Abstract
The bridge α-helix in the β′ subunit of RNA polymerase (RNAP) borders the active site and may have roles in catalysis and translocation. In Escherichia coli RNAP, a bulky hydrophobic segment near the N-terminal end of the bridge helix is identified (β′ 772-YFI-774; the YFI motif). YFI is located at a distance from the active center and adjacent to a glycine hinge (β′ 778-GARKG-782) involved in dynamic bending of the bridge helix. Remarkably, amino acid substitutions in YFI significantly alter intrinsic termination, pausing, fidelity and translocation of RNAP. F773V RNAP largely ignores the λ tR2 terminator at 200 µM NTPs and is strongly reduced in λ tR2 recognition at 1 µM NTPs. F773V alters RNAP pausing and backtracking and favors misincorporation. By contrast, the adjacent Y772A substitution increases fidelity and exhibits other transcriptional defects generally opposite to those of F773V. All atom molecular dynamics simulation revealed two separate functional connections emanating from YFI explaining the distinct effects of substitutions: Y772 communicates with the active site through the link domain in the β subunit, whereas F773 communicates through the fork domain in the β subunit. I774 interacts with the F-loop, which also contacts the glycine hinge of the bridge helix. These results identified negative and positive circuits coupled at YFI and employed for regulation of catalysis, elongation, termination and translocation.
Keywords: RNA polymerase, Bridge helix, Termination, Pausing, Transcriptional fidelity, Translocation
1. Introduction
The bridge α-helix (Escherichia coli (Ec) β′ 769–806 (Ec numbering is shown unless otherwise specified)) is a defining characteristic of multi-subunit RNAPs. The bridge helix approaches the RNAP active site and makes tight contacts to the mobile trigger loop (Ec β′ 913– 944 and 1134–1146, interrupted by a large sequence insertion SI3 in Ec RNAP (945–1133)). The trigger loop regulates the bond addition cycle by alternating between closed and open conformations [1,2]. The closed conformation is considered to be the catalytic form, participating in positioning of the incoming NTP in the active center and catalysis [3,4]. The open conformation may support release of the pyrophosphate by product generated from catalysis and may promote translocation of nucleic acids through RNAP [5–7]. One model for nucleic acid stepping through multi-subunit RNAPs posits that the bridge helix bends against the RNA/DNA hybrid helping to induce forward RNAP displacement [8,9].
Because the bridge helix contacts the active site and the trigger loop, mutations localized to the bridge may have large effects on catalytic activity, pausing and termination. Consistent with bridge helix bending associated with catalysis and translocation, some proline substitutions expected to induce bends result in transcriptional gain of function (i.e. rapid elongation) [10,11]. High throughput mutagenesis of the bridge helix has been reported for an archae on Methanocaldococcus jannaschii (Mj) RNAP [10,11]. From a combination of mutagenesis and molecular dynamics simulations, new models for bridge helix bending and dynamics in catalysis and translocation begin to emerge [11–15].
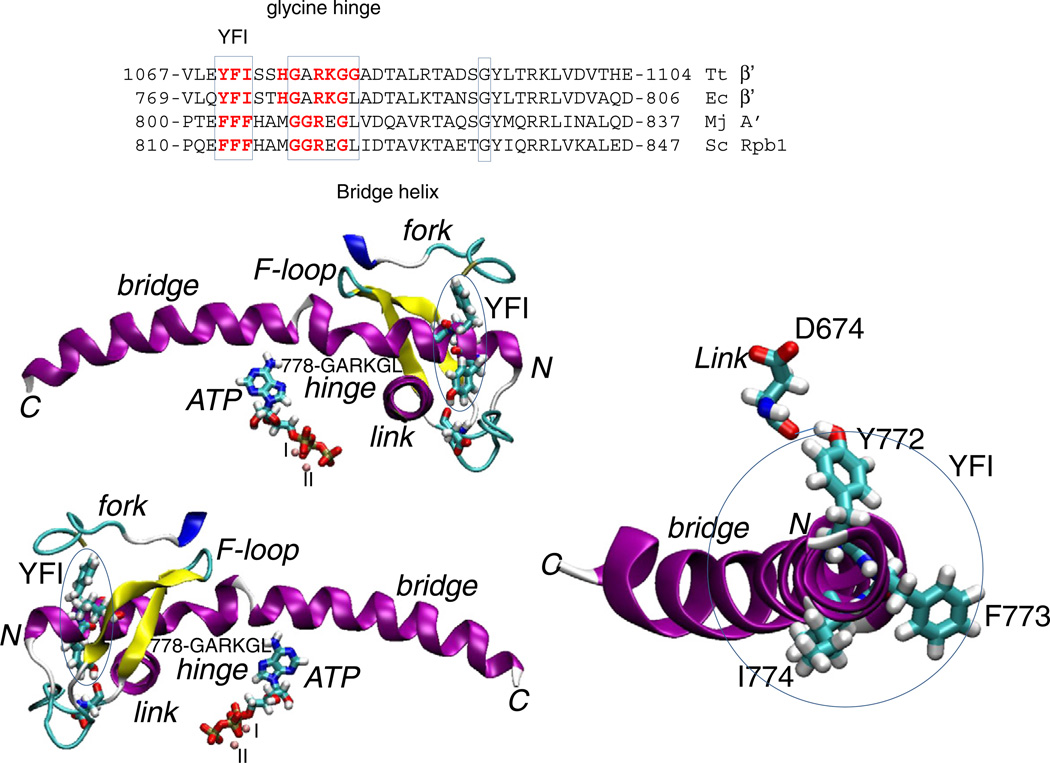
The amino-terminal end of the bridge helix contains a distinct and evolutionarily conserved but not identical triad of bulky hydrophobic amino acid residues (772-YFI-774 in Ec; FFF in Mj RNAP and Saccharomyces cerevisiae (Sc) RNAP II; described herein as the YFI motif) embedded into the protein domains called the link domain, the fork and the F-loop (Fig. 1). Near the N-terminal end of the bridge helix is the sequence β′ 778-GARKG-782 (Fig. 1). Flexibility at glycines (G778 and G782) may help to bend the bridge helix against the RNA–DNA hybrid [5,11,13]. YFI is just N-terminal to the glycine hinge and may form a brace against which the adjacent hinge can flex (Fig. 1). The hydroxyl group of Y772 forms a hydrogen bond to the main chain oxygen of β D674 within the link domain (β 666–685), which approaches the active site. Because tyrosine is substituted with phenylalanine in some organisms, this specific connection of the bridge helix and link domain is not necessarily maintained. F773 contacts the extended fork (β 540–570). YFI could potentially function in concert with surrounding protein to alter the dynamics and bending of the nearby β′ 778-GARKG-782 glycine hinge, which contacts the F-loop (β′ 736–756) [16] and the fork. In the catalytic RNAP ternary elongation complex (TEC), the trigger loop tightens over the accurately loaded NTP-Mg2+ substrate, so loading a NTP and closing the trigger loop stabilize the forward (post) translocation state of the ratchet [3,4].
Fig. 1.
The bridge helix YFI motif. On the right, an N-terminal end on view of the bridge helix is shown. Two images on the left indicate the RNAP regions surrounding YFI, the F-loop (β′ 736 to 770; Ec RNAP numbering), the fork (β 550 to 570) and the link domain approaching the active site (β 670 to 680). Blue ovals circumscribe YFI. The image is derived from PDB 2O5J [3] after molecular dynamics simulation. Secondary structures of RNAP chains are indicated: purple is α-helix; blue is 310 helix; yellow is β-sheet; white is coil; and cyan is turn. The image was drawn using Visual Molecular Dynamics [40]. A multiple sequence alignment of bridge helix sequences is shown to indicate the relationship of YFI and the adjacent, more N-terminal glycine hinge. Abbreviations: Tt, Thermus thermophilus; Ec, Escherichia coli; Mj, Methanocaldococcus jannaschii; Sc, Saccharomyces cerevisiae.
F773V was identified as a substitution that conferred dependence on the antibiotic CBR703 for E. coli cell survival [17]. To probe the roles of YFI in transcription, Ec RNAP β′ F773V was analyzed. The Y772A, I774A and 772-YFI → AAA-774 substitutions were also constructed. Mutant RNAPs were compared in elongation, termination, fidelity and translocation assays to gain insight into the functional connections of YFI, and amino acid substitutions were found to have dramatic effects using a number of transcription assays. The analysis identifies YFI as an important functional element of multi-subunit RNAPs integrating regulatory signals from the surrounding link, fork and flexible F-loop and apparently integrating signals between the active site and the glycine hinge on the bridge helix.
2. Materials and methods
2.1. RNAP mutagenesis
Mutants in rpoC residues (i.e. Y772A and F773V) were constructed in a pIA661 plasmid using the Phusion® Site-Directed Mutagenesis Kit (New England Biolabs) according to the manufacturer's protocol with minor modifications. A restriction digestion of the polymerase chain reaction (PCR) was performed with Dpn I to degrade the methylated template DNA and reduce the background of wild-type plasmids. The pIA661 plasmid contains the rpoC gene encoding the β′ subunit of RNAP under the control of an IPTG-inducible promoter. The mutation-containing Mva1269 I/Hind III fragment of pIA661 was subcloned into a pVS10 plasmid that contains the rpoA, B, C, and Z genes in a single polycistronic transcript under the control of the T7 RNAP promoter [16,18].
