− mice after prostate tumor initiation decreases telomere damage signaling, increases tumor weight and confers tumor invasiveness [78]. Additional models demonstrate that telomerase reactivation allows tumor progression in an Atm−/− mouse model of T-cell lymphoma with inducible telomerase expression [20]. Inhibiting telomerase expression after tumor initiation in these mice initially inhibits tumor growth but eventually pressures tumor cells to activate ALT pathways to facilitate tumor progression. ALT tumors were characterized by mitochondrial dysfunction, upregulation of mitochondrial regulators and sensitivity to inhibition of these regulators [20], suggesting that ALT cells are characterized by unique signaling alterations. These data show that telomerase enhances tumorigenesis in vivo, but that its inhibition can favor the activation of ALT. These data also show that ALT cells possess the potential for tumor progression.
Telomerase may offer some advantage in tumor progression, although metastatic potential is not mutually exclusive with ALT. Considering that only 2/17 primary human bone tumors but 11/13 corresponding metastases exhibit telomerase activity, it is perhaps not surprising that tumors lacking telomerase activity correlate with greater patient survival [79]. Soft tissue sarcomas also display a skewed incidence of telomere maintenance mechanisms in primary versus metastatic lesions: while 16/24 primary tumors display ALT characteristics by immunohistochemistry, only 9/27 metastases display ALT characteristics [80]. Similar results are observed in liposarcomas, where 4/19 primary tumors but 10/17 metastases express telomerase [81]. Immunohistochemical studies of TERT in a human lung squamous cell carcinoma and its metastatic lesions demonstrated differential telomerase: the primary lung tumor displayed no telomerase expression, while a hilar lymph node metastasis displayed robust telomerase expression [82]. These studies suggest that telomerase expression may offer a metastatic advantage to human tumor cells.
Current clinical data suggest that telomere maintenance mechanism does not necessarily impact overall clinical outcome despite its associations with tumor progression. This is true of diffuse malignant peritoneal mesothelioma (n=38) [83], osteosarcoma (n=58; median duration of patient follow-up was 28 months) and soft tissue sarcoma (n=101) [80]. However, some studies have associated telomerase expression with shorter disease-free survival and lower overall survival in osteosarcomas (n=44; median duration of patient follow-up was 46.8 months) [84]. Telomerase activity is also correlated with poor overall survival in pediatric osteosarcomas (n=17) and pediatric Ewing’s sarcomas (n=11) [79]. ALT-positive GBM tumors are associated with longer median survival (n=77) [40], better overall survival and a 3-fold longer median survival (n=32) [80] than tumors with telomerase expression.
Conversely, other studies have associated ALT with poor prognosis. ALT-positive liposarcomas are associated with a worse prognosis than liposarcomas expressing telomerase (n= 93) [90], as are ALT-positive malignant fibrous histiocytomas (n=43) [85]. Neuroblastomas expressing high telomerase activity correlate with a lower patient survival, however tumors without telomerase activity and with elongated telomeres (presumably ALT) also correlate with a lower patient survival in comparison to tumors with shorter telomeres [86]. These anecdotal and inconsistent data suggest complex contributions of telomere maintenance mechanisms to clinical outcome. Tumor types and other mutational or histopathological variables may affect outcome in some of these examples, although these variables are often not analyzed in many published studies. Limited patient data or tumor numbers as well as technical approaches may confound accurate assessment of telomere maintenance mechanism and outcomes.
5. Conclusions
Telomere maintenance is critical to the immortalization of tumor cells and tumor progression. Published work strongly supports the hypothesis that telomere dysfunction in the context of oncogenic mutations are early events in cancer initiation that permit neoplastic cells to bypass cell cycle checkpoints and grow in an unrestricted fashion. Telomere shortening and DNA damage signaling ensues, leading to the activation of telomere maintenance mechanisms. Without such activation, telomeres would shorten to a critical point and push cells to enter crisis, senesce and/or apoptose. Telomerase expression in normal somatic cells is tightly regulated, although neoplastic cells can bypass this repression, activate telomerase expression and enzymatically maintain telomere length. Alternatively, neoplastic cells may activate recombination-associated mechanisms to maintain telomere lengths. Mechanisms activating telomerase or ALT are complex, although specific mutations may favor the establishment of one mechanism or the other. Once activated, ALT uses telomere recombination and DNA repair proteins to maintain telomeres. Immortalization, tumor progression and metastatic potential are not equally supported by telomerase-positive and ALT-positive cells, suggesting therapeutic implications for characterizing telomere maintenance mechanisms in tumors and for developing better understanding of these mechanisms.
