Abstract
Autophagy-related 13 (Atg13) is a key early-acting factor in autophagy and the major locus for nutrient-dependent regulation of autophagy by Tor. The 2.3-Å resolution crystal structure of the N-terminal domain of Atg13 reveals a previously unidentified HORMA (Hop1p, Rev1p and Mad2) domain similar to that of the spindle checkpoint protein Mad2. Mad2 has two different stable conformations, O-Mad2 and C-Mad2, and the Atg13 HORMA structure corresponds to the C-Mad2 state. The Atg13 HORMA domain is required for autophagy and for recruitment of the phosphatidylinositol (PI) 3-kinase subunit Atg14 but is not required for Atg1 interaction or Atg13 recruitment to the preautophagosomal structure. The Atg13 HORMA structure reveals a pair of conserved Arg residues that constitute a putative phosphate sensor. One of the Arg residues is in the region corresponding to the “safety belt” conformational switch of Mad2, suggesting conformational regulation of phosphate binding. These two Arg residues are essential for autophagy, suggesting that the Atg13 HORMA domain could function as a phosphoregulated conformational switch.
Keywords: protein crystallography, protein degradation, protein structure, yeast genetics
Macroautophagy (henceforward, “autophagy”) is a conserved pathway for cell survival during starvation (1) and for the clearance of harmful materials from the cytosol (2, 3). Autophagy proceeds stepwise by the nucleation of a few high-curvature vesicles into a nascent double-membrane sheet; the growth of the sheet and its bending into the cup-shaped phagophore; the engulfment of cytosolic material by the cup; sealing of the cup to form the mature autophagosome; and finally, fusion of the autophagosome with the lysosome (4). Autophagy is mediated by a combination of general-purpose membrane trafficking proteins, including SNAREs and SNARE-interacting complexes, and 35 or so specialized Atg proteins (5).
In yeast, the initial nucleation of the phagophore occurs at a unique site in the cell, known as the phagophore assembly site or preautophagosomal structure (PAS) (6). The first vesicles to cluster at the phagophore contain the integral membrane protein Atg9 (7). These vesicles are apparently scaffolded by the earliest of all proteins to localize to the PAS, the Atg17–Atg31–Atg29 complex (6, 8). Atg17 and its partners are localized to the PAS in both fed and starved cells. The yeast PAS is present constitutively so as to facilitate the cytoplasm to vacuole targeting (Cvt) pathway. In the Cvt pathway, Atg11 serves as the functional counterpart of Atg17, and both are normally present in the PAS (9). Atg17 has a crescent-shaped fold reminiscent of the curvature-sensing and inducing Bin-Amphiphysin-Rvs (BAR) domain proteins (8). However, in its basal state, Atg17 does not bind directly to vesicles.
Upon starvation, the protein kinase Tor becomes inactive. The Tor substrate Atg13 becomes hypophosphorylated under these conditions (10, 11), thus gaining the ability to bind to Atg17 and to translocate to the PAS. Hypophosphorylated Atg13 also binds to the protein kinase Atg1 (12, 13), which it recruits to the PAS in yeast (14). Atg1 contains a C-terminal early autophagy targeting/tethering (EAT) domain that binds selectively to highly curved vesicles (8). The kinase activity of Atg1 is important in autophagy, although it seems to act on later steps in the pathway (15). The human orthologs of Atg1 are Unc-51-like kinase 1 (ULK1) and Unc-51-like kinase 2 (ULK2), and their interaction with human autophagy-related 13 (Atg13) seems to be regulated in a similar manner (16–19). The precise biochemical mechanisms for phosphoregulation of and by Atg1 and Atg13 are largely unknown.
The Atg1–Atg13–Atg17–Atg31–Atg29 complex assembles at the PAS with a 2:2:2:2:2 stoichiometry (20). It is thought that the two copies of the EAT domain belonging to each complex tether high-curvature vesicles together and help prime them for subsequent SNARE-dependent fusion (8). The generation of phosphatidylinositol (PI) 3-phosphate by the autophagy-specific class III PI 3-kinase complex is another key early event in autophagy (21). In yeast, this complex consists of the beclin ortholog Atg6/Vps30, the coiled-coil protein Atg14, the large kinase and WD40 repeat (WD40 repeats contain conserved Trp and Asp residues and consist of approximately 40 amino acid residues) protein Vps15, and the catalytic subunit Vps34 (5). Atg6, Vps15, and Vps34 also function in the endosomal class III PI 3-kinase complex, whereas Atg14 is uniquely specific to autophagy (5). The recruitment of Atg14 occurs immediately downstream of Atg13 localization to the PAS, but the molecular basis for the recruitment of Atg14 is unknown.
As part of an effort to understand the structural mechanisms of autophagy initiation at the PAS, we have been deconstructing the structure and function of Atg13. We and others have found that a predicted unstructured region in the center of Atg13 (Fig. 1 A and B) is necessary and sufficient for binding to both Atg1 and Atg17 (8, 22). This region is also the locus for most, although not all, of the reported regulatory phosphorylation sites in Atg13 (23). Yeast Atg proteins contain no known phosphopeptide binding domains, and the mechanism for readout of the phosphorylation signals in Atg13 is unknown. Moreover, it remains unclear what regions of Atg13 function in the recruitment of Atg14 (14) and the rest of the class III PI 3-kinase complex. In this study we decided to map the structure and function of the N-terminal domain of the Atg13. Comprising 260 residues, this is the only part of Atg13 predicted to have an ordered fold, yet its function is unknown, and it is not homologous to any domain of known function. Here we find that the Atg13 N-terminal domain is required for the recruitment of Atg14 but not for the localization of Atg13 itself. The 2.3-Å crystal structure of the first 260 residues of Lachancea thermotolerans Atg13 was solved and revealed a completely unexpected structural similarity to the spindle checkpoint protein Mad2 (24–28), which consists in its entirety of a HORMA (Hop1p, Rev1p, and Mad2) domain (29). We found that the Atg13 HORMA domain contains a functionally essential phosphate binding site. Together these findings illuminate how PI 3-kinase is recruited in early autophagy and suggest a role for the Atg13 HORMA domain in phosphoregulation of autophagy.
