Abstract
The Y box is an important sequence motif found in promoters and enhancers containing a CCAAT box – one of the few elements enriched in promoters of large sets of genes overexpressed in cancer. The search for the transcription factor(s) acting on it led to the biochemical purification of the nuclear factor Y (NF-Y) heterotrimer, and to the cloning – through the screening of expression libraries – of Y box-binding protein 1 (YB-1), an oncogene, overexpressed in aggressive tumors and associated with drug resistance. These two factors have been associated with Y/CCAAT-dependent activation of numerous growth-related genes, notably multidrug resistance protein 1. We review two decades of data indicating that NF-Y ultimately acts on Y/CCAAT in cancer cells, a notion recently confirmed by genome-wide data. Other features of YB-1, such as post-transcriptional control of mRNA biology, render it important in cancer biology.
Keywords: Y box, CCAAT box, NF-Y, YB-1
Facts
Precise sequence of the CCAAT element.
The nuclear factor Y (NF-Y) transcription factor (TF) binds to the CCAAT box.
Lack of solid evidence of DNA-binding specificity of the Y box-binding protein 1 (YB-1) oncogene.
Open Questions
How NF-Y turns on ‘cancer' genes with CCAAT boxes?
What is the exact role of YB-1 in the regulation of gene expression?
Is there an interplay between NF-Y and YB-1?
Y and CCAAT: Two Names, One Entity
The Y box – consensus CTGATTGGT/CT/C – was identified three decades ago as a DNA element conserved in promoters of major histocompatibility complex (MHC) class II genes.1 In vitro transcription, transfections and transgenic mice experiments with Y box-mutated promoters showed its crucial role, along with neighboring conserved sequences, for the coordinated and tissue-specific expression of these genes.2 In reality, the Y box contains an inverted CCAAT sequence – ATTGG underlined above – which had previously been identified in the globin and ovalbumin promoters along with the TATA box.3, 4 Importantly, the CCAAT/ATTGG pentanucleotide was shown to be required for transcriptional activation (TA).5, 6, 7, 8, 9, 10, 11 In essence, the Y and CCAAT boxes are functionally equivalent. Thereafter, the importance and widespread distribution of the Y/CCAAT box, as precisely defined by the initial genetic and biochemical experiments, has been substantiated by unbiased genomic studies. The exact sequence and frequency of Y/CCAAT in promoters was assessed using different bioinformatic tools: several labs reported the identification of Y/CCAAT as over-represented in human promoters and enhancers by searching with available matrices,12, 13, 14, 15, 16, 17, 18, 19 and two studies searching for unbiased ‘words' enriched within promoters identified Y/CCAAT and precise flanking motifs.20, 21
Y/CCAAT is Enriched in Promoters of ‘Cancer' Genes
Analysis of transcriptome profiles during cellular transformation identified the Y/CCAAT box as over-represented in promoters of genes overexpressed in diverse types of cancers, breast, colon, thyroid, prostate and leukemias.22, 23, 24, 25, 26, 27, 28, 29 Treating cells with cytotoxic drugs, or overexpressing growth suppressors, led to the downregulation of genes with CCAAT in their promoters.30, 31 However, it is important to remark that these exercises were performed with matrices included in TRANSFAC and JASPAR; hence, they could be highly biased, as the lists of transcription factor-binding sites (TFBS) present in databases certainly do not recapitulate all possible TF-binding sequences. More tellingly, de novo motif discovery methods that allow unbiased identification of sequence logos showed that promoters of genes, specifically overexpressed in tumors, are significantly enriched in Y/CCAAT elements.32, 33, 34 This indicates that Y/CCAAT is of importance in the overexpression of cancer genes in tumors. Therefore, the TFs recognizing this element are likely relevant for the process of cellular transformation.
The search for the TF(s) binding to Y/CCAAT started in the late 1980s, leading to the apparent identification of more than one activity.35 We will not discuss here C/EBP (CCAAT enhancer-binding proteins) and CTF/NF1 (CCAAT TF/nuclear factor 1), two bona fide sequence-specific TFs originally associated with CCAAT binding, as they were later unambiguously shown to have different sequence specificities.36, 37, 38 Two additional proteins have been ‘battling' over the Y/CCAAT ground for over two decades, NF-Y and YB-1. We review here the work of the past 20 years pertinent to the specific role of the two factors in Y/CCAAT activation.
NF-Y and CCAAT
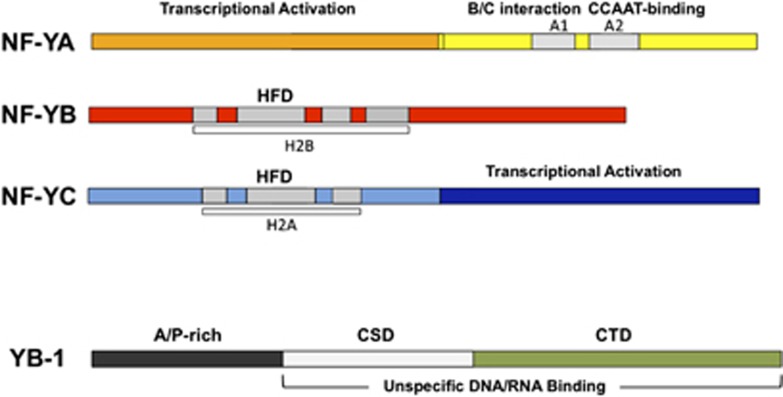
NF-Y was originally shown to bind to the Y box of the MHC class II Ea promoter using electrophoretic mobility shift assays (EMSAs).35, 39 Later, it became clear that it was identical to CBF (CCAAT-binding factor) shown to interact with collagen promoters,6 CP1 (CCAAT protein 1) binding to globin promoters7 and EFI binding to the Rous sarcoma virus long terminal repeat (RSV LTR).40 It was soon realized that this activity was ubiquitously expressed, composed of multiple subunits and conserved in yeast, where it is called HAP complex.41 The complex was biochemically purified using conventional affinity purification with oligomerized Y/CCAAT oligos. Initially, two subunits were characterized.42, 43, 44, 45 Eventually, NF-Y was shown to be a heterotrimer composed of NF-YA, NF-YB and NF-YC (Figure 1), whose genes are found in all eukaryotes (they are termed HAP2/3/5 in yeast). NF-YB and NF-YC have histone fold domains (HFDs) similar to core histones H2A/H2B, composed of three α-helices separated by short loop/strand regions;46 dimerization of HFDs is required for association with NF-YA, which provides sequence specificity to the complex.
Figure 1.
Scheme of NF-Y subunits and YB-1
Several lines of evidence indicate that NF-Y activates transcription through sequence-specific binding to Y/CCAAT:
Saturation mutagenesis studies on different Y/CCAAT boxes, using in vitro EMSAs, clearly showed that NF-Y binding is absolutely dependent on each of the five core nucleotides and pointed at important flanking nucleotides – 2 bp at the 5′ end and 3 bp at the 3′ end – as, indeed, found in the original Y box.6, 7, 35, 39, 40 Unbiased SELEX assays further confirmed the specificity of NF-Y for a 10 bp stretch.47
In vitro transcription and transfections of promoters mutated in the NF-Y-binding sites showed a perfect correlation between the decrease or abolition of NF-Y binding and the decrease of functional activity.6, 7, 8, 9, 10 In vitro transcription assays with purified NF-Y, antibodies and recombinant proteins showed an effect of NF-Y on transcriptional initiation, and re-initiation, in various promoters.6, 7, 11, 48, 49, 50, 51
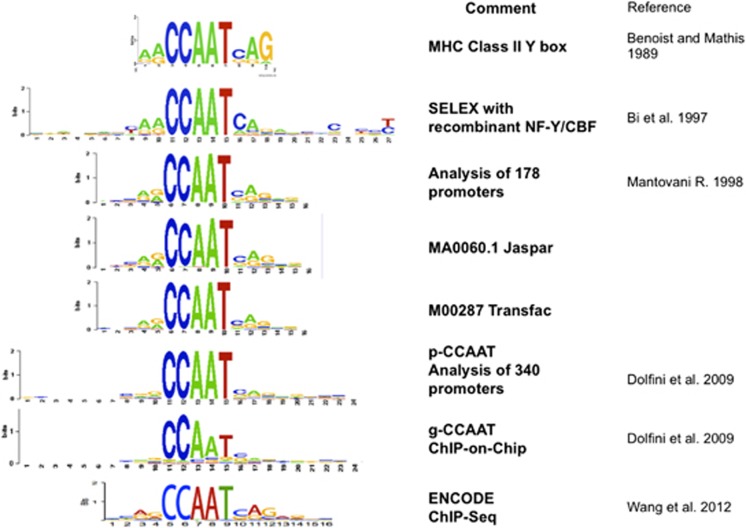
The development of specific antibodies, used in supershift EMSAs in vitro48 and in chromatin immunoprecipitation (ChIP) assays in vivo,52 enabled different labs to verify that the band observed in EMSAs, and the protein bound in cells, was indeed NF-Y. The initial in vitro experiments lead to the definition of a first NF-Y positional sequence frequency matrix (PSFM),53 which was soon incorporated into the TRANSFAC and JASPAR databases (Figure 2). Note that the bioinformatic analyses of motifs in promoters of cancer genes mentioned above also retrieved the NF-Y logo.
The use of dominant-negative NF-YA vectors54 and, more recently, the inactivation of NF-Y subunits by small interfering RNA (siRNA) or short hairpin RNA (shRNA) interference allowed the in vivo confirmation that a CCAAT promoter is regulated by NF-Y (reviewed in Dolfini et al.55).
