Abstract
Cells dying by apoptosis are normally cleared by phagocytes through mechanisms that can suppress inflammation and immunity. Molecules of the innate immune system, the pattern recognition receptors (PRRs), are able to interact not only with conserved structures on microbes (pathogen-associated molecular patterns, PAMPs) but also with ligands displayed by apoptotic cells. We reasoned that PRRs might therefore interact with structures on apoptotic cells – apoptotic cell-associated molecular patterns (ACAMPs) – that are analogous to PAMPs. Here we show that certain monoclonal antibodies raised against the prototypic PAMP, lipopolysaccharide (LPS), can crossreact with apoptotic cells. We demonstrate that one such antibody interacts with a constitutively expressed intracellular protein, laminin-binding protein, which translocates to the cell surface during apoptosis and can interact with cells expressing the prototypic PRR, mCD14 as well as with CD14-negative cells. Anti-LPS cross reactive epitopes on apoptotic cells colocalised with annexin V- and C1q-binding sites on vesicular regions of apoptotic cell surfaces and were released associated with apoptotic cell-derived microvesicles (MVs). These results confirm that apoptotic cells and microbes can interact with the immune system through common elements and suggest that anti-PAMP antibodies could be used strategically to characterise novel ACAMPs associated not only with apoptotic cells but also with derived MVs.
Keywords: apoptosis, innate immunity, pattern recognition, LPS, antibody
Apoptotic cells undergo multiple surface changes, the most renowned being the externalisation of the anionic phospholipid, phosphatidylserine (PS).1 Profound alterations in plasma-membrane topology also occur as a consequence of the apoptosis programme, involving protein, carbohydrate and nucleic acid moieties in addition to lipids (reviewed in Gregory and Pound2). Gross changes in apoptotic cells include the production of plasma-membrane-bound apoptotic bodies, blebs and microvesicles (MVs),3, 4 the latter serving functions in intercellular communication, including chemoattraction and activation of phagocytes.5, 6, 7, 8 Taken together, these changes permit a multitude of molecular interactions with phagocytes triggering apoptotic cell engulfment and additional phagocyte responses including immunomodulation. Detailed activities of underlying receptor–ligand interactions participating in these responses remain to be defined but it is clear that interference with these processes can have pathological consequences (see, Savill et al.,9 Lauber et al.,10 Ravichandran and Lorenz,11 Erwig and Henson,12 Elliott and Ravichandran13 and Gregory and Pound,14 for reviews).
Many of the host proteins, including CD14, C1q and collectins, that interact with apoptotic cells are molecules of the innate immune system that were first known for their involvement in host responses to microbes.15 The reverse relationship is also proven: brain angiogenesis inhibitor 1, characterised first in this context as a phagocyte receptor for apoptotic cells, was recently also shown to function as an immune receptor for bacterial binding and engulfment.16 All these molecules fall into the category of the so-called ‘pattern recognition receptors' (PRRs) that interact with evolutionarily conserved structures – ‘pathogen-associated molecular patterns' (PAMPs) – of microbes.17 For example, the prototypic PRR, CD14, interacts with, and regulates proinflammatory host responses to, the prototypic PAMP, lipopolysaccharide (LPS).18 Because of the involvement of PRRs in the molecular interactions of phagocytes with apoptotic cells, we and others previously suggested that apoptotic cells express host equivalents of PAMPs termed ‘apoptotic cell-associated molecular patterns' (ACAMPs).19, 20 This notion predicts that ACAMPs share spatial similarities with PAMPs that permit common PRR interactions. This led us to hypothesise that some antibodies raised against PAMPs could potentially crossreact with ACAMPs, thus providing an opportunity to use antibody approaches to probe the existence and nature of ACAMPs.
Here we test this hypothesis and demonstrate that certain anti-LPS antibodies indeed show strong crossreactive binding to apoptotic cells. We characterise a host epitope specified by one of these antibodies, demonstrating (1) its association with the intracellular protein, laminin-binding protein (LBP/p40), (2) its exposure on apoptotic cells and (3) its ability to bind to cells via both plasma-membrane CD14 (mCD14)-dependent and -independent mechanisms. These results provide further evidence supporting the notion that apoptotic cells and microbes share certain elements of immune recognition and suggest a strategy whereby anti-PAMP antibodies can be used to probe the molecular architecture of apoptotic cells and of derived MVs.
Results
Certain monoclonal anti-LPS antibodies crossreact with epitopes of apoptotic cells
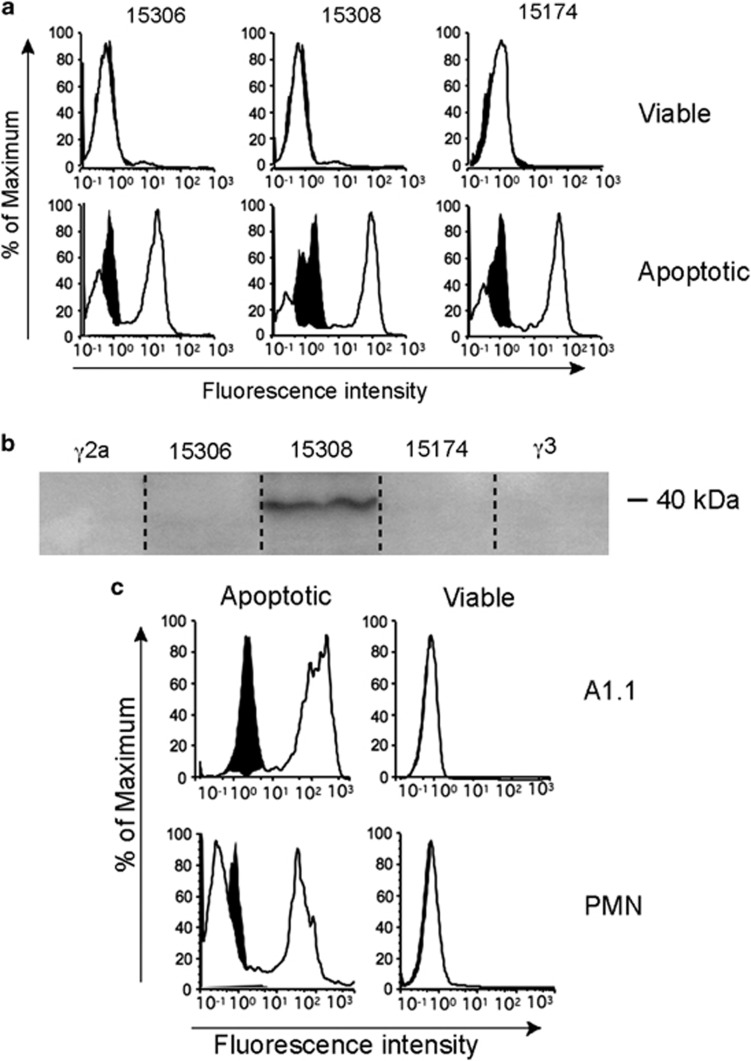
To test the hypothesis that certain anti-microbial antibodies can crossreact with ACAMPs, endogenous molecules exposed by cells undergoing apoptosis, a panel of monoclonal antibodies (mAbs) raised against LPS, was tested for reactivity towards apoptotic, compared with viable, cells. Initial screening was carried out by flow cytometry using a human lymphoma cell line. Antibodies were selected according to their previously characterised ability to react broadly across LPS species (Table 1). Three (out of 13 anti-LPS mAbs tested) showed clear binding to apoptotic, but not viable, cells (Figure 1a). Immunoblotting of apoptotic lymphoma cell lysates indicated that one of the three apoptotic cell-reactive anti-LPS mAbs, 15308, detected a protein-associated epitope, which, under reducing conditions, ran at around 40 kDa (Figure 1b); the remaining two apoptotic cell-reactive mAbs failed to detect the same species (Figure 1b) or any other protein epitopes under these conditions, indicating that additional, non-protein ACAMPs may be identified using anti-LPS mAbs.