Fig. 1.
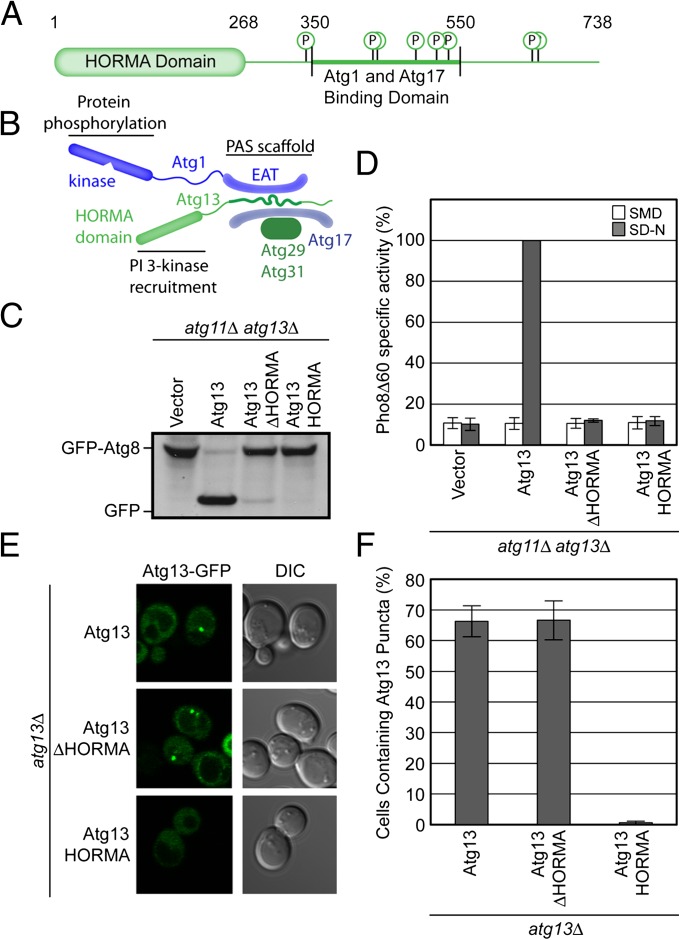
The HORMA Domain of Atg13 is required for autophagy but not for Atg13 localization. (A) Domain representation of S. cerevisiae Atg13. The HORMA domain is labeled and represented as a rounded rectangle. The unstructured region is represented as a straight line. Phosphorylation sites within the unstructured region are indicated. Amino acid numbers corresponding to the boundaries of the HORMA domain and the Atg1/Atg17 binding region are labeled. (B) Schematic of the organization of the Atg1 complex. This complex is a dimer of pentamers, with a single pentamer shown here. The loci of the previously identified PAS scaffolding and protein phosphorylation functions are indicated. The positioning of the HORMA domain identified in this study is highlighted and its PI 3-kinase recruitment activity annotated. (C) GFP-Atg8 processing assay as monitored by Western blot against GFP. The GFP-Atg8 and GFP bands are labeled. (D) Pho8Δ60 assay to monitor autophagy was performed in SMD (white) and SD-N (gray). Samples were normalized to the activity of Atg13 in starved cells. (E) Representative microscopy images of atg13Δ cells transformed with ATG13-GFP, ATG13ΔHORMA-GFP, and ATG13HORMA-GFP in the presence of rapamycin treatment. (F) Quantification of the Atg13 puncta from E. A total of three trials with 100 cells counted per trial. Error bars in D and F represent the SD of triplicate experiments.
Results
HORMA Domain of Atg13 Is Required for Autophagy.
The primary structure of Atg13 consists of a 260- to 270-residue N-terminal domain, which is conserved and predicted to have regular secondary structure, followed by 470 residues that are predicted to be intrinsically disordered (Fig. 1 A and B). As described below, structure determination reveals the N-terminal domain to belong to the HORMA family, and it will therefore be referred to as such. Upon starvation, Atg13 bridges Atg17, which resides at the PAS even under fed conditions, and Atg1, so as to recruit Atg1 to the PAS. This activity of Atg13 resides in the central portion of its primary structure, from residues 350 to 550 (Fig. 1A). Given the conservation of the HORMA domain of Atg13 from yeast to mammals, we inferred that it might be important for function. In the first instance, autophagic function was assessed by monitoring GFP-Atg8 processing (30) (Fig. 1C) and by the Pho8Δ60 assay (31) (Fig. 1D). During autophagy, Atg8 is attached to the membrane of the autophagosome and is subject to vacuolar proteolysis once the autophagosome matures and fuses with the vacuole. GFP is more resistant to proteolysis, and the presence of free GFP is a measure of autophagic function. Pho8Δ60 is an alkaline phosphatase whose activation requires its delivery to the vacuole. With its normal vacuolar sorting signal deleted, the Pho8Δ60 construct only reaches the vacuole and becomes enzymatically active as an autophagosomal passenger. Pho8Δ60 activity is thus a surrogate measure of the delivery of bulk cytosol to the vacuole via autophagosomes. To assay starvation-induced autophagy and avoid background activity due to the Atg11-dependent Cvt pathway, assays were conducted in atg11Δatg13Δ cells.
