Abstract
Mutations in Notch2, Jagged1 or homologs of the Hairy-related transcriptional repressor Hey2 cause congenital malformations involving the non-chamber atrioventricular canal (AVC) and inner curvature (IC) regions of the heart, but the underlying mechanisms have not been investigated. By manipulating signaling directly within the developing chick heart, we demonstrated that Notch2, Hey1 and Hey2 initiate a signaling cascade that delimits the non-chamber AVC and IC regions. Specifically, misactivation of Notch2 signaling, or misexpression of either Hey1 or Hey2, repressed Bmp2. Because Jagged (also known as Serrate in non-mammalian species) ligands were found to be present in prospective chamber myocardium, these data support the model that Notch2 and Hey proteins cause the progressive restriction of Bmp2 expression to within the developing AVC and IC, where it is essential for differentiation. Misactivation or inhibition of Notch2 specifically induced or inhibited Hey1, respectively, but these manipulations did not affect Hey2, implicating Hey1 as the direct mediator of Notch2. Bmp2 within the developing AVC and IC has been shown to induce Tbx2, and we found that Tbx2 misexpression inhibited the expression of both Hey1 and Hey2. Tbx2, therefore, is envisaged to constitute a feedback loop that sharpens the border with the developing AVC and IC by delimiting Hey gene expression to within prospective chamber regions. Analysis of the loss-of-function phenotype in mouse embryos homozygous for targeted disruption of Hey2 revealed an expanded AVC domain of Bmp2. Similarly, zebrafish gridlock (Hey2 homolog) mutant embryos showed ectopic expression of Bmp4, which normally marks AVC myocardium in this species. Thus, Hey pathway regulation of cardiac Bmp appears to be an evolutionarily conserved mechanism to delimit AVC and IC fate, and provides a potential mechanistic explanation for cardiac malformations caused by mutations in Serrate/Jagged1 and Notch signaling components.
Keywords: Notch, Hairy-related transcription factor, HRT, HES, Hey, Gridlock, Atrioventricular canal, T-box, Tbx
Introduction
During early embryogenesis, the linear heart [HH stage 10 in the chick (Hamburger and Hamilton, 1951), E8 in the mouse] develops atrial and ventricular chamber myocardium as the heart tube loops (HH stage 13-16 in chick; E8.5-E9.0 in mouse) (reviewed by Fishman and Chien, 1997; Srivastava and Olson, 2000). Chamber myocardium goes on to acquire high gap-junction density, high conduction velocity, and, in ventricular myocardium, pronounced trabeculae. By contrast, the atrioventricular canal (AVC) and inner curvature (IC) regions proliferate more slowly and preserve the automaticity, poor intercellular coupling and slow contraction characteristics of the embryonic myocardium of the tubular heart (de Jong et al., 1992; Delorme et al., 1997; Icardo and Fernandez-Teran, 1987). These distinctive structural and conduction characteristics are essential for heart function and approximately 5% of all congenital heart defects (CHDs) in the newborn are attributed to improper development of AVC and IC-derived tissue (reviewed by Moorman and Christoffels, 2003).
Models for specification of chamber versus AVC and IC myocardial fate have begun to emerge that depend on the expression of specific combinations of transcription factors (Abdelwahid et al., 2001; Chan-Thomas et al., 1993; Davis et al., 2001; Habets et al., 2002; Kupershmidt et al., 1999; Yamada et al., 2000). In particular, chamber-specific programs of gene expression depend on a cooperative interaction between Tbx5 and the cardiac transcription factor Nkx2.5, whereas Tbx2 inhibits Tbx5-dependent activation of chamber gene expression in the AVC and IC (Habets et al., 2002; Harrelson et al., 2004; Plageman and Yutzey, 2004; Stennard et al., 2003). Consequently, in the absence of Tbx2, the AVC and IC regions fail to constrict and express early chamber markers (Harrelson et al., 2004). Tbx20 also cooperates with Tbx5 and Tbx2 in the regulation of chamber fate (Brown et al., 2005; Cai et al., 2005; Singh et al., 2005; Stennard et al., 2005; Takeuchi et al., 2005). Emerging evidence indicates that localized expression of Bmp2 within the developing AVC and IC directs the spatiotemporal pattern of Tbx2 transcription within the tubular heart (Yamada et al., 2000). Very little information exists on the signals that establish the spatial patterns of transcription factor expression that distinguish chamber from non-chamber myocardium other than Bmp, and the upstream signals that control Bmp2 expression in the AVC and IC are largely unknown. Therefore, we tested whether Notch and Hey proteins are involved in the control of Bmp2 and, hence, AVC and IC identity.
Notch receptors encode transmembrane proteins that control numerous cell fate decisions during vertebrate embryogenesis through cell-cell interactions (for a review, see Artavanis-Tsakonas et al., 1999). In mammals, four Notch receptors (Notch1-Notch4) recognize two classes of transmembrane ligands (Lardelli et al., 1994; Uyttendaele et al., 1996; Weinmaster et al., 1991; Weinmaster et al., 1992): Serrate [Serrate1 and 2, commonly termed Jagged in mammals (Lindsell et al., 1995; Shawber et al., 1996)] and Delta [comprising Delta-like1, 3 and 4 (Bettenhausen et al., 1995; Dunwoodie et al., 1997; Shutter et al., 2000)]. For canonical Notch signaling, the extracellular binding of ligand induces a presenilin-dependent cleavage of Notch that releases its intracellular domain (NotchICD) into the cytoplasm, where it forms a complex with the DNA-binding protein CSL (CBF-1, Suppressor of Hairless, and Lag-2) (Honjo, 1996; Jarriault et al., 1995; Jeffries et al., 2002; Tamura et al., 1995) that in turn activates the transcription of target genes, such as the Hairy/Enhancer of Split (HES) family of transcriptional repressors, and the related Hey1 and Hey2 genes (see below).
Homozygous CSL−/− mutants (Oka et al., 1995) and presenilin1−/−, presenilin2−/− double mutants (Donoviel et al., 1999) die during mid-gestation with widespread cell death and underdeveloped hearts, in particular a thin, hypoplastic ventricular wall and impaired trabeculation. Overtly similar defects are seen in homozygous Notch2 hypomorphs (Hamada et al., 1999; McCright et al., 2001) and Notch1−/− (Conlon et al., 1995; Swiatek et al., 1994) embryos. Because CHDs often arise as consequences of malformations elsewhere in the developing embryo, the direct effects of Notch on myocardial development are not resolved by these systemic knockouts. Nonetheless, in humans, attenuated Notch activation causes Alagille syndrome (AGS), an autosomal dominant disorder characterized by CHDs, as well as cholestasis, vertebral and eye abnormalities, distinctive facial features, renal disease and growth retardation (Krantz et al., 1997). The CHDs range from mild to severe and can include tetralogy of Fallot, pulmonary artery stenosis, pulmonary atresia, truncus arteriosis, and ventricular and atrial septal defects. Jagged1 mutations leading to haploinsufficiency cause the syndrome in humans (Li et al., 1997; Oda et al., 1997) and AGS is phenocopied in the mouse by a reduction of Notch2 function in a heterozygous Jagged1+/null background (McCright et al., 2002).
Hey1 and Hey2 (also known as HRT2, CHF1, HESR2, HERP1 and gridlock) are highly expressed in the developing heart and vasculature (Chin et al., 2000; Kokubo et al., 1999; Leimeister et al., 1999; Nakagawa et al., 1999; Zhong et al., 2000). In mammals, these proteins are often referred to as Hairy-related transcriptional repressors (HRT1 and HRT2, respectively) and the zebrafish homolog of Hey2 is the product of gridlock. Hey proteins have conserved basic helix-loop-helix (bHLH) and orange domains, and a YRPW motif near the C terminus that differs from the WRPW motif found in the structurally related HES proteins (reviewed by Fischer and Gessler, 2003; Iso et al., 2003). Disruption of Hey2 in mice causes ventricular septal defects that are occasionally accompanied by other CHDs and cardiomyopathy (Donovan et al., 2002; Gessler et al., 2002; Sakata et al., 2002). Hey1−/− mice lack apparent cardiovascular phenotypes, but the spectrum of Hey2−/− anomalies is extended in double Hey1/Hey2 homozygous mutants to include a ubiquitous angiogenic remodeling deficit and strongly impaired arterial endothelial development, leading to death by E10.5 (Fischer et al., 2004).
Here, we show that Hey1 and Hey2 repress Bmp2 transcription in chick myocardium, thereby delimiting expression to within the developing AVC and IC as heart looping occurs. Hey-mediated repression of cardiac Bmp genes appears to be evolutionarily conserved in mice and zebrafish, as we observed ectopic cardiac Bmp transcription in mouse and zebrafish embryos with a disrupted Hey2 homolog. In addition, using the chick system, we provide evidence for a feedback mechanism whereby Tbx2, which is induced by Bmp2, inhibits both Hey1 and Hey2 expression, and we propose that this mechanism sharpens the border of Bmp2 transcription where chamber myocardium abuts the AVC and IC. Although both Hey1 and Hey2 suppress early cardiac Bmp2, we find that only Hey1 responds to Notch2 in chick hearts, thus leading to the model that segregation of prospective atrial and ventricular myocardium from AVC and IC regions by Hey proteins occurs in both a Notch-dependent and -independent manner.
Materials and Methods
Organ culture and electroporations
Chick eggs were obtained from commercial sources and were incubated at 38.5°C for 30-56 hours until they reached HH stage 10-16 [staging according to Hamburger and Hamilton (Hamburger and Hamilton, 1951)]. Embryos were explanted in Tyrode's solution, and the heart surgically removed with a sharpened Tungsten needle. Hearts were then placed in 1 × PBS with 200 ng/μl expression plasmid and transferred to a BTX372 electrode chamber (Genetronics). An ECM 830 Electro Square Porator was used to generate square pulses (2 × 20 V, pulse length 50 mseconds) before reversing electrode poles and repeating. Hearts were then incubated in 5% CO2 at 37°C for 24 hours in culture media [α-MEM (Gibco), supplemented with 15% donor calf serum (Gibco), 5% chick serum (Gibco), 75 U/ml penicillin/streptomycin (Gibco), 10 mM HEPES buffer (Omega Scientific), and 0.3% L-glutamine (Gibco)]. Hearts were then fixed in 4% paraformaldehyde for 2 hours at room temperature and stored in 100% methanol at −20°C until they were processed for histology.
