Abstract
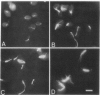
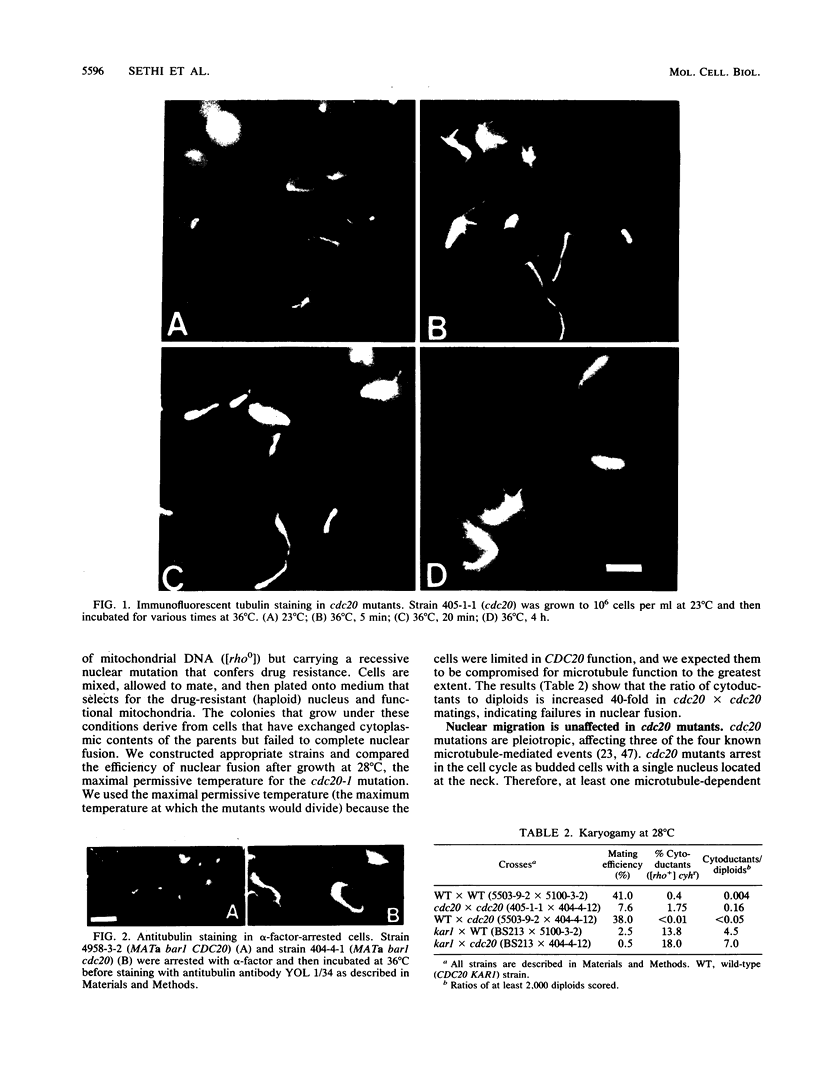
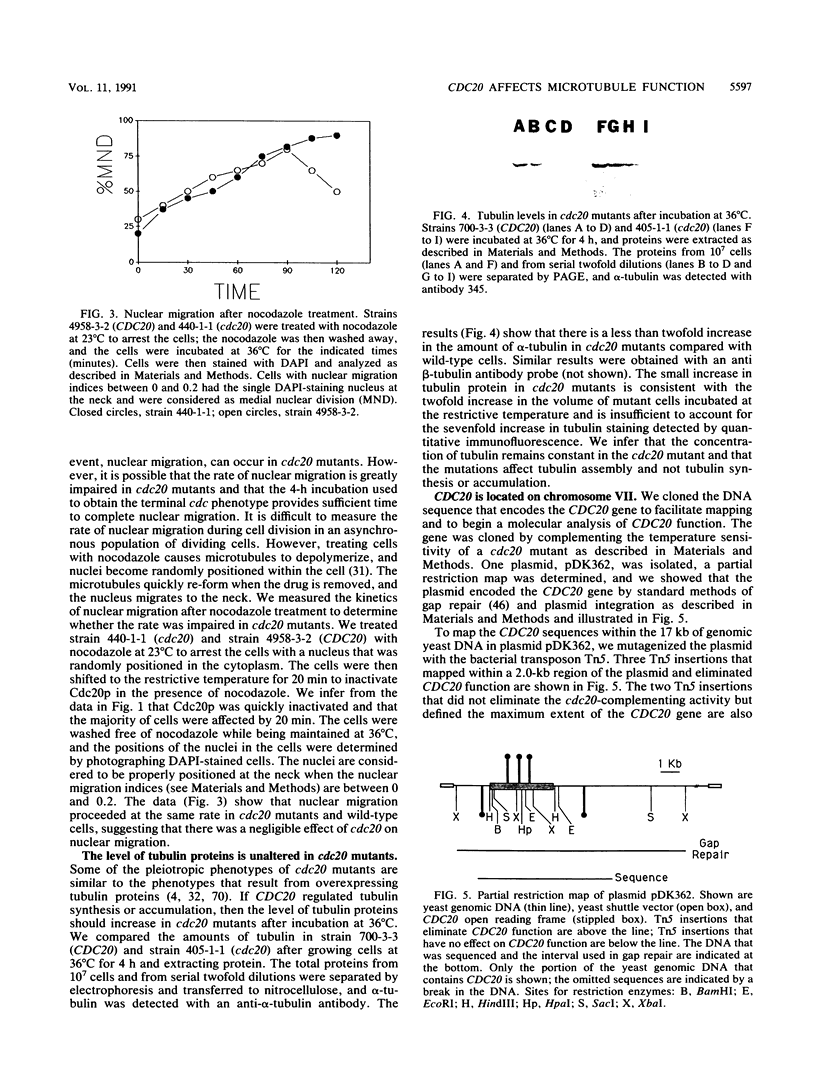
Previous analysis of cdc20 mutants of the yeast Saccharomyces cerevisiae suggests that the CDC20 gene product (Cdc20p) is required for two microtubule-dependent processes, nuclear movements prior to anaphase and chromosome separation. Here we report that cdc20 mutants are defective for a third microtubule-mediated event, nuclear fusion during mating of G1 cells, but appear normal for a fourth microtubule-dependent process, nuclear migration after DNA replication. Therefore, Cdc20p is required for a subset of microtubule-dependent processes and functions at multiple stages in the life cycle. Consistent with this interpretation, we find that cdc20 cells arrested by alpha-factor or at the restrictive temperature accumulate anomalous microtubule structures, as detected by indirect immunofluorescence. The anomalous microtubule staining patterns are due to cdc20 because intragenic revertants that revert the temperature sensitivity have normal microtubule morphologies. cdc20 mutants have a sevenfold increase in the intensity of antitubulin fluorescence in intranuclear spindles compared with spindles from wild-type cells, yet the total amount of tubulin is indistinguishable by Western immunoblot analysis. This result suggests that Cdc20p modulates microtubule structure in wild-type cells either by promoting microtubule disassembly or by altering the surface of the microtubules. Finally, we cloned and sequenced CDC20 and show that it encodes a member of a family of proteins that share homology to the beta subunit of transducin.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bilofsky H. S., Burks C., Fickett J. W., Goad W. B., Lewitter F. I., Rindone W. P., Swindell C. D., Tung C. S. The GenBank genetic sequence databank. Nucleic Acids Res. 1986 Jan 10;14(1):1–4. doi: 10.1093/nar/14.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond J. F., Fridovich-Keil J. L., Pillus L., Mulligan R. C., Solomon F. A chicken-yeast chimeric beta-tubulin protein is incorporated into mouse microtubules in vivo. Cell. 1986 Feb 14;44(3):461–468. doi: 10.1016/0092-8674(86)90467-8. [DOI] [PubMed] [Google Scholar]
- Botstein D., Segev N., Stearns T., Hoyt M. A., Holden J., Kahn R. A. Diverse biological functions of small GTP-binding proteins in yeast. Cold Spring Harb Symp Quant Biol. 1988;53(Pt 2):629–636. doi: 10.1101/sqb.1988.053.01.072. [DOI] [PubMed] [Google Scholar]
- Burke D., Gasdaska P., Hartwell L. Dominant effects of tubulin overexpression in Saccharomyces cerevisiae. Mol Cell Biol. 1989 Mar;9(3):1049–1059. doi: 10.1128/mcb.9.3.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers B., Goetsch L. Behavior of spindles and spindle plaques in the cell cycle and conjugation of Saccharomyces cerevisiae. J Bacteriol. 1975 Oct;124(1):511–523. doi: 10.1128/jb.124.1.511-523.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers B., Goetsch L. Duplication of spindle plaques and integration of the yeast cell cycle. Cold Spring Harb Symp Quant Biol. 1974;38:123–131. doi: 10.1101/sqb.1974.038.01.016. [DOI] [PubMed] [Google Scholar]