Expression of a HORMA domain deletion construct, ATG13ΔHORMA, failed to rescue function in atg11Δatg13Δ cells (Fig. 1 C and D). We then asked whether the HORMA domain was important for PAS localization of Atg13. Atg13ΔHORMA localized to the PAS, whereas the isolated HORMA domain (Atg13HORMA) of Atg13 did not (Fig. 1 E and F). This finding is consistent with the concept that Atg13 recruitment to the PAS depends on the presence of an intact Atg17-binding domain within the central region (8, 22) but not on the HORMA domain. It also shows that the isolated HORMA domain lacks an intrinsic PAS targeting activity. These data indicate that the HORMA domain of Atg13 has an essential function in autophagy but that this activity is not involved with the localization of Atg13 itself.
HORMA Domain of Atg13 Is Required for Atg14 Recruitment to the PAS.
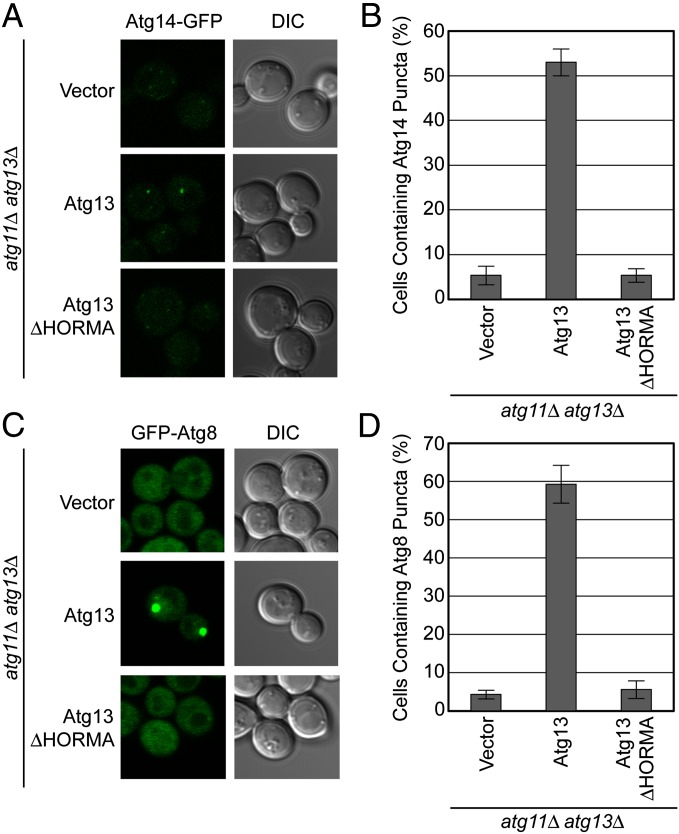
The hierarchy of Atg protein recruitment to the PAS has been elucidated (14), and the subunits of the autophagy-specific PI 3-kinase complex have been shown to translocate to the PAS immediately downstream of Atg13 (14, 15). We next tested whether the HORMA domain of Atg13 was involved in PI 3-kinase recruitment. Because the Atg6, Vps15, and Vps34 subunits of this complex have a dual function in endosome maturation and endolysosomal sorting (21), we focused on the only autophagy-specific subunit of the complex, Atg14. In atg11Δatg13Δ cells expressing wild-type Atg13 and treated with the Tor inhibitor rapamycin, Atg14-GFP forms puncta that fluoresce relatively weakly (Fig. 2 A and B), consistent with previous observations (15). In ATG13ΔHORMA atg11Δatg13Δ cells, the Atg14-GFP puncta are completely lost (Fig. 2 A and B), indicating that the Atg13 HORMA domain is required for recruitment of Atg14 to the PAS. Atg8 conjugation is the most downstream step in the hierarchy of protein recruitment to the PAS and depends on the presence of PI 3-kinase. Atg8 puncta formation is completely lost in ATG13ΔHORMA atg11Δatg13Δ cells (Fig. 2 C and D), consistent with the loss of Atg14 localization in these cells. Taken together, these data indicate that the HORMA domain of Atg13 is essential for PI 3-kinase recruitment and thus essential for autophagy.
Fig. 2.
The HORMA domain of Atg13 is necessary for the recruitment of downstream factors in autophagy. (A) Representative microscopy images of atg11Δ atg13Δ ATG14-GFP cells transformed with vector, ATG13, and ATG13ΔHORMA and treated with rapamycin. (B) Quantification of the Atg14 puncta from A using a brightness threshold of fourfold above the average background fluorescence. A total of three trials with 100 cells counted per trial were made. (C) Representative microscopy images of atg11Δ atg13Δ GFP-ATG8 cells transformed with vector, ATG13, and ATG13ΔHORMA and treated with rapamycin. (D) Quantification of the Atg8 puncta from C. A total of three trials with 100 cells counted per trial. Error bars in B and D represent the SD of triplicate experiments.
Structure of the N-Terminal Region of Atg13 Reveals a HORMA Fold.
