Abstract
We characterized intrinsic and extrinsic specification of progenitors in the lateral and medial ganglionic eminences (LGE and MGE). We identified seven genes whose expression is enriched or restricted in either the LGE [biregional cell adhesion molecule-related/downregulated by oncogenes binding protein (Boc), Frizzled homolog 8 (Fzd8), Ankrd43 (ankyrin repeat domain-containing protein 43), and Ikzf1 (Ikaros family zinc finger 1)] or MGE [Map3k12 binding inhibitory protein 1 (Mbip); zinc-finger, SWIM domain containing 5 (Zswim5); and Adamts5 [a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 5]]. Boc, Fzd8, Mbip, and Zswim5 are apparently expressed in LGE or MGE progenitors, whereas the remaining three are seen in the postmitotic mantle zone. Relative expression levels are altered and regional distinctions are lost for each gene in LGE or MGE cells propagated as neurospheres, indicating that these newly identified molecular characteristics of LGE or MGE progenitors depend on forebrain signals not available in the neurosphere assay. Analyses of Pax6Sey/Sey, Shh−/−, and Gli3XtJ/XtJ mutants suggests that LGE and MGE progenitor identity does not rely exclusively on previously established forebrain-intrinsic patterning mechanisms. Among a limited number of additional potential patterning mechanisms, we found that extrinsic signals from the frontonasal mesenchyme are essential for Shh- and Fgf8-dependent regulation of LGE and MGE genes. Thus, extrinsic and intrinsic forebrain patterning mechanisms cooperate to establish LGE and MGE progenitor identity, and presumably their capacities to generate distinct classes of neuronal progeny.
Keywords: forebrain, development, induction, interneuron, cell fate, explant, diversity, microarray
Introduction
Progenitors in the lateral and medial ganglionic eminences (LGE and MGE) produce postmitotic neurons with distinct migratory trajectories and fates: LGE progenitors produce striatal projection neurons and olfactory bulb interneurons, whereas MGE as well as caudal ganglionic eminence (CGE) progenitors produce cortical and hippocampal interneurons (Pleasure et al., 2000; Toresson and Campbell, 2001; Wichterle et al., 2001; Nery et al., 2002; Stenman et al., 2003; Xu et al., 2004; Butt et al., 2005; Tucker et al., 2006). Previously identified, regionally restricted, transcriptional regulators (Anderson et al., 1997; Sussel et al., 1999; Corbin et al., 2000; Toresson and Campbell, 2001; Stenman et al., 2003) may not fully account for these divergent capacities. Accordingly, we identified additional genes that distinguish LGE or MGE progenitors or progeny and determined the role of intrinsic and extrinsic signaling mechanisms in establishing LGE and MGE identity.
It is unclear whether LGE and MGE progenitors rely exclusively on local patterning mechanisms intrinsic to the forebrain neuroepithelium or require extrinsic signals to establish or retain molecular distinctions. Isolated neural progenitors from distinct forebrain regions, including the LGE and MGE, maintain some in vivo molecular identity when propagated as neurospheres (Hitoshi et al., 2002; Ostenfeld et al., 2002; Parmar et al., 2002). Thus, we asked whether novel molecular dimensions of LGE and MGE identity, once established in vivo, are maintained independent of local intrinsic and extrinsic forebrain signals. Patterned gene expression in the LGE and MGE depends on forebrain position, as well as local secreted signals and transcriptional regulators (Wilson and Rubenstein, 2000; Rallu et al., 2002b; Schuurmans and Guillemot, 2002; Campbell, 2003), including the transcription factor Pax6 (Stoykova et al., 1996; Götz et al., 1998; Stoykova et al., 2000; Toresson et al., 2000; Yun et al., 2001), the secreted signal sonic hedgehog (Shh) (Chiang et al., 1996; Fuccillo et al., 2004; Xu et al., 2005), and its transcriptional mediator Gli3 (Theil et al., 1999; Tole et al., 2000; Rallu et al., 2002a). Thus, we asked whether Pax6, Shh, or Gli3 regulate expression or patterning of the newly identified LGE- or MGE-enriched genes. In addition, signals from outside the forebrain may establish or reinforce LGE and MGE distinctions. The frontonasal mesenchyme (Fn:M), a source of inductive signals for the olfactory pathway during early forebrain development, is adjacent to the ventral forebrain neuroepithelium (LaMantia et al., 1993, 2000; Haskell and LaMantia, 2005). Thus, we asked whether this mesenchyme regulates expression or patterning of LGE- and MGE-enriched genes or modulates the activity of intrinsic signals thought to influence gene expression in the forebrain neuroepithelium.
Our results show that several molecular features of LGE and MGE progenitors are not selectively maintained in vitro, nor do they depend exclusively on intrinsic forebrain mechanisms previously thought to pattern the LGE and MGE. Instead, our in vitro observations suggest that local signaling via Shh and Fgf8 is modulated by signals from the frontonasal mesenchyme to establish or maintain multiple dimensions of LGE and MGE molecular identity.
Materials and Methods
Animals.
The University of North Carolina at Chapel Hill (UNC-CH) Division of Laboratory Animal Medicine maintained colonies of wild-type Institute of Cancer Research (ICR), small eye (Pax6Sey/+), extra-toe (XtJ/+), and Shh−/+ mice. Timed-pregnant litters [day of vaginal plug = embryonic day 0.5 (E0.5)] were generated by mating wild-type mice, or mice heterozygous for each mutation, independently. Timed-pregnant females were killed by rapid cervical dislocation, and embryos were collected for tissue culture and RNA isolation or fixed and embedded for in situ hybridization. All experimental procedures were reviewed and approved by the Institutional Animal Care and Use Committee at UNC-CH.
Forebrain microdissection and sample preparation.
Embryos were collected in ice-cold complete HBSS (Tucker et al., 2006) and dissected individually in a Sylgard dish containing ice-cold HBSS (Invitrogen) with 10% RNAlater (Ambion). Forebrains were microdissected as shown in Figure 1A under RNase-free conditions. After hemisecting the brain and removing the meninges, a microscalpel was used to separate the cortical and olfactory bulb rudiments from the basal forebrain. The basal forebrain was transected posterior to the interganglionic sulcus, defining the boundary of the lateral and caudal ganglionic eminences. The lateral and medial ganglionic eminences were separated along the interganglionic sulcus, and neighboring cortical and septal tissues were removed. The caudal ganglionic eminence was further separated from surrounding cortical and thalamic tissues. Samples of cortex, olfactory bulb, lateral, medial, and caudal ganglionic eminences were collected separately into RNAlater and pooled from a single litter comprised of 10 embryos. Tissue samples were subsequently solubilized in TRIzol (Invitrogen), homogenized through a graded series of hypodermic needles, and stored at −20°C.
Figure 1.
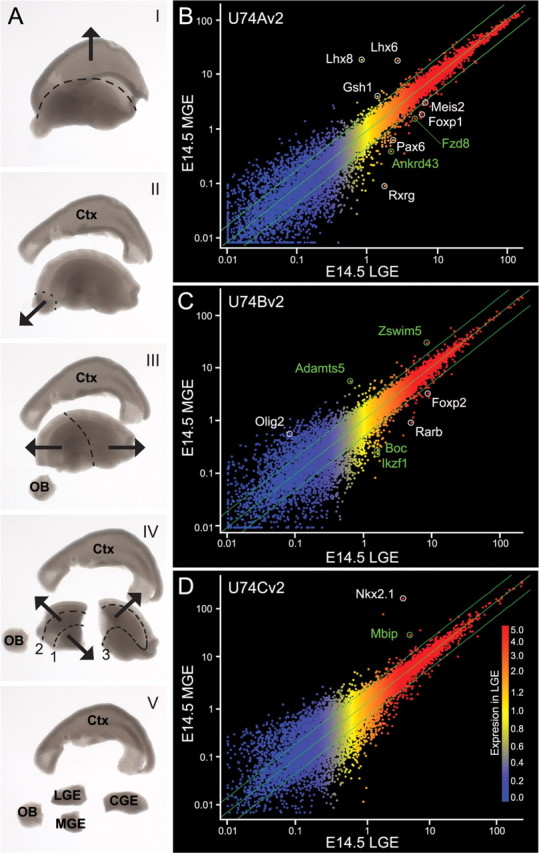
Affymetrix-based microarray screen identifies transcripts that are putatively enriched in the LGE or MGE at E14.5. A, Five-step (I–V) differential dissection of the E14.5 forebrain. Cortical (I) and olfactory bulb (II) rudiments were removed, the basal forebrain was bisected (III), and lateral, medial, and caudal ganglionic eminences were isolated (IV–V). B–D, MGE versus LGE relative gene expression determined from Affymetrix U74Av2 (B), U74Bv2 (C), and U74Cv2 (D) GeneChips. Intensity of expression in the LGE is indicated by color (blue, low; red, high). Green lines indicate twofold change in expression. Reference genes are in white; candidate genes are in green.
RNA was extracted from TRIzol samples, purified using RNeasy spin columns (Qiagen), and quantified using a ND-1000 spectrophotometer (NanoDrop Technologies). Fifteen micrograms of total RNA was supplied to the UNC Neuroscience Center quantitative expression core for cRNA synthesis and chip hybridization. For reverse transcription (RT)-PCR experiments, 2 μg of RNA was treated with 1 U of DNase I (Ambion) for 1 h at 37°C to eliminate contaminating genomic DNA. cDNA reactions were prepared with 1 μg of DNase I-treated RNA. Immediately before cDNA synthesis, template RNA was incubated with 0.05 μg of random primers (Invitrogen) for 5 min at 70°C and rapidly cooled on ice. This mixture was incubated with ImProm-II reverse transcriptase (Promega), 5 mm MgCl2, 0.5 mm dNTP, and RNasin ribonuclease inhibitor (Promega) for 10 min at 25°C followed by 1 h at 42°C. For negative controls, identical reactions were prepared without reverse transcriptase.
Quantitative RT-PCR.
Primers and dual-labeled probes (supplemental Table 1, available at www.jneurosci.org as supplemental material) were designed using Primer Express Software version 1.5 (Applied Biosystems), and where possible, their sequences spanned exon–exon junctions. Quantitative RT-PCRs (qRT-PCRs) were performed in 96-well optical plates on an Applied Biosystems 7500 Real-Time PCR System (Applied Biosystems). PCRs (50 μl) contained 1.25 U of HotStar TaqDNA Polymerase (Qiagen), 4.8 mm MgCl2, 50 μm dNTP (GE Healthcare), 1:500 Rox Dye (Invitrogen), forward and reverse primers (100 nm for Gapdh; 300 nm for all targets), 200 nm dual-labeled probe (5′ FAM reporter dye and 3′ TAMRA quencher dye), and 50 ng of cDNA in 1× Qiagen PCR buffer. PCR conditions were 95°C for 10 min, followed by 40 two-step cycles of 95°C for 15 s and 60°C for 1 min. For all experiments, Gapdh served as the endogenous control gene, and the LGE was used as the calibrator sample. Each primer–probe set amplified a single PCR product of predicted size as determined by agarose-gel electrophoresis and had approximately equal amplification efficiencies to Gapdh when validated with a serial dilution of representative cDNA. Relative quantification was determined according to the delta-delta CT method (Livak and Schmittgen, 2001). Mean fold change and SEM for each sample was log transformed before graphing.
Tissue preparation and in situ hybridization.
Wild-type (ICR) or mutant embryos and littermate controls were fixed overnight in RNase-free 4% paraformaldehyde in 1× PBS with 2 mm EGTA at 4°C. Embryos were rinsed in PBS, cryoprotected by immersion through sucrose (10, 20, and 30% sucrose in RNase-free 1× PBS; each step overnight at 4°C), embedded in freezing compound, rapidly frozen in liquid-nitrogen-cooled 2-methyl butane, and stored at −80°C. Serial cryosections were collected onto slides at 20 μm intervals and stored at −80°C before in situ hybridization. No qualitative differences in expression of candidate genes were observed between ICR and nonmutant littermate controls of Pax6 and Gli3 genotypes, suggesting no significant interstrain variability. Thus, ICR and nonmutant littermates were used interchangeably as control material in each in situ hybridization experiment. Approximately 10 mutants of each genotype were analyzed in the study. Mutant material was sequentially sectioned into five series; sets of five slides containing four mutants and one control were simultaneously processed under equivalent reaction conditions.
