Abstract
Catalytic DNA sequences (deoxyribozymes, DNA enzymes, or DNAzymes) have been identified by in vitro selection for various catalytic activities. Expanding the limits of DNA catalysis is an important fundamental objective and may facilitate practical utility of catalysts that can be obtained from entirely unbiased (random) sequence populations. In this study, we show that DNA can catalyze Zn2+-dependent phosphomonoester hydrolysis of tyrosine and serine side chains (i.e., exhibit phosphatase activity). The best deoxyribozyme decreases the half-life for phosphoserine hydrolysis from as high as >1010 y to <1 h. The phosphatase activity also occurs with nonpeptidic substrates but with reduced efficiency, indicating a preference for phosphopeptides. The newly identified deoxyribozymes can function with multiple turnover using free peptide substrates, have activity in the presence of human cell lysate or BSA, and catalyze dephosphorylation of a larger protein substrate, suggesting broader application of DNA catalysts as artificial phosphatases.
Development of catalysts is a major impetus for much of modern chemical research. Nature’s biomolecular protein and RNA catalysts are responsible for a wide range of chemical reactions, and protein enzymes in particular can achieve large rate enhancements (1, 2). Although DNA catalysts are unknown in nature, in vitro selection [first pioneered for RNA (3)] is readily applied to identify catalytically active artificial DNA sequences (4–6). Importantly, DNA (and RNA) catalysts can be identified by starting with entirely random sequence pools, whereas directed evolution of proteins typically requires a known, catalytically active starting point (7, 8). A growing range of chemical reactions has been shown to be catalyzed by DNA (4–6). For DNA phosphodiester hydrolysis, the uncatalyzed (spontaneous) half-life for P–O bond cleavage of ∼30 million y is reduced to as little as 0.5 min by a DNA catalyst (9, 10). However, in this reaction the DNA catalyst interacts with its DNA substrate by extensive Watson–Crick base pairing, and such an approach cannot be generalized to nonoligonucleotide substrates such as peptides and proteins. With the exception of the ribosome, the natural ribozymes catalyze RNA cleavage and ligation, and they generally have more modest rate enhancements (11–13), limited by the relatively high uncatalyzed half-lives of these reactions.
DNA catalysts that covalently modify peptide and protein substrates are fundamentally interesting and likely have practical value, especially for biologically relevant chemical modifications. We have initiated studies into DNA-catalyzed modifications of amino acid side chains of peptide substrates, such as nucleopeptide linkage formation involving tyrosine and serine (14–16). A major challenge in such studies is to achieve catalysis even though the DNA catalyst cannot engage in any preprogrammable Watson–Crick binding interactions with the substrate. In this report, we show that DNA can catalyze Zn2+-dependent hydrolysis of tyrosine and serine phosphomonoesters of peptide substrates (i.e., have phosphatase activity). The half-life for spontaneous phosphoserine hydrolysis at ∼37 °C is ∼4 × 1010 y as estimated on the basis of the half-life for methyl phosphate dianion (2), which emphasizes the difficulty of catalyzing this reaction. Our finding that Zn2+-dependent DNA catalysts can hydrolyze phosphoserine with half-life on the order of 1 h [or hydrolyze phosphotyrosine with half-life on the order of 1 min, versus spontaneous hydrolysis with half-life estimated as 2 × 104 y (2)] highlights the ability of DNA catalysts to achieve high rate enhancements by catalyzing otherwise very slow reactions. The new phosphatase DNA catalysts can function with multiple turnover using entirely free peptide substrates, which—unlike DNA substrates for DNA-catalyzed hydrolysis (9, 10)—inherently cannot interact with the DNA catalysts by Watson–Crick base pairing.
Results
In Vitro Selection Process.
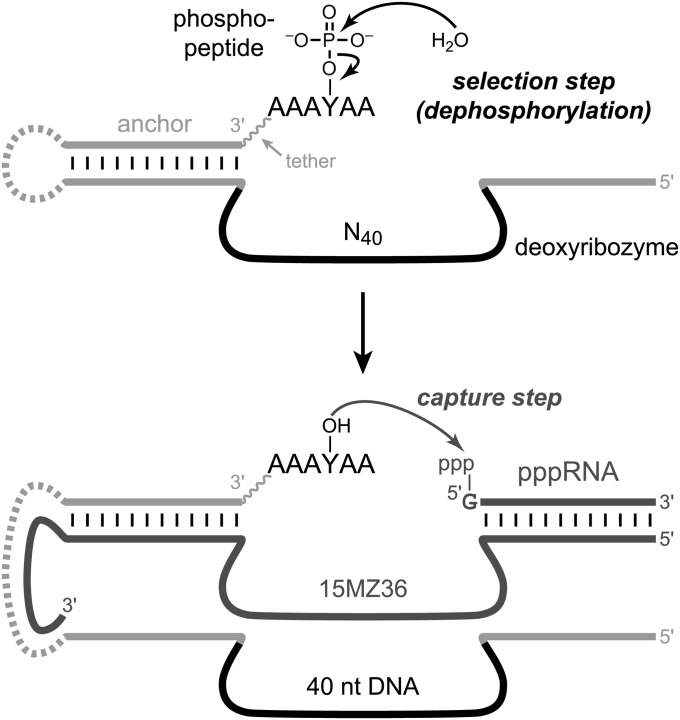
Our strategy to identify phosphatase deoxyribozymes by in vitro selection has key steps depicted in Fig. 1 (Experimental Procedures and SI Materials and Methods give a full description). A hexapeptide substrate AAAYPAA that incorporates an internal phosphotyrosine (YP) residue was covalently appended to a DNA anchor oligonucleotide (Fig. S1), which was itself ligated to the deoxyribozyme pool. After each key selection step that used incubation conditions of 70 mM Hepes (pH 7.5), 1 mM ZnCl2, 20 mM MnCl2, 40 mM MgCl2, and 150 mM NaCl at 37 °C for 14 h, active DNA catalyst sequences were “captured” with the aid of the 15MZ36 deoxyribozyme that selectively uses dephosphorylated tyrosine (YOH) as the nucleophile to attack 5′-triphosphorylated RNA, resulting in a PAGE shift for the corresponding deoxyribozyme sequence (Fig. S2) (17). The active DNA sequences were amplified by PCR and joined by T4 DNA ligase to the DNA-anchored hexapeptide substrate for entering the next selection round. After 14 rounds of selection, the phosphatase activity of the DNA pool reached 35% (Fig. S3), and individual deoxyribozymes were cloned and characterized.
