Abstract
Background & Aims
MicroRNAs (miRNAs) have been shown to be involved in many biological processes by affecting their target gene expression. miR-122 has been extensively studied in hepatocarcinogenesis. However, the role of miR-122 in liver fibrosis remains unknown.
Methods
The mRNA expression levels of miR-122, prolyl 4-hydroxylase subunit alpha-1 (P4HA1), and CCAAT/enhancer binding protein alpha (C/EBPα) were assessed by real-time PCR. The protein expression levels of P4HA1, C/EBPα and collagen, type I, alpha 1 (COL1A1) were analyzed by Western blot and immunofluorescence. MTT assay was used to assess cell proliferation. Chromatin immunoprecipitation (ChIP) assay was used to examine the binding activity of C/EBPα to miR-122 promoter.
Results
miR-122 expression was significantly reduced in transactivated HSCs and in the livers of mice treated with CCl4. Overexpression of miR-122 inhibited the proliferation of LX2 cells. We also demonstrated that P4HA1 was a target gene of miR-122. The mRNA expression level of PAHA1 inversely correlated with that of miR-122 in HSCs and in the mouse liver. Overexpression of miR-122 markedly attenuated the expression of P4HA1 via targeting a binding site located at 3′-UTR of P4HA1 mRNA. We further showed that miR-122 overexpression led to decreased collagen maturation and ECM production. Finally, the binding activity of C/EBPα to miR-122 promoter was significantly decreased in activated HSCs.
Conclusions
Our study suggests that miR-122 may play an important role in negatively regulating collagen production in HSCs and that targeted expression of miR-122 in HSCs may represent a new strategy for the treatment of liver fibrosis.
Keywords: miRNAs, miR-122, HSC, P4HA1, Collagen maturation, Liver fibrosis
Introduction
miRNAs are short ribonucleic acid (RNA) molecules found in eukaryotic cells, which regulate gene expression by binding to the 3′-untranslated region (3′-UTR) of the target gene mRNAs, to repress protein translation or induce mRNA degradation [1]. Several hundreds of miRNAs have been identified so far. They have been estimated to target about 60% of mammalian genes [2]. Studies on specific miRNAs in animal models have identified distinct roles of miRNAs in cell functions such as development, cell proliferation, differentiation, and apoptosis [3].
Liver cirrhosis is a major cause of morbidity and mortality worldwide, with limited therapeutic options. This advanced liver disease is the result of pathological response to initial liver injuries of various causes [4]. Upon liver damage, HSCs undergo an activation process from a fat storing quiescent phenotype to a highly proliferative myofibroblastic phenotype with significantly increased fibrogenic activity [5]. The build-up of excessive extracellular matrix (ECM) proteins leads to the fibrotic liver with impaired organ function [6]. The mechanisms of the pathogenesis and progression of liver fibrosis are complicated, and may vary with the underlying etiology. Increasing evidence suggests that certain miRNAs are critically involved in the different steps of liver fibrosis, including activation of HSCs [7], their proliferation [8] and production of ECM proteins [9]. The study by Mann et al. [7] showed that miR-132 was significantly suppressed during the transactivation of HSCs. miR-132 appeared to inhibit HSC activation by negatively regulating MeCP2 that encodes methyl-CpG binding protein 2. Increased activity of MeCP2 led to silencing of expression of PPAR-γ, a potent blocker of HSC transactivation. Roderburg et al. [9] reported that members of the miR-29 family were significantly downregulated in activated HSCs, in several mouse models of liver fibrosis. miR-29 members negatively regulated the expression of various fibrosis-related genes, including Col1A1, via targeting the 3′-UTR of their mRNAs [9,10]. They further showed that the activation of NF-κB and/or TGF-β signaling played an important role in reducing the expression of miR-29a in activated HSCs, suggesting that miR-29 acts as an important link between NF-κB/TGF-β signaling and the enhanced fibrogenic activity in activated HSCs [9]. In another study by Ji et al. [8], miRNA-27a and 27b were shown to be downregulated in activated HSCs and overexpression of both miRNAs in activated HSCs led to a partial reversion to quiescent phenotype.
miR-122 is the most abundant and liver-specific miRNA that accounts for 72% of total miRNA in the adult human liver [11]. It has been demonstrated to play a role in regulating hepatocyte growth development [12], differentiation [13], and metabolism [14]. miR-122 is decreased in advanced liver diseases, such as cirrhosis [15] and hepatocellular carcinoma [16]. Mice in which miR-122 was selectively deleted in hepatocytes, displayed altered lipid metabolism, hepatic inflammation, fibrosis, and a high incidence of hepatocellular carcinoma [17,18]. However, the role of miR-122 in HSCs and its implication in liver fibrosis are still unknown. In this study, we show that miR-122 is an important miRNA involved in regulation of collagen maturation by targeting P4HA1. miR-122 may also regulate the proliferation of HSCs. Our findings suggest that miR-122 may play an important role in negatively regulating collagen production in HSCs and that targeted expression of miR-122 in HSCs may represent a novel strategy for the treatment of liver fibrosis.