- Conde J., Fink G. R. A mutant of Saccharomyces cerevisiae defective for nuclear fusion. Proc Natl Acad Sci U S A. 1976 Oct;73(10):3651–3655. doi: 10.1073/pnas.73.10.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado M. A., Conde J. Benomyl prevents nuclear fusion in Saccharomyces cerevisiae. Mol Gen Genet. 1984;193(1):188–189. doi: 10.1007/BF00327435. [DOI] [PubMed] [Google Scholar]
- Dutcher S. K., Hartwell L. H. The role of S. cerevisiae cell division cycle genes in nuclear fusion. Genetics. 1982 Feb;100(2):175–184. doi: 10.1093/genetics/100.2.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong H. K., Amatruda T. T., 3rd, Birren B. W., Simon M. I. Distinct forms of the beta subunit of GTP-binding regulatory proteins identified by molecular cloning. Proc Natl Acad Sci U S A. 1987 Jun;84(11):3792–3796. doi: 10.1073/pnas.84.11.3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong H. K., Hurley J. B., Hopkins R. S., Miake-Lye R., Johnson M. S., Doolittle R. F., Simon M. I. Repetitive segmental structure of the transducin beta subunit: homology with the CDC4 gene and identification of related mRNAs. Proc Natl Acad Sci U S A. 1986 Apr;83(7):2162–2166. doi: 10.1073/pnas.83.7.2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilman A. G. G proteins: transducers of receptor-generated signals. Annu Rev Biochem. 1987;56:615–649. doi: 10.1146/annurev.bi.56.070187.003151. [DOI] [PubMed] [Google Scholar]
- Goebl M., Yanagida M. The TPR snap helix: a novel protein repeat motif from mitosis to transcription. Trends Biochem Sci. 1991 May;16(5):173–177. doi: 10.1016/0968-0004(91)90070-c. [DOI] [PubMed] [Google Scholar]
- Hartwell L. H. Macromolecule synthesis in temperature-sensitive mutants of yeast. J Bacteriol. 1967 May;93(5):1662–1670. doi: 10.1128/jb.93.5.1662-1670.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L. H., Mortimer R. K., Culotti J., Culotti M. Genetic Control of the Cell Division Cycle in Yeast: V. Genetic Analysis of cdc Mutants. Genetics. 1973 Jun;74(2):267–286. doi: 10.1093/genetics/74.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L. H., Smith D. Altered fidelity of mitotic chromosome transmission in cell cycle mutants of S. cerevisiae. Genetics. 1985 Jul;110(3):381–395. doi: 10.1093/genetics/110.3.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill J. E., Myers A. M., Koerner T. J., Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986 Sep;2(3):163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- Himmler A., Drechsel D., Kirschner M. W., Martin D. W., Jr Tau consists of a set of proteins with repeated C-terminal microtubule-binding domains and variable N-terminal domains. Mol Cell Biol. 1989 Apr;9(4):1381–1388. doi: 10.1128/mcb.9.4.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt M. A., Stearns T., Botstein D. Chromosome instability mutants of Saccharomyces cerevisiae that are defective in microtubule-mediated processes. Mol Cell Biol. 1990 Jan;10(1):223–234. doi: 10.1128/mcb.10.1.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker T. C., Hoyt M. A., Botstein D. Genetic analysis of the yeast cytoskeleton. Annu Rev Genet. 1987;21:259–284. doi: 10.1146/annurev.ge.21.120187.001355. [DOI] [PubMed] [Google Scholar]
