Abstract
Mutant human Cu/Zn superoxide dismutase 1 (SOD1) is associated with motor neuron toxicity and death in an inherited form of amyotrophic lateral sclerosis (ALS; Lou Gehrig disease). One aspect of toxicity in motor neurons involves diminished fast axonal transport, observed both in transgenic mice and, more recently, in axoplasm isolated from squid giant axons. The latter effect appears to be directly mediated by misfolded SOD1, whose addition activates phosphorylation of p38 MAPK and phosphorylation of kinesin. Here, we observe that several different oligomeric states of a fusion protein, comprising ALS-associated human G85R SOD1 joined with yellow fluorescent protein (G85R SOD1YFP), which produces ALS in transgenic mice, inhibited anterograde transport when added to squid axoplasm. Inhibition was blocked both by an apoptosis signal-regulating kinase 1 (ASK1; MAPKKK) inhibitor and by a p38 inhibitor, indicating the transport defect is mediated through the MAPK cascade. In further incubations, we observed that addition of the mammalian molecular chaperone Hsc70, abundantly associated with G85R SOD1YFP in spinal cord of transgenic mice, exerted partial correction of the transport defect, associated with diminished phosphorylation of p38. Most striking, the addition of the molecular chaperone Hsp110, in a concentration substoichiometric to the mutant SOD1 protein, completely rescued both the transport defect and the phosphorylation of p38. Hsp110 has been demonstrated to act as a nucleotide exchange factor for Hsc70 and, more recently, to be able to cooperate with it to mediate protein disaggregation. We speculate that it can cooperate with endogenous squid Hsp(c)70 to mediate binding and/or disaggregation of mutant SOD1 protein, abrogating toxicity.
Keywords: axoplasmic transport, SOD1-linked ALS
In a number of neurodegenerative diseases, aggregates of specific proteins are observed in particular affected neuronal populations. The nature of misfolding and aggregation of these proteins in an aging-related manner remains poorly understood, as does the dominant “gain of function” toxicity associated with them (loss of function remains a possibility in some cases). Among this collective of diseases is amyotrophic lateral sclerosis (ALS) or Lou Gehrig disease, in which a number of different mutationally altered proteins, including superoxide dismutase 1 (SOD1) (1, 2), TARDBP (3, 4), FUS (5, 6), optineurin (OPTN) (7), and ubiquilin 2 (UBQLN2) (8) have been associated with heritable forms. These proteins may in general be found in prominent cytosolic aggregates in motor neurons of affected individuals. SOD1 has perhaps been most amenable to study, because transgenic mice overproducing various mutant human versions of this normally dimeric superoxide–scavenging enzyme develop ALS after some months, with clinical signs resembling those of humans (9). A large number of different amino acid substitutions in human SOD1 have been associated with ALS, and many have been shown to destabilize the protein, disposing it to misfolding and aggregation (10, 11). The exact mechanisms of toxicity remain unclear, but development of disease is associated with alterations in motor neurons, including electrophysiological changes (12–14), mitochondrial defects (15), axonal transport deficiency (16–20), and neuromuscular junction dysfunction/retraction (21, 22). In addition, glia, which also express the mutant protein, play a significant role in progression of disease (23–26).
What forms of mutant SOD1 are toxic? How do they exert toxicity, and, particularly, can molecular chaperones, which specifically bind nonnative proteins, act to prevent a toxic action? Here we have tested these questions using a well-established system for study of fast axonal transport, the isolated axoplasm from the giant axon of the squid, Loligo pealei (27).
We find that added G85R mutant human SOD1 fused with yellow fluorescent protein (G85R SOD1YFP), a protein we previously associated with development of ALS in transgenic mice (28), produces inhibition of anterograde, kinesin-dependent, fast axonal transport in the isolated axoplasm, which is associated with activation of a MAPK cascade. By contrast, WT SOD1 fused with YFP exerts only a minor effect. We observe that addition of the cytosolic molecular chaperone, mammalian Hsc70, previously observed as the predominant protein associating with the G85R SOD1-YFP in spinal cord of transgenic mice (28), can partially reverse the transport defect. Strikingly, the molecular chaperone Hsp110, also associated with the mutant SOD1 in spinal cord (28) and established as a nucleotide exchange factor for Hsc70 (29, 30) that assists it in protein disaggregation (31, 32), completely reverses the transport defect when added at levels substoichiometric to the mutant protein. This establishes a role for molecular chaperones in potentially serving to bind and prevent the toxicity of disease-producing misfolded SOD1 species.
Results
G85R SOD1-YFP but Not WT SOD1-YFP Inhibits Anterograde Fast Axonal Transport in Squid Axoplasm.
Although deficiencies in axonal transport have been described in mouse models of ALS (16–20), the relative inaccessibility of mouse axons to biochemical manipulation led us to use axoplasm isolated from squid giant axon, a preparation free of the axonal membrane, to which it is possible to directly add purified proteins and small molecules and observe their effects on transport in real-time (27). Additionally, this system allows for recovery of the incubated axoplasm for biochemical and immunochemical analysis.
