Abstract
Suppressor of variegation 3–9 homolog 1 (SUV39H1), a histone methyltransferase, catalyzes histone 3 lysine 9 trimethylation and is involved in heterochromatin organization and genome stability. However, the mechanism for regulation of the enzymatic activity of SUV39H1 in cancer cells is not yet well known. In this study, we identified SET domain-containing protein 7 (SET7/9), a protein methyltransferase, as a unique regulator of SUV39H1 activity. In response to treatment with adriamycin, a DNA damage inducer, SET7/9 interacted with SUV39H1 in vivo, and a GST pull-down assay confirmed that the chromodomain-containing region of SUV39H1 bound to SET7/9. Western blot using antibodies specific for antimethylated SUV39H1 and mass spectrometry demonstrated that SUV39H1 was specifically methylated at lysines 105 and 123 by SET7/9. Although the half-life and localization of methylated SUV39H1 were not noticeably changed, the methyltransferase activity of SUV39H1 was dramatically down-regulated when SUV39H1 was methylated by SET7/9. Consequently, H3K9 trimethylation in the heterochromatin decreased significantly, which, in turn, led to a significant increase in the expression of satellite 2 (Sat2) and α-satellite (α-Sat), indicators of heterochromatin relaxation. Furthermore, a micrococcal nuclease sensitivity assay and an immunofluorescence assay demonstrated that methylation of SUV39H1 facilitated genome instability and ultimately inhibited cell proliferation. Together, our data reveal a unique interplay between SET7/9 and SUV39H1—two histone methyltransferases—that results in heterochromatin relaxation and genome instability in response to DNA damage in cancer cells.
Keywords: histone methylation, nonhistone posttranslational modifications
Heterochromatin is a specialized region of organized high-order chromosome structures that protects genome integrity and stability (1). When cells encounter DNA damage from various stresses, the structure of heterochromatin is modulated by unique mechanisms from those used by euchromatin (2). For example, the phosphorylation of histone H2A.x (H2AX) at Ser-139, which is a hallmark of DNA double-stranded breaks (DSBs), is refractory in heterochromatin but is enriched within euchromatin when cells encounter UV or ionizing irradiation (3). It is presumed that heterochromatin is less sensitive to DNA damage because heterochromatic DNA is covered and protected by chromatin-associated proteins, such as heterochromatin protein 1α and β (HP1α and HP1β) (4). However, once DNA damage occurs within heterochromatin, the local compacted architecture impedes recruitment of the related proteins to the DNA damage sites and thereby interferes with DNA repair. To overcome this disadvantage, the structure of heterochromatin is dynamically modulated in response to DNA damage. Generally, two mechanisms have been proposed for this process: ATP-dependent chromatin remodeling and covalent histone modifications (5, 6). These changes to the heterochromatin structure may lead to different cell fates in response to DNA damage. When the local chromatin adjacent to DNA damage sites is relaxed, the repair process is promoted, which is beneficial for cell survival. In contrast, if DNA damage persists for a long time, the relaxed structure of the heterochromatin may lead to further genome instability and ultimately cell death; this effect of relaxed heterochromatin may be useful in developing a therapeutic strategy for killing cancer cells (7).
Suppressor of variegation 3–9 homolog 1 (SUV39H1) is an evolutionarily conserved homolog of Drosophila Su(var)3–9, which functions as a histone methyltransferase that mainly catalyzes the trimethylation of histone 3 lysine 9 (H3K9) in vivo (8, 9). SUV39H1 is critical for the establishment and maintenance of heterochromatin structure through multiple mechanisms (10–13). Based on this property, SUV39H1 is thought to be essential for heterochromatin stability and integrity (14, 15). Evidence from transgenic mice reveals that the loss or overexpression of suv39h causes severe defects in growth and development and even genome instability and increased susceptibility to tumors (14, 16). Therefore, the regulation of SUV39H1 expression or activity is critical for the maintenance of heterochromatin stability. In fact, SUV39H1 activity is mainly modulated by posttranslational modifications (PTMs). For example, the phosphorylation of SUV39H1 during metaphase reinforces the association between SUV39H1 and the metaphase centromere (17). In addition, the deacetylation of SUV39H1 by silent mating type information regulation 2, homolog 1 (SIRT1) enhances SUV39H1 activity and facilitates heterochromatin formation (18), whereas the E3 ligase murine double minute 2, human homolog (MDM2)-mediated ubiquitination of SUV39H1 down-regulates SUV39H1 stability (19). Therefore, dissecting the potential PTMs of SUV39H1 in response to DNA damage may help us further understand the functions of SUV39H1.
SET domain-containing protein 7 (SET7/9) was initially identified as an H3K4 methyltransferase associated with gene expression (20, 21). SET7/9 also plays multiple roles in the DNA damage response by catalyzing the methylation of a series of nonhistone substrates such as p53, E2F transcription factor 1 (E2F1), and SIRT1 (22–26). Because SET7/9 and SUV39H1 exhibit similar properties in their responses to DNA damage, we were interested in investigating the possible coordination of these two histone methyltransferases in response to DNA damage.
In the present study, we showed that SET7/9 interacts with and methylates SUV39H1 at lysines 105 and 123 in response to DNA damage, resulting in the down-regulation of SUV39H1’s methyltransferase activity. In addition, the methylation of SUV39H1 induced heterochromatic relaxation by decreasing local H3K9 trimethylation and played a role in genome instability when methylation of SUV39H1 persisted. Together, our data reveal a unique link between SET7/9 and SUV39H1 in modulating heterochromatin structure and genome instability.
Results
Enhancement of the Interaction Between SET7/9 and SUV39H1 in Response to DNA Damage.
