Abstract
The Large-neutral Amino Acid Transporter 1 (LAT-1)—a sodium-independent exchanger of amino acids, thyroid hormones, and prescription drugs—is highly expressed in the blood–brain barrier and various types of cancer. LAT-1 plays an important role in cancer development as well as in mediating drug and nutrient delivery across the blood–brain barrier, making it a key drug target. Here, we identify four LAT-1 ligands, including one chemically novel substrate, by comparative modeling, virtual screening, and experimental validation. These results may rationalize the enhanced brain permeability of two drugs, including the anticancer agent acivicin. Finally, two of our hits inhibited proliferation of a cancer cell line by distinct mechanisms, providing useful chemical tools to characterize the role of LAT-1 in cancer metabolism.
Keywords: membrane transporter, polypharmacology, glioblastoma multiforme, solute carrier (SLC) transporter
Large-neutral Amino Acid Transporter 1 (LAT-1) is a sodium-independent exchanger found in the brain, testis, and placenta, where it mediates transport of large-neutral amino acids (e.g., tyrosine) and thyroid hormones (e.g., triiodothyronine) across the cell membrane (1). More specifically, LAT-1 is highly expressed in the blood- and brain-facing membranes of the blood–brain barrier (BBB) to supply the central nervous system (CNS) with essential nutrients and to help maintain the neural microenvironment (2). LAT-1 is also an important drug target because it transports several prescription drugs, such as the antiparkinsonian drug L-dopa and the anticonvulsant gabapentin, across the BBB, thereby enabling their pharmacologic effects (3, 4). This function at the BBB has made LAT-1 a target for drug delivery by modifying CNS-impermeable drugs such that they become LAT-1 substrates and have enhanced BBB penetration (5, 6).
In addition, LAT-1 expression levels are increased in many types of cancer, including non-small-cell lung cancer and glioblastoma multiforme (GBM) (7, 8). LAT-1 expression increases as cancers progress, leading to higher expression levels in high-grade tumors and metastases (9). In particular, LAT-1 plays a key role in cancer-associated reprogrammed metabolic networks by supplying growing tumor cells with essential amino acids that are used as nutrients to build biomass and signaling molecules to enhance proliferation by activating progrowth pathways such as the mammalian target of rapamycin (mTOR) pathway (10). Furthermore, inhibiting LAT-1 function reduces tumor cell proliferation, indicating that it may be a viable target for novel anticancer therapies (11–13). A cancer drug targeting LAT-1 can therefore be a LAT-1 inhibitor that deprives the cancer cells of nutrients or a cytotoxic LAT-1 substrate with an intracellular target (e.g., a metabolic enzyme).
LAT-1 is a large protein with 12 putative membrane-spanning helices (14). To transport solutes across the membrane, LAT-1 binds SLC3A2, a glycoprotein with a single membrane-spanning helix that serves as a chaperone for LAT-1 (14). The atomic structure of human LAT-1 is not known, but LAT-1 exhibits significant sequence similarity to prokaryotic transporters such as members of the amino acid/polyamine/organocation transporter (APC) family, whose representative structures have been recently determined by X-ray crystallography (15–19). Structures of the arginine:agmatine antiporter AdiC from Escherichia coli (15, 17, 18) and Salmonella enterica (20) in different conformations reveal an internal twofold pseudosymmetry, similar to the structures of the sodium- and chloride-dependent leucine transporter, LeuT (19, 21). These data, combined with structures of additional related transporters (22) and molecular dynamics (MD) simulations (23), suggest a common transport mechanism among the LAT-1 homologs and LeuT, in which the role of sodium in LeuT is proposed to be mimicked by a proton in some APC transporters (23). Thus, LAT-1 probably also transports ligands across the cell membrane via the alternating access transport mechanism (22, 24, 25).
In this study, we take an integrated computational and experimental approach to characterize previously unknown LAT-1 ligands. We construct structural models of LAT-1 based on structures of homologous APC family transporters from prokaryotic organisms and then perform virtual ligand screening of metabolite and prescription drug libraries against these models to predict small-molecule ligands. The top-scoring hits are tested experimentally for LAT-1 inhibition and transport by using cis-inhibition experiments and trans-stimulation assays, respectively. Furthermore, we characterize the effect of select validated ligands on cell proliferation. Finally, we describe the pharmacological implications of our results, including how the intended and unintended effects of the discovered ligands may be mediated by LAT-1 transport across the BBB as well as their potential use as chemical tools to characterize the role of LAT-1 in cancer.