Table 1. Reported reactivities of commercially available crossreactive anti-LPS mAbs used in these studies.
| mAb | LPS specificity | Species | Isotype |
|---|---|---|---|
| 15306 | Shigella sonnei Salmonella typhimurium Serratia marcescens Proteus mirabilis Proteus vulgaris Acinetobacter calcoaceticus | Mouse | IgG2a |
| 15308 | Enterobacter aerogenes Serratia marcescens Proteus mirabilis Acinetobacter calcoaceticus Pseudomonas aeruginosa | Mouse | IgG3 |
| 15174 | Chlamydia trachomatis | Mouse | IgG2a |
Antibodies were assessed by ELISA for binding to purified LPS. LPS of Chlamydia trachomatis lacks the O-polysaccharide region most distal to lipid A. Thus, although being marketed as anti-Chlamydia antibody, mAb 15174 was chosen for its potential reactivity towards conserved core regions of LPS.
Figure 1.
Anti-LPS antibodies crossreact with multiple epitopes of apoptotic cells. (a) Flow cytometric profiles of MUTU I BL cells triggered into apoptosis by ionomycin and stained using the indicated anti-LPS mAbs (white histograms), which were detected by fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse immunoglobulin G (IgG); isotype control binding (γ2a for mAbs 15174 and 15306, γ3 for mAb 15308) is indicated by the black histograms. (b) Immunoblotted SDS-PAGE of MUTU I lysates probed with the indicated anti-LPS mAbs or isotype controls demonstrating 40 kDa species specified by mAb 15308 but not 15174 or 15306. (c) Flow cytometric profiles of A1.1 hybridoma cells (upper panels) and human neutrophils (lower panels) after induction of apoptosis by ionomycin or serum deprivation, respectively. Cells were stained with mAb 15308 detected as above; isotype control binding shown in black. Apoptotic and viable cells were discriminated by light scatter as described6
We sought to characterise further the cellular reactivity of mAb 15308 and its cellular 40 kDa protein target. We first determined whether the cellular target(s) of mAb 15308 are conserved structures, as expected for ACAMPs,19, 20 by testing cells of different lineages and species. Figure 1c shows flow cytometric analysis of mAb 15308 reactivity towards primary human neutrophils and mouse thymoma cells. Our further studies showed wide reactivity across numerous cell lineages and species following induction of apoptosis (Supplementary Table 1) with reactivity having been found against all apoptotic cell types we have tested to date. By immunoblotting we have not demonstrated any qualitative changes in the antigen during apoptosis.
Specific binding of anti-LPS mAb 15308 to intracellular cytoskeletal sites within viable cells and to surface buds of apoptotic cells
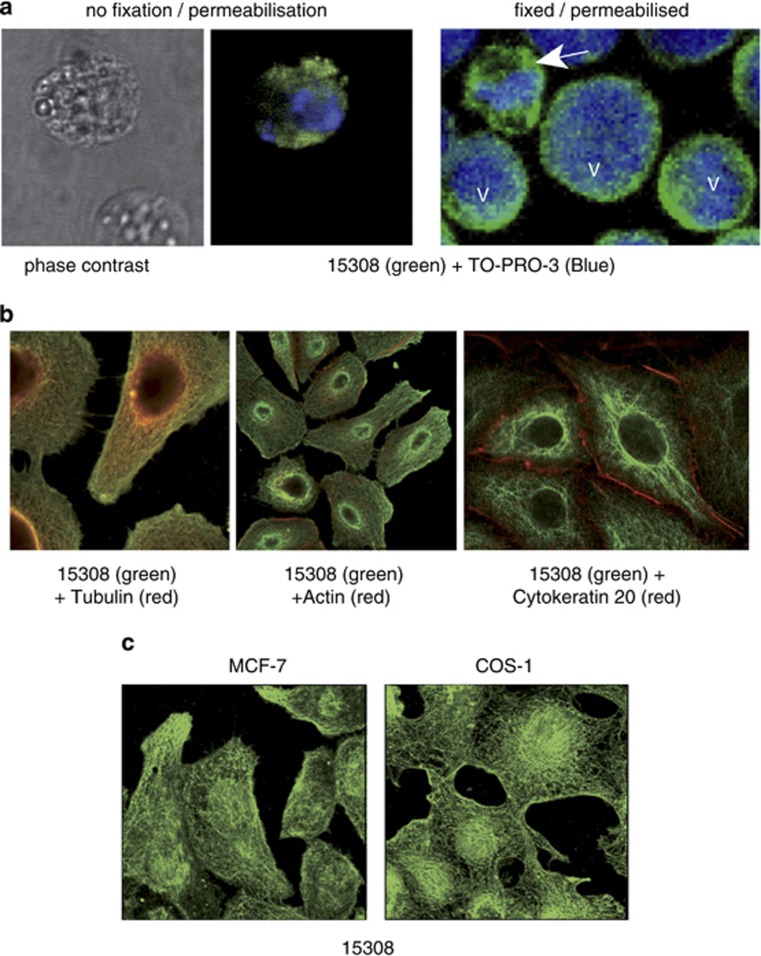
To determine whether the cellular targets of mAb 15308 were neoepitopes of apoptotic cells or, alternatively, intracellular epitopes exposed during apoptosis, we analysed the binding of mAb 15308 to cells that had been fixed and permeabilised in the absence of apoptosis induction. Permeabilised lymphoma cells displayed strong cytoplasmic mAb 15308 staining, comparable to that shown by plasma membrane-compromised apoptotic cells (Figure 2a and Supplementary Figure 1). To investigate the pattern of cytoplasmic staining further, a range of adherent cell lines were analysed in situ. Following fixation and permeabilisation of growing monolayers, mAb 15308 was found mainly to bind to intracytoplasmic filamentous structures (Figure 2b and c), colocalising with tubulin but not with other cytoskeletal elements such as actin or cytokeratin-20 (Figure 2b). All cell lines tested showed similar filamentous patterns of staining.
Figure 2.