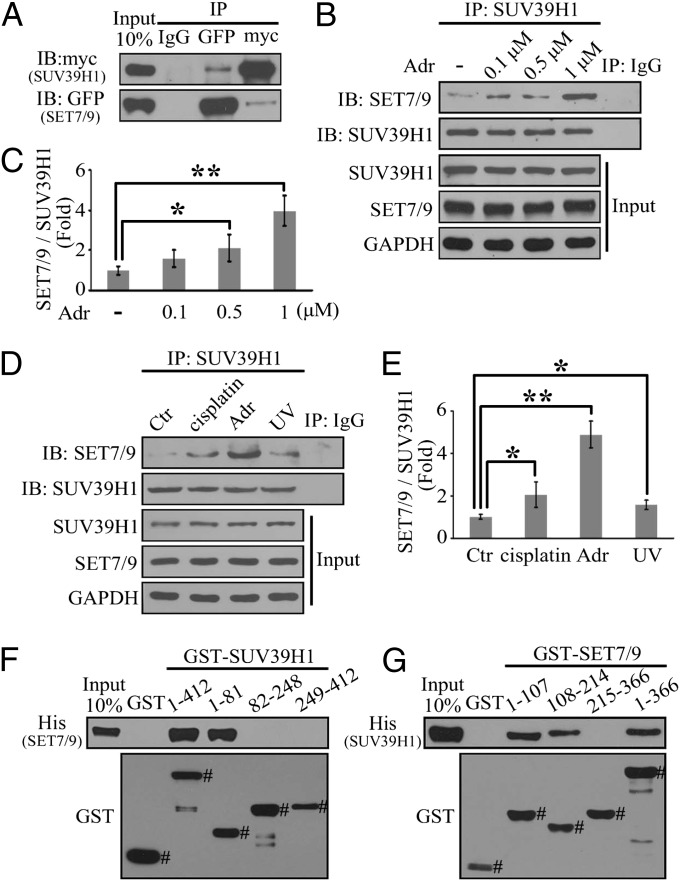
To investigate whether there is an interaction between SET7/9 and SUV39H1, we first performed a coimmunoprecipitation (Co-IP) assay in HEK293T cells with the overexpression of GFP–SET7/9 and myc–SUV39H1. As shown in Fig. 1A, the interaction between SET7/9 and SUV39H1 was obvious (the expression levels of exogenous SET7/9 and SUV39H1 are shown in Fig. S1). Notably, this interaction was specific, because SET7/9 did not associate with another H3K9 methyltransferase, G9A (Fig. S2 A and B). Next, to verify the existence of this interaction in physiological conditions, a Co-IP assay was performed by precipitating extracts of the human cancer cell line H1299 treated with or without adriamycin (Adr) with an anti-SUV39H1 antibody followed by blotting with an anti-SET7/9 antibody. As shown in Fig. 1 B and C, the interaction between SET7/9 and SUV39H1 was significantly enhanced by Adr treatment in a dose-dependent manner. We also performed a reciprocal Co-IP assay by precipitating with an anti-SET7/9 antibody, and again, we detected an enhanced interaction between SET7/9 and SUV39H1 upon Adr treatment (Fig. S2C). In addition, this interaction was enhanced by other DNA-damaging inducers, such as cisplatin and UV radiation (Fig. 1 D and E), suggesting that the enhanced interplay between SET7/9 and SUV39H1 is a general phenomenon of the DNA damage response. Furthermore, to address whether the interaction between SET7/9 and SUV39H1 was direct, a GST pull-down assay was performed by incubating the full-length (FL) or fragments of GST–SUV39H1 (FL, 1–412 aa; chromodomain-containing fragment, 1–81 aa; middle region fragment, 82–248 aa; and SET domain-containing fragment, 249–412 aa) with His–SET7/9. As shown in Fig. 1F, His–SET7/9 specifically interacted with the FL and the chromodomain-containing fragment of GST–SUV39H1, but not with GST alone. In addition, both the N terminus (1–39 aa) and chromodomain (40–81 aa) of SUV39H1 were associated with SET7/9 (Fig. S3), which was similar to the results reported in two cases (18, 27). To further map the region(s) of SET7/9 for SUV39H1 binding, we constructed and purified the FL and fragments of GST-SET7/9 (FL, 1–366 aa; N-terminal fragment, 1–107 aa; middle region fragment, 108–214 aa; and SET domain-containing fragment, 215–366 aa), and a GST pull-down assay showed that both the N-terminal and middle region fragments of SET7/9 bound to SUV39H1 (Fig. 1G). All of the above data indicate that SET7/9 directly interacts with SUV39H1 and that this interaction may be functionally associated with the cellular responses to DNA damage.
Fig. 1.
SET7/9 interacts with SUV39H1. (A) Whole-cell lysates of HEK293T cells transfected with GFP–SET7/9 and myc–SUV39H1 were precipitated with anti-GFP or anti-myc antibody, and then the interactive components were analyzed by Western blotting. (B and C) H1299 cells were treated with adriamycin (Adr) at a concentration of 0.1, 0.5, or 1 μM for 24 h. The cell extracts were then precipitated with anti-SUV39H1 antibody and probed with anti-SET7/9 antibody. The relative intensity of the interaction between SET7/9 and SUV39H1 was quantified and is shown as a histogram. (D and E) H1299 cells were treated with 10 μM cisplatin for 12 h, 1 μM Adr for 12 h, or UV-C (60 J/m2). A Co-IP assay was performed as described in B and C. Data are shown as means ± SD; n = 3. *P < 0.05; **P < 0.01 (C and E). (F) GST–SUV39H1 FL or fragments were incubated with His–SET7/9, and Western blotting was performed to detect the interaction with an anti-His antibody. (G) The reciprocal pull-down assay between GST–SET7/9 FL or fragments and His–SUV39H1. The relative quantity of GST–fusion proteins was measured by anti-GST antibody. # represents the specific bands in F and G.