- Huffaker T. C., Thomas J. H., Botstein D. Diverse effects of beta-tubulin mutations on microtubule formation and function. J Cell Biol. 1988 Jun;106(6):1997–2010. doi: 10.1083/jcb.106.6.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Icho T., Wickner R. B. The MAK11 protein is essential for cell growth and replication of M double-stranded RNA and is apparently a membrane-associated protein. J Biol Chem. 1988 Jan 25;263(3):1467–1475. [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs C. W., Adams A. E., Szaniszlo P. J., Pringle J. R. Functions of microtubules in the Saccharomyces cerevisiae cell cycle. J Cell Biol. 1988 Oct;107(4):1409–1426. doi: 10.1083/jcb.107.4.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz W., Weinstein B., Solomon F. Regulation of tubulin levels and microtubule assembly in Saccharomyces cerevisiae: consequences of altered tubulin gene copy number. Mol Cell Biol. 1990 Oct;10(10):5286–5294. doi: 10.1128/mcb.10.10.5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilmartin J. V., Adams A. E. Structural rearrangements of tubulin and actin during the cell cycle of the yeast Saccharomyces. J Cell Biol. 1984 Mar;98(3):922–933. doi: 10.1083/jcb.98.3.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilmartin J. V. Purification of yeast tubulin by self-assembly in vitro. Biochemistry. 1981 Jun 9;20(12):3629–3633. doi: 10.1021/bi00515a050. [DOI] [PubMed] [Google Scholar]
- Kirschner M. W. Biological implications of microtubule dynamics. Harvey Lect. 1987;83:1–20. [PubMed] [Google Scholar]
- Kirschner M., Mitchison T. Beyond self-assembly: from microtubules to morphogenesis. Cell. 1986 May 9;45(3):329–342. doi: 10.1016/0092-8674(86)90318-1. [DOI] [PubMed] [Google Scholar]
- Koshland D., Kent J. C., Hartwell L. H. Genetic analysis of the mitotic transmission of minichromosomes. Cell. 1985 Feb;40(2):393–403. doi: 10.1016/0092-8674(85)90153-9. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Meluh P. B., Rose M. D. KAR3, a kinesin-related gene required for yeast nuclear fusion. Cell. 1990 Mar 23;60(6):1029–1041. doi: 10.1016/0092-8674(90)90351-e. [DOI] [PubMed] [Google Scholar]
- Mitchison T., Kirschner M. Dynamic instability of microtubule growth. Nature. 1984 Nov 15;312(5991):237–242. doi: 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- Mortimer R. K., Schild D., Contopoulou C. R., Kans J. A. Genetic map of Saccharomyces cerevisiae, edition 10. Yeast. 1989 Sep-Oct;5(5):321–403. doi: 10.1002/yea.320050503. [DOI] [PubMed] [Google Scholar]
- Neff N. F., Thomas J. H., Grisafi P., Botstein D. Isolation of the beta-tubulin gene from yeast and demonstration of its essential function in vivo. Cell. 1983 May;33(1):211–219. doi: 10.1016/0092-8674(83)90350-1. [DOI] [PubMed] [Google Scholar]
- Noble M., Lewis S. A., Cowan N. J. The microtubule binding domain of microtubule-associated protein MAP1B contains a repeated sequence motif unrelated to that of MAP2 and tau. J Cell Biol. 1989 Dec;109(6 Pt 2):3367–3376. doi: 10.1083/jcb.109.6.3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmsted J. B. Microtubule-associated proteins. Annu Rev Cell Biol. 1986;2:421–457. doi: 10.1146/annurev.cb.02.110186.002225. [DOI] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W., Rothstein R. J. Genetic applications of yeast transformation with linear and gapped plasmids. Methods Enzymol. 1983;101:228–245. doi: 10.1016/0076-6879(83)01017-4. [DOI] [PubMed] [Google Scholar]