Expression plasmids
Each cDNA was cloned into pRS2, a modified pCS2 expression vector, in which the CMV promoter element was excised and an RSV promoter element cloned into the SalI/HindIII sites upstream of the multi-cloning site. Each cDNA was inserted into the pRS2 multi-cloning site as follows: pRS2-CSL-VP16, CSL-VP16-Myc, BamHI/XbaI (generous gift of J. Sklar, Brigham and Women's Hospital, MA, USA); pRS2-GFP (Myc-GFP, BamHI/XbaI); pRS2-Mfng, Man3HA, HindIII/XbaI (generous gift of T. Vogt, Merck Research Laboratories, West Point, PA, USA); pRS2-Hey2, FlagHrt2, HindIII/XhoI (generous gift of D. Srivastava, University of California, San Francisco, CA, USA); and pRS2-N2ICD, BamHI/StuI (generous gift of T. Maciag). To fuse PCR cloned mouse Hey1 to a 5′ Myc epitope in the pRS2 vector, R-GFP was cut with EcoRI and XbaI to excise GFP, and mouse Hey1 was inserted in frame at these sites.
To clone chick Hey2, HES and Notch cDNA, total RNA was prepared from HH stage 12–16 chick heart explants using TriZol reagent (Gibco) and the following degenerate primer pairs, where B=C,G,T; D=A,G,T; H=A,C,T; M=A,C; N=A,C,G,T; R=A,G; V=A,C,G; W=A,T;Y=C,T:
Hey2, 5′-CCCGCGGATCCAAYAGYTTGTCTGARCTG-3′ and 5′-CCCCCGCGGCCGCCCAAGGYCTRTAGGGYTT-3′;
Hes, 5′-CCCGCGGATCCAAYAGYTTGTCTGARCTG-3′ or 5′-CCC-GCGGATCCCARATGACWGTGGAYCAC-3′, and 5′-CCCCCGCGG-CCGCGTGRTCCACWGTCATYTG-3′ or 5′-CCCCCGCGGCCGCCC-AAGGYCTRTAGGGYTT-3′; and
Notch, 5′-TGATBCTYGGCHRCBMGVYTGGCHGT-3′ and 5′-TTM-ACVGCNGCNGCCCARTGNARDGC-3′.
To clone mouse Hey1, PCR was performed on the Mouse Brain Marathon-Ready cDNA library (Clontech). The 5′ primer (5′-ATG-AAGCGCCCTTGTGAGG-3′) includes the mouse Hey1 translational start codon, and the 3′ primer (5′-TTAAAAGGCTCCAACTTCTG-3′) includes the mouse Hey1 stop codon UAA.
In situ hybridization and epitope staining
Chick heart whole-mount in situ hybridization was performed as described previously (Levin et al., 1995), with the following modifications for combined in situ hybridization and immunohistochemistry. On day 1, after rehydration to PBT (PBS+0.1% TWEEN), hearts were incubated in RIPA wash with primary monoclonal antibodies against epitope tags for 45 minutes, followed by three 5-minute washes in PBT, a second 45-minute RIPA wash with Cy3-conjugated AffiniPure goat anti-mouse IgG diluted 1:100 (Jackson ImmunoResearch), and two 5-minute washes in PBT before fixation in 4% paraformaldehyde and 0.2% glutaraldehyde. Primary antibodies were anti-c-Myc 9E10 (Santa Cruz Biotechnology), anti-V5 antibody (Invitrogen), anti-HA HA.11 monoclonal antibody (Covance) and anti-FLAG M2 (Sigma), and each was diluted 1:100 in RIPA, with the exception of HA.11, which was diluted 1:1000 for immunohistochemistry.
Riboprobe templates for electroporated chick heart in situ hybridization were amplified from plasmids by PCR to eliminate the detection of plasmid DNA sequences. Each 3′ primer was made with an embedded T7 promoter element. Primer pairs were:
Bmp2, 5′-ATGTTCGGGCTGAAGC-3′ and 5′-GTAATACGACTCAC-TATAGGGCGATACACTCGCGGTG-3′;
Hey1, 5′-ATCATCGAGAAGCGCCGCCGCGACCGCATC-3′ and 5′-GAATTCTAATACGACTCACTATAGGGAGGACCGATCTCAGTCCC-3′;
Hey2, 5′-AACAGTTTGTCTGAGCTGAGGCGGCTGGTG-3′ and 5′-GAATTCTAATACGACTCACTATAGGGACACAGGAAGCAACGCTG-3′; and
Amhc, 5′-CGACGAGCGGGTCCAGCTTCTCCACTCC-3′ and 5′-GA-AAAATAATACGACTCACTATAGGGAGGCACCTTGACACGCCGC-3′.
Chick cDNAs for Cx42 (E. C. Beyer, University of Connecticut, Farmington, CT, USA) and Irx4 (C. Cepko, Harvard Medical School, Boston, MA, USA) were inserted in pBS, and Anf was PCR amplified from cDNA and inserted into pGEM T-easy. Both plasmids have T7 and T3 promoters flanking the inserts, so corresponding commercial primers were obtained for amplification of the template (IDT). All probes were transcribed using T7 RNA polymerase, with the exception of Cx42, which was transcribed with T3 RNA polymerase. Riboprobes for Serrate1, Serrate2 and Delta-like1 were made as described (Henrique et al., 1995; Laufer et al., 1997; Myat et al., 1996).
Mouse embryo whole-mount in situ hybridization was carried out as described previously (Leimeister et al., 1998). The riboprobe for mouse Bmp2 was generated from a PCR fragment (GCGGGATCCGTTTGGC-CTGAAGCAGAGAC, GCGGAATTCTGACGCTTTTCTCGTTTGTG) cloned into pCS2. Zebrafish whole-mount in situ hybridization was performed as described previously (Zhong et al., 2000). The Bmp4 riboprobe was as described (Chen et al., 1997).
Quantitative analyses
Quantification of the incidence of cells exhibiting altered gene expression after electroporation in each case reflects cumulative data over greater than three independent experiments. Identifying sample details were encoded prior to embedding and histological sections were scored by collaborators lacking knowledge of the experiments. Individual cells that expressed the electroporated transgenes (epitope-specific immunostain) were scored as having reduced, unchanged or elevated levels of the endogenous gene (in situ hybridization stain) relative to immediately neighboring cells. Thus, a cell in the midst of a patch of electroporated cells might show reduced expression relative to non-electroporated cells, but would be scored as unchanged relative to surrounding electroporated cells; thus, the analysis underestimates the magnitude of the effects observed.
The atrioventricular length of the Bmp2 expression domain in mouse E9.5-E10.0 embryos was performed using ImageJ (National Institutes of Health, http://rsb.info.nih.gov/ij). AVC length was determined as a straight line in the center of the AVC. Head length was measured along a line drawn from the anterior limit of the olfactory pit to the back surface of the head, intersecting the posterior limit of the mesencephalon.
In situ hybridization and epitope staining
Following culture, samples were processed for combined in situ hybridization, to detect endogenous gene expression, and immunohistochemistry, to detect the epitope-tagged proteins expressed from the electroporated pRSV construct. For immunohistochemistry, embryos were fixed in 4% paraformaldehyde for 30 minutes at room temperature. Rabbit polycolonal antibody against Notch2icd was used (Novus) with an alkaline phosphatase-conjugated secondary antibody, and detection was with BCIP/NBT a substrate.
Results
Dynamic expression patterns of Notch2, Serrate1 and Serrate2 demarcate the borders of the atrioventricular canal and inner curvature
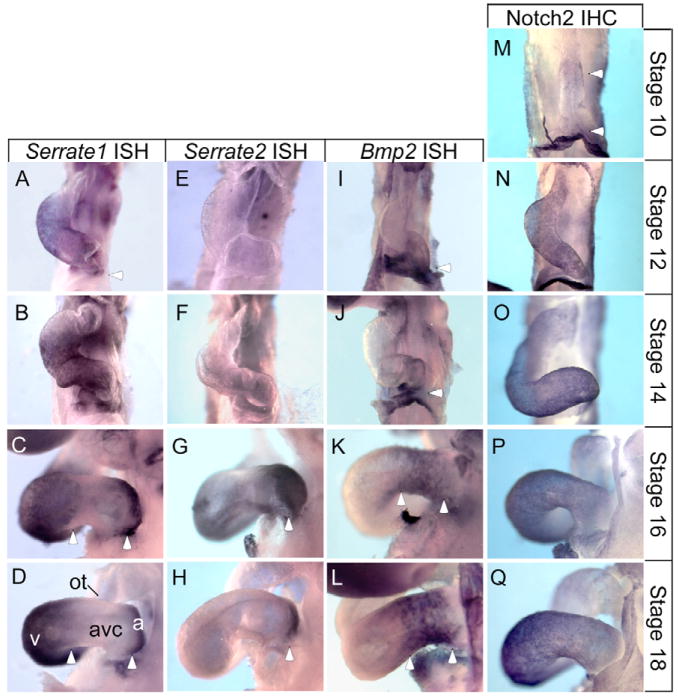
Three Notch ligands have been identified in the chick: Serrate1, Serrate2 (Hayashi et al., 1996; Myat et al., 1996) and Delta-like-1 (Henrique et al., 1995). Serrate1 mRNA was detected as early as HH stage 10 throughout the heart tube (not shown), but by stage 12 expression was absent in the developing atrium/sinus venosus region (Fig. 1A). Between stages 14 and 16, expression recedes from the AVC forming an abrupt boundary at both anterior and posterior borders, with Serrate1 expression continuing to mark mature myocardium of the ventricle and atrium (Fig. 1B,C). Notably, Serrate1 expression was less robust in the IC as well. Serrate1 maintained strong atrial and ventricular expression at all stages examined (up to stage 24, Fig. 1; data not shown). Serrate2 mRNA was detected in the developing heart from stage 16 (Fig. 1E-H), in the distal portion of the atrium overlapping the expression of Serrate1, where it remained at all stages examined. Delta-like1 was not detected at any stage surveyed (not shown).