We generated plasmids for riboprobe synthesis by cloning PCR products (supplemental Table 1, available at www.jneurosci.org as supplemental material) from E14.5 LGE or MGE cDNA into either pCRII-TOPO (Invitrogen) or pBluescript II KS (Stratagene). A 586 bp fragment of Pax6 containing 58 bases of 3′ untranslated region (UTR) and coding sequence through an internal BglII site was used to generate antisense Pax6 riboprobe. Nkx-2.1 riboprobe was synthesized from a plasmid containing a 603 bp SalI–NcoI fragment of 5′ UTR from an EST clone (IMAGE: 6416507). After proteinase K pretreatment and prehybridization, digoxigenin-labeled riboprobes were hybridized for 12–16 h at 55–60°C on cryosections or 65–70°C on explants. After rinsing, sections or explants were incubated with a 1:5000 dilution of alkaline phosphatase conjugated sheep anti-digoxigenin (Roche Diagnostics) in blocking buffer at 4°C for 14–16 h and developed in a nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate solution (20 μl/ml 5–10% polyvinyl alcohol; Roche Diagnostics) in the dark for 2–30 h. Fresh developing solution was exchanged every 8–12 h. After developing, material was rinsed, and alkaline phosphatase activity was quenched by fixation in 4% paraformaldehyde; sections were mounted in Aquatex (EM Science Harleco), and explants were stored at 4°C in a glycerine-azide buffer.
Neurosphere production and collection for RNA isolation.
LGE and MGE tissue was dissected from three to four E14.5 forebrains in ice-cold complete HBSS. The tissue was collected separately in supplemented DMEM/F12 medium (Invitrogen) containing 0.6% d-glucose (Sigma), 1× penicillin/streptomycin, a defined hormone and salt mixture (Reynolds et al., 1992), 2 mm glutamine, and 1× B27 (Invitrogen; supplemented medium) plus DNase I (0.01 mg/ml; Sigma). Trypsin (Invitrogen) was added to a final concentration of 0.25%, and tissue was digested for 15 min at 37°C. After enzymatic digestion, tissue was mechanically dissociated, and an equal volume of supplemented medium containing 5 mg/ml trypsin inhibitor (Invitrogen) was added to the cell suspensions. The enzyme-quenched cell suspensions were passed through 40 μm basket filters to remove large debris and centrifuged at 700 rpm for 15 min. Cell pellets were resuspended in 1 ml of supplemented medium, and cell densities were determined with a hemocytometer. Cells were diluted to a final concentration of 10 cells/ml in supplemented medium containing recombinant human epidermal growth factor (EGF) (20 ng/ml; Promega) and basic fibroblast growth factor (FGF) (20 ng/ml; Promega) and plated into six-well plates. Fresh media containing EGF and FGF were added on the third and sixth day of culture, and neurospheres were harvested on day 8. Large phase bright neurospheres were selected and transferred individually to 1.5 ml Eppendorf tubes containing RNAlater. Neurospheres were spun to pellet, homogenized in TRIzol, and processed for cDNA synthesis as above.
Forebrain explant cultures.
Forebrain epithelium (Fb:E)/frontonasal mesenchyme cultures were prepared as described previously (LaMantia et al., 2000; Tucker et al., 2006). Briefly, the E9.5 ventrolateral forebrain and adjacent frontonasal mass was microdissected, and the surface epithelium and mesenchyme was enzymatically digested and mechanically removed (forebrain epithelium only), or only the surface epithelium was removed (forebrain epithelium/frontonasal mesenchyme cocultures). Explants were grown floating on polycarbonate Nuclepore membranes (13 mm/8 μm pore; Whatman) for 3 d in DMEM containing 10% fetal calf serum. After 3 d, explants were briefly rinsed in RNase-free 1× PBS, fixed for 1 h at room temperature in RNase-free 4% paraformaldehyde in 1× PBS with 2 mm EGTA, rinsed three times for 5 min each in PTW (RNase-free 1× PBS with 2 mm EGTA and 0.1% Tween 20), and dehydrated through a 10, 20, 50, 75, 100% methanol series in PTW for 5 min each. Explants were rinsed and stored in a third exchange of 100% methanol at −20°C for up to 6 weeks before in situ hybridization.
Explant pharmacology.
Pairs of explanted forebrain epithelium and frontonasal mesenchyme cocultures were treated with 10 μm cyclopamine (kind gift from William Gaffield, Western Regional Research Center, Agricultural Research Service, United States Department of Agriculture, Albany, CA), 25 or 100 μm 4-diethylaminobenzaldehyde (DEAB; Sigma), 10 μm 3-[(3-(2-carboxyethyl)-4-methylpyrrol-2-yl)methylene]-2-indolinone (SU5402) (donated by Pfizer), or their respective vehicle controls. Stock solutions of cyclopamine (40 mm in 100% ethanol), DEAB (100 mm stock in DMSO), and SU5402 (40 mm in DMSO) were prepared aseptically, aliquoted, and stored at −20°C. Cultures were fed daily with medium containing drug or vehicle control and fixed after 72 h. For local delivery of Shh, retinoic acid (RA), and Fgf8, forebrain neuroepithelium explants were placed over individual agonist-soaked beads before positioning on membrane filters. Affi-Gel blue beads (75–150 μm wet diameter; Bio-Rad) were rinsed in sterile PBS, soaked in recombinant mouse Shh, N-term (0.3 mg/ml; R&D Systems) for 1 h at 37°C, and briefly rinsed in Leibovitz's L-15 medium (L-15) (Invitrogen). AG1-X2 resin beads (75–180 μm wet diameter; Bio-Rad) were converted to formate form, rinsed in PBS, soaked light protected in RA (10−3 m in DMSO; Sigma) for 40–60 min at room temperature, and rinsed three times for 5 min each in L-15 before use. Heparin acrylic beads (Sigma) were rinsed in PBS, incubated in a 5 μl droplet of recombinant mouse Fgf8b (1 mg/ml; R&D Systems) for 90 min at room temperature, and rinsed three times for 5 min each in L-15. Control beads were treated identically, but soaked in respective vehicles. Fgf8b was applied globally at 100 ng/ml in medium and replenished daily during the 72 h culture period.
Results
Microarray analysis identifies genes with enhanced LGE or MGE expression levels
We probed murine genome expression arrays with cDNAs from E14.5 LGE, MGE, CGE, neocortex (CTX), and olfactory bulb (OB) to identify novel molecular distinctions between LGE and MGE cells and those in related forebrain subdivisions (Fig. 1A) (see Materials and Methods). Samples from each of the five regions were microdissected and pooled from a single litter and then hybridized independently to Affymetrix U74Av2, U74Bv2, and U74Cv2 GeneChips. The quality of input RNA was examined before cRNA synthesis and after chip hybridization by examining 3′/5′ ratios of two endogenous control genes (β-actin and Gapdh). After hybridization and scanning, we used the Affymetrix GeneChip Operating Software to generate a list of apparently differentially expressed LGE and MGE genes.
LGE cells express 37.5% and MGE cells express 38.7% of the ∼36,000 transcripts represented as probe sets on the U74v2 GeneChips. Remarkably, most transcripts are equivalently expressed in the LGE and MGE (Fig. 1B–D); nevertheless, after removing nonmeaningful comparisons (i.e., nonrepresented transcripts or absent to absent calls), we found 951 differentially expressed transcripts. Most showed less than a twofold change in gene expression (Fig. 1B–D, green lines), and only 205 transcripts (∼22%) had a fold change greater than or equal to two. To facilitate the selection of candidates from this initial subset, we only considered transcripts present at a level of 2.8-fold or greater (signal log ratio of 1.5) between LGE and MGE, which decreased the number of differentially expressed transcripts to 71. By examining absolute detection signals across all tissue compartments for each of the 71 differentially expressed transcripts, we found that relatively few genes were uniquely expressed in either the LGE or MGE. Instead, 22 of 42 (52%) of the LGE-enriched transcripts were expressed at higher levels outside the LGE. Some LGE-enriched genes (Ngn2, Tbr1, and NeuroD6) are highly expressed in the cerebral cortex, and their presence may reflect an incomplete removal of lateral cortical tissue during sample preparation. Alternatively, as for Pax6, their LGE enrichment may reflect expression patterns that span the cortical rudiment–LGE boundary, thus resulting in enhanced LGE versus MGE expression. In contrast, MGE-enriched transcripts were less promiscuously expressed, because only 7 of 29 (24%) are expressed more abundantly elsewhere. The differentially expressed transcripts encode a wide assortment of proteins, including many nuclear, membrane-associated, extracellular, and cytosolic proteins (supplemental Table 2, available at www.jneurosci.org as supplemental material).
Seven candidate genes for restricted LGE or MGE expression
We selected seven candidate genes, whose apparent differential expression levels seemed likely to be matched with restricted cellular expression in the developing LGE and MGE, from the subset of 42 LGE and 29 MGE-enriched genes. We applied the following criteria to identify genes for detailed analysis: (1) the fold change had to be statistically significant at p < 0.00005; (2) the probability that the gene was present in its respective tissue had to be statistically significant [p < 0.005 for all genes except biregional cell adhesion molecule-related/downregulated by oncogenes (Cdon) binding protein (Boc), which was present at p < 0.05]; and (3) the function of the gene within the context of LGE/MGE molecular or cellular diversity had yet to be explored. Differential expression in additional forebrain compartments (CGE, OB, and CTX) was also considered in our evaluation of transcript abundance and possible LGE or MGE enrichment. Based on these criteria, we selected four genes apparently enriched in the LGE [ankyrin repeat domain-containing protein 43 (Ankrd43), Boc, Frizzled homolog 8 (Fzd8), and Ikaros family zinc finger 1 (Ikzf1; also known as Zfpn1a1)], and three genes apparently enriched in the MGE [a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 5 (Adamts5; also known as aggrecanase-2), Map3k12 binding inhibitory protein 1 (Mbip), and zinc-finger, SWIM domain containing 5 (Zswim5)] (Fig. 1B–D, Table 1).
Table 1.
Genes enriched in either the LGE or MGE as determined by Affymetrix microarray analysis
| Affymetrix ID | Gene symbol | Fold change | p value | Enrichment |
|---|---|---|---|---|
| Reference genes | ||||
| 92237_at | Rxrg | 19.7 | 0.000023 | LGE > MGE |
| 92271_at | Pax6 | 4.9 | 0.000001 | LGE > MGE |
| 104415_at | Foxp1 | 3.7 | 0.000002 | LGE > MGE |
| 105632_at | Rarb | 3.5 | 0.000322 | LGE > MGE |
| 164118_at | Foxp2 | 3.0 | 0.000000 | LGE > MGE |
| 97988_at | Meis2 (Mrg1) | 2.3 | 0.000001 | LGE > MGE |
| 134379_r_at | Nkx-2.1 (Titf1) | 36.8 | 0.000219 | MGE > LGE |
| 94754_at | Lhx8 | 18.4 | 0.000219 | MGE > LGE |
| 94176_at | Lhx6 | 6.1 | 0.000219 | MGE > LGE |
| 163516_at | Olig2 | 4.6 | 0.000068 | MGE > LGE |
| 101146_at | Gsh1 | 2.1 | 0.000099 | MGE > LGE |
| Candidate genes | ||||
| 112792_at | Ikzf1 | 4.9 | 0.000003 | LGE > MGE |
| 117276_at | Boc | 4.3 | 0.000031 | LGE > MGE |
| 103688_at | Ankrd43 | 3.5 | 0.000001 | LGE > MGE |
| 99415_at | Fzd8 | 2.8 | 0.000031 | LGE > MGE |
| 115173_at | Adamts5 | 4.9 | 0.004063 | MGE > LGE |
| 133340_f_at | Mbip | 3.7 | 0.001109 | MGE > LGE |
| 104778_at | Zswim5 | 2.8 | 0.000266 | MGE > LGE |
ID, Identification number.
The four LGE-enriched genes encode a variety of protein products. Ankrd43 encodes a 334 aa protein with 90% homology (299 of 334 aa) between human and mouse, including 96% homology within the distinctive ankyrin repeats located between residues 130–168 and 169–202. Informatic analysis of Ankrd43 predicts nuclear localization (56.5% probability determined by PSORT II) as well as DNA binding capacity [molecular function gene ontology (GO): 0003677; evidence inferred from electronic annotation (IEA)]. Boc encodes a type I transmembrane receptor containing four Ig repeats and three fibronectin type III repeats in its extracellular domain and no identifiable motifs in its intracellular domain. Boc regulates myogenic differentiation (Kang et al., 2002), is associated with Shh signaling (Okada et al., 2006; Tenzen et al., 2006), and is expressed in dorsal aspects of the brain and spinal cord during development (Mulieri et al., 2002; Tenzen et al., 2006). Fzd8 encodes a member of the Frizzled family of Wnt receptors and has been previously suggested to be differentially expressed in the developing forebrain (Kim et al., 2001). Ikzf1 encodes a zinc-finger transcription factor with a well established role in lymphocyte lineage specification (Georgopoulos et al., 1994), as well as regulation of growth hormone, prolactin, and pro-opiomelanocortin gene expression in the pituitary (Ezzat et al., 2005a,b). In addition, Ikzf1 transcript and protein localizes to the embryonic and postnatal striatum, where it appears to regulate the development of striatal enkephalinergic neurons (Agoston et al., 2007).