- Palmer R. E., Koval M., Koshland D. The dynamics of chromosome movement in the budding yeast Saccharomyces cerevisiae. J Cell Biol. 1989 Dec;109(6 Pt 2):3355–3366. doi: 10.1083/jcb.109.6.3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson J. B., Ris H. Electron-microscopic study of the spindle and chromosome movement in the yeast Saccharomyces cerevisiae. J Cell Sci. 1976 Nov;22(2):219–242. doi: 10.1242/jcs.22.2.219. [DOI] [PubMed] [Google Scholar]
- Pillus L., Solomon F. Components of microtubular structures in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1986 Apr;83(8):2468–2472. doi: 10.1073/pnas.83.8.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M. D., Fink G. R. KAR1, a gene required for function of both intranuclear and extranuclear microtubules in yeast. Cell. 1987 Mar 27;48(6):1047–1060. doi: 10.1016/0092-8674(87)90712-4. [DOI] [PubMed] [Google Scholar]
- Rose M. D., Novick P., Thomas J. H., Botstein D., Fink G. R. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60(2-3):237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- Ruggieri R., Tanaka K., Nakafuku M., Kaziro Y., Toh-e A., Matsumoto K. MSI1, a negative regulator of the RAS-cAMP pathway in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1989 Nov;86(22):8778–8782. doi: 10.1073/pnas.86.22.8778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sammak P. J., Gorbsky G. J., Borisy G. G. Microtubule dynamics in vivo: a test of mechanisms of turnover. J Cell Biol. 1987 Mar;104(3):395–405. doi: 10.1083/jcb.104.3.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz P. J., Georges G. E., Solomon F., Botstein D. Insertions of up to 17 amino acids into a region of alpha-tubulin do not disrupt function in vivo. Mol Cell Biol. 1987 Oct;7(10):3799–3805. doi: 10.1128/mcb.7.10.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz P. J., Pillus L., Grisafi P., Solomon F., Botstein D. Two functional alpha-tubulin genes of the yeast Saccharomyces cerevisiae encode divergent proteins. Mol Cell Biol. 1986 Nov;6(11):3711–3721. doi: 10.1128/mcb.6.11.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz P. J., Solomon F., Botstein D. Isolation and characterization of conditional-lethal mutations in the TUB1 alpha-tubulin gene of the yeast Saccharomyces cerevisiae. Genetics. 1988 Nov;120(3):681–695. doi: 10.1093/genetics/120.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze E., Kirschner M. Dynamic and stable populations of microtubules in cells. J Cell Biol. 1987 Feb;104(2):277–288. doi: 10.1083/jcb.104.2.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano L., Wandosell F., Avila J. Location of the regions recognized by five commercial antibodies on the tubulin molecule. Anal Biochem. 1986 Dec;159(2):253–259. doi: 10.1016/0003-2697(86)90340-4. [DOI] [PubMed] [Google Scholar]