Fig. 1. Serrate1 and Serrate2 gene expression patterns correspond to the borders of the AVC and indicate prospective atrial and ventricular chamber myocardium.
(A-D) Stage 12-18 embryos were hybridized with a Serrate1 antisense RNA probe. (A) Stage 12 embryos exhibit widespread Serrate1 expression throughout the posterior heart with the anterior boundary at the outflow tract. The posterior border stops at the presumptive atrium and sinus venosus, but may include primitive AVC tissue (white arrowhead). (B,C) Between stages 14 and 16 expression becomes restricted to the common ventricle and atria with boundaries at the AVC (white arrowheads, C). (D) At stage 18, expression is strong in the ventricles and atria, forming distinct boundaries at both the anterior and posterior AVC border (white arrowheads). (E-H) Stage 12-18 embryos were hybridized with a Serrate2 antisense RNA probe. (E,F) At early stages, 12 and 14, Serrate2 is not detected in the heart. (G) Serrate2 expression becomes apparent by stage 16 at the posterior-most region of the heart, corresponding to the distal atrium and sinus venosus (white arrowhead). (H) By stage 18, strong Serrate2 expression is observed in the atria and sinus venosus, overlapping that of Serrate1 (white arrowhead). (I-L) Stage 12-18 embryos were hybridized with the AVC marker Bmp2 for comparison with the Notch ligands. (I,J) Bmp2 is expressed in the posterior-most region of the heart, including the primitive AVC, atria and sinus venosus. The most anterior Bmp2 expression appears to correspond to the posterior border of Serrate1 expression (compare arrowheads in A,B and I,J). (J,K) Between stages 14 and 16, Bmp2 expression is restricted to the AVC. Again, Bmp2 expression appears complementary to that of Serrate1 in the primitive ventricle, and to both Serrate1 and Serrate2 in the atria (compare arrowheads in K with C,G). (L) Bmp2 mRNA persists in the AVC. (M-Q) Immunostaining against Notch2. (M) Notch2 is first detected in the linear heart tube in small patches in the ventral primitive ventricle and at the junction of the two horns of the primitive atria (white arrowheads). (N-Q) By Stage 12, Notch2 is present throughout the myocardium, where it remains at all stages examined. a, atrial region; avc, atrioventricular canal; ot, outflow tract; v, ventricular region.
By stage 16, the Serrate expression patterns become complementary to that of Bmp2, which marks the developing AVC and IC (compare Fig. 1K,L with Fig. 1C,D,G,H). Thus, Serrate might distinguish chamber from AVC and IC myocardium by controlling the spatial domain of Bmp2 expression.
Two Notch receptor homologs, Notch1 and Notch2, have been identified in chick (Hamada et al., 1999; Myat et al., 1996); however, only Notch2 was detected in the myocardium of the heart between HH stages 10 through 20 and is thus likely to mediate the Serrate signal. Low levels of Notch2 were first detected in the stage 10 linear heart tube, in particular within a smattering of cells in the ventral, presumptive ventricle and the center of the fusing, bilateral atria (Fig. 1M). As looping proceeds, prominent Notch2 expression encompasses the entire heart tube and persists through at least stage 20 (Fig. 1N-Q). Notch1 was not detected in the myocardium at these stages, as examined by immunohistochemistry, in situ hybridization or RT-PCR analysis (not shown).
Activation of the Notch pathway suppresses Bmp2 gene expression
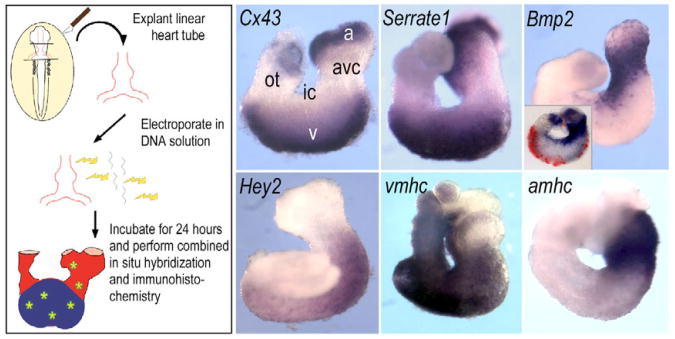
A whole-heart in vitro electroporation system was devised to study the effects of misexpressed Notch pathway components (Fig. 2). Explanted chick hearts from HH stage 12 embryos were electroporated with RSV promoter-based expression plasmids to direct production of exogenous proteins and then cultured for 24 hours to the point when age-matched embryos had developed to between HH stages 19 and 22. A panel of region-specific markers confirmed appropriate development of the hearts after the procedure. Exogenous protein was detected by immunohistochemistry against an epitope tag, and concurrent in situ hybridization revealed the effect on endogenous marker gene expression.
Fig. 2. Plasmid electroporation of linear heart tubes.
Stage 12 hearts were dissected, electroporated with RSV promoter cDNA constructs to direct production of epitope-tagged Notch pathway modulators or GFP (see Materials and methods). During the subsequent 24-hour culture period, the electroporated hearts looped and developed appropriate expression patterns of region-specific markers, including Cx43, vmhc and amhc in chamber myocardium, as well as Bmp2 in the AVC. In addition, Serrate1 and Hey2 showed characteristic expression patterns. Inset in Bmp2 panel shows Cy3 immunofluorescent detection of the Myc-epitope tag-labelled product of transfected eGFP gene. a, atrial region; avc, atrioventricular canal; ic, inner curvature; ot, outflow tract; v, ventricular region.
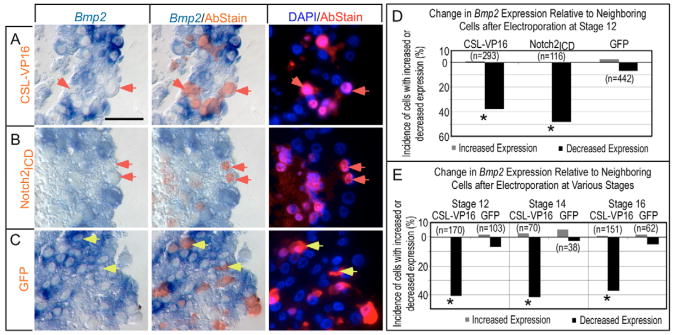
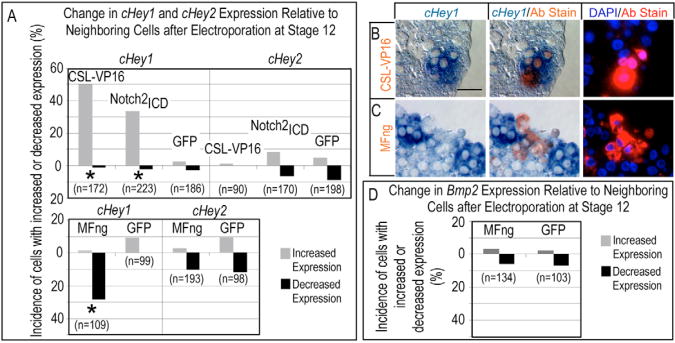
Paraffin sections of electroporated hearts revealed that ectopic Notch pathway activation in the AVC and IC suppressed Bmp2 expression. Either a Myc-tagged, human CSL DNA-binding region-VP16 transactivator (CSL-VP16) or a V5 epitope-tagged Notch2ICD (Small et al., 2003) cell-autonomously decreased the level of Bmp2 mRNA relative to that in surrounding cells in the AVC and IC (Fig. 3A,B). The response was quantified by scoring transfected cells as having either increased, decreased or similar levels of Bmp2 relative to their surrounding neighbors, revealing reduced expression in 37.9% and 48.3% of CSL-VP16- or Notch2ICD-expressing cells, respectively (Fig. 3D). By contrast, a control plasmid containing GFP fused to a Myc tag (GFP) did not elicit a reproducible change in Bmp2 expression (Fig. 3C,D). Notch activation suppressed Bmp2 with similar efficacy at stages 12, 14 and 16 (Fig. 3E), indicating that competence persists throughout this time window and might contribute to the progressive shift in endogenous Serrate and Bmp2 expression patterns.
Fig. 3. Notch pathway agonists inhibit Bmp2 expression cell autonomously in the AVC and IC.
(A-C) Histological sections demonstrating that misactivation of Notch signaling suppresses Bmp2. Immunohistochemical staining (red fluorescence) detected epitopes on expressed proteins from the electroporated cDNAs: CSL-VP16 (A), Notch2ICD (B) or GFP control (C). Brightfield images on the left show in situ hybridization (blue). Cy3 (red) and DAPI (blue) fluorescent images are shown on right. The center panels show brightfield images merged with the fluorescent Cy3 (red) staining of the epitope-tagged proteins. Red arrows indicate examples of cells with reduced Bmp2 relative to neighbors, whereas yellow arrows indicate examples of unchanged cells. Scale bar in A: 50 μM. (D,E) Quantification of cells scored for altered gene expression relative to neighboring cells after electroporation with the indicated cDNAs, scored from histological sections as in A-C. All sample identifiers were encoded prior to histological embedding so that tissue processing and scoring was performed blind (see Materials and methods). Data plotted are cumulative over greater than three trials. Note that Notch pathway activation suppresses Bmp2 expression (D) and that Bmp2 remains responsive throughout the period of heart looping (HH stages 12-16; E). Asterisks indicate statistical significance of the difference between the experimental modulation of Notch pathway and the control GFP (P<0.05, χ2 test).
It is important to emphasize that all samples in this and subsequent experiments were stripped of experimental identifiers and the identities encoded prior to embedding, so that histological preparation and scoring were performed blind. Moreover, our nearest neighbor analysis of single cells under-represented the magnitude of changes in gene expression, because cells in the midst of a cluster of transfected cells that all showed a similar response were scored as unchanged with respect to their neighbors, even though the entire cluster would be changed relative to untransfected cells.