Fig. 1.
In vitro selection strategy to identify DNA catalysts with phosphatase activity. Illustrated are the key selection and capture steps of each round. Each final deoxyribozyme has 40 particular nucleotides in its originally random N40 region. The dashed covalent loop on the left side was required for the selection process but was dispensable for catalysis by individual deoxyribozymes. The loop was absent for all assays of individual deoxyribozymes.
Five distinct deoxyribozyme sequences were identified (Fig. S4). Of these five DNA catalysts, one—named 14WM9—had substantial phosphatase activity not only with the initially used YP hexapeptide that is connected rather closely to the DNA anchor oligonucleotide, but also with a YP hexapeptide that is attached to the DNA anchor through a PEG-type tether that includes 60 ethylene glycol units (Fig. S5). The distinct 14WM27 deoxyribozyme also showed a small amount of activity with the PEG-linked YP hexapeptide. We focused our attention primarily on 14WM9, because its substantial activity with the PEG-tethered hexapeptide suggested the possibility of catalysis with entirely free peptides as well.
Single-Turnover Dephosphorylation by the 14WM9 Deoxyribozyme.
We found that 14WM9 catalyzes Zn2+-dependent single-turnover dephosphorylation of both phosphotyrosine (YP) and phosphoserine (SP) in DNA-anchored hexapeptide substrates (Fig. 2). The substrates were the same as those used during the selection process, but now the DNA-anchored hexapeptide was not covalently attached to the deoxyribozyme (the dashed loop on the far left side of Fig. 1 was absent). The identities of the phosphorylated and dephosphorylated DNA-anchored hexapeptides (i.e., substrates and products) were verified by MALDI MS analysis directly from the DNA-catalyzed reaction samples (all [M–H]–: YP substrate m/z calculated 6,705.6, found 6,708.3, Δ = +0.04%; YOH product m/z calculated 6,625.6, found 6,628.0, Δ = +0.04%; SP substrate m/z calculated 6,629.5, found 6,633.6, Δ = +0.06%; SOH product m/z calculated 6,549.5, found 6,554.3, Δ = +0.07%). MS was also used to verify the identities of separately synthesized standards (Table S1).
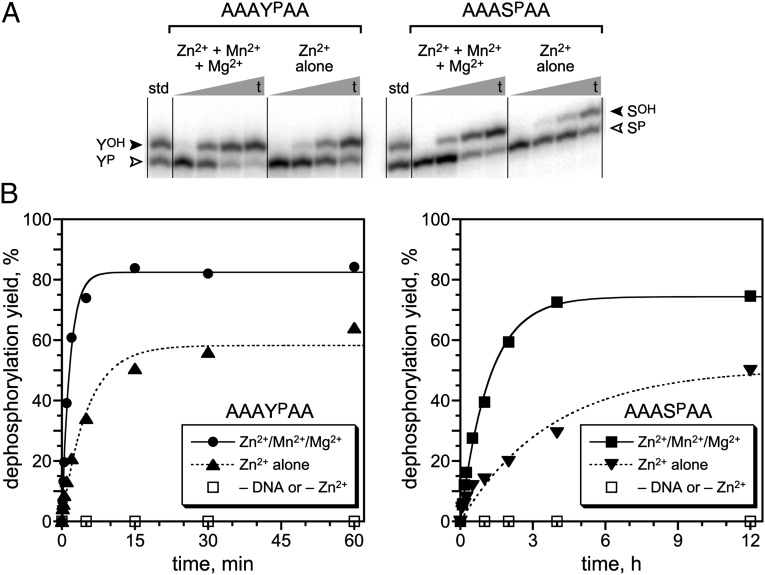
Fig. 2.
Single-turnover assays of the 14WM9 deoxyribozyme that dephosphorylates phosphotyrosine and phosphoserine side chains. (A) PAGE assays of the 14WM9-catalyzed reactions, using DNA-anchored hexapeptide substrates and showing representative time points. The open arrowhead marks the phosphopeptide substrate; the filled arrowhead marks the dephosphorylated product. Each hexapeptide substrate was joined via its N-terminal Ala residue to the 3′-end of the DNA anchor by reductive amination, as described in SI Materials and Methods. Assay conditions: 1 µM 14WM9, 12.5 nM 5′-32P-radiolabeled DNA-anchored hexapeptide, 70 mM Hepes (pH 7.5), 1 mM ZnCl2, 20 mM MnCl2, 40 mM MgCl2, and 150 mM NaCl at 37 °C. In the assays with Zn2+ alone, the Mn2+ and Mg2+ were omitted. Time points for YP substrate: 10 s, 1 min, 5 min, 1 h. Time points for SP substrate: 30 s, 15 min, 2 h, 12 h. (B) Kinetic plots for the assays. The negative controls without deoxyribozyme or without Zn2+ all showed no detectable dephosphorylation activity (<0.5%).
Substantial catalytic activity of 14WM9 with the YP substrate was observed not only using the initial selection conditions that included each of 1 mM Zn2+, 20 mM Mn2+, and 40 mM Mg2+ at pH 7.5 and 37 °C (single-turnover kobs = 0.65 ± 0.05 min–1; n = 3), but also with 1 mM Zn2+ alone, albeit with threefold reduction in rate constant (kobs = 0.19 ± 0.03 min–1; n = 3). Activity with the SP substrate was observed with Zn2+ in the presence or absence of Mn2+ and Mg2+, again with about threefold difference in rate constant (kobs = 0.79 ± 0.15 h–1 and 0.31 ± 0.06 h–1, respectively; n = 3). For both YP and SP substrates, no activity was observed in the absence of either the deoxyribozyme or Zn2+, and 1 mM Zn2+ was optimal; either decreasing or increasing the Zn2+ concentration reduced the activity. Using the YP substrate (SP was not tested in this regard), truncating the constant 5′-portion of 14WM9 (16-nt 5′-segment in Fig. 1) did not substantially disrupt catalytic function. The rate constant did not decrease (krel = 1.6 for 5′-truncated versus full-length deoxyribozyme), and the final yield was essentially unchanged. However, using a random DNA sequence in place of the 40-nt catalytic region—functionally equivalent to performing the first selection round with the N40 pool—abolished all catalytic activity, demonstrating that the mere presence of nonspecific DNA and Zn2+ is insufficient to support dephosphorylation.
