Abstract
Botulinum neurotoxin (BoNT) is produced by Clostridium botulinum and associates with nontoxic neurotoxin-associated proteins to form high-molecular weight progenitor complexes (PCs). The PCs are required for the oral toxicity of BoNT in the context of food-borne botulism and are thought to protect BoNT from destruction in the gastrointestinal tract and aid in absorption from the gut lumen. The PC can differ in size and protein content depending on the C. botulinum strain. The oral toxicity of the BoNT PC increases as the size of the PC increases, but the molecular architecture of these large complexes and how they contribute to BoNT toxicity have not been elucidated. We have generated 2D images of PCs from strains producing BoNT serotypes A1, B, and E using negative stain electron microscopy and single-particle averaging. The BoNT/A1 and BoNT/B PCs were observed as ovoid-shaped bodies with three appendages, whereas the BoNT/E PC was observed as an ovoid body. Both the BoNT/A1 and BoNT/B PCs showed significant flexibility, and the BoNT/B PC was documented as a heterogeneous population of assembly/disassembly intermediates. We have also determined 3D structures for each serotype using the random conical tilt approach. Crystal structures of the individual proteins were placed into the BoNT/A1 and BoNT/B PC electron density maps to generate unique detailed models of the BoNT PCs. The structures highlight an effective platform that can be engineered for the development of mucosal vaccines and the intestinal absorption of oral biologics.
Keywords: hemagglutinin, nontoxic nonhemagglutinin, sialic acid
Botulinum neurotoxin (BoNT) is produced by various strains of Clostridium botulinum, Clostridium butyricum, and Clostridium baratii and is the most potent toxin known. BoNTs are classified into seven serotypes (A–G) based upon serotype-specific antibody neutralization, and DNA sequencing has revealed multiple subtypes within each serotype (1). The BoNTs are synthesized as single-chain proteins that are proteolytically cleaved into di-chain proteins, consisting of a 50-kDa light chain (LC) and a 100-kDa heavy chain (HC) (2, 3). The HC binds receptors on the presynaptic neuron in the neuromuscular junction and directs the LC into the neuronal cell cytosol. The LC is a zinc metalloprotease that cleaves components of the synaptic membrane fusion complex and blocks neurotransmitter exocytosis (4). This inhibition of acetylcholine release in neuromuscular junctions results in the flaccid paralysis associated with botulism.
In adults, the most common form of botulism results from ingestion of BoNT contaminated food, whereas in infants, the disease typically results from C. botulinum colonization and in situ BoNT production in the gut (5). In the case of food contamination, BoNT has to survive passage through the low pH environment of the stomach. In both cases, BoNT must resist protease degradation in the intestine, cross the epithelial barrier of the digestive tract, and gain access to the nerve ending targets through the blood and lymph circulatory systems (6).
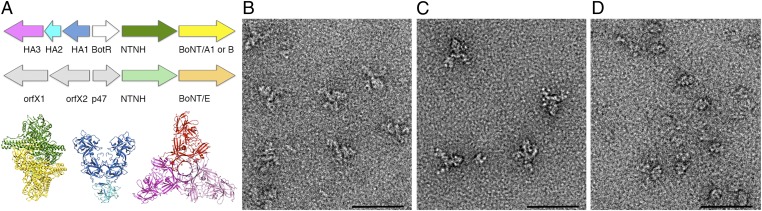
To maintain the activity of BoNT under these conditions, organisms that produce BoNT also produce one or more neurotoxin associated proteins (NAPs) that noncovalently associate with the neurotoxin to form progenitor complexes (PCs) (7). The PCs have been shown by ultracentrifugation to adopt three sizes: 12S (∼300 kDa), 16S (∼500 kDa), and 19S (∼900 kDa) (reviewed in ref. 7). The genes for the neurotoxin and its NAPs are located in the same locus (Fig. 1A) (8). The NAPs include three hemagglutinin (HA) proteins (HA1, HA2, and HA3, named for their capacity to agglutinate red blood cells) and a nontoxic, nonhemagglutinin protein (NTNH). Type A2, E, and F strains do not have the HA genes and only produce the 12S PC, a noncovalent complex of BoNT and NTNH. Types B, C, and D strains produce the 12S and 16S PCs. The 16S PC includes BoNT, NTNH, HA1, HA2, and HA3. Type A1 strains produce 12S, 16S, and 19S PCs. The 19S PC may represent a dimer of 16S complexes.
Fig. 1.
The BoNT PC is composed of the neurotoxin and NAPs. (A) The BoNT gene locus for representative strains expressing BoNT serotypes /A1, /B, and /E. Shown below are the crystal structures for the BoNT/A1-NTNH complex (3V0A), the BoNT/D (HA1)2-HA2 trimer (2E4M), and the BoNT/C HA3 trimer (4EN6). The structures are colored to match the colors in the gene locus such that BoNT/A1 is yellow, NTNH is green, HA1 is blue, HA2 is light blue, and HA3 is pink. The HA1, HA2, and HA3 proteins are also described in the literature as HA33, HA17, and HA70, respectively. (B) The BoNT/A1 PC has three arms projecting away from an ovoid body. (C) The BoNT/B PC has a similar structure to the /A1 complex. (D) The BoNT/E PC is smaller and resembles the ovoid body seen in the /A1 and /B PCs. (Scale bar in B–D, 50 nm.)