The pVS10-based constructs were expressed in an XJb(DE3) Autolysis™strain (Zymo Research) using the Overnight Express system for autoinduction (Novagen) and autolysis was induced with arabinose 1 h prior to cell collection. The lysate (in lysis buffer: 500 mM NaCl, 50 mM Tris–HCl pH 6.9, 5% glycerol, 0.2 mM β-mercaptoethanol) with 1× Complete EDTA-free protease inhibitors (Roche) was clarified by centrifugation and the histidine-tagged RNAP was bound to a HisTrap FF column (GE Healthcare). Columns were washed in lysis buffer and eluted with heparin column loading buffer (50 mM Tris–HCl, pH 6.9, 5% glycerol, 0.5 mM EDTA, 1 mM β-mercaptoethanol) containing 8 mM imidazole. The eluted RNAP was purified on a HiTrap Heparin HiTrap HP column (GE Healthcare) using the AKTA FPLC system (GE Life Sciences) and eluted with a 0 to 1.5 M NaCl gradient. RNAP-containing fractions were pooled and dialyzed into low salt buffer. The sample was loaded onto a Mono-Q anion exchange column (GE Life Sciences) and eluted with a 0 to 1.5 M NaCl gradient on the AKTA FPLC. The appropriate fractions were pooled and dialyzed into storage buffer (10 mM Tris pH 8.0, 100 mM NaCl, 50% glycerol, 0.1 mM EDTA). A typical yield was 10 mg mutant RNAP from a 4 liter culture of cells.
2.2. Preparation of TECs
TECs were prepared either by assembly in vitro [19,20] with Ec RNAP core or by initiation from a promoter with Ec RNAP holoenzyme (Ec RNAP core (α2ββ′ω) + σ70). Details are given with individual experiments.
2.3. Transcription elongation assays (Fig. 2)
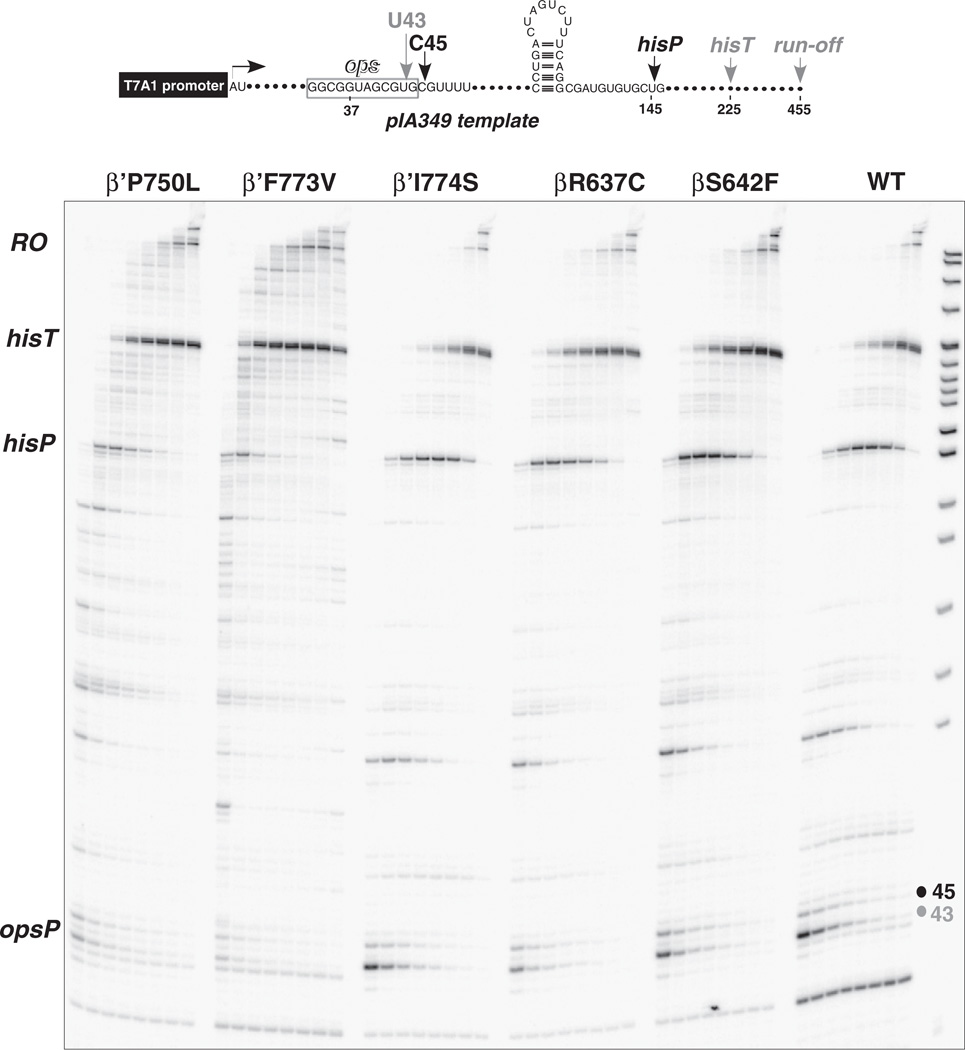
Fig. 2.
Elongation on a long DNA template comparing F773V and other RNAP mutants. The DNA template is indicated at the top of the figure. Elongation times are 10, 20, 30, 60, 120, 240, and 480 s. Mutants other than F773V are shown for comparison and are not discussed in detail.
For pause assays with RNAP variants, a linear DNA template generated by PCR amplification (30 nM), RNAP holoenzyme (40 nM), ApU (100 µM), and starting NTP subsets (1 µM CTP, 5 µM ATP and GTP, 10 µCi [α32P]-CTP, 3000 Ci/mmol) were mixed in 100 µl of 20 mM Tris-acetate, 20 mM Na-acetate, 2 mM Mg-acetate, 5% glycerol, 1 mM DTT, 0.1 mM EDTA, pH 7.9. Reactions were incubated for 15 min at 37 °C. Transcription was restarted by addition of NTPs (10 µM GTP, 150 µM ATP, CTP, and UTP) and rifapentine to 100 µg/ml at 37 °C. Samples were removed at various times, quenched by addition of an equal volume of STOP buffer (10 M urea, 20 mM EDTA, 45 mM Tris-borate; pH 8.3), and loaded on 8% denaturing urea/acrylamide (19:1) gels in 0.5× TBE.
2.4. Bulk elongation (Fig. 3)
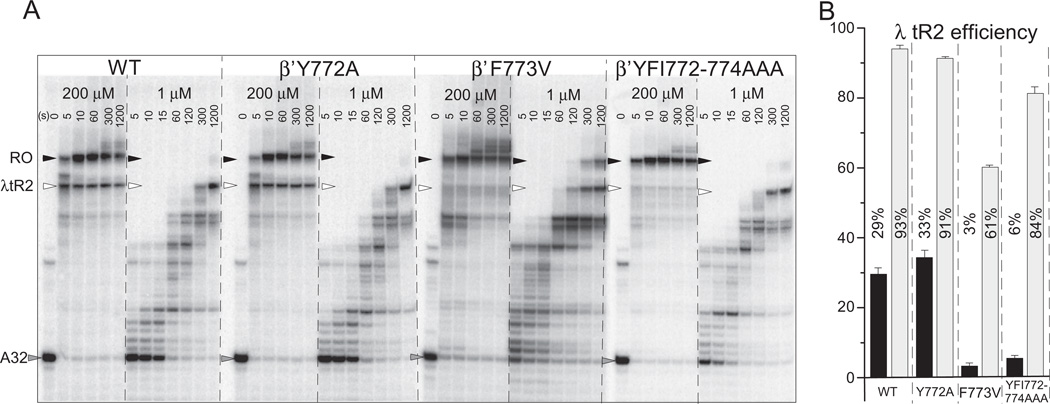
Fig. 3.
Bridge helix YFI substitutions affect λ tR2 intrinsic terminator recognition. A) 32P-labeled A32 RNA TECs were initiated from the bacteriophage T7 A1 promoter, walked to A32 and extended up to or through the λ tR2 terminator at 200 and 1 µM NTPs. B) λ tR2 termination efficiency. Triplicate samples at 30 s (200 µM NTPs; black bars) or 40 min (1 µM NTPs; gray bars) were done. Error bars indicate standard deviation.
Template DNA containing a T7A1 promoter and a λ tR2 terminator downstream was prepared by PCR amplification and purified using QiaQuick spin columns (QIAGEN). Transcription initiation was performed with RNAP-σ70 holoenzyme in the presence of 50 pmol template DNA, 10 µM ApUpC RNA primer, 50 µM GTP and ATP in transcription buffer (20 mM Tris HCl pH 7.9, 150 mM KCl, 5 mM MgCl2, 5% (wt/vol) glycerol, 0.003% Igepal CA-630 (Sigma), 10 µM ZnSO4, 1 mM 2-mercaptoethanol (Sigma)). 150 mM KCl was used for termination at λ tR2 because at 40 mM KCl λ tR2 termination was almost undetectable for F773V, even at 1 µM NTPs (data not shown). The stalled TECs were then immobilized on Ni2+-NTA agarose beads and washed with transcription buffer (the first wash containing 1 M KCl). The TECs were incubated with GTP, ATP, and [α-32P]-CTP (as described above), so that radioactively labeled NMP would be incorporated into the nascent RNA and transcription would stall at residue A32 due to lack of UTP. TECs were then washed and a chase mixture of complete nucleotides was added for the indicated times and stopped by 2× urea loading buffer.
Transcription using assembled RNAP TECs was done similarly. RNAP core was used rather than RNAP holoenzyme to assemble TECs in vitro. Precise protocols are indicated with the figures and in the text.
Samples were resolved on urea-polyacrylamide gels, exposed to the phosphorimaging screen, and scanned on the Typhoon phosphorimager (General Electric Healthcare). Quantification was performed using ImageQuant software (General Electric Healthcare).