Multiple-Turnover Dephosphorylation by the 14WM9 Deoxyribozyme.
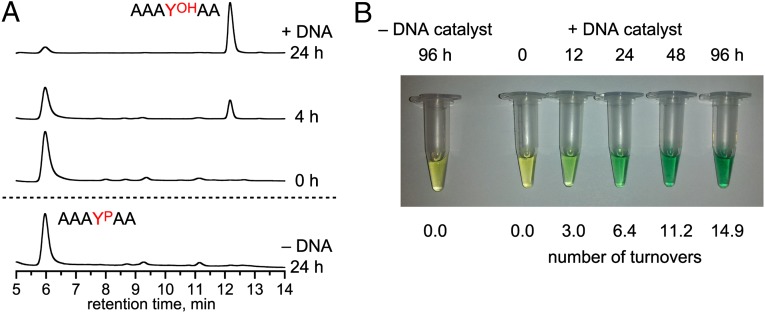
Although our in vitro selection strategy inherently requires a covalent tether between the peptide and the DNA anchor oligonucleotide (Fig. 1), we found that 14WM9 also catalyzes efficient multiple-turnover Zn2+-dependent dephosphorylation of the entirely free (untethered) AAAYPAA peptide substrate, as assayed both by HPLC of the peptide mixture (Fig. 3A) and by colorimetric detection of the released inorganic phosphate, Pi, using malachite green dye (Fig. 3B). Liquid chromatography-electrospray ionization MS of the reaction sample confirmed the identities of the phosphorylated substrate and dephosphorylated product hexapeptides (all [M+H]+: YP substrate m/z calculated 616.2, found 616.2; YOH product m/z calculated 536.3, found 536.2). These assays revealed 6 turnovers in 24 h and 15 turnovers in 96 h by 14WM9. The DNA anchor oligonucleotide, now unconnected to the free peptide substrate, was required for efficient reaction, presumably to sequester the corresponding deoxyribozyme binding arm (truncation of this 3′-binding arm led to 12-fold lower yield at 24 h). In all of these assays, the higher deoxyribozyme concentrations (100 µM or 10 µM, rather than 1 µM as in the single-turnover assays) required elevated Zn2+ concentration (6 mM or 2 mM, rather than 1 mM) for maximal activity, apparently to compensate for nonspecific chelation of Zn2+ by the increased amount of DNA. The Km for AAAYPAA was ≥1 mM (Fig. S6; the assays of Fig. 3 were performed at 0.5 mM peptide). The multiple-turnover activity of 14WM9 was not substantially inhibited by an excess of the dephosphorylated AAAYOHAA peptide product, but considerable inhibition by Pi was observed when over 2 equivalents was included (Fig. S7). The sequence-unrelated 14WM27 deoxyribozyme also exhibited multiple-turnover phosphatase activity with the free AAAYPAA substrate and Zn2+ (Fig. S8), demonstrating that DNA-catalyzed dephosphorylation of a free peptide substrate is not limited to a single deoxyribozyme.
Fig. 3.
Multiple-turnover assays of the 14WM9 deoxyribozyme with free phosphopeptide substrate. (A) HPLC assay. Conditions: 100 µM 14WM9, 125 µM DNA anchor oligonucleotide, 500 µM free AAAYPAA, 70 mM Hepes (pH 7.5), 6 mM ZnCl2, and 150 mM NaCl at 37 °C. Approximately five turnovers were observed in 24 h. (B) Malachite green dye assay, in which Pi is observed colorimetrically upon binding to ammonium molybdate and malachite green dye. Conditions: 10 µM 14WM9, 12.5 µM DNA anchor oligonucleotide, 500 µM free AAAYPAA, 70 mM Hepes (pH 7.5), 2 mM ZnCl2, and 150 mM NaCl at 37 °C. As calculated from A620 values, six turnovers in 24 h and 15 turnovers in 96 h were observed. In both the HPLC and malachite green assays, omission of Zn2+ rather than 14WM9 also led to no activity in 24 h.
Substrate Dependence of 14WM9.
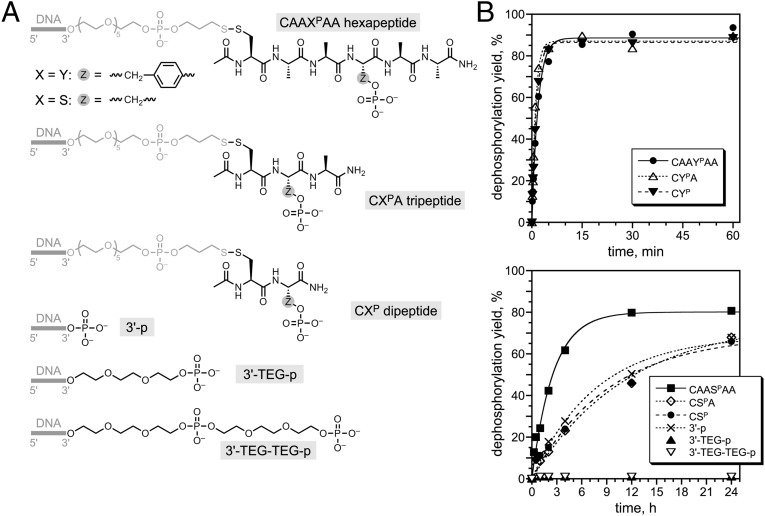
To gain insight into the interactions between the DNA catalyst and the substrate, we examined the dependence of 14WM9’s single-turnover catalytic activity on the chemical structure of its phosphorylated substrate. When the DNA-anchored hexapeptide was shortened to as little as a dipeptide (CYP or CSP, where the cysteine sulfhydryl was used to attach the dipeptide to the DNA anchor), 14WM9 retained full activity for YP and about fourfold lower kobs for SP (Fig. 4). Consistent with this finding, substrates with sequence CAXYPAA and CAAYPXA (where X = F, E, or K) were dephosphorylated to similar extents by 14WM9 (Fig. S9). In contrast, when the substrate was nonpeptidic and the phosphate group was instead connected to the DNA anchor through one or two triethylene glycol (TEG) units, 14WM9 had no detectable dephosphorylation activity (<0.5%; Fig. 4). When the phosphate group was attached directly to the 3′ hydroxyl of the DNA anchor, activity was comparable to that observed for the shorter CSP substrate, indicating that the flexibility introduced by the TEG linkers contributes to the lack of activity with those substrates (Fig. 4). From these data, we conclude that 14WM9 substantially prefers peptidic substrates, although only a relatively short peptide segment is required for optimal activity.