Anti-LPS antibody 15308 binding to intracellular epitopes of viable and apoptotic cells. (a) BL cells (MUTU I) were treated with ionomycin for 16 h to induce apoptosis. Binding of the anti-LPS antibody, 15308, was visualised using goat anti-mouse secondary antibody labelled with Alexa Fluor-488 (green). Cells were counterstained with the cell-impermeant dye, TO-PRO-3 (blue), for visualisation of nucleic acid and as a measure of membrane integrity. Unfixed, non-permeable viable cells were not stained at all (lower cell, left and centre panels), whereas apoptotic cells, which rapidly became permeable, displayed green cytoplasmic staining (upper cell, left and centre panels); the isotype-matched control antibody (γ3) exhibited no staining (not shown). When fixed and permeabilised, cells also exhibited cytoplasmic staining with mAb 15308 (green) whether viable or apoptotic (right panel). Viable cells displayed a large rounded nucleus (V, right panel), whereas the nuclei of apoptotic cells (arrow) were condensed and fragmented. (b) mAb 15308 associates with microtubules in adherent cell lines. Fixed and permeabilised A549 cells were double-stained with mAb 15308 (visualised green) and cytoskeletal markers (visualised red): anti-tubulin antibody (left), actin-binding phalloidin (centre) and cytokeratin 20 (right). (c) Filamentous patterns of 15308 staining in MCF-7 (left) and COS-1 (right) cells
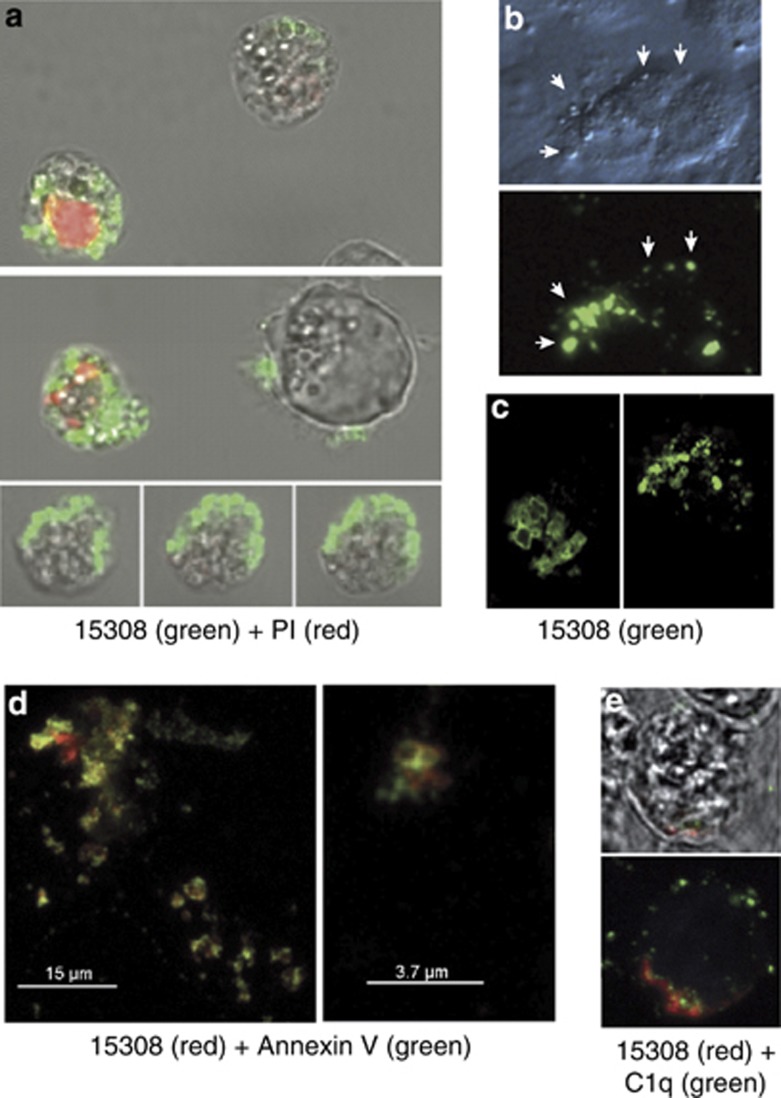
As surface architectural changes during apoptosis commonly result from translocation of molecules from intra- to extracellular loci, we next determined whether cellular epitopes of mAb 15308 were demonstrable on apoptotic cells with intact plasma membranes. Flow cytometric analyses initially suggested that this was not the case as only plasma membrane-compromised, propidium iodide (PI)-positive apoptosis-induced cells were reactive with mAb 15308. As apoptotic populations contain fragile cells, which become permeable during preparation for flow cytometry and/or during the flow procedure (our unpublished observations), it remained possible that mAb 15308 epitopes were displayed on the surface of fragile apoptotic cells. To investigate this, lymphoma cells and adherent cells (MCF-7) induced to undergo apoptosis were analysed by confocal microscopy following immunofluorescence and vital dye staining under gentle, preparatory conditions (as opposed to the high-shear steps required for flow cytometry). Strong mAb 15308 labelling could be observed on apoptosis-induced lymphoma cells whose membranes excluded PI (Figure 3a). In some cases, 15308 reactivity was seen to be associated with cells displaying only very weak PI fluorescence, indicative of low permeability and supporting the notion that mAb 15308 epitopes appeared on fragile apoptotic cells. Consistent with this, using flow cytometry we could detect low levels of mAb 15308 binding to 10–15% of apoptotic BL cells that were impermeable to PI (Supplementary Figure 2).
Figure 3.
Appearance of mAb 15308 reactivity on cell surface buds or blebs during apoptosis. (a) Confocal microscopy of exposure of mAb 15308 epitopes at the surface of MUTU I BL cells induced to undergo apoptosis with ionomycin. Binding of mAb 15308 was visualised using secondary antibody labelled with Alexa Fluor-488 (green); plasma membrane integrity was monitored using PI (red). Note that mAb 15308 binds to cells excluding PI as well as to those displaying weak or strong PI staining. (b) Microscopic analysis of mAb 15308 reactivity associated with surface buds (arrowheads) of PI-negative MCF-7 cells triggered to undergo apoptosis with etoposide. Top panel shows surface buds under transmitted light differential interference contrast. (c) Closer inspection by confocal microscopy of surface reactivity of two separate etoposide-treated MCF-7 cells. (d) Confocal fluorescence images of surface 15308 reactivity (visualised using secondary antibody labelled with Alexa Fluor-568, red) colocalised with AxV binding sites (most likely PS) visualised with streptavidin-labelled Alexa Fluor-488, green) on etoposide-treated MCF-7 cells. (e) Confocal microscopic analysis of mAb 15308 reactivity (red) and C1q binding (biotinylated C1q visualised with streptavidin–Alexa Fluor-488, green) at the surface of MUTU I BL cells induced to undergo apoptosis with ionomycin. No staining was observed when biotinylated IgG1 was used instead of C1q or when isotype control replaced mAb 15308 (not shown). Membrane integrity was monitored by exclusion of TO-PRO-3, which fluoresced blue in permeable cells (not shown). The upper panel is a bright field image overlaid with the fluorescence associated with a mid-way Z-section, and the lower panel shows total cell-associated fluorescence
Given its strong intracellular reactivity as well as its association with the cell surface during apoptosis, we wished to determine whether mAb 15308 showed preference for binding to immature glycoproteins via a carbohydrate ‘signature' present at the surface of apoptotic cells.8 As such immature glycoproteins are high in mannose residues, we sought inhibition of binding of mAb 15308 by free mannose and by the mannan-binding lectin, MBL. Free mannose failed to affect mAb 15308 binding and MBL caused only marginal inhibition (Supplementary Figure 3). Significantly, MBL did not bind to surfaces of apoptotic cells tested (Supplementary Figure 2). These results indicate that mAb 15308 is unlikely to bind to carbohydrate signatures on immature glycoproteins.
Further analyses revealed that mAb 15308 reactivity was predominantly associated with vesicular structures: small ‘buds' or ‘blebs' typical of the surface of apoptotic cells (Figure 3b and c). Similar observations were made with apoptotic lymphoma cells that grow in suspension culture (Supplementary Figure 4). To analyse this further, we investigated whether two additional known apoptotic cell surface-binding proteins, annexin V (AxV) and C1q associate with the same bleb-like surface structures as mAb 15308. Confocal microscopic analysis revealed co-distribution of mAb 15308 and AxV binding to isolated parts of cell surfaces and vesicular structures on apoptotic cells with intact plasma membranes (Figure 3d). Although both markers were generally colocalised to the same regions of the cell surface, coincident fluorescence was only occasionally observed, indicating that mAb 15308 and AxV target distinct molecular structures present in closely apposed regions of the plasma membrane. Similarly, regions that bound mAb 15308 colocalised with C1q binding (Figure 3e).
These results show that the anti-LPS mAb 15308 binds to intracellular, cytoskeleton-associated ligands in eukaryotic cells and strongly suggest that translocation of these binding sites to vesicular regions of the cell surface occurs during apoptosis. We next determined whether mAb 15308-reactive MVs were released from apoptotic cells.
MVs displaying protein ligands for mAb 15308 are released from cells undergoing apoptosis
Before examining the MVs released from apoptotic cells for mAb 15308 reactivity, we sought further characterisation of the cellular protein species identified by mAb 15308. Figure 4a illustrates the relative reactivity of mAb 15308 associated with cellular fractions isolated from human lymphoma cells by differential centrifugation. Immunoblotting with mAb 15308 revealed major differences among the four fractions, with the nuclear fraction negative and most reactivity being detected as a ∼40 kDa species found within the fraction (P1) containing large organelles and protein complexes. Lower levels of the 40 kDa band were observed in the insoluble membrane fraction (P2) and a faint higher-molecular-weight (∼65 kDa) species was seen in the soluble fraction.