Results
LAT-1 Predicted Structure and Ligand Binding.
LAT-1 was modeled based on the X-ray structure of the arginine/agmatine transporter AdiC from E. coli in the outward-occluded arginine-bound conformation (17) and the structure of the APC transporter ApcT from Methanococcus jannaschii in an inward-apo conformation (16) (Fig. S1 and SI Materials and Methods). The final LAT-1 model contains the whole transmembrane domain of the protein (i.e., the 12 transmembrane helices), including the residues constituting the predicted ligand-binding site. Comparative models were first scored by using Z-DOPE, a normalized atomic distance-dependent statistical potential based on known protein structures (26). The Z-DOPE scores of the top models were −0.3, suggesting that 60% of its Cα atoms are within 3.5 Å of their correct positions (27) (Table S1). Each model was also evaluated based on its ability to discriminate between known ligands and likely nonbinders (decoys), by using enrichment curves derived from ligand-docking calculations (28). The logAUC score for the final refined LAT-1 model was 31.9 (Table S1), suggesting that it is suitable for predicting ligands for experimental testing (28–30).
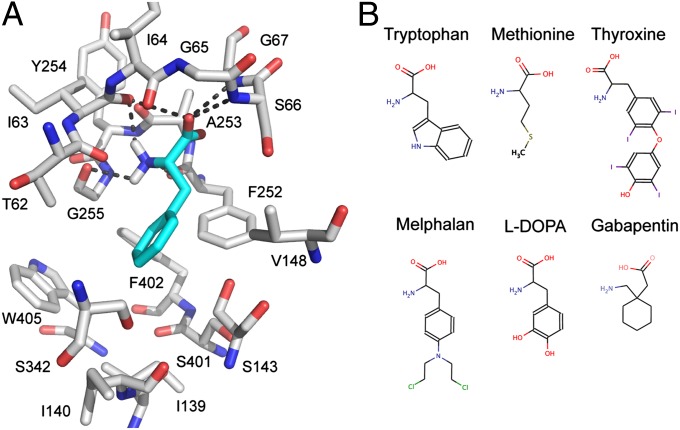
The model of LAT-1 interacting with phenylalanine indicates that the majority of the key polar interactions between LAT-1 and the carboxyl and amino group of the amino acid ligands are conserved between LAT-1 and the AdiC template structure (Fig. 1A and Fig. S1). For example, the backbone polar groups of LAT-1 residues T62, I63, I64, S66, G67, F252, A253, and G255 are predicted to form polar interactions with phenylalanine (Fig. 1). These residues correspond to A22, I23, M24, S26, G27, W202, S203, and I205 of AdiC, which make similar interactions with the carboxyl and amino groups of its ligand arginine (17). Because the carboxyl and amino groups are conserved among all other known LAT-1 ligands, such as thyroxine and gabapentin (Fig. 1B), we hypothesize that they make similar interactions with LAT-1.
Fig. 1.
Predicted LAT-1 structure and ligand-binding mode. (A) Predicted structure of the LAT-1–phenylalanine complex. LAT-1 (gray) and phenylalanine (cyan) are shown as the stick models; oxygen, nitrogen, and hydrogen atoms are depicted in red, blue, and white, respectively; key hydrogen bonds between phenylalanine and LAT-1 (involving residues Thr-62, Ile-63, Ile-64, Ser-66, Gly-67, Phe-252, Ala-253, and Gly-255) are shown as dotted gray lines. (B) Structures of representative LAT-1 substrates. Known LAT-1 substrates, including metabolites (tryptophan, methionine, and thyroxine) and prescription drugs (melphalan, L-dopa, and gabapentin) are shown using MarvinView 5.4.1.1 (Chemaxon).
Conversely, differences in the ligand preferences of LAT-1 and AdiC may be explained by two major differences in the binding sites of the LAT-1 model and the AdiC structure (Fig. S2). First, several residues with hydrophobic side chains (i.e., I139, V148, F252, F402, and W405) are located in the LAT-1 binding site, likely contributing to increased ligand-binding affinity of hydrophobic amino acids to LAT-1 via van der Waals interactions and the hydrophobic effect (e.g., the tryptophan indole ring). Some of these hydrophobic residues are replaced by nonhydrophobic residues in LAT-1 homologs, including the template structure AdiC and other SLC7 members. For instance, the aromatic residue W405 in LAT-1 corresponds to the polar T361 in AdiC. Second, several binding site residues in AdiC are replaced by residues with smaller side chains in LAT-1, creating a larger volume in LAT-1’s binding site that can accommodate larger amino acids. For instance, M104, I205, and W293 in AdiC correspond to the smaller V148, G255, and S342 in LAT-1 (Fig. 1A and Fig. S2).