Understanding how ALT processes facilitate cell survival during crisis and/or early events in tumor initiation or progression will illuminate therapeutic strategies to inhibit ALT. Telomerase inhibitors currently being tested in clinical trials may be more effective in some tumors than others, especially if some tumors are mosaic for telomere maintenance mechanisms. Genetically engineered mouse models of cancer demonstrate that if telomerase is repressed following telomerase-mediated tumor initiation, tumor cells activate ALT pathways [20], suggesting some flexibility with the commitment to a telomere maintenance mechanism. Tumor mosaicism for telomere maintenance mechanisms also can complicate therapeutic outcomes and may argue for combinations of ALT-directed and telomerase-directed therapies to treat some or all tumor types.
Highlights.
Neoplastic cells maintain telomeres by telomerase or ALT
Genetic mutations in p53, ATRX, DAXX or H3F3A may activate ALT
Many DNA repair proteins are involved in ALT
Tumor progression is favored by telomerase expression
Acknowledgements
The authors would like to acknowledge past and current members of the Groden laboratory and Drs. Samir Acharya, Gerard Nuovo and Hans Iwenofu for helpful discussions. We were supported by The Ohio State University Pelotonia Fellowship Program (AG), the National Science Foundation Graduate Research Fellowship 2011115452 (JH) and the National Institutes of Health CA-117898 (JG).
Abbreviations
- ALT
Alternative lengthening of telomeres
- APB
ALT-associated PML body
- ECTR
Extrachromosomal telomere repeat
- GBM
Glioblastoma multiforme
- MEN1
Multiple endocrine neoplasia type 1
- MRN
MRE11/RAD50/NBS1
- PanNET
Pancreatic neuroendocrine tumor
- TIF
Telomere dysfunction-induced focus
- TRAP
Telomere repeat amplification protocol
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest statement
The authors declare that they have no conflicts of interest that affect the presentation of these data and its discussion.
References
- 1.Moyzis RK, Buckingham JM, Cram LS, Dani M, Deaven LL, Jones MD, Meyne J, Ratliff RL, Wu JR. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc. Natl. Acad. Sci. 1988;85:6622–6626. doi: 10.1073/pnas.85.18.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 3.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 4.Harley CB, Futcher AB, Greider CW. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 5.Hastie ND, Dempster M, Dunlop MG, Thompson AM, Green DK, Allshire RC. Telomere reduction in human colorectal carcinoma and with aging. Nature. 1990;346:866–868. doi: 10.1038/346866a0. [DOI] [PubMed] [Google Scholar]
- 6.Lindsey J, McGill NI, Lindsey LA, Green DK, Cooke HJ. In vivo loss of telomeric repeats with age in humans. Mutat. Res. 1991;256:45–48. doi: 10.1016/0921-8734(91)90032-7. [DOI] [PubMed] [Google Scholar]
- 7.Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, Coviello GM, Wright WE, Weinrich SL, Shay JW. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 8.Wright WE, Piatyszek MA, Rainey WE, Byrd W, Shay JW. Telomerase activity in human germline and embryonic tissues and cells. Dev. Genet. 1996;18:173–179. doi: 10.1002/(SICI)1520-6408(1996)18:2<173::AID-DVG10>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 9.Stewart SA, Hahn WC, O’Connor BF, Banner EN, Lundberg AS, Modha P, Mizuno H, Brooks MW, Fleming M, Zimonjic DB, Popescu NC, Weinberg RA. Telomerase contributes to tumorigenesis by a telomere length-independent mechanism. Proc. Natl. Acad. Sci. USA. 2002;99:12606–12611. doi: 10.1073/pnas.182407599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bryan TM, Englezou A, Gupta J, Bacchetti S, Reddel RR. Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J. 1995;14:4240–4248. doi: 10.1002/j.1460-2075.1995.tb00098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Londono-Vallejo JA, Der-Sarkissian H, Cazes L, Bacchetti S, Reddel RR. Alternative lengthening of telomeres is characterized by high rates of telomeric exchange. Cancer Res. 2004;64:2324–2327. doi: 10.1158/0008-5472.can-03-4035. [DOI] [PubMed] [Google Scholar]