The three MGE-enriched genes also encode proteins with diverse functions. Adamts5 encodes a 930 aa zinc metalloproteinase implicated in aggrecan degradation in mouse models of osteoarthritis and inflammatory arthritis (Glasson et al., 2005; Stanton et al., 2005). Mbip encodes a 344 aa protein that binds Map3k12 through a pair of leucine zipper-like motifs and specifically inhibits the ability of Map3k12 to activate JNK (Fukuyama et al., 2000). Zswim5 encodes an 1188 aa protein of unknown function with 96% homology between mouse and human (1138 of 1188). Zswim5 protein is predicted to localize to the nucleus (60.9% by PSORT II), selectively bind zinc ions (molecular function GO: 0008270; evidence IEA), and interact with DNA or proteins in different contexts (Makarova et al., 2002). Thus, the MGE- and LGE-enriched genes identified in our microarray encode a diverse set of proteins with putative adhesive, signaling, and DNA binding and transcriptional functions.
qRT-PCR validates differential expression of LGE- and MGE-enriched genes
We first confirmed the reliability of our screen by quantifying expression levels of established LGE- and MGE-specific genes in parallel with those newly identified in our screen. Microarray analysis correctly detected differential expression of several established LGE-enriched genes, including Rxrg, Rarb, Meis2 (Mrg1), Foxp1, Foxp2, and Pax6, and MGE-enriched genes: Lhx6, Lhx8, Nkx-2.1, Gsh1, and Olig2 (Fig. 1B–D, Table 1). To validate the accuracy of our approach, we used quantitative PCR to compare expression levels of three control genes previously shown to be enriched in the LGE or MGE: Pax6 and Gsh2 (LGE) (Stoykova et al., 1996; Yun et al., 2001) and Nkx-2.1 (MGE) (Lazzaro et al., 1991; Price et al., 1992; Shimamura et al., 1995), with that of the seven newly identified LGE- or MGE-enriched genes. Expression levels of Pax6 (Fig. 2A) and Gsh2 (Fig. 2B) are significantly elevated in the LGE. Similarly, Nkx-2.1 (Titf1), which is predominantly localized to the MGE, is elevated by ∼50 times over the LGE (Fig. 2C). There was complete accord between the microarray and qRT-PCR estimates of differential LGE and MGE expression for two of these three control genes (Gsh2 is not represented in the U74v2 chip set), as well as all seven of our newly identified LGE- or MGE-enriched genes (Figs. 3A,D,G,J, 4A,D,G). Also, the distribution of mRNA abundance across additional forebrain regions (cortex and olfactory bulb) was consistent between microarray and qRT-PCR data for each control and candidate gene. Thus, our microarray data accurately reflect both the known expression patterns of LGE- and MGE-enriched genes and quantitative differences in LGE versus MGE mRNA levels determined by qRT-PCR.
Figure 2.
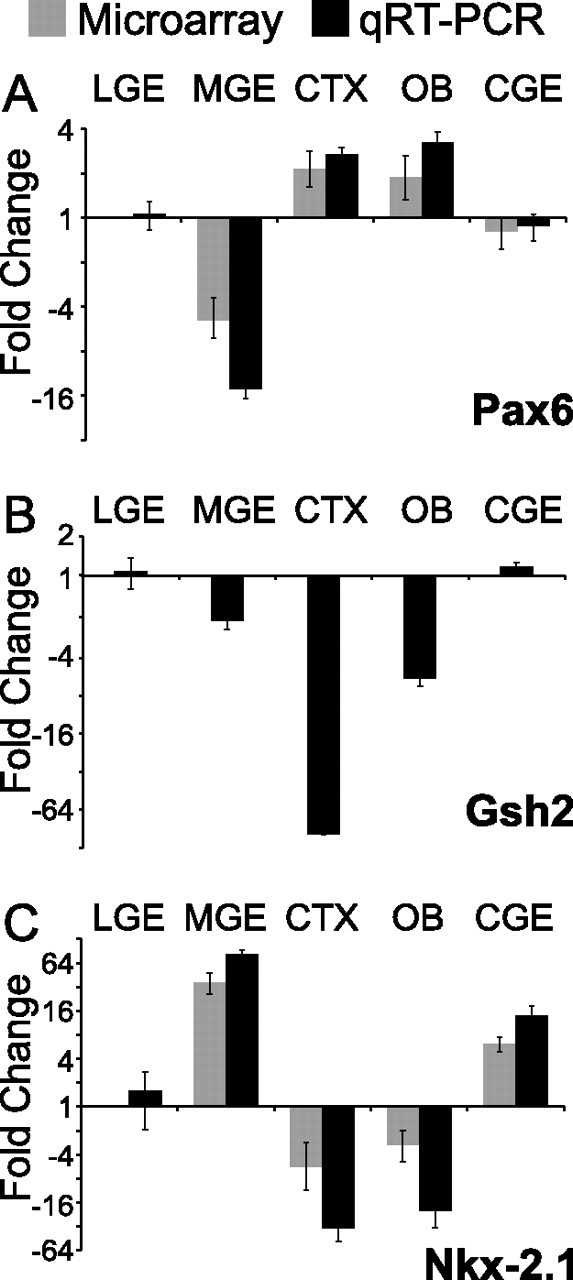
Quantitative RT-PCR of three independent reference genes confirms microarray data and validates differential dissection. Pax6 and Gsh2 are appropriately enriched in the LGE, whereas Nkx-2.1 is enriched in the MGE. Microarray (gray, signal log ratio ± signal log high/signal log low) and qRT-PCR (black, log mean fold change ± SEM; n = 3) data are comparable for Pax6 and Nkx-2.1; Gsh2-specific probes are not included on the U74v2 chipset. Gene expression in each tissue is relative to the LGE.
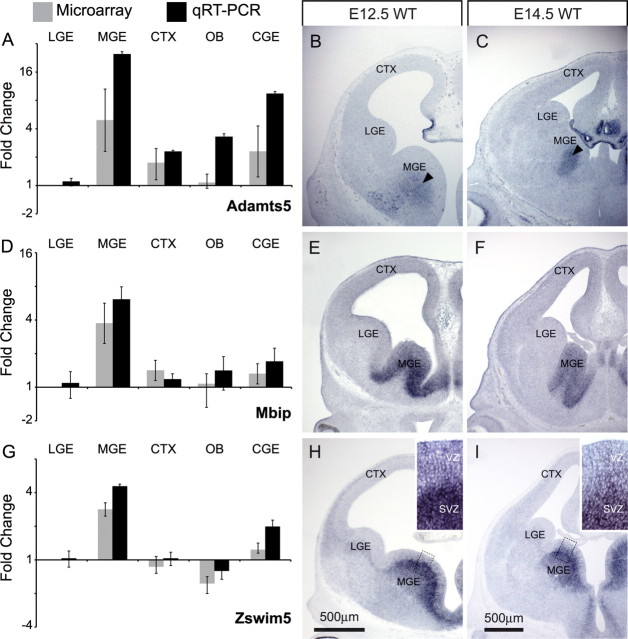
Figure 3.

qRT-PCR and in situ hybridization validates the expression of LGE-enriched candidate genes. A, D, G, J, qRT-PCR data (black) closely match microarray data (gray) for Boc (A), Fzd8 (D), Ankrd43 (G), and Ikzf1 (J). B, C, Boc is expressed in the ventricular zone of the cortex and LGE at E12.5 and E14.5. E, F, Fzd8 is expressed in the ventricular zone of the lateral cortex and the LGE at E12.5 and E14.5. H, I, Ankrd43 is weakly expressed in the LGE mantle at E12.5 (arrowhead). Ankrd43 message increases at E14.5 and localizes to cells within the LGE mantle (arrowhead), as well as cells within the cortical plate (arrows). K, L, Ikzf1 is absent from the E12.5 forebrain but is present in the LGE mantle at E14.5 (arrowhead). SC, Spinal cord; WT, wild type.
Figure 4.
qRT-PCR and in situ hybridization validates the expression of MGE-enriched candidate genes. A, D, G, qRT-PCR data (black) confirms microarray data (gray) for Adamts5 (A), Mbip (D), and Zswim5 (G). B, C, Adamts5 mRNA localizes to the lower SVZ/mantle of the MGE at E12.5 and E14.5 (arrowheads), as well as epithelial cells of the choroid plexus. E, F, Mbip is expressed in the VZ and SVZ of the MGE at E12.5 and E14.5. H, I, Zswim5 message localizes primarily to the SVZ of the MGE at E12.5 and E14.5. Insets, Higher magnification of boxed regions in H and I. Patchy expression is also seen in the MGE VZ at both time points.
Expression patterns demonstrate novel molecular diversity in the LGE and MGE
To determine whether quantitative differences in LGE and MGE mRNA levels were matched by distinct expression patterns, we examined the distribution of the seven newly identified LGE- or MGE-enriched genes by in situ hybridization. Of the LGE-enriched candidate genes, Boc and Fzd8 are easily detected at E12.5 (Fig. 3B,E) and remain through E14.5 (Fig. 3C,F). Boc extends throughout the LGE as well as cortical ventricular zone (VZ), but stops at the MGE border (Fig. 3B,C). Similarly, Fzd8 is found in the LGE VZ but not the MGE (Fig. 3E,F), and as previously reported, is also present in a lateral (high) to medial (low) gradient in the cortical VZ (Kim et al., 2001). Neither Ankrd43 nor Ikfz1 are robustly detected at E12.5 (Fig. 3H,K). At E14.5, however, Ankrd43 is seen in the LGE mantle, and to a lesser extent in the dorsal aspect of the LGE ventricular zone/subventricular zone (SVZ) (Fig. 3I). Ankrd43 transcripts are also found in the cortical marginal zone and to a lesser extent the cortical ventricular zone, as well as the nascent piriform cortex and marginal zone of the medial pallium (Fig. 3I). Similarly, Ikzf1 is seen in a restricted population of LGE mantle cells by E14.5, but is excluded from the MGE and neocortex (Fig. 3L). All three MGE-enriched genes can be detected by in situ hybridization in the E12.5 forebrain (Fig. 4). Adamts5 is restricted to the MGE mantle and nascent choroid plexus, whereas the other two genes are most prominent in the MGE, as well as the septal area and ventricular and subventricular zones (Fig. 3I); some transcripts, however, are detected at low levels in the LGE and cortex at E12.5. At E14.5, Adamts5 is seen at relatively low levels in the MGE SVZ and mantle, and more robustly the choroid plexus (Fig. 4B,C). Mbip, expressed in the VZ and SVZ (Fig. 4E,F), extends medially from a sharp boundary at the LGE through the MGE and into the septum. Zswim5 is limited to the MGE (Fig. 4H,I) and most strongly expressed in a narrow band of cells located at the boundary between the VZ and SVZ of the MGE (Fig. 4H,I, insets). Thus, cellular expression of LGE- and MGE-enriched genes identified by microarray and confirmed by qRT-PCR is either limited to or enhanced in the LGE or MGE. Boc, Fzd8, Mbip, and Zswim5 are restricted to the VZ/SVZ, where progenitors are found, whereas Ikzf1, Ankrd43, and Adamts5 are found in the mantle, where differentiating neurons are located.
Maintainence of LGE and MGE gene expression
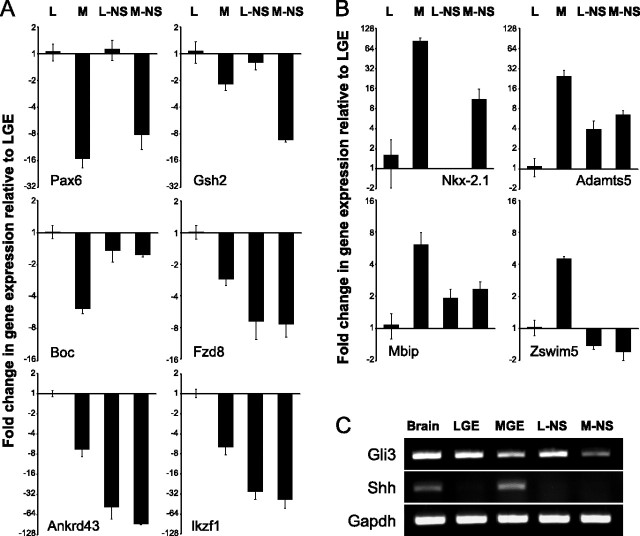
The expression of Boc, Fzd8, Mbip, and Zswim5 in the VZ/SVZ indicates that this subset of newly identified genes may be associated with LGE or MGE neural progenitors. Thus, we asked whether region-specific expression is maintained in progenitor cells isolated from the LGE and MGE and propagated in vitro as neurospheres. Some aspects of LGE and MGE progenitor identity are retained in neurospheres from the LGE, MGE, and cortex (Hitoshi et al., 2002; Ostenfeld et al., 2002; Parmar et al., 2002), even though the neurosphere assay represents highly selective, and clearly artificial, in vitro conditions for progenitor isolation and expansion. Pax6 and Gsh2, two established LGE-enriched genes, are selectively expressed in LGE-derived neurospheres, as is Nkx-2.1, an established MGE-enriched gene, in MGE-derived neurospheres (Parmar et al., 2002) (Fig. 5A,B). In contrast, regional specificity is not maintained for any of the newly identified LGE- or MGE-enriched genes. Expression of the LGE genes is diminished or absent in LGE as well as MGE neurospheres (Fig. 5A), whereas two of the MGE-enriched genes, Adamts5 (associated primarily with mantle rather than VZ/SVZ cells in vivo) and Mbip, are expressed at similar levels in MGE- as well as LGE-derived neurospheres (Fig. 5B).