- Serrano L., de la Torre J., Maccioni R. B., Avila J. Involvement of the carboxyl-terminal domain of tubulin in the regulation of its assembly. Proc Natl Acad Sci U S A. 1984 Oct;81(19):5989–5993. doi: 10.1073/pnas.81.19.5989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Shero J. H., Koval M., Spencer F., Palmer R. E., Hieter P., Koshland D. Analysis of chromosome segregation in Saccharomyces cerevisiae. Methods Enzymol. 1991;194:749–773. doi: 10.1016/0076-6879(91)94057-j. [DOI] [PubMed] [Google Scholar]
- Spencer F., Gerring S. L., Connelly C., Hieter P. Mitotic chromosome transmission fidelity mutants in Saccharomyces cerevisiae. Genetics. 1990 Feb;124(2):237–249. doi: 10.1093/genetics/124.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns T., Botstein D. Unlinked noncomplementation: isolation of new conditional-lethal mutations in each of the tubulin genes of Saccharomyces cerevisiae. Genetics. 1988 Jun;119(2):249–260. doi: 10.1093/genetics/119.2.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns T., Hoyt M. A., Botstein D. Yeast mutants sensitive to antimicrotubule drugs define three genes that affect microtubule function. Genetics. 1990 Feb;124(2):251–262. doi: 10.1093/genetics/124.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J. H., Neff N. F., Botstein D. Isolation and characterization of mutations in the beta-tubulin gene of Saccharomyces cerevisiae. Genetics. 1985 Dec;111(4):715–734. doi: 10.1093/genetics/111.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein B., Solomon F. Phenotypic consequences of tubulin overproduction in Saccharomyces cerevisiae: differences between alpha-tubulin and beta-tubulin. Mol Cell Biol. 1990 Oct;10(10):5295–5304. doi: 10.1128/mcb.10.10.5295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteway M., Hougan L., Dignard D., Thomas D. Y., Bell L., Saari G. C., Grant F. J., O'Hara P., MacKay V. L. The STE4 and STE18 genes of yeast encode potential beta and gamma subunits of the mating factor receptor-coupled G protein. Cell. 1989 Feb 10;56(3):467–477. doi: 10.1016/0092-8674(89)90249-3. [DOI] [PubMed] [Google Scholar]
- Williams F. E., Trumbly R. J. Characterization of TUP1, a mediator of glucose repression in Saccharomyces cerevisiae. Mol Cell Biol. 1990 Dec;10(12):6500–6511. doi: 10.1128/mcb.10.12.6500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams F. E., Varanasi U., Trumbly R. J. The CYC8 and TUP1 proteins involved in glucose repression in Saccharomyces cerevisiae are associated in a protein complex. Mol Cell Biol. 1991 Jun;11(6):3307–3316. doi: 10.1128/mcb.11.6.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson D. H., Fennell D. J. The use of fluorescent DNA-binding agent for detecting and separating yeast mitochondrial DNA. Methods Cell Biol. 1975;12:335–351. doi: 10.1016/s0091-679x(08)60963-2. [DOI] [PubMed] [Google Scholar]
- Yochem J., Byers B. Structural comparison of the yeast cell division cycle gene CDC4 and a related pseudogene. J Mol Biol. 1987 May 20;195(2):233–245. doi: 10.1016/0022-2836(87)90646-2. [DOI] [PubMed] [Google Scholar]
- de Bruijn F. J., Lupski J. R. The use of transposon Tn5 mutagenesis in the rapid generation of correlated physical and genetic maps of DNA segments cloned into multicopy plasmids--a review. Gene. 1984 Feb;27(2):131–149. doi: 10.1016/0378-1119(84)90135-5. [DOI] [PubMed] [Google Scholar]