The oral median lethal dose (MLD50) of BoNT decreases as the size of the PC increases, suggesting that the NAPs play a role in promoting BoNT passage through the gastrointestinal tract (9–11). A recent structure of a 12S complex shows how BoNT/A1 and NTNH/A1 bind each other in an interlocked complex that protects the BoNT from acidic and proteolytic degradation (Fig. 1A) (12). However, the role of the NAPs in promoting transport through epithelial cells is more controversial. Whereas the NAPs do not appear to effect the transport efficiency of iodinated BoNT in transcytosis experiments (13, 14), experiments in a guinea pig model suggest that only the 16S form of the BoNT/C PC is absorbed from the intestine and released into the serum (15). Mechanisms for how the HA proteins could enhance intestinal absorption involve their capacity to bind mucins (16, 17), bind intestinal microvilli (18), and disrupt the paracellular barrier (19). Crystal structures of HA1 and an HA3 trimer in complex with carbohydrates reveal sites where these molecules could bind mucins and sialylated gangliosides and/or glycoproteins of the epithelial cell surface (20, 21).
Despite these observations, there is limited information regarding the architecture of the BoNT 16S and 19S PCs. A 2D electron crystallography study of the BoNT/A1 19S PC suggested features with trigonal symmetry (22), and individual electron microscopy (EM) micrographs of the BoNT/D 16S PC suggested that the complex has three extended “arms” (23). Here, we have generated 3D reconstructions of the BoNT/A1, /B, and /E PCs, building on a series of 2D class averages obtained by single particle EM and using the random conical tilt (RCT) approach (24).
Results
Visualization of BoNT PCs by Negative Stain EM.
BoNT/A1, /B, and /E PC were adsorbed to carbon-coated glow-discharged grids and stained with uranyl formate. EM micrographs revealed monodisperse macromolecular assemblies suitable for additional structural analysis for all three serotypes. Examination of BoNT/A1 and /B PCs revealed particles composed of an ovoid-shaped “body” attached at one side to three extended arms (Figs. 1 B and C and 2A; see Fig. 4A). Although BoNT/E PC particles shared the same ovoid-shaped body, they lacked the extended arms (Fig. 1D). To more closely examine the organization of the PCs, image pairs of BoNT/A1, /B, and /E were collected so that the 3D structure of each serotype could be determined using the RCT approach.
Fig. 2.
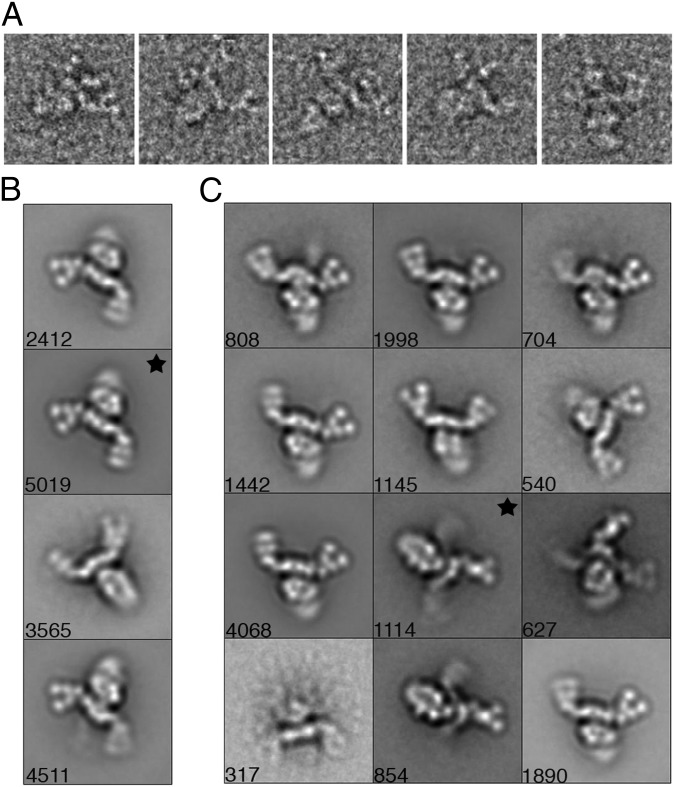
The BoNT/A1 PC is a flexible three-arm structure. (A) The three arms are evident in a gallery of single particles. (B) When 15,507 particles are sorted into four classes only two arms of the PCs’ three arms are visible. *“Flat” view class used for a 3D reconstruction. (C) BoNT/A1 PC class averages obtained by reference-based alignment. *Class used for an alternate “prong” view 3D reconstruction. Side length of panels, 420 Å.
Fig. 4.
The BoNT/B PC has a similar structure to the BoNT/A1 PC. (A) The three arms are evident in a gallery of single particles. (B) BoNT/B PC class averages obtained by reference-based alignment and classification. *Class used for the 3D reconstruction shown in C and D. Side length of panels in A and B, 420 Å. (C) Surface representation of the 3D reconstruction of BoNT/B PC. (D) Crystal structures (from Fig. 1A) placed into a mesh representation of the BoNT/B PC. (Scale bars in C and D, 50 Å.)
BoNT/A1 PC Is a Flexible Three-Armed Structure.
Image pairs of grids containing negatively stained BoNT/A1 PC were recorded at tilt angles of −55° and 0°. A total of 15,507 pairs of particles were selected, and images of the untilted specimens were classified into four class averages (Fig. 2B). Each class revealed a “pincher-like” feature at the end of the arms; however, unlike the raw images that clearly showed particles with three extended arms (Figs. 1B and 2A), only two arms were visible in the averages (Fig. 2B). To attempt to resolve the third arm apparent in our raw images, we expanded the number of classes to 50. (Fig. S1). As expected, when the particles were sorted into a larger number of classes, we began to observe averages containing three arms, although one of the three arms was always less resolved than the other two. In addition, from this group of 50 class averages we noticed that the particles adsorb to the carbon film in two distinct orientations. One is a “flat” orientation that is composed of an ovoid body with two well-defined arms (Fig. S1, black star) and the other is a “prong” orientation that contains an ovoid body with only one well-defined arm (Fig. S1, white star). A majority of the averages (containing ∼90% of the total particles) are in the flat orientation, whereas the remaining averages (containing ∼10% of the total particles) are in the prong orientation.