Figure 5.
Newly identified LGE- and MGE-enriched genes are not selectively maintained in primary neurosphere cultures. A, Pax6 and Gsh2 transcripts are expressed at higher levels in LGE-derived neurospheres (L-NS) than MGE-derived neurospheres (M-NS), reflecting a preservation of their in vivo expression profile. The selective expression of Boc, Fzd8, Ankrd43, and Ikzf1, however, is lost in L-NS. B, Nkx-2.1 is expressed in M-NS but absent from L-NS, mirroring its in vivo expression profile. Adamts5, Mbip, and Zswim5 are not selectively maintained by M-NS. L-NS display increased expression of Adamts5 and Mbip compared with native LGE levels. C, Semiquantitative RT-PCR indicates that Gli3 is selectively maintained in L-NS versus M-NS, whereas Shh expression is undetectable in either population. Brain, E14.5 brain; LGE and MGE, E14.5 LGE and MGE.
Because Shh-mediated signaling appears to differentially regulate MGE versus LGE gene expression in vivo (Rallu et al., 2002a), differences in maintenance of appropriate expression of the newly identified MGE- and LGE-enriched genes in neurospheres might reflect differences in expression or activity of Shh, perhaps via Gli3 transcriptional regulation in MGE- versus LGE-derived neurospheres. The partial maintenance of MGE genes in MGE-derived neurospheres, or loss of LGE genes in LGE-derived neurospheres, does not appear to reflect differential activity of Shh; Shh message is not detected in either LGE or MGE neurospheres (Fig. 5C). Similarly, the influence of Gli3 alone is unlikely to explain the partial maintenance of MGE or loss of LGE progenitor identity in vitro, because Gli3 is expressed in LGE and MGE neurospheres at levels comparable to those in the LGE and MGE in vivo (Fig. 5C). Accordingly, whereas the differential expression of Pax6, Gsh2, Nkx-2.1, and Gli3 persists in LGE versus MGE neurospheres, apparently independent of continued Shh availability, these distinctions are insufficient to establish or maintain region-specific expression of four newly identified genes that distinguish LGE and MGE progenitors. Apparently, the in vivo molecular identity of LGE and MGE progenitors is only partially retained in the conditions necessary to establish primary neurospheres from progenitors isolated from the LGE or MGE.
Regulation of LGE and MGE identity by Pax6, Gli3, and Shh
We next evaluated potential intrinsic, local context-dependent mechanisms that might regulate novel aspects of LGE and MGE progenitor identity. Pax6, Gli3, and Shh are major determinants of dorsal/ventral and medial/lateral forebrain patterning, including patterning of LGE and MGE gene expression (Chiang et al., 1996; Stoykova et al., 2000; Tole et al., 2000; Rallu et al., 2002a). The seven genes identified here may be influenced by these cardinal regulators in the context of morphogenetic axes (medial/lateral and dorsal/ventral) in the developing forebrain. Accordingly, their expression patterns or levels should change in response to mutations in Pax6, Gli3, or Shh, as is the case for other LGE- and MGE-associated genes, including Gsh2 and Nkx2.1 (Corbin et al., 2000; Pabst et al., 2000; Toresson et al., 2000; Yun et al., 2001; Rallu et al., 2002a). Alternatively, LGE and MGE patterning of some or all of the newly identified LGE and MGE genes may be independent of these established regulators of forebrain axes.
We assessed changes in expression of each of the seven novel genes, as well as the MGE-specific gene Nkx-2.1, in at least five E14.5 mutant embryos of each genotype. For all but two probes, there was absolute consistency in expression changes in each set of mutants analyzed. Multiple probes for Ankrd43 and Adamts5 had variability in both wild-type and mutant tissue, perhaps because of their relatively low abundance and restricted expression pattern. This technical issue diminished the number of reported observations in the expression analysis for these two genes.
There is a combination of stability and change in expression patterns and levels of the four LGE-enriched genes in E14.5 Pax6Sey/Sey, Shh−/−, or Gli3XtJ/XtJ mutant embryos. Boc expression patterns are not altered in homozygous Pax6 mutants (n = 5/5) (Fig. 6A). Expression levels, however, appear reduced in the forebrain (Fig. 6A), but not the spinal cord (n = 5/5) (Figs. 3C, 6A, compare insets). Loss of function of both Shh and Gli3 results in the expansion of Boc and Fzd8 expression in the forebrain (Fig. 6B,C). In Shh mutants, Boc is seen throughout the highly dysmorphic forebrain VZ (n = 5/5), and in the Gli3 mutant, Boc expands into the VZ of the MGE and septal region (n = 5/5) (Fig. 6B,C). Fzd8 expression also appears diminished in the Pax6Sey/Sey forebrain, without a noticeable change in pattern (n = 5/5) (Fig. 6D; supplemental Fig. 1A,B, available at www.jneurosci.org as supplemental material), and expands medially in Shh−/− (n = 5/5) (Fig. 6E; supplemental Fig. 1G,H, available at www.jneurosci.org as supplemental material) and Gli3XtJ/XtJ (n = 5/5) (Fig. 6F; supplemental Fig. 1M,N, available at www.jneurosci.org as supplemental material) embryos. Ankrd43 and Ikzf1 remain expressed in a lateral population of mantle cells in both Pax6 (n = 4/5; n = 5/5) and Gli3 (n = 3/5; n = 5/5) mutants (Fig. 6G,I,J,L); however, expression of both genes expands in Shh−/− embryos. Ankrd43 is seen throughout the entire forebrain mantle (n = 3/5) (Fig. 6H), whereas Ikzf1 expands throughout the ventral mantle region (n = 5/5) (Fig. 6K). Thus, expression of all four LGE-enriched genes apparently depends on Shh-mediated repression in the MGE. Moreover, for Boc and Fzd8, Gli3 but not Pax6 mediates this negative regulation (see Fig. 9).
Figure 6.
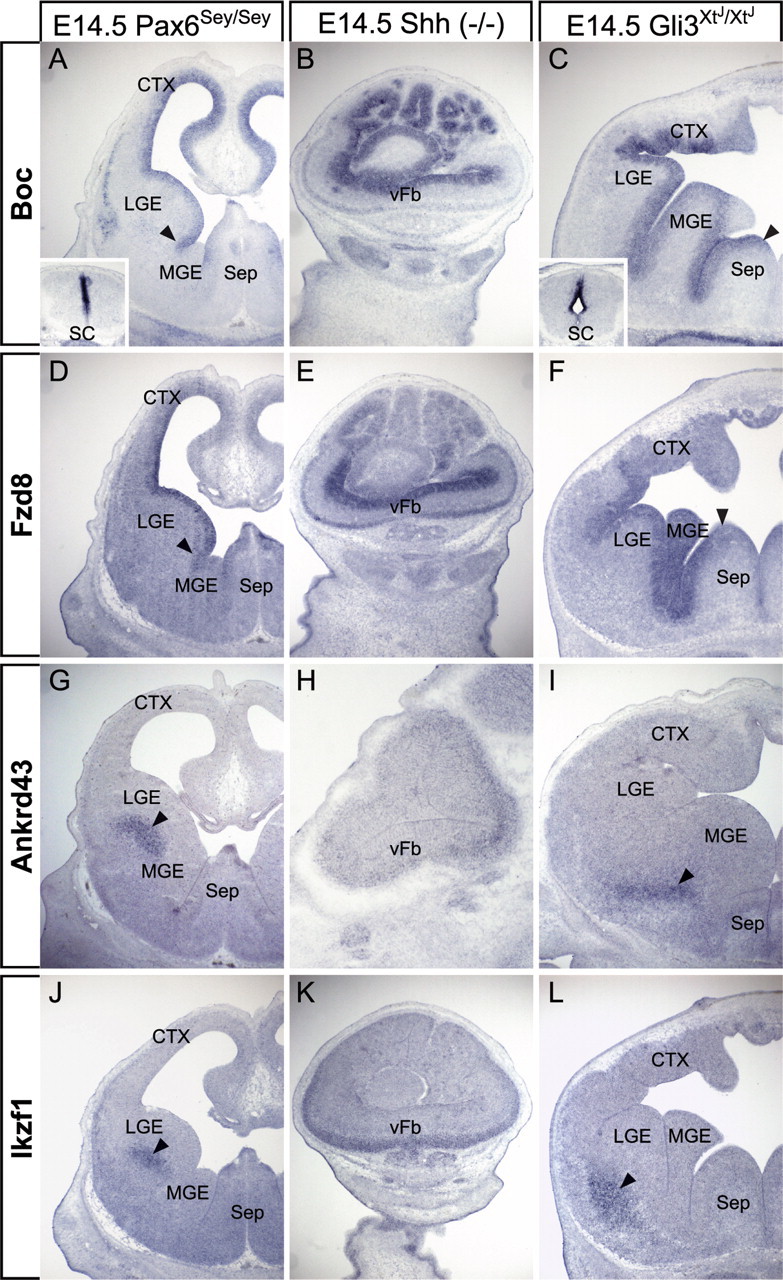
LGE-enriched gene expression is partially dependent on canonical axial patterning mechanisms. A, Boc expression is diminished in the Pax6 mutant forebrain, but remains appropriately patterned (arrowhead). Spinal cord (SC; inset) expression is less effected. B, Boc expression expands to the ventral forebrain (vFb) of the Shh mutant. C, Boc expression expands medially (arrowhead) in the Gli3 mutant forebrain. D, Pax6 mutation does not change patterning of Fzd8 expression (arrowhead). E, Fzd8 expression expands throughout the Shh−/− vFb. F, Loss of Gli3 function results in the medial expansion (arrowhead) of Fzd8 expression. G, Ankrd43 expression (arrowhead) is unaffected by Pax6 mutation. H, I, Its expression uniformly increases in the mantle region of the Shh mutant (H) and is shifted slightly ventromedially (arrowhead) in the Gli3 mutant (I). J–L, Ikzf1 remains appropriately patterned in the Pax6 mutant (J, arrowhead), expands throughout the ventral mantle region of the Shh mutant (K), and shifts somewhat ventrolaterally (arrowhead) in the Gli3 mutant (L). Sep, Septum or septal area.
Figure 9.

Frontonasal mesenchyme regulates gene expression in the ventral forebrain by modulating the activities of Shh and Fgf8. A, Fn:M/Fb:E cocultures treated with vehicle control and DEAB (equivalent results at 25 and 100 μm) display normal MGE (Mbip and Nkx-2.1) and LGE (Fzd8 and Pax6) patterning, indicating that a local source of retinoic acid in the mesenchyme does not significantly impact ventral forebrain gene expression. MGE gene induction is blocked by cyclopamine (Cyclop; 10 μm), whereas LGE gene expression expands uniformly. SU5402 (10 μm), a selective inhibitor of Fgfr tyrosine kinase activity (Mohammadi et al., 1997), prevents Mbip but not Nkx-2.1 induction, while expanding LGE gene expression. B, Bead (1 mg/ml) or bath (100 ng/ml) application of Fgf8b enhances Mbip but not Nkx-2.1 expression in Fb:E cultures. Asterisks indicate placement of heparin acrylic bead.