Cardiac expression of Hey1 and Hey2 suppresses Bmp2
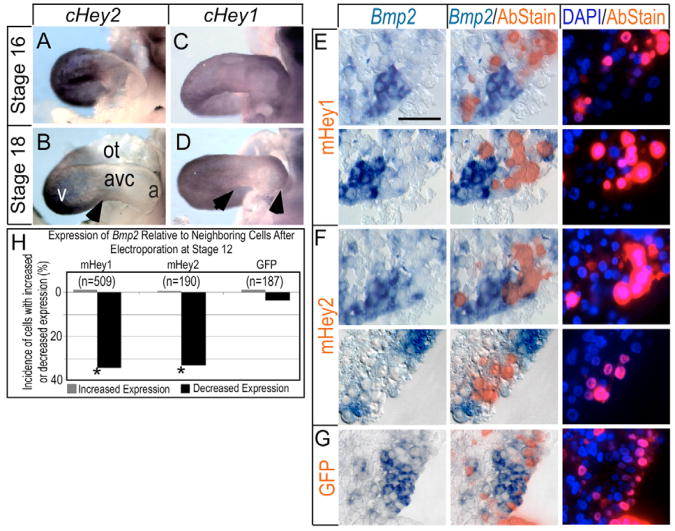
Endogenous Hey1 and Hey2 transcripts are localized to overlapping regions of Serrate1 and Serrate2 expression in the developing chick heart (compare Fig. 1 with Fig. 4) and are absent in the intervening AVC. Hey2 expression was limited to the myocardium of the developing ventricle at all stages examined, in a similar pattern to that described in mouse (Fig. 4A,B). Hey1 was expressed not only in the atrium, as in the mouse, but also in the myocardial wall of the ventricle (Fig. 4C,D). We were unable to detect other members of the HES family of bHLH proteins in the developing myocardium using RT-PCR and specific and degenerate oligonucleotide primer sets (not shown), and are unaware of reports of other HES family members in the myocardium of the linear heart. We therefore examined whether Hey proteins mediate Notch2 function in AVC and IC patterning. Misexpression of N-terminal Myc-tagged mouse Hey1 by electroporation into HH stage 12 hearts, as before, and cultured for 24 hours, repressed Bmp2 in 34.4% of the cells relative to neighbors (Fig. 4E,H). Hey1 and Hey2 show a 51.3% sequence identity at the protein level, with greater similarity in the bHLH and orange domains. Both proteins act as transcriptional repressors and recognize similar DNA motifs (Chin et al., 2000; Fischer and Gessler, 2003). As for Hey1, electroporation of Flag-tagged mouse Hey2 reduced Bmp2 expression in 33.2% of transfected cells, relative to neighbors (Fig. 4F,H). Thus, both Hey1 and Hey2 suppress Bmp2.
Fig. 4. Ectopic Hey1 and Hey2 suppress Bmp2 cell autonomously in the AVC and IC.
(A-D) Examples of endogenous Hey2 and Hey1 mRNA expression in stage 16 and 18 embryos. Arrowheads denote the borders of mRNA expression. (A,B) Hey2 expression is restricted to the developing ventricles at all stages examined, similar to patterns reported for the mouse. (C) Low levels of Hey1 are detected throughout the myocardium of stage 16 chick hearts. (D) By stage 18, Hey1 is detected in atrial and ventricular myocardium, but not AVC, as for expression of Serrate1 (see Fig. 1). Arrowheads indicate the borders of Hey gene expression with the AVC. (E-G)Bmp2 suppression by exogenously provided Myc-tagged mouse Hey1 and Flag-tagged mouse Hey2. Immunohistochemical staining (red fluorescence) detected epitopes on expressed proteins from the electroporated cDNAs, as in Fig. 3: Hey1 (E), Hey2 (F) or GFP control (G). Scale bar in E: 50 μM. (H) Quantification was performed as for Fig. 3. Asterisk indicates statistical significance of a difference between the effect of Hey proteins and GFP control (P<0.05, χ2 test).
Notch directly activates Hey1, but not Hey2
Electroporation of HH stage 12 hearts with Notch pathway agonists CSL-VP16 and Notch2ICD induced strong activation of endogenous Hey1 in the AVC and IC myocardium, with relative increases occurring in 50.0% and 33.6% of cells, respectively (Fig. 5A,B), implicating Notch signaling in direct activation of Hey1. Upregulation was also apparent in the ventricular region over endogenous expression levels, implying a dose-response mechanism (not shown). As before, misexpression of the control plasmid containing GFP had no effect (Fig. 5A). Surprisingly, endogenous Hey2 transcripts were not induced by Notch pathway agonists (Fig. 5A). To test the effect of inhibiting Serrate/Notch2 signaling cell autonomously, we next introduced the Manic Fringe glycosyltransferase, which inhibits responsiveness to Serrate ligands in a highly selective and cell-autonomous manner (Shimizu et al., 2001; Weinmaster and Kintner, 2003). HA-tagged mouse Mfng expressed following electroporation into stage 12 hearts significantly reduced the level of endogenous Hey1 transcripts in chamber myocardium (28.4% of transfected cells exhibited lowered levels of expression relative to nearest neighbors), whereas GFP misexpression had no effect (Fig. 5A,C). Hey2 was unaffected by Notch pathway antagonists (Fig. 5A). These experiments indicate differential regulation of Hey1 and Hey2 in the linear heart tube.
Fig. 5. Differential effects of Notch on Hey1, Hey2 and Bmp2.
Stage 12 hearts were electroporated to misexpress modulators of the Notch pathway signaling, cultured for 24 hours and analyzed for effects on endogenous gene expression, as described in Fig. 3. (A) Activation of the Notch pathway with Myc-tagged CSL-VP16 or Notch2icd increased the number of cells that expressed endogenous Hey1, relative to neighboring cells, and attenuation of Notch with HA-tagged Mfng caused a decrease in Hey1-positive cells. By contrast, Notch pathway agonists and antagonists did not affect Hey2 expression. (B,C) Histological sections demonstrating that misactivation of the Notch pathway (B) or inhibition of endogenous Notch responsiveness by Mfng (C) elicited reciprocal and cell-autonomous effects on endogenous Hey1. Scale bar: 25 μm. (D) Attenuation of Notch signaling with Mfng did not cause a cell-autonomous effect on the expression of Bmp2, consistent with the observations that Hey2 is not regulated directly by Notch (A) yet suppresses Bmp2 (see Fig. 4). Asterisks in A,D indicate statistical significance of the difference between the Notch pathway modulators and the GFP control (P<0.05, χ2 test).
From the preceding results, we predicted that loss of Notch alone should not cause the Bmp2 expression domain to expand into the ventricular myocardium because of persistent Hey2 expression. Accordingly, Mfng, which suppressed Hey1, failed to induce ectopic myocardial Bmp2 transcripts (Fig. 5D). We conclude, therefore, that Notch2 signaling involving Hey1, as well as Notch2-independent signaling involving Hey2, delimit the AVC and IC domain of Bmp2 transcription.
Notch and Hey proteins do not regulate the identity of chamber myocardium directly
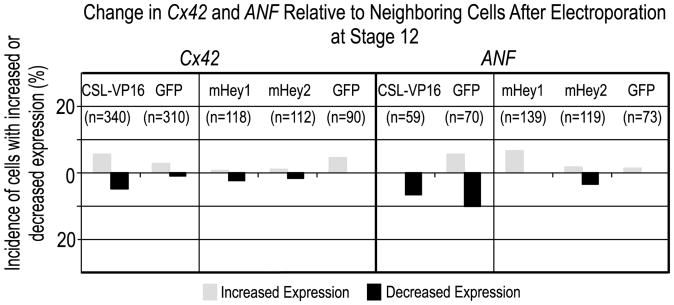
Lineage choice by Notch can occur by suppressing and/or promoting differentiation, depending on context. Two early chamber markers, Cx42 and Anf, were examined to determine whether Notch2-mediated suppression of AVC and IC fate was accompanied by a reciprocal stimulation of chamber identity, which would be apparent by cell-autonomous, ectopic activation of these genes in the AVC and IC region. Neither ectopic Notch pathway activation, nor ectopic mouse Hey1 or Hey2 affected either gene (Fig. 6). Thus, Notch2 in the myocardium suppresses AVC and IC fate, but another signal must specify chamber identity.
Fig. 6. Ectopic Notch2 signaling does not cause expansion of early chamber marker expression into the developing AVC or IC.
Neither Anf nor Cx42 was induced following ectopic Notch pathway activation by misexpression of Myc-tagged CSL-VP16 in stage 12 hearts or by Flag-tagged mouse Hey1 and mouse Hey2. Analysis was as for preceding experiments.
Repression of Hey genes by Tbx2
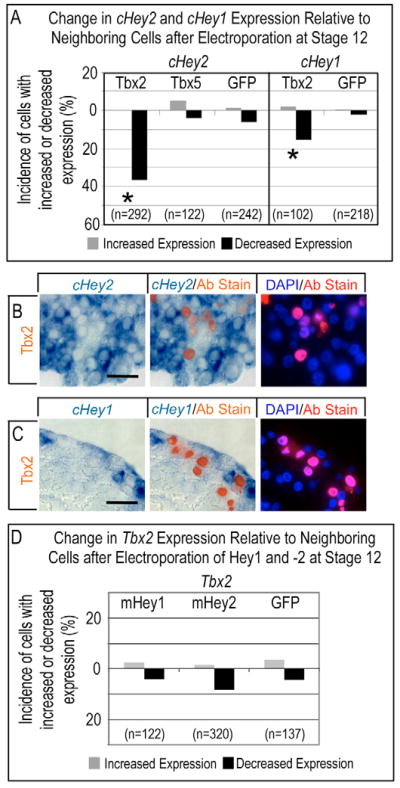
Several targeted disruption transgenic studies in mice indicate Tbx genes regulate Hey1 and Hey2 transcription. For instance, disruption of Tbx5 caused a reduction in ventricular Hey2 expression (Bruneau et al., 2001), and disruption of Tbx20 caused an expansion of the Tbx2 and a reduction of the Hey1 (Stennard et al., 2005) domain. As Bmp2 induces Tbx2 (Yamada et al., 2000), repression of Hey by this factor would help sharpen the border of Hey transcription where chamber myocardium abuts the AVC and IC. To directly test this model, we misexpressed Myc-tagged Tbx2 and Tbx5 proteins in tubular hearts. The efficacy of our Myc-tagged Tbx constructs was verified by showing reciprocal effects on endogenous Cx42 (the chick homolog of Cx43). As expected (e.g. Bruneau et al., 2001; Habets et al., 2002), electroporated Tbx5 induced Cx42, whereas Tbx2 repressed Cx42 expression (not shown). Myc-tagged Tbx2 introduced into the ventricular myocardium of HH stage 12 hearts caused a robust and cell-autonomous inhibition of Hey2, and a less pronounced but significant reduction of Hey1, as indicated by a 36.6% and 15.7% incidence of reduced expression relative to neighboring cells, respectively (Fig. 7A-C). By contrast, Serrate1 was unaffected by Tbx2 (not shown), thus suggesting that the point of regulation lies downstream. We did not find a consensus T-box binding element (TBE) to be conserved within 10 Kb upstream of the start sites of transcription of the mouse, human or chick Hey1 and Hey2 genes, or in the first intron, nor did we find a consensus Nkx element (NKE), such as is often located near a TBE in Tbx-responsive cardiac genes (e.g. Habets et al., 2002; Small and Krieg, 2003). The electroporation data, together with the lack of conserved T-box response elements, strongly indicate that Tbx regulation of Hey genes involves an unknown intermediary, consistent with the finding that Myc-tagged Tbx5 protein did not activate either Hey gene (Fig. 7A). Thus, we conclude that the activation of Tbx2 by Bmp2 is likely to suppress Hey1 and Hey2 in the AVC and IC, and might constitute a feedback circuit to sharpen the border of Hey gene expression.