Genomic analysis by ChIP-on-Chip56, 57, 58, 59 and ChIP-Seq60 confirmed that NF-Y binds to Y/CCAAT in vivo. These experiments further refined the NF-Y PSFM (Figure 2). In summary, a very robust set of data leads to the accepted conclusion that NF-Y regulates gene expression through specific binding of the Y/CCAAT box.
Figure 2.
Y/CCAAT ‘evolution' over time
YB-1 and CCAAT?
YB-1 was identified through screening of a phage expression library using a multimerized Y box oligo from the MHC class II DRa promoter.61 DRa is the human homolog of the mouse Ea, which was the starting point for the biochemical identification of NF-Y.35 A similar strategy was reported in the cloning of rat EFIa, using a CCAAT oligo from the RSV LTR,62 Xenopus FRG-163 and chicken YB-1.64 This technique identifies phages producing a single polypeptide, and it could not have been used for NF-Y, as all three of its subunits are required for DNA binding. YB-1 was shown to be a protein with a known nucleic acid-binding domain, termed CSD (cold-shock domain), which is highly conserved in eukaryotes and prokaryotes (Figure 1 and Mihailovich et al.65).
Intriguingly, two cloning manuscripts published in the same period were at odds with the interpretation that YB-1 was a classic sequence-specific TF: (i) YB-1 was identified in screenings of expression libraries with a completely unrelated oligo, the W box, also from the DRa promoter, but sharing no sequence similarity to Y/CCAAT;66 these authors first provided evidence that YB-1 binds well to single-stranded (ssDNA) and, surprisingly, to abasic DNA. The chicken YB-1, subsequently cloned, also showed DNA binding with little dependence on the presence of a Y/CCAAT box.64 (ii) The Xenopus FRG-1 gene was cloned with expression libraries67probed with antibodies directed against p54/p56,68 one of the subunits of the mRNA-binding complex biochemically identified in the 1970s, and widely studied for its role in mRNA translation.68 Thereafter, research on YB-1 proceeded, by and large, in two parallel fields: its role in the control of mRNA biology (splicing, stability, translation), in which it has taken a center-stage position (Bouvet and Wolffe69; reviewed in Evdokimova et al.70), and its role in the control of transcriptional initiation, which has been more controversial.
NF-Y is the Sequence-Specific CCAAT Factor
We list below a comparison of features supporting NF-Y and YB-1 as bona fide sequence-specific TFs.
DNA affinity and sequence specificity
When recombinant YB-1 was tested in in vitro binding assays, which are more sensitive and specific than the Southwestern blots used in the initial cloning papers, the protein was shown to bind to RNA and DNA oligos.71 With SELEX and Chip methods, it was established that the strings of nucleic acids preferred by YB-1 are GGGG (ssDNA), CACC/T (double-stranded DNA (dsDNA)) and AACAUC (RNA). None of these motifs resemble to a canonical Y/CCAAT box (Bouvet et al.,72 Zasedateleva et al.73 and Ray et al.74; reviewed by Eliseeva et al.71). In vitro, YB-1 prefers RNA and ssDNA – KD in the order of 10−9 – with respect to dsDNA. The appetite of NF-Y for DNA is much higher, with a KD of 10−11.7, 39 As for the specificity, single-nucleotide substitutions within the CCAAT and flanking nucleotides of Y/CCAAT drop affinity by one/two log levels.6, 10, 35 Typically, nanograms of NF-Y give a robust EMSA shift,47, 75 as opposed to micrograms of recombinant YB-1.76 Methylation interference and orthophenanthroline footprinting confirmed that the bases contacted by NF-Y are centered on Y/CCAAT.6, 7, 35, 39, 40, 47 The only such data available for YB-1 is relative to a site in the MMP-2 promoter – CTGCTGGGCAAG – which lacks a Y/CCAAT sequence.77
In summary, in vitro biochemical analyses indicate that the affinity and specificity of NF-Y for Y/CCAAT is superior to that of YB-1.
Mechanisms of TA
Sequence-specific TFs are known to be modular proteins, composed of a minimum of two domains: a DNA-binding domain (DBD) and a TA domain. Different types of DBDs and TAs have been described. NF-Y has two large Q-rich TAs over150 amino acids in length, in the NF-YA and NF-YC subunits. These TAs function when fused to heterologous DBDs, such as those of yeast GAL4 or bacterial LexA.49, 78, 79 Importantly, removal of the NF-YA (CBF-B) Q-rich domain generates a dominant-negative mutant, which affects the activity of CCAAT promoters upon transfection.80 Co-transfections of the NF-Y trimer with TFs binding the loci neighboring CCAAT boxes in mammalian and Drosophila cells synergistically activate transcription of CCAAT promoters.52, 81, 82 Finally, transfections of recombinant proteins in which NF-YA was linked to a cell-penetrating TAT peptide activate endogenous CCAAT genes but not CCAAT-less units.83, 84 All these features are quite standard for TFs: sequence specificity, modularity of DBD and TA, and synergistic activation with neighboring TFs.
YB-1 has two domains flanking the central CSD: the A/P domain, and the C-terminal domain, in none of which is a typical TA apparent; most importantly, there are no data showing that these domains can function as TAs, in hybrids, or within the intact protein. The inclusion of YB-1 among TFs is based on changes in the expression of many genes by overexpression or functional inactivation of YB-1 by siRNA or shRNA.71 However, it should be remembered that northern blots, RT-PCR, qRT-PCR and microarray profiles all measure mRNA steady-state levels and not RNA Pol II transcription rates. In general, a troublesome issue in the manuscripts showing an effect of YB-1 in transcription is that they do not take into account its known role in mRNA biology: one needs to be sure that the reported changes in mRNA levels upon overexpression or inactivation are really due to transcriptional initiation events, rather than to post-transcriptional effects on mRNA stability. This consideration is particularly relevant for experiments measuring enzymatic activities of reporter genes, such as chloramphenicol acetyltransferase or Luciferase.
Nuclear run-on assays can provide evidence for a role in transcriptional initiation events, but to the best of our knowledge, no such experiments have been described for overexpression or inactivation of YB-1, in contrast to the situation for NF-Y-dependent transcriptional units.85, 86, 87, 88, 89, 90 Analogously, many in vitro transcription assays suggest a role for NF-Y in the formation of the pre-initiation complex and in re-loading of the RNA Pol II machinery.6, 7, 11, 48, 49, 50, 51 The only report showing in vitro transcription data on YB-1 used the heat-shock 70 kDa protein (HSP70) and thymidine kinase (TK) promoters with an unpurified bacterially produced protein.63 It is worth noting that the HSP70 CCAAT element was subsequently investigated as a canonical NF-Y target by the same group.91, 92
As to the mechanistic details of TA, NF-Y has been shown to (i) promote the DNA binding of neighboring activators, (ii) make contacts with multiple general transcription factors, including TBP (TATA-binding protein) and TAFs (TBP-associated factors), (iii) mediate recruitment of Pol II and (iv) bind multiple coactivators – p300, PCAF, MLL – and be post-translationally modified by some of them.93 The role of YB-1 in activation appears to be related to ssDNA binding in promoter regions (Stein et al.94; reviewed in Eliseeva et al.71).
The targets
YB-1 has been shown to regulate a number of genes. Among them, the multidrug resistance protein 1 (MDR1) promoter was analyzed in detail, due to the paramount importance that this gene has in the mechanisms of drug resistance. Overexpression of this efflux pump as a result of TA is a normal response to the treatment of cells to many chemicals, including cytotoxic drugs. Upon administration of anticancer compounds, cancer cells often acquire resistance to pharmacologic doses of drugs through the overexpression of MDR1, rendering antitumor regimens ineffective. The transcriptional control of MDR1 has been the object of many studies, as preventing its overexpression could be highly desirable.95 The MDR1 promoter contains a crucial Y/CCAAT element, and a number of contradictory reports concerning the identity of the activator were published. Several papers suggested that YB-1 activates MDR1 through the Y/CCAAT box.96, 97, 98, 99 The identification of YB-1 as the CCAAT activator relied on EMSAs challenged with anti-YB-1 antibodies and on overexpression of YB-1 and inactivation by an antisense YB-1 transcript. Other investigators later showed that overexpression of YB-1 has no effect on MDR1 transcription,100 and that inactivation by different siRNAs and shRNAs, which completely obliterates YB-1 expression, have no effect on MDR1 basal, or activated expression.101 The MDR1 Y/CCAAT is a perfect NF-Y site – CTGATTGGCT – located in the NF-Y canonical position, at −70 from the TSS. Indeed, several labs showed that NF-Y is the activator, both under basal and under multiple inducing conditions.102, 103, 104, 105, 106, 107, 108 In EMSAs performed with the Y/CCAAT box, these authors detected NF-Y as the only DNA-binding complex. Specifically, Scotto's lab showed that a NF-YA dominant-negative mutant affects MDR1 expression.104 Finally, in vivo ChIPs reported NF-Y binding to MDR1.109
Recent experiments reported interactions of YB-1 with APE1, a protein originally characterized for its role in base excision repair (BER), and shown to be important in the specific system to recruit Pol II by association with p300 and YB-1. APE1 is also known as redox effector factor 1 (Ref1), as it affects the redox status of, among other proteins, many TFs. APE1/Ref1 and YB-1 directly interact,110 they bind the MDR1 core promoter and removal of APE1 leads to decreased YB-1, Pol II and p300 promoter association in ChIP assays.111 This led the authors to propose that the YB-1/CCAAT interaction mediates the recruitment of the APE1/p300/Pol II complex onto the promoter. The redox potential of NF-Y affects directly its DNA-binding capacity, acting on three conserved cysteines of the NF-YB HFD, and Ref1 is an important regulator.112 Therefore, the above data are consistent with an alternative explanation in which APE1/Ref1 acts on NF-Y/CCAAT interactions to activate MDR1 transcription.