2.5. Fast kinetics and fidelity assays
G9 TECs (5′-32P-labeled RNA) were assembled using the TDS45 DNA template and a complementary non-template DNA strand [20,21]. Using a KinTek Rapid Chemical Quench Flow Instrument (RQF-3), G9 TECs were incubated with GTP and ATP for 0.002–0.5 at 25 °C. The reactions were quenched by addition of 1 M HCl or 0.25 M EDTA final concentrations.
2.6. Competition fidelity assays
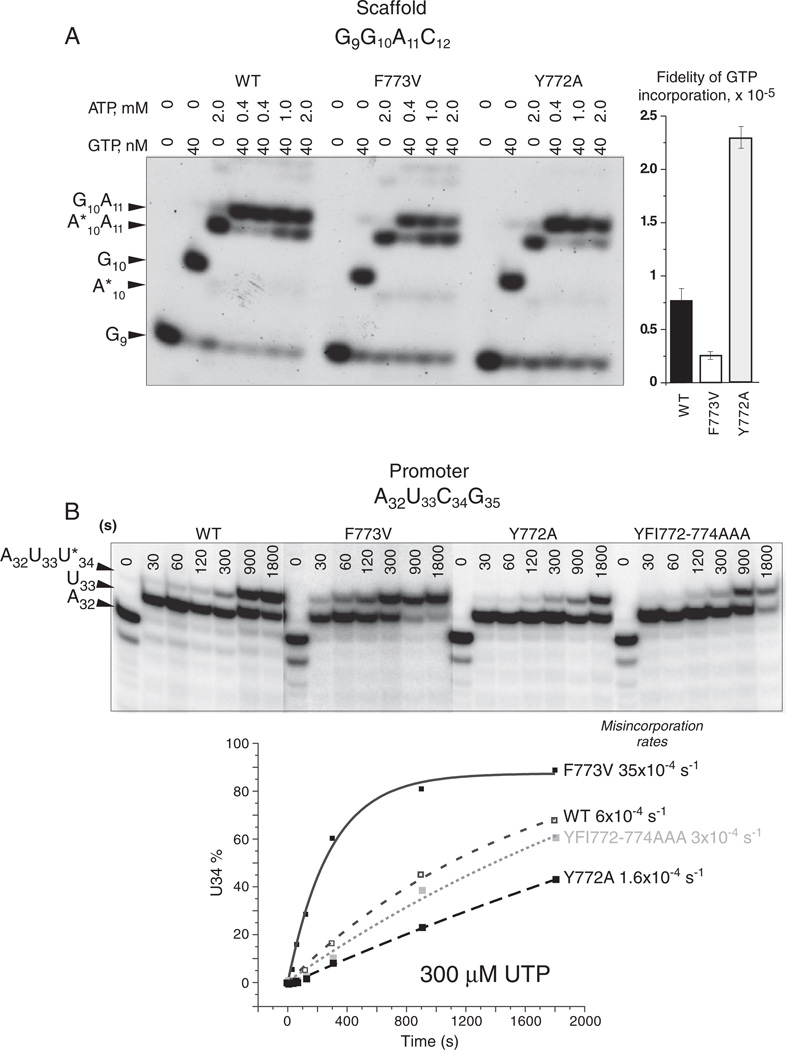
The competition transcriptional fidelity assay is done by extension of TEC G9 at a high concentration of ATP (the incorrect substrate for G10 incorporation) and a low concentration of GTP (the accurate substrate for G10 incorporation) [5,22]. Because of the short length of transcripts, inaccurate (A*10A11 transcripts; asterisk indicates misincorporation) are resolved from accurate (G10A11 transcripts) on gels (Fig. 7A). Reactions with incorrect substrates were stopped manually by gel-loading buffer (5 M urea; 25 mM EDTA final concentrations). RNA products were resolved on 20% polyacrylamide-7 M urea gels. Fidelity is calculated as: fidelity = (correct product %/misincorporation products %)*([non-cognate NTP]/[cognate NTP]).
Fig. 7.
YFI substitutions Y772A and F773V strongly and conversely affect transcriptional fidelity. A) Normalized AMP (from ATP) misincorporation for GMP (from GTP) done as an ATP versus GTP competition experiment in the presence of high ATP and low GTP. A*10A11 indicates misincorporation of AMP for GMP at A*10 followed by accurate incorporation at A11. Phosphorimager quantification is shown at the right. Error bars indicate standard deviation. B) Misincorporation rates for UMP (from UTP) in place of CMP (with no CTP added). Phosphorimager quantification is shown at the bottom of the figure.
2.7. Exonuclease III footprinting assay
Exo III mapping was done as previously described [20]. Templates for Exo III RNAP downstream border footprinting were done on assembled G9 TECs, immobilized on Ni2+-NTA agarose beads, walked to G10 with 5 µM GTP and washed free of GTP. G9 RNA (AUCGAGAGG) was 5′-32P-labeled. The DNA non-template strand was also 5′-32P-labeled for detection of TEC boundaries by Exo III digestion. The template DNA strand included a 3′-thio-G to block digestion by Exo III. Ec Exo III (New England Biolabs) mapping (4 units/µl, except as noted) was at 25 °C. The reaction buffer was 20 mM Tris–HCl pH 7.9, 40 mM KCl, 5 mM MgCl2, 5% glycerol, 0.003% IGEPAL CA-630 (Sigma), 10 µM ZnS04 and 1 mM β-mercaptoethanol. 5 µM ATP and CTP were added with Exo III to extend the RNA chain to C14 and to back up the stall position in the case of RNAP-mediated RNA hydrolysis. To some reactions, GMPcPP was added as a GTP analog (templated at G15 and G16). RNAs and DNAs were analyzed on 20% or 6% polyacrylamide-7 M urea gels and quantified using a phosphorimager (GE Health; Typhoon 9200).
2.8. Molecular dynamics simulations
Molecular images generated in this work were derived from all atom molecular dynamics simulations of Thermus thermophilus (Tt) RNAP TECs done in explicit water with counter-ions, as previously described [5,13]. Details of simulation protocols are given in Supplementary Materials.
3. Results
3.1. F773V RNAP has altered pausing
F773 is one of the three clustered, bulky hydrophobic residues found within the bridge helix N-terminal segment (β′ 772-YFI-774) (Fig. 1). We posit that the YFI motif forms a hydrophobic brace against which the bridge helix bends, particularly at the adjacent 778-GARKG-782 glycine hinge [5]. We further propose that these conformational changes influence surrounding regions including the link domain, the extended fork, the F-loop and the active site.
In bulk elongation, β′ F773V reaches the runoff position faster than wild type RNAP and some other β and β′ RNAP mutants (Fig. 2) [18]. F773V RNAP, however, is not uniformly “fast” in elongation and appears to pause more at some sites but less at others, compared to wild type. Depending on the sequence context and the NTP concentration, F773V has complex effects on elongation and pausing (Figs. 2 and S1). Interestingly, F773V appears substantially faster than the wild type at lower but not at higher NTP concentrations probably because F773V has modified pause site selection (Fig. S1). On the other hand, pause recognition by the Y772A mutant was more similar to the wild type on both templates and at different NTP concentrations (Figs. 2 and S1).
As reported previously [18], F773V suppresses RNAP pausing at the opsP and hisP pause sites (Fig. 2). F773V recognizes the strong hisT intrinsic terminator, although we show here that F773V, 772-YFI → AAA-774 and I774A are defective in recognition of bacteriophage λ tR2, a somewhat weaker intrinsic terminator than hisT. Read through of hisT involves formation of alternate secondary structures blocking formation of the long hisT terminator stem (see Fig. S2B) [23]. Different responses to λ tR2 and the observation that F773V and neighboring Y772A have unusual and often opposing transcriptional properties compared to wild type and other bridge helix substitutions, led us to analyze YFI as a functional feature of the bridge helix.
3.2. Termination efficiency at λ tR2
Because YFI substitutions are altered for pause site preference and elongation rate, and because pausing enhances termination, the efficiency of termination for YFI substitutions was tested at the λ tR2 terminator. λ tR2 is a moderately strong intrinsic terminator consisting of an 8 bp stem-8 nucleotide loop followed by a U-track (Fig. S2B) [24,25]. Transcription was initiated from the bacteriophage T7 A1 promoter, which is recognized by Ec RNAP-σ70 holoenzyme. Transcripts were extended from A32 (a 32 nucleotide RNA ending in 3′-AMP). Transcription was monitored up to and/or through the λ tR2 terminator (Fig. 3). Supplemental Fig. S2A shows a longer gel on which most RNA bands can be distinguished. As expected, λ tR2 termination occurs primarily at U68 within the terminator U-stretch 62-UUUUUAUUU-70 [25]. Fig. S2B compares predicted RNA secondary structures for λ tR2 and hisT, showing why λ tR2 is expected to be a weaker intrinsic terminator than hisT.