To generate better resolved prong averages, we selected 12 class averages from our analysis using 50 classes (Fig. S1, black dots) and used these for a further round of reference-based alignment (Fig. 2C). In an effort to generate averages containing an ovoid body with three well-resolved arms, the particles were sorted into groups of 100, 200, or 300 classes. When divided into 300 classes, the third arm is visible in a third of the class averages; however, each class contains only a small number of particles making the averages noisy and not suitable for 3D reconstructions. When comparing distinct classes composed of particles either in the flat or prong view, it is clear there is significant flexibility in the orientation of the pinchers relative to the arms and the orientation of the arms relative to the ovoid body (Movies S1 and S2) and suggests that flexibility is an intrinsic feature of the BoNT/A1 PC.
The conformational heterogeneity made it difficult to generate well-populated classes containing three well-resolved arms. Therefore, we opted to calculate two 3D reconstructions using tilted images either associated with a class average in the flat orientation (Fig. 2B, black star) or associated with a class average in the prong orientation (Fig. 2C, black star) using the RCT approach. Both reconstructions led to well-defined 3D structures that contained features seen in the 2D averages (Fig. 3 A and B), suggesting that the 3D reconstructions were successful. The overall features of the structure generated from the flat-view particles (Fig. 3A) are better resolved than the features seen in the structure generated from the prong-view particles (Fig. 3B). This is due to the larger number of images in the flat projection average (5,019) than in the prong projection average (1,114). We aligned our two 3D reconstructions (flat and prong structures) to produce a 3D model of the three-armed BoNT/A1 PC (Fig. 3C and Movie S3). Our model shows a triangular plate and V-shaped pinchers assembled at one end of an ovoid-shaped body and thus agrees well with the three-armed complex we observed at the single particle level in our EM micrographs (Fig. 2A).
Fig. 3.
The molecular organization of the BoNT/A1 PC. (A) Surface representations of 3D reconstructions of the BoNT/A1 “flat” view and (B) “prong” view in negative stain. (C) An alignment of the two BoNT/A1 3D views reproduces the three-armed complex observed in our EM micrographs. (D) Crystal structures (from Fig. 1A) placed into a mesh representation of the flat view reconstruction. (E) Class averages of the BoNT/A1 PC with an antibody specific to the neurotoxin’s catalytic domain reveal specific binding at the end of the ovoid body furthest away from the HA proteins (black arrows). (Scale bars in A–D, 50 Å.)
The observed geometric features of ovoid body, triangular plate, and V-shape pinchers correlate with the shapes observed in the high-resolution crystal structures of the BoNT/A1-NTNH complex (12), the BoNT/C HA3 trimer (18), and the BoNT/D (HA1)2-HA2 trimer (16), respectively (Figs. 1A and 3D and Movie S3). Whereas the EM structure is not at a resolution that would permit a detailed docking analysis, the BoNT/D (HA1)2-HA2 trimer could easily be placed into the pinchers and the BoNT/C HA3 trimer was placed into the triangular plate of the electron density map. The orientation of the BoNT/C HA3 trimer was guided by the fact that the BoNT/C HA3 proteins, when viewed from the side, form a shallow “W” shape consistent with features we observe in our structure (Fig. S2). Although it was clear that the remaining ovoid body corresponded to the BoNT/A1-NTNH complex, there were two possible ways to orient the crystal structure of the complex within our density map. The genes encoding BoNT/A1 and NTNH are thought to have emerged from a gene duplication event, and the structures of these two molecules are similar (12). In one scenario, the BoNT/A1 LC would make contact with the HA3 triangular plate, whereas the LC equivalent of the NTNH (nLC) would form the base of the ovoid body. In the other scenario, the nLC would be in contact with the HA3 trimer, whereas the BoNT LC was at the distal end of the complex. To distinguish between these two possibilities, we labeled the PC with a BoNT/A1 LC-specific monoclonal antibody (4LCA) (25). We observed consistent and efficient labeling of the distal end of the ovoid body (Fig. 3E and Fig. S3) thus confirming the placement where the nLC interacts with the HA3 trimer. The placement of the crystal structures (Fig. 1A) into the two-arm and three-arm structures of the BoNT/A1 PC is shown in Fig. 3D and Fig. S4, respectively.
BoNT/B PC Has a Similar Structure to the BoNT/A1 PC and Was Observed as a Population of Assembly/Disassembly Intermediates.
Negative stain EM micrographs of the BoNT/B PC show a macromolecular assembly that closely resembles the BoNT/A1 PC (Figs. 1C and 4A), although the sample appeared to be more heterogeneous with PC assembly or disassembly intermediates evident as single particles. Image pairs were collected at tilt angles of −55° and 0° and a total of 11,765 particles were selected. The untilted particles were initially sorted into 75 classes and averaged (Fig. S5). In addition to a flat view, like we observed with the BoNT/A1 PC, we observed complexes where one or more of the components seemed to be missing (Fig. S5, colored boxes). A total of 19 classes representing the range of complexes observed in the sample was selected for multireference alignment (Fig. S5, black dots). A class containing 2,614 particles that clearly showed the main body with two of the three arms well resolved was selected for 3D reconstruction (Fig. 4B, black star). The face view of the density map closely resembles the projection map, suggesting a successful 3D reconstruction (Fig. 4C). The overall BoNT/B PC reconstruction resembles that of the BoNT/A1 PC. Independent placement of the crystal structures (Fig. 4D) suggests that the BoNT/A1 and BoNT /B PCs have a similar structural organization.