Virtual Screening of Drugs and Metabolites.
We computationally screened filtered libraries of 6,436 and 12,730 small molecules from the Kyoto Encyclopedia of Genes and Genomes (KEGG) DRUG and KEGG LIGAND COMPOUND databases (28), respectively, against two LAT-1 models (Fig. 2 and Table S1). Some of the top-scoring hits were shown previously to be LAT-1 ligands, increasing our confidence in the binding site model. For example, the known substrate L-Trp was ranked 50th in the docking screen of KEGG LIGAND COMPOUND. The 200 (3.1%) KEGG DRUG and 500 (3.9%) KEGG COMPOUND top-scoring hits against our top two models were analyzed manually. A compound was selected for experimental testing based on three criteria: (i) similarity between its docking pose and those of known ligands in complex with LAT-1 (28); (ii) the chemical novelty of its scaffold, especially if it occurred frequently among the top-scoring compounds; and (iii) its pharmacological effect (28).
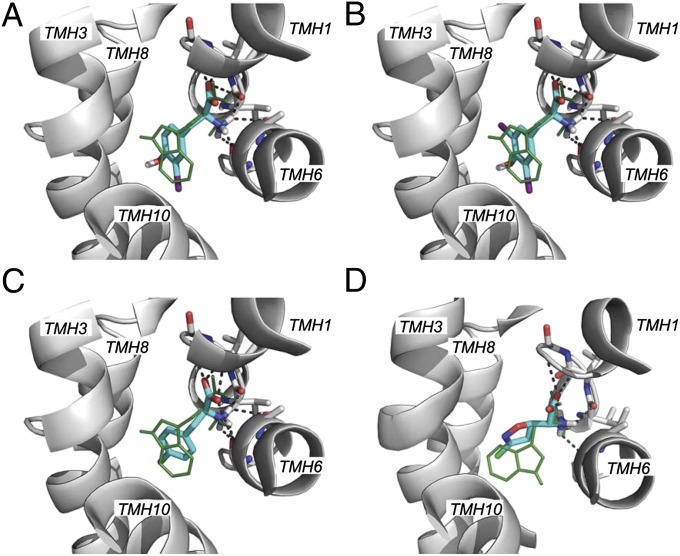
Fig. 2.
Predicted binding modes for LAT-1 ligands. Predicted binding modes of the known substrate tryptophan (green lines) and four ligands discovered in the docking screen. Residues making polar interactions with the ligand are illustrated with sticks; carbon atoms are colored in white, nitrogen atoms in blue, and oxygen atoms in red; hydrogen bonds are represented by dotted gray lines. The predicted pose of a known LAT-1 ligand, tryptophan, is shown with green lines. The compounds depicted are 3-Iodo-L-tyrosine (A), 3,5-diiodo-L-tyrosine (B), fenclonine (C), and acivicin (D). Halogen atoms in the discovered ligands are colored in purple (iodine) and green (chlorine).
Experimental Validation of Predicted Ligands.