Fig. 4.
Dependence of 14WM9 catalytic activity on the chemical structure of the phosphorylated substrate. (A) Structures of the evaluated substrates. Note that for these experiments the hexapeptide was connected via an N-terminal Cys, rather than an N-terminal Ala as in Fig. 2. (B) Single-turnover assays, under conditions of Fig. 2 (Zn2+/Mn2+/Mg2+). kobs values from PAGE assays, top to bottom as listed in each plot: upper plot 0.54, 0.93, and 0.75 min–1; first four data sets in lower plot 0.39, 0.087, 0.11, and 0.13 h–1 (i.e., 6.5 × 10−3, 1.5 × 10−3, 1.8 × 10−3, and 2.2 × 10−3 min–1).
Catalysis by 14WM9 Under More Biologically Relevant Conditions.
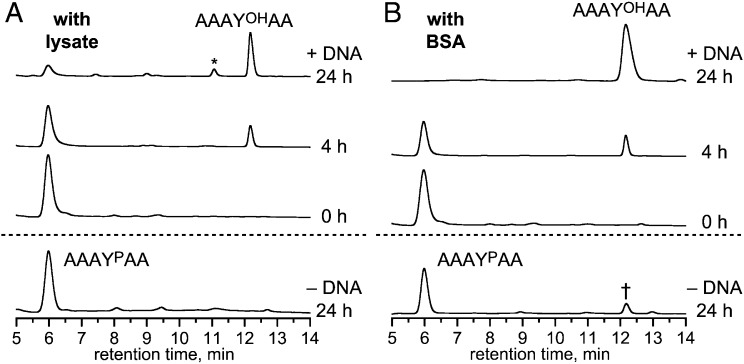
To investigate the ability of 14WM9 to function under more biologically representative conditions, its activity was assayed in the presence of human cell lysate or BSA, using the free AAAYPAA substrate. When up to 160 µg/mL of H1299 human non-small-cell lung carcinoma lysate protein was supplemented into the same buffer and metal ion components as used for the previous experiments, 14WM9 retained multiple-turnover activity (Fig. 5A). Separately, when 14WM9 was assayed in the presence of BSA, substantial activity was maintained up to 20 mg/mL BSA (Fig. 5B and Fig. S10). Multiple-turnover behavior for the 14WM27 deoxyribozyme was also observed in the presence of lysate or BSA (Fig. S11). Collectively, these findings establish that robust DNA-catalyzed phosphatase activity is compatible with high concentrations of nonspecific proteins and other native cellular components.
Fig. 5.
Activity of the 14WM9 deoxyribozyme in the presence of human cell lysate and BSA. (A) HPLC multiple-turnover assay with free AAAYPAA peptide in the presence of human cell lysate (160 µg/mL of lysate protein), under conditions of Fig. 3A. The asterisk denotes the pentapeptide AAYOHAA formed by nonspecific peptidase activity in the lysate, as validated by electrospray ionization MS ([M+H]+ m/z calculated 465.2, found 465.2). (B) HPLC multiple-turnover assay in the presence of 20 mg/mL BSA, under conditions of Fig. 3A except 10 mM Zn2+. Substantial activity was also observed at 6 and 8 mM Zn2+, but reduced presumably due to nonspecific chelation of Zn2+ by BSA (Fig. S10). The dagger denotes a small amount (∼14%) of dephosphorylation product formed in 24 h in the absence of 14WM9. In both panels, omission of Zn2+ rather than 14WM9 led to no activity in 24 h. This finding suggests for B that Zn2+ activates nonspecific phosphatases present in the BSA, and inclusion of the DNA suppresses this phosphatase activity.
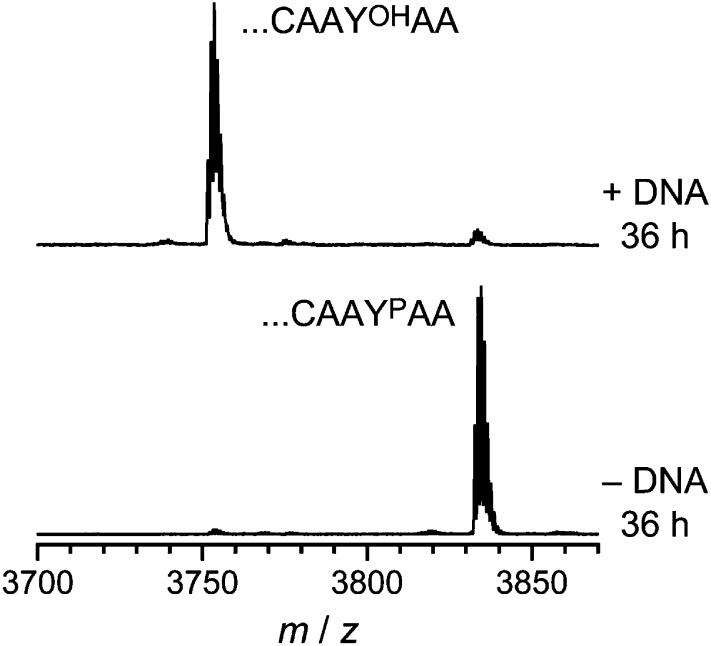
Finally, we assessed the ability of 14WM9 to dephosphorylate a YP residue when presented as part of a large 91-mer protein substrate rather than merely on a short hexapeptide. For this purpose, we used as substrate a 66-mer fragment of prochlorosin ProcA2.8 (18), fused to an N-terminal His6 tag and joined at its C terminus by native chemical ligation to the CAAYPAA hexapeptide (Experimental Procedures). The resulting 91-mer protein was incubated with 14WM9 and analyzed by MALDI MS, revealing substantial dephosphorylation (∼94% in 36 h; Fig. 6).