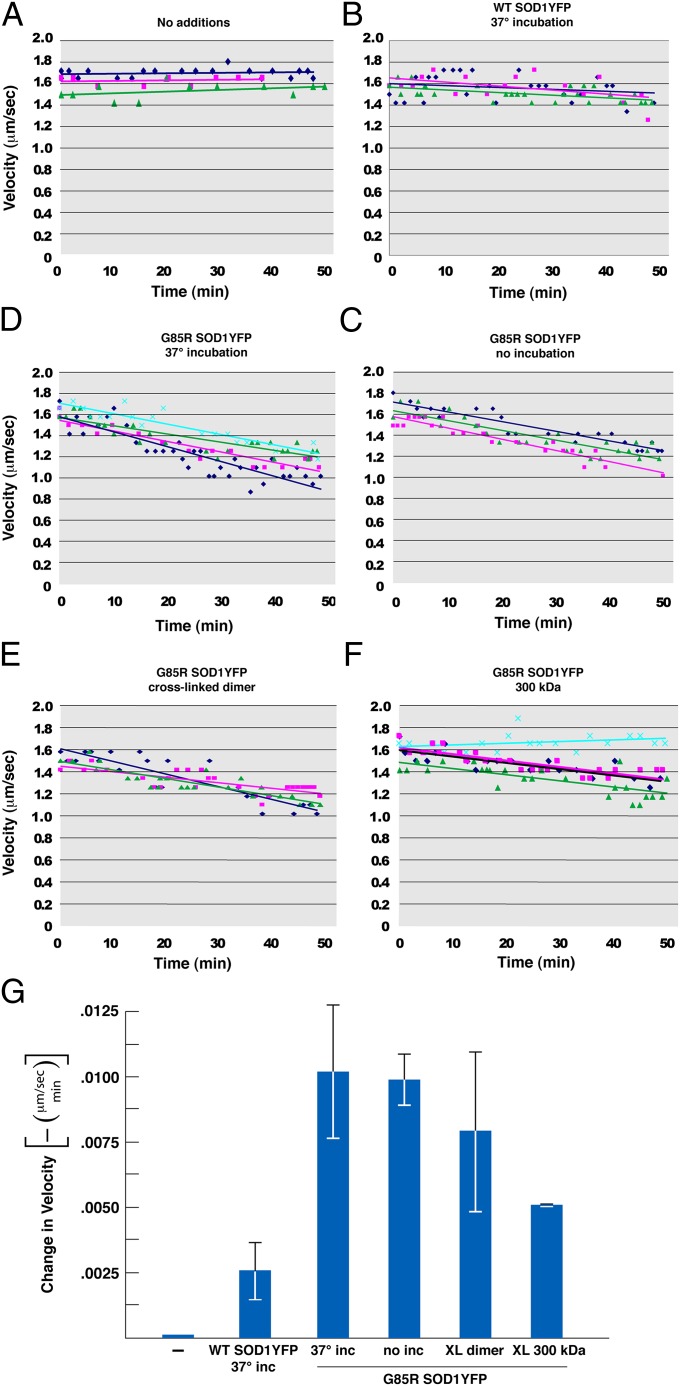
To provide proteins for measuring effects on axoplasmic transport, we overexpressed both WT and ALS-associated G85R mutant forms of human SOD1 fused to YFP bearing a C-terminal hexahistidine tag in Escherichia coli and purified the soluble protein (WT SOD1YFP and G85R SOD1YFP, respectively; Methods). The purified mutant protein behaved mainly as a monomer on gel filtration, with some earlier-eluting material reflecting misfolded or larger molecular size species (Fig. S1A). To increase production of such species, the mutant protein was incubated at 37 °C for 24 h, which indeed expanded the earlier-eluting species (Fig. S1B). We observed from rechromatography studies, however, that the apparent size of the larger material was dynamic. Therefore, to stabilize individual species, we carried out disuccinimidyl suberate (DSS) crosslinking after 37 °C incubation, followed by gel filtration and isolation of specific size fractions (Fig. S1C; Fig. S2 for SDS gel analysis of the crosslinked fractions). These various mutant species as well as 37 °C-incubated WT dimer (Fig. S1D) were then mixed with the perfusion buffer surrounding the intact axoplasm extruded from a squid giant axon, rapidly diffusing through the axoplasm as observed by fluorescence microscopy. The effect of such addition on the rate of transport of vesicular structures (∼20–200 nm diameter) was measured in real-time by video-enhanced contrast differential interference contrast (DIC) microscopy (27).
In all cases, the mutant G85R SOD1YFP species produced progressive slowing of anterograde axonal transport (Fig. 1, compare C–F with A). In contrast, there was no specific effect of the mutant on retrograde transport (Fig. S3). Whereas the mutant species all produced relatively similar slowing of anterograde transport, the WT dimer exhibited only a small effect (Fig. 1B). Note that the same microgram amount of each mutant species was added so that, for example, the molarity of the 300-kDa species is reduced relative to that of the dimer fraction.
Fig. 1.
Fast anterograde axoplasmic transport measured by video-enhanced contrast DIC microscopy is inhibited by G85R SOD1YFP. Instantaneous rates of anterograde vesicular movement (μm/sec) as a function of incubation time in axoplasms incubated in 25 µL perfusion buffer containing (A) no additions; (B) 4.6 µM WT SOD1YFP (final concentration) previously incubated at 37 °C for 24 h; (C) 4.6 µM G85R SOD1YFP, without prior incubation; (D) 4.6 µM G85R SOD1YFP, incubated for 24 h at 37 °C; and (E and F), two gel filtration fractions of G85R SOD1YFP, which had been incubated and crosslinked (XL) with DSS, corresponding to dimeric SOD1YFP (E) and multimeric species of ∼300 kDa (F) (see Fig. S1 and Methods for additional detail), both in final concentrations equivalent to 4.6 µM monomer. In each panel, the individual points are the rate of anterograde movement at the specified time after addition of SOD1YFP protein to the perfusion buffer surrounding the axoplasm, with each color representing a single axoplasm. The corresponding colored lines are linear best fits to the corresponding points. (G) The negative of the slope from the respective sets of fit lines in panels A–F are plotted, showing the mean and SE for the three or four experiments under each condition. Although the WT exerted a small slowing of anterograde transport, the various forms of G85R SOD1YFP all produced significantly greater slowing of transport.
These results parallel those reported by Bosco et al. (33), in which either a different human mutant SOD1 (H46R) or hydrogen peroxide–treated WT human SOD1 had similar inhibitory effects specifically on anterograde transport in the axoplasm assay, whereas WT human SOD1 had little or no effect. The present data suggest that a variety of oligomeric forms of mutant SOD1 species may be toxic to transport, potentially including monomeric forms, but the dynamic nature of the noncrosslinked material precludes conclusions about which species might be most toxic in that context.
G85R SOD1YFP Activates p38 MAPK in Squid Axoplasm.