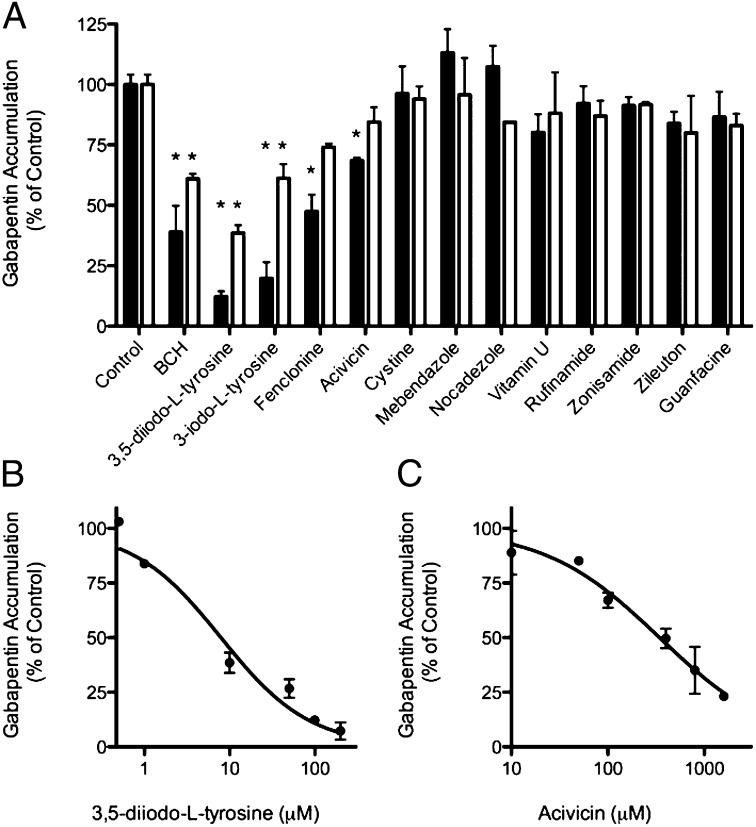
A LAT-1–overexpressing cell line was generated by stably transfecting HEK cells with human LAT-1 cDNA. HEK-LAT1 cells expressed 20-fold higher levels of LAT-1 mRNA relative to HEK-EV cells and demonstrated LAT-1–specific uptake of the established system L substrates, gabapentin and L-leucine (Fig. S3 A–D). Twelve of the top-scoring molecules were selected for experimental testing by cis-inhibition assay (Table 1, Table S2, and Fig. 2). Each molecule was tested as a LAT-1 ligand by determining its ability to inhibit transport of a known LAT-1 substrate in HEK-LAT1 cells at concentrations of 10 and 100 μM (Fig. 3 and Fig. S3E). The known LAT-1 inhibitor 2-aminobicyclo-(2,2,1)-heptane-2-carboxylic acid (BCH) was also included as a positive control. At 100 μM, inhibition of intracellular gabapentin accumulation ranged from 88% (3,5-diiodo-L-tyrosine) to <10% (cystine, mebendazole, and nocadezole), with the metabolites 3,5-diiodo-L-tyrosine and 3-iodo-L-tyrosine, as well as the tryptophan hydroxylase inhibitor fenclonine and the anticancer agent acivicin, demonstrating significant inhibition of gabapentin and L-leucine transport (Fig. 3A and Fig. S3E). Acivicin also obtained a dissimilarity score of 0.74 using the JCDissimilarityCFTanimoto score, which calculates dissimilarities among molecules based on chemical fingerprints, indicating that it is a chemically novel LAT-1 ligand (Table 1 and Materials and Methods).
Table 1.
Small-molecule ligands confirmed experimentally
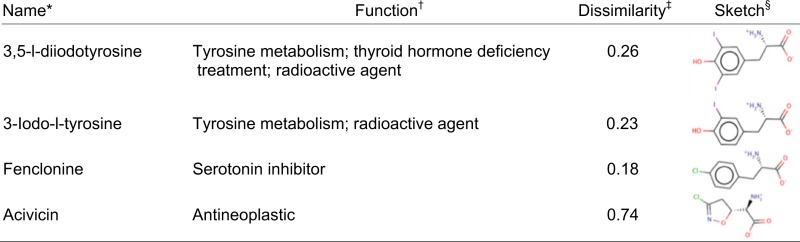 |
Generic or chemical name of the molecule.
Pharmacological function of the drug or the physiological function of the metabolites, when applicable.
Dissimilarity measure calculated with the Chemaxon fingerprints. Dissimilarity values of >0.7 suggest that the molecule is chemically different from all known LAT-1 ligands.
A 2D sketch of the molecule is shown.
Fig. 3.
Experimental validation of predicted LAT-1 ligands. Predicted LAT-1 ligands were validated by cis-inhibition of gabapentin uptake. (A) Cells were coincubated with 12 predicted ligands and a positive control (BCH) at either 100 μM (filled bars) or 10 μM (open bars) concentrations and gabapentin (1 μM unlabeled and 10 nM radiolabeled). Each bar depicts the mean of two to four separate experiments; error bars represent SEM. (B and C) Dose-dependent inhibition of gabapentin (1 μM unlabeled with 10 nM radiolabeled) accumulation by 3,5-diiodo-L-tyrosine (IC50 = 7.9 μM) and acivicin (IC50 = 340 μM), respectively. Each point is the mean of two or three separate experiments; error bars represent SEM. Statistical analysis in A was by one-way ANOVA and Dunnett’s multiple comparison test. *P < 0.05.