Materials and methods
Animals and HSC isolation
Male Sprague-Dawley rats from Charles River Laboratories (Wilmington, MA) were used for HSC isolation. All procedures were reviewed and approved by the Institutional Animal Care and Use Committee of the University of Pittsburgh. Primary rat HSCs were isolated as previously described and used 3 days after isolation as quiescent cells. The purity of isolated HSCs was >90% [10]. HSCs were cultured for 7 days to allow transactivation. A mouse model of liver fibrosis was established via intraperitoneal injection of carbon tetrachloride (CCl4; Merck; 0.6 ml/kg of body weight), twice a week for six weeks. Common bile duct ligation (CBDL) was performed according to the literature [9]. Sham-operated rats served as controls.
miRNA isolation and stem-loop real-time RT-PCR
All kits and reagents were obtained from Applied Biosystems (Foster City, CA). Total RNA enriched with miRNAs was isolated from HSCs using the miRVana miR-NA isolation kit. To examine the expression of miRNA-122, stem-loop real-time RT-PCR (SLqRT-PCR) was performed according to the manufacturer’s instructions. The relative miRNA expression was calculated from three different experiments.
Cell culture and miRNA transfection
LX-2, an immortalized human HSC line [24], was kindly provided by Dr. S.L. Friedman, Mount Sinai Medical School, NY. LX-2 cells were cultured and transfected as reported by us previously [10].
Cell proliferation
LX-2 cells were seeded on 96-well plates and were transfected with miRNAs. Cells were assayed for proliferation and cell survival using the MTT (Roche Diagnostics, Indianapolis, IN), according to the manufacturer’s instructions.
Bioinformatics analysis
The miR-122 sequence was analyzed for its predicted target genes using Target-scan 4.1, miRNADA, and Microcosm.
RNA isolation and qRT-PCR
Total RNA was isolated from HSCs using TRIzol, and cDNA was synthesized according to manufacturer’s instructions (Invitrogen, San Diego, CA). The primers for P4HA1, collagen 1A1, Bcl-w, IGFR-1, HNF4, C/EBPα, and GAPDH were obtained from MWG Biotech. First-strand cDNA was synthesized, followed by SYBR green qRT-PCR, using ABI Prism 7300 (Applied Biosystems, Foster City, CA). The relative expression was analyzed, and data were collected from three different experiments, as reported by us previously [10].
Western blot analysis
Western blots were performed using anti-COL1A1, anti-P4HA1, anti-C/EBPα, and anti-β-actin antibodies (Santa Cruz Biotechnology) following our published method [10].
Immunofluorescence
LX-2 cells were grown on chamber slides and miR-122 transfection was carried out as described before. Thirty-six hours following the transfection, LX-2 cells were fixed in 4% paraformaldehyde/PBS for 15 min at room temperature and washed with PBS three times. Cells were blocked with 2% bovine serum albumin in PBS for 1 h followed by incubation with anti-COL1A1 antibody (Santa Cruz Biotechnology) (1:200) for 16 h at 4 °C. After washing with PBS, a FITC-labeled secondary antibody (1:1000) was applied and incubated for 60 min. After an additional washing, cells were mounted and analyzed by fluorescence microscopy.
Chromatin immunoprecipitation (ChIP) assay
ChIP assay was performed as previously described [10]. Primer sequences used to amplify a fragment that contains a putative C/EBPα binding site on miR-122 promoter are shown in Supplementary Table 1. Briefly, quiescent and activated cells were treated with 1% formaldehyde for 10 min at 37 °C. After lysis, lysate was sonicated to shear DNA to lengths between 200 and 1000 bp. The sheared chromatin was used for immunoprecipitation with either normal rabbit IgG (negative control) or 5 μg of anti-C/EBPα (14AA, Santa Cruz Biotechnology). Immunoprecipitates were washed, eluted, and decrosslinked. After proteins and RNA were degraded by treatment with proteinase K and RNase A, DNA was purified using QIAquick PCR Purification Kit (QIAGEN). Real-time PCR was performed by using SYBR Green Master Mix (Applied Biosystems) on a 7300 real-time PCR system (Applied Biosystems).
Statistical analysis
All the experiments were performed in triplicate and at least three times. Data were expressed as means ± SEM and calculated using variance analysis and the Newman–Keuls test for multiple comparisons among groups. p <0.05 was considered as statistically significant.
Results
miR-122 is downregulated in activated HSCs and the fibrotic liver
miR-122 is the most abundant miRNA in the liver and its expression in the liver was decreased in advanced liver diseases, such as primary biliary cirrhosis (PBC) and hepatocellular carcinoma (HCC) [14,15]. To further study the role of miR-122 in liver fibrosis, we examined the expression level of miR-122 in mice treated with CCl4 for 6 weeks, a protocol that reproducibly produces liver fibrosis. Fig. 1A shows that repeated CCl4 treatment led to significant downregulation of miR-122 in the mouse liver. This result was similar to that observed in human patients with PBC [14].
Fig. 1. miR-122 is downregulated in activated HSC and the fibrotic liver.
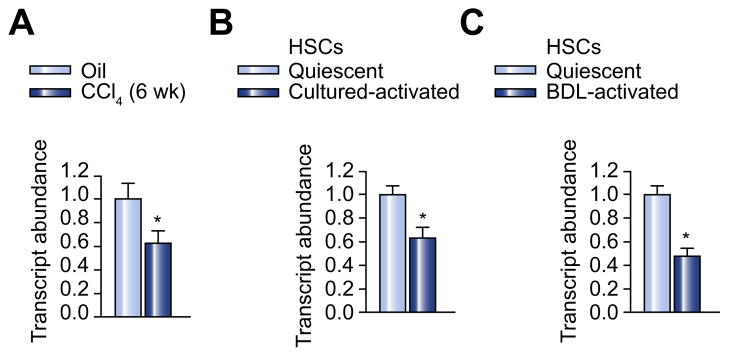
(A) Expression of miR-122 was decreased in the liver of mice treated with CCl4. (B) Expression of miR-122 was decreased in culture-activated rat HSCs. (C) Expression of miR-122 was decreased in CBDL-activated rat HSCs. miR-122 expression was determined by qPCR. n = 6 per group. Results are expressed as means ± SE. p <0.05.