In general, we find it extremely surprising that in the reports pointing to YB-1 as the MDR1 CCAAT activator, NF-Y was either not observed or not recognized as such in EMSAs, given its superior affinity for the Y/CCAAT sequence. Technical considerations are perhaps unlikely to account for this, as NF-Y is readily observed in nuclear extracts of all growing cells, regardless of extraction protocols employed, incubation and EMSA conditions. In fact, it has been detected in EMSAs in hundreds of manuscripts reporting CCAAT binding in disparate promoters (Dolfini et al.55 and references therein). Essentially, the MDR1 system is apparently a rare exception to the rule that a Y/CCAAT binding complex detected in vitro contains NF-Y. This confusion has been replicated with the related MRP2 promoter, suggested to be either activated by NF-Y113 or YB-1.114
The same argument applies to other Y/CCAAT promoters, which YB-1 was shown to activate, including DNA Pola,115 cyclin A, cyclin B1,116 where many reports have also demonstrated an NF-Y dependence (Farina et al.117; reviewed in Gurtner et al.118). For other YB-1 targets, such as EGFR, PIK3CA, MET and CD44,119, 120, 121, 122 the Y/CCAAT, as it has been characterized genetically and bioinformatically, is absent.
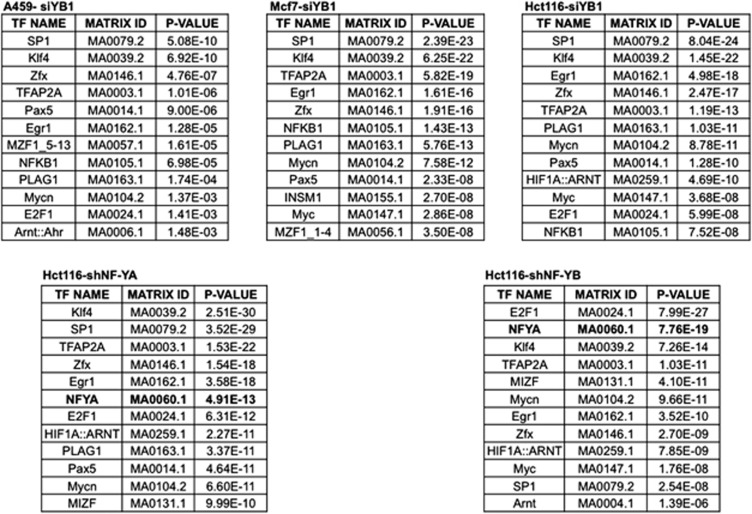
Focusing on the global picture, transcription profiling analysis after functional inactivation is available both for YB-1123 and NF-Y.124 In YB-1-regulated genes, an abundance of E2F sites was reported: we re-examined these data with bioinformatic tools and indeed confirm this point, but neither the Y/CCAAT logo nor related variants are over-represented (Figure 3). On the other hand, inactivation of single NF-Y subunits led to different phenotypes and sets of regulated genes,124, 125 with the enrichment of the Y/CCAAT logo in downregulated genes a common feature (Figure 3).
Figure 3.
Enriched TFBS in YB-1- or NF-Y-regulated genes. Data of gene expression profilings of different cell lines, reported by Lasham et al.,123 were analyzed by pscan (upper panels). In the lower panels, a similar analysis is reported on gene expression profiles of HCT116 cells inactivated with shNF-YA or shNF-YB (Benatti et al.124)
The genomic loci
The genome-wide identification of TFBSs through ChIP-on-Chip and, more recently, ChIP-Seq has vastly advanced our understanding. For NF-Y, ChIP-on-Chip studies performed on CpG islands, promoters and oligo tiling arrays led to the conclusion that the protein is bound not only to promoters but also to many enhancers. The presence of Y/CCAAT in these regions was consistent, although not all loci contained the pentanucleotide. ChIP-Seq experiments performed in the framework of the ENCODE Project (Wang et al.60) provide far higher precision. The data are clearcut: almost all peaks do contain the Y/CCAAT consensus, essentially identical to the original NF-Y matrix (Figure 2). Moreover, the CCAAT-less sites are variation of one nucleotide in the core sequence that also harbor optimal flanking sequences. The technique is so spectacularly powerful and precise that it is possible to discriminate the exact area bound by NF-YA – the CCAAT pentanucleotide – from the immediately flanking nucleotides bound by NF-YB, in perfect accordance with the in vitro biochemical data.
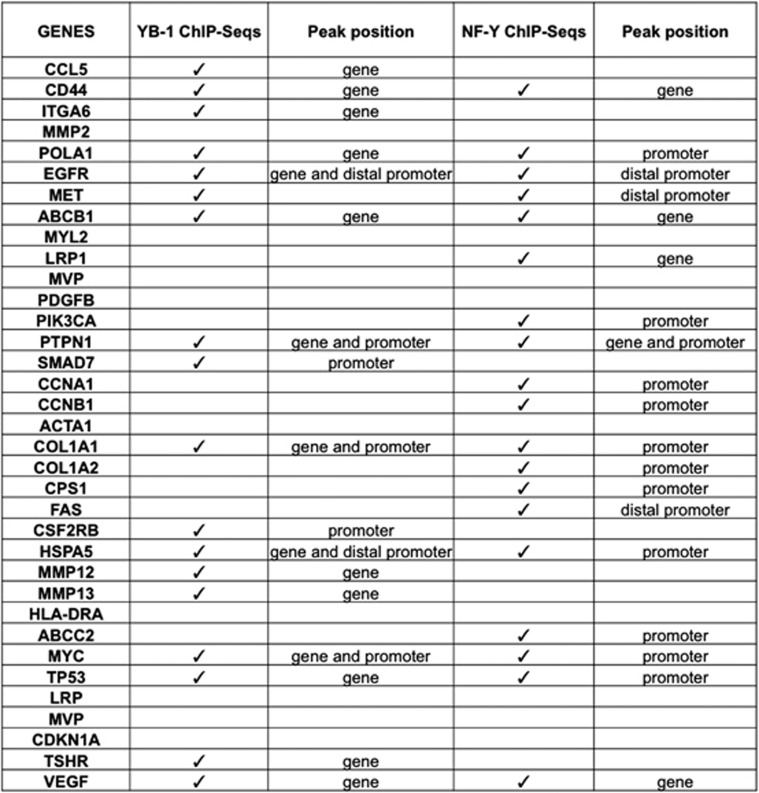
ChIP-on-Chip and ChIP-Seq experiments for YB-1 were also reported recently.121, 126 We analyzed the data for >30 000 ChIP-Seq peaks in three cancer cell types and could not identify a Y/CCAAT sequence, either searching for known TFBS or with de novo motif discovery tools.127 This is uncommon for a sequence-specific TF, whose logo is usually recognizable within the bound peaks, but not unheard of:60 in many cases, one can identify either a new logo or one of those characterized for other TFs. Of the 35 YB-1-targeted genes previously analyzed by various means, including reporter assays (summarized in Eliseeva et al.71), 5 are associated with YB-1 peaks in BT747 cells, 15 in HR5 and 13 in HR6 cells: collectively, 18 genes are targeted (Figure 4). A minority (seven) of the YB-1 peaks is in promoters and even fewer are at the exact sites described in the functional studies. Obviously, these results can be heavily influenced by the dissimilar cellular contexts used in the functional and location assays. However, a similar NF-Y ChIP-Seq analysis performed on the same 35 genes, also from disparate cells, showed positivity for 19 units, with all but 4 peaks being present in promoters: this leads to the somewhat paradoxical notion that, in unbiased experiments, NF-Y targets more ‘bona fide' YB-1 sites in vivo than YB-1 itself.
Figure 4.
Presence of YB-1 and NF-Y peaks in YB-1-regulated genes. YB-1-regulated genes, as summarized by Eliseeva et al.,71 were analyzed for the presence of YB-1 peaks in the YB-1 ChIP-Seq data reported by Astanehe et al.,126 and for NF-Y peaks present in ENCODE data of K562, HeLa-S3 and GM12878 cells (Wang et al.60 and Fleming and Struhl, submitted for publication)
Cellular localization
Intuitively, TFs exert their function in the nucleus, where their genomic targets are located. In this respect, NF-Y is prototypical, with NF-YA and NF-YB being found exclusively in the nucleus, NF-YC being in part in the cytoplasm and traveling to the nucleus with the NF-YB HFD partner.128 YB-1 is fundamentally cytoplasmic in normal cells, and nuclear in transformed cells, or after specific stimuli. Many inducible TFs are found in the cytoplasm or membrane bound, and are transferred to the nucleus only after a specific stimulus. To explain the activation of G1/S cell-cycle-regulated promoters, such as DNA Pol α and cyclin A by YB-1, it was proposed that there is a transient nuclearization of the protein at the G1/S boundary.116 Even so, it not clear how YB-1 could activate transcription of the many proposed target genes that are constitutively expressed, or of the G2/M-specific cyclin B, in normal cells.