BoNT/E PC Resembles the Ovoid Body Observed in BoNT/A1 and BoNT/B Complexes.
Negative stain EM micrographs of the BoNT/E PC show a macromolecular assembly that closely resembles the ovoid-shaped body observed in the BoNT/A1 and BoNT/B PCs (Fig. 1D). The smaller particles are consistent with the description of BoNT/E PC as a 12S complex and the fact that the BoNT/E gene locus does not have the HA operon. Image pairs were collected at tilt angles of −55° and 0° and a total of 7,792 particles was selected. The untilted particles were sorted and averaged in 10 classes (Fig. S6) that revealed classes resembling the ovoid body of the BoNT/A1-NTNH/A1 complex and classes that likely represent dissociated BoNT/E or NTNH/E monomers. Four classes were selected for multireference alignment (Fig. S6, black dots). A class containing 4,518 particles that clearly showed the ovoid body was selected for a 3D reconstruction (Fig. 5A, black star). The face view of the density map resembles the projection map, suggesting a successful 3D reconstruction (Fig. 5B). We could not unambiguously place the BoNT/A1 12S PC crystal structure into the 3D reconstruction of the BoNT/E 12S PC, because we lacked an antibody that could be used to determine which end corresponded to the BoNT/E LC and which end corresponded to the nLC of NTNH/E. However, the symmetry of the BoNT/E 12S structure (Fig. 5B) and the similarity with the ovoid bodies observed in the structures of the BoNT/A1 (Fig. 3 A and C) and BoNT/B (Fig. 4C) PCs suggest that BoNT/E is capable of binding NTNH/E in a conformation similar to how BoNT/A1 binds NTNH/A.
Fig. 5.
The BoNT/E PC resembles the ovoid body observed in the /A1 and /B PCs. (A) BoNT/E class averages obtained by reference-based alignment and classification. *Class used for the 3D reconstruction shown in B. Side length of panels, 420 Å. (B) Surface representation of the 3D reconstruction of the BoNT/E PC. (Scale bar, 50 Å.)
Discussion
Our study provides unique 3D descriptions of the BoNT/A1 16S PC, the BoNT/B 16S PC and the BoNT/E 12S PC. Our data suggest that the BoNT/A1 PC adopts a flexible three-armed structure with a BoNT:NTNH:HA1:HA2:HA3 stoichiometry of 1:1:6:3:3. A BoNT/A1 PC with this stoichiometry has a calculated molecular weight of ∼760 kDa, halfway between the estimates provided by centrifugation studies (500 kDa for the 16S PC and 900 kDa for the 19S PC). The BoNT/A1 PC structure (Fig. 3 A and D) resembles the BoNT/B PC structure (Fig. 4 C and D) as well as the model put forth by Hasegawa et al. (23) based on electron micrographs of the BoNT/D PC. We interpret this similarity to mean that the BoNT/A1 PC structure is in the 16S form. No evidence of dimers or larger complexes was observed in our analysis of the BoNT/A1 PC sample and could reflect dissociation of the 19S PC at low concentrations (26).
In addition to dissociation of the 19S complex, there are reports that document the reversible assembly and disassembly of components in the smaller complexes (27, 28). We observed this in our analysis of the BoNT/B PC sample, which had structures resembling the (i) BoNT-NTNH ovoid body, (ii) BoNT-NTNH bound to an HA3 trimer, or (iii) BoNT-NTNH-(HA3)3 associated with either 1, 2, or 3 (HA1)2-HA2 assemblies (Fig. S5). In vitro, the complexes are known to be pH sensitive, with disassembly occurring at pH ≥ 7.5 (29–31). What occurs in the context of the small intestine is less clear. Some reports suggest that the BoNT will be fully released from the NAPs in the small intestine (the pH of the duodenum is ∼7.0), and there is evidence documenting the capacity of the BoNT to cross the epithelial barrier in the absence of the NAPs (13, 14, 32). Experiments in pH 7 rat intestinal juices, however, indicate that the PC does not dissociate (33). Whole PCs are capable of being absorbed from the intestine into the lymphatics in a rat ligated duodenum loop assay (34), consistent with the hypothesis that the NAPs contribute to absorption in the intestine (35).
There are now strong data to support the role of NTNH in protecting the BoNT from acidic and proteolytic degradation in the digestive tract (12). These data are consistent with the 10- to 20-time enhancement in oral MLD50 when the 12S PC is compared with the neurotoxin alone (9). We know that the addition of the HA proteins to form the larger 16S PC enhances the oral MLD50 even further (the BoNT/A1 16S PC is 1.6 times more potent than the BoNT/A1 12S PC and the BoNT/B 16S PC is 733 times more potent than the BoNT/B 12S PC) (9). Our structures of the BoNT/A1 and BoNT/B 16S PCs indicate that the HAs have no direct contact with the BoNTs and suggest that the HAs do not play a “protective” role but instead contribute to intestinal absorption.