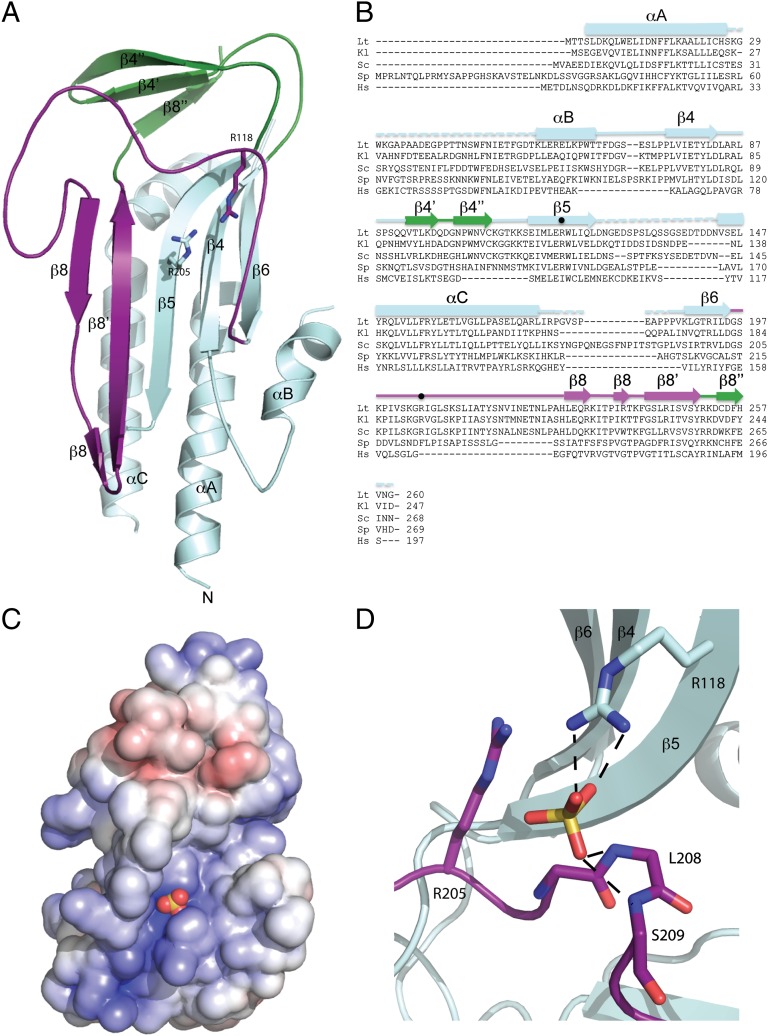
The HORMA domain of L. thermotolerans Atg13 consisting of residues 1–260 was crystallized and its structure determined at 2.3-Å resolution (Fig. 3 A and B, Fig. S1, and Table S1). The domain has an α/β fold centered on a five-stranded antiparallel β-sheet. Four helices pack against one side of the β-sheet, and a smaller three-stranded antiparallel β-sheet sits on one end of the structure. The longest loop in the structure connects the end of β6, which is at the right side of the sheet as seen in Fig. 3A, with the start of β8. A sulfate ion from the crystallization medium is bound by Arg118 of β5 and the main chain amide groups of Leu208 and Ser209 of the β6-β8 loop. Together with nearby Arg205, these residues form an electropositive focal point near the center of a long groove in the protein surface (Fig. 3 C and D). Although Arg205 is not in direct contact with the sulfate ion, its close proximity suggests it contributes to binding through its effect on the local electrostatic potential. Furthermore, this Arg would only need to move approximately 1 Å to directly engage the sulfate.
Fig. 3.
Crystal structure of the Atg13 HORMA domain. (A) Ribbon diagram of the Atg13 HORMA domain. Regions structurally related to MAD2 are colored in cyan except the safety belt region, which is highlighted in purple. β4’, β4’’, and β8’’ of the Atg13 HORMA domain, which are unique to Atg13, are colored in green. (B) Sequence alignment of the Atg13 HORMA domain with the secondary structure elements colored as in A. The two arginine residues located within the sulfate binding site are marked by dots. Species are abbreviated as follows: L. thermotolerans (Lt), Kluyveromyces lactis (Kl), S. cerevisiae (Sc), Schizosaccharomyces pombe (Sp), and Homo sapiens (Hs). (C) Surface representation of the HORMA domain colored according to electrostatic potential, with saturating blue and red at ±7.5 kT/e. The sulfate ion is represented as spheres. (D) Close-up view of the sulfate binding site. The sulfate is coordinated by the side chain of R118 and backbone NH of L208 and L209. Hydrogen bonds are represented as dashed lines. R205 contributes to the overall positive charge of the binding pocket but is not within hydrogen bonding distance.