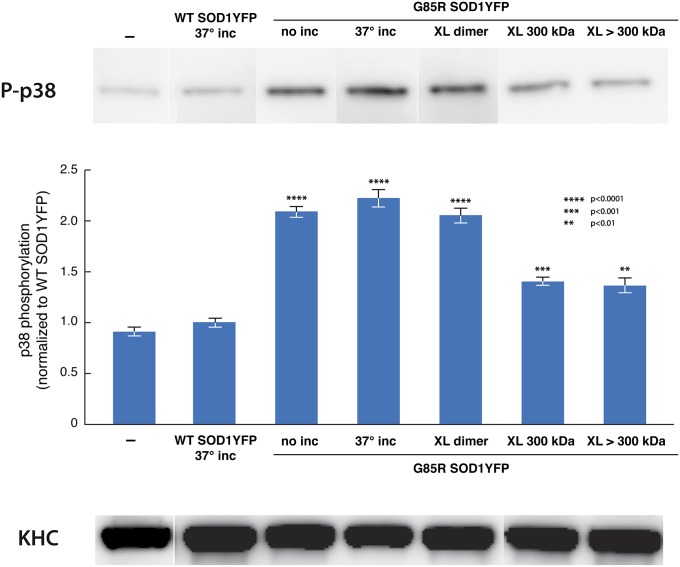
Studies of fast axonal transport in squid axoplasm in the presence of H46R SOD1 and oxidized SOD1, as well as other neurodegeneration-associated proteins, have associated transport defects with activation of various kinase cascades (34). In the case of SOD1, such transport defects are associated with phosphorylation of p38 MAPK, which stimulates its kinase activity (33). To assess whether the presence of G85R SOD1YFP could likewise lead to increased phosphorylation of p38, axoplasm was incubated with WT SOD1YFP or various forms of G85R SOD1YFP, and phosphorylation of p38 was assayed by Western blot analysis with an anti-phospho p38 antibody. As shown in Fig. 2, whereas WT SOD1YFP did not detectably activate phosphorylation of the kinase relative to axoplasm alone, the mutant G85R SOD1YFP forms produced increases ranging from 1.5-fold for 300 kDa and >300 kDa to greater than twofold for nonincubated, 37 °C-incubated, and crosslinked dimer. This extent of phosphorylation of p38 in the presence of the various mutant species correlates well with the inhibition of fast anterograde transport. The specific activity of phosphorylation of p38 may be roughly similar for all of the G85R SOD1YFP species on a molar basis.
Fig. 2.
An increase of p38 MAPK phosphorylation correlates with the inhibitory effect of G85R SOD1YFP on anterograde transport. (Top) Axoplasms were incubated as in Fig. 1 for 50 min, and aliquots were electrophoresed and immunoblotted with anti-phospho p38 antibody. Lanes from a representative blot are shown. (Middle) Quantitation of phosphorylation by densitometry from three individual experiments of each type, with mean, SEs, and P values relative to no additions as indicated. (Bottom) An internal loading control, endogenous kinesin heavy chain (KHC), determined by immunoblotting the same aliquot of each axoplasm as in the top panel. Phosphorylation was greatest in the presence of G85R SOD1YFP (whether incubated at 37 °C or not) as well as with DSS-crosslinked dimer (purified by gel filtration). Because equal amounts of protein were added in each case, the relatively lower level of phosphorylation in the presence of 300 kDa and >300 kDa, relative to nonfractionated G85R SOD1YFP, could reflect that their “specific activity” for activation of p38 phosphorylation is relatively similar to that of the other mutant species.
Inhibition of Anterograde Fast Axonal Transport Is Abolished by Either p38 MAPK Inhibitor MW069 or ASK1 (MAPKKK) Inhibitor NQDI-1.
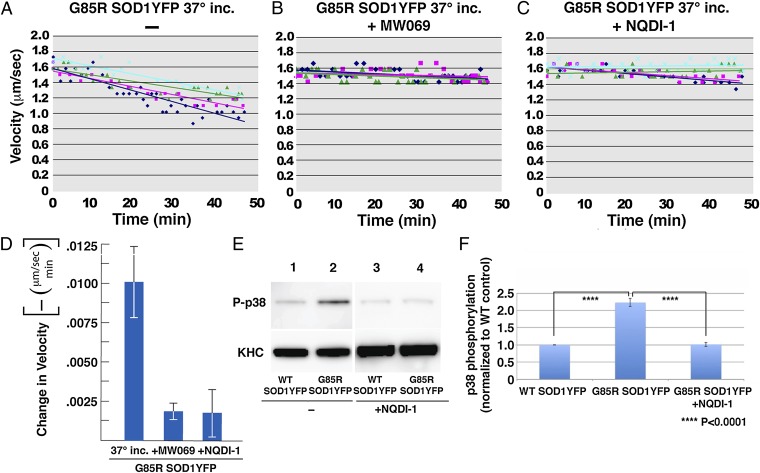
We asked whether there was a causal relationship between phosphorylation (activation) of p38 in the presence of G85R SOD1YFP and the slowing of anterograde transport. The p38 kinase inhibitor MW069 (35) was added to an axoplasmic transport assay containing 37 °C-incubated G85R SOD1YFP, and we observed that it completely normalized the rate of anterograde transport (Fig. 3 A and B). This indicates a proximate involvement of p38 kinase activity in the effects on anterograde transport. We then asked whether the entire MAPK cascade was involved. Several inhibitors at the level of MAPKKK were tested, and here we observed that the inhibitor NQDI-1, which inhibits the MAPKKK apoptosis signal-regulating kinase 1 (ASK1) (36), also corrected the anterograde transport defect (Fig. 3 A, C, and D). As expected, addition of NQDI-1 led to a block of the downstream phosphorylation of p38 (Fig. 3 E and F).
Fig. 3.
Inhibitors of the MAPK cascade reverse transport inhibition by G85R SOD1YFP, associated with inhibition of the increased phosphorylation of p38. G85R SOD1YFP that had been incubated at 37 °C for 24 h was added to axoplasm (A) in the absence of inhibitor; (B) in the presence of the MAPK inhibitor, MW069; or (C) in the presence of the ASK1 MAPKKK inhibitor NQDI-1. Transport was assayed as in Fig. 1; (A) is a duplication of Fig. 1D. (D) Negatives of the slopes of the fitted lines in A–C, showing means and SEs. Both MW069 and NQDI-1 block the inhibition of transport by G85R SOD1YFP. The same effects were obtained when unincubated G85R SOD1YFP was used (not shown). (E) Immunoblot for phospho-p38 in axoplasms incubated with WT or mutant SOD1YFP in the absence (left two lanes) or presence (right two lanes) of ASK inhibitor NQDI-1. Each phosphorylation experiment was conducted three times, with a representative experiment shown. (F) Quantitation of the three immunoblots corresponding to E, with means and SDs shown; P values are for the comparison indicated by the brackets. ASK inhibitor NQDI-1, by blocking the ASK1 MAPKKK, blocks the downstream activation of phosphorylation of p38 in the presence of G85R SOD1YFP (compare lane 4 with lane 2).
Hsc70 Partially Inhibits Slowing of the Fast Axonal Transport Defect of G85R SOD1YFP, Whereas Addition of Hsp110 (HSPA4L) Completely Blocks Slowing.