All reported studies on miR-122 in liver diseases have examined its expression in either the intact liver or cultured hepatocytes. Little is known about the expression of miR-122 in HSCs and its implication in chronic liver diseases such as liver fibrosis. Therefore, we examined the expression of miR-122 in isolated, quiescent rat HSCs and compared it with that in isolated rat hepatocytes. miR-122 expression was also examined in LX2 cells, an immortalized human hepatic stellate cell line. Our data show that miR-122 was constitutively expressed in both human and rat HSCs, although expression levels were relatively lower than those in hepatocytes (Supplementary Fig. 1). However, the expression of miR-122 was significantly downregulated in rat HSCs upon culture-induced activation (Fig. 1B). The expression level of miR-122 was also significantly decreased in HSCs isolated from rats, 3 weeks following CBDL (Fig. 1C).
Overexpression of miR-122 suppresses proliferation of HSCs
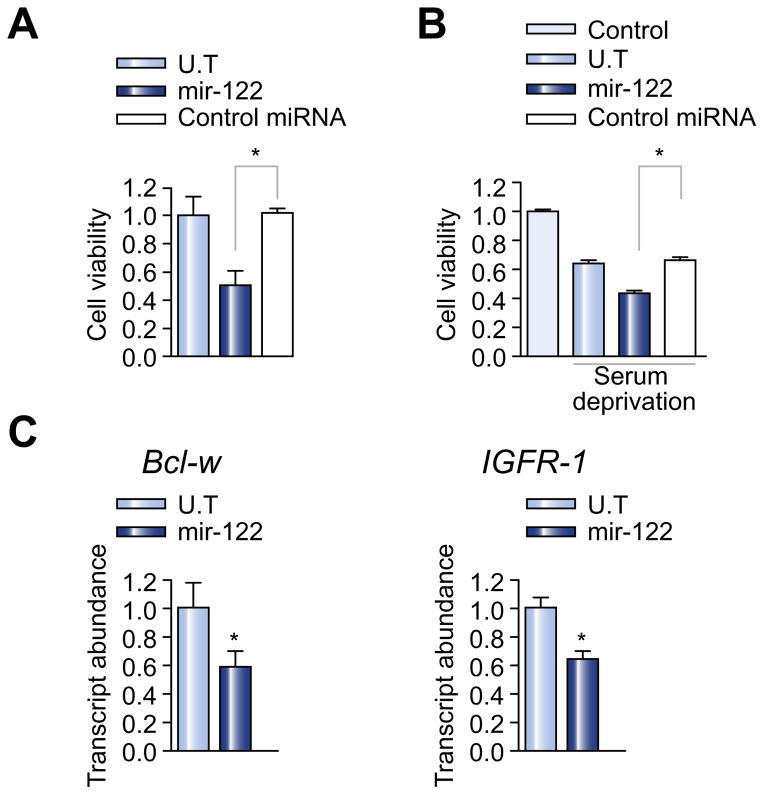
In light of the decreased expression of miR-122 in activated HSCs, we next evaluated the direct effect of miR-122 on LX2 cells. LX2 cells were transiently transfected with miR-122 or a control miRNA. Five days after transfection, cell proliferation was examined by MTT assay. As shown in Fig. 2A, overexpression of miR-122 led to a moderate inhibition of cell proliferation in comparison with the control group. A similar cell growth inhibition was observed when this experiment was performed in serum deprivation condition (Fig. 2B).
Fig. 2. Overexpression of miR-122 suppresses proliferation of HSCs.
(A) LX2 cells were transfected with miR-122 or control miRNA. 5 days after transfection, MTT assay was used to examine cell proliferation. (B) Cells were similarly transfected as described above, followed by serum deprivation. Seventy-two hours after serum deprivation, cell proliferation was determined by MTT assay. (C) Bcl-w and IGFR-1 mRNA expression in miR-122-transfected LX-2 cells was determined by qPCR. Values are means ± SE of three independent experiments (p <0.05). U.T, untreated.
Overexpression of members of the Bcl-2 family and enhanced IGF-1 signaling have been shown to play a role in the high proliferation rate of activated stellate cells [19,20]. Bcl-w and IGFR-1 are two reported target genes of miR-122 [21,22]. Therefore, we examined the effect of miR-122 on the mRNA expression levels of Bcl-w and IGFR-1. Fig. 2C shows that treatment of LX2 cells with miR-122 led to significant downregulation of mRNA expression levels of both genes, suggesting a potential role of Bcl-w and IGFR-1 downregulation in miR-122-mediated growth inhibition.