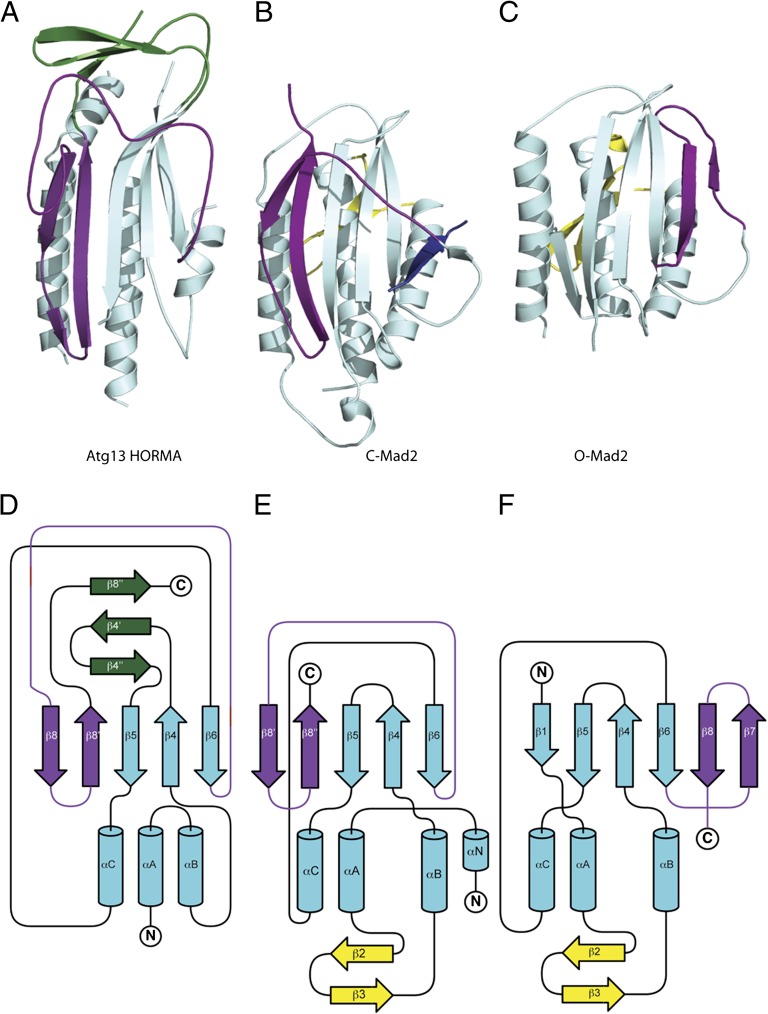
A search of the protein data bank revealed an unexpected structural similarity with the spindle checkpoint protein Mad2 (24–28). Mad2 consists in its entirety of a HORMA domain. Roughly half of the ordered portion (120 residues) of the Atg13 HORMA domain can be superimposed upon the structure of Mad2 with an rmsd of 2.1 Å, leaving no doubt that this region is a bona fide HORMA domain. The secondary structure elements shown in Fig. 3 A and B are numbered to correspond with those of Mad2. Mad2 is a remarkable protein in that it has two different folds, known as C-Mad2 and O-Mad2 (Fig. 4) (27). C-Mad2 is the conformation involved in binding other proteins at the spindle checkpoint. The O-Mad2 structure superimposes on just 95 residues of Atg13 with an rmsd of 2.9 Å, consistent with the clearer visual similarity between Atg13 HORMA and C-Mad2 (Fig. 4). The α-helices and β-strands 4–6 form the fixed core of Mad2, whereas β-strands 7–8’ and the long β6-β8 loop rearrange from one side of the sheet to the other. The β6-β8 loop of Mad2 has a special role in conformation-dependent ligand binding. The loop covers ligands in the C-Mad2 state and holds them securely in place. The entire region encompassing the loop and the conformationally switchable β-strands 7–8’ is termed the “safety belt” (27) (Fig. 4).
Fig. 4.
Unexpected structural homology between Atg13 and C-Mad2. (A) Ribbon diagram of the HORMA domain of Atg13 colored as is shown in Fig. 3A. (B) Ribbon diagram of the closed state of Mad2 oriented identical to Atg13 in A, with the Mad1 binding peptide shown in dark blue. Regions structurally related to the HORMA domain of Atg13 are colored in cyan except the safety belt region, which is highlighted in purple. Regions unique to Mad2 are colored in yellow. Coordinates taken from Protein Data Bank (PDB) ID 1GO4. (C) Ribbon diagram of the open state of Mad2 oriented identical to Atg13 in A and colored exactly as in B. Coordinates taken from PDB ID 2V64. (D–F) Topology diagrams for the Atg13 HORMA domain (D), closed state of Mad2 (E), and open state of Mad2 (F) colored exactly as in A–C.
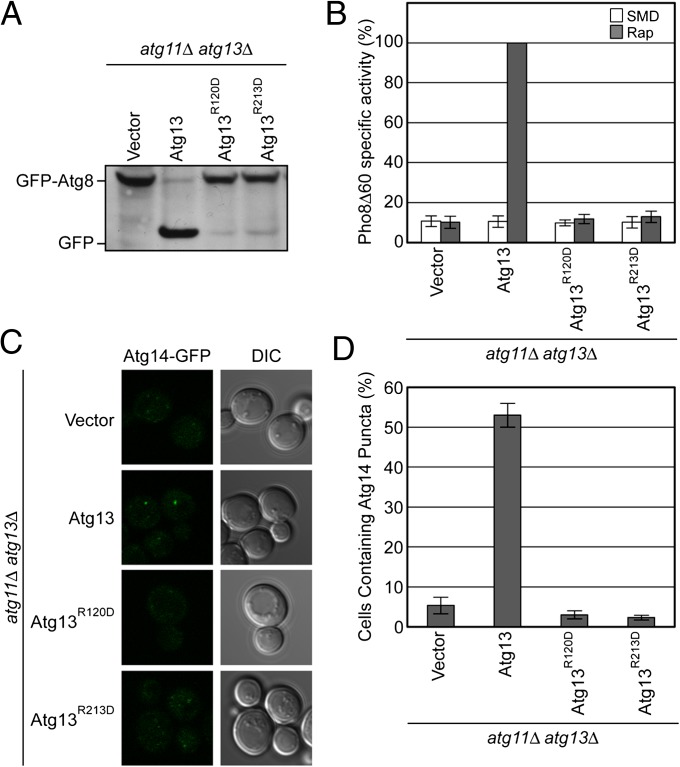
Bound sulfate ions are often markers for biologically relevant binding sites for phosphoryl moieties of phosphoproteins, nucleotides, phospholipids, or other phosphate-containing molecules. We hypothesized that Arg118 and Arg205 of L. thermotolerans Atg13 might comprise a biologically important phosphate sensor. To determine whether the putative phosphate sensor residues were important for function, the relevant mutants were assayed. In Saccharomyces cerevisiae Atg13, the putative phosphate ligands of the L. thermotolerans structure correspond to Arg120 and Arg213. The mutants R120D and R213D were constructed and found to express at levels identical to wild type (Table S2 and Fig. S2). Expression of ATG13R120D or ATG13R213D failed to rescue autophagy in atg11Δatg13Δ cells as measured by the GFP-Atg8 processing and Pho8Δ60 assays (Fig. 5 A and B). Therefore, we infer that the two Arg residues are directly involved in autophagic function. We went on to assess whether the Arg residues were involved in Atg14 localization to the PAS and found that they were required (Fig. 5 C and D).