Because pausing contributes to intrinsic termination, λ tR2 is expected to be a more efficient terminator at lower NTP concentrations. Termination efficiency, therefore, was determined at 200 and at 1 µM NTPs. Fig. 3A shows a time course of TECs encountering λ tR2. Fig. 3B shows quantification of triplicate determinations at a single time point judged to be long enough that U68 transcripts are terminated rather than paused. Wild type and Y772A RNAPs recognize λ tR2 reasonably efficiently at 200 µM NTPs (29 and 33% efficiency) and very strongly at 1 µM NTPs (93 and 91% efficiency). By contrast, F773V and 772-YFI → AAA-774 RNAPs only weakly recognize λ tR2 at 200 µM NTPs (3 and 6% efficiency). As expected, F773V and 772-YFI → AAA-774 RNAPs recognize λ tR2 more efficiently at 1 µM NTPs (61 and 84% efficiency) but not as well as wild type and Y772A RNAPs (93 and 91% efficiency). F773V and 772-YFI → AAA-774 substitutions, therefore, are defective in recognition of λ tR2 under these conditions (150 mM KCl), but Y772A is not. In Fig. S3 (Supplementary Material), a comparison between wild type and I774A RNAP is shown. I774A RNAP is defective in λ tR2 recognition at 200 µM NTPs but not at 1 µM NTPs. Because 772-YFI → AAA-774 and I774A RNAPs recognize λ tR2 very similarly, the I774A substitution may be sufficient to explain the results with the triple alanine substitution.
3.3. Catalysis and stable NTP-Mg2+ sequestration
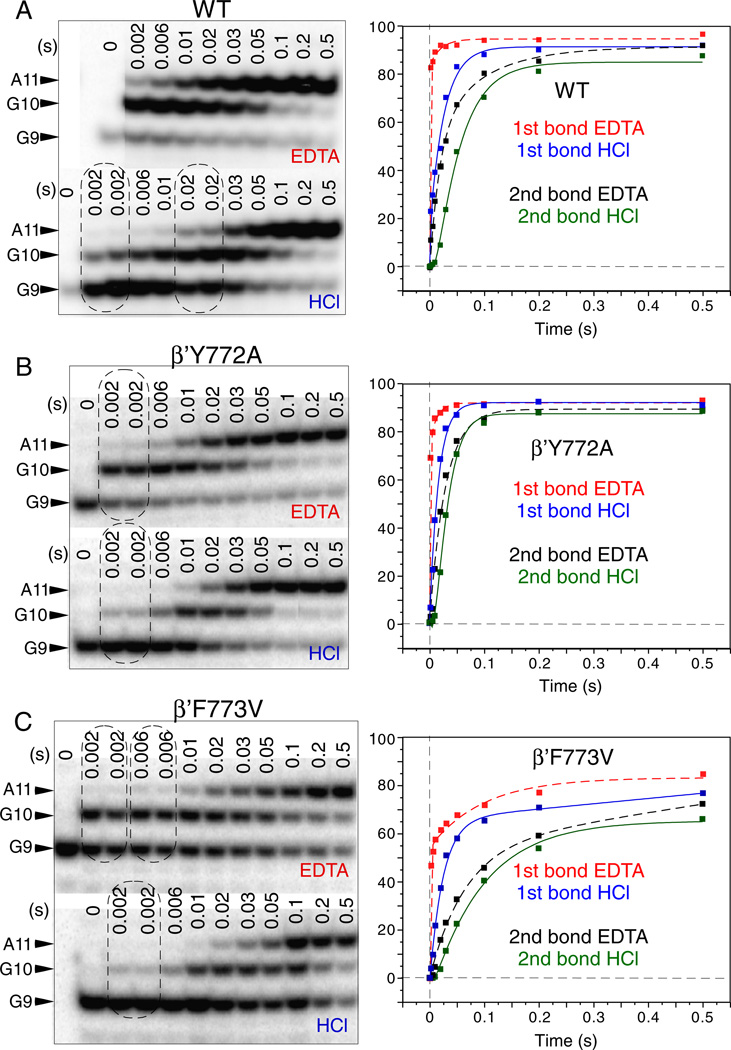
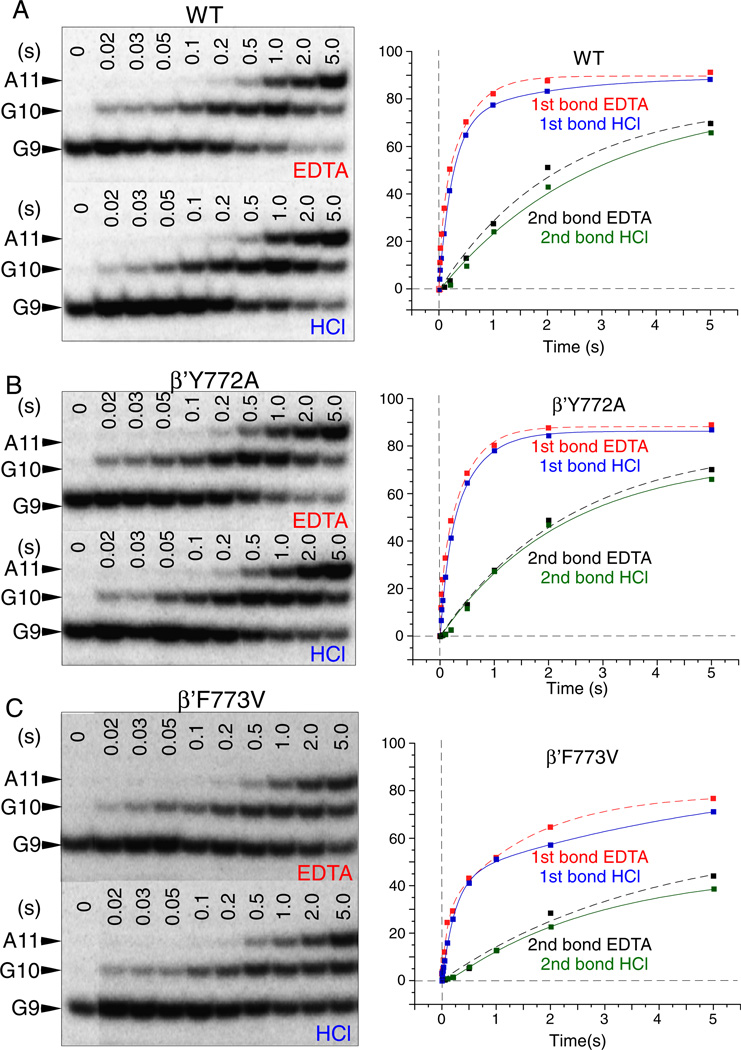
A two bond, double quench rapid chemical quench flow assay is a very sensitive technique for measurement of stable NTP-Mg2+ commitment and phosphodiester bond addition by RNAP [21]. Because TECs can go offline at a stall position, this assay also gives insight into transcriptional pausing. Tracking two consecutive bonds in a single reaction provides information about the translocation of RNAP, which must occur after synthesis of the first bond and before fully stabilized NTP loading for synthesis of the subsequent bond. In the assays represented in Figs. 4–6, transcription was started from TEC G9 assembled in vitro on a synthetic RNA/DNA scaffold directing synthesis of G10 followed by A11. Y772A, F773V and wild type RNAPs show differences in the rates of GTP-Mg2+ sequestration (for G10 synthesis) (EDTA quench), ATP-Mg2+ sequestration (for A11 synthesis) (EDTA quench) and two phosphodiester bond formations (G10 and A11) (HCl quench). Elongation from G9 was analyzed at 200 µM (Figs. 4 and 6) and 1 µM (Figs. 5 and 6B) GTP + ATP.
Fig. 4.
Millisecond phase kinetic analysis of YFI substitutions. 200 µM GTP and ATP were added to G9 RNAP TECs assembled in vitro. Reactions were quenched in a KinTek RQF-3 instrument with EDTA or HCl. A) Wild type RNAP. B) Y772A RNAP. C) F773V RNAP. Data were fit with double or single phase exponential curves (Fig. 6B).
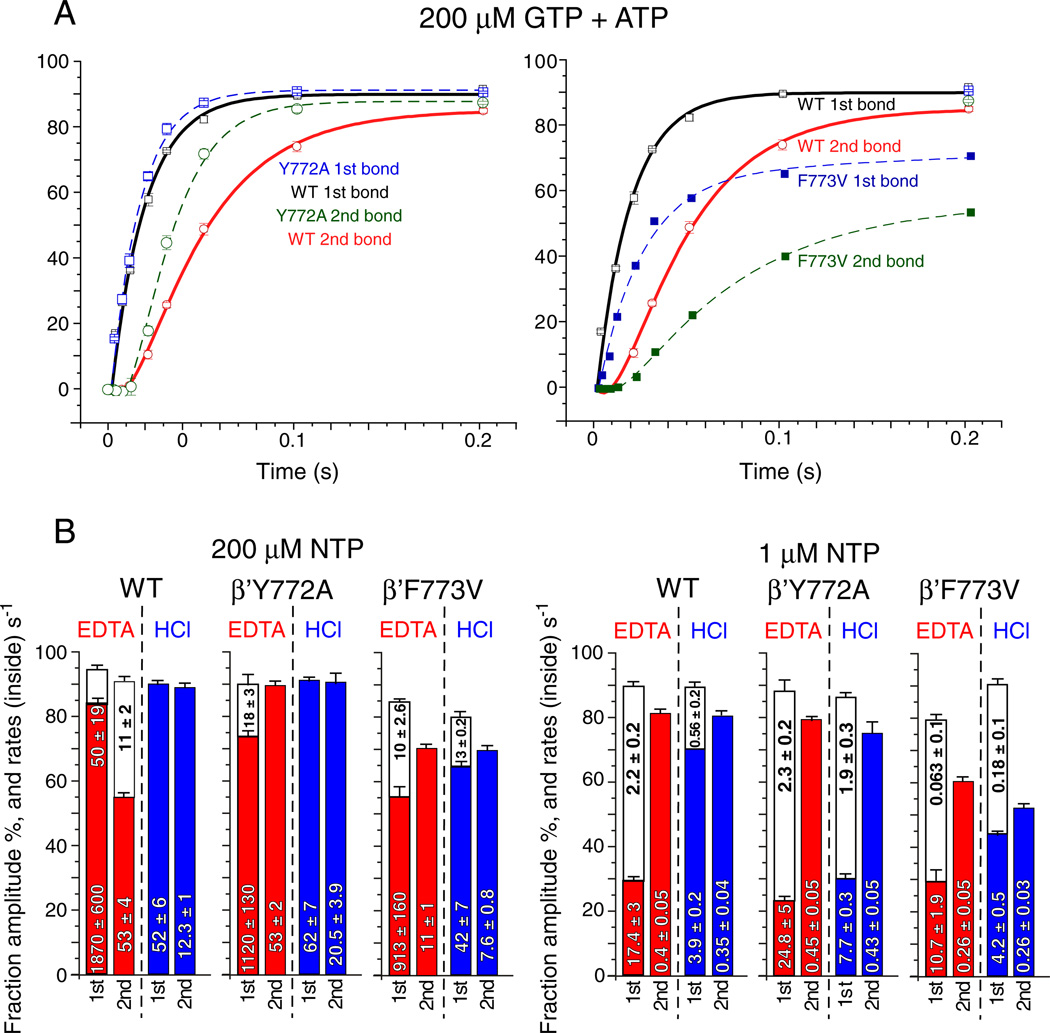
Fig. 6.