Figure 4.
mAb 15308 reveals a 40 kDa protein associated with apoptotic cell-derived microvesicles. (a) SDS-PAGE and immunoblots of lysate from MUTU I BL cells fractionated by sequential centrifugation into 1000 × g (N), 27 000 × g (P1), 100 000 × g (P2) pellets and remaining supernatant (S). (b) Flow cytometric analysis of mAb 15308 reactivity with MV produced by MUTU I BL cells with (left panel) and without (right) induction of apoptosis by UV irradiation for 16 h. Black histogram indicates isotype control binding. (c) Immunoblotting of mAb 15308 reactivity with MVs produced by MUTU I BL cells after induction of apoptosis by UV irradiation for 16 h. (d) Quantification by protein assay of MVs liberated by 10 × 106 BL2 cells and Bcl-2-overexpressing BL2 cells (BL2-Bcl-2) following induction of apoptosis by staurosporine. One-tailed unpaired Mann–Whitney test; *P≤0.05. By 4 h, >70% of staurosporine-treated BL2 cells bound AxV, compared with 5–25% BL2-Bcl-2 cells. (e) Flow cytometric analyses of mAb 15308-stained MVs isolated from BL2 and BL2-Bcl-2 cells following apoptosis induction by staurosporine for 3 h. Histograms of representative samples of MVs isolated from 3 h staurosporine-treated BL2 (left panel) and BL2-Bcl-2 (middle) cells are shown. Percentages of 15308-positive MVs are indicated on the histograms and mean percentages±S.E.M (n=3 independent experiments for each cell type) are shown (right). One-tailed unpaired Mann–Whitney test; *P≤0.05
Flow cytometric analysis of MVs isolated by ultracentrifugation of supernatants from lymphoma cells induced to undergo apoptosis by UV irradiation indicated strong staining with mAb 15308 (Figure 4b) and immunoblotting confirmed the 40 kDa species associated with these MVs (Figure 4c). MVs from untreated lymphoma cells also stained positively for mAb 15308 but at lower levels than those undergoing apoptosis (Figure 4b), possibly reflecting the low level of spontaneous apoptosis in the untreated cells. To investigate further the ‘15308 phenotype' of MVs produced by apoptotic cells, we induced a lymphoma line, BL2, into apoptosis using staurosporine and analysed the resultant MVs, comparing them with MVs from BL2-Bcl-2 cells, which were largely protected from staurosporine-induced apoptosis. We observed an increase in MV production over time by apoptotic BL2 cells but not by the apoptosis-protected BL2-Bcl-2 cells (Figure 4d). It is worth noting that the low-level production of MVs by BL2-Bcl-2 cells may relate to the low-level apoptosis induced in these cells (Figure 4) as well as representing MVs from viable cells. As shown in Figure 4e, in addition to lower levels of MVs being released from Bcl-2-protected BL2 cells, their capacity to bind mAb 15308 was markedly lower (∼40% were 15308 positive) than the MVs released from overtly apoptotic BL2 cells (∼70% 15308 positive). These results indicate that mAb 15308 binding sites are preferentially released in association with MVs produced by apoptotic cells.
Cellular targets of mAb 15308 include LBP/p40
MALDI-TOF-mass spectrometry was used to characterise further the main (∼40 kDa) protein species observed in immunoblots reactive with mAb 15308. Three polypeptide species were initially identified: β-actin, LBP/p40 and the septin, NEDD5 (neuronally expressed, developmentally downregulated-5; Supplementary Figure 5). β-Actin was not considered as a candidate mAb 15308-reactive protein as (a) distinct patterns of intracellular localisation were seen when phalloidin staining was compared directly with immunocytochemical labelling by mAb 15308 (Figure 2b), and (b) immunoblotting of fractionated cell lysates had shown actin to be present in different relative proportions to the mAb 15308-binding species (Figure 4a).
We next undertook molecular cloning of LBP/p40 and NEDD5 using mRNA from lymphoma cells. Lysates of HEK293T cells expressing exogenous V5-tagged LBP/p40 were immunoblotted both with a V5-specific mAb and with mAb 15308. Exogenously expressed LBP/p40 was observed as two species of ∼40 kDa and of 50–70 kDa as detected using the anti-V5 mAb antibody (Figure 5a, left). This is consistent with both the faint band seen in fraction S of lymphoma cell lysates (Figure 4a) and the previously reported 67 kDa form of this protein.21 Lysates of cells transfected with V5-tagged exogenous NEDD5 displayed the NEDD5 polypeptide (∼42 kDa) in immunoblots using anti-V5 but mAb 15308 failed to bind NEDD5 (Figure 5b). Furthermore, mAb 15308 failed to detect any bands in addition to the endogenous 40 kDa (LBP/p40) species found in lysates of mock-transfected cells.
Figure 5.
Detection of LBP/p40 by mAb 15308 and expression of exogenous LBP/p40 on the surface of apoptotic cells. (a) Exogenous expression of the LBP/p40 cDNA (pcDNA3.1D/LBP/p40-V5-His) in lysates from transiently transfected HEK293T cells (lane: p40) and detection of 40 and 50–70 kDa species by anti-V5 antibody and by mAb 15308 but not by isotype control antibody (γ3) or anti-actin antibody. Lysates from mock-transfected cells are shown for comparison (lane: M). (b) Exogenous expression of the NEDD5 cDNA (pcDNA3.1D/NEDD5-V5-His) in lysates from transiently transfected HEK293T cells (lane: N5) and detection by anti-V5 antibody but not by mAb 15308. NEDD5 was not detected in mock-transfected cells (lane: M) and mAb 15308 detected endogenous 40 kDa bands in lysates from both NEDD5- and mock-transfected cells. Note that the differences in signal intensity between anti-V5 and mAb 15308 can be explained by the exquisitely high affinity of anti-V5 for its epitope. (c) Immunofluorescence microscopic analysis of MCF-7 cells transiently transfected with pcDNA3.1D/LBP/p40-V5-His and double-stained with anti-V5 followed by Alexa Fluor-488-tagged secondary antibody (green, left panel), and mAb 15308 followed by Alexa Fluor 568-tagged secondary antibody (red, centre panel). Right panel shows overlay of red/green fluorescence and arrows indicate an example of an area where coincident fluorescence was observed. (d) Surface exposure of exogenous V5-tagged LBP/p40 stably expressed in MCF-7 cells. Confocal microscopy of MCF-7/V5-LBP/p40 transfectants after 48 h etoposide treatment. Exogenous LBP/p40 was detected using anti-V5 and secondary antibody tagged with Alexa Fluor-488 (green). Plasma membrane integrity was monitored by PI (red) exclusion. Z-sections of rare cells revealed the presence of the V5 epitope on surface blebs with plasma membranes intact (arrows, upper panels) as opposed to membrane compromised cells (arrows, lower panels), which stained green throughout the cell and were positive for PI
To determine whether the intracellular distribution of the exogenous LBP/p40 reflected that of the reactivity of mAb 15308, immunofluorescence double-labelling of MCF-7 cells by anti-V5 and 15308 mAbs was performed. Anti-V5 and mAb 15308 labelling were generally independent, with only occasional coincident fluorescence, especially in the perinuclear region (Figure 5c). Similar results were seen using a GFP-LBP/p40 construct in place of the V5 expression system (not shown). It seems likely that mAb 15308 binds multiple intracellular epitopes and that only a small subset is represented by LBP/p40. Notably, exogenous LBP/p40 expression failed to generate detectable increases in mAb 15308 binding.
Given our earlier observations that mAb 15308 reactivity could be detected on the surface of apoptotic cells, we next sought surface expression of exogenous LBP/p40 during apoptosis. Following induction of exogenous V5-tagged LBP/p40-expressing MCF-7 cells into apoptosis, it was possible to detect by confocal microscopy small amounts of the V5 epitope at the surface of cells with intact plasma membranes (Figure 5d). Perhaps unsurprisingly given the observations above (Figure 5c), such display of exogenously exposed LBP/p40 was a rare event, observed on <2% of the cells.