- 12.Dunham MA, Neumann AA, Fasching CL, Reddel RR. Telomere maintenance by recombination in human cells. Nat. Genet. 2000;26:447–450. doi: 10.1038/82586. [DOI] [PubMed] [Google Scholar]
- 13.Muntoni A, Neumann AA, Hills M, Reddel RR. Telomere elongation involves intra-molecular DNA replication in cells utilizing alternative lengthening of telomeres. Hum. Mol. Genet. 2009;18:1017–1027. doi: 10.1093/hmg/ddn436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cesare AJ, Griffith JD. Telomeric DNA in ALT cells is characterized by free telomeric circles and heterogeneous t-loops. Mol. Cell Biol. 2004;24:9948–9957. doi: 10.1128/MCB.24.22.9948-9957.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bryan TM, Englezou A, Dalla-Pozza L, Dunham MA, Reddel RR. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat. Med. 1997;3:1271–1274. doi: 10.1038/nm1197-1271. [DOI] [PubMed] [Google Scholar]
- 16.Grobelny JV, Godwin AK, Broccoli D. ALT-associated PML bodies are present in viable cells and are enriched in cells in the G2/M phase of the cell cycle. J. Cell Sci. 2000;113:4577–4585. doi: 10.1242/jcs.113.24.4577. [DOI] [PubMed] [Google Scholar]
- 17.Murnane JP, Sabatier L, Marder BA, Morgan WF. Telomere dynamics in an immortal human cell line. EMBO J. 1994;13:4953–4962. doi: 10.1002/j.1460-2075.1994.tb06822.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perrem K, Colgin LM, Neumann AA, Yeager TR, Reddel RR. Coexistence of alternative lengthening of telomeres and telomerase in hTERT-transfected GM847 cells. Mol. Cell Biol. 2001;21:3862–3875. doi: 10.1128/MCB.21.12.3862-3875.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yeager TR, Neumann AA, Englezou A, Huschtscha LI, Noble JR, Reddel RR. Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body. Cancer Res. 1999;59:4175–4179. [PubMed] [Google Scholar]
- 20.Hu J, Hwang SS, Liesa M, Gan B, Sahin E, Jaskelioff M, Ding Z, Ying H, Boutin AT, Zhang H, Johnson S, Ivanova E, Kost-Alimova M, Protopopov A, Wang YA, Shirihai OS, Chin L, DePinho RA. Antitelomerase therapy provokes ALT and mitochondrial adaptive mechanisms in cancer. Cell. 2012;148:651–663. doi: 10.1016/j.cell.2011.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henson JD, Reddel RR. Assaying and investigating alternative lengthening of telomeres activity in human cells and cancers. FEBS Lett. 2010;584:3800–3811. doi: 10.1016/j.febslet.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 22.Heaphy CM, Subhawong AP, Hong SM, Goggins MG, Montgomery EA, Gabrielson E, Netto GJ, Epstein JI, Lotan TL, Westra WH, Shih IM, Iacobuzio-Donahue CA, Maitra A, Li QK, Eberhart CG, Taube JM, Rakheja D, Kurman RJ, Wu TC, Roden RB, Argani P, De Marzo AM, Terracciano L, Torbenson M, Meeker AK. Prevalence of the alternative lengthening of telomeres telomere maintenance mechanism in human cancer subtypes. Am. J. Pathol. 2011;179:1608–1615. doi: 10.1016/j.ajpath.2011.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zimmermann S, Voss M, Kaiser S, Kapp U, Waller CF, Martens UM. Lack of telomerase activity in human mesenchymal stem cells. Leukemia. 2003;17:1146–1149. doi: 10.1038/sj.leu.2402962. [DOI] [PubMed] [Google Scholar]
- 24.Serakinci N, Hoare SF, Kassem M, Atkinson SP, Keith WN. Telomerase promoter regprogramming and interaction with general transcription factors in the human mesenchymal stem cell. Regen. Med. 2006;1:125–131. doi: 10.2217/17460751.1.1.125. [DOI] [PubMed] [Google Scholar]