All three MGE-enriched genes are sensitive to homozygous loss of Pax6, Shh, or Gli3 function. Homozygous Pax6 mutation has a similar influence on Mbip and Zswim5 expression. Mbip expression is diminished in the MGE SVZ, but expands laterally into the medial aspect of the LGE (n = 4/5) (Fig. 7A; supplemental Fig. 1C,D, available at www.jneurosci.org as supplemental material). Zswim5 remains expressed near the VZ/SVZ border, but its boundary shifts laterally into the LGE (n = 5/5) (Fig. 7D; supplemental Fig. 1E,F, available at www.jneurosci.org as supplemental material). This lateral expansion across the MGE/LGE border is also seen for Nkx-2.1, as previously reported (supplemental Fig. 2A–A″, available at www.jneurosci.org as supplemental material) (Stoykova et al., 2000). Adamts5 is diminished in both abundance and frequency in the Pax6Sey/Sey ventral forebrain. When detected, the remaining Adamts5 message is displaced ventrolaterally from its normal position at the SVZ/mantle border of the MGE (n = 3/5) (Fig. 7G); however, choroid plexus expression consistently appears unaltered (n = 4/4) (Fig. 7G). Shh has a more uniform influence; there is no detectable expression of any of the three MGE-enriched genes in the Shh−/− forebrain (n = 5/5 for each) (Fig. 7B,E,H; supplemental Fig. 1I–L, available at www.jneurosci.org as supplemental material). In contrast, Gli3 loss of function has distinct effects on each gene. Mbip becomes restricted to the VZ of the most medial aspect of the MGE and septum (n = 5/5) (Fig. 7C; supplemental Fig. 1O,P, available at www.jneurosci.org as supplemental material). This domain of Mbip expression corresponds to an analogous domain of Nkx-2.1 expression in Gli3 mutants, which also appears shifted medially from its normal boundary at the LGE border (n = 4/4) (supplemental Fig. 2B–E, available at www.jneurosci.org as supplemental material). Zswim5 is diminished in the VZ/SVZ of the MGE, but unlike Mbip and Nkx-2.1, expands laterally throughout the LGE and cortex (n = 5/5) (Fig. 7F; supplemental Fig. 1Q,R, available at www.jneurosci.org as supplemental material). Adamts5 cannot be detected in the MGE of Gli3XtJ/XtJ embryos, and because Gli3 mutants lack a choroid plexus in the lateral ventricle (Theil et al., 1999), Adamts5 message is absent from the remaining regions of the mutant forebrain (n = 5/5) (Fig. 7I). Thus, like the newly identified LGE genes, MGE genes are all regulated through Shh-dependent mechanisms. Nevertheless, there is divergent dependence on Gli3: expanded expression of Zswim5 suggests regulation by Gli3-mediated repression of Shh signaling, whereas the repressor activity of Gli3 alone is unlikely to account for patterned expression of Mbip and Adamts5 in the MGE (see Fig. 9).
Figure 7.
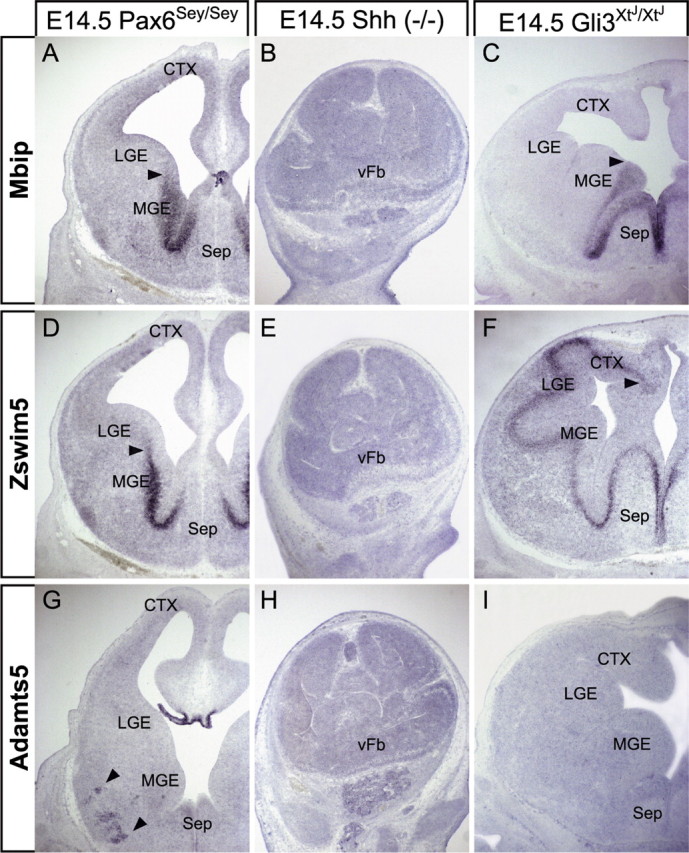
MGE-enriched gene expression is differentially regulated by mutations that disrupt axial patterning in the forebrain. A, Mbip expression is expanded laterally (arrowhead) in the Pax6 mutant MGE. B, E, H, Mbip expression (B) is lost in the Shh−/− vFb, as is the expression of Zswim5 (E) and Adamts5 (H). C, Mbip expression is retracted medially (arrowhead) in the Gli3 mutant forebrain. D, Similar to Mbip, Zswim5 expression expands laterally (arrowhead) into the Pax6 mutant LGE. F, Zswim5, unlike Mbip, expands laterally (arrowhead) throughout much of the Gli3 mutant forebrain. G, Adamts5 expression is reduced and displaced ventrolaterally (arrowhead) in the Pax6 mutant MGE, but is present in the choroid plexus. I, Adamts5 expression is lost entirely in the Gli3 mutant forebrain.
Extrinsic patterning of LGE and MGE genes
Together, the results of the neurosphere and mutant analyses indicate that the molecular identity of LGE and MGE progenitors, particularly that defined by expression of the four newly identified progenitor markers, depends on additional mechanisms beyond those that establish or maintain intrinsic axes in the forebrain. Potential candidates are limited: the major tissue adjacent to the ventral forebrain between E9.5 and E12.5 (when LGE and MGE identity emerges) is the frontonasal mesenchyme. This mesenchyme is a source of inductive signals for the nascent olfactory epithelium (LaMantia et al., 1993, 2000); therefore, we used an in vitro assay (Fig. 8A) to determine whether this mesenchyme also influences regional differences in LGE and MGE gene expression in the forebrain neuroepithelium. The tissue used for our in vitro mesenchyme/epithelium (M/E) interaction assay is microdissected from the forebrain and frontonasal mass at E9.5. Thus, our assay begins with ventral forebrain tissue in which there is no indication of MGE/LGE patterning. The assay period extends for 3 d in vitro, thus providing a culture period that parallels the initial patterning of LGE and MGE domains that occurs between E9.5 and E12.5 in vivo.
Figure 8.
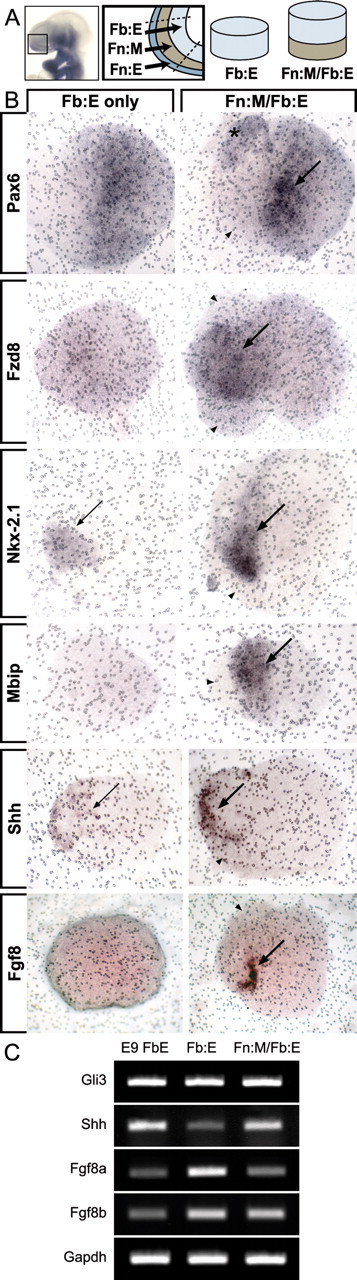
Frontonasal mesenchyme enhances and patterns gene expression in the developing forebrain. A, At E9.5, Fn:M directly apposes the nascent telencephalon at the rostrolateral pole of the prosencephalic vesicle. By microdissecting and culturing either Fb:E alone or the Fn:M and the Fb:E paired (Fn:M/Fb:E), we assessed the influence of the Fn:M on LGE and MGE gene expression. B, Pax6 mRNA is present in Fb:E-only cultures, but enriched (arrow) and predictably patterned (asterisk) in the Fb:E of Fn:M/Fb:E cocultures. Fzd8, Nkx-2.1, and Mbip expression show significant enhancement (arrows) and patterning of expression in Fn:M/Fb:E cultures. In addition, the Fn:M influences the inductive signaling molecules Shh and Fgf8 by enhancing and refining their expression domains in the overlying Fb:E. C, Semiquantitative RT-PCR indicates that Shh transcripts increase, whereas Gli3 transcript levels appear unchanged between Fb:E only and Fn:M/Fb:E cultures. Also, mesenchymal signals selectively reduce Fgf8a (inactive splice isoform) and maintain Fgf8b (active splice isoform) expression levels in the forebrain neuroepithelium. E9 FbE, Uncultured forebrain neuroepithelium. Arrowheads point to frontonasal mesenchyme in Fn:M/Fb:E cultures.
We examined six genes expressed in either LGE or MGE progenitors by E12.5: Pax6, Fzd8, and Boc for the LGE, and Nkx-2.1, Mbip, and Zswim5 for the MGE. Frontonasal mesenchyme apparently enhances expression levels and pattern of Pax6 (n = 4/4) and Fzd8 (n = 8/8) (Fig. 8B, top two panels). The frontonasal mesenchyme also influences MGE genes: Nkx-2.1 expression and pattern is enhanced by frontonasal mesenchyme (n = 4/4), and the mesenchyme both induces and patterns Mbip (n = 9/9) (Fig. 8B, middle two panels). Influence of the frontonasal mesenchyme on induction and patterning is not universal, however, because neither Boc (n = 3/3) nor Zswim5 (n = 4/4) appear to be significantly regulated by this tissue (data not shown).
Inductive signaling intrinsic to the ventromedial forebrain is also regulated by frontonasal mesenchyme. Thus frontonasal mesenchymal signals influence expression and pattern of Shh and Fgf8. In the absence of frontonasal mesenchyme, Shh is detected in the presumed ventral aspect of the isolated Fb:E; however, the in situ signal is attenuated, and its expression domain is relatively broad. In the presence of frontonasal mesenchyme, Shh expression appears enhanced, and the expression domain boundaries are sharpened and refined (n = 8/9) (Fig. 8B, bottom panel). Fgf8 message localizes to a small but intensely labeled stripe of forebrain neuroepithelial cells found only in the presence of frontonasal mesenchyme (n = 2/5) (Fig. 8B); in situ detection of Fgf8 was variable, however, and we were unable to localize Fgf8 transcripts in all explant pairs. Consistent with a difference in the expression pattern and intensity of Shh, there is an apparent difference in the abundance of Shh transcripts in isolated forebrain epithelium versus paired frontonasal mesenchyme/forebrain epithelium explants. Shh transcripts appear attenuated in the isolated epithelium, whereas Gli3 levels are indistinguishable in Fb:E versus Fn:M/Fb:E explants (Fig. 8C). Fgf8a, a splice variant thought to be an endogenous antagonist of Fgf8 signaling (Lee et al., 1997; Liu et al., 1999; Sato et al., 2001), declines in the presence of mesenchyme (Fig. 8C), whereas Fgf8b (the primary active Fgf8 isoform) levels appear equivalent between Fb:E and Fn:M/Fb:E explant cultures (Fig. 8C). Thus, signals from the frontonasal mesenchyme establish or maintain expression or pattern of several genes associated with either LGE or MGE progenitors as well as the inductive signals Shh and Fgf8 in the ventral forebrain neuroepithelium.
M/E interaction regulates Shh and Fgf signaling in the ventral forebrain
Our results suggest that MGE as well as LGE patterning relies on signals from the lateral frontonasal mesenchyme; however, ventral forebrain patterning is thought to depend primarily on Shh and Fgf8 from the neuroepithelium itself (Chiang et al., 1996; Ohkubo et al., 2002; Storm et al., 2003, 2006; Fuccillo et al., 2004; Gutin et al., 2006). Because Shh expression levels and pattern are modulated by frontonasal mesenchyme (Fig. 8B,C), we asked whether M/E interaction facilitates Shh-mediated patterning in the ventral forebrain. Pharmacological disruption of Shh in the presence of frontonasal mesenchyme leads to loss of MGE gene expression and enhancement of LGE gene expression; Mbip (n = 6/6) and Nkx-2.1 (n = 5/5) expression levels are consistently reduced, whereas Fzd8 (n = 5/5) and Pax6 (n = 4/5) expression domains are expanded (Fig. 9A, second row). Nevertheless, Shh alone, in the absence of mesenchyme, is not sufficient to enhance Nkx2.1 expression (n = 9/9), nor induce Mbip expression (n = 8/8) (supplemental Fig. 3A, available at www.jneurosci.org as supplemental material). Apparently, Shh-mediated MGE gene expression depends critically on signals from the lateral frontonasal mesenchyme. Previous observations show that RA, which is known to regulate Shh at several sites, including the facial primordial (Schneider et al., 2001), is produced by the frontonasal mesenchyme, and signals to subsets of forebrain precursor cells (Haskell and LaMantia, 2005). Nevertheless, pharmacological loss of RA signaling does not influence initial expression or patterning of LGE (Fzd8, n = 7/8; Pax6, n = 6/7) or MGE (Mbip, n = 7/7; Nkx-2.1, n = 8/8) genes (Fig. 9A, third row), nor does gain of RA signaling induce or enhance LGE (Fzd8, n = 6/7; Pax6, n = 6/7) or MGE (Mbip, 5 = 5; Nkx-2.1 = 3/3) gene expression in absence of mesenchyme (supplemental Fig. 3B, available at www.jneurosci.org as supplemental material). Efficacy of DEAB in reducing RA-sensitive gene expression was independently confirmed in whole embryos by qRT-PCR and in situ hybridization (data not shown). Thus, mesenchymal influence on the LGE and MGE genes we have evaluated is unlikely to reflect RA regulation of Shh or other local forebrain signals.