miR-122 downregulates the expression of P4HA1 via targeting the 3′-UTR of P4HA1 mRNA
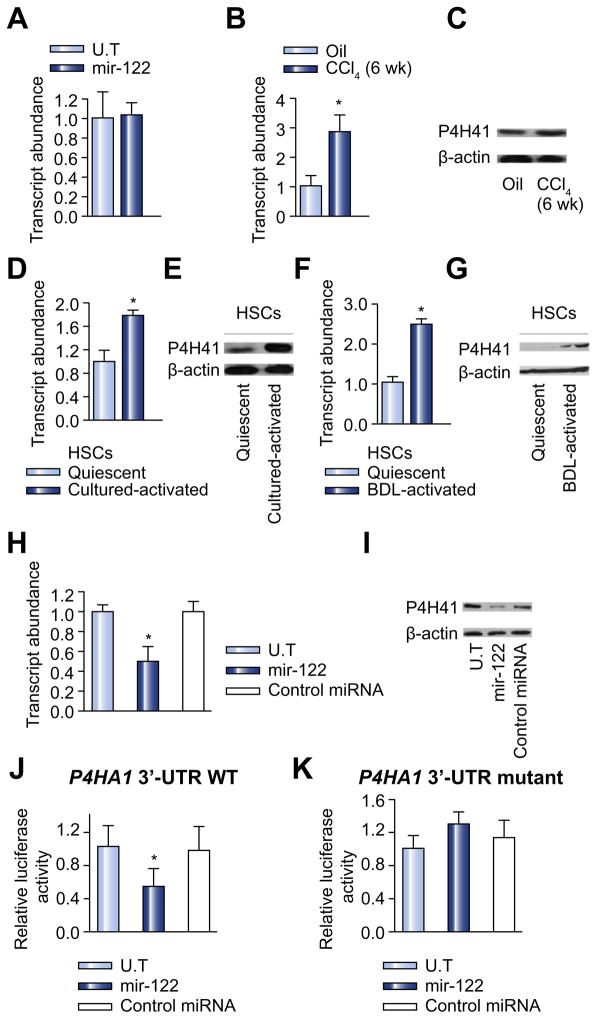
After demonstrating the growth inhibitory effect of miR-122 on HSCs, we investigated whether miR-122 also affects the fibrogenic activity in activated HSCs. A preliminary study showed that overexpression of miR-122 in LX-2 cells had no effect on the mRNA expression of a number of fibrosis-related genes, including Col1A1 (Fig. 3A). These data were consistent with the results of a bioinformatics analysis with multiple algorithms (Targetscan 4.1, MiRnada and Microcosm) showing that none of them was a predicted target gene of miR-122. However, P4HA1, which encodes a component of prolyl 4-hydroxylase, a key enzyme in collagen maturation, was identified to be a putative target gene of miR-122. This prompted us to examine the expression levels of P4HA1 in the fibrotic liver and activated HSCs, and to correlate their expression levels with those of miR-122. Fig. 3B and C show that the expression of P4HA1 in the mouse liver was significantly increased at both mRNA (Fig. 3B) and protein (Fig. 3C) levels following CCl4 treatment. The expression level of P4HA1 in the mouse fibrotic liver was inversely correlated with that of miR-122 (Fig. 1A). A similar result was observed for the mRNA (Fig. 3D) and protein expression (Fig. 3E) of P4HA1 in primary rat HSCs upon culture-induced activation. P4HA1 expression was also significantly upregulated at both mRNA (Fig. 3F) and protein (Fig. 3G) levels in HSCs isolated from rats, 3 weeks following CBDL. To investigate whether P4HA1 expression is, indeed, regulated by miR-122, we examined P4HA1 expression at both mRNA and protein levels before and after transfection with miR-122. As shown in Fig. 3H, overexpression of miR-122 significantly reduced the mRNA level of P4HA1 in LX-2 cells. In contrast, the control miRNA had no effect on P4HA1 mRNA expression. miR-122 also inhibited the expression of P4HA1 at protein level in a sequence-specific manner (Fig. 3I).miRNAs regulate the expression of target genes by binding to the 3′-UTR of specific mRNAs and triggering mRNA degradation or translational repression. Bioinformatics analysis suggested the presence of a putative target site for miR-122 in the 3′-UTR of P4HA1 mRNA. To confirm whether P4HA1 is, indeed, a direct target of miR-122 in HSCs, a ~1000-bp fragment of the P4HA1 3′-UTR containing the putative target sequence, was cloned into a luciferase reporter vector (pmiR-122-wt) (Supplementary Fig. 2). A control reporter plasmid, in which the putative target site was deleted, was similarly constructed (Supplementary Fig. 2). The effect of miR-122 on the luciferase activity from each construct in CV-1 cells was then examined. As shown in Fig. 3J, miR-122 suppressed the luciferase activity of the pmiR-122-wt by about 50%. miR-122 inhibitor did not show significant effect on reporter expression possibly due to a very low level of endogenous miR-122 expression in CV-1 cells. The inhibitory effect of miR-122 on the reporter gene expression was essentially abolished when the putative binding site of miR-122 was deleted from the cloned P4HA1 3′-UTR fragment (Fig. 3K). These data strongly suggest that P4HA1 is a direct target of miR-122.
Fig. 3. miR-122 downregulates the expression of P4HA1 via targeting the 3′- UTR of P4HA1 mRNA.
(A) mRNA expression of collagen 1A1 was examined in miR-122 transfected cells by qPCR. n = 6 per group. (B and C) mRNA (B) and protein (C) expression levels of P4HA1 in livers of CCl4-treated or oil-treated CD-1 mice were analyzed by qPCR and Western blot. n = 6 per group. (D and E) mRNA (D) and protein (E) expression levels of P4HA1 in cultured-activated HSCs were analyzed by qPCR and Western blot. n = 6 per group. (F and G) mRNA (F) and protein (G) expression levels of P4HA1 in CBDL-activated HSCs were analyzed by qPCR and Western blot. n = 6 per group. (H and I) HSCs were transfected with miR-122 mimic or non-specific control miRNA mimic. P4HA1 mRNA expression was analyzed by qPCR, 24 h later (H); protein expression levels of P4HA1 were determined by Western blot, 36 h following the treatment (I). n = 3. p <0.05 (vs. control miRNA). (J) HSCs were transfected with a luciferase construct with P4HA1-3′-UTR, in the presence of miR-122 mimic or non-specific control miRNA mimic. Luciferase assay was performed 24 h after transfection. (K) HSCs were transfected with a P4HA1-3′-UTR luciferase construct with miR-122 binding site deletion in the presence of miR-122 mimic or non-specific control miRNA mimic. Luciferase assay was performed 24 h after transfection. Data shown in the panels represent mean ± SE. n = 3; p <0.05 (vs. control miRNA). U.T, untreated.