Fig. 7. Tbx2 represses Hey1 and Hey2 cell autonomously.
(A) Misexpression of Myc-tagged Tbx2 in stage 12 hearts caused a cell-autonomous reduction in the incidence of Hey1- and Hey2-positive cells. Tbx5, by contrast, had no effect. Asterisks indicate statistical significance of the difference between electroporated Tbx2 and the GFP control (P<0.05, χ2 test). (B,C) Histological sections demonstrating the cell-autonomous suppression of endogenous Hey1 (B) and Hey2 (C) by electroporated Tbx2. (D) Flag-tagged mouse Hey1 and mouse Hey2 did not alter Tbx2 expression. Brightfield, merged and fluorescent images are as in Fig. 3. Scale bar: 50 μm.
Because Tbx2 suppressed both Hey1 and Hey2, we next investigated whether either Hey protein cell-autonomously suppresses Tbx2, constituting a mutually repressive interaction. As seen in Fig. 7D, Tbx2 was unaffected by cell-autonomous expression of either Hey protein, indicating that Hey proteins suppress AVC and IC fate through the inhibition of Bmp2 rather than by regulation of Tbx2. In addition, Tbx2 is insufficient to maintain Bmp2 expression cell autonomously, as cells expressing Hey proteins are always negative for Bmp2.
Expansion of the Bmp2 domain in AVC/IC of mouse and zebrafish embryos lacking Hey2 homologs
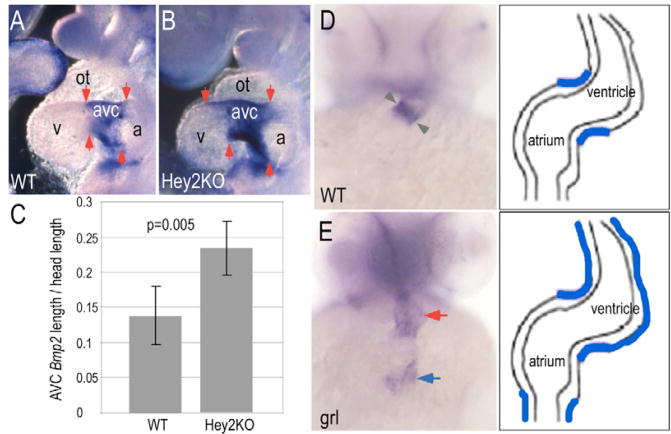
Having demonstrated that Hey proteins suppress Bmp2 in chick AVC/IC, we tested whether loss of Hey function would cause a corresponding increase. As discussed above, Hey2 (also known as HRT2 in mammals) is the only Hey homolog transcribed in mouse ventricular myocardium (Hey1 is atrial) and targeted disruption has not been correlated with early AVC defects. When E9.5 embryos were analyzed by in situ hybridization, however, we found a distinct increase in the Bmp2 expression domain relative to that of wild-type littermates (Fig. 8A,B). The linear length of the domain along the atrioventricular axis of the heart was expanded approximately 1.7-fold, measured relative to either the head length of the embryo (Fig. 8C, P=0.005) or the maximal width of the mandibular arch (not shown, P=0.005) in order to normalize for natural variation in embryo size within a litter. Zebrafish gridlock (grl) is the only Hey homolog expressed in the dorsal aorta and the early looping heart, where transcripts are localized to ventricular and atrial myocardium (Fischer et al., 2002; Zhong et al., 2000) (H.J. and T.P.Z., unpublished). We examined the expression of Bmp genes in the early looping zebrafish heart at 48 hours postfertilization (hpf) and found that Bmp4, rather than Bmp2, shows AVC expression (Fig. 8D). grl mutants showed ectopic inflow tract expression of Bmp4 and diffuse ventricular myocardial expression (Fig. 8E). Neither species showed measurable expansion of Tbx2 homologs outside of the normal AVC expression domain (not shown), possibly reflecting overriding repression by Tbx20 in ventricular myocardium (Brown et al., 2005; Cai et al., 2005; Singh et al., 2005; Stennard et al., 2005; Takeuchi et al., 2005).
Fig. 8. Expansion of the cardiac Bmp expression domain in the absence of Hey2 homologs in mouse and zebrafish embryos.
(A,B) Bmp2 expression marks the AVC and IC of E9.5-E10.0 mouse wild-type (WT) embryos (A), and this region is expanded in homozygous Hey2 mutant siblings (B). Arrows indicate the borders of Bmp2 expression. (C) Measurements of the atrioventricular length of the Bmp2 expression domain relative to head length revealed a 1.7-fold expansion in mutants (n=5) relative to wild type (n=5; P<0.005, Student's t-test), compiled from two litters. Error bars correspond to standard deviation. (D,E)Bmp4 marked the developing AVC (gray arrowheads) in 48 hpf wild-type zebrafish embryos (D), whereas gridlock(grl) mutant embryos (E), which lack the Hey2 homolog, showed diffusely distributed Bmp4 in the AVC and ventricular regions (red arrow), and strong ectopic expression in the inflow tract (blue arrow), as diagrammed in schematics to the right.
Discussion
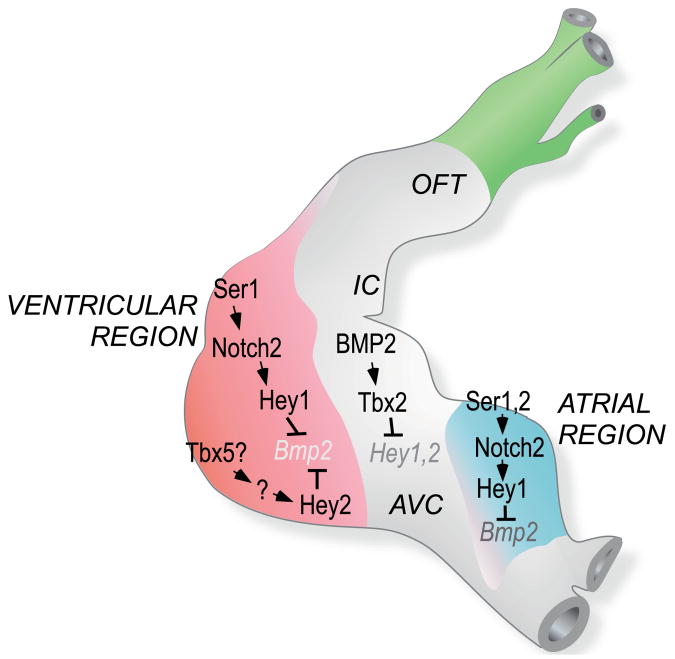
Recent evidence suggests that Tbx5 in the developing chambers of the linear to early looping heart cooperates with the more broadly expressed transcription factors Nkx2.5 and Gata4 to induce expression of Anf and Cx43 (which encodes a gap junction protein homologous to Cx42 in chick) (Bruneau et al., 2001; Habets et al., 2002). The transcriptional repressor Tbx2 (possibly together with Tbx3) disrupts these chamber-specific transcription complexes (Habets et al., 2002; Harrelson et al., 2004) and might operate in the AVC and IC to antagonize chamber-specific differentiation and morphogenesis (Yamada et al., 2000). Therefore, Bmp2 plays a pivotal role in AVC and IC identity by inducing Tbx2. The upstream signals that regulate Bmp2 were previously unidentified. We showed that Notch2 and the Hey proteins provide evolutionarily conserved and essential patterning cues for normal spatial localization of Bmp2 (Bmp4 in zebrafish) in the looping stage heart, and we propose that this mechanism demarcates the border that separates chamber from AVC and IC myocardium. The genetic hierarchy is summarized in Fig. 9 for chick heart development, showing Notch2-dependent Hey1 expression in the prospective ventricular and atrial regions that express Serrate1 and both Serrate1 and Serrate2, respectively. By contrast, Hey2 is induced independently of Notch in the chick. The expression of Hey proteins in chamber myocardium suppresses Bmp2 expression, constraining Bmp2 to the AVC and IC, where it induces Tbx2. Furthermore, our finding that Tbx2 inhibits Hey gene expression suggests a feedback circuit capable of driving the initially graded expression of Hey genes in the early looping heart into a sharp border at the juncture of chamber with AVC and IC myocardium.
Fig. 9. Diagram of the proposed interactions between Notch2, Hey proteins and Tbx2 in the regulation of Bmp in the chick heart.