Fgf8, presumably from the anterior neural ridge of the telencephalic vesicle (the “rostral patterning center”), also appears to be critical for MGE morphogenesis and gene expression (Storm et al., 2006). Because frontonasal mesenchyme influences expression and proportions of active versus inactive Fgf8 isoforms in the forebrain neuroepithelium (Fig. 8B,C, available at www.jneurosci.org as supplemental material), the mesenchyme may regulate ventral forebrain patterning by modulating Fgf8 activity. In the presence of frontonasal mesenchyme, pharmacological disruption of Fgfr tyrosine kinase activity does not alter mesenchymally regulated Nkx-2.1 expression or pattern (n = 5/5); Mbip induction, however, is substantially attenuated (n = 5/5) (Fig. 9A, fourth row). LGE gene expression is also disrupted: Fzd8 (n = 5/5) and Pax6 (n = 4/4) expand to the entire explant neuroepithelium (Fig. 9A, fourth row). Apparently, Fgf signaling influences LGE patterning independent of the establishment and maintenance of restricted Nkx2.1 expression. To determine whether Fgf8 regulates ventral forebrain gene expression directly, we exposed isolated forebrain epithelium to either a local or global source of Fgf8. Fgf8b beads do not enhance Nkx-2.1 (n = 5/5), whereas Mbip appears to be modestly enhanced (n = 4/6) (Fig. 9B, top row). Similarly, bath application of Fgf8b upregulates Mbip expression (n = 3/3), without altering Nkx-2.1 expression (n = 2/2) (Fig. 9B, bottom row). Apparently, Fgf8 positively regulates at least one MGE gene, Mbip, and this regulation occurs independently of Nkx2.1. Together, these observations indicate that Shh and Fgf8 are maximally effective in patterning MGE and LGE gene expression in the context of M/E interactions between the frontonasal mesenchyme and the forebrain neuroepithelium.
Discussion
We defined novel dimensions of LGE and MGE progenitor identity and specification. Incomplete maintenance of LGE and MGE genes in neurospheres, coupled with divergent changes in expression patterns of LGE and MGE genes in Pax6Sey/Sey, Shh−/−, and Gli3XtJ/XtJ mutants, suggests that progenitor identity depends on mechanisms beyond Pax6- or Gli3-mediated Shh signaling. Instead, integration of signals from extrinsic tissues and forebrain neuroepithelium is critical for establishing LGE and MGE identity. The frontonasal mesenchyme, adjacent to the rudimentary ventral forebrain, is a major source of extrinsic signals that modulate expression and activity of Shh and Fgf8 in the ventral forebrain, as well as induction and patterning of LGE and MGE genes. Thus, LGE and MGE progenitor identity depends on forebrain position and likely reflects a balance of extrinsic signals from the frontonasal mesenchyme and intrinsic signals from neighboring forebrain cells.
Novel dimensions of LGE and MGE molecular diversity
Most previously identified LGE- or MGE-restricted genes encode transcription factors (Campbell, 2003; Takahashi and Liu, 2006; Wonders and Anderson, 2006). Of seven LGE- or MGE-enriched genes identified here, only one, Ikzf1, which is not expressed in progenitors, is an established transcription factor. Boc, enhanced in LGE progenitors, is implicated in myogenesis and axon guidance (Kang et al., 2002; Mulieri et al., 2002; Kang et al., 2003; Connor et al., 2005; Okada et al., 2006). Boc initiates adhesive signaling by binding Cdon, which our microarray data suggest is expressed uniformly in LGE, MGE, and cortex. Thus, Boc may limit Cdon-dependent signaling to LGE and cortical progenitors. Fzd8, a Wnt receptor also enhanced in LGE precursors, may regulate progenitor or neuroblast polarity and motility through activation of canonical or noncanonical signaling pathways (Veeman et al., 2003; Kikuchi et al., 2007). Mbip, limited to MGE progenitors, selectively inhibits Map3k12 induction of JNK/SAPK activity (Fukuyama et al., 2000). Because Map3k12 and JNK apparently regulate cortical radial migration (Hirai et al., 2002, 2006; Kawauchi et al., 2003), Mbip inhibition of Map3k12 signaling may prohibit radial and support tangential migration of MGE cells. Zswim5, also in the MGE, has DNA and protein binding capacity, but no known function (Makarova et al., 2002). Ankrd43, in the LGE mantle, has no known function and remains expressed in apparent adult striatal projection neurons (Allan Brain Atlas, Ankrd43). Adamts5 in the MGE mantle, as well as in the choroid plexus like another family member, Adamts9 (Jungers et al., 2005), may expose MGE cells to additional metalloprotease activity at the ventricular surface, perhaps modifying the local extracellular environment. Thus, our data suggests that adhesion and signaling are selectively regulated in LGE and MGE progenitors, as well as their postmitotic progeny.
Limited maintenance of LGE and MGE progenitor identity
Several aspects of LGE and MGE identity are not maintained when progenitors are isolated in vitro. Loss of four novel progenitor-associated genes either reflects incomplete retention or in vitro respecification of endogenous precursor identity. There is conflicting evidence for retention of neural progenitor identity in vitro: some reports suggest it is preserved (Hitoshi et al., 2002; Ostenfeld et al., 2002; Parmar et al., 2002), whereas others suggest such distinctions are lost (Gabay et al., 2003; Santa-Olalla et al., 2003; Hack et al., 2004). Neurosphere formation likely selects against the expression of late progenitor markers, such as Zswim5, heavily enriched in the MGE SVZ, as well as early differentiation markers of the striatal mantle, including Ankrd43 and Ikzf1. Nevertheless, some regional identity remains intact: Pax6 and Gsh2 in LGE, and Nkx-2.1 in MGE neurospheres. These factors alone, however, do not maintain expression of additional LGE or MGE genes, perhaps because of selection of EGF- and FGF-responsive progenitors with more generic identity (Gabay et al., 2003; Hack et al., 2004). Indeed, the altered milieu in the neurosphere assay leads to anomalous expression of LGE and MGE genes, including Boc, Mbip, and Adamts5, as well as loss of Shh. These results, together with previous comparisons of molecular identity of embryonic and adult forebrain neurospheres (Hack et al., 2004; Haskell and LaMantia, 2005; Councill et al., 2006), suggest in vivo progenitor identities are not completely maintained in vitro. Maintenance of LGE and MGE progenitor identity defined here, in contrast to that associated with Pax6, Gsh2, or Nkx-2.1, must rely on systemic signals or local cell–cell interactions.
Pax6-, Shh-, and Gli3-dependent and -independent mechanisms pattern the LGE and MGE
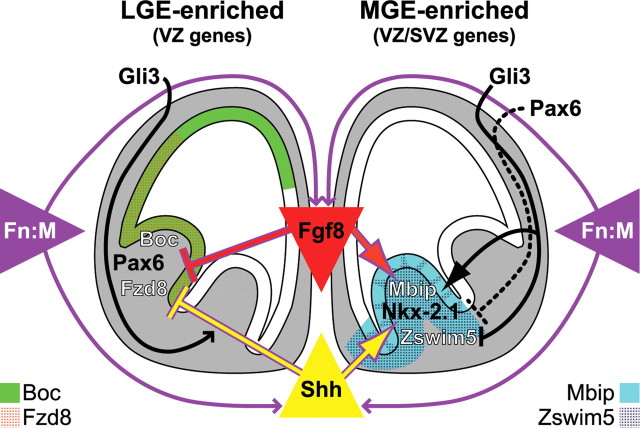
Patterned expression of LGE- or MGE-enriched genes depends on Shh signaling; nevertheless, this regulation is not limited to Gli3-dependent repression. LGE genes expand in the Shh−/− forebrain, indicating Shh repression; however, Boc and Fzd8 also expand in the Gli3XtJ/XtJ forebrain (Fig. 10). Apparently, Shh-dependent repression of Boc and Fzd8 in the MGE depends on Gli3, because Shh remains available in the Gli3XtJ/XtJ MGE (Theil et al., 1999; Tole et al., 2000). Neither Boc nor Fzd8 are lost in the Gli3XtJ/XtJ LGE. Thus, Gli3 must not regulate Boc and Fzd8 by repressing Shh signaling. Instead, Gli3-dependent Shh repression of Boc and Fzd8 in the MGE may depend on full-length Gli3 activator function (Sasaki et al., 1999; Wang et al., 2000), present in the Shh-rich MGE (Fotaki et al., 2006). Gli3-activator function regulates ventral spinal cord progenitor segregation and neuronal differentiation (Bai et al., 2004; Lei et al., 2004). In the ventral forebrain, Shh-dependent Gli3-mediated transcriptional activation presumably upregulates inhibitors of Boc and Fzd8 expression in MGE cells, thereby establishing an MGE/LGE expression boundary.
Figure 10.
A balance of intrinsic and extrinsic signaling regulates molecular specification of the LGE and MGE. Gli3 regulates novel aspects of ventral forebrain patterning by differentially influencing gene expression at the LGE/MGE boundary, whereas Pax6 reinforces LGE identity by apparently regulating expression levels in the LGE and bounding MGE gene expression at the LGE border. Extrinsic signals from the Fn:M are critical positive regulators of Shh and Fgf8 activity in the forebrain. This novel molecular mechanism of ventral forebrain induction and patterning indicates that initial specification of the LGE and MGE reflects mesenchymal influence on forebrain signaling sources as well as response capacity of the forebrain neuroepithelium, and subsequently limits or refines region-specific gene expression in LGE or MGE progenitors or their postmitotic progeny.
Gli3 loss of function also highlights key distinctions in Shh and Gli3 regulation of MGE gene expression, reinforcing the hypothesis that Shh-dependent maintenance of ventral forebrain patterning involves more than Gli3-mediated repression of Shh target genes. Mbip and Zswim5 have opposing changes in Gli3XtJ/XtJ mutants (Fig. 10). Mbip, like Nkx-2.1, is limited to a medial domain, whereas Zswim5 expands laterally throughout the dorsal telencephalon. This suggests differential requirements for Gli3-mediated repression in defining the lateral MGE boundary. Zswim5 depends on Gli3-mediated repression of Shh activity to reinforce the MGE/LGE border; Mbip does not. Moreover, because Mbip and Nkx-2.1 appear diminished and restricted medially in the Gli3XtJ/XtJ ventral forebrain, their normal expression at the LGE border may depend on Gli3 activator function or repression of inhibitory mechanisms in the lateral MGE. Alternatively, Gli3-mediated patterning of LGE and MGE gene expression may reflect currently uncharacterized Shh-independent signaling mechanisms.
Pax6 loss of function has distinct effects on the newly identified LGE- and MGE-enriched genes. Pax6 apparently influences Boc and Fzd8 expression levels, but not pattern. In contrast, Pax6 regulates Mbip and Zswim5 patterning at the MGE/LGE border. Pax6 loss of function results in medial to lateral expansion of Mbip and Zswim5, consistent with a similar shift in Shh, Nkx-2.1, and Lhx6 (Stoykova et al., 2000). This regulation is likely indirect; however, Pax6 and Nkx-2.1 expression converge in the early ventral telencephalon (Corbin et al., 2003), perhaps allowing Pax6 to influence the lateral MGE. Apparently, Pax6 operates in parallel with Gli3 to define or maintain the MGE/LGE boundary. Unlike Gli3, however, Pax6 maintains bounded expression of both Mbip and Zswim5 (Fig. 10). Thus, the influences of three canonical forebrain patterning genes, Pax6, Shh, and Gli3, are not uniform when assessed within multiple dimensions of LGE and MGE molecular diversity.
Extrinsic regulation of LGE and MGE progenitor identity
We have shown that frontonasal mesenchyme patterns the prosencephalon (LaMantia et al., 1993; Anchan et al., 1997) and promotes migratory capacity of early forebrain neuroblasts (Tucker et al., 2006). We now demonstrate that frontonasal mesenchyme influences LGE and MGE progenitor gene expression. Frontonasal mesenchyme facilitates Mbip induction and patterning and enhances or restricts Fzd8, Pax6, and Nkx-2.1 in forebrain neuroepithelium. Moreover, this mesenchyme promotes and refines Shh and Fgf8 expression and regulates active versus inactive Fgf8 isoforms. Indeed, Shh and Fgf regulation of MGE- and LGE-enriched genes depends on mesenchymal signaling (Fig. 10). Induction and patterning of Mbip and Nkx-2.1 requires Shh, yet their requirement for Fgf signaling differs: presumed global inhibition of Fgfr tyrosine kinase activity blocks Mbip induction without altering Nkx-2.1. Moreover, Fgf8 directly activates Mbip, whereas our data, along with previous observations (Shimamura and Rubenstein, 1997; Crossley et al., 2001), indicate that Fgf8 alone does not induce expression of Nkx-2.1 in the early ventral forebrain. Loss of Nkx-2.1 expression in Fgf8 (Storm et al., 2006) as well as Fgfr1−/−; Fgfr2−/− (Gutin et al., 2006) mutants may therefore reflect failed MGE development rather than direct Fgf8 regulation of Nkx-2.1 expression. In addition, Shh and Fgf signaling are necessary to maintain mesenchyme-mediated patterning of LGE genes: Fzd8 and Pax6 expand when Shh and Fgf signaling is blocked, despite the presence of mesenchyme. Nevertheless, Shh and Fgf8 alone, without mesenchymal signals, appear insufficient to insure normal MGE and LGE gene expression. The integration of extrinsic and intrinsic signaling mechanisms may be essential for initial definition of LGE and MGE progenitor identity, thus establishing distinct migratory and differentiation capacities of neurons derived from these forebrain subdivisions.