- 25.Sun B, Huang Q, Liu S, Chen M, Hawks CL, Wang L, Zhang C, Hornsby PJ. Progressive loss of malignant behavior in telomerase-negative tumorigenic adrenocortical cell and restoration of tumorigenicity by human telomerase reverse transcriptase. Cancer Res. 2004;64:6144–6151. doi: 10.1158/0008-5472.CAN-04-1376. [DOI] [PubMed] [Google Scholar]
- 26.Farazi PA, Glickman J, Jiang S, Yu A, Rudolph KL, DePinho RA. Differential impact of telomere dysfunction on initiation and progression of hepatocellular carcinoma. Cancer Res. 2003;63:5021–5027. [PubMed] [Google Scholar]
- 27.Chin L, Artandi SE, Shen Q, Tam A, Lee S-L, Gottlieb GJ, Greider CW, DePinho RA. p53 deficiency rescues the adverse effects of telomere loss and cooperates with telomere dysfunction to accelerate carcinogenesis. Cell. 1999;97:527–538. doi: 10.1016/s0092-8674(00)80762-x. [DOI] [PubMed] [Google Scholar]
- 28.Artandi SE, Chang S, Lee S-L, Alson S, Gottlieb GJ, Chin L, DePinho RA. Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature. 2000;406:641–645. doi: 10.1038/35020592. [DOI] [PubMed] [Google Scholar]
- 29.O’Hagan RC, Chang S, Maser RS, Mohan R, Artandi SE, Chin L, DePinho RA. Telomere dysfunction provokes regional amplification and deletion in cancer genomes. Cancer Cell. 2002;2:149–155. doi: 10.1016/s1535-6108(02)00094-6. [DOI] [PubMed] [Google Scholar]
- 30.Suram A, Kaplunov J, Patel PL, Ruan H, Cerutti A, Boccardi V, Fumagalli M, Di Micco R, Mirani N, Gurung RL, Hande MP, d’Adda di Fagagna F, Herbig U. Oncogene-induced telomere dysfunction enforces cellular senescence in human cancer precursor lesions. EMBO J. 2012;31:2839–2851. doi: 10.1038/emboj.2012.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kyo S, Takakura M, Fujiwara T, Inoue M. Understanding and exploiting hTERT promoter regulation for diagnosis and treatment of human cancers. Cancer Sci. 2008;99:1528–1538. doi: 10.1111/j.1349-7006.2008.00878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu J, Zhao Y, Wang S. Chromatin and epigenetic regulation of the telomerase reverse transcriptase gene. Protein Cell. 2010;1:22–32. doi: 10.1007/s13238-010-0014-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sengupta S, Harris CC. p53: traffic cop at the crossroads of DNA repair and recombination. Nat. Rev. Mol. Cell. Biol. 2005;6:44–55. doi: 10.1038/nrm1546. [DOI] [PubMed] [Google Scholar]
- 34.Bertrand P, Rouillard D, Boulet A, Levalois C, Soussi T, Lopez BS. Increase of spontaneous intrachromosomal homologous recombination in mammalian cells expressing a mutant p53 protein. Oncogene. 1997;14:1117–1122. doi: 10.1038/sj.onc.1200931. [DOI] [PubMed] [Google Scholar]
- 35.Razak ZR, Varkonyi RJ, Kulp-McEliece M, Caslini C, Testa JR, Murphy ME, Broccoli D. p53 differentially inhibits cell growth depending on the mechanism of telomere maintenance. Mol. Cell. Biol. 2004;24:5967–5976. doi: 10.1128/MCB.24.13.5967-5977.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Akyüz N, Boehden GS, Süsse S, Rimek A, Preuss U, Scheidtmann KH, Wiesmüller L. DNA substrate dependence of p53-mediated regulation of double strand break repair. Mol. Cell. Biol. 2002;22:6306–6317. doi: 10.1128/MCB.22.17.6306-6317.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Farazi PA, Glickman J, Horner J, DePinho RA. Cooperative interactions of p53 mutation, telomere dysfunction, and chronic liver damage in hepatocellular carcinoma progression. Cancer Res. 2006;66:4766–4773. doi: 10.1158/0008-5472.CAN-05-4608. [DOI] [PubMed] [Google Scholar]
- 38.Perera SA, Maser RS, Xia H, McNamara K, Protopopov A, Chen L, Hezel AF, Kim CF, Bronson RT, Castrillon DH, Chin L, Bardeesy N, DePinho RA, Wong K-K. Telomere dysfunction promotes genome instability and metastatic potential in a K-ras p53 mouse model of lung cancer. Carcinogenesis. 2008;29:747–753. doi: 10.1093/carcin/bgn050. [DOI] [PubMed] [Google Scholar]