Fig. 6.
Dephosphorylation of a larger protein substrate by 14WM9. The 91-mer protein derived from prochlorosin ProcA2.8 and ending with ...CAAYPAA at its C terminus was dephosphorylated by 14WM9, digested in-gel by Lys-C, and analyzed by MALDI MS (Experimental Procedures gives details). Dephosphorylation conditions: 100 µM 14WM9, 125 µM DNA anchor oligonucleotide, 20 µM protein, 70 mM Hepes (pH 7.5), 6 mM ZnCl2, 40 mM MgCl2, 20 mM MnCl2, and 150 mM NaCl at 37 °C for 36 h. YOH product Lys-C fragment [M+H]+ m/z calculated 3,751.725, found 3,751.699, Δ = –6.9 ppm. Difference between product and substrate due to dephosphorylation Δ(m/z) calculated 79.966, found 79.993, Δ = +0.033%.
Discussion
In this study, we have expanded the scope of DNA catalysis to include the challenging reaction of phosphomonoester hydrolysis in the context of free peptide substrates. We observed multiple-turnover behavior even in the presence of human cell lysate or high concentrations of BSA, as well as substantial phosphatase activity with a larger protein substrate. We previously demonstrated DNA-catalyzed DNA phosphodiester hydrolysis, that is, nuclease activity (9, 10). Both reactions involve “phosphoester” substrates, but a phosphomonoester as used here is substantially more difficult to hydrolyze than is a phosphodiester, considering the >3 orders of magnitude difference in uncatalyzed half-lives [4 × 1010 y for an aliphatic phosphomonoester dianion (2), versus 3 × 107 y for a phosphodiester (19)]. Regardless of the uncatalyzed reaction rate constants, the peptide substrate for phosphomonoester dephosphorylation cannot be bound by the DNA catalyst using any Watson–Crick base pairs, whereas the DNA substrate for phosphodiester hydrolysis inherently interacts with the deoxyribozyme by extensive preprogrammed base pairing. This means that the phosphatase deoxyribozyme has a much greater challenge to interact well with its substrate. The DNA-catalyzed phosphatase activity has a rate constant as fast as 0.7 min–1 for YP dephosphorylation, a value commensurate with many other in vitro selected deoxyribozymes for various catalytic activities (4–6).
From a fundamental perspective, we wish to understand the mechanism of DNA-catalyzed dephosphorylation, in the context of broader studies of biological phosphoryl transfer (20). Such mechanistic studies will benefit greatly from future high-resolution structural information that is currently unavailable for any deoxyribozyme of any kind (21). Natural protein phosphatases (2, 22, 23) function either by a double-displacement mechanism [cysteine-dependent phosphatases (24, 25) and haloacid dehalogenase (HAD) superfamily aspartate-dependent phosphatases (26–28)] or via direct metal-bound water attack using cofactors such as Mn2+ and Fe2+ (29). Both phosphomonoester dianion (30) and monoanion (31, 32) substrates have been implicated (33). Although we can speculate that 14WM9, 14WM27, and the other newly identified phosphatase deoxyribozymes activate Zn2+ for direct hydrolysis of the phosphorylated side chains, determining their detailed mechanisms requires considerable further experiments, especially without a sound structural basis for the catalysis.
The marked preference of the 14WM9 deoxyribozyme for a peptidic substrate suggests direct interaction of this deoxyribozyme with some portion of the substrate’s polyamide backbone, although the efficient dephosphorylation of a dipeptide substrate means that any such contact is not very extensive. Expanding the examination of peptide substrate sequences may provide DNA catalysts that have sequence selectivity to discriminate among various phosphopeptides. As one potential application with nonpeptide substrates, we anticipate that with further development, DNA-catalyzed dephosphorylation of small molecules (rather than peptides) could be used for controlled prodrug activation, noting that many prodrugs are phosphorylated versions of small-molecule compounds (34).
Experimental Procedures
Oligonucleotides.
DNA oligonucleotides were obtained from Integrated DNA Technologies or prepared by solid-phase synthesis on an ABI 394 instrument using reagents from Glen Research. The 5′-triphosphorylated RNA oligonucleotides were prepared by in vitro transcription using synthetic DNA templates and T7 RNA polymerase (35). All oligonucleotides were purified by 7 M urea denaturing PAGE with running buffer 1× TBE [89 mM each Tris and boric acid and 2 mM EDTA (pH 8.3)] as described previously (36, 37).
Synthesis of Peptides and DNA-Anchored Peptides.
Peptides were prepared by solid-phase synthesis on Fmoc Rink amide MBHA resin (AAPPTec). Each peptide was coupled to the DNA anchor oligonucleotide via either the N-terminal α-amino group (linkage created by reductive amination) or the N-terminal cysteine side chain (linkage created by disulfide formation). SI Materials and Methods gives full procedures.
In Vitro Selection Procedures.
The selection procedure, cloning, and initial analysis of individual clones were performed essentially as described previously (36, 38), but with a different ligation step as recently reported (17). An overview of the key selection and capture steps of each round is shown in Fig. 1, and full procedures are given in SI Materials and Methods.
Single-Turnover Assay Procedure Using PAGE.
The DNA-anchored phosphopeptide substrate was 5′-32P-radiolabeled using γ-[32P]ATP and Optikinase (USB), which lacks the 3′-phosphatase activity that we have found also dephosphorylates tyrosine and serine side chains. A 10-µL sample containing 0.25 pmol of 5′-32P-radiolabeled DNA-anchored phosphopeptide substrate and 20 pmol of deoxyribozyme was annealed in 5 mM Hepes (pH 7.5), 15 mM NaCl, and 0.1 mM EDTA by heating at 95 °C for 3 min and cooling on ice for 5 min. The DNA-catalyzed dephosphorylation reaction was initiated by bringing the sample to a 20-µL total volume containing 70 mM Hepes (pH 7.5), 1 mM ZnCl2, 20 mM MnCl2, 40 mM MgCl2, and 150 mM NaCl (Mn2+ and Mg2+ were omitted in assays that used Zn2+ alone). The sample was incubated at 37 °C. At appropriate time points, 2-µL aliquots were quenched with 5 µL of stop solution (80% formamide, 1× TBE [89 mM each Tris and boric acid and 2 mM EDTA (pH 8.3)], 50 mM EDTA, 0.025% bromophenol blue, and 0.025% xylene cyanol). Samples were separated by 20% PAGE and quantified with a PhosphorImager. Values of kobs were obtained by fitting the yield versus time data directly to first-order kinetics; that is, yield = Y•(1 – e–kt), where k = kobs and Y is the final yield. Each kobs value is reported with error calculated as the SD from the indicated number of independent determinations.