The potencies of selected active ligands were further established by determining the IC50 values for inhibiting gabapentin accumulation in the HEK-LAT1 cells. IC50 values ranged from 7.9 μM (3,5-diiodo-l-tyrosine; Fig. 3B) to 340 μM (acivicin; Fig. 3C). At 10 μM, inhibition of gabapentin accumulation ranged from 61% (3,5 diiodo-l-tyrosine) to <10%, with 3,5-diiodo-l-tyrosine and 3-iodo-l-tyrosine significantly inhibiting gabapentin transport (Fig. 3A). Interestingly, 3,5-diiodo-l-tyrosine is a stronger inhibitor than the positive control BCH. In summary, one-third (4 of 12) of the top-scoring molecules selected for experimental testing are LAT-1 ligands capable of inhibiting gabapentin and L-leucine transport in HEK-LAT1 cells.
Identification of LAT-1 Substrates.
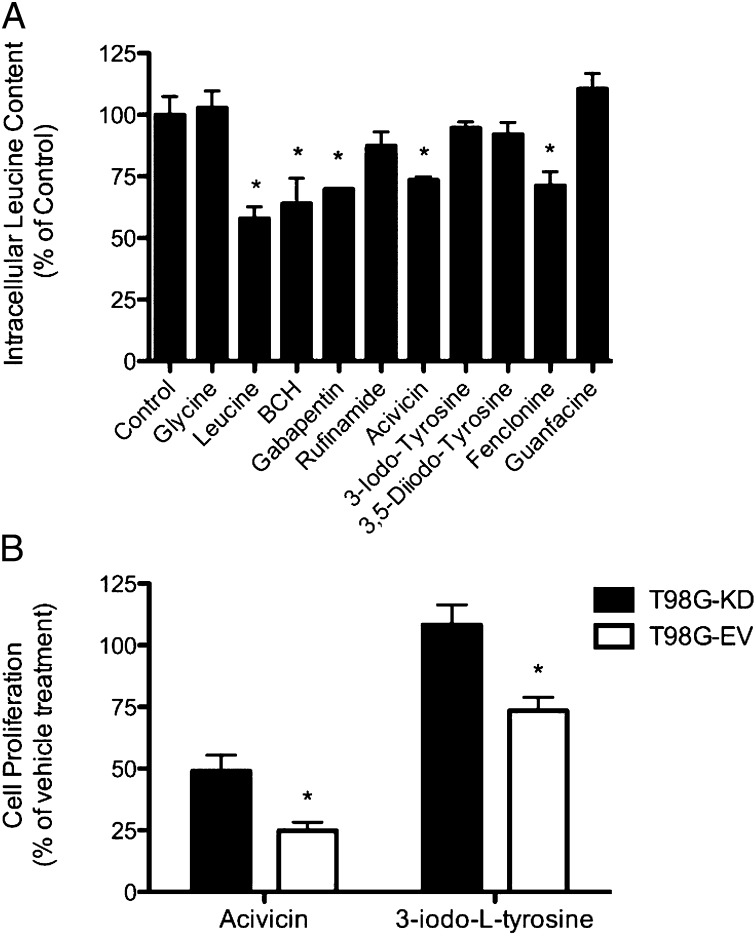
The four molecules found to significantly inhibit gabapentin accumulation in the HEK-LAT1 cells were further analyzed as putative substrates by trans-stimulation assay. This assay takes advantage of LAT-1’s obligatory exchange mechanism of transport by exchanging intracellular L-leucine from preloaded HEK-LAT1 cells with an extracellular molecule only if it is a LAT-1 substrate. Three known LAT-1 substrates served as positive controls and were able to induce L-leucine efflux from the HEK-LAT1 cells, including L-leucine (43%), gabapentin (36%), and BCH (30%) (Fig. 4A). In contrast, glycine was used as a negative control because it is known not to be a LAT-1 substrate and did not induce any L-leucine efflux. Two of the four inhibitors confirmed in our cis-inhibition assay also induced L-leucine efflux. Acivicin and fenclonine induced L-leucine efflux by 27% and 29%, respectively, indicating that they are transported by LAT-1. These results indicate that the drug-like molecules acivicin and fenclonine, which both have pharmacodynamic effects in the CNS, are likely LAT-1 substrates. Surprisingly, both of the more potent LAT-1 inhibitors, the metabolites 3,5-diiodo-L-tyrosine and 3-iodo-L-tyrosine, were only able to induce 7.9% and 5.4% L-leucine efflux, respectively, suggesting that they are inhibitors that only bind to, but are not transported by, LAT-1. Finally, guanfacine and rufinamide were also studied, and neither induced significant L-leucine efflux.