Hey1 and Hey2 are proposed to function cell autonomously within prospective chamber myocardium to inhibit Bmp2 expression (Bmp4 in zebrafish), and thereby suppress AVC and IC identity. Our data indicate that Notch regulates Hey1 but not Hey2, suggesting a Notch-independent input into Hey2. Although loss of ventricular Hey2 expression is seen in Tbx5-deficient mouse embryos (Bruneau et al., 2001), Tbx5 is unable to induce Hey2 (see Fig. 7), suggesting that any influence on Hey2 is indirect (as indicated by ‘?’). An important finding from the present study is that Tbx2 feeds back to suppress Hey1 and Hey2 in the AVC and IC region (Fig. 7), and this mechanism is postulated to sharpen the border with prospective chamber myocardium.
Our data implicate both Hey1 and Hey2 as inhibitors of Bmp2, but only Hey1 as a direct effector of Notch2 in the chick. Both Hey genes can be Notch regulated: reduction of Hey1 and Hey2 mRNA occurs in Notch1−/− (Leimeister et al., 2000) and Dll3−/− (Dunwoodie et al., 2002) mutant mice, and both Hey1 and Hey2 genes have binding sites for CSL and can be upregulated in response to Notch pathway activators such as N1icd (Iso et al., 2001; Maier and Gessler, 2000; Nakagawa et al., 2000). Nonetheless, Notch-independent transcription of Hey1 and Hey2 genes has also been reported (Iso et al., 2001; Rones et al., 2002), pointing to the existence of tissue-specific factors that modulate Hey gene responsiveness to CSL and the Notch pathway.
How might Hey genes contribute to congenital cardiac defects such as those present in AGS? Jagged1 haploinsufficient AGS patients can exhibit atrial and ventricular septal defects and tetralogy of Fallot (Li et al., 1997; McCright et al., 2002; Oda et al., 1997). Bmp2, Bmp4 and Tbx2 regulate differentiation of the AVC, IC and outflow tract (OFT) regions, so that chamber alignment and development of the valves, septa and conduction system can proceed normally, probably by sustaining the AVC, IC and OFT regions in what has been characterized as an immature, non-chamber state (Moorman and Christoffels, 2003). Our misexpression experiments in chicks, combined with our analyses of mouse Hey2 and zebrafish grl mutants, suggest that attenuation of ventricular Hey2 or atrial Hey1 function in human patients would cause an expansion of Bmp2 into ventricular and atrial myocardium, respectively, which we predict would have patterning and/or morphogenetic consequences for structures that form at the border of the AVC and IC with chamber myocardium. Interestingly, neither Hey2 mutant mice nor zebrafish gridlock mutants showed a corresponding alteration in Tbx2 and Tbx2a or Tbx2b, respectively, pointing to the existence of other factors, such as Tbx20, that modulate responsiveness downstream of Bmp. The morphological consequences of potential genetic interactions between such factors and the genetic cascade identified here would be best characterized using mouse models of loss of Notch2, Hey and Tbx gene function. Recent studies have demonstrated Notch-dependent endocardial effects on myocardial development (Noseda et al., 2004; Raya et al., 2003; Timmerman et al., 2004), and paracrine effects from endocardial and epicardial sources are well established (Baliga et al., 1999; Ford et al., 1999; Ozcelik et al., 2002; Zhao et al., 1998), underscoring the importance of using tissue-specific misexpression strategies to distinguish intramyocardial effects from confounding indirect effects of Notch components. Accordingly, we are investigating the myocardial role of Notch2 in the control of Bmp2 and Hey genes, as well as potential downstream Tbx, Iroquois and Hand genes, through the use of specific myocardial conditional mutants that should overcome the severe cardiac anomalies that are present in mouse embryos that have systemic deletions of Notch2 or Notch signaling components.
In conclusion, our results indicate that Hey proteins play an unforeseen role in constraining Bmp expression to the AVC and IC during cardiac looping. The data also provide a novel role for Notch2 in this process directly within the developing myocardium that complements previous studies illustrating Notch function in endocardium. AVC malformations are particularly common, and mutations in Jagged/Serrate, Notch2 and both Hey proteins might occur in affected individuals. Indeed, mutations in Hey1 and Hey2 have been observed recently in patients with congenital heart disease (Deepak Srivastava, personal communication).
Acknowledgments
We regret the passing of Tom Maciag, who provided reagents and advice. We thank Derek Kedra for database searching for TBEs and NKEs; and Eric Beyer, Connie Cepko, Jeff Sklar, Deepak Srivastava, Cliff Tabin, Katherine Yutzey and Tom Vogt for providing cDNAs. The authors are grateful to Rolf Bodmer, Seigo Izumo, Fred Levine, Tom Schultheiss, Cliff Tabin, Malcolm Whitman and Ramon Diaz Trelles, and members of the Mercola lab, for many helpful discussions. This research was supported by grants from NIH (R01HL067079, R01HL059502 and R01HL083463) and Exelixis to M.M., the DFG (Ge539/9) to M.G., and NIH (RO1HL073348), March of Dimes and American Heart Association to T.P.Z.
References
- Abdelwahid E, Rice D, Pelliniemi LJ, Jokinen E. Overlapping and differential localization of Bmp-2, Bmp-4, Msx-2 and apoptosis in the endocardial cushion and adjacent tissues of the developing mouse heart. Cell Tissue Res. 2001;305:67–78. doi: 10.1007/s004410100399. [DOI] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- Baliga RR, Pimental DR, Zhao YY, Simmons WW, Marchionni MA, Sawyer DB, Kelly RA. NRG-1-induced cardiomyocyte hypertrophy. Role of PI-3-kinase, p70(S6K), and MEK-MAPK-RSK. Am J Physiol. 1999;277:H2026–H2037. doi: 10.1152/ajpheart.1999.277.5.H2026. [DOI] [PubMed] [Google Scholar]
- Bettenhausen B, Hrabe de Angelis M, Simon D, Guenet JL, Gossler A. Transient and restricted expression during mouse embryogenesis of Dll1, a murine gene closely related to Drosophila Delta. Development. 1995;121:2407–2418. doi: 10.1242/dev.121.8.2407. [DOI] [PubMed] [Google Scholar]
- Brown DD, Martz SN, Binder O, Goetz SC, Price BM, Smith JC, Conlon FL. Tbx5 and Tbx20 act synergistically to control vertebrate heart morphogenesis. Development. 2005;132:553–563. doi: 10.1242/dev.01596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruneau BG, Nemer G, Schmitt JP, Charron F, Robitaille L, Caron S, Conner DA, Gessler M, Nemer M, Seidman CE, et al. A murine model of Holt-Oram syndrome defines roles of the T-box transcription factor Tbx5 in cardiogenesis and disease. Cell. 2001;106:709–721. doi: 10.1016/s0092-8674(01)00493-7. [DOI] [PubMed] [Google Scholar]
- Cai CL, Zhou W, Yang L, Bu L, Qyang Y, Zhang X, Li X, Rosenfeld MG, Chen J, Evans S. T-box genes coordinate regional rates of proliferation and regional specification during cardiogenesis. Development. 2005;132:2475–2487. doi: 10.1242/dev.01832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan-Thomas PS, Thompson RP, Robert B, Yacoub MH, Barton PJ. Expression of homeobox genes Msx-1 (Hox-7) and Msx-2 (Hox-8) during cardiac development in the chick. Dev Dyn. 1993;197:203–216. doi: 10.1002/aja.1001970305. [DOI] [PubMed] [Google Scholar]
- Chen JN, van Eeden JM, Warren KS, Chin A, Nusslein-Volhard C, Haffter P, Fishman MC. Left-right pattern of cardiac BMP4 may drive asymmetry of the heart in zebrafish. Development. 1997;124:4373–4382. doi: 10.1242/dev.124.21.4373. [DOI] [PubMed] [Google Scholar]
- Chin MT, Maemura K, Fukumoto S, Jain MK, Layne MD, Watanabe M, Hsieh CM, Lee ME. Cardiovascular basic helix loop helix factor 1, a novel tanscriptional repressor expressed preferentially in the developing and adult cardiovascular system. J Biol Chem. 2000;275:6381–6387. doi: 10.1074/jbc.275.9.6381. [DOI] [PubMed] [Google Scholar]
- Conlon RA, Reaume AG, Rossant J. Notch1 is required for the coordinate segmentation of somites. Development. 1995;121:1533–1545. doi: 10.1242/dev.121.5.1533. [DOI] [PubMed] [Google Scholar]
- Davis DL, Edwards AV, Juraszek AL, Phelps A, Wessels A, Burch JB. A GATA-6 gene heart-region-specific enhancer provides a novel means to mark and probe a discrete component of the mouse cardiac conduction system. Mech Dev. 2001;108:105–119. doi: 10.1016/s0925-4773(01)00500-7. [DOI] [PubMed] [Google Scholar]
- de Jong F, Opthof T, Wilde AA, Janse MJ, Charles R, Lamers WH, Moorman AF. Persisting zones of slow impulse conduction in developing chicken hearts. Circ Res. 1992;71:240–250. doi: 10.1161/01.res.71.2.240. [DOI] [PubMed] [Google Scholar]
- Delorme B, Dahl E, Jarry-Guichard T, Briand JP, Willecke K, Gros D, Theveniau-Ruissy M. Expression pattern of connexin gene products at the early developmental stages of the mouse cardiovascular system. Circ Res. 1997;81:423–437. doi: 10.1161/01.res.81.3.423. [DOI] [PubMed] [Google Scholar]