SET7/9 Methylates SUV39H1 at Lysines 105 and 123 in Vitro and in Vivo.
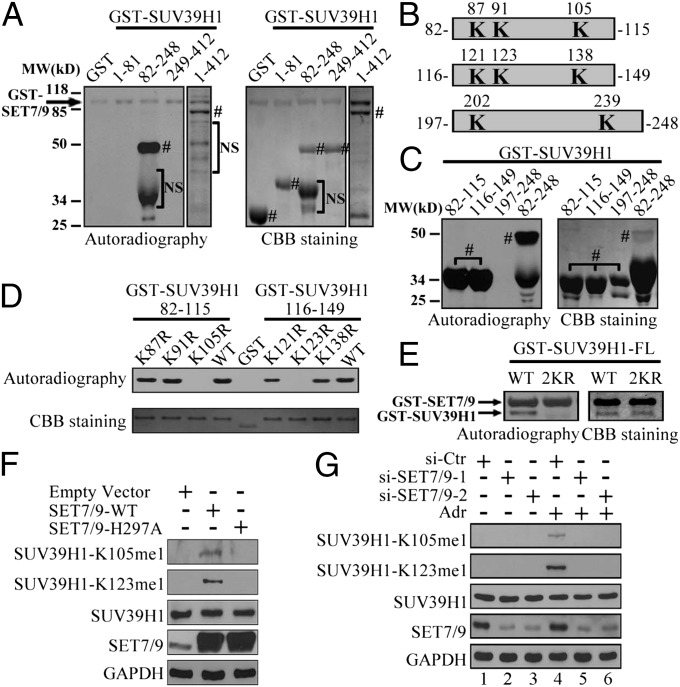
Given that several nonhistone proteins have been identified as substrates for SET7/9 methylation, we investigated whether SUV39H1 was also methylated by SET7/9. To this end, the FL and three fragments of GST-SUV39H1 were purified, and an in vitro methylation assay was performed by incubating SET7/9 with 3H-labeled S-adenosyl methionine (3H-SAM). As shown in Fig. 2A, the FL and middle region fragment (82–248 aa) of SUV39H1 were methylated by SET7/9. Next, we further constructed three subfragments within the middle region fragment (82–115, 116–149, and 197–248 aa) (Fig. 2B) and performed an in vitro methylation assay. As shown in Fig. 2C, the methylation signals were detected within the 82- to 115-aa and 116- to 149-aa fragments. To delineate the exact methylation site(s) that were catalyzed by SET7/9, we conducted a site-directed mutagenesis assay by changing each lysine to arginine within the 82- to 115-aa and 116- to 149-aa fragments. The methylation signal completely disappeared when lysine 105 or 123 was replaced with arginine within the 82- to 115-aa or 116- to 149-aa fragments, respectively (Fig. 2D). In addition, we performed an in vitro methylation assay with FL SUV39H1–WT or –K105/123R (2KR) as the substrate, and we found that lysines 105 and 123 were the only sites methylated by SET7/9 (Fig. 2E). Although one to three methyl groups could be added to the lysine residues, in this study, only the monomethylation of SUV39H1 was detected by mass spectrometry analysis (Fig. S4). These data indicate that SUV39H1 is a unique substrate of SET7/9 in vitro.
Fig. 2.
SET7/9 methylates SUV39H1 in vitro and in vivo. (A) GST–SUV39H1 FL or fragments were incubated with SET7/9 and 3H–SAM for 1 h at 30 °C. The samples were subsequently separated by SDS/PAGE, stained by Coomassie brilliant blue (CBB), or exposed by autoradiography. The arrow indicates automethylation of GST-SET7/9. # represents specific protein bands; NS refers to nonspecific bands. (B) A schematic diagram of the subfragments of SUV39H1. The localization of lysines is in bold. (C) GST–SUV39H1 (82–115 aa) or three subfragments mentioned in B were catalyzed by SET7/9, and autoradiography or CBB staining was performed as indicated in A. # represents specific protein bands. (D) Indicated GST–SUV39H1 fragments with or without individual lysine mutations were catalyzed by SET7/9 and analyzed by autoradiography or CBB staining. (E) FL GST–SUV39H1–WT or –2KR was catalyzed by SET7/9 and analyzed as described in A. (F) Empty vector, flag–SET7/9–WT, or –H297A was transfected into H1299 cells for 48 h. Western blotting was then performed with indicated antibodies. (G) siRNA control (si-Ctr) or two SET7/9 siRNA fragments were transfected into H1299 cells for 48 h with or without 1 μM Adr for the last 24 h. Western blotting was subsequently performed by using the indicated antibodies.
To determine whether SUV39H1 can be methylated by SET7/9 in vivo, we generated antibodies specific to SUV39H1–K105me1 or –K123me1. The efficiency and specificity of these antibodies were evaluated, and they proved to be sufficient for subsequent experiments (Fig. S5). Next, a flag–SET7/9–WT or –H297A (catalytic defect) expressing plasmid was transfected into H1299 cells, and the total cell lysate was analyzed with the anti-SUV39H1–K105me1 or –K123me1 antibody. As shown in Fig. 2F, both SUV39H1–K105me1 and –K123me1 were obviously enhanced upon overexpression of SET7/9–WT. However, SET7/9–H297A overexpression did not have such an effect on the methylation of SUV39H1. In addition, SUV39H1–K105me1 and –123me1 were significantly induced in H1299 cells in response to Adr treatment (Fig. 2G). However, depletion of SET7/9 by siRNA efficiently blocked SUV39H1 methylation in response to Adr treatment (Fig. 2G, lanes 5 and 6 vs. 4). Together, these data clearly indicate that SET7/9 methylates SUV39H1 at lysines 105 and 123 in vivo, and the methylation of these lysines is significantly enhanced in response to DNA damage.