Protein structure considerations
The modality of CCAAT recognition by NF-Y is now fully understood, as the crystal structure of the complex bound to a CCAAT oligo has been solved.129 NF-Y contacts DNA in a non-sequence-specific manner via the HFD subunits, which bind the double helix in a way that is essentially identical to H2A/H2B within nucleosomes, with sequence specificity imparted by minor groove binding to the CCAAT, via an α helix (A2) and a novel motif of NF-YA. This modality of DNA recognition is unprecedented among TFs. Overall, a high number of amino-acid residues (46) contact the DNA over a 25–28 bp area, which helps to explain the extraordinary affinity of the trimer for DNA. The domain of YB-1 required for nucleic acid binding is the CSD, composed of RNP1 and RNP2: the structure of this domain is known, both from X-ray crystallography and NMR studies,62 but the interactions with RNA or DNA are not detailed and modeling exercises could not provide compelling reasons why YB-1 should bind to double-stranded Y/CCAAT with any type of specificity.
Altogether, from the most reductionistic in vitro assays to in vivo approaches, a large set of data strongly argues against YB-1 being a direct regulator of transcriptional initiation by binding to the Y/CCAAT sequence, or variations of it. Because of the widespread and profound influence played by YB-1 on mRNA biology, likely impacting on results obtained in overexpression or functional inactivation experiments, it is even debatable as to whether it has any role in direct promotion of transcriptional initiation events. On the contrary, NF-Y's credentials as a paradigmatic sequence-specific TF are impeccable.
The – Apparent – Paradox of Y/CCAAT, NF-Y and YB-1 in Cancer
To the best of our knowledge, NF-Y subunits are neither consistently mutated – as for p53 – nor generally overexpressed – as for MYC – in human tumors. However, changes in the expression of NF-Y subunits, often NF-YA, have indeed been reported,93 and this phenomenon should be studied in a quantitatively credible and statistically significant manner. Moreover, the links of NF-Y with activation of cancer pathways mediated by mutant p53 and E2Fs are well established.130 More recently, strong connections emerged from ChIP-Seq experiments with JNKs,131 the PRAME oncogene,132 and a specific group of oncogenic TFs in ENCODE data (Fleming and Struhl, submitted for publication).
There is little doubt that YB-1 expression changes dramatically, at both the mRNA and protein levels, in transformed cells; the correlation is so impressive that in some tumors it is considered a prognostic marker (for a review see Eliseeva et al.71 and Costessi et al.133). Furthermore, the localization of YB-1 becomes strongly nuclear in tumor cells. The changes are maximal in aggressive tumors resistant to drugs, and in advanced stages of cancer. It is also undisputed that a large set of genetic experiments point to YB-1 as a protein promoting transformation, epithelial–mesenchymal transition and growth of metastatic cancer cells: in essence, YB-1 is a powerful oncogene.134, 135, 136
Largely because of the alleged role of YB-1 on MDR1 transcription, two syllogisms emerged in the literature. First, Y/CCAAT is enriched in cancer genes, YB-1 is a Y/CCAAT binding TF enriched in cancer cells; hence, YB-1 is responsible for the activation of CCAAT cancer genes in cancer cells.137 Second, MDR1 expression is induced by and responsible for resistance of cancer cells to cytotoxic drugs, YB-1 is responsible for MDR1 overexpression (and it is overexpressed in cancers); hence, YB-1 mediates cancer resistance by enhancing MDR1 expression. Flaws in the antithesis, as explained above, lead to an at least partially incorrect synthesis.
So how could YB-1 be mediating cancer progression and resistance to drugs, if not by binding directly to the Y/CCAAT boxes of MDR1 or other overexpressed genes? Reviews of available literature point to many possible hypotheses, the most likely of which highlight the control of various aspects of RNA metabolism, such as stability, splicing and translation.71, 135 The mechanisms of YB-1 mRNA regulation were studied in reliable reconstituted in vitro systems, and they are now relatively well understood, and consistent with specific binding of the protein to RNA. Interestingly, the preferred target logo, derived from the analysis of ChIP-Seq experiments, is consistent with the RNA-binding features of YB-1, and indeed resembles Kozak sequences.127
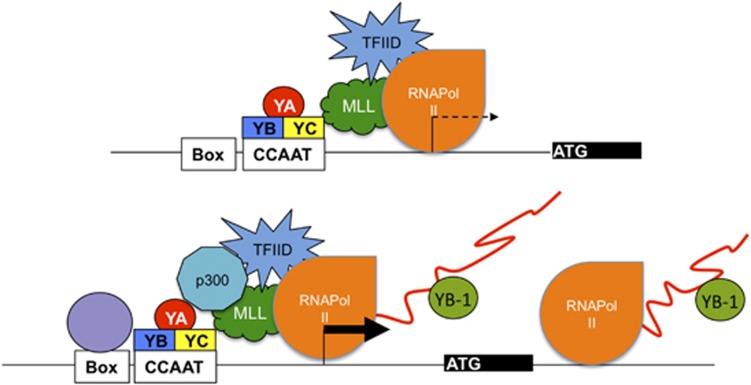
Thus, we offer an alternative explanation for the many reports of YB-1 binding in ChIP experiments, and indeed ChIP-Seq, in transformed cells:126 YB-1 could be transitorily located in an area physically ‘close' to promoters, or other important regulatory regions, where transcriptional initiation decisions are made, but loaded on partially synthesized primary RNAs (Figure 5). One finding consistent with this interpretation is that chromatin association of YB-1 is apparently lost upon treatment with ribonucleases, which destroys pre-mRNAs.138 It is well established that coactivators, which do not bind to DNA directly, can be crosslinked efficiently to DNA, resulting in peaks in ChIPs and ChIP-Seq. It is also known that promoter structures and specific TF combinations have an impact on the loading and composition of the mRNA splicing apparatus.139, 140 Thus, we propose that rather than being enemies battling over the same DNA sequence, NF-Y and YB-1 take on different tasks, cooperating to alter gene expression in cancer cells: transcriptional initiation through Y/CCAAT sequence-specific binding the former, and post-transcriptional mechanisms through RNA binding the latter.
Figure 5.
Scheme of mechanisms of gene expression control by NF-Y and YB-1
Acknowledgments
We thank the members of the lab for helpful discussions. Our special thanks to David Horner for his patience and dedication in reviewing the manuscript. The work is supported by the Regione Lombardia NEPENTE Grant.
Glossary
- NF-Y
nuclear factor Y
- YB-1
Y box-binding protein 1
- MDR1
multidrug resistance protein 1
- TFBS
transcription factor-binding sites
- TF
transcription factor
- CBF
CCAAT-binding factor
- MHC
major histocompatibility complex
- EMSA
electrophoretic mobility shift assay
- HFDs
histone fold domains
- RSV LTR
Rous sarcoma virus long terminal repeat
- ChIP
chromatin immunoprecipitation
- HSP70
heat-shock 70 kDa protein
- PSFM
positional sequence frequency matrix
- siRNA
small interfering RNA
- shRNA
short hairpin RNA
- CSD
cold-shock domain
- ssDNA
single-stranded DNA
- dsDNA
double-stranded DNA
- DBD
DNA-binding domain
- TA
transcriptional activation
The authors declare no conflict of interest.