The HA proteins could contribute to intestinal absorption by enhancing adhesion of the BoNT PC to mucins or the cell surface, disrupting the epithelial barrier, or facilitating transcytosis (35). The HAs bind a variety of sialo- and asialooligosaccharides. Binding studies using a glycoconjugate microarray indicate that HA3/C preferentially recognizes α2–3 and α2–6 sialylated sugars, and the binding site for these sugars has been elucidated by X-ray crystallography (21) (Fig. 6B). N-acetylneuraminic acid, N-acetylgalactosamine (GalNAc), and galactose block the binding of HA1/C to mucins, and crystallography has revealed a common site where these sugars can bind (20). Galactose and GalNAc are also capable of binding a second site on HA1/C (20, 36). These binding sites could represent points of contact between the BoNT/C PC and mucins, glycolipids, or glycoproteins within the intestine. Whereas cocrystal structures of the HAs from /A1 and /B strains with sugars are not yet available, the second sugar binding site in HA1/C has been implicated in the sugar binding properties of HA1/A (36, 37). Importantly, each binding site is accessible in the models we have generated of the BoNT/A1 and BoNT/B 16S PC (Fig. 6 A, D, and E).
Fig. 6.
16S PCs have multiple sugar binding sites. (A) Two N-acetylgalactosamine binding sites have been identified in the structure of HA1/C (3AH2 and 3AJ6), and the sugars have been superimposed on the BoNT/D (HA1)2-HA2 trimer structure (blue) and depicted as red sticks. (B) The HA3 trimer (pink) was crystallized with alpha 2–3-sialyllactose, which is depicted in red sticks. (C) The GT1b binding site (with GT1b shown in red sticks) is located between BoNT/A1 (gold) and NTNH (green). Superposition of the BoNT/B receptor-binding domain (HCR)-SytII structure (2NM1) onto the BoNT/A1 HCR suggests that the SytII binding site will be accessible in the BoNT/B PC structure (SytII depicted as a red α-helix). (D) Model of a “flat” view BoNT/A1 PC shown with three arms. (E) The BoNT PC rotated 180° to the “prong” view reveals the accessibility of the GT1b binding site and the SytII binding site.
The BoNTs are also known to interact with sialic acid containing sugars, and the interaction with GT1b gangliosides is known to be important in the dual-receptor model, in which BoNT uses a ganglioside and protein to mediate interaction with the neuronal cell surface (38). The GT1b binding pocket has been defined by X-ray crystallography (39) and is accessible in the context of the 16S PC structure (Fig. 6 C and E). Because GT1b gangliosides are present in epithelial cells of the small intestine, this interaction could contribute to adhesion as well. At the neuromuscular junction, synaptic vesicle protein (SV2) and synaptotagmin II (SytII) are the presynaptic protein receptors for BoNT/A1 and BoNT/B, respectively (40–42), but the relevance of these receptors in the intestine is not clear. We note that the SytII binding site for BoNT/B (43, 44) is fully accessible in the context of the 16S PC structure (Fig. 6 C and E).
Our structural studies have shown that the BoNT/A1 and BoNT/B 16S PCs are flexible three-armed structures. The fact that the HA proteins do not interact directly with the BoNT suggests that they are not required for toxin protection but may be important for multivalent binding interactions with mucin and/or the epithelial cell surface. It is tempting to speculate that the flexibility we observed in the analysis of the BoNT/A1 PC (Movies S1 and S2) allows the complex to effectively “sample” the intestinal surface and improve the chances of absorption of the toxin before high pH dissociation of the complex and proteolytic degradation of the neurotoxin. Our description of the BoNT PCs and the provisional model whereby HA proteins contribute to the delivery of BoNT to the intestinal epithelium provide a platform for optimizing the intestinal delivery of orally administered polypeptide vaccines and therapeutics.
Methods
Specimen Preparation and EM.
The BoNT/A1, /B, and /E PCs were purified by List Biological Laboratories and stored in lyophilized form. The purity of the BoNT/A1 and /B PC at the time of purification is indicated by SDS and native gels (Fig. S7) The samples were hydrated with water to yield solutions that were 0.1 mg/mL protein, 20 mM Hepes, pH 6.8, and 1.25% (wt/vol) lactose. The samples were further diluted to 50 μg/mL in 20 mM Hepes, pH 6.8. Uranyl formate [0.7% (wt/vol)] was used for conventional negative staining as previously described (45). Images of the BoNT/A1, /B, and /E PCs were recorded using a Tecnai F20 electron microscope (FEI) equipped with a field emission electron source and operated at an acceleration voltage of 200 kV. Images were taken under low-dose conditions at a magnification of 67,000× using a defocus value of −1.5 μm. Images were recorded on a Gatan 4K × 4K CCD camera. Images were converted to mixed raster content format, and binned by a factor of 2, yielding final images with 3.5 Å/pixel. Images of the tilted and untilted BoNT/A1, /B, and /E PC particles were selected and analyzed using SPIDER and the associated display program WEB (46).
Random Conical Tilt Reconstruction of Negatively Stained BoNT/A1, /B, and /E PCs.
Micrograph tilt pairs of BoNT/A1, /B, and /E PCs were recorded at −55° and 0°. Particle pairs (BoNT/A1-PC: 15,507; BoNT/B-PC: 11,765; and BoNT/E-PC: 7,792) were selected interactively from the tilted and untilted images using WEB and windowed into 120 × 120 pixel images (3.5 Å/pixel). The untilted images were rotationally and translationally aligned and subjected to 10 cycles of multireference alignment and K-means classification using no designated initial reference.