SET7/9 Negatively Regulates SUV39H1 Methyltransferase Activity.
Generally, SUV39H1 is responsible for establishing and maintaining H3K9 trimethylation (8, 9); therefore, we initially investigated the effect of SUV39H1 methylation on the total trimethyl H3K9 level in cells. To this end, a myc–SUV39H1–WT or –2KR and a flag–SET7/9 were cotransfected into H1299 cells, and H3K9 methylation was measured by Western blotting. As shown in Fig. 3 A and B, coexpression of SET7/9 largely alleviated SUV39H1–WT-induced, but not SUV39H1–2KR-induced, H3K9 trimethylation (lane 2 vs. 5; lane 3 vs. 6). Accordingly, as the preferential substrate of SUV39H1 in vivo (8, 9), monomethyl H3K9 had a reverse change relative to that of trimethyl H3K9. However, the total level of dimethyl H3K9 was not changed in our study. In addition, we noticed that the catalytic activity of SET7/9 is essential for its effect on H3K9 trimethylation; overexpression of a catalytically defective SET7/9 had no effect on SUV39H1–WT-induced H3K9 trimethylation (Fig. S6A). Moreover, SUV39H1 and SET7/9 had no regulatory effect on H3K4 mono, di-, or trimethylation (Fig. S6B), indicating that there was no cross-talk between H3K4 and H3K9 methylation in our study.
Fig. 3.
SET7/9 negatively regulates SUV39H1 methyltransferase activity. (A) myc–SUV39H1–WT or –2KR was cotransfected with or without flag–SET7/9 into H1299 cells for 48 h. Cells were lysed with acid extract buffer for histone modification analysis or Nonidet P-40 buffer for soluble protein analysis. Western blotting was performed with the indicated antibodies. (B) Quantitative analysis of H3K9me3 in A. Data are shown as means ± SD (n = 3). **P < 0.01. (C and D) A UAS–tk–luciferase reporter and a pRL–renilla control reporter were cotransfected with other indicated plasmids into HEK293T cells for 24 h. The luciferase activity was measured and is shown as relative fold changes. Data are shown as means ± SD (n = 3). *P < 0.05; **P < 0.01. (E) Schematic diagram showing the process of purification of methylated or unmethylated SUV39H1. (F) An increased amount of purified methylated or unmethylated SUV39H1 was incubated with recombinant FL histone H3 and SAM for 30 min at 30 °C. The products were subjected to Western blotting and probed with the indicated antibodies. The acid extract was set as a quality control for each antibody. (G) The relative quantification of H3K9me1 in F. Data are shown as means ± SD (n = 3). *P < 0.05; **P < 0.01.
Based on the above data, we hypothesized that SET7/9 may decrease SUV39H1 methyltransferase activity by methylating SUV39H1. To test this hypothesis, a Gal4–upstream activating sequence (UAS)–thymidine kinase (tk)–luciferase system was used to measure the transcriptional repression activity of SUV39H1 because the transcriptional repression activity of SUV39H1 is directly correlated to its methyltransferase activity (28). As shown in Fig. 3C, overexpression of SUV39H1–WT significantly decreased the relative luciferase activity driven by the Gal4–VP16 (lane 5 vs. 2–4). In addition, the coexpression of SET7/9–WT, but not of the catalytically defective mutant, restored the SUV39H1–WT-mediated decrease of the relative luciferase activity in a dose-dependent manner (Fig. 3C, lanes 6 and 7 vs. 8 and 9). Furthermore, SET7/9 antagonized SUV39H1–WT-mediated repression activity alone, but had no such effect on SUV39H1–2KR-mediated repression (Fig. 3D, lanes 1–3 vs. 4–6), which supports our hypothesis. To further verify our results, we purified methylated and unmethylated SUV39H1 from HEK293T cells (Fig. 3E) and performed an in vitro methylation assay using the recombinant FL histone H3 or the monomethyl H3K9 peptide (1–21 aa) as substrate. As shown in Fig. 3 F and G and Fig. S7A, unmethylated SUV39H1 exhibited stronger methyltransferase activity than did methylated SUV39H1. In addition, we noticed that only monomethyl H3K9 was detected when using the recombinant H3 as the substrate (Fig. 3F), even though the monomethyl H3K9 peptide can be trimethylated (Fig. S7A), which may indicate a substrate preference of SUV39H1 in vitro (9). To account for the possible effect of the single-point mutation (K to R) on SUV39H1 activity, we also prepared methylated SUV39H1 by a direct in vitro methylation reaction (Fig. S7B), and it was clear that unmethylated SUV39H1 had more activity than that of methylated SUV39H1 (Fig. S7 C and D). Together, the above data demonstrate that SET7/9 negatively regulates SUV39H1 methyltransferase activity by methylating SUV39H1 at lysines 105 and 123.
SET7/9-Mediated SUV39H1 Methylation Induces Heterochromatin Relaxation in Response to DNA Damage.