Earlier studies have indicated that the molecular chaperone Hsc70 interacts with mutant human SOD1 protein in the spinal cord of transgenic mice that develop ALS (37, 38). This interaction also occurred in mouse strains transgenic for G85R SOD1YFP (28). We have also observed the association of G85R SOD1YFP with the nucleotide exchange factors for Hsc70 known collectively as Hsp110s (three related proteins, Hspa4, Hspa4L, and Hsph1; 28). By contrast, no such chaperone interactions were observed in spinal cords of WT SOD1YFP transgenic mice. Notably, Hsc70/Hsp70 chaperone interactions occur with extended segments of nonnative proteins that expose hydrophobic side chains (39, 40). Such features are presented both by newly made proteins and by misfolded ones, which in some cases are associated with neurodegenerative processes. The latter interactions may be relatively persistent compared with the interactions with proteins that can proceed to the native state. Such chaperone interactions with mutant SOD1s may comprise the most proximate protein interactions.
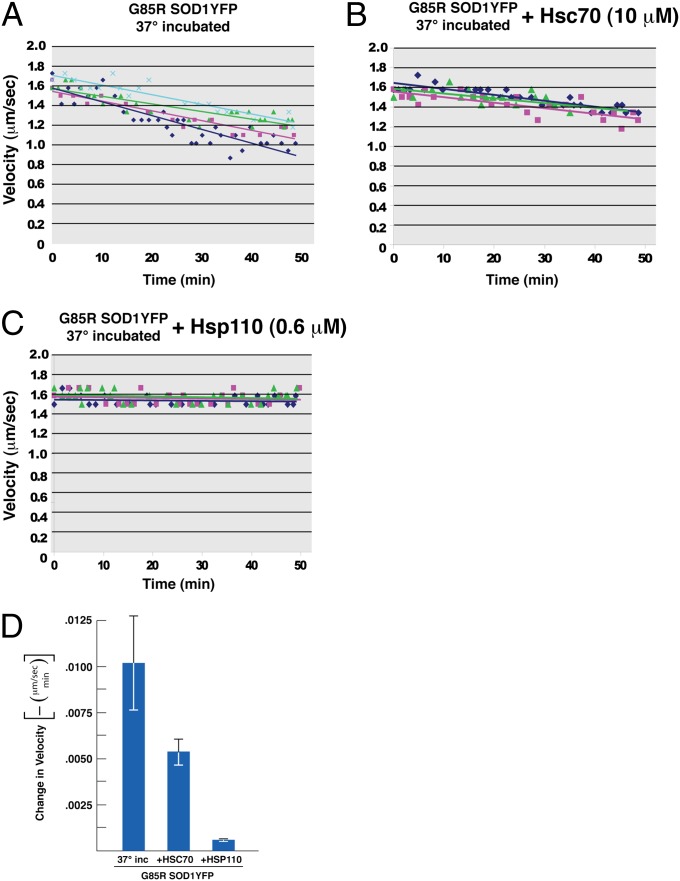
We conjectured that exogenously added Hsc70 or Hsp110 might provide additional protection against MAPK activation beyond that afforded by endogenous squid chaperones and thus reverse the slowing of anterograde axonal transport. Addition of purified bovine Hsc70 in twofold molar excess over G85R SOD1YFP (10 μM vs. 4.6 μM) indeed reduced the G85R SOD1YFP–induced transport defect by ∼50% (Fig. 4B). Even more strikingly, addition of substoichiometric amounts of Hsp110 (0.6 μM of human HSPA4L), completely corrected the transport defect produced by G85R SOD1YFP (Fig. 4C). The substoichiometric action exerted by Hsp110 would be consistent with its cooperation with an endogenous squid component(s), particularly endogenous Hsc70 or Hsp70, and likely a DnaJ homolog, to mediate masking of a toxic element of G85R SOD1YFP or to drive dissociation of a toxic protein–protein interaction. For example, if misfolded monomer associates with other proteins to produce toxicity, then the addition of Hsp110 and its cooperation with squid Hsp70 (and DnaJ) components might dissociate such oligomeric species and thus detoxify the mutant protein. Notably, recent studies have indicated a role for Hsp110 in disaggregation, functioning via its nucleotide exchange activity with Hsc70 (32; see also ref. 31, in which direct binding of substrate protein by Hsp110 is also indicated). Importantly, the chaperone effects observed here were not associated with proteolytic turnover of G85R SOD1YFP, because Western blot analysis of the incubated squid axoplasm did not reveal an alteration in the level of the mutant protein (Fig. S4). Finally, incubation of the Hsc70 and Hsp110 chaperones together exerted the same corrective effect on transport as Hsp110 alone.
Fig. 4.
Addition of molecular chaperones Hsc70 or Hsp110 reduces and reverses, respectively, the effect of G85R SOD1YFP on anterograde axoplasmic transport. Transport assays were carried out and plotted as described in Fig. 1, with 4.6 µM final concentration of G85R SOD1YFP that had been incubated at 37 °C for 24 h. (A) Reproduction of Fig. 1D. (B) 10 µM Hsc70 (final concentration) was added to the axoplasm mixture at the same time as G85R SOD1YFP; (C) 0.6 µM Hsp110 (final concentration) was added at the same time as G85R SOD1YFP. (D) The negative of the slopes with means and SEs from the transport data taken from three experiments for each condition are plotted. Whereas Hsc70 reduced the inhibition of transport by ∼50%, the addition of Hsp110 completely blocked inhibition of transport by G85R SOD1YFP. Note that addition of Hsp110 to axoplasm in the absence of added G85R SOD1YFP did not affect the rate of anterograde transport (not shown).
The nature of the interactions in the squid axoplasm between the endogenous chaperones and exogenously added Hsc70 and Hsp110 is not clear. We have, however, used anti-YFP affinity chromatography to capture G85R SOD1YFP from the squid axoplasm assay after incubation without exogenous chaperones. Proteolysis/HPLC-MS readily identified two Hsp70-related squid proteins as the dominant species captured with mutant SOD1YFP itself, along with two DnaJ homologs (Table S1). Interestingly, there was also selective capture of cytoskeletal and synaptic vesicle proteins as well as both kinesin and dynein. By contrast, much lower levels of chaperone species were captured from axoplasm that had been incubated with WT SOD1YFP (Table S1). Notably, in neither case was either of two predicted endogenous squid Hsp110 proteins detected.