Reproducibility and model-independent analysis of the fast kinetic experiment. A) HCl quench data at 200 µM GTP + ATP (as in Fig. 4) was obtained in triplicate for wild type and Y772A RNAPs and compared to F773V RNAP. Error bars indicate standard deviation of three replicates. B) Model-independent analysis. Apparent rates (s−1) and amplitudes (% of TECs) based on double or single phase exponential curve fitting. Error bars indicate standard error.
Fig. 5.
Elongation of G9 RNAP TECs at 1 µM GTP and ATP (as in Fig. 4). A) Wild type RNAP. B) Y772A RNAP. C) F773V RNAP.
Model-independent analysis of rate curves is shown in Fig. 6B. Apparent rates and amplitudes (A1 and A2) were determined by fitting to two phase (y = y0 + A1*(1 − e^(− t*k1)) + A2*(1 − e^(− t*k2))) or single phase (y = y0 + A1*(1 − e^(− t*k1))) exponential curves, as appropriate. A1 and A2 are amplitudes (% of TECs); k1 and k2 are rates (s−1). For many of these rate curves, single-phase exponential fits are inadequate. Those rate curves fit with a double phase exponential equation could not be adequately fit using a single phase exponential equation [26,27].
Of particular interest is the very fast rate of stable GTP-Mg2+ sequestration for G10 synthesis (k ~ 1870 s−1; EDTA quench) at 200 µM GTP + ATP for the wild-type RNAP (Figs. 4 and 6B). Comparing the interval between the HCl quench for G10 synthesis and the EDTA quench for A11 synthesis provides insight into the processive transition between G10 phosphodiester bond synthesis (HCl quench) and stable ATP-Mg2+ loading for A11 synthesis (EDTA quench). From structural and mechanistic studies, this interval is expected to include trigger loop relaxation, pyrophosphate release and translocation [21,28]. Comparing this interval at 200 (Fig. 4) and 1 µM (Fig. 5) GTP + ATP, it appears that, at 1 µM GTP + ATP, this transition becomes surprisingly slow (Fig. 6B; k ~ 0.4 s−1 (EDTA quench for A11 synthesis)). This result may indicate an ATP-dependent step between G10 phosphodiester bond synthesis (HCl quench) and EDTA-resistant ATP-Mg2+ loading for A11 synthesis (EDTA quench) that may not be directly related to ATP loading to the RNAP i + 1 catalytic site [29–32].
Another way to view this issue is that, for G10 synthesis, there are faster and slower rates at 1 µM GTP, indicating two processes for stable GTP loading (Figs. 5 and 6B). By contrast, at 1 µM ATP, for ATP loading for A11 synthesis, only the much slower rate is observed, indicating the loss of the mechanism underlying the faster rate seen for G10 synthesis. The interpretation is that the stalled G9 TEC has faster (k ~ 17.4 s−1) and slower (k ~ 2.2 s−1) mechanisms to acquire a stably bound GTP, but only a much slower mechanism for stable ATP binding (k ~ 0.4 s−1) is available to newly synthesized G10 TECs at 1 µM ATP. Very slow kinetics for A11 synthesis, therefore, may not be consistent with ATP loading being the only ATP-dependent event for A11 synthesis. For instance, the strong ATP dependence of the G10 → A11 interval could be considered evidence for ATP-stimulated trigger loop opening, translocation and/or pyrophosphate release [21,29,31–33].
Fig. 6A presents a direct comparison between Y772A, F773V and wild type RNAP for G10 and A11 phosphodiester bond synthesis (HCl quench) at 200 µM GTP + ATP. Y772A is slightly faster than wild type RNAP for G10 and A11 synthesis (left panel). F773V is slower than wild type RNAP for G10 and A11 synthesis, at least in part because F773V stalls more strongly at the G9 TEC position (right panel). For G10 addition, Y772A and F773V show opposite effects relative to wild type RNAP. However, as is also shown in Fig. S1, F773V is not a “fast” RNAP at every template position under all circumstances. For instance, F773V RNAP was significantly faster than the wild type in a bulk elongation assay at 1 µM NTPs (Fig. S1) and also poorly recognized the opsP and hisP pause sites (Fig. 2) [18]. G9 appears to be a position at which F773V is slower than wild type and Y772A and is also more prone to stalling. It appears that F773V suppresses pausing at many template positions causing F773V to appear rapid in elongation, particularly at lower NTP concentrations. However, F773V recognizes some pause sites that are less strongly selected by wild type and Y772A RNAP, which can cause F773V to appear slower than wild type and Y772A RNAP at other template positions. These differences in pause site recognition and response to NTPs can cause F773V to either appear faster or slower than wild type RNAP in elongation (Fig. S1).
The biphasic nature of the EDTA quench curves might be explained by equilibration of RNAP between a catalytically-active post-translocation state and an inactive pre-translocation counterpart, which is incapable of stable NTP-Mg2+ binding. Then, the fast phase of the curve (also called the EDTA burst) corresponds to rapid binding and sequestration of a GTP (G10, k ~ 1870 s−1 for wild type) in the vacant active center in the post-translocated G9 TEC. The slower phase (G10, k ~ 50 s−1) derives from the pre-translocated fraction of the same complex, which requires forward translocation to vacate the active center in the enzyme for NTP binding. Therefore, the ~50 s−1 rate observed in TEC G9 for GMP incorporation might reflect the forward translocation rate of the G9 TEC. Notably, there is a ~5-fold difference in the slow rate for G10 (EDTA quench, 200 µMNTPs) between the wild type RNAP (k~50 s−1) and F773V mutant (k ~ 10 s−1) (Fig. 6B). This difference suggests that the F773V substitution may slow forward translocation at G9 [15].
The apparent biphasic nature of HCl quench rate curves is more difficult to explain, although similar observations have been reported by others [27]. The interpretation of the HCl quench is that this is the timing for phosphodiester bond formation, but fitting biphasic kinetics to catalysis is challenging using a linear pathway kinetic model, which would be expected to result in monophasic kinetics for the HCl quench data. Biphasic kinetics for phosphodiester bond formation might be more consistent with a branched elongation mechanism. We infer from this observation that the RNAP mechanism, which includes conformational changes, translocation, NTP loading, NTP sequestration, bond formation and pyrophosphate release, may be more complex than a simple linear model. In order to fit RNAP elongation to an adequate kinetic model, additional experiments will be necessary.
3.4. Transcriptional fidelity
Because Y772A and F773V have altered pausing, termination and transcriptional properties, YFI substitutions were tested in transcriptional fidelity assays (Fig. 7) [5,20,27,33]. In Fig. 7A, a competition fidelity assay was done in the presence of high ATP (the non-cognate substrate for G10 synthesis) and low GTP (the cognate substrate for G10 synthesis) [22]. Fidelity normalized for ATP and GTP concentrations was calculated as described in Experimental Procedures. The competition assay has some advantages over other determinations of fidelity, because tight error bars indicate high reproducibility and because inspection of the gel provides a visual recognition of misincorporation products compared to accurate products (Fig. 7A; compare G10A11 to A*10A11) [22]. Fidelity measurements (right panel) indicate that, at equal ATP and GTP concentrations, wild type RNAP would make about 1 error (AMP misincorporation) per 75,000 nucleotide additions at the G10 position. Significantly, F773V RNAP has decreased fidelity (about 1 error in 25,000 bond additions) and Y772A has enhanced fidelity (about 1 error in 225,000 bond additions) compared to wild type RNAP. F773V and Y772A, therefore, appear to have opposite effects on fidelity relative to wild type RNAP. Interestingly, fidelities of YFI substitutions correlate with time-resolved Exo III mapping experiments that indicate that F773V (reduced fidelity) has a restrained and Y772A (elevated fidelity) has a hyper-active translocation ratchet [15].
Consistent with the results shown in Fig. 7A, Fig. 7B shows UMP (from UTP) misincorporation rates at the C34 position. These misincorporation rates are not normalized to accurate elongation rates, but the bulk elongation rates of YFI mutants are very similar to those of the wild type RNAP (Figs. 2 and S1), so UMP for CMP misincorporation rates likely reflect the fidelity of NTP recognition. Remarkably, the results in Fig. 7B are qualitatively the same as those in Fig. 7A for normalized AMP for GMP misincorporation. For UMP for CMP misincorporation, F773V appears error-prone, Y772A makes few errors and 772-YFI → AAA-774 appears similar to wild type RNAP (Fig. 7B). Interestingly, the result with the triple alanine substitution indicates that its effects on transcriptional fidelity may be a cancelation of the opposite effects of the Y772A (increased fidelity) and F773V (error prone) substitutions.