Based on the above observations, we hypothesized that, in response to DNA damage, SET7/9 induces a structural change in heterochromatin through the methylation of SUV39H1. To verify this hypothesis, we selectively evaluated the transcripts of centromeric satellite repeats, satellite 2 (Sat2) and α-satellite (α-Sat), as markers for the alteration of heterochromatin structure. Generally, Sat2 and α-Sat are transcribed by polymerase II at a low level; however, when the centromeric region is relaxed, the transcripts of both Sat2 and α-Sat are increased (12, 29). First, H1299 cells were treated with 1 μM Adr for the times indicated in Fig. 4A, and total RNA was later extracted and analyzed by real-time PCR. As shown in Fig. 4A, the relative expression of Sat2 or α-Sat was significantly elevated in a time-dependent manner. Accordingly, a quantitative ChIP (qChIP) assay also revealed that H3K9 trimethylation was down-regulated in both Sat2 and α-Sat loci in response to Adr treatment (Fig. 4B). These data suggest that DNA damage induces heterochromatin relaxation, which correlates with the loss of H3K9 trimethylation in heterochromatic loci.
Fig. 4.
SET7/9-mediated SUV39H1 methylation induces heterochromatin relaxation in response to DNA damage. (A) H1299 cells were treated with 1 μM Adr for the indicated times. The relative expression levels of Sat2 and α-Sat were measured by real-time PCR. (B) H1299 cells were treated with 1 μM Adr for 24 h. The relative occupancy of H3K9me1, H3K9me2, and H3K9me3 within the region of Sat2 and α-Sat was analyzed by real-time PCR. (C) A si-RNA control (si-Ctr) or SET7/9 siRNA fragment was transfected into H1299 cells for 48 h with or without 1 μM Adr for the last 24 h. The relative expression of Sat2 or α-Sat was measured by real-time PCR. (D) H1299 cells were treated as in C, and the enrichment of H3K9me3 in the region of Sat2 or α-Sat was analyzed by real-time PCR. (E) A fragment of siRNA against SET7/9 or SUV39H1 was transfected into H1299 cells, alone or together, for 48 h, with 1 μM Adr for the last 24 h. The relative expression of Sat2 or α-Sat was measured by real-time PCR. (F) H1299 cells were treated as in E, and the enrichment of H3K9me3 within the region of Sat2 or α-Sat was measured by real-time PCR. *P < 0.05; **P < 0.01; NS, no significant difference.
To confirm that SET7/9 is involved in DNA-damage-induced heterochromatin relaxation, we depleted SET7/9 in H1299 cells using siRNA and subsequently treated the cells with or without Adr (Fig. S8A). Real-time PCR analysis indicated that SET7/9 knockdown (KD) compromised the Adr-induced expression of Sat2 and α-Sat. (Fig. 4C, lane 3 vs. 4; lane 7 vs. 8). In addition, a qChIP assay also revealed that the depletion of SET7/9 by siRNA restored the Adr-induced decrease of H3K9 trimethylation in the Sat2 and α-Sat loci (Fig. 4D, lane 7 vs. 8; lane 15 vs. 16). These data indicate that SET7/9 participates in DNA-damage-induced heterochromatin relaxation.
Next, we investigated whether SUV39H1 methylation is necessary for SET7/9-induced heterochromatin relaxation in response to DNA damage. First, we transfected siRNAs into H1299 cells against SUV39H1, SET7/9, or both to deplete the expression of SUV39H1 or SET7/9, separately or together. We then treated the cells with Adr for 24 h (Fig. S8B). As shown in Fig. 4E, SET7/9 KD alone blocked the expression of Sat2 and α-Sat in response to DNA damage (lane 1 vs. 2; lane 5 vs. 6); however, the depletion of SUV39H1 compromised the effect of the SET7/9 KD on the expression of Sat2 and α-Sat (lane 3 vs. 4; lane 7 vs. 8). In addition, when SUV39H1 and SET7/9 were depleted together, we observed no effect of SET7/9 KD on H3K9 trimethylation in Sat2 and α-Sat loci in response to DNA damage, as demonstrated by a qChIP assay (Fig. 4F, lane 7 vs. 8; lane 15 vs. 16). The similar results were also seen in Set7/9−/− mouse embryonic fibroblast (MEF) cells (Fig. S8 C and D). All of the above data indicate that SET7/9-mediated SUV39H1 methylation plays a role in heterochromatin relaxation in response to DNA damage.
Methylation of SUV39H1 Induces Genome Instability.
The relaxation of heterochromatin may result in genome instability if it persists. To investigate the role of SUV39H1 methylation in genome instability, we established four stable H1299 cell lines expressing control shRNA, SUV39H1 shRNA, SUV39H1 shRNA plus RNAi-resistant SUV39H1–WT or –2KR plasmid, separately (hereafter these cell lines are referred to as sh–Ctr, KD, re–WT, or re–2KR; Fig. S9A). The expression of SUV39H1 in these cell lines was measured by Western blot (Fig. S9B). Noticeably, ectopic re–SUV39H1–WT, but not re–SUV39H1–2KR, was methylated in physiological conditions, which was suitable for measuring the long-term function of methylated SUV39H1 on genome instability (Fig. S9B). First, genomic instability was evaluated by a micrococcal nuclease (MNase) sensitivity assay. As shown in Fig. 5A, KD cells were very sensitive to MNase digestion. Although cells with re–SUV39H1–WT or –2KR were able to reverse this phenomenon, re–WT cells were relatively less efficient in resisting MNase digestion than re–2KR cells, suggesting that methylated SUV39H1 compromised genomic stability. Similar results were also found in Set7/9−/− MEF cells; SUV39H1 was unmethylated, and the cells were more resistant to Adr-induced genome instability (Fig. 5B, lane 4 vs. 2). In addition, KD cells showed severe genomic abnormalities, such as micronuclei and abnormal mitoses (Fig. 5 C and D). Although re–SUV39H1–WT and –2KR rescued the cells from genomic abnormalities, the rescue efficiency was more pronounced in re–2KR cells (Fig. 5 C and D). The structural changes in the chromosomes were further evaluated by performing a fluorescence in situ hybridization (FISH) assay. Cells were synchronized in metaphase by using colchicine, an inhibitor of microtubule organization, and then the chromosomes were spread and subsequently detected by using human-specific pan-centromere peptide nucleic acid (PNA) probes. As shown in Fig. 5E, KD cells developed significant centromere defects, such as acentric or multiple-centric chromosome. Although cells with re–SUV39H1–WT or –2KR were able to recover centromere structure, cells with re–SUV39H1–2KR were more efficient than cells with re–SUV39H1–WT. Consequently, re–WT cells proliferated slower than re–2KR cells (Fig. 5F). These data demonstrate that SET7/9-mediated SUV39H1 methylation indeed plays a key role in inducing genome instability and suggest that SUV39H1 methylation-induced genome instability may be related to tumor growth inhibition.