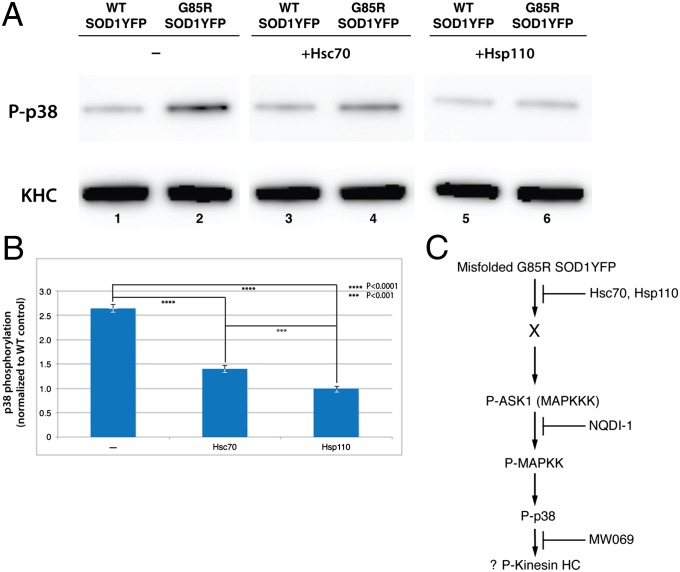
Hsc70 Inhibits Phosphorylation of p38 in the Presence of G85R SOD1YFP, and Hsp110 Completely Blocks Phosphorylation.
Consistent with the placement of MAPK activation distal to chaperone-mediated action, addition of either Hsc70 or Hsp110 (or both) to the G85R SOD1YFP–containing axoplasm was able to inhibit phosphorylation of p38 (Fig. 5). The action of Hsc70 (10 μM) was partial (compare lanes 2 and 4), corresponding to the degree of its effect on transport (Fig. 4B). In contrast, the action of Hsp110 (0.6 μM) was complete, with phosphorylation reduced to the level seen with WT SOD1YFP (compare lanes 2, 5, and 6). This effect corresponds to the complete correction of the slowing of anterograde transport (Fig. 4B).
Fig. 5.
Addition of the molecular chaperones Hsc70 or Hsp110 inhibits activation of p38 phosphorylation by G85R SOD1YFP to a degree that parallels that of blocking the inhibitory effect of the mutant protein on anterograde transport. (A) Phosphorylation of p38, detected by immunoblotting with anti-phospho-p38 antibody, in axoplasm incubated with WT SOD1YFP (lanes 1, 3, and 5) or G85R SOD1YFP (lanes 2, 4, and 6), without additions (lanes 1 and 2) or in the presence of either Hsc70 (lanes 3 and 4) or Hsp110 (lanes 5 and 6). (B) Quantitation of the inhibition of p38 phosphorylation, from three independent experiments as in (A), displayed as mean ± SE, with P values for indicated comparisons. Whereas Hsc70 partially inhibits activation of p38 phosphorylation, Hsp110 completely abolishes activation of p38 phosphorylation, reducing it to the basal level (compare lane 6 with lane 1). (C). Model for action of G85R SOD1YFP in inhibiting anterograde axonal transport. The mutant protein is proposed to activate the MAPK cascade via an interaction with a component(s), designated X, that activates ASK1 that, in turn, inhibits anterograde transport via p38-mediated phosphorylation of kinesin heavy chain (KHC). Such phosphorylation has been detected in another study of a mutant SOD1 in squid axoplasm (34). Notably, the phosphorylation of KHC is not detectable on immunoblots, requiring instead addition of [32P]ATP to the axoplasm, followed by immunoprecipitation (G. Morfini, Y.S. and S.T.B., unpublished). The effects of the molecular chaperones, Hsc70 and Hsp110, are suggested to lie upstream of these phosphorylation-dependent actions, forestalling cascade activation presumably by cooperative binding and release of the mutant protein in the presence of ATP (here added to all axoplasms at 2–5 mM final concentration), mediating its disaggregation or unfolding and preventing the toxic effect of G85R SOD1YFP to activate the MAPK cascade.
Discussion
The studies here of effects of a mutant human SOD1 on anterograde fast transport in squid axoplasm likely have relevance to interactions that produce neurotoxicity in the mammalian setting (Fig. 5C). The misfolded mutant protein apparently exposes epitopes that are sufficiently extended and hydrophobic to recruit endogenous Hsp(c)70 in axoplasm, as occurs in vivo in mouse spinal cord (Table S1; 28, 37, 38). Notably, Hsc70 is the most abundant chaperone in mouse motor neurons, as revealed by laser capture microdissection and proteolysis/MS. Such physical interaction of both squid endogenous Hsp(c)70 and DnaJ proteins, as well as exogenously added bovine Hsc70, with mutant SOD1, can apparently partially mitigate the toxic effects of the mutant protein that lead to MAPK activation and inhibition of anterograde axonal transport (Figs. 4 and 5). More striking is the ability of the exogenously added human chaperone/nucleotide exchange factor, Hsp110, to mediate complete inhibition of this downstream toxic pathway at a concentration substoichiometric to G85R SOD1YFP (Figs. 4 and 5). This action, because it can occur with Hsp110 addition alone, is likely accomplished in cooperation with endogenous squid Hsp(c)70 and DnaJ, and might reflect the augmentation of what is otherwise a rate limiting step in a disaggregation reaction. Because the added human Hsp110 was recovered from the axoplasm with the affinity-captured mutant SOD1YFP as observed by MS, it appears that the chaperone has the ability, in this context, either to stably bind directly to the mutant SOD1 or to stably associate with the mutant SOD1 via endogenous Hsp(c)70, which is also recovered in this sample. As mentioned, it is unclear whether the action of Hsp110/Hsp(c)70 (and putatively DnaJ) disrupts a homomeric interaction between misfolded G85R SOD1YFP proteins or a heteromeric one between the mutant SOD1 protein and a squid protein(s), designated X in Fig. 5C, that somehow mediates the toxic effect. Potentially, the same surfaces in the mutant protein that interact with the molecular chaperones are able, when left unbound, to interact with endogenous protein(s) in the axoplasm to produce the gain of toxic function that here results in activation of the MAPK cascade.
It seems possible that G85R SOD1YFP directly interacts with ASK1 to activate its kinase activity and the downstream cascade. Interaction of WT SOD1 with two casein kinases has been recently reported in the context of a signaling cascade that controls cellular respiration (41). There is also a report that WT SOD1 can bind directly to Rac1 and affect its activity (42). However, whether the misfolded state could mediate either such association is currently unknown.