Footnotes
Edited by G Melino
References
- Mathis DJ, Benoist CO, Williams VE, Kanter MR, McDevitt HO. The murine E alpha immune response gene. Cell. 1983;32:745–754. doi: 10.1016/0092-8674(83)90060-0. [DOI] [PubMed] [Google Scholar]
- Benoist C, Mathis D. Regulation of major histocompatibility complex class-II genes: X, Y and other letters of the alphabet. Annu Rev Immunol. 1990;8:681–715. doi: 10.1146/annurev.iy.08.040190.003341. [DOI] [PubMed] [Google Scholar]
- Efstratiadis A, Posakony JW, Maniatis T, Lawn RM, O'Connell C, Spritz RA, et al. The structure and evolution of the human beta-globin gene family. Cell. 1980;21:653–668. doi: 10.1016/0092-8674(80)90429-8. [DOI] [PubMed] [Google Scholar]
- Grosveld GC, Shewmaker CK, Jat P, Flavell RA. Localization of DNA sequences necessary for transcription of the rabbit beta-globin gene in vitro. Cell. 1981;25:215–226. doi: 10.1016/0092-8674(81)90246-4. [DOI] [PubMed] [Google Scholar]
- Myers RM, Tilly K, Maniatis T. Fine structure genetic analysis of a beta-globin promoter. Science. 1986;232:613–618. doi: 10.1126/science.3457470. [DOI] [PubMed] [Google Scholar]
- Maity SN, Golumbek PT, Karsenty G, de Crombrugghe B. Selective activation of transcription by a novel CCAAT binding factor. Science. 1988;241:582–585. doi: 10.1126/science.3399893. [DOI] [PubMed] [Google Scholar]
- Kim CG, Sheffery M. Physical characterization of the purified CCAAT transcription factor, alpha-CP1. J Biol Chem. 1990;265:13362–13369. [PubMed] [Google Scholar]
- Greuel BT, Sealy L, Majors JE. Transcriptional activity of the Rous sarcoma virus long terminal repeat correlates with binding of a factor to an upstream CCAAT box in vitro. Virology. 1990;177:33–43. doi: 10.1016/0042-6822(90)90457-3. [DOI] [PubMed] [Google Scholar]
- Viville S, Jongeneel V, Koch W, Mantovani R, Benoist C, Mathis D. The E alpha promoter: a linker-scanning analysis. J Immunol. 1991;146:3211–3217. [PubMed] [Google Scholar]
- Tronche F, Rollier A, Sourdive D, Cereghini S, Yaniv M. NFY or a related CCAAT binding factor can be replaced by other transcriptional activators for co-operation with HNF1 in driving the rat albumin promoter in vivo. J Mol Biol. 1991;222:31–43. doi: 10.1016/0022-2836(91)90735-o. [DOI] [PubMed] [Google Scholar]
- Zeleznik-Le NJ, Azizkhan JC, Ting JP. Affinity-purified CCAAT-box-binding protein (YEBP) functionally regulates expression of a human class II major histocompatibility complex gene and the herpes simplex virus thymidine kinase gene. Proc Natl Acad Sci USA. 1991;88:1873–1877. doi: 10.1073/pnas.88.5.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Tsunoda T, Sese J, Taira H, Mizushima-Sugano J, Hata H, et al. Identification and characterization of the potential promoter regions of 1031 kinds of human genes. Genome Res. 2001;11:677–684. doi: 10.1101/gr.164001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Yamashita R, Shirota M, Sakakibara Y, Chiba J, Mizushima-Sugano J, et al. Sequence comparison of human and mouse genes reveals a homologous block structure in the promoter regions. Genome Res. 2004;14:1711–1718. doi: 10.1101/gr.2435604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkon R, Linhart C, Sharan R, Shamir R, Shiloh Y. Genome-wide in silico identification of transcriptional regulators controlling the cell cycle in human cells. Genome Res. 2003;13:773–780. doi: 10.1101/gr.947203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linhart C, Elkon R, Shiloh Y, Shamir R. Deciphering transcriptional regulatory elements that encode specific cell cycle phasing by comparative genomics analysis. Cell Cycle. 2005;4:1788–1797. doi: 10.4161/cc.4.12.2173. [DOI] [PubMed] [Google Scholar]
- Zhu Z, Shendure J, Church GM. Discovering functional transcription–factor combinations in the human cell cycle. Genome Res. 2005;15:848–855. doi: 10.1101/gr.3394405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grskovic M, Chaivorapol C, Gaspar-Maia A, Li H, Ramalho-Santos M. Systematic identification of cis-regulatory sequences active in mouse and human embryonic stem cells. PLoS Genet. 2007;3:e145. doi: 10.1371/journal.pgen.0030145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halperin Y, Linhart C, Ulitsky I, Shamir R. Allegro: analyzing expression and sequence in concert to discover regulatory programs. Nucleic Acids Res. 2009;37:1566–1579. doi: 10.1093/nar/gkn1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häkkinen A, Healy S, Jacobs HT, Ribeiro AS. Genome wide study of NF-Y type CCAAT boxes in unidirectional and bidirectional promoters in human and mouse. J Theor Biol. 2011;281:74–83. doi: 10.1016/j.jtbi.2011.04.027. [DOI] [PubMed] [Google Scholar]
- Mariño-Ramírez L, Spouge JL, Kanga GC, Landsman D. Statistical analysis of over-represented words in human promoter sequences. Nucleic Acids Res. 2004;32:949–958. doi: 10.1093/nar/gkh246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald PC, Shlyakhtenko A, Mir AA, Vinson C. Clustering of DNA sequences in human promoters. Genome Res. 2004;14:1562–1574. doi: 10.1101/gr.1953904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabach Y, Milyavsky M, Shats I, Brosh R, Zuk O, Yitzhaky A, et al. The promoters of human cell cycle genes integrate signals from two tumor suppressive pathways during cellular transformation. Mol Syst Biol. 2005;1:0022. doi: 10.1038/msb4100030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvatore G, Nappi TC, Salerno P, Jiang Y, Garbi C, Ugolini C, et al. A cell proliferation and chromosomal instability signature in anaplastic thyroid carcinoma. Cancer Res. 2007;67:10148–10158. doi: 10.1158/0008-5472.CAN-07-1887. [DOI] [PubMed] [Google Scholar]
- Scafoglio C, Ambrosino C, Cicatiello L, Altucci L, Ardovino M, Bontempo P, et al. Comparartive gene expression profiling reveals partially overlapping but distinct genomic actions of different antiestrogens in human breast cancer cells. J Cell Biochem. 2006;98:1163–1184. doi: 10.1002/jcb.20820. [DOI] [PubMed] [Google Scholar]
- Niida A, Smith AD, Imoto S, Tsutsumi S, Aburatani H, Zhang MQ, et al. Integrative bioinformatics analysis of transcriptional regulatory programs in breast cancer cells. BMC Bioinform. 2008;9:404. doi: 10.1186/1471-2105-9-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomassen M, Tan Q, Kruse TA. Gene expression meta-analysis identifies metastatic pathways and transcription factors in cancer. BMC Cancer. 2008;8:394. doi: 10.1186/1471-2407-8-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum R, Gupta R, Burger PE, Ontiveros CS, Salm SN, Xiong X, et al. Molecular signatures of prostate stem cells reveal novel signaling pathways and provide insights into prostate cancer. PLoS One. 2009;4:e5722. doi: 10.1371/journal.pone.0005722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo A, Perez-Stable C, Segura V, Catena R, Guruceaga E, Nguewa P, et al. Molecular characterization of the Ggamma-globin-Tag transgenic mouse model of hormone refractory prostate cancer: comparison to human prostate cancer. Prostate. 2010;70:630–645. doi: 10.1002/pros.21097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsberg EC, Passegué E, Prohaska SS, Wagers AJ, Koeva M, Stuart JM, et al. Molecular signatures of quiescent, mobilized and leukemia-initiating hematopoietic stem cells. PLoS One. 2010;5:e8785. doi: 10.1371/journal.pone.0008785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum R, Elkon R, Yaari S, Zundelevich A, Jacob-Hirsch J, Rechavi G, et al. Gene expression signature of human cancer cell lines treated with the ras inhibitor salirasib (S-farnesylthiosalicylic acid) Cancer Res. 2007;67:3320–3328. doi: 10.1158/0008-5472.CAN-06-4287. [DOI] [PubMed] [Google Scholar]
- Yamanaka K, Mizuarai S, Eguchi T, Itadani H, Hirai H, Kotani H. Expression levels of NF-Y target genes changed by CDKN1B correlate with clinical prognosis in multiple cancers. Genomics. 2009;94:219–227. doi: 10.1016/j.ygeno.2009.06.003. [DOI] [PubMed] [Google Scholar]
- Goodarzi H, Elemento O, Tavazoie S. Revealing global regulatory perturbations across human cancers. Mol Cell. 2009;36:900–911. doi: 10.1016/j.molcel.2009.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Barrette TR, Ghosh D, Chinnaiyan AM. Mining for regulatory programs in the cancer transcriptome. Nat Genet. 2005;37:579–583. doi: 10.1038/ng1578. [DOI] [PubMed] [Google Scholar]
- Sinha S, Adler AS, Field Y, Chang HY, Segal E. Systematic functional characterization of cis-regulatory motifs in human core promoters. Genome Res. 2008;18:477–488. doi: 10.1101/gr.6828808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn A, Bollekens J, Staub A, Benoist C, Mathis D. A multiplicity of CCAAT box-binding proteins. Cell. 1987;50:863–672. doi: 10.1016/0092-8674(87)90513-7. [DOI] [PubMed] [Google Scholar]