These results indicate that LBP/p40 is a component, albeit minor, of the cellular crossreactivity of mAb 15308 and demonstrate that this protein is translocated from its intracellular location in viable cells to be exposed on the surface of cells undergoing apoptosis.
Interaction of recombinant LBP/p40 with cells via mCD14-dependent and -independent mechanisms
To investigate further the possibility that LBP/p40 constitutes an ACAMP, we next determined its capacity for PRR interaction, nominating mCD14, the prototypic PRR as a candidate. For these studies, recombinant LBP/p40 was produced in Escherichia coli, in human K562 and MCF-7 cells and in insect sf9 cells. Immunoblots of transformed E. coli lysates and of derived nickel affinity-purified preparations probed with mAb 15308 revealed three protein species that were absent from non-transformed lysates, the main bands being ∼40 and ∼65 kDa (Figure 6a and b). The latter species were readily detected with the anti-V5 mAb and also by an antibody against the 67 kDa laminin receptor (Lam-R; Figure 6b). These results indicate that eukaryotic processing is unnecessary for binding of LBP/p40 to mAb 15308. Recombinant LBP/p40 purified from MCF-7 and K562 transfectants displayed similar major species of ∼40 and 50–70 kDa (Figure 6b), although K562 material was only visible in blots using the sensitive anti-V5 antibody, reflecting the relatively low level of recombinant protein produced by these cells (Supplementary Figure 6). Using the insect system, high levels of expression were obtained and mAb 15308 reactivity was almost entirely associated in immunoblots of cell lysates or nickel-purified protein with the 50–70 kDa species (Figure 6c), although a ∼40 kDa band could also be observed upon overexposure (not shown).
Figure 6.
Production of recombinant LBP/p40 in different expression systems and reactivity with mCD14. (a) Presence of recombinant protein in LBP/p40 (p40)-transformed or non-transformed (NT) E. coli lysates was detected by probing immunoblots of total protein with mAb 15308. (b) LBP/p40 was expressed as an intracellular protein in E. coli and MCF-7 cells or as a secreted protein in K562 cells. Immunoblots of nickel affinity-purified LBP/p40 preparations were probed with mAb 15308 (left panel), Lam-R (a commercially available anti-LBP/p40 antibody; middle panel) or anti-V5 (right panel). (c) Expression of recombinant LBP/p40 in baculovirus-infected sf9 insect cells detected by probing immunoblots of total protein from non-infected (NI) or recombinant-LBP/p40 (p40) baculovirus-infected cells with mAb 15308. Nickel metal affinity-purified LBP/p40 (p40/F) is shown for comparison. In all immunoblots, equivalent amounts of total protein were included in each lane. (d) Flow cytometric analysis of binding of purified preparations of recombinant LBP/p40 produced in E. coli to wild-type K562 cells (CD14 negative) or to K562 cells overexpressing CD14 (CD14 positive). Bound recombinant protein was detected with anti-V5 antibody followed by anti-mouse secondary antibody labelled with FITC (GAM-FITC). (e) Failure of contaminating LPS to mediate binding of recombinant LBP/p40 to CD14. Purified preparations of LPS-free, eukaryotic LBP/p40, prepared from MCF-7 or K562 expression systems were incubated with CD14 negative or CD14 positive K562 cells and binding visualised as in (d). Binding of recombinant protein preparations were assessed in the presence or absence of LPS in concentration equivalent to that found contaminating the bacterial preparation used in (d). (f) LBP/p40 derived from a baculovirus insect expression system fails to bind preferentially to mCD14-expressing cells. Purified preparations of insect-derived LBP/p40 were tested for binding to mCD14 negative and positive cells as described for (d) and (e). Numbers at the top right of each histogram represent the total protein concentration of recombinant LBP/p40 incubated with cells (μg/ml). Surface expression of CD14 in K562 (d–f) cells was monitored by staining with mAb 63D3 (anti-CD14)
Recombinant LBP/p40 proteins from bacterial, human and insect cells were tested in flow cytometric assays for their ability to bind to K562 cells, which are constitutively CD14 negative, and to K562 transfectants stably expressing mCD14 (Figures 6d–f). Recombinant protein binding to cells was visualised by indirect immunofluorescence using anti-V5 mAb as primary. Preferential binding of bacterial LBP/p40 preparations to mCD14-expressing cells was observed (Figure 6d). Given the role of CD14 as an LPS receptor, it is conceivable that LPS contamination of the bacterial LBP/p40 preparations aided mCD14 binding. However, similar concentrations of LPS found in preparations of LBP/p40 derived from bacteria failed to enhance binding of eukaryotic, LPS-free preparations either secreted by K562 transfectants or purified from lysates of MCF-7 transfectants (Figure 6e). While it is possible that the levels of purified LBP/p40 in these preparations were too low for detection in these preparations, this seems unlikely as binding activity (albeit low) was consistently observed, especially with the K562-derived protein (Figure 6e, bottom right). When higher concentrations of eukaryotic protein were tested, as was possible using the insect expression system (Figure 6f), significant levels of binding to both mCD14-negative and -positive cells were observed. Differences in binding patterns between recombinant LBP/p40 produced in prokaryotic versus different eukaryotic cells may be due to the differential processing of the protein in these different contexts.
These results indicate that, while recombinant LBP/p40 (at least when produced in bacteria) may show propensity to bind mCD14, this is not an absolute requirement and CD14-independent binding pathways are also evident.
Discussion
To clear apoptotic cells efficiently, the innate immune system deploys multiple receptors, including PRRs that are renowned for their roles in host responses to microbes. Notwithstanding the well-established, opposed inflammatory responses of the host to apoptotic cells versus microbial invaders, the use of common receptors to engage with apoptotic cell- and pathogen-borne ligands has suggested that molecular patterns sharing three-dimensional structure are displayed by both microbes and apoptotic cells, leading to the proposed parallels between PAMPs and ACAMPs. Here we investigated a natural extension of this concept of innate immunity, rationalising that common structural features of PAMPs and ACAMPs should be definable on the basis of crossreactivity of derivatives of adaptive immune receptors, antibodies. We predicted that certain antibodies raised against PAMPs would crossreact with ACAMPs. The results of these studies show that, in a small sample of anti-LPS mAbs, just under a quarter crossreacted with cells undergoing apoptosis. The findings of the initial screening suggested that such crossreactivities encompassed both protein and non-protein molecular species, including a cytoskeleton-associated protein, LBP/p40, which carries epitopes recognised by the anti-LPS mAb, 15308.