Fig. 4.
Substrate determination and cytotoxicity characterization of predicted ligands. Predicted LAT-1 ligands validated in cis-inhibition assays were subjected to substrate determination by trans-stimulation of L-leucine efflux (1 μM unlabeled and 10 nM radiolabeled). (A) Cells were preloaded with L-leucine, and efflux was induced by subsequent addition of each test compound at a concentration of 1 mM. Gabapentin, L-leucine, and BCH were included as positive controls, and glycine and guanfacine were included as negative controls. (B) The cytotoxic effects of acivicin (100 μM) and 3-iodo-L-tyrosine (1 mM) against T98G glioblastoma cells stably expressing an shRNA against LAT-1 (T98G-KD; filled bars) or EV (T98G-EV; open bars) are depicted. Cell proliferation for both cell lines and treatment conditions were normalized to cell density at treatment day 0 and then to the vehicle control treatment at 48 h. Each bar represents the mean of three or four separate experiments, and error bars represent the SEM. Statistical analysis in A was by one-way ANOVA and Dunnett’s multiple comparison test and in B was by two-way ANOVA and Bonferroni correction for multiple testing. *P < 0.05.
Inhibition of LAT-1–Dependent Cell Proliferation.
LAT-1 is highly expressed in various cancer cells, providing them with nutrients and signaling molecules for growth. Thus, a drug targeting LAT-1 in cancer can be an inhibitor that deprives the cancer cells from nutrients or a cytotoxic substrate with an intracellular target. We therefore investigated the antiproliferative effects of select validated LAT-1 ligands, including the LAT-1 substrate acivicin and the inhibitor 3-iodo-L-tyrosine, by cell proliferation assay in the high LAT-1–expressing GBM cell line, T98G (8). The LAT-1–specific effects of each ligand on cell growth were determined in control cells (T98G-EV) and cells with LAT-1 expression (Fig. S4A) and function (Fig. S4B) knocked down (T98G-KD). The anticancer drug acivicin was a more potent growth inhibitor of T98G-EV (75% growth reduction) than T98G-KD (51% growth reduction) (Fig. 4B). Similarly, 3-iodo-L-tyrosine had a more potent effect on T98G-EV cells, reducing their growth by 27%, whereas having no effect on T98G-KD (Fig. 4B Right). These results suggest that both 3-iodo-L-tyrosine and acivicin are capable of inhibiting cancer cell proliferation in a LAT-1–dependent manner by means of two distinct mechanisms, including nutrient deprivation and cytotoxicity, respectively.
Discussion
Three key findings emerge from our study. First, two drug-like molecules that interact with different proteins in the CNS are also substrates of LAT-1. This finding may explain the mechanism by which these drugs penetrate the BBB to reach their targets in the CNS. It also provides a starting point for optimizing the two drugs for better BBB permeability. Second, two of the discovered LAT-1 ligands, including one inhibitor and one substrate, inhibit proliferation of cancer cells. This result indicates that LAT-1 can be targeted for cancer therapy by means of different mechanisms and reveals chemical tools for further characterizing the role of LAT-1 in cancer. Third, the identified LAT-1 ligands achieve their pharmacological effect (positive or negative) on the CNS or cancer by interacting with multiple targets. This finding suggests that effective therapy can be obtained by applying modeling and docking approaches to whole systems, including pathways and networks. We take each of the three key findings in turn.
LAT-1–Mediated BBB Drug Permeability.
Passive diffusion has long been thought of as the primary mechanism by which most drugs cross the BBB to permeate the CNS (31). The contribution of carrier-mediated transport to this process is assumed to be minimal, even though different classes of membrane transporters have been shown to restrict and/or facilitate access of drugs, nutrients, and toxins to the CNS (32–34). LAT-1 is one such influx transporter known to transport nutrients and xenobiotics across the BBB. In this study, we identified two previously unknown LAT-1 substrates, including acivicin and fenclonine, which may also cross the BBB via LAT-1–mediated transport. Both were found to be likely LAT-1 substrates in trans-stimulation studies (Fig. 4), and both are known to have pharmacodynamic effects in the CNS. Even though previous studies have used trans-stimulation to establish whether or not a specific transporter can transport different compounds (35–37), this assay provides indirect evidence that a compound may be a substrate for a specific transporter. Nevertheless, acivicin was assessed in a clinical trial for treating various solid tumors that did not involve the CNS but failed these trials due to CNS-related toxic side effects (e.g., lethargy and confusion) (38). Furthermore, these side effects were reversed when acivicin was concomitantly administered with a mixture of amino acids, including the prototypical LAT-1 substrate, L-leucine. These observations highly implicate LAT-1 in mediating acivicin’s CNS permeability in humans. The second molecule, fenclonine, is an irreversible tryptophan hydroxylase inhibitor used to deplete CNS serotonin levels in animal models of human disease (39). Together with our results, LAT-1 likely mediates the effects in the CNS by transporting fenclonine across the BBB. Therefore, influx transporters such as LAT-1 may be important mediators of drug efficacy and toxicity in the CNS and have a greater contribution to drug penetration across the BBB than previously thought.