Fig. 5.
Methylation of SUV39H1 induces genome instability. (A) Indicated cells were digested with MNase for 2 min at 37 °C, and the genomic DNA was subsequently extracted and separated by a 1.2% agarose gel. The intensity of each lane was consecutively quantified by using Quantity One software (Bio-Rad). N1–N5 in Right indicate the number of nucleosomes in each oligonucleosome. (B) Set7/9–WT (Set7/9+/+) or –KO (Set7/9−/−) MEFs were transfected with myc–SUV39H1–WT and then treated with 1 μM Adr for 3 h. The MNase sensitivity assay was performed as described in A. (C and D) Indicated cells were fixed and stained with DAPI. Micronuclei or abnormal metaphase chromosomes were measured from interval (n = 500) or anaphase (n = 30) cells, respectively. The arrow indicates individual abnormalities. *P < 0.05; **P < 0.01. (E) Metaphase chromosomes were labeled by using a pan-centromere probe (green). Chromosomes were counterstained by DAPI (blue). The yellow arrow represents acentric chromosomes, and the pink arrow represents multiple-centric chromosomes. The centromere aberrances were statistically analyzed (n = 50). *P < 0.05; **P < 0.01. (F) Approximately 500 of the indicated cells were cultured in a six-well plate for 2 wk, and then the cloning formation was analyzed by using a crystal violet staining. Data are shown as means ± SD (n = 3). **P < 0.01. (G) A hypothetical model showing how SET7/9-mediated SUV39H1 methylation participates in regulating heterochromatin relaxation and genome instability.
Discussion
In the present study, we identified SET7/9 as a unique regulator of SUV39H1 activity. In light of our data, we present a model to hypothesize how SET7/9-mediated SUV39H1 methylation plays a role in heterochromatin relaxation and genome instability (Fig. 5G). In response to DNA damage, SET7/9 interacts with and methylates SUV39H1 at lysines 105 and 123, which, in turn, compromises the methyltransferase activity of SUV39H1. Consequently, the level of H3K9 trimethylation in heterochromatin is significantly decreased, and, thus, the structure of heterochromatin becomes relaxed further, inducing genome instability and inhibiting cell proliferation.
PTMs have emerged as a key step in the regulation of the functions of nonhistone proteins. In this study, we revealed that SUV39H1 can be methylated at specific lysine residues that are functionally different from other previously reported PTMs in SUV39H1. For example, cellular SUV39H1 is acetylated at Lys-266 within the SET domain, which reduces the activity of SUV39H1 in vivo. However, SIRT1 is able to reverse this modification and elevate SUV39H1 activity by facilitating the interaction between the SET domain and the post-SET domain of SUV39H1 (18). In addition, SIRT1 inhibits MDM2-mediated ubiquitination and the subsequent degradation of SUV39H1 during oxidative stress, which, in turn, increases the turnover of SUV39H1 in heterochromatin and protects genome stability (19). In comparison with the acetyl group or the ubiquitin molecule, the methyl group is neutral and relatively smaller; therefore, methylation may influence SUV39H1 by other mechanisms. Although we did not see changes in the half-life and localization of SUV39H1 (Fig. S10), the methylation of SUV39H1 significantly reduced its own methyltransferase activity (Fig. 3 F and G; Fig. S7). Noticeably, the methylation sites identified here were close to the chromodomain of SUV39H1, which is a key region for the interaction between chromatin and SUV39H1 (15). Therefore, we hypothesized that methylation of SUV39H1 induces a conformational change of SUV39H1 and thus disrupts the enzyme–substrate affinity, thereby down-regulating SUV39H1 activity.
The dynamics of H3K9 methylation may be implicated in the DNA damage response. In both yeast and human cells, DNA damage induced by UV irradiation resulted in a significant increase of H3K9 acetylation at damage sites and a modest increase genome-wide (30, 31), which may also imply the loss of H3K9 methylation, because acetylation and methylation of H3K9 are mutually exclusive. In particular, H3K9 trimethylation is enriched in mammalian heterochromatin and is mainly kept in balance by methyltransferase and demethylase activities (32, 33). Evidence from Caenorhabditis elegans indicates that the depletion of H3K9/K36 trimethyl lysine demethylase JMJD2A contributed to the delay of DNA repair (32). These data partially suggest that a decrease in H3K9 trimethylation is important for the DNA damage response. However, there is still no evidence that these demethylases are involved in the DNA-damage-related decline of H3K9 trimethylation in mammalian cells. Our study indicates that the methylation of the H3K9 methyltransferase SUV39H1 contributed to the down-regulation of H3K9 trimethylation in mammalian heterochromatin in response to DNA damage and resulted in a relaxation of heterochromatin structure. This phenomenon suggests that the loss of H3K9 trimethylation in heterochromatin is a stress-dependent, passive progress—or, more specifically, that the trimethylation of H3K9 is not maintained because of a decrease in SUV39H1 activity.