Fig. 5.
The putative phosphate sensor of the Atg13 HORMA domain is required for autophagy and PI 3-kinase recruitment. (A) GFP-Atg8 processing assay of atg11Δ atg13Δ cells transformed with ATG13 wild type and mutants designed to disrupt the putative phosphate binding site. GFP-Atg8 processing was monitored by Western blot against GFP. The GFP-Atg8 and GFP bands are labeled. (B) Pho8Δ60 assay to monitor autophagy was performed in SMD (white) and SD-N (gray) using atg11Δatg13Δ cells transformed with the same ATG13 constructs as in A. Samples were normalized to the activity of Atg13 in starved cells. (C) Representative images of atg11Δ atg13Δ ATG14-GFP cells transformed with vector, and ATG13 (as shown also in Fig. 2A) and mutant ATG13 alleles, and treated with rapamycin. (D) Quantification of the Atg14 puncta from C using the procedure described in Fig. 2. A total of three trials with 100 cells counted per trial. Error bars in B and C represent the SD of triplicate experiments.
Discussion
Taking the results described above together with other recent reports, the division of labor among different regions of Atg13 is becoming clearer. Here we established that the N-terminal region of Atg13 has a HORMA domain fold and is specifically involved in Atg14 recruitment. The first finding makes a striking and completely unexpected link between Atg13 and the unusual HORMA domain family. At least for Mad2, the HORMA domain is capable of refolding into two dramatically different conformations, suggesting the same could be true for Atg13. The second clarifies the division of labor between different regions of Atg13.
The putative phosphate sensor on the Atg13 HORMA domain is intriguing because the binding site can only be formed when Atg13 is in the C-conformation. It is not known whether Atg13 possesses an O-conformation. If an O-state were to exist, the change in position of the safety belt would be incompatible with phosphate binding at this site, because the C-to-O conformational change would move Arg205 out of position for binding. This suggests that the Atg13 HORMA domain could be a phosphoregulated conformational switch and/or a conformation-dependent phosphate sensor.
The concept of a phosphoregulated switch begs the question as to the identity of the phosphorylated ligand. The two key Arg residues line the walls of a long electropositive groove that runs from southwest to northeast as seen in the view in Fig. 3C. The groove is strongly suggestive of a binding site for a phosphorylated peptide segment. One obvious potential phosphorylated ligand for the Atg13 HORMA domain would be a phosphorylated segment within the central or C-terminal region of the selfsame Atg13 molecule. Tor is the best-characterized, although not the only, kinase involved in Atg13 phosphorylation. Mutation of all eight of the Tor phosphorylation sites in Atg13 to Ala leads to constitutive autophagy (23). We found that mutation of the putative phosphate sensor in the HORMA domain has no such constitutive autophagy phenotype. This observation is inconsistent with a role for the HORMA phosphate sensor in the autoinhibition of Tor phosphorylated Atg13. Binding to a cis-autoactivation sequence phosphorylated by some kinase other than Tor remains a possibility.
The HORMA domain recruits Atg14, but it remains to be seen whether the domain binds directly to the Atg14-containing autophagy-specific PI 3-kinase complex. One important future direction will be to reconstitute this complex and to determine whether it or other bridging factors bind the HORMA domain to drive recruitment. In conclusion, the unexpected findings that Atg13 contains a HORMA domain, that the HORMA domain is required for PI 3-kinase recruitment, and that the HORMA domain in turn contains a functionally required putative phosphate sensor, casts a key facet of autophagy initiation in a new light. The full ramifications of these discoveries will be an important goal of further investigation.
Materials and Methods
Autophagy Assays.
For the Pho8Δ60 assay, MJR20 cells containing yCplac-111, ATG13-yCPLAC111 ATG13ΔHORMA-yCPLAC111, ATG13HORMA-yCPLAC111, ATG13R120D-yCPLAC111, and ATG13R213D-yCPLAC111 were grown to mid log phase in SMD [0.67% (wt/vol) yeast nitrogen base with ammonium sulfate, 2% (wt/vol) glucose, supplemented with the appropriate amino acids]. For starvation, cells were harvested, resuspended in SD-N [0.17% (wt/vol) yeast nitrogen base without ammonium sulfate and amino acids, 2% glucose] and incubated for 4 h at 30 °C shaking. The Pho8Δ60 assay was performed on nonstarved and starved cells as previously described (31).
For the GFP-Atg8 processing assays MJR17 cells containing yCplac-111, ATG13-yCPLAC111 ATG13ΔHORMA-yCPLAC111, ATG13HORMA-yCPLAC111, ATG13R120D-yCPLAC111, and ATG13R213D-yCPLAC111 were grown to mid log phase in SMD. Cells were harvested and resuspended in SD-N and incubated for 4 h at 30 °C. Cells were harvested, resuspended in 50 mM Tris (pH 8.0), 1% (wt/vol) SDS, 6 M urea, and 1 mM EDTA, and lysed by the addition of a half volume of 425- to 600-mm glass beads (Sigma, G8772) using the Mini-Beadbeater-16 (Biospec Products). Cell lysates were cleared by centrifugation at 16,000 × g for 5 min at 4 °C and subjected to Western blot analysis using an anti-GFP antibody (Santa Cruz, sc9996).