BoNT/A1 PC particles were grouped into 4, 5, 25, 50, 75, 100, 200, and 300 classes. An initial 3D reconstruction was done using the tilted particles (2,550) associated with one average from the five group classification (Fig. S8, black star). The initial 3D reconstruction was done by back projection using the in-plane rotation angles determined by rotational alignment and the preselected tilt angle of 55° implemented in the processing package SPIDER. To increase the number of particles included in the 3D reconstruction, 5,019 tilted images plus 500 nontilted images associated with a class average in the 4-group alignment (Fig. 2B) were used to further refine the structure using angular refinement in SPIDER. Using a Fourier shell correlation (FSC) = 0.5 criterion the resolution is ∼15 Å (Fig. S9A); however, the lack of any secondary structural details in our map suggests that the resolution more likely falls closer to 20 Å.
Twelve classes were selected from the class average of 50 for additional multireference alignment (Fig. S1, black dots). A class of 1,114 particles showing a prong orientation (Fig. 2C, black star) was used to calculate an initial 3D reconstruction by back projection using the same variables described above. The resulting density map was used for the back projection and angular refinement in SPIDER. Ten percent of the untilted particles were included in the data set (115 particle images) and angular refinement was repeated. Using a FSC = 0.5 criterion, the resolution is ∼15 Å (Fig. S9B); however, examination of structural features in the density map suggests the resolution is closer to 20 Å or higher.
BoNT/B PC particles were grouped into 75 classes. Nineteen classes were selected from the class average of 75 for additional multireference alignment (Fig. S5, black dots). A class of 2,614 particles (Fig. 4B, black star) was used to calculate an initial 3D reconstruction by back projection using the same variables described above. The resulting density map was used for the back projection and angular refinement in SPIDER. Ten percent of the untilted particles were included in the data set (260 particle images) and angular refinement was repeated. Using a FSC = 0.5 criterion, the resolution is ∼15 Å (Fig. S9C); however, the lack of any secondary structural details in our map suggests that the resolution more likely falls closer to 20 Å. Although the FSC curves for BoNT/A1 and /B PCs initially approach 0 correlation as is expected, at the highest resolution the curves once again begin approaching 0.5 correlation or higher. We attribute this to artifacts created by the large amount of noise generated by the flexible third arm that could not be resolved in the maps.
BoNT/E PC particles were grouped into 10 classes using reference-free alignment. Four classes were selected as references for an additional round of multireference alignment (Fig. S6, black dots). A 3D structure was calculated using back projection from the tilted images associated with a class of 692 particles from the initial alignment (Fig. S6, black star) as an initial model for angular refinement of 4,518 tilted images plus 450 nontilted images from a class generated in the reference-based alignment (Fig. 5, black star). Using a FSC = 0.5 criterion, the resolution is ∼15 Å (Fig. S9D); however, the lack of any secondary structural details in our map suggests that the resolution more likely falls closer to 20 Å.
The contouring threshold for all structures was chosen so that the volume of the structure was continuous. For display purposes, structures were filtered in Chimera (47) using the “Hide Dust” command to diminish “salt and pepper” noise from the maps by removing single voxels that were unconnected to the main volume of the 3D density.
Negative Staining of an Antibody-BoNT/A1 PC complex.
The 4LCA antibody was purified as previously described (25). For labeling, the BoNT/A1 PC and 4LCA proteins were mixed in a ∼1:10 molar ratio and incubated at room temperature for 3 h before grid preparation. Grids were prepared as described (45). A total of 403 particles of BoNT/A1 PC bound to 4LCA was selected interactively using WEB and windowed into 150 × 150 pixel images (3.5 Å/pixel). The images were rotationally and translationally aligned and subjected to 10 cycles of multireference alignment and K-means classification with no initial references designated.
Supplementary Material
Acknowledgments
We thank Melissa Chambers, Taylor Jones, Rory Pruitt, and Yoshimasa Takizawa for technical assistance. This project was supported by Public Health Service Grant R01 AI075259 from the National Institutes of Health (to D.B.L.) and R01 AI065967 (to S.K.D.).
Footnotes
Conflict of interest statement: S.K.D. owns equity in a start-up company (Immunome, Inc.) that has licensed the 4LCA antibody. N.S. is an employee of List Biologicals, Inc.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1222139110/-/DCSupplemental.