3.5. Exonuclease III mapping
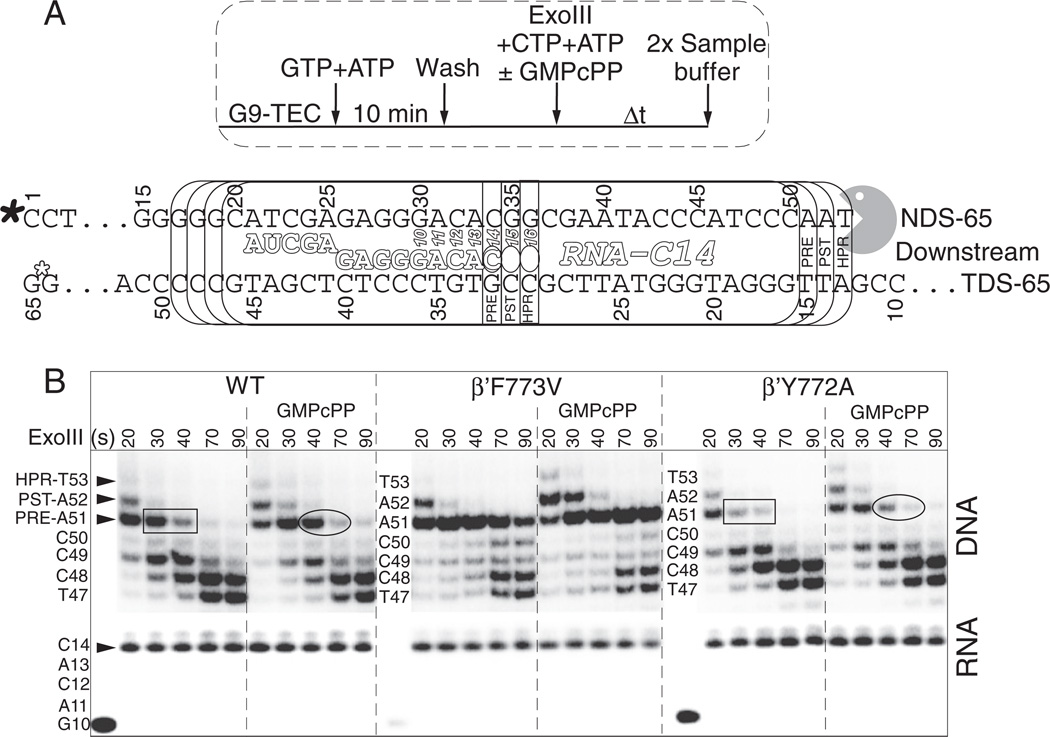
Exo III was used to map the downstream border of the C14 TEC and to gain insight into the resting translocation distribution of Ec RNAP (Fig. 8) [5,15,20,34,35]. DNA band assignments and the inferred active site RNA positions are indicated in Fig. 8A. The post-translocated C14 TEC results in Exo III digestion 18 nucleotides downstream from the RNA 3′ end (counting from the next DNA nucleotide downstream from the RNA 3′ end). For the pre-translocated TEC, the Exo III cleavage site is 17 nucleotides downstream of the RNA 3′ end. Transient backtracking by one or two nucleotides can result in Exo III digestion to 16 and 15 nucleotides downstream from the RNA 3′ end. Details and interpretations of the Exo III experiment and mapping from both the upstream and downstream RNAP borders have recently been published from our laboratory, and these studies provide additional information about the Y772A and F773V substitutions [15].
Fig. 8.
RNAP C14 TEC downstream border Exo III mapping indicating the slow post → pre and pre → backtracked transitions. A) Protocol and schematic. In vitro assembled Ec RNAP G9 TECs were extended to G10 and washed. After releasing TECs from beads, ATP and CTP were added together with Exo III to extend the TEC to C14 and to map the downstream RNAP border, as indicated. Filled asterisk indicates the position of the 5′ DNA radiolabel. Open asterisk indicates that the template DNA strand is blocked from Exo III digestion using a thio linkage. B) Exo III mapping (20–90 s incubation at 25 °C) and RNA gel data. 500 µM GMPcPP was added to the indicated reactions as a non-incorporatable GTP analog to stabilize the hyper-, post- and pre-translocated TEC Exo III gel bands. Rectangles and ovals highlight apparent differences in the stability of DNA A51 for wild type and Y772A RNAPs.
To form C14 TECs, G9 TECs were assembled in vitro. Immobilized G9 TECs were walked to G10, washed and released from beads. ATP and CTP (10 µM each) were added together with Exo III for 20 to 90 s, so, during Exo III digestion, the TECs were stalled at C14. GMPcPP, which is accurately templated at G15 and G16, is expected to stabilize the post-translocated TEC (Exo III cleavage to produce DNA A52). GMPcPP, therefore, can preserve the digestion fragment that represents the pre-translocated TEC (A51), by binding to and stabilizing the post-translocated TEC. Furthermore, GMPcPP is expected to reduce the time the TEC dwells in the pre-translocated register, from which it could backtrack, resulting in Exo III digestion to DNA C50, C49 and shorter lengths.
Because Exo III digests DNA 3′ → 5′ and because a DNA that is digested by Exo III remains shortened (even when RNAP translocates forward), band patterns do not represent the distribution of translocation states. Rather, Exo III provides a record of the most extreme (in this case upstream) translocation excursions made by RNAP during Exo III digestion, assuming DNA hydrolysis by Exo III at functional excess is reasonably efficient. As a result, with downstream RNAP border mapping, GMPcPP stabilizes both the post- and pre-translocation states (Exo III bands DNA A52 and A51).
The post-translocated RNAP TEC at C14 appears to be long-lived, indicating that RNAP may primarily reside in the post-translocation state [15]. The post-translocated TEC is apparently detected for 20–30 s of Exo III mapping (at Exo III functional excess) in the absence of GMPcPP and for 20–40 s of mapping in the presence of GMPcPP. Because these TECs may not have accessed the pre-translocated register during this extended time interval, the post-translocated register appears to be very stable, and the C14 post → pre rate appears therefore to be slow (k ~ 0.4 to 0.03 s−1; t1/2 ~ 2–25 s). In Fig. S4, a very consistent Exo III downstream border mapping experiment of an Ec RNAP A39 TEC is shown.
The results of Exo III mapping reveal that wild type and Y772A RNAP are similar to one another but startlingly distinct from F773V. As expected, GMPcPP stabilizes the post-translocation registers in all three RNAPs. Compared to wild type and Y772A RNAPs, forward translocation of F773V is strongly stimulated by addition of GMPcPP or GMP added as a non-incorporatable GTP analog (Figs. 8 and S4). Because F773V RNAP appears to efficiently load and/or to retain GMPcPP or GMP as a GTP analog, F773V might be expected to be error-prone, as was shown to be the case in Fig. 7. Notably, F773V RNAP appeared more resistant to backtracking at the C14 position with and without GMPcPP present (Fig. 8B). The resistance of F773V to backtracking might indicate stiffness of the bridge helix acting as a translocation ratchet. This conclusion is supported in reference [15], in which time-resolved Exo III mapping is shown from the downstream and upstream RNAP borders for wild type, Y772A and F773V RNAPs. In those experiments, in the absence of an incoming NTP analog, Y772A appears to have hyper-active and F773V appears to have restrained translocation [15]. A restrained translocation ratchet for F773V is consistent with its reduced intrinsic termination, suppressed pausing and lowered fidelity. Somewhat subtle differences between wild type and Y772A RNAP, consistent with a more active translocation ratchet in Y772A, are highlighted in Fig. 8.
4. Discussion
4.1. Bridge helix dynamics
Time-resolved Exo III mapping of RNAP TECs from both upstream and downstream boundaries provides insight into bridge helix dynamics [15]. In the absence of incoming NTPs or analogs, F773V RNAP is observed to restrain hyper-translocation, when mapped from the upstream border, and to restrict backtracking, when mapped from the downstream border (Fig. 8) [15]. Y772A, by contrast, appears more prone to both backtracking (Fig. 8) and hyper-translocation than wild type RNAP [15]. Because Y772A and F773V are bridge helix substitutions that affect translocation, we interpret these results to indicate that, in the absence of NTP addition, F773V substitution appears to decrease and the Y772A substitution appears to increase bridge helix bending and flexibility. We also consider these results with YFI substitutions in Exo III mapping to be evidence that bridge helix bending has a central role in RNAP translocation.
4.2. Properties of YFI mutants
Ec RNAP F773V is surprisingly different from wild type and Y772A RNAP in a broad spectrum of transcriptional elongation properties. Compared to Y772A and wild type RNAPs, F773V is strongly defective in recognition of the λ tR2 intrinsic terminator (Fig. 3) but not the stronger hisT terminator (Fig. 2) [23]. F773V also has a distinct elongation pattern, suppressing pausing at many sites but enhancing pausing at some other sites relative to wild type RNAP (Figs. 2 and S1). F773V is an error-prone enzyme, whereas Y772A is more accurate in transcription than wild type RNAP (Fig. 7). Compared to Y772A and wild type RNAP, F773V is more responsive to GMP and GMPcPP acting as GTP analogs to stabilize the post-translocation register (Figs. 8 and S4). Because F773V resists backtracking (Fig. 8), valine substitution may result in a stiffer bridge helix to reduce dynamics and bending [15]. Bridge helix dynamics, therefore, appears to be coupled to pausing, backtracking, NTP sequestration, catalysis, termination, fidelity, NTP selection and forward translocation.