Multiple-Turnover Assay Procedure Using HPLC.
A 20-µL sample containing 2 nmol (100 µM) of deoxyribozyme, 2.5 nmol (125 µM) of free DNA anchor oligonucleotide, and 10 nmol (500 µM) of free AAAYPAA hexapeptide in 70 mM Hepes (pH 7.5), 6 mM ZnCl2, and 150 mM NaCl was incubated at 37 °C. At an appropriate time point (0–24 h), the sample was quenched with 5 µL of 50 mM EDTA (pH 8.0) (250 nmol, versus 120 nmol Zn2+ present) and frozen at –80 °C until further processing. To remove the deoxyribozyme, a filtration step was performed (this step was confirmed not to distort substantially the ratio of phosphorylated to dephosphorylated peptide). An Amicon Ultra-0.5 mL 3-kDa centrifugal filter was washed with 300 µL of water by centrifugation at 14,000 × g for 10 min. The sample was diluted with water to 300 µL and transferred to the filter, which was centrifuged at 14,000 × g for 15 min, and the filtrate was collected. Another 300 µL of water was passed through the filter. The combined filtrates were evaporated to dryness, redissolved in 25 µL of water, and analyzed by HPLC. Analysis was performed on a Beckman System Gold instrument with a Beckman Ultrasphere C18 column (5 µm, 2 × 150 mm). Samples were analyzed using a gradient of 1% solvent A (acetonitrile) and 99% solvent B [0.1% trifluoroacetic acid (TFA) in water] at 0 min to 21:79 A:B at 25 min at a flow rate 0.5 mL/min.
For assays in the presence of H1299 human non-small-cell lung carcinoma lysate, the lysate (40 mg/mL of lysate protein) was a gift prepared by E. Parkinson in the P. Hergenrother laboratory (University of Illinois, Urbana, IL) [lysis in radioimmunoprecipitation assay lysis buffer containing protease inhibitor, with cell debris removed by centrifugation at 16,000 × g for 5 min (39) and quantification by Bradford and bicinchoninic acid assays]. The 20-µL sample additionally contained 1/250 by volume of lysate (160 µg/mL of lysate protein). For assays in the presence of BSA, the 20-µL sample additionally contained 20 mg/mL of BSA (A4503; Sigma), and the Zn2+ concentration was 10 mM.
Multiple-Turnover Assay Procedure Using Malachite Green Dye.
A 20-µL sample containing 200 pmol (10 µM) of deoxyribozyme, 250 pmol (12.5 µM) of free DNA anchor oligonucleotide, and 10 nmol (500 µM) of free AAAYPAA hexapeptide in 70 mM Hepes (pH 7.5), 2 mM ZnCl2, and 150 mM NaCl was incubated at 37 °C. At an appropriate time point (0–96 h), the sample was quenched with 5 µL of 50 mM EDTA (pH 8.0) (250 nmol) and frozen at –80 °C until further processing. To the sample was added 75 µL of water and 20 µL of malachite green assay solution (POMG-25H; BioAssay Systems). After incubation at room temperature for 30 min, the sample was diluted with 180 µL of water, and the absorbance at 620 nm was measured (NanoDrop 2000c; Thermo Scientific). Standard curves for Pi were obtained using solutions containing known amounts of Pi and all other assay components except the free hexapeptide. Numbers of turnovers were calculated directly from the ratio of mole amounts of released Pi and deoxyribozyme.
MS of DNA-Anchored Hexapeptides.
The DNA-anchored AAAYPAA substrate was prepared by the reductive amination procedure (SI Materials and Methods). The DNA-anchored AAAYOHAA dephosphorylation product was prepared from a 10-µL sample containing 50 pmol of DNA-anchored AAAYPAA substrate and 80 pmol of 14WM9 deoxyribozyme, which was annealed in 5 mM Hepes (pH 7.5), 15 mM NaCl, and 0.1 mM EDTA by heating at 95 °C for 3 min and cooling on ice for 5 min. The DNA-catalyzed dephosphorylation reaction was initiated by bringing the sample to a 20-µL total volume containing 70 mM Hepes (pH 7.5), 1 mM ZnCl2, 20 mM MnCl2, 40 mM MgCl2, and 150 mM NaCl. The sample was incubated at 37 °C for 5 min, quenched with 5 µL of 50 mM EDTA (pH 8.0) (250 nmol), desalted by Millipore C18 ZipTip, and analyzed by MALDI MS (matrix 3-hydroxypicolinic acid).
MS of Free Hexapeptides.
The free AAAYOHAA dephosphorylation product was prepared from 10 nmol of the free AAAYPAA substrate using 2 nmol of the 14WM9 deoxyribozyme and the multiple-turnover procedure described above (24-h incubation). Analysis was performed on a Waters 2795 LC-MS instrument with a Beckman Ultrasphere C18 column (5 µm, 2 × 150 mm) and detection by electrospray ionization MS (Q-Tof Ultima API in positive ion scan mode using the manufacturer’s suggested parameters). Samples were analyzed using a gradient of 1% solvent A (0.1% formic acid in acetonitrile) and 99% solvent B (0.1% formic acid in water) at 0 min to 21:79 A:B at 25 min.
Preparation of Larger Protein and Analysis of Dephosphorylation by MS.