- Donovan J, Kordylewska A, Jan YN, Utset MF. Tetralogy of fallot and other congenital heart defects in Hey2 mutant mice. Curr Biol. 2002;12:1605–1610. doi: 10.1016/s0960-9822(02)01149-1. [DOI] [PubMed] [Google Scholar]
- Donoviel DB, Hadjantonakis AK, Ikeda M, Zheng H, Hyslop P, Bernstein A. Mice lacking both presenilin genes exhibit early embryonic patterning defects. Genes Dev. 1999;13:2801–2810. doi: 10.1101/gad.13.21.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunwoodie SL, Henrique D, Harrison SM, Beddington RS. Mouse Dll3: a novel divergent Delta gene which may complement the function of other Delta homologues during early pattern formation in the mouse embryo. Development. 1997;124:3065–3076. doi: 10.1242/dev.124.16.3065. [DOI] [PubMed] [Google Scholar]
- Dunwoodie SL, Clements M, Sparrow DB, Sa X, Conlon RA, Beddington RS. Axial skeletal defects caused by mutation in the spondylocostal dysplasia/pudgy gene Dll3 are associated with disruption of the segmentation clock within the presomitic mesoderm. Development. 2002;129:1795–1806. doi: 10.1242/dev.129.7.1795. [DOI] [PubMed] [Google Scholar]
- Fischer A, Gessler M. Hey genes in cardiovascular development. Trends Cardiovasc Med. 2003;13:221–226. doi: 10.1016/s1050-1738(03)00082-3. [DOI] [PubMed] [Google Scholar]
- Fischer A, Leimeister C, Winkler C, Schumacher N, Klamt B, Elmasri H, Steidl C, Maier M, Knobeloch KP, Amann K, et al. Hey bHLH factors in cardiovascular development. Cold Spring Harb Symp Quant Biol. 2002;67:63–70. doi: 10.1101/sqb.2002.67.63. [DOI] [PubMed] [Google Scholar]
- Fischer A, Schumacher N, Maier M, Sendtner M, Gessler M. The Notch target genes Hey1 and Hey2 are required for embryonic vascular development. Genes Dev. 2004;18:901–911. doi: 10.1101/gad.291004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman MC, Chien KR. Fashioning the vertebrate heart: earliest embryonic decisions. Development. 1997;124:2099–2117. doi: 10.1242/dev.124.11.2099. [DOI] [PubMed] [Google Scholar]
- Ford BD, Loeb JA, Fischbach GD. Neuregulin stimulates DNA synthesis in embryonic chick heart cells. Dev Biol. 1999;214:139–150. doi: 10.1006/dbio.1999.9394. [DOI] [PubMed] [Google Scholar]
- Gessler M, Knobeloch KP, Helisch A, Amann K, Schumacher N, Rohde E, Fischer A, Leimeister C. Mouse gridlock: no aortic coarctation or deficiency, but fatal cardiac defects in Hey2 −/− mice. Curr Biol. 2002;12:1601–1604. doi: 10.1016/s0960-9822(02)01150-8. [DOI] [PubMed] [Google Scholar]
- Habets PE, Moorman AF, Clout DE, van Roon MA, Lingbeek M, van Lohuizen M, Campione M, Christoffels VM. Cooperative action of Tbx2 and Nkx2.5 inhibits ANF expression in the atrioventricular canal: implications for cardiac chamber formation. Genes Dev. 2002;16:1234–1246. doi: 10.1101/gad.222902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada Y, Kadokawa Y, Okabe M, Ikawa M, Coleman JR, Tsujimoto Y. Mutation in ankyrin repeats of the mouse Notch2 gene induces early embryonic lethality. Development. 1999;126:3415–3424. doi: 10.1242/dev.126.15.3415. [DOI] [PubMed] [Google Scholar]
- Hamburger V, Hamilton HL. A series of normal stages in the development of the chick embryo. J Morphol. 1951;88:49–92. [PubMed] [Google Scholar]
- Harrelson Z, Kelly RG, Goldin SN, Gibson-Brown JJ, Bollag RJ, Silver LM, Papaioannou VE. Tbx2 is essential for patterning the atrioventricular canal and for morphogenesis of the outflow tract during heart development. Development. 2004;131:5041–5052. doi: 10.1242/dev.01378. [DOI] [PubMed] [Google Scholar]
- Hayashi H, Mochii M, Kodama R, Hamada Y, Mizuno N, Eguchi G, Tachi C. Isolation of a novel chick homolog of Serrate and its coexpression with C-Notch-1 in chick development. Int J Dev Biol. 1996;40:1089–1096. [PubMed] [Google Scholar]
- Henrique D, Adam J, Myat A, Chitnis A, Lewis J, Ish-Horowicz D. Expression of a Delta homologue in prospective neurons in the chick. Nature. 1995;375:787–790. doi: 10.1038/375787a0. [DOI] [PubMed] [Google Scholar]
- Honjo T. The shortest path from the surface to the nucleus: RBP-J kappa/Su(H) transcription factor. Genes Cells. 1996;1:1–9. doi: 10.1046/j.1365-2443.1996.10010.x. [DOI] [PubMed] [Google Scholar]
- Icardo JM, Fernandez-Teran A. Morphologic study of ventricular trabeculation in the embryonic chick heart. Acta Anat Basel. 1987;130:264–274. doi: 10.1159/000146455. [DOI] [PubMed] [Google Scholar]
- Iso T, Sartorelli V, Chung G, Shichinohe T, Kedes L, Hamamori Y. HERP, a new primary target of Notch regulated by ligand binding. Mol Cell Biol. 2001;21:6071–6079. doi: 10.1128/MCB.21.17.6071-6079.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iso T, Kedes L, Hamamori Y. HES and HERP families: multiple effectors of the Notch signaling pathway. J Cell Physiol. 2003;194:237–255. doi: 10.1002/jcp.10208. [DOI] [PubMed] [Google Scholar]
- Jarriault S, Brou C, Logeat F, Schroeter EH, Kopan R, Israel A. Signalling downstream of activated mammalian Notch. Nature. 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- Jeffries S, Robbins DJ, Capobianco AJ. Characterization of a high-molecular-weight Notch complex in the nucleus of Notch(ic)-transformed RKE cells and in a human T-cell leukemia cell line. Mol Cell Biol. 2002;22:3927–3941. doi: 10.1128/MCB.22.11.3927-3941.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokubo H, Lun Y, Johnson RL. Identification and expression of a novel family of bHLH cDNAs related to Drosophila hairy and enhancer of split. Biochem Biophys Res Commun. 1999;260:459–465. doi: 10.1006/bbrc.1999.0880. [DOI] [PubMed] [Google Scholar]
- Krantz ID, Piccoli DA, Spinner NB. Alagille syndrome. J Med Genet. 1997;34:152–157. doi: 10.1136/jmg.34.2.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupershmidt S, Yang T, Anderson ME, Wessels A, Niswender KD, Magnuson MA, Roden DM. Replacement by homologous recombination of the minK gene with lacZ reveals restriction of minK expression to the mouse cardiac conduction system. Circ Res. 1999;84:146–152. doi: 10.1161/01.res.84.2.146. [DOI] [PubMed] [Google Scholar]
- Lardelli M, Dahlstrand J, Lendahl U. The novel Notch homologue mouse Notch 3 lacks specific epidermal growth factor-repeats and is expressed in proliferating neuroepithelium. Mech Dev. 1994;46:123–136. doi: 10.1016/0925-4773(94)90081-7. [DOI] [PubMed] [Google Scholar]
- Laufer E, Dahn R, Orozco OE, Yeo CY, Pisenti J, Henrique D, Abbott UK, Fallon JF, Tabin C. Expression of Radical fringe in limb-bud ectoderm regulates apical ectodermal ridge formation. Nature. 1997;386:366–373. doi: 10.1038/386366a0. [DOI] [PubMed] [Google Scholar]
- Leimeister C, Bach A, Gessler M. Developmental expression patterns of mouse sFRP genes encoding members of the secreted frizzled related protein family. Mech Dev. 1998;75:29–42. doi: 10.1016/s0925-4773(98)00072-0. [DOI] [PubMed] [Google Scholar]
- Leimeister C, Externbrink A, Klamt B, Gessler M. Hey genes: a novel subfamily of hairy- and enhancer of split related genes specifically expressed during mouse embryogenesis. Mech Dev. 1999;85:173–177. doi: 10.1016/s0925-4773(99)00080-5. [DOI] [PubMed] [Google Scholar]
- Leimeister C, Dale K, Fischer A, Klamt B, Hrabe de Angelis M, Radtke F, McGrew MJ, Pourquie O, Gessler M. Oscillating expression of c-Hey2 in the presomitic mesoderm suggests that the segmentation clock may use combinatorial signaling through multiple interacting bHLH factors. Dev Biol. 2000;227:91–103. doi: 10.1006/dbio.2000.9884. [DOI] [PubMed] [Google Scholar]
- Levin M, Johnson RL, Stern CD, Kuehn M, Tabin C. A molecular pathway determining left-right asymmetry in chick embryogenesis. Cell. 1995;82:803–814. doi: 10.1016/0092-8674(95)90477-8. [DOI] [PubMed] [Google Scholar]
- Li L, Krantz ID, Deng Y, Genin A, Banta AB, Collins CC, Qi M, Trask BJ, Kuo WL, Cochran J, et al. Alagille syndrome is caused by mutations in human Jagged1, which encodes a ligand for Notch1. Nat Genet. 1997;16:243–251. doi: 10.1038/ng0797-243. [DOI] [PubMed] [Google Scholar]
- Lindsell CE, Shawber CJ, Boulter J, Weinmaster G. Jagged: a mammalian ligand that activates Notch1. Cell. 1995;80:909–917. doi: 10.1016/0092-8674(95)90294-5. [DOI] [PubMed] [Google Scholar]
- Maier MM, Gessler M. Comparative analysis of the human and mouse Hey1 promoter: Hey genes are new Notch target genes. Biochem Biophys Res Commun. 2000;275:652–660. doi: 10.1006/bbrc.2000.3354. [DOI] [PubMed] [Google Scholar]
- McCright B, Gao X, Shen L, Lozier J, Lan Y, Maguire M, Herzlinger D, Weinmaster G, Jiang R, Gridley T. Defects in development of the kidney, heart and eye vasculature in mice homozygous for a hypomorphic Notch2 mutation. Development. 2001;128:491–502. doi: 10.1242/dev.128.4.491. [DOI] [PubMed] [Google Scholar]