4.3. F773, Y772 and I774 support distinct communication links
In several respects, F773V and Y772A appear to have opposing effects on transcription compared to wild type RNAP. This result seems reasonable because F773 and Y772 project from the bridge helix in different directions to form distinct contacts with RNAP domains. As shown in Figs. 4 and 6A, at 200 µM NTPs F773V is slower and Y772A is faster than wild type RNAP for G10 and A11 syntheses. Furthermore, F773V appears to have a more restrained and Y772A appears to have a more active translocation ratchet than wild type RNAP, judging from upstream border Exo III mapping [15]. In transcriptional fidelity, F773V is error prone and Y772A makes fewer errors than wild type RNAP (Fig. 7). At 40 mM KCl, F773V is strongly resistant and Y772A is more efficient than wild type RNAP at λ tR2 terminator recognition (data not shown). Differences between Y772A and wild type RNAP in λ tR2 recognition are suppressed at 150 mM KCl (Fig. 3).
F773 primarily contacts the fork domain, and Y772 primarily interacts with the link domain including a specific hydrogen bond to β D674 that is broken in Y772A (Figs. 1 and S5–S7) [14,36]. The F773V substitution is expected to affect bridge helix bending and dynamics and to weaken coupling of the bridge helix to the fork. A more complete and detailed discussion of Y772 and F773 atomic contacts and the anticipated effects of substitutions at these positions is found in Supplementary Materials (Figs. S5–S7). I774 interacts with the F-loop, which contacts the 778-GARKG-782 glycine hinge (Fig. S5). The base of the F-loop contacts the link domain near the active site through an ion pair between β′ R744 and β D675 (Fig. S5).
4.4. Translocation and fidelity
Bridge helix bending against the RNA/DNA hybrid has been hypothesized to induce translocation [8,9]. The F773 residue of the bulky bridge helix 772-YFI-774 appears to be very important in synchronizing the motions and dynamics of the bridge helix, the link domain, the fork and the F-loop. Both the link domain and the extended fork approach the RNAP active site, potentially coupling motions at YFI on the bridge to active site function. We posit, therefore, that YFI and the 778-GARKG-782 glycine hinge respond to the stage of the nucleotide addition cycle through these remote connections between the bridge helix and the active site (Figs. 1 and S5–S7) [5,11]. The F-loop makes contacts to I774 and to the 778-GARKG-782 glycine hinge.
The strong effects of the F773V substitution on intrinsic termination, pausing, fidelity, NTP selection and translocation are very consistent with a model in which bending of the bridge helix 778-GARKG-782 glycine hinge is strongly coupled to motions of YFI and surrounding protein. A strong stimulation of the NTP-stabilized post-translocated state of the RNAP TEC by F773V substitution (Fig. 8B) suggests that the wild type F773 residue enhances flexibility of the adjacent hinge probably by interaction with the fork. By reducing fork contact, F773V substitutionmay delay forward translocation and decrease isomerization of the active center into the closed conformation after NTP binding. This hypothesis may explain a significant stimulation of forward translocation of F773V by the incoming NTP (Figs. 8 and S4) and reduced F773V fidelity (Fig. 7). By these two properties, F773V appears very similar to the previously characterized Sc RNAP II Rpb1 E1103G trigger loop hinge substitution, which also exhibits enhanced forward translocation in the presence of the next NTP and reduced transcriptional fidelity [20].
4.5. RNAP may stall primarily in the post-translocated state
The majority of X-ray crystal structures of RNAP TECs appear to be primarily post-translocated [3,4,37,38]. Using fluorescence changes to monitor Ec RNAP translocation, Malinen et al. also concluded that RNAP rests primarily in the post-translocated state [39]. From Exo III mapping of the Ec RNAP TEC downstream border, the post → pre transition appears to be very slow at least for some DNA sequences, requiring 15–40 s to complete, even in the absence of a NTP analog (Figs. 8 and S4). Based on these results, it appears that the Ec RNAP TEC may stall in a primarily post-translocated position. Recent work from our laboratory on time-resolved Exo III footprinting of Ec RNAP TECs also supports this conclusion [15].
4.6. Bridge helix flexibility in fidelity
Transcriptional fidelity is proposed to depend on NTP orientation and to be enhanced by active site dehydration through trigger loop closing during or just after phosphodiester bond synthesis [5]. Here, we indicate that bridge helix bending and dynamics may also be important because F773V, which appears to restrict bridge helix bending (Fig. 8), is reduced in transcriptional fidelity (Fig. 7). Y772A, which has enhanced transcriptional fidelity, appears to have enhanced bridge helix dynamics, which might result in more rapid NTP exchange when a NTP or analog is inaccurately loaded [15]. Alternatively, F773V and Y772A may alter fidelity by influencing NTP orientation through unknown mechanisms. Sc RNAP II Rpb1 E1103G, which locates to the more C-terminal hinge at the base of the trigger loop (designated hinge H2), may reduce fidelity by limiting bridge helix bending by increasing the flexibility of the H2 trigger loop hinge [5], because increased flexibility in the trigger loop in E1103G may limit the loop's capacity to bend the bridge helix. Consistent with this interpretation, deletion of the trigger loop also appears to stiffen the bridge helix [15]. As a simple working model, therefore, we posit that a more flexible bridge helix hinge may enhance fidelity (Y772A) by increasing the likelihood of NTP exchange, and a less flexible bridge helix hinge (F773V, Sc Rpb1 E1103G) may decrease fidelity by causing NTPs in the active site to be less exchangeable. Currently, we are attempting to test this correlation using more extensive Exo III and fidelity analyses. Models for YFI interaction with the 778-GARKG-782 hinge, surrounding protein and the active site will be challenged by molecular dynamics simulations of F773V, Y772A, I774A and wild type RNAP in TECs with open and closed trigger loop conformations.
4.7. YFI
From molecular dynamics simulations [5], the β′ 772-YFI-774 motif appears to move as part of the surrounding N-terminal segment (β′ 769-VLQYFISTH-777) of the bridge helix leading up to the glycine hinge β′ 778-GARKG-782 at which bending mostly occurs. In simulations, the C-terminal segment of the bridge helix (β′ 783 to 806) also appears to move as an intact helical unit. Depending on whether the trigger loop is in a closed (catalytic) or an open (translocating) conformation and depending on bending at the bridge helix hinge, motions of the C-terminal bridge helix segment are either highly correlated or highly anti-correlated with those of the N-terminal segment including YFI. This result indicates that the N- and C-terminal helical segments of the bridge helix tend to remain helical, whereas the hinge is much more dynamic and mobile [5,13]. Combined with functional studies, these simulations indicate that hinge bending and dynamics regulate bridge helix function in catalysis, pausing, termination, fidelity and translocation. During simulations, N- and C-terminal helical segments of the bridge remain intact and bending appears restricted to the glycine hinge (Fig. S6).
Detailed connections described in this paper for bacterial RNAPs are somewhat different in other RNAPs. For instance, YFI is FFF in Sc RNAP II and Mj RNAP (Fig. 1). The Y → F substitution prevents hydrogen bonding to the link domain through YFI. The glycine hinge in Sc RNAP II and Mj RNAPs is GGREG and in some RNAP IIIs and RNAP Is GREG rather than GARKG, indicating that the glycine hinge is maintained but hinge action is regulated and integrated somewhat differently in different RNAPs. In archaeal RNAPs and RNAP II, the F loop reaches further than the bacterial F loop to contact the link domain and the active site, so a remote connection is made between the bridge helix and active site that cannot form in bacterial RNAPs. Differences in the YFI/FFF motif and the glycine hinge, therefore, might potentially be exploited in RNAP inhibitor development, for instance, in designing new antibiotics.
Supplementary Material
Acknowledgements
This work was supported by the National Science Foundation MCB-1050867 (70%) (to ZFB (PI) and RIC (co-I)) and the National Institutes of Health R01 GM 092949 (30%) to Michael Feig (PI) and ZFB (co-I). This work was supported by National Institutes of Health grant R01 GM58750 to EN. ZFB receives support from Michigan State University, the Michigan State University Agricultural Experiment Station, and the Michigan State University College of Osteopathic Medicine. The contents of this publication do not necessarily reveal the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. government.
Abbreviations
- Ec
Escherichia coli
- Sc
Saccharomyces cerevisiae
- Mj
Methanocaldococcus jannaschii
- Tt
Thermus thermophilus
- Exo III
exonuclease III
- GMPcPP
α,β-methylene guanosine triphosphate
- NTP
nucleoside triphosphate
- PPi
pyrophosphate (diphosphate)
- RNAP
RNA polymerase
- RNA G9
9 nucleotide RNA ending in 3′-GMP
- TEC
ternary elongation complex (RNAP, DNA, nascent RNA)
Footnotes
Appendix A. Supplementary data
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.bbagrm.2012.11.005.