- 39.Hakin-Smith V, Jellinek DA, Levy D, Carroll T, Teo M, Timperley WR, McKay MJ, Reddel RR, Royds JA. Alternative lengthening of telomeres and survival in patients with glioblastoma multiforme. Lancet. 2003;361:836–838. doi: 10.1016/s0140-6736(03)12681-5. [DOI] [PubMed] [Google Scholar]
- 40.Chen Y, Hakin-Smith V, Teo M, Xinarianos GE, Jellinek DA, Carroll T, McDowell D, MacFarlane MR, Boet R, Baguley BC, Braithwaite AW, Reddel RR, Royds JA. Association of mutant TP53 with alternative lengthening of telomeres and favorable prognosis in glioma. Cancer Res. 2006;66:6473–6576. doi: 10.1158/0008-5472.CAN-06-0910. [DOI] [PubMed] [Google Scholar]
- 41.Olivier M, Hollstein M, Hainaut P. TP53 mutations in human cancers: origins, consequences, and clinical utility. Cold Spring Harb. Perspect. Biol. 2010;2:a001008. doi: 10.1101/cshperspect.a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tang J, Wu S, Liu H, Stratt R, Barak OG, Shiekhattr R, Picketts DJ, Yang X. A novel transcription regulatory complex containing death domain-associated protein and the ATR-X syndrome protein. J. Biol. Chem. 2004;279:20369–20377. doi: 10.1074/jbc.M401321200. [DOI] [PubMed] [Google Scholar]
- 43.Law MJ, Lower KM, Voon HPJ, Hughes JR, Garrick D, Viprakasit V, Mitson M, de Gobbi M, Marra M, Morris A, Abbott A, Wilder SP, Taylor S, Santos GM, Cross J, Ayyub H, Jones S, Ragoussis J, Rhodes D, Dunham I, Higgs DR, Gibbons RJ. ATR-X syndrome protein targets tandem repeats and influences allele-specific expression in a size-dependent manner. Cell. 2010;143:367–378. doi: 10.1016/j.cell.2010.09.023. [DOI] [PubMed] [Google Scholar]
- 44.Lewis P, Elsaesser S, Noh K, Stadler S, Allis CD. DAXX is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc. Natl. Acad. Sci. USA. 2010;107:14075–14080. doi: 10.1073/pnas.1008850107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goldberg AD, Banaszynski LA, Noh K, Lewis PW, Elsaesser SJ, Stadler S, Dewell S, Law M, Guo X, Li X, Wen D, Chapgier A, DeKelver RC, Miller JC, Lee Y, Boydston EA, Holmes MC, Gregory PD, Greally JM, Rafii S, Yang C, Scambler PJ, Garrick D, Gibbons RJ, Higgs DR, Cristea IM, Urnov FD, Zheng D, Allis CD. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140:678–691. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ahmad K, Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell. 2002;9:1191–2000. doi: 10.1016/s1097-2765(02)00542-7. [DOI] [PubMed] [Google Scholar]
- 47.Wong L, Ren H, Williams E, McGhie J, Ahn S, Sim M, Tam A, Earle E, Anderson M, Mann J, Choo KHA. Histone H3.3 incorporation provides a unique and functionally essential telomeric chromatin in embryonic stem cells. Genome Res. 2009;19:404–414. doi: 10.1101/gr.084947.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Heaphy CM, de Wilde RF, Jiao Y, Klein AP, Edil BH, Shi C, Bettegowda C, Rodriguez FJ, Eberhart CG, Hebbar S, Offerhaus GJ, McLendon R, Rasheed BA, He Y, Yan H, Bigner DD, Oba-Shinjo SM, Marie SKN, Riggins GJ, Kinzler KW, Vogelstein B, Hruban RH, Maitra A, Papadopoulos N, Meeker AK. Altered telomeres in tumors with ATRX and DAXX mutations. Science. 2011;333:6041. doi: 10.1126/science.1207313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thakker RV. Multiple endocrine neoplasia type 1 (MEN1) Best Pract. Res. Clin. Endocrinol. Metab. 2010;24:355–370. doi: 10.1016/j.beem.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 50.de Wilde RF, Heaphy CM, Maitra A, Meeker AK, Edil BH, Wolfgan CL, Ellison TA, Schulick RD, Molenaar IQ, Valk GD, Vriens MR, Borel Rinkes IHM, Offerhaus GJA, Hruban RH, Matsukuma KE. Loss of ATRX or DAXX expression and concomitant acquisition of the alternative lengthening of telomeres phenotype are late events in a small subset of MEN-1 syndrome pancreatic neuroendocrine tumors. Mod. Pathology. 