The 85-mer His6-ProcA2.8(1–66) mercaptoethanesulfonic acid (MESNA) thioester was overexpressed and partially purified as described in SI Materials and Methods. A 100-µL sample containing 6 mg of crude protein thioester (∼0.6 mM thioester; most of the mass is salts) and 5 mM CAAYPAA hexapeptide in 100 mM Hepes (pH 7.5), 1 mM tris(2-carboxyethyl)phosphine hydrochloride, and 10 mM MESNA was incubated at 4 °C for 16 h. To alkylate the cysteine side chain, 50 µL of 1 M iodoacetamide was added, and the sample was incubated at 25 °C for 45 min in the dark. The reaction mixture was acidified by adding 3 µL of 5% TFA to a final concentration of 0.1% TFA. The ligated protein was purified by HPLC on a Beckman System Gold instrument with a Grace Vydac C4 column (5 µm, 4.6 × 250 mm) using a gradient of 2% solvent A (80% acetonitrile and 0.1% TFA in water) and 98% solvent B (0.1% TFA in water) at 0 min to 100% solvent B at 45 min with flow rate of 1 mL/min. The ligated protein eluted at 29 min and was collected, lyophilized, and redissolved in water. MALDI MS [M+H]+ calculated 9,681.5, found 9,681.7, Δ = +0.002% (matrix 2,5-dihydroxybenzoic acid). The final 91-mer protein sequence was GSSHHHHHHSSGLVPRGSHMSEEQLKAFLTKVQADTSLQEQLKIEGADVVAIAK^AAGFSITTEDLNSHRQNLSDDELEGVAGGAACAAYPAA, where the caret marks the Lys-C cleavage site closest to the YP position, and the single cysteine has been alkylated by iodoacetamide.
To assay 14WM9-catalyzed dephosphorylation of the 91-mer protein, a 10-µL sample containing 1 nmol (100 µM) of deoxyribozyme, 1.25 nmol (125 µM) of free DNA anchor oligonucleotide, and 200 pmol (20 µM) of protein in 70 mM Hepes (pH 7.5), 6 mM ZnCl2, 40 mM MgCl2, 20 mM MnCl2, and 150 mM NaCl was incubated at 37 °C. The deoxyribozyme was omitted in the control experiment. After 36 h, 5 µL of the sample was added to 10 µL of solution containing 60 mM Tris (pH 6.8), 2% SDS, 0.1% bromophenol blue, and 10% glycerol. The sample was separated by SDS/PAGE [1 M Tris (pH 8.45), 0.1% SDS, 5% acrylamide stacking, 15% acrylamide + 10% glycerol resolving], run at 120 V for 75 min. The gel was stained with Imperial Protein Stain (24615; Pierce), and the band corresponding to the protein was excised. In-gel Lys-C digestion was performed as reported (40), except omitting treatment of the gel slice with DTT and iodoacetamide. For the Lys-C digestion step, the gel slice was treated with 40 µL of a solution containing 100 mM Tris (pH 8.0), 40 mM DTT, and 0.6 ng/µL Lys-C (11420429001; Roche). The acidic extraction step used 5% formic acid in acetonitrile. The sample was concentrated by SpeedVac to 10 µL, desalted by Millipore C18 ZipTip, and analyzed by MALDI MS (matrix 2,5-dihydroxybenzoic acid). Monoisotopic m/z values were calculated using the PeptideMass calculator at ExPASy.
Supplementary Material
Acknowledgments
We thank E. Parkinson for providing the human cell lysate, C. Thibodeaux for assistance with protein expression, and numerous members of the W. van der Donk laboratory for experimental advice. This research was supported by National Institutes of Health Grant R01GM065966 (to S.K.S.), Defense Threat Reduction Agency Grant HDTRA1-09-1-0011 (to S.K.S.), and National Science Foundation Grant CHE0842534 (to S.K.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1221946110/-/DCSupplemental.
References
- 1.Radzicka A, Wolfenden R. A proficient enzyme. Science. 1995;267(5194):90–93. doi: 10.1126/science.7809611. [DOI] [PubMed] [Google Scholar]
- 2.Lad C, Williams NH, Wolfenden R. The rate of hydrolysis of phosphomonoester dianions and the exceptional catalytic proficiencies of protein and inositol phosphatases. Proc Natl Acad Sci USA. 2003;100(10):5607–5610. doi: 10.1073/pnas.0631607100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Joyce GF. Forty years of in vitro evolution. Angew Chem Int Ed. 2007;46(34):6420–6436. doi: 10.1002/anie.200701369. [DOI] [PubMed] [Google Scholar]
- 4.Silverman SK. Deoxyribozymes: Selection design and serendipity in the development of DNA catalysts. Acc Chem Res. 2009;42(10):1521–1531. doi: 10.1021/ar900052y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schlosser K, Li Y. Biologically inspired synthetic enzymes made from DNA. Chem Biol. 2009;16(3):311–322. doi: 10.1016/j.chembiol.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 6.Silverman SK. DNA as a versatile chemical component for catalysis, encoding, and stereocontrol. Angew Chem Int Ed. 2010;49(40):7180–7201. doi: 10.1002/anie.200906345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dalby PA. Strategy and success for the directed evolution of enzymes. Curr Opin Struct Biol. 2011;21(4):473–480. doi: 10.1016/j.sbi.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 8.Keefe AD, Szostak JW. Functional proteins from a random-sequence library. Nature. 2001;410(6829):715–718. doi: 10.1038/35070613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chandra M, Sachdeva A, Silverman SK. DNA-catalyzed sequence-specific hydrolysis of DNA. Nat Chem Biol. 2009;5(10):718–720. doi: 10.1038/nchembio.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiao Y, Wehrmann RJ, Ibrahim NA, Silverman SK. Establishing broad generality of DNA catalysts for site-specific hydrolysis of single-stranded DNA. Nucleic Acids Res. 2012;40(4):1778–1786. doi: 10.1093/nar/gkr860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doudna JA, Cech TR. The chemical repertoire of natural ribozymes. Nature. 2002;418(6894):222–228. doi: 10.1038/418222a. [DOI] [PubMed] [Google Scholar]
- 12.Emilsson GM, Nakamura S, Roth A, Breaker RR. Ribozyme speed limits. RNA. 2003;9(8):907–918. doi: 10.1261/rna.5680603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Breaker RR, et al. A common speed limit for RNA-cleaving ribozymes and deoxyribozymes. RNA. 2003;9(8):949–957. doi: 10.1261/rna.5670703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pradeepkumar PI, Höbartner C, Baum DA, Silverman SK. DNA-catalyzed formation of nucleopeptide linkages. Angew Chem Int Ed. 2008;47(9):1753–1757. doi: 10.1002/anie.200703676. [DOI] [PubMed] [Google Scholar]