Targeting LAT-1 for Cancer Therapy.
Changes in cell metabolism are strongly associated with cancer. Membrane transporters have been shown to play a key role in such reprogrammed metabolic networks by providing nutrients to transforming cells. For example, the glucose transporter (GLUT1; SLC2A1) is up-regulated in various cancers to provide glucose as a carbon source to accommodate an increased rate of anabolic cellular reactions and to maintain a microecosystem favorable for cancer cells (40). Moreover, LAT-1 imports essential amino acids that serve as nutrients and pro-proliferative signaling molecules by exporting glutamine brought into cancer cells via the glutamine transporter ASCT2 (10). Thus, therapeutics targeting LAT-1 can be (i) inhibitors that selectively block transport by LAT-1 and/or ASCT2, depriving the cancer cell of nutrients required for proliferation, or (ii) cytotoxic substrates that are delivered into the cell via LAT-1 and/or ASCT2 to act on an intracellular target. LAT-1 ligands that act through each of these mechanisms were discovered in our screen (Figs. 2–4 and Table 1).
First, 3-iodo-L-tyrosine is a thyroid hormone derivative typically used to treat hormone deficiencies and as a radioactive agent. Here, cis-inhibition and cell-proliferation experiments identified 3-iodo-L-tyrosine as a potent LAT-1 inhibitor (Fig. 3A) that reduces proliferation of T98G glioblastoma cells (Fig. 4B), possibly by starving these cells of nutrients supplied by LAT-1. Our results suggest that, in addition to its putative anticancer applications, 3-iodo-L-tyrosine may be useful as a diagnostic imaging agent to identify tumors and other disease states associated with LAT-1 up-regulation (41).
Second, acivicin is a cytotoxic agent with antitumor activity that targets glutamine-dependent amidotransferases in the biosynthesis of purines and pyrimidines (42). Trans-stimulation and cell-proliferation experiments indicated that acivicin is likely a LAT-1 substrate (Fig. 4), suggesting that LAT-1 can be targeted for acivicin delivery into tumor cells. Interestingly, acivicin failed in various clinical trials (e.g., for advanced solid malignancies) because of CNS-related toxic side effects (38) or insufficient efficacy (43). Thus, targeting LAT-1 in a tumor with a drug that is a LAT-1 substrate may not be a rational therapeutic strategy because LAT-1 would also facilitate entry of the drug into the CNS. However, design of other cytotoxic substrates of LAT-1, which are not associated with deleterious CNS effects, may represent a viable drug development strategy for cancer. Although cell proliferation experiments indicate that multiple transporters may mediate acivicin accumulation in cells, the significant difference in sensitivity to acivicin between T98G-KD and -EV cells clearly indicates a LAT-1–specific effect on cell proliferation, most likely by mediating acivicin uptake.
Targeting Biological Systems Using a Combined Structural Pharmacology Approach.
Although polypharmacology can be exploited to improve the treatment of various nervous system disorders and cancers, it may also lead to toxicity. Virtual screening against the LAT-1 model identifies ligands that likely achieve their pharmacological effect by interacting with multiple proteins. Current efforts to design reagents, including drugs or chemical tools, for treating complex diseases include optimizing binding affinities of one or more molecules against more than one target. Recent advances in comparative modeling and molecular docking for ligand discovery, coupled with the determination of a number of membrane protein structures including transporters, enables us to target multiple components of a single pathway (e.g., mTOR) or organ (e.g., the BBB) by using structure-based ligand discovery. Importantly, some of these newly determined structures represent different protein conformations, allowing in silico screens of small molecules against comparative models of different conformations to suggest chemically distinct ligands. For example, a structure-based approach predicted that molecules binding to a model for the outward-facing conformation of the GABA transporter 2 were chemically distinct from those predicted to bind an occluded model (44). Thus, as more structures of LAT-1 homologs are discovered, our results can be refined to identify novel LAT-1 ligands for effective therapy and the study of CNS diseases and cancer.