Expression of ATG13, ATG13ΔHORMA, ATG13HORMA, ATG13R120D, and ATG13R213D was confirmed by transforming atg13Δ yeast cells with ATG13-GFP, ATG13ΔHORMA-GFP, ATG13HORMA-GFP, ATG13R120D-GFP, or ATG13R213D-GFP alleles subcloned into pJK59. Cells were grown to mid log phase in SMD. Cells were subsequently harvested, resuspended in 50 mM Tris (pH 8.0), 1% (wt/vol) SDS, 6 M urea, and 1 mM EDTA and lysed by the addition of a half volume of 425- to 600-mm glass beads (Sigma, G8772) using the Mini-Beadbeater-16 (Biospec Products). Cell lysates were cleared by centrifugation at 16,000 × g for 5 min at 4 °C and subjected to Western blot analysis using an anti-GFP antibody (Santa Cruz, sc9996).
Microscopy.
For analysis of Atg13 localization, atg13Δ yeast cells (Invitrogen) were transformed with ATG13-GFP, ATG13ΔHORMA-GFP, and ATG13HORMA-GFP alleles subcloned into pJK59. For analysis of Atg14 puncta, HCY142 (atg11Δ atg13Δ ATG14-GFP) cells were transformed with yCPLAC111, ATG13-yCPLAC111, ATG13ΔHORMA-yCPLAC111, ATG13R120D- yCPLAC111, and ATG13R213D- yCPLAC111. For analysis of Atg8 puncta, MJR17 (atg11Δ atg13Δ GFP-ATG8) cells were transformed with yCPLAC111, ATG13-yCPLAC111, and ATG13ΔHORMA-yCPLAC111. Cells were grown to mid log phase in SMD [0.67% (wt/vol) yeast nitrogen base with ammonium sulfate, 2% (wt/vol) glucose, supplemented with the appropriate amino acids] and subsequently treated with 0.2 µg mL−1 rapamycin. After rapamycin treatment, cells were incubated at 30 °C shaking for 1 h and then visualized. For analysis of Atg13 and Atg14 puncta, a stack of 11 to 12 z-sections was collected at an interval of 0.44 µm. Atg8 puncta are bright and were therefore observed in a single confocal section. Atg8 and Atg13 puncta were unambiguously identified by inspection. An unbiased method was used to quantitate Atg14 puncta. A maximum intensity projection was performed on the z-sections using ZEN (Carl Zeiss Microscopy), and the images were imported into ImageJ. The fluorescent GFP background in cells was determined for each image by averaging the background within a 30 × 30 pixel zone in five cells. Atg14 puncta were quantitated on the basis of a threshold fourfold brighter than the background. All microscopy was performed on an LSM780 scanning confocal microscope (Carl Zeiss Microscopy) with a 100× oil immersion objective. For all puncta quantitation, 100 cells were counted per trial, and the final experiment is the result of three independent trials.
Crystallization and Structure Determination
Native and selenomethionyl (SeMet) protein were crystallized by hanging drop vapor diffusion using equal volumes of protein and well solution containing 0.2 M AmoSO4, 0.1 M Mes (pH 6.5), and 20% wt/vol Peg 8K at 21 °C. Crystals were cryoprotected by stepwise addition of glycerol to 20% (vol/vol), followed by flash freezing in liquid nitrogen. SeMet-1 contained the mutation (L123M) residue and data were collected at Diamond Light Source on I02 beamline (Oxfordshire, United Kingdom). SeMet-2 contained two additional methionines (L116M/L123M), and the data were collected at the Advanced Photon Source on the SER-CAT 22-BM beamline (Chicago, IL). Data were processed with HKL2000 (HKL Research), and the crystals are in the C2 space group with two copies in the asymmetric unit. The structure was initially solved by single anomalous diffraction (SAD) phasing using AutoSol (32) with the SeMet-1 dataset; however, the phasing power was weak, and the initial experimental maps were of low resolution. Higher-resolution phases were determined by SAD with AutoSol using the SeMet-2 data. An initial backbone trace of the protein was built manually in Coot (33), and then this was used for combination with the SeMet-1 SAD phases to determine the structure in Autosol. Autobuild (32) was used to build the model, followed by several cycles of manual model building with COOT and refinement with Refmac (34). The final structure was refined to an Rwork/Rfree of 0.22/0.27.
Supplementary Material
Acknowledgments
We thank A. Toulmay and W. Prinz for advice, N. Noinaj and P. Lukacik for data collection at the Diamond Light Source, and D. J. Klionksy and R. Youle for yeast strains. Crystallographic data were collected at Southeast Regional Collaborative Access Team 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory and at the Diamond Light Source, Oxfordshire, United Kingdom. Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38. This work was supported by the Intramural Program of the National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases (J.H.H.), a fellowship from the National Institute of General Medical Sciences Postdoctoral Research Associate Program (C.C.J.), a Damon Runyon Cancer Research Fellowship (R.E.S.), and by Ruth Kirschtein National Research Service Award Fellowship GM099319 (to M.J.R.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4J2G).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1220306110/-/DCSupplemental.