The identity of a relevant component associating with mutant SOD1 might be intimated by its recovery among the proteins bound to G85R SOD1YFP captured via anti-YFP antibodies when G85R SOD1YFP alone was added to axoplasm but not when exogenously added Hsp110 was also present (see Table S2 for a list of such selectively captured components). Prominent in this list are cytoskeletal proteins and cytoskeletal-modifying components as well as synaptic vesicle-associated proteins, but no obvious candidate for protein X is apparent.
The possibility also has to be considered that the mutant SOD1 protein could be depleting limiting amounts of squid Hsc70 and/or Hsp110, leading somehow to MAPK cascade activation. The nature of such a mechanism is unclear. Finally, it is conceivable that the complex of mutant SOD1 and X itself is the toxic species that has gained the ability to activate the ASK1 kinase cascade. It will be crucial to identify the activating component(s), regardless of whether the same kinase cascade is operative to produce toxicity in mammals, because its identity may provide a window into the nature of a protein species that can interact with a neurodegeneration-associated protein to activate a kinase cascade as a gain of function.
Methods
Antibodies and Reagents.
The following antibodies were used: anti-kinesin heavy chain (H2 clone; 1:100,000) (43) and anti-phospho p38 (Cell Signaling 9215; 1:500). The following secondary antibodies were used: Jackson 111-035-045 HRP-conjugated goat anti-rabbit and Jackson 115-035-146 HRP-conjugated goat anti-mouse IgG (1:20,000). Immunoblotting was carried out as in Bosco et al. (33). MW01-2-069SRM (MW069) is a small-molecule CNS drug developed at Northwestern University and described in Munoz et al. (35). NQDI-1 (R & D Systems) is a selective inhibitor of ASK1.
Protein Preparations.
Both G85R SOD1YPFHis and WT SOD1YFPHis were produced in E. coli BL21/DE3 cells by overexpression from pET vectors. The former transformant was induced at low temperature in 50 µM isopropyl β-D-1-thiogalactopyranoside to optimize the fraction that remained soluble (∼2%). Both mutant and WT fusion proteins were purified on Talon resin, and the eluted material further purified by chromatography on MonoQ 10/10, eluting at 0.1–0.15 M NaCl. Bovine Hsc70 was overexpressed in E. coli and purified by anion exchange chromatography on Q Sepharose Fast Flow, followed by ATP agarose chromatography (Sigma/Fluka/02065). Human Hsp110 (HSPA4L) was produced as a 6His-SUMO-2G-HSPA4L fusion in E. coli and purified on Talon resin. The eluted protein was treated with purified ULP1-His to cleave the SUMO moiety, and the HSPA4L was recovered free of both His-SUMO and ULP1-His by passage through Talon resin (32, 44).
Vesicle Motility Assays in Isolated Axoplasm.
Intact axoplasms were extruded from giant axons of the squid L. pealei (Marine Biological Laboratory) as described previously (45). Recombinant proteins and pharmacological inhibitors were diluted into X/2 buffer (175 mM potassium aspartate, 65 mM taurine, 35 mM betaine, 25 mM glycine, 10 mM Hepes, 6.5 mM MgCl2, 5 mM EGTA, 1.5 mM CaCl2, 0.5 mM glucose, pH 7.2) supplemented with 2–5 mM ATP, and 25 µL were perfused into chambers holding membrane-free axoplasms. The final concentration of the SOD1-YFP was 4.6 µM with respect to the fusion monomer; the final concentration of Hsc70 was 10 µM, and that of Hsp110 was 0.6 µM. Axoplasms were visualized on a Zeiss Axiomat microscope with a 100×, 1.3 n.a. objective and DIC optics used. Images were acquired with a Hamamatsu Model 2400 CCD camera and Argus 20 interface, and organelle velocities were measured with a Photonics Microscopy C2117 video manipulator (Hamamatsu) as described previously (46). The rate measurements obtained by this method reflect a sampling of vesicle movements in and out of the plane of focus, so the average velocity correlates with both the rate and number of vesicles moving in a given treatment [i.e., high rates also mean high numbers of vesicles, whereas low rates reflect reduced numbers as well as slower mean velocities (46–48)].
Statistical Analyses.
All experiments were repeated at least three times. The data were typically analyzed by pooled t test of µ1–µ2 using DataDesk statistical software. Quantitative data were expressed as mean ± SEM unless otherwise stated; significance was determined as P values as indicated.
Squid Transcriptome and Proteomic Analyses.
RNA.
Optic lobes were dissected from freshly caught squid and frozen in liquid nitrogen. Two lobes (∼0.2 g each) from the same animal were ground separately in a stainless steel mortar and pestle under liquid nitrogen, then thawed/dispersed in lysis buffer (PureLink Mini kit, Invitrogen) with homogenization. The kit protocols for column application, washing, and elution were followed, with the inclusion of on-column DNase I treatment. The yield was 2.5–3.0 µg total RNA per optic lobe. mRNA was purified from 500 ng total RNA with oligo-dT beads (Invitrogen) and sheared in SuperScript III reverse transcription buffer (Invitrogen) by incubation at 94 °C for 2 min. First-strand synthesis by SuperScript III was primed with random primers; the second-strand synthesis reaction contained UTP instead of TTP so that uracil DNA-glycosylase (New England Biolabs) could be used later to remove uracil residues and produce a strand-specific sequencing library. The cDNA library was end-repaired and A-tailed, and a custom adapter containing Illumina sequencing primer binding sites was ligated to both ends of the insert. The 400–500 bp fraction of the library was recovered from a 2% (wt/vol) agarose gel, PCR amplified, and quantified by quantitative RT-PCR. The library was sequenced on a version 2 Illumina MiSeq using paired-end 250-bp reads. Adapter sequences, empty reads, and low-quality sequences were removed; and the first 15 and last 50 (read 1) or last 100 (read 2) nucleotides were trimmed to remove low quality bases using the FASTX toolkit (http://hannonlab.cshl.edu/fastx_toolkit/index.html). Putative transcripts were assembled using the Trinity method (49).
Protein.
The transcripts were conceptually translated within the Trinity package, and all predicted ORFs >50 amino acids long were compiled in a FASTA-formatted file. This file was used as the database against which the MS data (Affinity capture and MS) were searched with Sequest (Thermo Scientific) and DTASelect (50). Identified protein sequences were searched against the nonredundant protein database (National Center for Biotechnology Information; January 12, 2013) using BLAST 2.2.27+, running locally. The best “hit” reported for each sequence was curated manually to ensure a significant level of similarity, remove duplicates, and resolve isoforms when possible.