The above data suggest that activation of HSCs is associated with downregulation of miR-122 and upregulation of P4HA1. TGF-β signaling is known to play a key role in HSC activation. We then examined the effect of TGF-β treatment on the expression of miR-122 and P4HA1 in quiescent rat HSCs. Consistent with previous reports, TGF-β treatment resulted in a significant increase in the mRNA expression level of Col1A1 (Supplementary Fig. 3A). Treatment of HSCs with TGF-β also led to decreased expression of miR-122 with a concomitant upregulation of P4HA1 expression (Supplementary Fig. 3B and C), suggesting a potential role of TGF-β signaling in regulating the expression of miR-122.
Overexpression of miR-122 led to decreased collagen maturation
P4HA1 encodes a component of prolyl 4-hydroxylase, an enzyme that is critically involved in collagen post-translational modification (maturation). As shown in Fig. 3A, overexpression of miR-122 had no effect on the mRNA level of Col1A1. We then investigated whether miR-122 affected the synthesis of mature Col1A1 in HSCs. Fig. 4A shows that the protein expression of mature Col1A1 was significantly decreased in miR-122 transfected cells in comparison with the control group. Immunofluorescence staining of extracellular type I collagen revealed that the staining patterns differed significantly between miR-122-overexpressing cells and control miRNA-treated cells (Fig. 4B). Type I collagen fibrils secreted from miR-122 transfected cells were sparse and weakly stained compared with those secreted from cells in the control group (Fig. 4B). These results indicated that miR-122 overexpression inhibited the maturation of collagen in HSCs.
Fig. 4. Overexpression of miR-122 decreases collagen maturation.
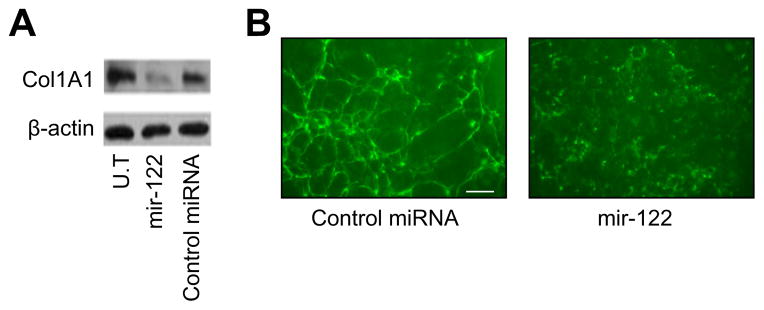
HSCs were transfected with miR-122 mimic or non-specific control miRNA mimic. Thirty-six hours after transfection, (A) mature Col1A1 expression was analyzed by Western blot, and (B) immunofluorescence staining of type I collagen was carried out without permeabilization. Bar: 50 μm. U.T, untreated.
Potential role of decreased expression of HNF4 and C/EBPα in the downregulation of miR-122 expression in the fibrotic liver and activated HSCs
It has been reported that miR-122 is regulated by several liverenriched transcription factors, including C/EBPα and hepatocyte nuclear factor 4 (HNF4) [12]. As an initial approach to the understanding of the potential mechanism responsible for the down-regulation of miR-122 expression, we examined the expression levels of C/EBPα and HNF4 in the mouse fibrotic liver and activated rat HSCs. As shown in Fig. 5A and B, mRNA levels of both transcription factors were significantly decreased in the mouse liver following CCl4 treatment. The mRNA expression of C/EBPα was also decreased in culture-activated rat HSCs (Fig. 5C). Furthermore, the expression of C/EBPα was significantly down-regulated at both mRNA (Fig. 5D) and protein (Fig. 5E) levels in CBDL-activated HSCs. HNF4 expression was below the detection limit in quiescent rat HSCs and remained undetectable when cells were transactivated (data not shown).
Fig. 5. Decreased expression of C/EBPa in activated HSCs may lead to lower expression of miR-122.
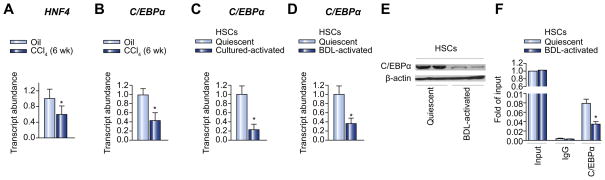
(A and B) mRNA expression levels of HNF-4 (A) and C/EBPα (B), in livers of CCl4-treated or oil-treated CD-1 mice, were analyzed by qPCR. n = 6 per group. (C and D) mRNA expression levels of C/EBPα in culture-activated HSCs (C) and CBDLactivated HSCs (D) were determined by qPCR. n = 6 per group. Results are expressed as means SE. *p <0.05. (E) Protein expression levels of C/EBPα in CBDL-activated HSCs were examined by Western blot. n = 6 per group. (F) The binding activity of C/EBPα to miR-122 promoter in CBDL-activated rat HSCs was analyzed by ChIP assay. The fragment containing a putative C/EBPα binding site was analyzed by qPCR. Results are expressed as means SE. *p <0.05.
To further establish a role of decreased transcription activity of C/EBPα in the downregulation of miR-122, we examined the binding activity of C/EBPα to miR-122 promoter in both quiescent and activated rat HSCs. As shown in Fig. 5F, significantly decreased binding activity of C/EBPα to miR-122 promoter was shown in activated HSCs compared to quiescent HSCs, which may contribute to the downregulation of miR-122 expression in activated HSCs.