An underlying principle of apoptosis – the translocation of intracellular structures to the dying-cell surface – has been demonstrated for a variety of molecules, most famously PS, which flops from the inner to the outer plasma membrane leaflet and engages with phagocytes via multiple PS receptors.1, 22, 23, 24, 25, 26 Nucleic acid27 and a diverse range of proteins, including calreticulin,28 annexin 1,29, 30 the large subunit of the eukaryotic translation initiation factor 331 and the long pentraxin PTX3,32 have also been shown to move to the surface of apoptotic cells from intracellular locations via as yet unknown mechanisms to facilitate phagocytic clearance. Recently in Drosophila, two proteins of the endoplasmic reticulum, Pretaporter and DmCaBP1, have been observed to become externalised in apoptosis aiding phagocytosis mediated by Draper, a fly hemocyte receptor for apoptotic cells that is homologous to CED-1.33, 34 We established that LBP/p40 translocates from its intracellular location to the plasma membrane surface during apoptosis. In light of our observations (e.g. Figure 5c) mAb 15308-reactive epitopes are most probably not limited to LBP/p40. Regardless of their molecular species, however, they associate with exposed PS and C1q binding sites, appear on the blebs of apoptotic cells and constitute components of released MVs. As changes in plasma membrane topology associated with apoptosis are likely to form the molecular basis for establishing the apoptotic cell side of the phagocytic synapse,9, 35, 36 we speculate that ACAMPs, as defined according to the principle demonstrated here, are present within an assortment of molecular structures that, together with additional changes such as PS exposure, endow the apoptotic cell with discrete regions through which to interact effectively with phagocyte receptors. Concentrating here on mCD14, a well-defined PRR that interacts both with LPS and with apoptotic cells, our results were unable to show, at least for eukaryotic cell-derived LBP/p40, preferential binding to this receptor. A further PRR, MBL, also failed to interact with surface ACAMPs (Supplementary Figure 2). It will be important to determine not only the PRR preferences of ‘LPS-like' ACAMPs but also their functional attributes in engagement with phagocytes. In relation to the latter, we undertook preliminary investigations of possible involvement of LPS-like ACAMPs in interactions between apoptotic cells and macrophages under conditions in which a role for CD14 was demonstrable using an antagonistic CD14 mAb. However, none of the three crossreactive anti-LPS mAbs tested here was observed to antagonise macrophage/apoptotic cell interactions in these assays (Supplementary Figure 7). Further work is warranted to clarify functional effects of LPS-like ACAMPs.
In conclusion, the present work demonstrates that anti-PAMP mAbs can be used to probe the molecular architecture of the apoptotic cell surface, which remains poorly defined to date. Although we have restricted these initial investigations to anti-LPS antibodies, we believe that more widespread cross-reactivities with ACAMPs defined by antibodies specifying alternative PAMPs will be defined in the future. We propose that anti-PAMP mAbs constitute useful and readily accessible tools, which will help not only to improve our understanding of the structural and functional properties of apoptotic cell surfaces, but also to accelerate the acquisition of much-needed knowledge of the phenotypes and functions of apoptotic cell-derived MVs. Although our present reasoning favours PAMP-like epitopes as ultimately activating non-phlogistic responses to apoptotic cells in the ACAMPs model, it is also conceivable that such epitopes could play roles in proinflammatory, immunogenic responses to apoptosis, thereby behaving as DAMPs – damage-associated molecular patterns.37, 38 Such responses may mirror the classical proinflammatory effects of PAMPs acting via PRRs.
Materials and Methods
Cells
Adherent cells (MCF-7, A549, COS-1 and 293T) were cultured in Dulbecco's modified Eagle's medium (Invitrogen, Paisley, UK) supplemented with 10% (v/v) Serum Supreme (Lonza, Slough, UK) or 10% foetal bovine serum (PAA Laboratories Ltd, Yeovil, UK), 100 U/ml penicillin (PAA Laboratories), 100 μg/ml streptomycin (PAA Laboratories) and 2 mM ℒ-glutamine (PAA Laboratories). Suspension cells (MUTU I, BL2, K562, A1.1 and BJAB) were maintained in RPMI 1640 (Invitrogen) supplemented with 10% (v/v) Serum Supreme, 100 U/ml penicillin, 100 μg/ml streptomycin and 2 mM ℒ-glutamine. Primary polymorphonuclear leucocytes were isolated as described.39 Sf9 insect cells were cultured at 27 °C in Sf-900 II medium (Invitrogen).
Antibodies and Proteins
Antibodies were from QED Biosciences (San Diego, CA, USA; accession numbers 15174, 15306 and 15308), Invitrogen (anti-V5; goat-anti-mouse IgG Alexa Fluor conjugates), Sigma-Aldrich (Poole, UK; goat-anti-mouse IgG-FITC; isotype controls IgG1, IgG2a and IgG3; anti-β-actin (AC15)), AbD Serotec (Kidlington, UK; IgG3 isotype control), Neomarkers (Runcorn, UK; anti-β-tubulin (DM-1B)), DAKO M7901 (Carpinteria, CA, USA; anti-cytokeratin-20 (KS20.8)), Santa Cruz Biotechnology (Santa Cruz, CA, USA; LamR (F-18)) and Amersham Biosciences (Little Chalfont, UK; goat-anti-mouse IgG-HRP). Additional anti-LPS antibodies (non-crossreactive with apoptotic cells) were produced in-house by Universities of Birmingham and Edinburgh (UK). 63D3 anti-CD14 was obtained as described.40 Oligomeric recombinant mannan binding lectin (rMBL), MW 700 kDa, was a generous gift from Prof. JC Jensenius University of Aarhus (Aarhus, Denmark) and was biotinylated by standard methods. Alexa Fluor-488-conjugated streptavidin was from Invitrogen.
Apoptosis induction and measurement
Apoptosis was induced by 1 μM ionomycin (Calbiochem, San Diego, CA, USA) or UV-B irradiation (200 mJ/cm2) for up to 16 h for MUTU I, 1 μM staurosporine (Calbiochem) for BL2 and BL2-Bcl-2 cells, 100 μM etoposide (Sigma) for MCF-7 for 48–72 h, 125 ng/ml TRAIL (Affiniti Research Products, Exeter, UK) with 10 μg/ml cycloheximide (Sigma) for K562 for 8 h, or aged in the absence of serum (PMNs, 16–24 h). Levels of apoptosis were measured using TO-PRO-3 (3 μM; Invitrogen) by assessing condensation of chromatin by fluorescence microscopy (Axioskop 2; Zeiss, Feldbach, Switzerland) or by flow cytometric analysis of Alexa Fluor-488 conjugated AxV (Invitrogen) binding and PI (Sigma) uptake as described.6
Molecular cloning, recombinant protein expression and purification
Reverse transcriptase primers were designed and PCR was used to amplify the complete coding sequence of LBP/p4041 and NEDD542 (accession numbers X61156.1 and BC014455.1, respectively) from mRNA prepared from MUTU I cells using TRI reagent. Primer sequences, designed using the Amplify 1.0 software (University of Winconsin, WI, USA), were forward, 5′-CACCATGTCCGGAGCCCTTGAT-3′ and reverse, 5′-AGACCAGTCAGTGGTTGCTCCTAC3′-for LBP/p40; and forward, 5′-CACCATGTCTAAGCAACAGCCAACTCAG-3′ and reverse, 5′-CACGTGGTGCCCGAGAGCCCCG-3′ for NEDD5. DNA sequences were amplified using Pfu Polymerase (Qiagen, Crawley, UK). Blunt-ended PCR products were cloned into pcDNA3.1D/V5-His-TOPO vector using the pcDNA3.1 Directional TOPO % expression kit (Invitrogen) using the manufacturer's protocol. HEK293T, MCF7 and K562 cells were transfected by electroporation. DNA encoding LBP/p40 was subcloned into pTrcHis2C (Invitrogen) for subsequent expression in E. coli by IPTG. LBP/p40 was expressed in Spodeoptera frugiperda Sf9 insect cells using the Bac-to-Bac expression system (Invitrogen). His-tagged proteins were purified by His-Trap columns (Amersham Biosciences) and Fc-fusion proteins by Hitrap protein G column (Amersham Biosciences), and fractions containing eluted protein were identified using a modified Bradford Reagent assay (Bio-Rad Laboratories, Hercules, CA, USA) according to the manufacturer's instructions.
MV isolation
Cells were cultured at 3 × 106/ml (for flow cytometry analysis, unless otherwise stated) or 10 × 106/ml (for immunoblotting) in 0.22 μm filtered RPMI 1640 supplemented with 2 mM ℒ-glutamine, 100 U/ml penicillin and 100 μg/ml streptomycin, with or without 0.5% bovine serum albumin (low endotoxin; Sigma-Aldrich). Apoptosis was induced by exposure of cells to 1 μM staurosporine. MVs were isolated by sequential centrifugation; cell culture supernatants were obtained by centrifugation at 300 × g for 5 min followed by 750 × g for 10 min to remove cells and large debris, respectively. Supernatants were then centrifuged at either 14 000 × g for 5 min (for flow cytometry analysis, unless otherwise stated) or at 100 000 × g for 30 min (for immunoblotting) at 4°C and resuspended in 0.22 μm double-filtered Dulbecco's PBS without Ca2+ and Mg2+ (PAA). Quantification of MV proteins was by the Bradford method (Bio-Rad).