Footnotes
This work was supported by a National Institute on Deafness and Other Communication Disorders postdoctoral fellowship (DC007047) to E.S.T., a National Alliance for Research on Schizophrenia and Depression Young Investigator Award to F.P., National Institute of Neurological Disorders and Stroke (NINDS) Grant NS047701 to F.P., National Institute of Child Health and Human Development Grants HD029178 and HD042182 and National Institute of Mental Health Grant MH64065 to A.-S.L, and NINDS Grant NS031768, which provides support for the University of North Carolina Neuroscience Center Expression Profiling and In Situ Hybridization core facilities. We thank Daniel Meechan for advice and guidance in qRT-PCR experiments, Tom Maynard for assistance in construct/primer design and digital art production, Thomas Sugimoto for technical assistance in histological sample preparation, Cliff Heindel for mouse care and technical support, and Amanda Peters for laboratory management.
References
- Agoston DV, Szemes M, Dobi A, Palkovits M, Georgopoulos K, Gyorgy A, Ring MA. Ikaros is expressed in developing striatal neurons and involved in enkephalinergic differentiation. J Neurochem. 2007;102:1805–1816. doi: 10.1111/j.1471-4159.2007.04653.x. [DOI] [PubMed] [Google Scholar]
- Anchan RM, Drake DP, Haines CF, Gerwe EA, LaMantia AS. Disruption of local retinoid-mediated gene expression accompanies abnormal development in the mammalian olfactory pathway. J Comp Neurol. 1997;379:171–184. [PubMed] [Google Scholar]
- Anderson SA, Eisenstat DD, Shi L, Rubenstein JL. Interneuron migration from basal forebrain to neocortex: dependence on Dlx genes. Science. 1997;278:474–476. doi: 10.1126/science.278.5337.474. [DOI] [PubMed] [Google Scholar]
- Bai CB, Stephen D, Joyner AL. All mouse ventral spinal cord patterning by hedgehog is Gli dependent and involves an activator function of Gli3. Dev Cell. 2004;6:103–115. doi: 10.1016/s1534-5807(03)00394-0. [DOI] [PubMed] [Google Scholar]
- Butt SJ, Fuccillo M, Nery S, Noctor S, Kriegstein A, Corbin JG, Fishell G. The temporal and spatial origins of cortical interneurons predict their physiological subtype. Neuron. 2005;48:591–604. doi: 10.1016/j.neuron.2005.09.034. [DOI] [PubMed] [Google Scholar]
- Campbell K. Dorsal-ventral patterning in the mammalian telencephalon. Curr Opin Neurobiol. 2003;13:50–56. doi: 10.1016/s0959-4388(03)00009-6. [DOI] [PubMed] [Google Scholar]
- Chiang C, Litingtung Y, Lee E, Young KE, Corden JL, Westphal H, Beachy PA. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature. 1996;383:407–413. doi: 10.1038/383407a0. [DOI] [PubMed] [Google Scholar]
- Connor RM, Allen CL, Devine CA, Claxton C, Key B. BOC, brother of CDO, is a dorsoventral axon-guidance molecule in the embryonic vertebrate brain. J Comp Neurol. 2005;485:32–42. doi: 10.1002/cne.20503. [DOI] [PubMed] [Google Scholar]
- Corbin JG, Gaiano N, Machold RP, Langston A, Fishell G. The Gsh2 homeodomain gene controls multiple aspects of telencephalic development. Development. 2000;127:5007–5020. doi: 10.1242/dev.127.23.5007. [DOI] [PubMed] [Google Scholar]
- Corbin JG, Rutlin M, Gaiano N, Fishell G. Combinatorial function of the homeodomain proteins Nkx2.1 and Gsh2 in ventral telencephalic patterning. Development. 2003;130:4895–4906. doi: 10.1242/dev.00717. [DOI] [PubMed] [Google Scholar]
- Councill JH, Tucker ES, Haskell GT, Maynard TM, Meechan DW, Hamer RM, Lieberman JA, LaMantia AS. Limited influence of olanzapine on adult forebrain neural precursors in vitro. Neuroscience. 2006;140:111–122. doi: 10.1016/j.neuroscience.2006.02.014. [DOI] [PubMed] [Google Scholar]
- Crossley PH, Martinez S, Ohkubo Y, Rubenstein JL. Coordinate expression of Fgf8, Otx2, Bmp4, and Shh in the rostral prosencephalon during development of the telencephalic and optic vesicles. Neuroscience. 2001;108:183–206. doi: 10.1016/s0306-4522(01)00411-0. [DOI] [PubMed] [Google Scholar]
- Ezzat S, Mader R, Yu S, Ning T, Poussier P, Asa SL. Ikaros integrates endocrine and immune system development. J Clin Invest. 2005a;115:1021–1029. doi: 10.1172/JCI22486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezzat S, Yu S, Asa SL. The zinc finger Ikaros transcription factor regulates pituitary growth hormone and prolactin gene expression through distinct effects on chromatin accessibility. Mol Endocrinol. 2005b;19:1004–1011. doi: 10.1210/me.2004-0432. [DOI] [PubMed] [Google Scholar]
- Fotaki V, Yu T, Zaki PA, Mason JO, Price DJ. Abnormal positioning of diencephalic cell types in neocortical tissue in the dorsal telencephalon of mice lacking functional Gli3. J Neurosci. 2006;26:9282–9292. doi: 10.1523/JNEUROSCI.2673-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuccillo M, Rallu M, McMahon AP, Fishell G. Temporal requirement for hedgehog signaling in ventral telencephalic patterning. Development. 2004;131:5031–5040. doi: 10.1242/dev.01349. [DOI] [PubMed] [Google Scholar]
- Fukuyama K, Yoshida M, Yamashita A, Deyama T, Baba M, Suzuki A, Mohri H, Ikezawa Z, Nakajima H, Hirai S, Ohno S. MAPK upstream kinase (MUK)-binding inhibitory protein, a negative regulator of MUK/dual leucine zipper-bearing kinase/leucine zipper protein kinase. J Biol Chem. 2000;275:21247–21254. doi: 10.1074/jbc.M001488200. [DOI] [PubMed] [Google Scholar]
- Gabay L, Lowell S, Rubin LL, Anderson DJ. Deregulation of dorsoventral patterning by FGF confers trilineage differentiation capacity on CNS stem cells in vitro. Neuron. 2003;40:485–499. doi: 10.1016/s0896-6273(03)00637-8. [DOI] [PubMed] [Google Scholar]
- Georgopoulos K, Bigby M, Wang JH, Molnar A, Wu P, Winandy S, Sharpe A. The Ikaros gene is required for the development of all lymphoid lineages. Cell. 1994;79:143–156. doi: 10.1016/0092-8674(94)90407-3. [DOI] [PubMed] [Google Scholar]
- Glasson SS, Askew R, Sheppard B, Carito B, Blanchet T, Ma HL, Flannery CR, Peluso D, Kanki K, Yang Z, Majumdar MK, Morris EA. Deletion of active ADAMTS5 prevents cartilage degradation in a murine model of osteoarthritis. Nature. 2005;434:644–648. doi: 10.1038/nature03369. [DOI] [PubMed] [Google Scholar]
- Götz M, Stoykova A, Gruss P. Pax6 controls radial glia differentiation in the cerebral cortex. Neuron. 1998;21:1031–1044. doi: 10.1016/s0896-6273(00)80621-2. [DOI] [PubMed] [Google Scholar]
- Gutin G, Fernandes M, Palazzolo L, Paek H, Yu K, Ornitz DM, McConnell SK, Hébert JM. FGF signalling generates ventral telencephalic cells independently of SHH. Development. 2006;133:2937–2946. doi: 10.1242/dev.02465. [DOI] [PubMed] [Google Scholar]
- Hack MA, Sugimori M, Lundberg C, Nakafuku M, Götz M. Regionalization and fate specification in neurospheres: the role of Olig2 and Pax6. Mol Cell Neurosci. 2004;25:664–678. doi: 10.1016/j.mcn.2003.12.012. [DOI] [PubMed] [Google Scholar]
- Haskell GT, LaMantia AS. Retinoic acid signaling identifies a distinct precursor population in the developing and adult forebrain. J Neurosci. 2005;25:7636–7647. doi: 10.1523/JNEUROSCI.0485-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai S, Kawaguchi A, Hirasawa R, Baba M, Ohnishi T, Ohno S. MAPK-upstream protein kinase (MUK) regulates the radial migration of immature neurons in telencephalon of mouse embryo. Development. 2002;129:4483–4495. doi: 10.1242/dev.129.19.4483. [DOI] [PubMed] [Google Scholar]
- Hirai S, Cui DF, Miyata T, Ogawa M, Kiyonari H, Suda Y, Aizawa S, Banba Y, Ohno S. The c-Jun N-terminal kinase activator dual leucine zipper kinase regulates axon growth and neuronal migration in the developing cerebral cortex. J Neurosci. 2006;26:11992–12002. doi: 10.1523/JNEUROSCI.2272-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitoshi S, Tropepe V, Ekker M, van der Kooy D. Neural stem cell lineages are regionally specified, but not committed, within distinct compartments of the developing brain. Development. 2002;129:233–244. doi: 10.1242/dev.129.1.233. [DOI] [PubMed] [Google Scholar]
- Jungers KA, Le Goff C, Somerville RP, Apte SS. Adamts9 is widely expressed during mouse embryo development. Gene Expr Patterns. 2005;5:609–617. doi: 10.1016/j.modgep.2005.03.004. [DOI] [PubMed] [Google Scholar]
- Kang JS, Mulieri PJ, Hu Y, Taliana L, Krauss RS. BOC, an Ig superfamily member, associates with CDO to positively regulate myogenic differentiation. EMBO J. 2002;21:114–124. doi: 10.1093/emboj/21.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JS, Feinleib JL, Knox S, Ketteringham MA, Krauss RS. Promyogenic members of the Ig and cadherin families associate to positively regulate differentiation. Proc Natl Acad Sci U S A. 2003;100:3989–3994. doi: 10.1073/pnas.0736565100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawauchi T, Chihama K, Nabeshima Y, Hoshino M. The in vivo roles of STEF/Tiam1, Rac1 and JNK in cortical neuronal migration. EMBO J. 2003;22:4190–4201. doi: 10.1093/emboj/cdg413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi A, Yamamoto H, Kishida S. Multiplicity of the interactions of Wnt proteins and their receptors. Cell Signal. 2007;19:659–671. doi: 10.1016/j.cellsig.2006.11.001. [DOI] [PubMed] [Google Scholar]
- Kim AS, Lowenstein DH, Pleasure SJ. Wnt receptors and Wnt inhibitors are expressed in gradients in the developing telencephalon. Mech Dev. 2001;103:167–172. doi: 10.1016/s0925-4773(01)00342-2. [DOI] [PubMed] [Google Scholar]
- LaMantia AS, Colbert MC, Linney E. Retinoic acid induction and regional differentiation prefigure olfactory pathway formation in the mammalian forebrain. Neuron. 1993;10:1035–1048. doi: 10.1016/0896-6273(93)90052-s. [DOI] [PubMed] [Google Scholar]
- LaMantia AS, Bhasin N, Rhodes K, Heemskerk J. Mesenchymal/epithelial induction mediates olfactory pathway formation. Neuron. 2000;28:411–425. doi: 10.1016/s0896-6273(00)00121-5. [DOI] [PubMed] [Google Scholar]
- Lazzaro D, Price M, de Felice M, Di Lauro R. The transcription factor TTF-1 is expressed at the onset of thyroid and lung morphogenesis and in restricted regions of the foetal brain. Development. 1991;113:1093–1104. doi: 10.1242/dev.113.4.1093. [DOI] [PubMed] [Google Scholar]
- Lee SM, Danielian PS, Fritzsch B, McMahon AP. Evidence that FGF8 signalling from the midbrain-hindbrain junction regulates growth and polarity in the developing midbrain. Development. 1997;124:959–969. doi: 10.1242/dev.124.5.959. [DOI] [PubMed] [Google Scholar]
- Lei Q, Zelman AK, Kuang E, Li S, Matise MP. Transduction of graded Hedgehog signaling by a combination of Gli2 and Gli3 activator functions in the developing spinal cord. Development. 2004;131:3593–3604. doi: 10.1242/dev.01230. [DOI] [PubMed] [Google Scholar]