- Osada S, Matsubara T, Daimon S, Terazu Y, Xu M, Nishihara T, et al. Expression, DNA-binding specificity and transcriptional regulation of nuclear factor 1 family proteins from rat. Biochem J. 1999;342:189–198. [PMC free article] [PubMed] [Google Scholar]
- Osada S, Yamamoto H, Nishihara T, Imagawa M. DNA binding specificity of the CCAAT/enhancer-binding protein transcription factor family. J Biol Chem. 1996;271:3891–3896. doi: 10.1074/jbc.271.7.3891. [DOI] [PubMed] [Google Scholar]
- Roulet E, Busso S, Camargo AA, Simpson AJ, Mermod N, Bucher P. High-throughput SELEX SAGE method for quantitative modeling of transcription-factor binding sites. Nat Biotechnol. 2002;20:831–835. doi: 10.1038/nbt718. [DOI] [PubMed] [Google Scholar]
- Hooft van Huijsduijnen RA, Bollekens J, Dorn A, Benoist C, Mathis D. Properties of a CCAAT box-binding protein. Nucleic Acids Res. 1987;15:7265–7282. doi: 10.1093/nar/15.18.7265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber M, Sealy L. Rous sarcoma virus enhancer factor I is a ubiquitous CCAAT transcription factor highly related to CBF and NF-Y. J Biol Chem. 1990;265:22243–22254. [PubMed] [Google Scholar]
- Chodosh LA, Olesen J, Hahn S, Baldwin AS, Guarente L, Sharp PA. A yeast and a human CCAAT-binding protein have heterologous subunits that are functionally interchangeable. Cell. 1988;53:25–35. doi: 10.1016/0092-8674(88)90484-9. [DOI] [PubMed] [Google Scholar]
- Hatamochi A, Golumbek PT, Maity SN, de Crombrugghe B. A CCAAT DNA binding factor consisting of two different components that are both required for DNA binding. J Biol Chem. 1988;263:5940–5947. [PubMed] [Google Scholar]
- Vuorio T, Maity SN, de Crombrugghe B. Purification and molecular cloning of the ‘A' chain of a rat heteromeric CCAAT-binding protein. Sequence identity with the yeast HAP3 transcription factor. J Biol Chem. 1990;265:22480–22486. [PubMed] [Google Scholar]
- Maity SN, Vuorio T, de Crombrugghe B. The B subunit of a rat heteromeric CCAAT-binding transcription factor shows a striking sequence identity with the yeast Hap2 transcription factor. Proc Natl Acad Sci USA. 1990;87:5378–5382. doi: 10.1073/pnas.87.14.5378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooft van Huijsduijnen R, Li XY, Black D, Matthes H, Benoist C, Mathis D. Co-evolution from yeast to mouse: cDNA cloning of the two NF-Y (CP-1/CBF) subunits. EMBO J. 1990;9:3119–3127. doi: 10.1002/j.1460-2075.1990.tb07509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romier C, Cocchiarella F, Mantovani R, Moras D. The NF-YB/NF-YC structure gives insight into DNA binding and transcription regulation by CCAAT factor NF-Y. J Biol Chem. 2003;278:1336–1345. doi: 10.1074/jbc.M209635200. [DOI] [PubMed] [Google Scholar]
- Bi W, Wu L, Coustry F, de Crombrugghe B, Maity SN. DNA binding specificity of the CCAAT-binding factor CBF/NF-Y. J Biol Chem. 1997;272:26562–26572. doi: 10.1074/jbc.272.42.26562. [DOI] [PubMed] [Google Scholar]
- Mantovani R, Pessara U, Tronche F, Li XY, Knapp AM, Pasquali JL, et al. Monoclonal antibodies to NF-Y define its function in MHC class II and albumin gene transcription. EMBO J. 1992;11:3315–3322. doi: 10.1002/j.1460-2075.1992.tb05410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coustry F, Maity SN, de Crombrugghe B. Studies on transcription activation by the multimeric CCAAT-binding factor CBF. J Biol Chem. 1995;270:468–475. doi: 10.1074/jbc.270.1.468. [DOI] [PubMed] [Google Scholar]
- Coustry F, Hu Q, de Crombrugghe B, Maity SN. CBF/NF-Y functions both in nucleosomal disruption and transcription activation of the chromatin-assembled topoisomerase IIalpha promoter. Transcription activation by CBF/NF-Y in chromatin is dependent on the promoter structure. J Biol Chem. 2001;276:40621–40630. doi: 10.1074/jbc.M106918200. [DOI] [PubMed] [Google Scholar]
- Coustry F, Maity SN, Sinha S, de Crombrugghe B. The transcriptional activity of the CCAAT-binding factor CBF is mediated by two distinct activation domains, one in the CBF-B subunit and the other in the CBF-C subunit. J Biol Chem. 2006;271:14485–14491. doi: 10.1074/jbc.271.24.14485. [DOI] [PubMed] [Google Scholar]
- Bennett MK, Osborne TF. Nutrient regulation of gene expression by the sterol regulatory element binding proteins: increased recruitment of gene-specific coregulatory factors and selective hyperacetylation of histone H3 in vivo. Proc Natl Acad Sci USA. 2000;97:6340–6344. doi: 10.1073/pnas.97.12.6340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani R. A survey of 178 NF-Y binding CCAAT boxes. Nucleic Acids Res. 1998;26:1135–1143. doi: 10.1093/nar/26.5.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani R, Li XY, Pessara U, Hooft van HR, Benoist C, Mathis D. Dominant negative analogs of NF-YA. J Biol Chem. 1994;269:20340–20346. [PubMed] [Google Scholar]
- Dolfini D, Zambelli F, Pavesi G, Mantovani R. A perspective of promoter architecture from the CCAAT box. Cell Cycle. 2009;8:4127–4137. doi: 10.4161/cc.8.24.10240. [DOI] [PubMed] [Google Scholar]
- Testa A, Donati G, Yan P, Romani F, Huang TH, Viganò MA, et al. Chromatin immunoprecipitation (ChIP) on chip experiments uncover a widespread distribution of NF-Y binding CCAAT sites outside of core promoters. J Biol Chem. 2005;280:13606–13615. doi: 10.1074/jbc.M414039200. [DOI] [PubMed] [Google Scholar]
- Ceribelli M, Alcalay M, Vigano MA, Mantovani R. Repression of new p53 targets revealed by ChIP on chip experiments. Cell Cycle. 2006;5:1102–1110. doi: 10.4161/cc.5.10.2777. [DOI] [PubMed] [Google Scholar]
- Ceribelli M, Dolfini D, Merico D, Gatta R, Viganò AM, Pavesi G, et al. The histone-like NF-Y is a bifunctional transcription factor. Mol Cell Biol. 2008;28:2047–2058. doi: 10.1128/MCB.01861-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed BD, Charos AE, Szekely AM, Weissman SM, Snyder M. Genome-wide occupancy of SREBP1 and its partners NFY and SP1 reveals novel functional roles and combinatorial regulation of distinct classes of genes. PLoS Genet. 2008;4:e1000133. doi: 10.1371/journal.pgen.1000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhuang J, Iyer S, Lin X, Whitfield TW, Greven MC, et al. Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors. Genome Res. 2012;22:1798–1812. doi: 10.1101/gr.139105.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didier DK, Schiffenbauer J, Woulfe SL, Zacheis M, Schwartz BD. Characterization of the cDNA encoding a protein binding to the major histocompatibility complex class II Y box. Proc Natl Acad Sci USA. 1988;85:7322–7326. doi: 10.1073/pnas.85.19.7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozer J, Faber M, Chalkley R, Sealy L. Isolation and characterization of a cDNA clone for the CCAAT transcription factor EFIA reveals a novel structural motif. J Biol Chem. 1990;265:22143–22152. [PubMed] [Google Scholar]
- Tafuri SR, Wolffe AP. Xenopus Y-box transcription factors: molecular cloning, functional analysis and developmental regulation. Proc Natl Acad Sci USA. 1990;87:9028–9032. doi: 10.1073/pnas.87.22.9028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant CE, Deeley RG. Cloning and characterization of chicken YB-1: regulation of expression in the liver. Mol Cell Biol. 1993;13:4186–4196. doi: 10.1128/mcb.13.7.4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihailovich M, Militti C, Gabaldón T, Gebauer F. Eukaryotic cold shock domain proteins: highly versatile regulators of gene expression. Bioessays. 2010;32:109–118. doi: 10.1002/bies.200900122. [DOI] [PubMed] [Google Scholar]
- Hasegawa SL, Doetsch PW, Hamilton KK, Martin AM, Okenquist SA, Lenz J, et al. DNA binding properties of YB-1 and dbpA: binding to double-stranded, single-stranded, and abasic site containing DNAs. Nucleic Acids Res. 1991;19:4915–4920. doi: 10.1093/nar/19.18.4915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray MT, Schiller DL, Franke WW. Sequence analysis of cytoplasmic mRNA-binding proteins of Xenopus oocytes identifies a family of RNA-binding proteins. Proc Natl Acad Sci USA. 1992;89:11–15. doi: 10.1073/pnas.89.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minich WB, Ovchinnikov LP. Role of cytoplasmic mRNP proteins in translation. Biochimie. 1992;74:477–483. doi: 10.1016/0300-9084(92)90088-v. [DOI] [PubMed] [Google Scholar]
- Bouvet P, Wolffe AP. A role for transcription and FRGY2 in masking maternal mRNA within Xenopus oocytes. Cell. 1994;77:931–941. doi: 10.1016/0092-8674(94)90141-4. [DOI] [PubMed] [Google Scholar]
- Evdokimova V, Ovchinnikov LP, Sorensen PH. Y-box binding protein 1: providing a new angle on translational regulation. Cell Cycle. 2006;5:1143–1147. doi: 10.4161/cc.5.11.2784. [DOI] [PubMed] [Google Scholar]
- Eliseeva IA, Kim ER, Guryanov SG, Ovchinnikov LP, Lyabin DN. Y-box-binding protein 1 (YB-1) and its functions. Biochemistry. 2011;76:1402–1433. doi: 10.1134/S0006297911130049. [DOI] [PubMed] [Google Scholar]