Flow cytometry
Fluorescence staining and flow cytometric analysis of cells was carried out as described.6 Binding of biotinylated rMBL was detected using Alexa Fluor-488-conjugated streptavidin. For MV staining, MVs from 6 × 106 cells were isolated as described above. Washing and incubation with antibodies was in 0.22 μm filtered PBS containing 5% normal goat serum (Biosera, Ringmer, UK) at 14 000 × g. After two washes, MVs were stained with primary antibody or isotype control for 40 min at 4 °C. After washing two times, MVs were stained with Alexa Fluor-488-conjugated goat anti-mouse IgG secondary antibody for 15 min at 4 °C, followed by two washes and resuspension in 400 μl washing buffer for analysis. Cell and MV samples were analysed using an Epics-XL-MCL flow cytometer (Beckman Coulter, High Wycombe, UK). Electronic gates for MVs were set using fluorescence-labelled 1 μm beads (Polysciences, Eppelheim, Germany). Analysis was performed using FlowJo (Treestar Inc., Ashland, OR, USA).
Immunofluorescence microscopy
Cells were seeded onto 4-well Teflon-coated slides 24 h before analysis. Cells were fixed and permeabilized in PBS containing 3% formaldehyde/5% sucrose/0.2% Triton X-100 (Bio-Rad). Excess free aldehyde groups were quenched with 50 mM NH4Cl in PBS. Cells were sequentially incubated with specified primary followed by conjugated secondary antibody. Slides were air-dried and mounted using Mowiol (Calbiochem) and viewed using confocal laser scanning microscope systems (Leica Microsystems, Milton Keynes, UK and Zeiss) equipped with Ti:Sapphire lasers. Formal microscopic quantification was not carried out prospectively throughout these studies but when scored, the incidences of mAb 15308-positive cells as a percentage of the total were found to be closely equivalent to those reported by flow cytometry (data not shown).
Immunoblotting
Cell lysates were prepared by sonication in 10 mM HEPES (Sigma), pH 7.4, 1 mM EDTA and 1% Triton X-100 with mammalian protease inhibitor cocktail (Sigma) and used in immunoblotting as described.6
Mass spectrometry
Protein bands of interest were excised from SDS-PAGE gels and analysed by peptide mass fingerprinting by MALDI-TOF Mass Spectrometry (Applied Biosystems, Warrington, UK; Voyager DE STR MALDI-TOF mass spectrometer). These masses were submitted to database search engines (SwissProt and Mascott) to identify a list of potential identities.
Statistics
A one-tailed unpaired Mann–Whitney test was applied to calculate P-values. All statistics were performed using the GraphPad Prism 5 Software (GraphPad Software, San Diego, CA, USA).
Acknowledgments
This work was supported by Leukaemia and Lymphoma Research and the Medical Research Council (UK).
Glossary
- ACAMP
apoptotic cell-associated molecular pattern
- AxV
annexin V
- LBP
laminin binding protein
- LPS
lipopolysaccharide
- mAb
monoclonal antibody
- MV
microvesicle
- PAMP
pathogen-associated molecular pattern
- PI
propidium iodide
- PRR
pattern recognition receptor
- rMBL
mannan binding lectin
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by S Nagata
Supplementary Material
References
- Fadok VA, Voelker DR, Campbell PA, Cohen JJ, Bratton DL, Henson PM. Exposure of phosphatidylserine on the surface of apoptotic lymphocytes triggers specific recognition and removal by macrophages. J Immunol. 1992;148:2207–2216. [PubMed] [Google Scholar]
- Gregory CD, Pound JD. Microenvironmental influences of apoptosis in vivo and in vitro. Apoptosis. 2010;15:1029–1049. doi: 10.1007/s10495-010-0485-9. [DOI] [PubMed] [Google Scholar]
- Kerr JF, Wyllie AH, Currie AR. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer. 1972;26:239–257. doi: 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majno G, Joris I. Apoptosis, oncosis, and necrosis. An overview of cell death. Am J Pathol. 1995;146:3–15. [PMC free article] [PubMed] [Google Scholar]
- Segundo C, Medina F, Rodriguez C, Martinez-Palencia R, Leyva-Cobian F, Brieva JA. Surface molecule loss and bleb formation by human germinal center B cells undergoing apoptosis: role of apoptotic blebs in monocyte chemotaxis. Blood. 1999;94:1012–1020. [PubMed] [Google Scholar]
- Truman LA, Ford CA, Pasikowska M, Pound JD, Wilkinson SJ, Dumitriu IE, et al. CX3CL1/fractalkine is released from apoptotic lymphocytes to stimulate macrophage chemotaxis. Blood. 2008;112:5026–5036. doi: 10.1182/blood-2008-06-162404. [DOI] [PubMed] [Google Scholar]
- Torr EE, Gardner DH, Thomas L, Goodall DM, Bielemeier A, Willetts R, et al. Apoptotic cell-derived ICAM-3 promotes both macrophage chemoattraction to and tethering of apoptotic cells. Cell Death Differ. 2012;19:671–679. doi: 10.1038/cdd.2011.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilyy RO, Shkandina T, Tomin A, Munoz LE, Franz S, Antonyuk V, et al. Macrophages discriminate glycosylation patterns of apoptotic cell-derived microparticles. J Biol Chem. 2012;287:496–503. doi: 10.1074/jbc.M111.273144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savill J, Dransfield I, Gregory C, Haslett C. A blast from the past: clearance of apoptotic cells regulates immune responses. Nat Rev Immunol. 2002;2:965–975. doi: 10.1038/nri957. [DOI] [PubMed] [Google Scholar]
- Lauber K, Blumenthal SG, Waibel M, Wesselborg S. Clearance of apoptotic cells: getting rid of the corpses. Mol Cell. 2004;14:277–287. doi: 10.1016/s1097-2765(04)00237-0. [DOI] [PubMed] [Google Scholar]
- Ravichandran KS, Lorenz U. Engulfment of apoptotic cells: signals for a good meal. Nat Rev Immunol. 2007;7:964–974. doi: 10.1038/nri2214. [DOI] [PubMed] [Google Scholar]
- Erwig LP, Henson PM. Clearance of apoptotic cells by phagocytes. Cell Death Differ. 2008;15:243–250. doi: 10.1038/sj.cdd.4402184. [DOI] [PubMed] [Google Scholar]
- Elliott MR, Ravichandran KS. Clearance of apoptotic cells: implications in health and disease. J Cell Biol. 2010;189:1059–1070. doi: 10.1083/jcb.201004096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory CD, Pound JD. Cell death in the neighbourhood: direct microenvironmental effects of apoptosis in normal and neoplastic tissues. J Pathol. 2011;223:177–194. doi: 10.1002/path.2792. [DOI] [PubMed] [Google Scholar]
- Gregory CD, Devitt A. The macrophage and the apoptotic cell: an innate immune interaction viewed simplistically. Immunology. 2004;113:1–14. doi: 10.1111/j.1365-2567.2004.01959.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Owen KA, Ly KT, Park D, Black SG, Wilson JM, et al. Brain angiogenesis inhibitor 1 (BAI1) is a pattern recognition receptor that mediates macrophage binding and engulfment of Gram-negative bacteria. Proc Natl Acad Sci USA. 2011;108:2136–2141. doi: 10.1073/pnas.1014775108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R, Janeway CA. Innate immunity: the virtues of a nonclonal system of recognition. Cell. 1997;91:295–298. doi: 10.1016/s0092-8674(00)80412-2. [DOI] [PubMed] [Google Scholar]