Based on its weak activity in the nucleosome, SET7/9 is thought to function mainly as a nonhistone modifier. SET7/9 dynamically interacts with diverse substrates in response to various stresses. For example, when the colon cancer cell line HCT116 suffers DNA damage caused by Adr, the association between SIRT1 and SET7/9 is dramatically enhanced (26). In addition, the proportion of SET7/9 that binds with the NF-κB subunit Rel-A is elevated in a time-dependent manner upon TNF-α stimulation in the HEK293T cell line (34). Similarly, our study identified that the interaction between SET7/9 and SUV39H1 was highly dependent on Adr treatment. In addition, the methylation level of SUV39H1 at lysines 105 and 123 was specifically enhanced in response to DNA damage, which was SET7/9-dependent (Fig. 2G). Our data suggest that the interaction between SET7/9 and SUV39H1 and the accompanying SUV39H1 methylation are critical for the cellular response to DNA damage. Consistent with previous reports, SET7/9 plays important roles in this process through multiple pathways. For example, SET7/9 elevates p21 expression and induces cell-cycle arrest by directly methylating p53 or disrupting p53–SIRT1 interaction (22, 26). In addition, SET7/9 regulates the cell cycle or apoptosis by mediating retinoblastoma tumor suppressor protein (pRB) or E2F1 methylation (25, 35). As such, SET7/9 seems to be a potential DNA damage transducer that signals and assists the cellular response to DNA damage. Here we further characterized the functions of SET7/9 in response to DNA damage by observing the heterochromatin relaxation and genome instability that was induced by SET7/9-mediated SUV39H1 methylation. Our findings suggest that SET7/9 not only plays a role in DNA-damage-induced cell-cycle arrest or apoptosis but also participates in modulating heterochromatin structure. In addition, persistent methylation of SUV39H1 by SET7/9 induces genome instability and ultimately cell death, which may indicate an approach for designing an anticancer therapy by targeting cancer chromatin.
Materials and Methods
Cell culture and treatment, plasmid construction, Co-IP, GST pull-down, ChIP, RNA interference, real-time PCR, luciferase assay, immunofluorescence, and antibody generation are described in detail in SI Materials and Methods.
In Vitro Methylation Assay.
For enzyme SET7/9, GST–SUV39H1 was incubated with SET7/9 and 1 μCi 3H–SAM (PerkinElmer) in methylation buffer І (50 mM Tris⋅HCl, pH 9.0, 0.5 mM DTT, 1 mM PMSF) for 1 h at 30 °C. For enzyme SUV39H1, 0.2 or 1 μg of purified methylated/unmethylated SUV39H1 was incubated with 1 μg of core histone H3 (NEB) or H3K9me1 peptide (Millipore) and 1 mM SAM (NEB) in methylation buffer II (50 mM Tris⋅HCl, pH 8.5, 5 mM MgCl2, 4 mM DTT) for 30 min at 30 °C. The products were separated by SDS/PAGE and detected by autoradiography or indicated antibodies.
FISH Assay.
Cells were synchronized into metaphase by treatment with colchicine for 2.5 h. Individual chromosomes were spread following incubation with a centromere-specific PNA probe (Panagene) and then observed with a fluorescent microscope.
Supplementary Material
Acknowledgments
We thank Drs. Peter Atadja and Jidong Zhu at Novartis (Shanghai) for kindly donating Set7/9−/− MEF cells. We also thank Dr. Zhiqi Xiong and Zengqiang Yuan for technical help. This work was supported by National Key Basic Research Program of China Grant 2011CB504200; National Natural Science Foundation of China Grants 90919030, 31070691, 30921140311, and 31261140372; Minister of Education of China “111 Project”; and Ministry of Education of China Academic Award for Young Excellent Doctoral Students 8910021370228001 (to D.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.L.W. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1216596110/-/DCSupplemental.
References
- 1.Hennig W. Heterochromatin. Chromosoma. 1999;108(1):1–9. doi: 10.1007/s004120050346. [DOI] [PubMed] [Google Scholar]
- 2.Cann KL, Dellaire G. Heterochromatin and the DNA damage response: the need to relax. Biochem Cell Biol. 2011;89(1):45–60. doi: 10.1139/O10-113. [DOI] [PubMed] [Google Scholar]
- 3.Kim JA, Kruhlak M, Dotiwala F, Nussenzweig A, Haber JE. Heterochromatin is refractory to gamma-H2AX modification in yeast and mammals. J Cell Biol. 2007;178(2):209–218. doi: 10.1083/jcb.200612031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Falk M, Lukásová E, Kozubek S. Chromatin structure influences the sensitivity of DNA to gamma-radiation. Biochim Biophys Acta. 2008;1783(12):2398–2414. doi: 10.1016/j.bbamcr.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 5.Goodarzi AA, Kurka T, Jeggo PA. KAP-1 phosphorylation regulates CHD3 nucleosome remodeling during the DNA double-strand break response. Nat Struct Mol Biol. 2011;18(7):831–839. doi: 10.1038/nsmb.2077. [DOI] [PubMed] [Google Scholar]
- 6.Sun Y, et al. Histone H3 methylation links DNA damage detection to activation of the tumour suppressor Tip60. Nat Cell Biol. 2009;11(11):1376–1382. doi: 10.1038/ncb1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lord CJ, Ashworth A. The DNA damage response and cancer therapy. Nature. 2012;481(7381):287–294. doi: 10.1038/nature10760. [DOI] [PubMed] [Google Scholar]
- 8.Peters AH, et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell. 2003;12(6):1577–1589. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- 9.Rice JC, et al. Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol Cell. 2003;12(6):1591–1598. doi: 10.1016/s1097-2765(03)00479-9. [DOI] [PubMed] [Google Scholar]