- Bouvet P, Matsumoto K, Wolffe AP. Sequence-specific RNA recognition by the Xenopus Y-box proteins. An essential role for the cold shock domain. J Biol Chem. 1995;270:28297–28303. doi: 10.1074/jbc.270.47.28297. [DOI] [PubMed] [Google Scholar]
- Zasedateleva OA, Krylov AS, Prokopenko DV, Skabkin MA, Ovchinnikov LP, Kolchinsky A, et al. Specificity of mammalian Y-box binding protein p50 in interaction with ss and ds DNA analyzed with generic oligonucleotide microchip. J Mol Biol. 2002;324:73–87. doi: 10.1016/s0022-2836(02)00937-3. [DOI] [PubMed] [Google Scholar]
- Ray D, Kazan H, Chan ET, Peña Castillo L, Chaudhry S, Talukder S, et al. Rapid and systematic analysis of the RNA recognition specificities of RNA-binding proteins. Nat Biotechnol. 2009;27:667–670. doi: 10.1038/nbt.1550. [DOI] [PubMed] [Google Scholar]
- Liberati C, di Silvio A, Ottolenghi S, Mantovani R. NF-Y binding to twin CCAAT boxes: role of Q-rich domains and histone fold helices. J Mol Biol. 1999;285:1441–1455. doi: 10.1006/jmbi.1998.2384. [DOI] [PubMed] [Google Scholar]
- Valadão AF, Fantappie MR, LoVerde PT, Pena SD, Rumjanek FD, Franco GR. Y-box binding protein from Schistosoma mansoni: interaction with DNA and RNA. Mol Biochem Parasitol. 2002;125:47–57. doi: 10.1016/s0166-6851(02)00210-4. [DOI] [PubMed] [Google Scholar]
- Mertens PR, Harendza S, Pollock AS, Lovett DH. Glomerular mesangial cell-specific transactivation of matrix metalloproteinase 2 transcription is mediated by YB-1. J Biol Chem. 1997;272:22905–22912. doi: 10.1074/jbc.272.36.22905. [DOI] [PubMed] [Google Scholar]
- Li XY, Hooft van Huijsduijnen R, Mantovani R, Benoist C, Mathis D. Intron–exon organization of the NF-Y genes. Tissue-specific splicing modifies an activation domain. J Biol Chem. 1992;267:8984–8990. [PubMed] [Google Scholar]
- di Silvio A, Imbriano C, Mantovani R. Dissection of the NF-Y transcriptional activation potential. Nucleic Acids Res. 1999;27:2578–2584. doi: 10.1093/nar/27.13.2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q, Maity SN. Stable expression of a dominant negative mutant of CCAAT binding factor/NF-Y in mouse fibroblast cells resulting in retardation of cell growth and inhibition of transcription of various cellular genes. J Biol Chem. 2000;275:4435–4444. doi: 10.1074/jbc.275.6.4435. [DOI] [PubMed] [Google Scholar]
- Caretti G, Cocchiarella F, Sidoli C, Villard J, Peretti M, Reith W, et al. Dissection of functional NF-Y-RFX cooperative interactions on the MHC class II Ea promoter. J Mol Biol. 2000;302:539–552. doi: 10.1006/jmbi.2000.4028. [DOI] [PubMed] [Google Scholar]
- Ge Y, Jensen TL, Matherly LH, Taub JW. Synergistic regulation of human cystathionine-beta-synthase-1b promoter by transcription factors NF-YA isoforms and Sp1. Biochim Biophys Acta. 2002;1579:73–80. doi: 10.1016/s0167-4781(02)00509-2. [DOI] [PubMed] [Google Scholar]
- Domashenko AD, Danet-Desnoyers G, Aron A, Carroll MP, Emerson SG. TAT-mediated transduction of NF-Ya peptide induces the ex vivo proliferation and engraftment potential of human hematopoietic progenitor cells. Blood. 2010;116:2676–2683. doi: 10.1182/blood-2010-03-273441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolfini D, Minuzzo M, Pavesi G, Mantovani R. The short isoform of NF-YA belongs to the ES cell transcription factor circuitry. Stem Cells. 2012;30:2450–2459. doi: 10.1002/stem.1232. [DOI] [PubMed] [Google Scholar]
- Song B, Young CS. Functional analysis of the CAAT box in the major late promoter of the subgroup C human adenoviruses. J Virol. 1998;72:3213–3220. doi: 10.1128/jvi.72.4.3213-3220.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibinga NE, Wang H, Perrella MA, Endege WO, Patterson C, Yoshizumi M, et al. Interferon-gamma-mediated inhibition of cyclin A gene transcription is independent of individual cis-acting elements in the cyclin A promoter. J Biol Chem. 1999;274:12139–12146. doi: 10.1074/jbc.274.17.12139. [DOI] [PubMed] [Google Scholar]
- Nenoi M, Ichimura S, Mita K, Yukawa O, Cartwright IL. Regulation of the catalase gene promoter by Sp1, CCAAT-recognizing factors, and a WT1/Egr-related factor in hydrogen peroxide-resistant HP100 cells. Cancer Res. 2001;61:5885–5894. [PubMed] [Google Scholar]
- Grimm M, Spiecker M, De Caterina R, Shin WS, Liao JK. Inhibition of major histocompatibility complex class II gene transcription by nitric oxide and antioxidants. J Biol Chem. 2002;277:26460–26467. doi: 10.1074/jbc.M110538200. [DOI] [PubMed] [Google Scholar]
- Lagor WR, de Groh ED, Ness GC. Diabetes alters the occupancy of the hepatic 3-hydroxy-3-methylglutaryl-CoA reductase promoter. J Biol Chem. 2005;280:36601–36608. doi: 10.1074/jbc.M504346200. [DOI] [PubMed] [Google Scholar]
- Leclerc GJ, Leclerc GM, Kinser TT, Barredo JC. Analysis of folylpoly-gamma-glutamate synthetase gene expression in human B-precursor ALL and T-lineage ALL cells. BMC Cancer. 2006;6:132. doi: 10.1186/1471-2407-6-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landsberger N, Wolffe AP. Role of chromatin and Xenopus laevis heat shock transcription factor in regulation of transcription from the X. laevis hsp70 promoter in vivo. Mol Cell Biol. 1995;15:6013–6024. doi: 10.1128/mcb.15.11.6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Herrler M, Landsberger N, Kaludov N, Ogryzko VV, Nakatani Y, et al. Xenopus NF-Y pre-sets chromatin to potentiate p300 and acetylation-responsive transcription from the Xenopus hsp70 promoter in vivo. EMBO J. 1998;17:6300–6315. doi: 10.1093/emboj/17.21.6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolfini D, Gatta R, Mantovani R. NF-Y and the transcriptional activation of CCAAT promoters. Crit Rev Biochem Mol Biol. 2012;47:29–49. doi: 10.3109/10409238.2011.628970. [DOI] [PubMed] [Google Scholar]
- Stein U, Jürchott K, Walther W, Bergmann S, Schlag PM, Royer HD. Hyperthermia-induced nuclear translocation of transcription factor YB-1 leads to enhanced expression of multidrug resistance-related ABC transporters. J Biol Chem. 2001;276:28562–28569. doi: 10.1074/jbc.M100311200. [DOI] [PubMed] [Google Scholar]
- Scotto KW. Transcriptional regulation of ABC drug transporters. Oncogene. 2003;22:7496–7511. doi: 10.1038/sj.onc.1206950. [DOI] [PubMed] [Google Scholar]
- Asakuno K, Kohno K, Uchiumi T, Kubo T, Sato S, Isono M, et al. Involvement of a DNA binding protein, MDR-NF1/YB-1, in human MDR1 gene expression by actinomycin D. Biochem Biophys Res Commun. 1994;199:1428–1435. doi: 10.1006/bbrc.1994.1390. [DOI] [PubMed] [Google Scholar]
- Ohga T, Koike K, Ono M, Makino Y, Itagaki Y, Tanimoto M, et al. Role of the human Y box-binding protein YB-1 in cellular sensitivity to the DNA-damaging agents cisplatin, mitomycin C, and ultraviolet light. Cancer Res. 1996;56:4224–4228. [PubMed] [Google Scholar]
- Ohga T, Uchiumi T, Makino Y, Koike K, Wada M, Kuwano M, et al. Direct involvement of the Y-box binding protein YB-1 in genotoxic stress-induced activation of the human multidrug resistance 1 gene. J Biol Chem. 1998;273:5997–6000. doi: 10.1074/jbc.273.11.5997. [DOI] [PubMed] [Google Scholar]
- Bargou RC, Jurchott K, Wagener C, Bergmann S, Metzner S, Bommert K, et al. Nuclear localization and increased levels of transcription factor YB-1 in primary human breast cancers are associated with intrinsic MDR1 gene expression. Nat Med. 1997;3:447–450. doi: 10.1038/nm0497-447. [DOI] [PubMed] [Google Scholar]
- Vaiman AV, Stromskaya TP, Rybalkina EY, Sorokin AV, Guryanov SG, Zabotina TN, et al. Intracellular localization and content of YB-1 protein in multidrug resistant tumor cells. Biochemistry. 2006;71:146–154. doi: 10.1134/s0006297906020052. [DOI] [PubMed] [Google Scholar]
- Kaszubiak A, Kupstat A, Müller U, Hausmann R, Holm PS, Lage H. Regulation of MDR1 gene expression in multidrug-resistant cancer cells is independent from YB-1. Biochem Biophys Res Commun. 2007;357:295–301. doi: 10.1016/j.bbrc.2007.03.145. [DOI] [PubMed] [Google Scholar]
- Goldsmith ME, Madden MJ, Morrow CS, Cowan. KH. A Y-box consensus sequence is required for basal expression of the human multidrug resistance (mdr1) gene. J Biol Chem. 1993;268:5856–5860. [PubMed] [Google Scholar]
- Sundseth R, MacDonald G, Ting J, King AC. DNA elements recognizing NF-Y and Sp1 regulate the human multidrug-resistance gene promoter. Mol Pharmacol. 1997;51:963–971. doi: 10.1124/mol.51.6.963. [DOI] [PubMed] [Google Scholar]
- Jin S, Scotto KW. Transcriptional regulation of the MDR1 gene by histone acetyltransferase and deacetylase is mediated by NF-Y. Mol Cell Biol. 1998;18:4377–4384. doi: 10.1128/mcb.18.7.4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Jin S, Scotto KW. Transcriptional activation of the MDR1 gene by UV irradiation. Role of NF-Y and Sp1. J Biol Chem. 2000a;275:2979–2985. doi: 10.1074/jbc.275.4.2979. [DOI] [PubMed] [Google Scholar]