- Pugin J, Heumann D, Tomasz A, Kravchenko VV, Akamatsu Y, Nishijima M, et al. CD14 is a pattern recognition receptor. Immunity. 1994;1:509–516. doi: 10.1016/1074-7613(94)90093-0. [DOI] [PubMed] [Google Scholar]
- Franc NC, White K, Ezekowitz RAB. Phagocytosis and development: back to the future. Curr Opin Immunol. 1999;11:47–52. doi: 10.1016/s0952-7915(99)80009-0. [DOI] [PubMed] [Google Scholar]
- Gregory CD. CD14-dependent clearance of apoptotic cells: relevance to the immune system. Curr Opin Immunol. 2000;12:27–34. doi: 10.1016/s0952-7915(99)00047-3. [DOI] [PubMed] [Google Scholar]
- Ardini E, Tagliabue E, Magnifico A, Buto S, Castronovo V, Colnaghi MI, et al. Co-regulation and physical association of the 67-kDa monomeric laminin receptor and the alpha6beta4 integrin. J Biol Chem. 1997;272:2342–2345. doi: 10.1074/jbc.272.4.2342. [DOI] [PubMed] [Google Scholar]
- Martin SJ, Reutelingsperger CPM, McGahon AJ, Rader JA, van Schie RCAA, LaFace DM, et al. Early redistribution of plasma membrane phosphatidylserine is a general feature of apoptosis regardless of the initiating stimulus: inhibition by overexpression of Bcl-2 and Abl. J Exp Med. 1995;182:1545–1556. doi: 10.1084/jem.182.5.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratton DL, Fadok VA, Richter DA, Kailey JM, Guthrie LA, Henson PM. Appearance of phosphatidylserine on apoptotic cells requires calcium- mediated nonspecific flip–flop and is enhanced by loss of the aminophospholipid translocase. J Biol Chem. 1997;272:26159–26165. doi: 10.1074/jbc.272.42.26159. [DOI] [PubMed] [Google Scholar]
- Matsura T, Serinkan BF, Jiang J, Kagan VE. Phosphatidylserine peroxidation/externalization during staurosporine-induced apoptosis in HL-60 cells. FEBS Lett. 2002;524:25–30. doi: 10.1016/s0014-5793(02)02990-3. [DOI] [PubMed] [Google Scholar]
- Kagan VE, Gleiss B, Tyurina YY, Tyurin VA, Elenstrom-Magnusson C, Liu SX, et al. A role for oxidative stress in apoptosis: oxidation and externalization of phosphatidylserine is required for macrophage clearance of cells undergoing Fas-mediated apoptosis. J Immunol. 2002;169:487–499. doi: 10.4049/jimmunol.169.1.487. [DOI] [PubMed] [Google Scholar]
- Zhou Z. New phosphatidylserine receptors: clearance of apoptotic cells and more. Dev Cell. 2007;13:759–760. doi: 10.1016/j.devcel.2007.11.009. [DOI] [PubMed] [Google Scholar]
- Palaniyar N, Nadesalingam J, Clark H, Shih MJ, Dodds AW, Reid KB. Nucleic acid is a novel ligand for innate, immune pattern recognition collectins surfactant proteins A and D and mannose-binding lectin. J Biol Chem. 2004;279:32728–32736. doi: 10.1074/jbc.M403763200. [DOI] [PubMed] [Google Scholar]
- Gardai SJ, McPhillips KA, Frasch SC, Janssen WJ, Starefeldt A, Murphy-Ullrich JE, et al. Cell-surface calreticulin initiates clearance of viable or apoptotic cells through trans-activation of LRP on the phagocyte. Cell. 2005;123:321–334. doi: 10.1016/j.cell.2005.08.032. [DOI] [PubMed] [Google Scholar]
- Arur S, Uche UE, Rezaul K, Fong M, Scranton V, Cowan AE, et al. Annexin I is an endogenous ligand that mediates apoptotic cell engulfment. Dev Cell. 2003;4:587–598. doi: 10.1016/s1534-5807(03)00090-x. [DOI] [PubMed] [Google Scholar]
- Blume KE, Soeroes S, Waibel M, Keppeler H, Wesselborg S, Herrmann M, et al. Cell surface externalization of annexin A1 as a failsafe mechanism preventing inflammatory responses during secondary necrosis. J Immunol. 2009;183:8138–8147. doi: 10.4049/jimmunol.0902250. [DOI] [PubMed] [Google Scholar]
- Nakai Y, Shiratsuchi A, Manaka J, Nakayama H, Takio K, Zhang JT, et al. Externalization and recognition by macrophages of large subunit of eukaryotic translation initiation factor 3 in apoptotic cells. Exp Cell Res. 2005;309:137–148. doi: 10.1016/j.yexcr.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Jaillon S, Jeannin P, Hamon Y, Fremaux I, Doni A, Bottazzi B, et al. Endogenous PTX3 translocates at the membrane of late apoptotic human neutrophils and is involved in their engulfment by macrophages. Cell Death Differ. 2009;16:465–474. doi: 10.1038/cdd.2008.173. [DOI] [PubMed] [Google Scholar]
- Kuraishi T, Nakagawa Y, Nagaosa K, Hashimoto Y, Ishimoto T, Moki T, et al. Pretaporter, a Drosophila protein serving as a ligand for Draper in the phagocytosis of apoptotic cells. EMBO J. 2009;28:3868–3878. doi: 10.1038/emboj.2009.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada R, Nagaosa K, Kuraishi T, Nakayama H, Yamamoto N, Nakagawa Y, et al. Apoptosis-dependent externalization and involvement in apoptotic cell clearance of DmCaBP1, an endoplasmic reticulum protein of Drosophila. J Biol Chem. 2012;287:3138–3146. doi: 10.1074/jbc.M111.277921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadok VA, Bratton DL, Henson PM. Phagocyte receptors for apoptotic cells: recognition, uptake, and consequences. J Clin Invest. 2001;108:957–962. doi: 10.1172/JCI14122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimsley C, Ravichandran KS. Cues for apoptotic cell engulfment: eat-me, don't eat-me and come-get-me signals. Trends Cell Biol. 2003;13:648–656. doi: 10.1016/j.tcb.2003.10.004. [DOI] [PubMed] [Google Scholar]
- Rubartelli A, Lotze MT. Inside, outside, upside down: damage-associated molecular-pattern molecules (DAMPs) and redox. Trends Immunol. 2007;28:429–436. doi: 10.1016/j.it.2007.08.004. [DOI] [PubMed] [Google Scholar]
- Garg AD, Krysko DV, Verfaillie T, Kaczmarek A, Ferreira GB, Marysael T, et al. A novel pathway combining calreticulin exposure and ATP secretion in immunogenic cancer cell death. EMBO J. 2012;31:1062–1079. doi: 10.1038/emboj.2011.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dransfield I, Stocks SC, Haslett C. Regulation of cell adhesion molecule expression and function associated with neutrophil apoptosis. Blood. 1995;85:3264–3273. [PubMed] [Google Scholar]
- Flora PK, Gregory CD. Recognition of apoptotic cells by human macrophages − inhibition by a monocyte/macrophage-specific monoclonal antibody. Eur J Immunol. 1994;24:2625–2632. doi: 10.1002/eji.1830241109. [DOI] [PubMed] [Google Scholar]
- Yow HK, Wong JM, Chen HS, Lee CG, Davis S, Steele GD, et al. Increased mRNA expression of a laminin-binding protein in human colon carcinoma: complete sequence of a full-length cDNA encoding the protein. Proc Natl Acad Sci USA. 1988;85:6394–6398. doi: 10.1073/pnas.85.17.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strausberg RL, Feingold EA, Grouse LH, Derge JG, Klausner RD, Collins FS, et al. Generation and initial analysis of more than 15 000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci USA. 2002;99:16899–16903. doi: 10.1073/pnas.242603899. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.