- 10.Bannister AJ, et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410(6824):120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 11.Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410(6824):116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 12.Lehnertz B, et al. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol. 2003;13(14):1192–1200. doi: 10.1016/s0960-9822(03)00432-9. [DOI] [PubMed] [Google Scholar]
- 13.Vaute O, Nicolas E, Vandel L, Trouche D. Functional and physical interaction between the histone methyl transferase Suv39H1 and histone deacetylases. Nucleic Acids Res. 2002;30(2):475–481. doi: 10.1093/nar/30.2.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peters AH, et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107(3):323–337. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- 15.Melcher M, et al. Structure-function analysis of SUV39H1 reveals a dominant role in heterochromatin organization, chromosome segregation, and mitotic progression. Mol Cell Biol. 2000;20(10):3728–3741. doi: 10.1128/mcb.20.10.3728-3741.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Czvitkovich S, et al. Over-expression of the SUV39H1 histone methyltransferase induces altered proliferation and differentiation in transgenic mice. Mech Dev. 2001;107(1-2):141–153. doi: 10.1016/s0925-4773(01)00464-6. [DOI] [PubMed] [Google Scholar]
- 17.Aagaard L, Schmid M, Warburton P, Jenuwein T. Mitotic phosphorylation of SUV39H1, a novel component of active centromeres, coincides with transient accumulation at mammalian centromeres. J Cell Sci. 2000;113(Pt 5):817–829. doi: 10.1242/jcs.113.5.817. [DOI] [PubMed] [Google Scholar]
- 18.Vaquero A, et al. SIRT1 regulates the histone methyl-transferase SUV39H1 during heterochromatin formation. Nature. 2007;450(7168):440–444. doi: 10.1038/nature06268. [DOI] [PubMed] [Google Scholar]
- 19.Bosch-Presegué L, et al. Stabilization of Suv39H1 by SirT1 is part of oxidative stress response and ensures genome protection. Mol Cell. 2011;42(2):210–223. doi: 10.1016/j.molcel.2011.02.034. [DOI] [PubMed] [Google Scholar]
- 20.Wang H, et al. Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol Cell. 2001;8(6):1207–1217. doi: 10.1016/s1097-2765(01)00405-1. [DOI] [PubMed] [Google Scholar]
- 21.Nishioka K, et al. Set9, a novel histone H3 methyltransferase that facilitates transcription by precluding histone tail modifications required for heterochromatin formation. Genes Dev. 2002;16(4):479–489. doi: 10.1101/gad.967202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chuikov S, et al. Regulation of p53 activity through lysine methylation. Nature. 2004;432(7015):353–360. doi: 10.1038/nature03117. [DOI] [PubMed] [Google Scholar]
- 23.Campaner S, et al. The methyltransferase Set7/9 (Setd7) is dispensable for the p53-mediated DNA damage response in vivo. Mol Cell. 2011;43(4):681–688. doi: 10.1016/j.molcel.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 24.Lehnertz B, et al. p53-dependent transcription and tumor suppression are not affected in Set7/9-deficient mice. Mol Cell. 2011;43(4):673–680. doi: 10.1016/j.molcel.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 25.Kontaki H, Talianidis I. Lysine methylation regulates E2F1-induced cell death. Mol Cell. 2010;39(1):152–160. doi: 10.1016/j.molcel.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 26.Liu X, et al. Methyltransferase Set7/9 regulates p53 activity by interacting with Sirtuin 1 (SIRT1) Proc Natl Acad Sci USA. 2011;108(5):1925–1930. doi: 10.1073/pnas.1019619108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sewalt RG, et al. Selective interactions between vertebrate polycomb homologs and the SUV39H1 histone lysine methyltransferase suggest that histone H3-K9 methylation contributes to chromosomal targeting of Polycomb group proteins. Mol Cell Biol. 2002;22(15):5539–5553. doi: 10.1128/MCB.22.15.5539-5553.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nielsen SJ, et al. Rb targets histone H3 methylation and HP1 to promoters. Nature. 2001;412(6846):561–565. doi: 10.1038/35087620. [DOI] [PubMed] [Google Scholar]
- 29.Zhu Q, et al. BRCA1 tumour suppression occurs via heterochromatin-mediated silencing. Nature. 2011;477(7363):179–184. doi: 10.1038/nature10371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo R, Chen J, Mitchell DL, Johnson DG. GCN5 and E2F1 stimulate nucleotide excision repair by promoting H3K9 acetylation at sites of damage. Nucleic Acids Res. 2011;39(4):1390–1397. doi: 10.1093/nar/gkq983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu Y, Teng Y, Liu H, Reed SH, Waters R. UV irradiation stimulates histone acetylation and chromatin remodeling at a repressed yeast locus. Proc Natl Acad Sci USA. 2005;102(24):8650–8655. doi: 10.1073/pnas.0501458102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whetstine JR, et al. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006;125(3):467–481. doi: 10.1016/j.cell.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 33.Klose RJ, et al. The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature. 2006;442(7100):312–316. doi: 10.1038/nature04853. [DOI] [PubMed] [Google Scholar]
- 34.Yang XD, et al. Negative regulation of NF-kappaB action by Set9-mediated lysine methylation of the RelA subunit. EMBO J. 2009;28(8):1055–1066. doi: 10.1038/emboj.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carr SM, Munro S, Kessler B, Oppermann U, La Thangue NB. Interplay between lysine methylation and Cdk phosphorylation in growth control by the retinoblastoma protein. EMBO J. 2011;30(2):317–327. doi: 10.1038/emboj.2010.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.