Affinity capture and MS.
After vesicle transport experiments in the presence of WT or mutant SOD1YFP, with or without added Hsp110, the axoplasm and its surrounding perfusion buffer were recovered, briefly homogenized with a pipet tip, and centrifuged at 15,000 × g for 5 min in a microcentrifuge. The supernatant was recovered, and 10 µL of Ultralink-bound anti-YFP antibody was added (prior experiments indicated that this quantity of resin was more than sufficient to bind all of the 4.6 µg of SOD1YFP added to each axoplasm). The resin was washed and eluted as detailed previously (28), and the recovered proteins were modified and proteolyzed with trypsin as described, except that chloroacetamide was used to block sulfhydryl groups. The peptide mixtures from two identical transport experiments were combined and subjected to MudPIT HPLC/MS analysis as described (28) on an LTQ Orbitrap XL mass spectrometer (Thermo Scientific).
Supplementary Material
Acknowledgments
The authors thank G. Morfini for valuable comments regarding the biochemistry of MAPK in axoplasm and A. Apetri, K. Furtak, and K. Kowalik for technical assistance. This work was supported by the Howard Hughes Medical Institute and by grants from the ALS Association and the National Institutes of Health (to S.B.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1303279110/-/DCSupplemental.
References
- 1.Rosen DR, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362(6415):59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 2.Hart PJ. Pathogenic superoxide dismutase structure, folding, aggregation and turnover. Curr Opin Chem Biol. 2006;10(2):131–138. doi: 10.1016/j.cbpa.2006.02.034. [DOI] [PubMed] [Google Scholar]
- 3.Neumann M, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314(5796):130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 4.Sreedharan J, et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319(5870):1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vance C, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323(5918):1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kwiatkowski TJ, Jr, et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323(5918):1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- 7.Maruyama H, et al. Mutations of optineurin in amyotrophic lateral sclerosis. Nature. 2010;465(7295):223–226. doi: 10.1038/nature08971. [DOI] [PubMed] [Google Scholar]
- 8.Deng HX, et al. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature. 2011;477(7363):211–215. doi: 10.1038/nature10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bruijn LI, Miller TM, Cleveland DW. Unraveling the mechanisms involved in motor neuron degeneration in ALS. Annu Rev Neurosci. 2004;27:723–749. doi: 10.1146/annurev.neuro.27.070203.144244. [DOI] [PubMed] [Google Scholar]
- 10.Lindberg MJ, Byström R, Boknäs N, Andersen PM, Oliveberg M. Systematically perturbed folding patterns of amyotrophic lateral sclerosis (ALS)-associated SOD1 mutants. Proc Natl Acad Sci USA. 2005;102(28):9754–9759. doi: 10.1073/pnas.0501957102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Valentine JS, Doucette PA, Zittin Potter S. Copper-zinc superoxide dismutase and amyotrophic lateral sclerosis. Annu Rev Biochem. 2005;74:563–593. doi: 10.1146/annurev.biochem.72.121801.161647. [DOI] [PubMed] [Google Scholar]
- 12.Kuo JJ, Siddique T, Fu R, Heckman CJ. Increased persistent Na(+) current and its effect on excitability in motoneurones cultured from mutant SOD1 mice. J Physiol. 2005;563(Pt 3):843–854. doi: 10.1113/jphysiol.2004.074138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Zundert B, et al. Neonatal neuronal circuitry shows hyperexcitable disturbance in a mouse model of the adult-onset neurodegenerative disease amyotrophic lateral sclerosis. J Neurosci. 2008;28(43):10864–10874. doi: 10.1523/JNEUROSCI.1340-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meehan CF, et al. Intrinsic properties of lumbar motor neurones in the adult G127insTGGG superoxide dismutase-1 mutant mouse in vivo: Evidence for increased persistent inward currents. Acta Physiol (Oxf) 2010;200(4):361–376. doi: 10.1111/j.1748-1716.2010.02188.x. [DOI] [PubMed] [Google Scholar]
- 15.Pasinelli P, Brown RH. Molecular biology of amyotrophic lateral sclerosis: Insights from genetics. Nat Rev Neurosci. 2006;7(9):710–723. doi: 10.1038/nrn1971. [DOI] [PubMed] [Google Scholar]
- 16.Zhang B, Tu P, Abtahian F, Trojanowski JQ, Lee VM. Neurofilaments and orthograde transport are reduced in ventral root axons of transgenic mice that express human SOD1 with a G93A mutation. J Cell Biol. 1997;139(5):1307–1315. doi: 10.1083/jcb.139.5.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Borchelt DR, et al. Axonal transport of mutant superoxide dismutase 1 and focal axonal abnormalities in the proximal axons of transgenic mice. Neurobiol Dis. 1998;5(1):27–35. doi: 10.1006/nbdi.1998.0178. [DOI] [PubMed] [Google Scholar]
- 18.Williamson TL, Cleveland DW. Slowing of axonal transport is a very early event in the toxicity of ALS-linked SOD1 mutants to motor neurons. Nat Neurosci. 1999;2(1):50–56. doi: 10.1038/4553. [DOI] [PubMed] [Google Scholar]
- 19.Warita H, Itoyama Y, Abe K. Selective impairment of fast anterograde axonal transport in the peripheral nerves of asymptomatic transgenic mice with a G93A mutant SOD1 gene. Brain Res. 1999;819(1-2):120–131. doi: 10.1016/s0006-8993(98)01351-1. [DOI] [PubMed] [Google Scholar]
- 20.De Vos KJ, et al. Familial amyotrophic lateral sclerosis-linked SOD1 mutants perturb fast axonal transport to reduce axonal mitochondria content. Hum Mol Genet. 2007;16(22):2720–2728. doi: 10.1093/hmg/ddm226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fischer LR, et al. Amyotrophic lateral sclerosis is a distal axonopathy: Evidence in mice and man. Exp Neurol. 2004;185(2):232–240. doi: 10.1016/j.expneurol.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 22.Pun S, Santos AF, Saxena S, Xu L, Caroni P. Selective vulnerability and pruning of phasic motoneuron axons in motoneuron disease alleviated by CNTF. Nat Neurosci. 2006;9(3):408–419. doi: 10.1038/nn1653. [DOI] [PubMed] [Google Scholar]