2012;25:1033–1039. doi: 10.1038/modpathol.2012.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lovejoy CA, Li W, Reisenweber S, Thongthip S, Bruno J, de Lange T, De S, Petrini JH, Sung PA, Jasin M, Rosenbluh J, Zwang Y, Weir BA, Hatton C, Ivanova E, Macconaill L, Hanna M, Hahn WC, Lue NF, Reddel RR, Jiao Y, Kinzler K, Vogelstein B, Papadopoulos N, Meeker AK. Loss of ATRX, genome instability, and an altered DNA damage response are hallmarks of the alternative lengthening of telomeres pathway. PLoS Genet. 2012;8:e1002772. doi: 10.1371/journal.pgen.1002772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schwartzentruber J, Korshunov A, Liu X-Y, Jones DTW, Pfaff E, Jacob K, Sturm D, Fontebasso AM, Quang D, Tönjes M, Hovestadt V, Albrecht S, Kool M, Nantel A, Konermann C, Lindroth A, Jäger N, Rausch T, Ryzhova M, Korbel JO, Hielscher T, Hauser P, Garami M, Klekner A, Bognar L, Ebinger M, Schuhmann MU, Scheurlen W, Pekrun A, Frühwald MC, Roggendorf W, Kramm C, Dürken M, Atkinson J, Lepage P, Montpetit A, Zakrzewska M, Zakrzewski K, Liberski PP, Dong Z, Siegel P, Kulozik AE, Zapatka M, Guha A, Malkin D, Felsberg J, Reifenberger G, von Deimling A, Ichimura K, Collins VP, Witt H, Milde T, Witt O, Zhang C, Castelo-Branco P, Lichter P, Faury D, Tabori U, Plass C, Majewski J, Pfister SM, Jabado N. Driver mutations in histone H3.3 and chromatin remodeling genes in pediatric glioblastoma. Nature. 2012;484:130. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 53.Lafferty-Whyte K, Cairney CJ, Will MB, Serakinci N, Daidone M-G, Zaffaroni N, Bilsland A, Keith WN. A gene expression signature classifying telomerase and ALT immortalization reveals an hTERT regulatory network and suggests a mesenchymal stem cell origin for ALT. Oncogene. 2009;28:3765–3774. doi: 10.1038/onc.2009.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lafferty-Whyte K, Bilsland A, Hoare SF, Burns S, Zaffaroni N, Cairney CJ, Keith WN. TCEAL7 inhibition of c-MYC activity in alternative lengthening of telomeres regulates hTERT expression. Neoplasia. 2010;12:405–414. doi: 10.1593/neo.10180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu KJ, Grandori C, Amacker M, Simon-Vermot N, Polack A, Lingner J, Dalla-Favera R. Direct activation of TERT transcription by c-MYC. Nat. Genet. 1999;21:220–224. doi: 10.1038/6010. [DOI] [PubMed] [Google Scholar]
- 56.Gocha ARS, Nuovo G, Iwenofu OH, Groden J. Human sarcomas are mosaic for telomerase-dependent and –independent telomere maintenance mechanisms: implications for telomere-based therapies. Am. J. Pathol. (in review) doi: 10.1016/j.ajpath.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jiang WQ, Zhong ZH, Henson JD, Neumann AA, Chang AC, Reddel RR. Suppression of alternative lengthening of telomeres by Sp100-mediated sequestration of the MRE11/RAD50/NBS1 complex. Mol. Cell. Biol. 2005;25:2708–2721. doi: 10.1128/MCB.25.7.2708-2721.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhong ZH, Jiang WQ, Cesare AJ, Neumann AA, Wadhwa R, Reddel RR. Disruption of telomere maintenance by depletion of the MRE11/RAD50/NBS1 complex in cells that use alternative lengthening of telomeres. J. Biol. Chem. 2007;282:29314–29322. doi: 10.1074/jbc.M701413200. [DOI] [PubMed] [Google Scholar]
- 59.Compton SA, Choi JH, Cesare AJ, Ozgür S, Griffith JD. Xrcc3 and Nbs1 are required for the production of extrachromosomal telomeric circles in human alternative lengthening of telomere cells. Cancer Res. 2007;67:1513–1519. doi: 10.1158/0008-5472.CAN-06-3672. [DOI] [PubMed] [Google Scholar]