References
- 1.Zhang J, Palangat M, Landick R. Role of the RNA polymerase trigger loop in catalysis and pausing. Nat. Struct. Mol. Biol. 2010;17:99–104. doi: 10.1038/nsmb.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nudler E. RNA polymerase active center: the molecular engine of transcription. Annu. Rev. Biochem. 2009;78:335–361. doi: 10.1146/annurev.biochem.76.052705.164655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vassylyev DG, Vassylyeva MN, Zhang J, Palangat M, Artsimovitch I, Landick R. Structural basis for substrate loading in bacterial RNA polymerase. Nature. 2007;448:163–168. doi: 10.1038/nature05931. [DOI] [PubMed] [Google Scholar]
- 4.Wang D, Bushnell DA, Westover KD, Kaplan CD, Kornberg RD. Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis. Cell. 2006;127:941–954. doi: 10.1016/j.cell.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kireeva ML, Opron K, Seibold SA, Domecq C, Cukier RI, Coulombe B, Kashlev M, Burton ZF. Molecular dynamics and mutational analysis of the catalytic and translocation cycle of RNA polymerase. BMC Biophys. 2012;5:11. doi: 10.1186/2046-1682-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feig M, Burton ZF. RNA polymerase II with open and closed trigger loops: active site dynamics and nucleic acid translocation. Biophys. J. 2010;99:2577–2586. doi: 10.1016/j.bpj.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feig M, Burton ZF. RNA polymerase II flexibility during translocation from normal mode analysis. Proteins. 2010;78:434–446. doi: 10.1002/prot.22560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 A resolution. Science. 2001;292:1876–1882. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- 9.Bar-Nahum G, Epshtein V, Ruckenstein AE, Rafikov R, Mustaev A, Nudler E. A ratchet mechanism of transcription elongation and its control. Cell. 2005;120:183–193. doi: 10.1016/j.cell.2004.11.045. [DOI] [PubMed] [Google Scholar]
- 10.Tan L, Wiesler S, Trzaska D, Carney HC, Weinzierl RO. Bridge helix and trigger loop perturbations generate superactive RNA polymerases. J. Biol. 2008;7:40. doi: 10.1186/jbiol98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weinzierl RO. The nucleotide addition cycle of RNA polymerase is controlled by two molecular hinges in the Bridge Helix domain. BMC Biol. 2010;8:134. doi: 10.1186/1741-7007-8-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heindl H, Greenwell P, Weingarten N, Kiss T, Terstyanszky G, Weinzierl RO. Cation-pi interactions induce kinking of a molecular hinge in the RNA polymerase bridge-helix domain. Biochem. Soc. Trans. 2011;39:31–35. doi: 10.1042/BST0390031. [DOI] [PubMed] [Google Scholar]
- 13.Seibold SA, Singh BN, Zhang C, Kireeva M, Domecq C, Bouchard A, Nazione AM, Feig M, Cukier RI, Coulombe B, Kashlev M, Hampsey M, Burton ZF. Conformational coupling, bridge helix dynamics and active site dehydration in catalysis by RNA polymerase. Biochim. Biophys. Acta. 2010;1799:575–587. doi: 10.1016/j.bbagrm.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weinzierl RO. The BridgeHelix of RNA polymerase acts as a central nanomechanical switchboard for coordinating catalysis and substrate movement. Archaea. 2011;2011:608385. doi: 10.1155/2011/608385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nedialkov YA, Nudler E, Burton ZF. RNA polymerase stalls in a post-translocated register and can hyper-translocate. Transcription. 2012;3:260–269. doi: 10.4161/trns.22307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miropolskaya N, Artsimovitch I, Klimasauskas S, Nikiforov V, Kulbachinskiy A. Allosteric control of catalysis by the F loop of RNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 2009;106:18942–18947. doi: 10.1073/pnas.0905402106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Artsimovitch I, Chu C, Lynch AS, Landick R. A new class of bacterial RNA polymerase inhibitor affects nucleotide addition. Science. 2003;302:650–654. doi: 10.1126/science.1087526. [DOI] [PubMed] [Google Scholar]
- 18.Svetlov V, Belogurov GA, Shabrova E, Vassylyev DG, Artsimovitch I. Allosteric control of the RNA polymerase by the elongation factor RfaH. Nucleic Acids Res. 2007;35:5694–5705. doi: 10.1093/nar/gkm600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kireeva ML, Komissarova N, Waugh DS, Kashlev M. The 8-nucleotide-long RNA:DNA hybrid is a primary stability determinant of the RNA polymerase II elongation complex. J. Biol. Chem. 2000;275:6530–6536. doi: 10.1074/jbc.275.9.6530. [DOI] [PubMed] [Google Scholar]
- 20.Kireeva ML, Nedialkov YA, Cremona GH, Purtov YA, Lubkowska L, Malagon F, Burton ZF, Strathern JN, Kashlev M. Transient reversal of RNA polymerase II active site closing controls fidelity of transcription elongation. Mol. Cell. 2008;30:557–566. doi: 10.1016/j.molcel.2008.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kireeva M, Nedialkov YA, Gong XQ, Zhang C, Xiong Y, Moon W, Burton ZF, Kashlev M. Millisecond phase kinetic analysis of elongation catalyzed by human, yeast, and Escherichia coli RNA polymerase. Methods. 2009;48:333–345. doi: 10.1016/j.ymeth.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walmacq C, Cheung AC, Kireeva ML, Lubkowska L, Ye C, Gotte D, Strathern JN, Carell T, Cramer P, Kashlev M. Mechanism of translesion transcription by RNA polymerase II and its role in cellular resistance to DNA damage. Mol. Cell. 2012;46:18–29. doi: 10.1016/j.molcel.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnston HM, Barnes WM, Chumley FG, Bossi L, Roth JR. Model for regulation of the histidine operon of Salmonella. Proc. Natl. Acad. Sci. U. S. A. 1980;77:508–512. doi: 10.1073/pnas.77.1.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Epshtein V, Cardinale CJ, Ruckenstein AE, Borukhov S, Nudler E. An allosteric path to transcription termination. Mol. Cell. 2007;28:991–1001. doi: 10.1016/j.molcel.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 25.Gusarov I, Nudler E. The mechanism of intrinsic transcription termination. Mol. Cell. 1999;3:495–504. doi: 10.1016/s1097-2765(00)80477-3. [DOI] [PubMed] [Google Scholar]
- 26.Foster JE, Holmes SF, Erie DA. Allosteric binding of nucleoside triphosphates to RNA polymerase regulates transcription elongation. Cell. 2001;106:243–252. doi: 10.1016/s0092-8674(01)00420-2. [DOI] [PubMed] [Google Scholar]
- 27.Yuzenkova Y, Bochkareva A, Tadigotla VR, Roghanian M, Zorov S, Severinov K, Zenkin N. Stepwise mechanism for transcription fidelity. BMC Biol. 2010;8:54. doi: 10.1186/1741-7007-8-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kireeva M, Kashlev M, Burton ZF. Translocation by multi-subunit RNA polymerases. Biochim. Biophys. Acta. 2010;1799:389–401. doi: 10.1016/j.bbagrm.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 29.Kennedy SR, Erie DA. Templated nucleoside triphosphate binding to a noncatalytic site on RNA polymerase regulates transcription. Proc. Natl. Acad. Sci. U. S. A. 2011;108:6079–6084. doi: 10.1073/pnas.1011274108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Larson MH, Zhou J, Kaplan CD, Palangat M, Kornberg RD, Landick R, Block SM. Trigger loop dynamics mediate the balance between the transcriptional fidelity and speed of RNA polymerase II. Proc. Natl. Acad. Sci. U. S. A. 2012;109:6555–6560. doi: 10.1073/pnas.1200939109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson RS, Strausbauch M, Carraway JK. Rapid pyrophosphate release from transcriptional elongation complexes appears to be coupled to a nucleotide-induced conformational change in E. coli core polymerase. J. Mol. Biol. 2011;412:849–861. doi: 10.1016/j.jmb.2011.05.023. [DOI] [PubMed] [Google Scholar]
- 32.Johnson RS, Strausbauch M, Cooper R, Register JK. Rapid kinetic analysis of transcription elongation by Escherichia coli RNA polymerase. J. Mol. Biol. 2008;381:1106–1113. doi: 10.1016/j.jmb.2008.06.089. [DOI] [PubMed] [Google Scholar]
- 33.Holmes SF, Santangelo TJ, Cunningham CK, Roberts JW, Erie DA. Kinetic investigation of Escherichia coli RNA polymerase mutants that influence nucleotide discrimination and transcription fidelity. J. Biol. Chem. 2006;281:18677–18683. doi: 10.1074/jbc.M600543200. [DOI] [PubMed] [Google Scholar]
- 34.Hein PP, Palangat M, Landick R. RNA transcript 3′-proximal sequence affects translocation bias of RNA polymerase. Biochemistry. 2011;50:7002–7014. doi: 10.1021/bi200437q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Toulokhonov I, Zhang J, Palangat M, Landick R. A central role of the RNA polymerase trigger loop in active-site rearrangement during transcriptional pausing. Mol. Cell. 2007;27:406–419. doi: 10.1016/j.molcel.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 36.Weinzierl RO. Nanomechanical constraints acting on the catalytic site of cellular RNA polymerases. Biochem. Soc. Trans. 2011;38:428–432. doi: 10.1042/BST0380428. [DOI] [PubMed] [Google Scholar]
- 37.Vassylyev DG, Vassylyeva MN, Perederina A, Tahirov TH, Artsimovitch I. Structural basis for transcription elongation by bacterial RNA polymerase. Nature. 2007;448:157–162. doi: 10.1038/nature05932. [DOI] [PubMed] [Google Scholar]
- 38.Brueckner F, Cramer P. Structural basis of transcription inhibition by alpha-amanitin and implications for RNA polymerase II translocation. Nat. Struct. Mol. Biol. 2008;15:811–818. doi: 10.1038/nsmb.1458. [DOI] [PubMed] [Google Scholar]
- 39.Malinen AM, Turtola M, Parthiban M, Vainonen L, Johnson MS, Belogurov GA. Active site opening and closure control translocation of multisubunit RNA polymerase. Nucleic Acids Res. 2012;40:7442–7451. doi: 10.1093/nar/gks383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. (27–38) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.