Discussion
In this study, we have identified a novel miR-122 regulatory circuit in HSCs that may be implicated in the pathogenesis and progression of liver fibrosis. Our data suggest that miR-122 negatively regulates fibrogenesis through downregulation of the expression of P4HA1 that encodes a component of prolyl 4- hydroxylase. In addition, miR-122 may indirectly inhibit liver fibrosis through inhibition of the proliferation of activated HSCs.
Various miRNAs have been reported to regulate liver fibrosis through different mechanisms. Some miRNAs inhibit liver fibrosis through inhibiting the transactivation of HSCs (e.g., miR-27a, b [8]) or promoting apoptosis of activated HSCs (e.g., miR-15, 16 [23]), while others modulate liver fibrosis by directly regulating the mRNA expression levels of various ECM genes [10]. For example, members of the miR-29 family inhibit liver fibrosis through targeting the 3′-UTRs of mRNAs of a number of fibrosis-related genes [10]. Our study revealed a new mechanism by which miRNA-122 controls liver fibrosis via targeting prolyl 4-hydroxylase that is involved in the process of collagen maturation.
Synthesis of collagen, the major component of the ECM, starts with translation of collagen mRNA on ribosomes of the rough endoplasmic reticulum and hydroxylation of its proline residue to hydroxyproline by prolyl 4-hydroxylase [24]. After several modifications, procollagen with a triple helix is formed, and is transported to the Golgi complex for extracellular secretion in secretory granules [25]. Proline hydroxylation plays an important role in stabilizing the triple helix of the collagen molecule. Inhibition of prolyl 4-hydroxylase produces unstable collagen associated with decreased collagen production [26]. Several inhibitors of prolyl 4-hydroxylase have been developed and examined as new therapeutics for the treatment of liver fibrosis [27,28]. The expression of prolyl 4-hydroxylase is regulated by various proinflammatory molecules, including several cytokines [29]. However, little is known about the role of miRNAs in the regulation of prolyl 4-hydroxylase. Our data suggest miR-122 negatively regulates the expression of prolyl 4-hydroxylase in HSCs. The mRNA expression of P4HA1 was significantly increased in the mouse fibrotic liver and rat activated HSCs, which was inversely correlated with that of miR-122. Overexpression of miR-122 in HSCs led to significant downregulation of P4HA1 expression at both mRNA and protein levels. Reporter assay suggested that miR-122 was targeted to the 3′-UTR of P4HA1 mRNA. We have further shown that overexpression of miR-122 led to significant inhibition of the production of mature Col1A1 as determined by Western analysis. Immunofluorescence staining of extracellular type I collagen revealed that type I collagen fibrils secreted from miR-122 transfected cells were sparse and weakly stained compared with those secreted from control group cells. Taken together, our data strongly support the notion that miR-122 plays a role in blocking the maturation process of collagen via targeting P4HA1 mRNA.
The mechanism involved in the regulation of miR-122 expression in HSCs is not known at present. However, studies in hepatocytes have shown that a number of transcriptional factors are involved in the regulation of miR-122 including C/EBPα, HNF1α, FoxA2, and HNF4α [22,30]. To investigate whether these transcriptional factors are similarly involved in the regulation of miR-122 expression in HSCs, we examined the expression levels of C/EBPα and HNF4 in the mouse fibrotic liver and activated rat HSCs. Our data showed that HNF4 expression was below the detection limit in quiescent rat HSCs and remained undetectable when cells were transactivated. This is in agreement with the notion that HNF4 is a hepatocyte-specific transcriptional factor. However, C/EBPα is constitutively expressed in quiescent HSCs and its expression is significantly downregulated in activated HSCs. ChIP assay shows that C/EBPα binds to a putative binding site(s) in the miR-122 gene promoter in both rat primary HSCs (Fig. 5) and human LX2 cells (data not shown). More importantly, the binding activity of C/EBPα to the miR-122 promoter was significantly decreased in activated HSCs compared to quiescent cells. These data suggest a potential role for decreased transcriptional activity of C/EBPα in the downregulation of miR-122, during the activation of HSCs. It has been shown that TGF-β inhibits the expression of C/EBPα and PPAR-γ in adipocytes [31]. Our data showed that TGF-β treatment led to downregulation of miR-122 expression (Supplementary Fig. 3). These results suggest a likely role of TGF-β/C/EBPα signaling in the negative regulation of miR-122 in hepatic stellate cells, which require more studies in the future.
In addition to blocking collagen maturation, miR-122 also showed a moderate effect in inhibiting the proliferation of activated HSCs. As an initial approach to understand the mechanism, we examined the effect of miR-122 on the expression of Bcl-w and IGFR-1, because enhanced Bcl-2 and IGFR signaling is implicated in the rapid proliferation of activated HSCs, and Bcl-w and IGFR-1 are two target genes of miR-122. Our results showed that miR-122 treatment led to a significant downregulation of the expression of both genes. A similar growth inhibitory effect of miR-122 has previously been demonstrated in hepatoma cells by regulating several tumorigenesis-related proteins, including ADAM10, IGF1R, CCNG1 and ADAM17 [32–34]. More studies are needed to further define the mechanism of miR-122-mediated growth inhibition of HSCs. Nonetheless, this may add favorably to the overall antifibrotic effect of miR-122, as induction of apoptosis of activated HSCs is one of the strategies that are effective in reversing or decreasing fibrotic changes in the liver [35].