- 60.Potts PR, Yu H. The SMC5/6 complex maintains telomere length in ALT cancer cells through SUMOylation of telomere-binding proteins. Nat. Struct. Mol. Biol. 2007;14:581–590. doi: 10.1038/nsmb1259. [DOI] [PubMed] [Google Scholar]
- 61.Bhattacharyya S, Keirsey J, Russell B, Kavecansky J, Lillard-Wetherell K, Tahmaseb K, Turchi JJ, Groden J. Telomerase-associated protein 1, HSP90, and topoisomerase IIα associate directly with the BLM helicase in immortalized cells using ALT and modulate its helicase activity using telomeric DNA substrates. J. Biol. Chem. 2009;284:14966–14977. doi: 10.1074/jbc.M900195200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Temime-Smaali N, Guittat L, Wenner T, Bayart E, Douarre C, Gomez D, Giraud-Panis MJ, Londono-Vallejo A, Gilson E, Amor-Guéret M, Riou JF. Topoisomerase IIIα is required for normal proliferation and telomere stability in alternative lengthening of telomeres. EMBO J. 2008;27:1513–1524. doi: 10.1038/emboj.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gocha ARS, Acharya S, Groden J. A variable requirement for the WRN helicase in human cell lines using ALT suggests multiple mechanisms of telomere maintenance in the absence of telomerase, in preparation [Google Scholar]
- 64.Fasching CL, Bower K, Reddel RR. Telomerase-independent telomere length maintenance in the absence of alternative lengthening of telomeres-associated promyelocytic leukemia bodies. Cancer Res. 2005;65:2722–2729. doi: 10.1158/0008-5472.CAN-04-2881. [DOI] [PubMed] [Google Scholar]
- 65.Zeng S, Xiang T, Pandita TK, Gonzalez-Suarez I, Gonzalo S, Harris CC, Yang Q. Telomere recombination requires the MUS81 endonuclease. Nat. Cell Biol. 2009;11:616–623. doi: 10.1038/ncb1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Grudic A, Jul-Larsen A, Haring SJ, Wold MS, Lønning PE, Bjerkvig R, Bøe SO. Replication protein A prevents accumulation of single-stranded telomeric DNA in cells that use alternative lengthening of telomeres. Nucleic Acids Res. 2007;35:7267–7278. doi: 10.1093/nar/gkm738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li B, Reddy S, Comai L. Depletion of Ku70/80 reduces the levels of extrachromosomal telomeric circles and inhibits proliferation of ALT cells. Aging. 2011;3:395–406. doi: 10.18632/aging.100308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Saharia A, Stewart SA. FEN1 contributes to telomere stability in ALT-positive tumor cells. Oncogene. 2009;28:1162–1167. doi: 10.1038/onc.2008.458. [DOI] [PubMed] [Google Scholar]
- 69.Fan Q, Zhang F, Barrett B, Ren K, Andreassen PR. A role for monoubiquitinated FANCD2 at telomeres in ALT cells. Nucleic Acids Res. 2009;37:1740–1754. doi: 10.1093/nar/gkn995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tarsounas M, Muñoz P, Claas A, Smiraldo PG, Pittman DL, Blasco MA, West SC. Telomere maintenance requires the RAD51D recombination/repair protein. Cell. 2004;117:337–347. doi: 10.1016/s0092-8674(04)00337-x. [DOI] [PubMed] [Google Scholar]
- 71.Jiang WQ, Zhong ZH, Henson JD, Reddel RR. Identification of candidate alternative lengthening of telomeres genes by methionine restriction and RNA interference. Oncogene. 2007;26:4635–4647. doi: 10.1038/sj.onc.1210260. [DOI] [PubMed] [Google Scholar]
- 72.Stagno D’Alcontres M, Mendez-Bermudez A, Foxon JL, Royle NJ, Salomoni P. Lack of TRF2 in ALT cells causes PML-dependent p53 activation and loss of telomeric DNA. J. Cell Biol. 2007;179:855–867. doi: 10.1083/jcb.200703020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nabetani A, Ishikawa F. Alternative lengthening of telomeres pathway: recombination-mediated telomere maintenance mechanism in human cells. J. Biochem. 2011;149:5–14. doi: 10.1093/jb/mvq119. [DOI] [PubMed] [Google Scholar]