- McCright B, Lozier J, Gridley T. A mouse model of Alagille syndrome: Notch2 as a genetic modifier of Jag1 haploinsufficiency. Development. 2002;129:1075–1082. doi: 10.1242/dev.129.4.1075. [DOI] [PubMed] [Google Scholar]
- Moorman AF, Christoffels VM. Cardiac chamber formation: development, genes, and evolution. Physiol Rev. 2003;83:1223–1267. doi: 10.1152/physrev.00006.2003. [DOI] [PubMed] [Google Scholar]
- Myat A, Henrique D, Ish-Horowicz D, Lewis J. A chick homologue of Serrate and its relationship with Notch and Delta homologues during central neurogenesis. Dev Biol. 1996;174:233–247. doi: 10.1006/dbio.1996.0069. [DOI] [PubMed] [Google Scholar]
- Nakagawa O, Nakagawa M, Richardson JA, Olson EN, Srivastava D. HRT1, HRT2, and HRT3: a new subclass of bHLH transcription factors marking specific cardiac, somitic, and pharyngeal arch segments. Dev Biol. 1999;216:72–84. doi: 10.1006/dbio.1999.9454. [DOI] [PubMed] [Google Scholar]
- Nakagawa O, McFadden DG, Nakagawa M, Yanagisawa H, Hu T, Srivastava D, Olson EN. Members of the HRT family of basic helix-loop-helix proteins act as transcriptional repressors downstream of Notch signaling. Proc Natl Acad Sci USA. 2000;97:13655–13660. doi: 10.1073/pnas.250485597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noseda M, McLean G, Niessen K, Chang L, Pollet I, Montpetit R, Shahidi R, Dorovini-Zis K, Li L, Beckstead B, et al. Notch activation results in phenotypic and functional changes consistent with endothelial-to-mesenchymal transformation. Circ Res. 2004;94:910–917. doi: 10.1161/01.RES.0000124300.76171.C9. [DOI] [PubMed] [Google Scholar]
- Oda T, Elkahloun AG, Pike BL, Okajima K, Krantz ID, Genin A, Piccoli DA, Meltzer PS, Spinner NB, Collins FS, et al. Mutations in the human Jagged1 gene are responsible for Alagille syndrome. Nat Genet. 1997;16:235–242. doi: 10.1038/ng0797-235. [DOI] [PubMed] [Google Scholar]
- Oka C, Nakano T, Wakeham A, de la Pompa JL, Mori C, Sakai T, Okazaki S, Kawaichi M, Shiota K, Mak TW, et al. Disruption of the mouse RBP-J kappa gene results in early embryonic death. Development. 1995;121:3291–3301. doi: 10.1242/dev.121.10.3291. [DOI] [PubMed] [Google Scholar]
- Ozcelik C, Erdmann B, Pilz B, Wettschureck N, Britsch S, Hubner N, Chien KR, Birchmeier C, Garratt AN. Conditional mutation of the ErbB2 (HER2) receptor in cardiomyocytes leads to dilated cardiomyopathy. Proc Natl Acad Sci USA. 2002;99:8880–8885. doi: 10.1073/pnas.122249299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plageman TF, Jr, Yutzey KE. Differential expression and function of Tbx5 and Tbx20 in cardiac development. J Biol Chem. 2004;279:19026–19034. doi: 10.1074/jbc.M314041200. [DOI] [PubMed] [Google Scholar]
- Raya A, Koth CM, Buscher D, Kawakami Y, Itoh T, Raya RM, Sternik G, Tsai HJ, Rodriguez-Esteban C, Izpisua-Belmonte JC. Activation of Notch signaling pathway precedes heart regeneration in zebrafish. Proc Natl Acad Sci USA. 2003;100(1):11889–11895. doi: 10.1073/pnas.1834204100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rones MS, Woda J, Mercola M, McLaughlin KA. Isolation and characterization of Xenopus Hey-1: A downstream mediator of Notch signaling. Dev Dyn. 2002;225:554–560. doi: 10.1002/dvdy.10192. [DOI] [PubMed] [Google Scholar]
- Sakata Y, Kamei CN, Nakagami H, Bronson R, Liao JK, Chin MT. Ventricular septal defect and cardiomyopathy in mice lacking the transcription factor CHF1/Hey2. Proc Natl Acad Sci USA. 2002;99:16197–16202. doi: 10.1073/pnas.252648999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shawber C, Boulter J, Lindsell CE, Weinmaster G. Jagged2: a serrate-like gene expressed during rat embryogenesis. Dev Biol. 1996;180:370–376. doi: 10.1006/dbio.1996.0310. [DOI] [PubMed] [Google Scholar]
- Shimizu K, Chiba S, Saito T, Kumano K, Takahashi T, Hirai H. Manic fringe and lunatic fringe modify different sites of the Notch2 extracellular region, resulting in different signaling modulation. J Biol Chem. 2001;276:25753–25758. doi: 10.1074/jbc.M103473200. [DOI] [PubMed] [Google Scholar]
- Shutter JR, Scully S, Fan W, Richards WG, Kitajewski J, Deblandre GA, Kintner CR, Stark KL. Dll4, a novel Notch ligand expressed in arterial endothelium. Genes Dev. 2000;14:1313–1318. [PMC free article] [PubMed] [Google Scholar]
- Singh MK, Christoffels VM, Dias JM, Trowe MO, Petry M, Schuster-Gossler K, Burger A, Ericson J, Kispert A. Tbx20 is essential for cardiac chamber differentiation and repression of Tbx2. Development. 2005;132:2697–2707. doi: 10.1242/dev.01854. [DOI] [PubMed] [Google Scholar]
- Small D, Kovalenko D, Soldi R, Mandinova A, Kolev V, Trifonova R, Bagala C, Kacer D, Battelli C, Liaw L, et al. Notch activation suppresses fibroblast growth factor-dependent cellular transformation. J Biol Chem. 2003;278:16405–16413. doi: 10.1074/jbc.M300464200. [DOI] [PubMed] [Google Scholar]
- Small EM, Krieg PA. Transgenic analysis of the atrialnatriuretic factor (ANF) promoter: Nkx2-5 and GATA-4 binding sites are required for atrial specific expression of ANF. Dev Biol. 2003;261:116–131. doi: 10.1016/s0012-1606(03)00306-3. [DOI] [PubMed] [Google Scholar]
- Srivastava D, Olson EN. A genetic blueprint for cardiac development. Nature. 2000;407:221–226. doi: 10.1038/35025190. [DOI] [PubMed] [Google Scholar]
- Stennard FA, Costa MW, Elliott DA, Rankin S, Haast SJ, Lai D, McDonald LP, Niederreither K, Dolle P, Bruneau BG, et al. Cardiac T-box factor Tbx20 directly interacts with Nkx2-5, GATA4, and GATA5 in regulation of gene expression in the developing heart. Dev Biol. 2003;262:206–224. doi: 10.1016/s0012-1606(03)00385-3. [DOI] [PubMed] [Google Scholar]
- Stennard FA, Costa MW, Lai D, Biben C, Furtado MB, Solloway MJ, McCulley DJ, Leimena C, Preis JI, Dunwoodie SL, et al. Murine T-box transcription factor Tbx20 acts as a repressor during heart development, and is essential for adult heart integrity, function and adaptation. Development. 2005;132:2451–2462. doi: 10.1242/dev.01799. [DOI] [PubMed] [Google Scholar]
- Swiatek PJ, Lindsell CE, Franco del Amo F, Weinmaster G, Gridley T. Notch1 is essential for postimplantation development in mice. Genes Dev. 1994;8:707–719. doi: 10.1101/gad.8.6.707. [DOI] [PubMed] [Google Scholar]
- Takeuchi JK, Mileikovskaia M, Koshiba-Takeuchi K, Heidt AB, Mori AD, Arruda EP, Gertsenstein M, Georges R, Davidson L, Mo R, et al. Tbx20 dose-dependently regulates transcription factor networks required for mouse heart and motoneuron development. Development. 2005;132:2463–2474. doi: 10.1242/dev.01827. [DOI] [PubMed] [Google Scholar]
- Tamura K, Taniguchi Y, Minoguchi S, Sakai T, Tun T, Furukawa T, Honjo T. Physical interaction between a novel domain of the receptor Notch and the transcription factor RBP-J kappa/Su(H) Curr Biol. 1995;5:1416–1423. doi: 10.1016/s0960-9822(95)00279-x. [DOI] [PubMed] [Google Scholar]
- Timmerman LA, Grego-Bessa J, Raya A, Bertran E, Perez-Pomares JM, Diez J, Aranda S, Palomo S, McCormick F, Izpisua-Belmonte JC, et al. Notch promotes epithelial-mesenchymal transition during cardiac development and oncogenic transformation. Genes Dev. 2004;18:99–115. doi: 10.1101/gad.276304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyttendaele H, Marazzi G, Wu G, Yan Q, Sassoon D, Kitajewski J. Notch4/int-3, a mammary proto-oncogene, is an endothelial cell-specific mammalian Notch gene. Development. 1996;122:2251–2259. doi: 10.1242/dev.122.7.2251. [DOI] [PubMed] [Google Scholar]
- Weinmaster G, Kintner C. Modulation of Notch signaling during somitogenesis. Annu Rev Cell Dev Biol. 2003;19:367–395. doi: 10.1146/annurev.cellbio.19.111301.115434. [DOI] [PubMed] [Google Scholar]
- Weinmaster G, Roberts VJ, Lemke G. A homolog of Drosophila Notch expressed during mammalian development. Development. 1991;113:199–205. doi: 10.1242/dev.113.1.199. [DOI] [PubMed] [Google Scholar]
- Weinmaster G, Roberts VJ, Lemke G. Notch2: a second mammalian Notch gene. Development. 1992;116:931–941. doi: 10.1242/dev.116.4.931. [DOI] [PubMed] [Google Scholar]
- Yamada M, Revelli JP, Eichele G, Barron M, Schwartz RJ. Expression of chick Tbx-2, Tbx-3, and Tbx-5 genes during early heart development: evidence for BMP2 induction of Tbx2. Dev Biol. 2000;228:95–105. doi: 10.1006/dbio.2000.9927. [DOI] [PubMed] [Google Scholar]
- Zhao YY, Sawyer DR, Baliga RR, Opel DJ, Han X, Marchionni MA, Kelly RA. Neuregulins promote survival and growth of cardiac myocytes Persistence of ErbB2 and ErbB4 expression in neonatal and adult ventricular myocytes. J Biol Chem. 1998;273:10261–10269. doi: 10.1074/jbc.273.17.10261. [DOI] [PubMed] [Google Scholar]
- Zhong TP, Rosenberg M, Mohideen MAPK, Weinstein B, Fishman MC. gridlock, an HLH gene required for assembly of the aorta in zebrafish. Science. 2000;287:1820–1824. doi: 10.1126/science.287.5459.1820. [DOI] [PubMed] [Google Scholar]