- Tanaka H, Ohshima N, Ikenoya M, Komori K, Katoh F, Hidaka H. HMN-176 an active metabolite of the synthetic antitumor agent HMN-214, restores chemosensitivity to multidrug-resistant cells by targeting the transcription factor NF-Y. Cancer Res. 2003;63:6942–6947. [PubMed] [Google Scholar]
- Okamura H, Yoshida K, Sasaki E, Morimoto H, Haneji T. Transcription factor NF-Y regulates mdr1 expression through binding to inverted CCAAT sequence in drug-resistant human squamous carcinoma cells. Int J Oncol. 2004;25:1031–1037. [PubMed] [Google Scholar]
- Huo H, Magro PG, Pietsch EC, Patel BB, Scotto KW. Histone methyltransferase MLL1 regulates MDR1 transcription and chemoresistance. Cancer Res. 2010;70:8726–8735. doi: 10.1158/0008-5472.CAN-10-0755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Jiang Z, Yin P, Li Q, Liu J. Role for class I histone deacetylases in multidrug resistance. Exp Cell Res. 2012;318:177–186. doi: 10.1016/j.yexcr.2011.11.010. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay R, Das S, Maiti AK, Boldogh I, Xie J, Hazra TK, et al. Regulatory role of human AP-endonuclease (APE1/Ref-1) in YB-1-mediated activation of the multidrug resistance gene MDR1. Mol Cell Biol. 2008;28:7066–7080. doi: 10.1128/MCB.00244-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S, Mantha AK, Mitra S, Bhakat KK. Human AP endonuclease (APE1/Ref-1) and its acetylation regulate YB-1-p300 recruitment and RNA polymerase II loading in the drug-induced activation of multidrug resistance gene MDR1. Oncogene. 2011;30:482–493. doi: 10.1038/onc.2010.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakshatri H, Bhat-Nakshatri P, Currie RA. Subunit association and DNA binding activity of the heterotrimeric transcription factor NF-Y is regulated by cellular redox. J Biol Chem. 1996;271:28784–28791. doi: 10.1074/jbc.271.46.28784. [DOI] [PubMed] [Google Scholar]
- Kauffmann HM, Vorderstemann B, Schrenk D. Basal expression of the rat, but not of the human, multidrug resistance protein 2 (MRP2) gene is mediated by CBF/NF-Y and Sp1 promoter-binding sites. Toxicology. 2001;167:25–35. doi: 10.1016/s0300-483x(01)00455-3. [DOI] [PubMed] [Google Scholar]
- Geier A, Mertens PR, Gerloff T, Dietrich CG, En-Nia A, Kullak-Ublick GA, et al. Constitutive rat multidrug-resistance protein 2 gene transcription is down-regulated by Y-box protein 1. Biochem Biophys Res Commun. 2003;309:612–618. doi: 10.1016/j.bbrc.2003.08.041. [DOI] [PubMed] [Google Scholar]
- En-Nia A, Yilmaz E, Klinge U, Lovett DH, Stefanidis I, Mertens PR. Transcription factor YB-1 mediates DNA polymerase alpha gene expression. J Biol Chem. 2005;280:7702–7711. doi: 10.1074/jbc.M413353200. [DOI] [PubMed] [Google Scholar]
- Jurchott K, Bergmann S, Stein U, Walther W, Janz M, Manni I, et al. YB-1 as a cell cycle-regulated transcription factor facilitating cyclin A and cyclin B1 gene expression. J Biol Chem. 2003;278:27988–27996. doi: 10.1074/jbc.M212966200. [DOI] [PubMed] [Google Scholar]
- Farina A, Manni I, Fontemaggi G, Tiainen M, Cenciarelli C, Bellorini M, et al. Down-regulation of cyclin B1 gene transcription in terminally differentiated skeletal muscle cells is associated with loss of functional CCAAT-binding NF-Y complex. Oncogene. 1999;18:2818–2827. doi: 10.1038/sj.onc.1202472. [DOI] [PubMed] [Google Scholar]
- Gurtner A, Manni I, Fuschi P, Mantovani R, Guadagni F, Sacchi A, et al. Requirement for down-regulation of the CCAAT-binding activity of the NF-Y transcription factor during skeletal muscle differentiation. Mol Biol Cell. 2003;14:2706–2715. doi: 10.1091/mbc.E02-09-0600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Lee C, Yokom D, Jiang H, Cheang MC, Yorida E, et al. Disruption of the Y-box binding protein-1 results in suppression of the epidermal growth factor receptor and HER-2. Cancer Res. 2006;66:4872–4879. doi: 10.1158/0008-5472.CAN-05-3561. [DOI] [PubMed] [Google Scholar]
- Astanehe A, Finkbeiner MR, Hojabrpour P, To K, Fotovati A, Shadeo A, et al. The transcriptional induction of PIK3CA in tumor cells is dependent on the oncoprotein Y-box binding protein-1. Oncogene. 2009;28:2406–2418. doi: 10.1038/onc.2009.81. [DOI] [PubMed] [Google Scholar]
- Finkbeiner MR, Astanehe A, To K, Fotovati A, Davies AH, Zhao Y, et al. Profiling YB-1 target genes uncovers a new mechanism for MET receptor regulation in normal and malignant human mammary cells. Oncogene. 2009;28:1421–1431. doi: 10.1038/onc.2008.485. [DOI] [PubMed] [Google Scholar]
- To K, Fotovati A, Reipas KM, Law JH, Hu K, Wang J, et al. Y-box binding protein-1 induces the expression of CD44 and CD49f leading to enhanced self-renewal, mammosphere growth, and drug resistance. Cancer Res. 2010;70:2840–2851. doi: 10.1158/0008-5472.CAN-09-3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasham A, Samuel W, Cao H, Patel R, Mehta R, Stern JL, et al. YB-1, the E2F pathway, and regulation of tumor cell growth. J Natl Cancer Inst. 2012;104:133–146. doi: 10.1093/jnci/djr512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benatti P, Dolfini D, Viganò MA, Ravo M, Weisz A, Imbriano C. Specific inhibition of NF-Y subunits triggers different cell proliferation defects. Nucleic Acids Res. 2011;39:5356–5368. doi: 10.1093/nar/gkr128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benatti P, Basile V, Merico D, Fantoni LI, Tagliafico E, Imbriano C. A balance between NF-Y and p53 governs the pro- and anti-apoptotic transcriptional response. Nucleic Acids Res. 2008;36:1415–1428. doi: 10.1093/nar/gkm1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astanehe A, Finkbeiner MR, Krzywinski M, Fotovati A, Dhillon J, Berquin IM, et al. MKNK1 is a YB-1 target gene responsible for imparting trastuzumab resistance and can be blocked by RSK inhibition. Oncogene. 2012;31:4434–4436. doi: 10.1038/onc.2011.617. [DOI] [PubMed] [Google Scholar]
- Dolfini D, Mantovani R.YB-1 (YBX1) does not bind to the Y/CCAAT box in vivo Oncogene 2012. e-pub ahead of print 19 November 2012; doi: 10.1038/onc.2012.521 [DOI]
- Frontini M, Imbriano C, Manni I, Mantovani R. Cell cycle regulation of NF-YC nuclear localization. Cell Cycle. 2004;3:217–222. [PubMed] [Google Scholar]
- Nardini M, Gnesutta N, Donati G, Gatta R, Forni C, Fossati A, et al. NF-Y is a sequence-specific transcription factor displaying histone-like DNA binding and H2B-like ubiquitination. Cell. 2013;152:132–143. doi: 10.1016/j.cell.2012.11.047. [DOI] [PubMed] [Google Scholar]
- Davies AH, Dunn SE. YB-1 drives preneoplastic progression: insight into opportunities for cancer prevention. Oncotarget. 2011;2:401–406. doi: 10.18632/oncotarget.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbriano C, Gnesutta N, Mantovani R. The NF-Y/p53 liaison: well beyond repression. BBA Rev Cancer. 2012;1825:131–139. doi: 10.1016/j.bbcan.2011.11.001. [DOI] [PubMed] [Google Scholar]
- Tiwari VK, Stadler MB, Wirbelauer C, Paro R, Schübeler D, Beisel C. A chromatin-modifying function of JNK during stem cell differentiation. Nat Genet. 2011;44:94–100. doi: 10.1038/ng.1036. [DOI] [PubMed] [Google Scholar]
- Costessi A, Mahrour N, Tijchon E, Stunnenberg R, Stoel MA, Jansen PW, et al. The tumour antigen PRAME is a subunit of a Cul2 ubiquitin ligase and associates with active NFY promoters. EMBO J. 2011;30:3786–3798. doi: 10.1038/emboj.2011.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evdokimova V, Tognon C, Ng T, Ruzanov P, Melnyk N, Fink D, et al. Translational activation of snail1 and other developmentally regulated transcription factors by YB-1 promotes an epithelial–mesenchymal transition. Cancer Cell. 2009;15:402–415. doi: 10.1016/j.ccr.2009.03.017. [DOI] [PubMed] [Google Scholar]
- Evdokimova V, Tognon C, Ng T, Sorensen PH. Reduced proliferation and enhanced migration: two sides of the same coin? Molecular mechanisms of metastatic progression by YB-1. Cell Cycle. 2009;8:2901–2906. doi: 10.4161/cc.8.18.9537. [DOI] [PubMed] [Google Scholar]
- Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, et al. The epithelial–mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jürchott K, Kuban RJ, Krech T, Blüthgen N, Stein U, Walther W, et al. Identification of Y-box binding protein 1 as a core regulator of MEK/ERK pathway-dependent gene signatures in colorectal cancer cells. PLoS Genet. 2010;6:e1001231. doi: 10.1371/journal.pgen.1001231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soop T, Nashchekin D, Zhao J, Sun X, Alzhanova-Ericsson AT, Bjorkroth B, et al. J Cell Sci. 2003. pp. 1493–1503. [DOI] [PubMed]
- Kornblihtt AR. Promoter usage and alternative splicing. Curr Opin Cell Biol. 2005;17:262–268. doi: 10.1016/j.ceb.2005.04.014. [DOI] [PubMed] [Google Scholar]
- Allemand E, Batsché E, Muchardt C. Splicing, transcription, and chromatin: a ménage à trois. Curr Opin Genet Dev. 2008;18:145–151. doi: 10.1016/j.gde.2008.01.006. [DOI] [PubMed] [Google Scholar]