In summary, we have shown that the expression level of miR-122 is significantly decreased in activated HSCs, which may contribute to the development of liver fibrosis through upregulating the expression of prolyl 4-hydroxylase and the subsequent production of overly cross-linked collagen. It should be noted that miR-122 is downregulated in the livers of patients with primary biliary cirrhosis [15] as well as in the livers of mice treated with CCl4 [36]. We propose that targeted delivery of miR-122 to the liver, particularly HSCs, may be a novel therapeutic approach for the treatment of liver fibrosis.
Supplementary Material
Acknowledgments
Financial support
An NIH grant HL091828 and a grant from the University of Pittsburgh Central Research Development Fund.
We thank Dr S.L. Friedman, Mount Sinai School of Medicine, NY, for his generous gift of LX2 cells.
Abbreviations
- miR
microRNA
- P4HA1
prolyl 4-hydroxylase subunit alpha-1
- C/ EBPα
CCAAT/enhancer binding protein alpha
- COL1A1
collagen, type I, alpha 1
- HSCs
hepatic stellate cells
- CCl4
carbon tetrachloride
- 3′-UTR
3′-untranslated region
- ECM
extracellular matrix
- PBC
primary biliary cirrhosis
- HCC
hepatocellular carcinoma
- HNF4
hepatocyte nuclear factor 4
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.jhep.2012.11.011.
Conflict of interest
The authors who have taken part in this study declared that they do not have anything to disclose regarding funding or conflict of interest with respect to this manuscript.
References
- 1.Valencia-Sanchez MA, Liu JD, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 2.Friedman RC, Farh KKH, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 4.Olaso E, Friedman SL. Molecular regulation of hepatic fibrogenesis. J Hepatol. 1998;29:836–847. doi: 10.1016/s0168-8278(98)80269-9. [DOI] [PubMed] [Google Scholar]
- 5.Friedman SL. Hepatic stellate cells: protean, multifunctional, and enigmatic cells of the liver. Physiol Rev. 2008;88:125–172. doi: 10.1152/physrev.00013.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiao JJ, Friedman SL, Aloman C. Hepatic fibrosis. Curr Opin Gastroenterol. 2009;25:223–229. doi: 10.1097/mog.0b013e3283279668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mann J, Chu DCK, Maxwell A, Oakley F, Zhu N, Tsukamoto H, et al. MeCP2 controls an epigenetic pathway that promotes myofibroblast transdifferentiation and fibrosis. Gastroenterology. 2010;138:705–714. doi: 10.1053/j.gastro.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ji JL, Zhang JS, Huang GC, Qian J, Wang XQ, Mei S. Over-expressed microRNA-27a and 27b influence fat accumulation and cell proliferation during rat hepatic stellate cell activation. FEBS Lett. 2009;583:759–766. doi: 10.1016/j.febslet.2009.01.034. [DOI] [PubMed] [Google Scholar]
- 9.Roderburg C, Urban GW, Bettermann K, Vucur M, Zimmermann H, Schmidt S, et al. Micro-RNA profiling reveals a role for miR-29 in human and murine liver fibrosis. Hepatology. 2011;53:209–218. doi: 10.1002/hep.23922. [DOI] [PubMed] [Google Scholar]
- 10.Li J, Zhang YF, Kuruba R, Gao X, Gandhi CR, Xie W, et al. Roles of microRNA-29a in the antifibrotic effect of farnesoid X receptor in hepatic stellate cells. Mol Pharmacol. 2011;80:191–200. doi: 10.1124/mol.110.068247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 12.Xu H, He JH, Xiao ZD, Zhang QQ, Chen YQ, Zhou H, et al. Liver-enriched transcription factors regulate microRNA-122 that targets CUTL1 during liver development. Hepatology. 2010;52:1431–1442. doi: 10.1002/hep.23818. [DOI] [PubMed] [Google Scholar]
- 13.Kim N, Kim H, Jung I, Kim Y, Kim D, Han YM. Expression profiles of miRNAs in human embryonic stem cells during hepatocyte differentiation. Hepatol Res. 2011;41:170–183. doi: 10.1111/j.1872-034X.2010.00752.x. [DOI] [PubMed] [Google Scholar]
- 14.Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, et al. MiR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 15.Padgett KA, Lan RY, Leung PC, Lleo A, Dawson K, Pfeiff J, et al. Primary biliary cirrhosis is associated with altered hepatic microRNA expression. J Autoimmun. 2009;32:246–253. doi: 10.1016/j.jaut.2009.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bai S, Nasser MW, Wang B, Hsu SH, Datta J, Kutay H, et al. MicroRNA-122 inhibits tumorigenic properties of hepatocellular carcinoma cells and sensitizes these cells to sorafenib. J Biol Chem. 2009;284:32015–32027. doi: 10.1074/jbc.M109.016774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsu SH, Wang B, Kota J, Yu J, Costinean S, Kutay H, et al. Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. J Clin Invest. 2012;122:2871–2883. doi: 10.1172/JCI63539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsai WC, Hsu SD, Hsu CS, Lai TC, Chen SJ, Shen R, et al. MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J Clin Invest. 2012;122:2884–2897. doi: 10.1172/JCI63455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saile B, DiRocco P, Dudas J, El-Armouche H, Sebb H, Eisenbach C, et al. IGF-I induces DNA synthesis and apoptosis in rat liver hepatic stellate cells (HSC) but DNA synthesis and proliferation in rat liver myofibroblasts (rMF) Lab Invest. 2004;84:1037–1049. doi: 10.1038/labinvest.3700116. [DOI] [PubMed] [Google Scholar]