- 74.Bednarek A, Budunova I, Slaga TJ, Aldaz CM. Increased telomerase activity in mouse skin premalignant progression. Cancer Res. 1995;55:4566–4569. [PubMed] [Google Scholar]
- 75.Blasco MA, Rizen M, Greider CW, Hanahan D. Differential regulation of telomerase activity and telomerase RNA during multi-stage tumorigenesis. Nat. Genet. 1996;12:200–204. doi: 10.1038/ng0296-200. [DOI] [PubMed] [Google Scholar]
- 76.Greenberg RA, Chin L, Femino A, Lee K-H, Gottlieb GJ, Singer RH, Greider CW, DePinho RA. Short dysfunctional telomeres impair tumorigenesis in the INK4aΔ2/3 cancer-prone mouse. Cell. 1999;97:515–525. doi: 10.1016/s0092-8674(00)80761-8. [DOI] [PubMed] [Google Scholar]
- 77.Chang S, Khoo CM, Naylor ML, Maser RS, DePinho RA. Telomere-based crisis: functional differences between telomerase activation and ALT in tumor progression. Genes Dev. 2003;17:88–100. doi: 10.1101/gad.1029903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ding Z, Wu C-J, Jaskelioff M, Ivanova E, Kost-Alimova M, Protopopov A, Chu GC, Wang G, Lu X, Labrot ES, Hu J, Wang W, Xiao Y, Zhang H, Zhang J, Zhang J, Gan B, Perry SR, Jiang S, Li L, Homer JW, Wang YA, Chin L, DePinho RA. Telomerase reactivation following telomere dysfunction yields murine prostate tumors with bone metastases. Cell. 2012;148:1–12. doi: 10.1016/j.cell.2012.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sotillo-Piñeiro E, Sierrasesúmaga L, Patiño-García A. Telomerase activity and telomere length in primary and metastatic tumors from pediatric bone cancer patients. Pediatr. Res. 2004;55:231–235. doi: 10.1203/01.PDR.0000102455.36737.3C. [DOI] [PubMed] [Google Scholar]
- 80.Henson JD, Hannay JA, McCarthy SW, Royds JA, Yeager TR, Robinson RA, Wharton S, Jellinek DA, Arbuckle SM, Yoo J, Robinson BG, Learoyd DL, Stalley PD, Bonar SF, Yu D, Pollock RE, Reddel RR. A robust assay for alternative lengthening of telomeres in tumors shows the significance of alternative lengthening of telomeres in sarcomas and astrocytomas. Clin. Cancer Res. 2005;11:217–225. [PubMed] [Google Scholar]
- 81.Costa A, Daidone MG, Daprai L, Villa R, Cantù S, Pilotti S, Mariani L, Gronchi A, Henson JD, Reddel RR, Zaffaroni N. Telomere maintenance mechanisms in liposarcomas: association with histologic subtypes and disease progression. Cancer Res. 2006;66:8918–8924. doi: 10.1158/0008-5472.CAN-06-0273. [DOI] [PubMed] [Google Scholar]
- 82.Hiyama E, Hiyama K, Yokoyama T, Shay JW. Immunohistochemical detection of telomerase (hTERT) protein in human cancer tissues and a subset of cells in normal tissues. Neoplasia. 2001;3:17–26. doi: 10.1038/sj.neo.7900134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Villa R, Daidone MG, Motta R, Venturini L, De Marco C, Vannelli A, Kusamura S, Baratti D, Deraco M, Costa A, Reddel RR, Zaffaroni N. Multiple mechanisms of telomere maintenance exist and differentially affect clinical outcome in diffuse malignant peritoneal mesothelioma. Clin. Cancer Res. 2008;14:4134–4140. doi: 10.1158/1078-0432.CCR-08-0099. [DOI] [PubMed] [Google Scholar]
- 84.Sanders RP, Drissi R, Billups CA, Daw NC, Valentine MB, Dome JS. Telomerase expression predicts unfavorable outcome in osteosarcoma. J. Clin. Oncol. 2004;22:3790–3797. doi: 10.1200/JCO.2004.03.043. [DOI] [PubMed] [Google Scholar]
- 85.Matsuo T, Shay JW, Wright WE, Hiyama E, Shimose S, Kubo T, Sugita T, Yasunaga Y, Ochi M. Telomere-maintenance mechanisms in soft-tissue malignant fibrous histiocytomas. J. Bone Joint Surg. Am. 2009;91:928–937. doi: 10.2106/JBJS.G.01390. [DOI] [PubMed] [Google Scholar]
- 86.Onitake Y, Hiyama E, Kamei N, Yamaoka H, Sueda T, Hiyama K. Telomere biology in neuroblastoma: telomere binding proteins and alternative strengthening of telomeres. J. Pediatr. Surg. 2009;44:2258–2266. doi: 10.1016/j.jpedsurg.2009.07.046. [DOI] [PubMed] [Google Scholar]