References
- 1.Peck MW. Biology and genomic analysis of Clostridium botulinum. Adv Microb Physiol. 2009;55:183–265, 320. doi: 10.1016/S0065-2911(09)05503-9. [DOI] [PubMed] [Google Scholar]
- 2.Montecucco C, Schiavo G. Structure and function of tetanus and botulinum neurotoxins. Q Rev Biophys. 1995;28(4):423–472. doi: 10.1017/s0033583500003292. [DOI] [PubMed] [Google Scholar]
- 3.Lacy DB, Tepp W, Cohen AC, DasGupta BR, Stevens RC. Crystal structure of botulinum neurotoxin type A and implications for toxicity. Nat Struct Biol. 1998;5(10):898–902. doi: 10.1038/2338. [DOI] [PubMed] [Google Scholar]
- 4.Schiavo G, Rossetto O, Benfenati F, Poulain B, Montecucco C. Tetanus and botulinum neurotoxins are zinc proteases specific for components of the neuroexocytosis apparatus. Ann N Y Acad Sci. 1994;710:65–75. doi: 10.1111/j.1749-6632.1994.tb26614.x. [DOI] [PubMed] [Google Scholar]
- 5.Arnon SS. Human Tetanus and Human Botulism. In: Rood JI, McClane BA, Songer JG, Titball RW, editors. The Clostridia: Molecular Biology and Pathogenesis. San Diego: Academic; 1997. pp. 95–115. [Google Scholar]
- 6.Simpson LL. Identification of the major steps in botulinum toxin action. Annu Rev Pharmacol Toxicol. 2004;44:167–193. doi: 10.1146/annurev.pharmtox.44.101802.121554. [DOI] [PubMed] [Google Scholar]
- 7.Sakaguchi G. Clostridium botulinum toxins. Pharmacol Ther. 1982;19(2):165–194. doi: 10.1016/0163-7258(82)90061-4. [DOI] [PubMed] [Google Scholar]
- 8.Raffestin S, Marvaud JC, Cerrato R, Dupuy B, Popoff MR. Organization and regulation of the neurotoxin genes in Clostridium botulinum and Clostridium tetani. Anaerobe. 2004;10(2):93–100. doi: 10.1016/j.anaerobe.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 9.Ohishi I, Sugii S, Sakaguchi G. Oral toxicities of Clostridium botulinum toxins in response to molecular size. Infect Immun. 1977;16(1):107–109. doi: 10.1128/iai.16.1.107-109.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohishi I, Sakaguchi G. Oral toxicities of Clostridium botulinum type C and D toxins of different molecular sizes. Infect Immun. 1980;28(2):303–309. doi: 10.1128/iai.28.2.303-309.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sakaguchi G, Sakaguchi S. Oral toxicities of Clostridium botulinum type E toxins of different forms. Jpn J Med Sci Biol. 1974;27(4):241–244. doi: 10.7883/yoken1952.27.241. [DOI] [PubMed] [Google Scholar]
- 12.Gu S, et al. Botulinum neurotoxin is shielded by NTNHA in an interlocked complex. Science. 2012;335(6071):977–981. doi: 10.1126/science.1214270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maksymowych AB, Simpson LL. Binding and transcytosis of botulinum neurotoxin by polarized human colon carcinoma cells. J Biol Chem. 1998;273(34):21950–21957. doi: 10.1074/jbc.273.34.21950. [DOI] [PubMed] [Google Scholar]
- 14.Park JB, Simpson LL. Inhalational poisoning by botulinum toxin and inhalation vaccination with its heavy-chain component. Infect Immun. 2003;71(3):1147–1154. doi: 10.1128/IAI.71.3.1147-1154.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujinaga Y, et al. The haemagglutinin of Clostridium botulinum type C progenitor toxin plays an essential role in binding of toxin to the epithelial cells of guinea pig small intestine, leading to the efficient absorption of the toxin. Microbiology. 1997;143(Pt 12):3841–3847. doi: 10.1099/00221287-143-12-3841. [DOI] [PubMed] [Google Scholar]
- 16.Nishikawa A, et al. The receptor and transporter for internalization of Clostridium botulinum type C progenitor toxin into HT-29 cells. Biochem Biophys Res Commun. 2004;319(2):327–333. doi: 10.1016/j.bbrc.2004.04.183. [DOI] [PubMed] [Google Scholar]
- 17.Uotsu N, et al. Cell internalization and traffic pathway of Clostridium botulinum type C neurotoxin in HT-29 cells. Biochim Biophys Acta. 2006;1763(1):120–128. doi: 10.1016/j.bbamcr.2005.11.014. [DOI] [PubMed] [Google Scholar]
- 18.Fujinaga Y, et al. Identification and characterization of functional subunits of Clostridium botulinum type A progenitor toxin involved in binding to intestinal microvilli and erythrocytes. FEBS Lett. 2000;467(2–3):179–183. doi: 10.1016/s0014-5793(00)01147-9. [DOI] [PubMed] [Google Scholar]
- 19.Matsumura T, et al. The HA proteins of botulinum toxin disrupt intestinal epithelial intercellular junctions to increase toxin absorption. Cell Microbiol. 2008;10(2):355–364. doi: 10.1111/j.1462-5822.2007.01048.x. [DOI] [PubMed] [Google Scholar]
- 20.Nakamura T, et al. Sugar-binding sites of the HA1 subcomponent of Clostridium botulinum type C progenitor toxin. J Mol Biol. 2008;376(3):854–867. doi: 10.1016/j.jmb.2007.12.031. [DOI] [PubMed] [Google Scholar]
- 21.Yamashita S, et al. Carbohydrate recognition mechanism of HA70 from Clostridium botulinum deduced from X-ray structures in complexes with sialylated oligosaccharides. FEBS Lett. 2012;586(16):2404–2410. doi: 10.1016/j.febslet.2012.05.055. [DOI] [PubMed] [Google Scholar]
- 22.Burkard F, Chen F, Kuziemko GM, Stevens RC. Electron density projection map of the botulinum neurotoxin 900-kilodalton complex by electron crystallography. J Struct Biol. 1997;120(1):78–84. doi: 10.1006/jsbi.1997.3910. [DOI] [PubMed] [Google Scholar]
- 23.Hasegawa K, et al. A novel subunit structure of Clostridium botulinum serotype D toxin complex with three extended arms. J Biol Chem. 2007;282(34):24777–24783. doi: 10.1074/jbc.M703446200. [DOI] [PubMed] [Google Scholar]