References
- 1.Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: Lessons from yeast. Nat Rev Mol Cell Biol. 2009;10(7):458–467. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- 2.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451(7182):1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132(1):27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rubinsztein DC, Shpilka T, Elazar Z. Mechanisms of autophagosome biogenesis. Curr Biol. 2012;22(1):R29–R34. doi: 10.1016/j.cub.2011.11.034. [DOI] [PubMed] [Google Scholar]
- 5.Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 6.Suzuki K, Ohsumi Y. Current knowledge of the pre-autophagosomal structure (PAS) FEBS Lett. 2010;584(7):1280–1286. doi: 10.1016/j.febslet.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 7.Mari M, et al. An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. J Cell Biol. 2010;190(6):1005–1022. doi: 10.1083/jcb.200912089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ragusa MJ, Stanley RE, Hurley JH. Architecture of the Atg17 complex as a scaffold for autophagosome biogenesis. Cell. 2012;151(7):1501–1512. doi: 10.1016/j.cell.2012.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yorimitsu T, Klionsky DJ. Atg11 links cargo to the vesicle-forming machinery in the cytoplasm to vacuole targeting pathway. Mol Biol Cell. 2005;16(4):1593–1605. doi: 10.1091/mbc.E04-11-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scott SV, et al. Apg13p and Vac8p are part of a complex of phosphoproteins that are required for cytoplasm to vacuole targeting. J Biol Chem. 2000;275(33):25840–25849. doi: 10.1074/jbc.M002813200. [DOI] [PubMed] [Google Scholar]
- 11.Funakoshi T, Matsuura A, Noda T, Ohsumi Y. Analyses of APG13 gene involved in autophagy in yeast, Saccharomyces cerevisiae. Gene. 1997;192(2):207–213. doi: 10.1016/s0378-1119(97)00031-0. [DOI] [PubMed] [Google Scholar]
- 12.Reggiori F, Tucker KA, Stromhaug PE, Klionsky DJ. The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev Cell. 2004;6(1):79–90. doi: 10.1016/s1534-5807(03)00402-7. [DOI] [PubMed] [Google Scholar]
- 13.Chang YY, Neufeld TP. An Atg1/Atg13 complex with multiple roles in TOR-mediated autophagy regulation. Mol Biol Cell. 2009;20(7):2004–2014. doi: 10.1091/mbc.E08-12-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suzuki K, Kubota Y, Sekito T, Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007;12(2):209–218. doi: 10.1111/j.1365-2443.2007.01050.x. [DOI] [PubMed] [Google Scholar]
- 15.Cheong H, Nair U, Geng JF, Klionsky DJ. The Atg1 kinase complex is involved in the regulation of protein recruitment to initiate sequestering vesicle formation for nonspecific autophagy in Saccharomyces cerevisiae. Mol Biol Cell. 2008;19(2):668–681. doi: 10.1091/mbc.E07-08-0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mercer CA, Kaliappan A, Dennis PB. A novel, human Atg13 binding protein, Atg101, interacts with ULK1 and is essential for macroautophagy. Autophagy. 2009;5(5):649–662. doi: 10.4161/auto.5.5.8249. [DOI] [PubMed] [Google Scholar]
- 17.Jung CH, et al. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20(7):1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hosokawa N, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20(7):1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ganley IG, et al. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem. 2009;284(18):12297–12305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kabeya Y, et al. Characterization of the Atg17-Atg29-Atg31 complex specifically required for starvation-induced autophagy in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2009;389(4):612–615. doi: 10.1016/j.bbrc.2009.09.034. [DOI] [PubMed] [Google Scholar]
- 21.Backer JM. The regulation and function of Class III PI3Ks: Novel roles for Vps34. Biochem J. 2008;410(1):1–17. doi: 10.1042/BJ20071427. [DOI] [PubMed] [Google Scholar]
- 22.Kabeya Y, et al. Atg17 functions in cooperation with Atg1 and Atg13 in yeast autophagy. Mol Biol Cell. 2005;16(5):2544–2553. doi: 10.1091/mbc.E04-08-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamada Y, et al. Tor directly controls the Atg1 kinase complex to regulate autophagy. Mol Cell Biol. 2010;30(4):1049–1058. doi: 10.1128/MCB.01344-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo XL, et al. Structure of the Mad2 spindle assembly checkpoint protein and its interaction with Cdc20. Nat Struct Biol. 2000;7(3):224–229. doi: 10.1038/73338. [DOI] [PubMed] [Google Scholar]
- 25.Mapelli M, Massimiliano L, Santaguida S, Musacchio A. The Mad2 conformational dimer: Structure and implications for the spindle assembly checkpoint. Cell. 2007;131(4):730–743. doi: 10.1016/j.cell.2007.08.049. [DOI] [PubMed] [Google Scholar]
- 26.Mapelli M, et al. Determinants of conformational dimerization of Mad2 and its inhibition by p31comet. EMBO J. 2006;25(6):1273–1284. doi: 10.1038/sj.emboj.7601033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sironi L, et al. Crystal structure of the tetrameric Mad1-Mad2 core complex: Implications of a ‘safety belt’ binding mechanism for the spindle checkpoint. EMBO J. 2002;21(10):2496–2506. doi: 10.1093/emboj/21.10.2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chao WCH, Kulkarni K, Zhang ZG, Kong EH, Barford D. Structure of the mitotic checkpoint complex. Nature. 2012;484(7393):208–213. doi: 10.1038/nature10896. [DOI] [PubMed] [Google Scholar]
- 29.Aravind L, Koonin EV. The HORMA domain: A common structural denominator in mitotic checkpoints, chromosome synapsis and DNA repair. Trends Biochem Sci. 1998;23(8):284–286. doi: 10.1016/s0968-0004(98)01257-2. [DOI] [PubMed] [Google Scholar]
- 30.Klionsky DJ, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4(2):151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klionsky DJ. Monitoring autophagy in yeast: The Pho8Δ60 assay. Methods Mol Biol. 2007;390:363–371. doi: 10.1007/978-1-59745-466-7_24. [DOI] [PubMed] [Google Scholar]
- 32.Terwilliger TC, et al. Decision-making in structure solution using Bayesian estimates of map quality: The PHENIX AutoSol wizard. Acta Crystallogr D Biol Crystallogr. 2009;65(Pt 6):582–601. doi: 10.1107/S0907444909012098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murshudov GN, Vagin AA, Lebedev A, Wilson KS, Dodson EJ. Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr D Biol Crystallogr. 1999;55(Pt 1):247–255. doi: 10.1107/S090744499801405X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.