- 15.Sachdeva A, Silverman SK. DNA-catalyzed serine side chain reactivity and selectivity. Chem Commun. 2010;46(13):2215–2217. doi: 10.1039/b927317d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wong OY, Pradeepkumar PI, Silverman SK. DNA-catalyzed covalent modification of amino acid side chains in tethered and free peptide substrates. Biochemistry. 2011;50(21):4741–4749. doi: 10.1021/bi200585n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sachdeva A, Chandra M, Chandrasekar J, Silverman SK. Covalent tagging of phosphorylated peptides by phosphate-specific deoxyribozymes. ChemBioChem. 2012;13(5):654–657. doi: 10.1002/cbic.201200048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li B, et al. Catalytic promiscuity in the biosynthesis of cyclic peptide secondary metabolites in planktonic marine cyanobacteria. Proc Natl Acad Sci USA. 2010;107(23):10430–10435. doi: 10.1073/pnas.0913677107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schroeder GK, Lad C, Wyman P, Williams NH, Wolfenden R. The time required for water attack at the phosphorus atom of simple phosphodiesters and of DNA. Proc Natl Acad Sci USA. 2006;103(11):4052–4055. doi: 10.1073/pnas.0510879103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lassila JK, Zalatan JG, Herschlag D. Biological phosphoryl-transfer reactions: understanding mechanism and catalysis. Annu Rev Biochem. 2011;80:669–702. doi: 10.1146/annurev-biochem-060409-092741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nowakowski J, Shim PJ, Prasad GS, Stout CD, Joyce GF. Crystal structure of an 82-nucleotide RNA-DNA complex formed by the 10-23 DNA enzyme. Nat Struct Biol. 1999;6(2):151–156. doi: 10.1038/5839. [DOI] [PubMed] [Google Scholar]
- 22.Barford D, Das AK, Egloff MP. The structure and mechanism of protein phosphatases: insights into catalysis and regulation. Annu Rev Biophys Biomol Struct. 1998;27:133–164. doi: 10.1146/annurev.biophys.27.1.133. [DOI] [PubMed] [Google Scholar]
- 23.Jackson MD, Denu JM. Molecular reactions of protein phosphatases—Insights from structure and chemistry. Chem Rev. 2001;101(8):2313–2340. doi: 10.1021/cr000247e. [DOI] [PubMed] [Google Scholar]
- 24.Denu JM, Dixon JE. A catalytic mechanism for the dual-specific phosphatases. Proc Natl Acad Sci USA. 1995;92(13):5910–5914. doi: 10.1073/pnas.92.13.5910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tonks NK. Protein tyrosine phosphatases: From genes, to function, to disease. Nat Rev Mol Cell Biol. 2006;7(11):833–846. doi: 10.1038/nrm2039. [DOI] [PubMed] [Google Scholar]
- 26.Allen KN, Dunaway-Mariano D. Phosphoryl group transfer: Evolution of a catalytic scaffold. Trends Biochem Sci. 2004;29(9):495–503. doi: 10.1016/j.tibs.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 27.Allen KN, Dunaway-Mariano D. Markers of fitness in a successful enzyme superfamily. Curr Opin Struct Biol. 2009;19(6):658–665. doi: 10.1016/j.sbi.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seifried A, Schultz J, Gohla A. Human HAD phosphatases: structure, mechanism, and roles in health and disease. FEBS J. 2013;280(2):549–571. doi: 10.1111/j.1742-4658.2012.08633.x. [DOI] [PubMed] [Google Scholar]
- 29.Shi Y. Serine/threonine phosphatases: Mechanism through structure. Cell. 2009;139(3):468–484. doi: 10.1016/j.cell.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 30.Nikolic-Hughes I, Rees DC, Herschlag D. Do electrostatic interactions with positively charged active site groups tighten the transition state for enzymatic phosphoryl transfer? J Am Chem Soc. 2004;126(38):11814–11819. doi: 10.1021/ja0480421. [DOI] [PubMed] [Google Scholar]
- 31.Bone R, Frank L, Springer JP, Atack JR. Structural studies of metal binding by inositol monophosphatase: Evidence for two-metal ion catalysis. Biochemistry. 1994;33(32):9468–9476. doi: 10.1021/bi00198a012. [DOI] [PubMed] [Google Scholar]
- 32.Choe J-Y, Fromm HJ, Honzatko RB. Crystal structures of fructose 1,6-bisphosphatase: mechanism of catalysis and allosteric inhibition revealed in product complexes. Biochemistry. 2000;39(29):8565–8574. doi: 10.1021/bi000574g. [DOI] [PubMed] [Google Scholar]
- 33.Kamerlin SCL, Wilkie J. The role of metal ions in phosphate ester hydrolysis. Org Biomol Chem. 2007;5(13):2098–2108. doi: 10.1039/b701274h. [DOI] [PubMed] [Google Scholar]
- 34.Rautio J, et al. Prodrugs: Design and clinical applications. Nat Rev Drug Discov. 2008;7(3):255–270. doi: 10.1038/nrd2468. [DOI] [PubMed] [Google Scholar]
- 35.Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15(21):8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Flynn-Charlebois A, et al. Deoxyribozymes with 2′-5′ RNA ligase activity. J Am Chem Soc. 2003;125(9):2444–2454. doi: 10.1021/ja028774y. [DOI] [PubMed] [Google Scholar]
- 37.Wang Y, Silverman SK. Characterization of deoxyribozymes that synthesize branched RNA. Biochemistry. 2003;42(51):15252–15263. doi: 10.1021/bi0355847. [DOI] [PubMed] [Google Scholar]
- 38.Kost DM, Gerdt JP, Pradeepkumar PI, Silverman SK. Controlling the direction of site-selectivity and regioselectivity in RNA ligation by Zn2+-dependent deoxyribozymes that use 2′,3′-cyclic phosphate RNA substrates. Org Biomol Chem. 2008;6(23):4391–4398. doi: 10.1039/b813566e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bair JS, Palchaudhuri R, Hergenrother PJ. Chemistry and biology of deoxynyboquinone, a potent inducer of cancer cell death. J Am Chem Soc. 2010;132(15):5469–5478. doi: 10.1021/ja100610m. [DOI] [PubMed] [Google Scholar]
- 40.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc. 2006;1(6):2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.