- Liu A, Losos K, Joyner AL. FGF8 can activate Gbx2 and transform regions of the rostral mouse brain into a hindbrain fate. Development. 1999;126:4827–4838. doi: 10.1242/dev.126.21.4827. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Makarova KS, Aravind L, Koonin EV. SWIM, a novel Zn-chelating domain present in bacteria, archaea and eukaryotes. Trends Biochem Sci. 2002;27:384–386. doi: 10.1016/s0968-0004(02)02140-0. [DOI] [PubMed] [Google Scholar]
- Mohammadi M, McMahon G, Sun L, Tang C, Hirth P, Yeh BK, Hubbard SR, Schlessinger J. Structures of the tyrosine kinase domain of fibroblast growth factor receptor in complex with inhibitors. Science. 1997;276:955–960. doi: 10.1126/science.276.5314.955. [DOI] [PubMed] [Google Scholar]
- Mulieri PJ, Kang JS, Sassoon DA, Krauss RS. Expression of the boc gene during murine embryogenesis. Dev Dyn. 2002;223:379–388. doi: 10.1002/dvdy.10063. [DOI] [PubMed] [Google Scholar]
- Nery S, Fishell G, Corbin JG. The caudal ganglionic eminence is a source of distinct cortical and subcortical cell populations. Nat Neurosci. 2002;5:1279–1287. doi: 10.1038/nn971. [DOI] [PubMed] [Google Scholar]
- Ohkubo Y, Chiang C, Rubenstein JL. Coordinate regulation and synergistic actions of BMP4, SHH and FGF8 in the rostral prosencephalon regulate morphogenesis of the telencephalic and optic vesicles. Neuroscience. 2002;111:1–17. doi: 10.1016/s0306-4522(01)00616-9. [DOI] [PubMed] [Google Scholar]
- Okada A, Charron F, Morin S, Shin DS, Wong K, Fabre PJ, Tessier-Lavigne M, McConnell SK. Boc is a receptor for sonic hedgehog in the guidance of commissural axons. Nature. 2006;444:369–373. doi: 10.1038/nature05246. [DOI] [PubMed] [Google Scholar]
- Ostenfeld T, Joly E, Tai YT, Peters A, Caldwell M, Jauniaux E, Svendsen CN. Regional specification of rodent and human neurospheres. Brain Res Dev Brain Res. 2002;134:43–55. doi: 10.1016/s0165-3806(01)00291-7. [DOI] [PubMed] [Google Scholar]
- Pabst O, Herbrand H, Takuma N, Arnold HH. NKX2 gene expression in neuroectoderm but not in mesendodermally derived structures depends on sonic hedgehog in mouse embryos. Dev Genes Evol. 2000;210:47–50. doi: 10.1007/pl00008188. [DOI] [PubMed] [Google Scholar]
- Parmar M, Skogh C, Björklund A, Campbell K. Regional specification of neurosphere cultures derived from subregions of the embryonic telencephalon. Mol Cell Neurosci. 2002;21:645–656. doi: 10.1006/mcne.2002.1204. [DOI] [PubMed] [Google Scholar]
- Pleasure SJ, Anderson S, Hevner R, Bagri A, Marin O, Lowenstein DH, Rubenstein JL. Cell migration from the ganglionic eminences is required for the development of hippocampal GABAergic interneurons. Neuron. 2000;28:727–740. doi: 10.1016/s0896-6273(00)00149-5. [DOI] [PubMed] [Google Scholar]
- Price M, Lazzaro D, Pohl T, Mattei MG, Rüther U, Olivo JC, Duboule D, Di Lauro R. Regional expression of the homeobox gene Nkx-2.2 in the developing mammalian forebrain. Neuron. 1992;8:241–255. doi: 10.1016/0896-6273(92)90291-k. [DOI] [PubMed] [Google Scholar]
- Rallu M, Machold R, Gaiano N, Corbin JG, McMahon AP, Fishell G. Dorsoventral patterning is established in the telencephalon of mutants lacking both Gli3 and Hedgehog signaling. Development. 2002a;129:4963–4974. doi: 10.1242/dev.129.21.4963. [DOI] [PubMed] [Google Scholar]
- Rallu M, Corbin JG, Fishell G. Parsing the prosencephalon. Nat Rev Neurosci. 2002b;3:943–951. doi: 10.1038/nrn989. [DOI] [PubMed] [Google Scholar]
- Reynolds BA, Tetzlaff W, Weiss S. A multipotent EGF-responsive striatal embryonic progenitor cell produces neurons and astrocytes. J Neurosci. 1992;12:4565–4574. doi: 10.1523/JNEUROSCI.12-11-04565.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santa-Olalla J, Baizabal JM, Fregoso M, del Carmen Cárdenas M, Covarrubias L. The in vivo positional identity gene expression code is not preserved in neural stem cells grown in culture. Eur J Neurosci. 2003;18:1073–1084. doi: 10.1046/j.1460-9568.2003.02824.x. [DOI] [PubMed] [Google Scholar]
- Sasaki H, Nishizaki Y, Hui C, Nakafuku M, Kondoh H. Regulation of Gli2 and Gli3 activities by an amino-terminal repression domain: implication of Gli2 and Gli3 as primary mediators of Shh signaling. Development. 1999;126:3915–3924. doi: 10.1242/dev.126.17.3915. [DOI] [PubMed] [Google Scholar]
- Sato T, Araki I, Nakamura H. Inductive signal and tissue responsiveness defining the tectum and the cerebellum. Development. 2001;128:2461–2469. doi: 10.1242/dev.128.13.2461. [DOI] [PubMed] [Google Scholar]
- Schneider RA, Hu D, Rubenstein JL, Maden M, Helms JA. Local retinoid signaling coordinates forebrain and facial morphogenesis by maintaining FGF8 and SHH. Development. 2001;128:2755–2767. doi: 10.1242/dev.128.14.2755. [DOI] [PubMed] [Google Scholar]
- Schuurmans C, Guillemot F. Molecular mechanisms underlying cell fate specification in the developing telencephalon. Curr Opin Neurobiol. 2002;12:26–34. doi: 10.1016/s0959-4388(02)00286-6. [DOI] [PubMed] [Google Scholar]
- Shimamura K, Rubenstein JL. Inductive interactions direct early regionalization of the mouse forebrain. Development. 1997;124:2709–2718. doi: 10.1242/dev.124.14.2709. [DOI] [PubMed] [Google Scholar]
- Shimamura K, Hartigan DJ, Martinez S, Puelles L, Rubenstein JL. Longitudinal organization of the anterior neural plate and neural tube. Development. 1995;121:3923–3933. doi: 10.1242/dev.121.12.3923. [DOI] [PubMed] [Google Scholar]
- Stanton H, Rogerson FM, East CJ, Golub SB, Lawlor KE, Meeker CT, Little CB, Last K, Farmer PJ, Campbell IK, Fourie AM, Fosang AJ. ADAMTS5 is the major aggrecanase in mouse cartilage in vivo and in vitro. Nature. 2005;434:648–652. doi: 10.1038/nature03417. [DOI] [PubMed] [Google Scholar]
- Stenman J, Toresson H, Campbell K. Identification of two distinct progenitor populations in the lateral ganglionic eminence: implications for striatal and olfactory bulb neurogenesis. J Neurosci. 2003;23:167–174. doi: 10.1523/JNEUROSCI.23-01-00167.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storm EE, Rubenstein JL, Martin GR. Dosage of Fgf8 determines whether cell survival is positively or negatively regulated in the developing forebrain. Proc Natl Acad Sci U S A. 2003;100:1757–1762. doi: 10.1073/pnas.0337736100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storm EE, Garel S, Borello U, Hebert JM, Martinez S, McConnell SK, Martin GR, Rubenstein JL. Dose-dependent functions of Fgf8 in regulating telencephalic patterning centers. Development. 2006;133:1831–1844. doi: 10.1242/dev.02324. [DOI] [PubMed] [Google Scholar]
- Stoykova A, Fritsch R, Walther C, Gruss P. Forebrain patterning defects in small eye mutant mice. Development. 1996;122:3453–3465. doi: 10.1242/dev.122.11.3453. [DOI] [PubMed] [Google Scholar]
- Stoykova A, Treichel D, Hallonet M, Gruss P. Pax6 modulates the dorsoventral patterning of the mammalian telencephalon. J Neurosci. 2000;20:8042–8050. doi: 10.1523/JNEUROSCI.20-21-08042.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussel L, Marin O, Kimura S, Rubenstein JL. Loss of Nkx2.1 homeobox gene function results in a ventral to dorsal molecular respecification within the basal telencephalon: evidence for a transformation of the pallidum into the striatum. Development. 1999;126:3359–3370. doi: 10.1242/dev.126.15.3359. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Liu FC. Genetic patterning of the mammalian telencephalon by morphogenetic molecules and transcription factors. Birth Defects Res C Embryo Today. 2006;78:256–266. doi: 10.1002/bdrc.20077. [DOI] [PubMed] [Google Scholar]
- Tenzen T, Allen BL, Cole F, Kang JS, Krauss RS, McMahon AP. The cell surface membrane proteins Cdo and Boc are components and targets of the Hedgehog signaling pathway and feedback network in mice. Dev Cell. 2006;10:647–656. doi: 10.1016/j.devcel.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Theil T, Alvarez-Bolado G, Walter A, Rüther U. Gli3 is required for Emx gene expression during dorsal telencephalon development. Development. 1999;126:3561–3571. doi: 10.1242/dev.126.16.3561. [DOI] [PubMed] [Google Scholar]
- Tole S, Ragsdale CW, Grove EA. Dorsoventral patterning of the telencephalon is disrupted in the mouse mutant extra-toes(J) Dev Biol. 2000;217:254–265. doi: 10.1006/dbio.1999.9509. [DOI] [PubMed] [Google Scholar]
- Toresson H, Campbell K. A role for Gsh1 in the developing striatum and olfactory bulb of Gsh2 mutant mice. Development. 2001;128:4769–4780. doi: 10.1242/dev.128.23.4769. [DOI] [PubMed] [Google Scholar]
- Toresson H, Potter SS, Campbell K. Genetic control of dorsal-ventral identity in the telencephalon: opposing roles for Pax6 and Gsh2. Development. 2000;127:4361–4371. doi: 10.1242/dev.127.20.4361. [DOI] [PubMed] [Google Scholar]
- Tucker ES, Polleux F, LaMantia AS. Position and time specify the migration of a pioneering population of olfactory bulb interneurons. Dev Biol. 2006;297:387–401. doi: 10.1016/j.ydbio.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Veeman MT, Axelrod JD, Moon RT. A second canon. Functions and mechanisms of beta-catenin-independent Wnt signaling. Dev Cell. 2003;5:367–377. doi: 10.1016/s1534-5807(03)00266-1. [DOI] [PubMed] [Google Scholar]
- Wang B, Fallon JF, Beachy PA. Hedgehog-regulated processing of Gli3 produces an anterior/posterior repressor gradient in the developing vertebrate limb. Cell. 2000;100:423–434. doi: 10.1016/s0092-8674(00)80678-9. [DOI] [PubMed] [Google Scholar]
- Wichterle H, Turnbull DH, Nery S, Fishell G, Alvarez-Buylla A. In utero fate mapping reveals distinct migratory pathways and fates of neurons born in the mammalian basal forebrain. Development. 2001;128:3759–3771. doi: 10.1242/dev.128.19.3759. [DOI] [PubMed] [Google Scholar]
- Wilson SW, Rubenstein JL. Induction and dorsoventral patterning of the telencephalon. Neuron. 2000;28:641–651. doi: 10.1016/s0896-6273(00)00171-9. [DOI] [PubMed] [Google Scholar]
- Wonders CP, Anderson SA. The origin and specification of cortical interneurons. Nat Rev Neurosci. 2006;7:687–696. doi: 10.1038/nrn1954. [DOI] [PubMed] [Google Scholar]
- Xu Q, Cobos I, De La Cruz E, Rubenstein JL, Anderson SA. Origins of cortical interneuron subtypes. J Neurosci. 2004;24:2612–2622. doi: 10.1523/JNEUROSCI.5667-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Wonders CP, Anderson SA. Sonic hedgehog maintains the identity of cortical interneuron progenitors in the ventral telencephalon. Development. 2005;132:4987–4998. doi: 10.1242/dev.02090. [DOI] [PubMed] [Google Scholar]
- Yun K, Potter S, Rubenstein JL. Gsh2 and Pax6 play complementary roles in dorsoventral patterning of the mammalian telencephalon. Development. 2001;128:193–205. doi: 10.1242/dev.128.2.193. [DOI] [PubMed] [Google Scholar]