- 20.Saile B, Knittel T, Matthes N, Schott P, Ramadori G. CD95/CD95L-mediated apoptosis of the hepatic stellate cell. A mechanism terminating uncontrolled hepatic stellate cell proliferation during hepatic tissue repair. Am J Pathol. 1997;151:1265–1272. [PMC free article] [PubMed] [Google Scholar]
- 21.Lin CJ, Gong HY, Tseng HC, Wang WL, Wu JL. MiR-122 targets an antiapoptotic gene, Bcl-w, in human hepatocellular carcinoma cell lines. Biochem Biophys Res Commun. 2008;375 (3):315–320. doi: 10.1016/j.bbrc.2008.07.154. [DOI] [PubMed] [Google Scholar]
- 22.Zeng C, Wang R, Li D, Lin XJ, Wei QK, Yuan Y, et al. A novel GSK-3 beta-C/EBP alpha-miR-122-insulin-like growth factor 1 receptor regulatory circuitry in human hepatocellular carcinoma. Hepatology. 2010;52 (5):1702–1712. doi: 10.1002/hep.23875. [DOI] [PubMed] [Google Scholar]
- 23.Guo CJ, Pan Q, Li DG, Sun H, Liu BW. MiR-15b and miR-16 are implicated in activation of the rat hepatic stellate cell: an essential role for apoptosis. J Hepatol. 2009;50:766–778. doi: 10.1016/j.jhep.2008.11.025. [DOI] [PubMed] [Google Scholar]
- 24.Myllyharju J. Prolyl 4-hydroxylases, the key enzymes of collagen biosynthesis. Matrix Biol. 2003;22:15–24. doi: 10.1016/s0945-053x(03)00006-4. [DOI] [PubMed] [Google Scholar]
- 25.Bonfanti L, Mironov AA, Jr, Martínez-Menárguez JA, Martella O, Fusella A, et al. Procollagen traverses the golgi stack without leaving the lumen of cisternae: evidence for cisternal maturation. Cell. 1998;95:993–1003. doi: 10.1016/s0092-8674(00)81723-7. [DOI] [PubMed] [Google Scholar]
- 26.Sakaida I, Matsumura Y, Kubota M, Kayano K, Takenaka K, Okita K. The prolyl 4-hydroxylase inhibitor HOE 077 prevents activation of Ito cells, reducing procollagen gene expression in rat liver fibrosis induced by choline-deficient L-amino acid-defined diet. Hepatology. 1996;23:755–763. doi: 10.1053/jhep.1996.v23.pm0008666329. [DOI] [PubMed] [Google Scholar]
- 27.Bickel M, Baringhaus KH, Gerl M, Günzler V, Kanta J, Schmidts L, et al. Selective inhibition of hepatic collagen accumulation in experimental liver fibrosis in rats by a new prolyl 4-hydroxylase inhibitor. Hepatology. 1998;28:404–411. doi: 10.1002/hep.510280217. [DOI] [PubMed] [Google Scholar]
- 28.Fujiwara K, Ogata I, Ohta Y, Hayashi S, Mishiro S, Takatsuki K, et al. Decreased collagen accumulation by a prolyl hydroxylase inhibitor in pig serum-induced fibrotic rat liver. Hepatology. 1988;8:804–807. doi: 10.1002/hep.1840080418. [DOI] [PubMed] [Google Scholar]
- 29.Li L, Zhang K, Cai XJ, Feng M, Zhang Y, Zhang M. Adiponectin upregulates prolyl-4-hydroxylase α1 expression in interleukin 6-stimulated human aortic smooth muscle cells by regulating ERK 1/2 and Sp1. PLoS One. 2011;6:e22819. doi: 10.1371/journal.pone.0022819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laudadio I, Manfroid I, Achouri Y, Schmidt D, Wilson MD, Cordi S, et al. A feedback loop between the liver-enriched transcription factor network and mir-122 controls hepatocyte differentiation. Gastroenterology. 2012;142:119–129. doi: 10.1053/j.gastro.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 31.Zamani N, Brown CW. Emerging roles for the transforming growth factor- {beta} superfamily in regulating adiposity and energy expenditure. Endocr Rev. 2011;32:387–403. doi: 10.1210/er.2010-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsai WC, Hsu PWC, Lai TC, Chau GY, Lin CW, Chen CM, et al. MicroRNA-122, a tumor suppressor microRNA that regulates intrahepatic metastasis of hepatocellular carcinoma. Hepatology. 2009;49:1571–1582. doi: 10.1002/hep.22806. [DOI] [PubMed] [Google Scholar]
- 33.Gramantieri L, Ferracin M, Fornari F, Veronese A, Sabbioni S, Liu CG, et al. Cyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res. 2007;67:6092–6099. doi: 10.1158/0008-5472.CAN-06-4607. [DOI] [PubMed] [Google Scholar]
- 34.Zhang G, Wang Q, Xu R. Therapeutics based on microRNA: a new approach for liver cancer. Curr Genomics. 2010;11:311–325. doi: 10.2174/138920210791616671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gressner AM. The cell biology of liver fibrogenesis – an imbalance of proliferation, growth arrest and apoptosis of myofibroblasts. Cell Tissue Res. 1998;292:447–452. doi: 10.1007/s004410051073. [DOI] [PubMed] [Google Scholar]
- 36.Marquez RT, Bandyopadhyay S, Wendlandt EB, Keck K, Hoffer BA, Icardi MS, et al. Correlation between microRNA expression levels and clinical parameters associated with chronic hepatitis C viral infection in humans. Lab Invest. 2010;90:1727–1736. doi: 10.1038/labinvest.2010.126. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.