- 23.Clement AM, et al. Wild-type nonneuronal cells extend survival of SOD1 mutant motor neurons in ALS mice. Science. 2003;302(5642):113–117. doi: 10.1126/science.1086071. [DOI] [PubMed] [Google Scholar]
- 24.Di Giorgio FP, Carrasco MA, Siao MC, Maniatis T, Eggan K. Non-cell autonomous effect of glia on motor neurons in an embryonic stem cell-based ALS model. Nat Neurosci. 2007;10(5):608–614. doi: 10.1038/nn1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagai M, et al. Astrocytes expressing ALS-linked mutated SOD1 release factors selectively toxic to motor neurons. Nat Neurosci. 2007;10(5):615–622. doi: 10.1038/nn1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haidet-Phillips AM, et al. Astrocytes from familial and sporadic ALS patients are toxic to motor neurons. Nat Biotechnol. 2011;29(9):824–828. doi: 10.1038/nbt.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brady ST, Lasek RJ, Allen RD. Fast axonal transport in extruded axoplasm from squid giant axon. Science. 1982;218(4577):1129–1131. doi: 10.1126/science.6183745. [DOI] [PubMed] [Google Scholar]
- 28.Wang J, et al. Progressive aggregation despite chaperone associations of a mutant SOD1-YFP in transgenic mice that develop ALS. Proc Natl Acad Sci USA. 2009;106(5):1392–1397. doi: 10.1073/pnas.0813045106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dragovic Z, Broadley SA, Shomura Y, Bracher A, Hartl FU. Molecular chaperones of the Hsp110 family act as nucleotide exchange factors of Hsp70s. EMBO J. 2006;25(11):2519–2528. doi: 10.1038/sj.emboj.7601138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raviol H, Sadlish H, Rodriguez F, Mayer MP, Bukau B. Chaperone network in the yeast cytosol: Hsp110 is revealed as an Hsp70 nucleotide exchange factor. EMBO J. 2006;25(11):2510–2518. doi: 10.1038/sj.emboj.7601139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shorter J. The mammalian disaggregase machinery: Hsp110 synergizes with Hsp70 and Hsp40 to catalyze protein disaggregation and reactivation in a cell-free system. PLoS ONE. 2011;6(10):e26319. doi: 10.1371/journal.pone.0026319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rampelt H, et al. Metazoan Hsp70 machines use Hsp110 to power protein disaggregation. EMBO J. 2012;31(21):4221–4235. doi: 10.1038/emboj.2012.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bosco DA, et al. Wild-type and mutant SOD1 share an aberrant conformation and a common pathogenic pathway in ALS. Nat Neurosci. 2010;13(11):1396–1403. doi: 10.1038/nn.2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morfini GA, et al. Axonal transport defects in neurodegenerative diseases. J Neurosci. 2009;29(41):12776–12786. doi: 10.1523/JNEUROSCI.3463-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Munoz L, et al. A novel p38 alpha MAPK inhibitor suppresses brain proinflammatory cytokine up-regulation and attenuates synaptic dysfunction and behavioral deficits in an Alzheimer’s disease mouse model. J Neuroinflammation. 2007;4:21. doi: 10.1186/1742-2094-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Volynets GP, et al. Identification of 3H-naphtho[1,2,3-de]quinoline-2,7-diones as inhibitors of apoptosis signal-regulating kinase 1 (ASK1) J Med Chem. 2011;54(8):2680–2686. doi: 10.1021/jm200117h. [DOI] [PubMed] [Google Scholar]
- 37.Watanabe Y, et al. Adherent monomer-misfolded SOD1. PLoS ONE. 2008;3(10):e3497. doi: 10.1371/journal.pone.0003497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zetterström P, Graffmo KS, Andersen PM, Brännström T, Marklund SL. Proteins that bind to misfolded mutant superoxide dismutase-1 in spinal cords from transgenic amyotrophic lateral sclerosis (ALS) model mice. J Biol Chem. 2011;286(23):20130–20136. doi: 10.1074/jbc.M111.218842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Flynn GC, Pohl J, Flocco MT, Rothman JE. Peptide-binding specificity of the molecular chaperone BiP. Nature. 1991;353(6346):726–730. doi: 10.1038/353726a0. [DOI] [PubMed] [Google Scholar]
- 40.Zhu X, et al. Structural analysis of substrate binding by the molecular chaperone DnaK. Science. 1996;272(5268):1606–1614. doi: 10.1126/science.272.5268.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reddi AR, Culotta VC. SOD1 integrates signals from oxygen and glucose to repress respiration. Cell. 2013;152(1-2):224–235. doi: 10.1016/j.cell.2012.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harraz MM, et al. SOD1 mutations disrupt redox-sensitive Rac regulation of NADPH oxidase in a familial ALS model. J Clin Invest. 2008;118(2):659–670. doi: 10.1172/JCI34060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.DeBoer SR, et al. Conventional kinesin holoenzymes are composed of heavy and light chain homodimers. Biochemistry. 2008;47(15):4535–4543. doi: 10.1021/bi702445j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Andréasson C, Fiaux J, Rampelt H, Mayer MP, Bukau B. Hsp110 is a nucleotide-activated exchange factor for Hsp70. J Biol Chem. 2008;283(14):8877–8884. doi: 10.1074/jbc.M710063200. [DOI] [PubMed] [Google Scholar]
- 45.Brady ST, Lasek RJ, Allen RD. Video microscopy of fast axonal transport in extruded axoplasm: A new model for study of molecular mechanisms. Cell Motil. 1985;5(2):81–101. doi: 10.1002/cm.970050203. [DOI] [PubMed] [Google Scholar]
- 46.Morfini G, Szebenyi G, Elluru R, Ratner N, Brady ST. Glycogen synthase kinase 3 phosphorylates kinesin light chains and negatively regulates kinesin-based motility. EMBO J. 2002;21(3):281–293. doi: 10.1093/emboj/21.3.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brady ST, Pfister KK, Bloom GS. A monoclonal antibody against kinesin inhibits both anterograde and retrograde fast axonal transport in squid axoplasm. Proc Natl Acad Sci USA. 1990;87(3):1061–1065. doi: 10.1073/pnas.87.3.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brady ST, Richards BW, Leopold PL. Assay of vesicle motility in squid axoplasm. Methods Cell Biol. 1993;39:191–202. doi: 10.1016/s0091-679x(08)60171-5. [DOI] [PubMed] [Google Scholar]
- 49.Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29(7):644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tabb DL, McDonald WH, Yates JR., 3rd DTASelect and Contrast: Tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res. 2002;1(1):21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.