- 24.Radermacher M, Wagenknecht T, Verschoor A, Frank J. Three-dimensional reconstruction from a single-exposure, random conical tilt series applied to the 50S ribosomal subunit of Escherichia coli. J Microsc. 1987;146(Pt 2):113–136. doi: 10.1111/j.1365-2818.1987.tb01333.x. [DOI] [PubMed] [Google Scholar]
- 25.Adekar SP, et al. Neutralization of botulinum neurotoxin by a human monoclonal antibody specific for the catalytic light chain. PLoS ONE. 2008;3(8):e3023. doi: 10.1371/journal.pone.0003023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eisele KH, Fink K, Vey M, Taylor HV. Studies on the dissociation of botulinum neurotoxin type A complexes. Toxicon. 2011;57(4):555–565. doi: 10.1016/j.toxicon.2010.12.019. [DOI] [PubMed] [Google Scholar]
- 27.Kitamura M, Sakaguchi G. Dissociation and reconstitution of 12-S toxin of Clostridium botulinum type E. Biochim Biophys Acta. 1969;194(2):564–571. doi: 10.1016/0005-2795(69)90118-4. [DOI] [PubMed] [Google Scholar]
- 28.Miyazaki S, Kozaki S, Sakaguchi S, Sakaguchi G. Comparison of progenitor toxins of nonproteolytic with those of proteolytic Clostridium botulinum Type B. Infect Immun. 1976;13(3):987–989. doi: 10.1128/iai.13.3.987-989.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wagman J. Isolation and sedimentation study of low molecular weight forms of type A Botulins toxin. Arch Biochem Biophys. 1954;50(1):104–112. doi: 10.1016/0003-9861(54)90013-2. [DOI] [PubMed] [Google Scholar]
- 30.Wagman J. Low molecular weight forms of type A botulinum toxin. II. Action of pepsin on intact and dissociated toxin. Arch Biochem Biophys. 1963;100:414–421. doi: 10.1016/0003-9861(63)90106-1. [DOI] [PubMed] [Google Scholar]
- 31.Wagman J, Bateman JB. The behavior of the bolulinus toxins in the ultracentrifuge. Arch Biochem Biophys. 1951;31(3):424–430. doi: 10.1016/0003-9861(51)90158-0. [DOI] [PubMed] [Google Scholar]
- 32.Couesnon A, Pereira Y, Popoff MR. Receptor-mediated transcytosis of botulinum neurotoxin A through intestinal cell monolayers. Cell Microbiol. 2008;10(2):375–387. doi: 10.1111/j.1462-5822.2007.01051.x. [DOI] [PubMed] [Google Scholar]
- 33.Sugii S, Ohishi I, Sakaguchi G. Correlation between oral toxicity and in vitro stability of Clostridium botulinum type A and B toxins of different molecular sizes. Infect Immun. 1977;16(3):910–914. doi: 10.1128/iai.16.3.910-914.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sugii S, Ohishi I, Sakaguchi G. Intestinal absorption of botulinum toxins of different molecular sizes in rats. Infect Immun. 1977;17(3):491–496. doi: 10.1128/iai.17.3.491-496.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fujinaga Y. Interaction of botulinum toxin with the epithelial barrier. J Biomed Biotechnol. 2010;2010:974943. doi: 10.1155/2010/974943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakamura T, et al. Molecular diversity of the two sugar-binding sites of the β-trefoil lectin HA33/C (HA1) from Clostridium botulinum type C neurotoxin. Arch Biochem Biophys. 2011;512(1):69–77. doi: 10.1016/j.abb.2011.05.012. [DOI] [PubMed] [Google Scholar]
- 37.Inoue K, et al. Structural analysis by X-ray crystallography and calorimetry of a haemagglutinin component (HA1) of the progenitor toxin from Clostridium botulinum. Microbiology. 2003;149(Pt 12):3361–3370. doi: 10.1099/mic.0.26586-0. [DOI] [PubMed] [Google Scholar]
- 38.Montecucco C. How do tetanus and botulinum neurotoxins bind to neuronal membranes? Trends Biochem Sci. 1986;11:314–317. [Google Scholar]
- 39.Stenmark P, Dupuy J, Imamura A, Kiso M, Stevens RC. Crystal structure of botulinum neurotoxin type A in complex with the cell surface co-receptor GT1b-insight into the toxin-neuron interaction. PLoS Pathog. 2008;4(8):e1000129. doi: 10.1371/journal.ppat.1000129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dong M, et al. SV2 is the protein receptor for botulinum neurotoxin A. Science. 2006;312(5773):592–596. doi: 10.1126/science.1123654. [DOI] [PubMed] [Google Scholar]
- 41.Dong M, et al. Synaptotagmins I and II mediate entry of botulinum neurotoxin B into cells. J Cell Biol. 2003;162(7):1293–1303. doi: 10.1083/jcb.200305098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nishiki T, et al. The high-affinity binding of Clostridium botulinum type B neurotoxin to synaptotagmin II associated with gangliosides GT1b/GD1a. FEBS Lett. 1996;378(3):253–257. doi: 10.1016/0014-5793(95)01471-3. [DOI] [PubMed] [Google Scholar]
- 43.Chai Q, et al. Structural basis of cell surface receptor recognition by botulinum neurotoxin B. Nature. 2006;444(7122):1096–1100. doi: 10.1038/nature05411. [DOI] [PubMed] [Google Scholar]
- 44.Jin R, Rummel A, Binz T, Brunger AT. Botulinum neurotoxin B recognizes its protein receptor with high affinity and specificity. Nature. 2006;444(7122):1092–1095. doi: 10.1038/nature05387. [DOI] [PubMed] [Google Scholar]
- 45.Ohi M, Li Y, Cheng Y, Walz T. Negative staining and image classification: Powerful tools in modern electron microscopy. Biol Proced Online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Frank J, et al. SPIDER and WEB: Processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116(1):190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 47.Pettersen EF, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.