In summary, we constructed structural models for LAT-1 based on atomic structures of distantly related prokaryotic homologs. Two small organic molecule libraries containing endogenous metabolites and prescription drugs were then virtually screened against these models. Select top-ranked docking hits were tested experimentally, and four previously unknown LAT-1 ligands were identified: 3,5-diiodo-L-tyrosine, 3-iodo-L-tyrosine, fenclonine, and acivicin. Furthermore, acivicin and 3-iodo-L-tyrosine were found to have LAT-1–mediated antiproliferative effects in a GBM cancer cell line. These findings provide chemical tools to elucidate the role of membrane transporters as potential drug targets and in mediating tissue permeability to small organic molecules. Future studies are needed to further elucidate the mechanism by which these ligands interact with LAT-1.
Materials and Methods
Comparative Model Construction.
LAT-1 was modeled by using MODELLER-9v11 based on the X-ray structure of the arginine/agmatine transporter AdiC from E. coli in the outward-occluded arginine-bound conformation [Protein Data Bank (PDB) ID code 3L1L] (17). We also modeled LAT-1 based on the structure of ApcT from M. jannaschii in an inward-apo conformation (PDB ID code 3GI9) (ref. 16; SI Materials and Methods). We relied on a published alignment (15), as well as alignments obtained from the Promals3D server (45), where gaps were present primarily, but not only, in the predicted extracellular loops and were manually refined (SI Materials and Methods and Fig. S1). For each template structure and alignment, 100 models were generated by using the standard “automodel” routine of MODELLER-9v11 (46). The initial models were assessed by using Z-DOPE, a normalized atomic distance-dependent statistical potential based on known protein structures (26). For selected LAT-1 models, the binding site was refined by repacking the side chains on a fixed backbone using Scwrl4 (47). The final models for virtual screening were selected based on their ability to discriminate known ligands from decoy compounds using enrichment curves derived from ligand-docking calculations (SI Materials and Methods) (28, 29, 48). These final models were also evaluated based on residue hydrophobicity (49) and evolutionary conservation profiles (SI Materials and Methods and Fig. S5) (50).
Virtual Screening and Ligand Docking.
Virtual screening against the LAT-1 models was performed by using a semiautomatic docking procedure (29) relying on DOCK 3.5.54 (51). The docking poses of the database molecules were ranked by DOCK score, which is a sum of van der Waals, Poisson–Boltzmann finite-difference electrostatics, and ligand desolvation penalty terms. Poses of the 500 highest-ranked compounds from each of the docking screens were inspected by eye to prioritize compounds for experimental testing (SI Materials and Methods) (28, 48).
Chemical Similarity Calculations.
The chemical novelty of the top hits was first evaluated by using Instant JChem (Version 5.7.0; ChemAxon). Specifically, we calculated the chemical dissimilarity measure JCDissimilarityCFTanimoto among the top small-molecule hits and the 44 known LAT-1 ligands from the databases ChEMBL (52) and UniProt (53), as well as from the literature (1); predicted ligands with values of >0.7 were classified as chemically novel.
Cell Lines.
Stably transfected HEK 293 cells were created by transfecting pcDNA5/FRT (Invitrogen) vector containing the full-length human LAT-1 cDNA (HEK-LAT1) and the empty vector (HEK-EV) by using Lipofectamine 2000 (Invitrogen) per the manufacturer’s instructions. Transfected cells were maintained in DMEM-H21 containing 10% (vol/vol) FBS, 100 units/mL penicillin, 100 µg/mL streptomycin, and 200 µg/mL hygromycin B at 37 °C and 5% CO2. Stable LAT-1 knockdown cells were created by infecting 2 × 105 T98G GBM cells with lentivirus produced by the University of California, San Francisco (UCSF) Lentiviral RNAi core (54) carrying a pSicoR vector expressing green fluorescent protein (GFP) and either an anti–LAT-1 shRNA (T98G-KD; Table S3) or empty vector (T98G-EV) at a multiplicity of infection equal to 10. One week after infection, GFP+ cells were isolated by using fluorescence-activated cell sorting (FACS) analysis by the Laboratory for Cell Analysis at the UCSF Comprehensive Cancer Center. GFP+ T98G-KD and T98G-EV cells were validated for LAT-1 RNA and functional knockdown as described in SI Materials and Methods. T98G, T98G-KD, and T98G-EV cells were maintained in DMEM-H21 containing 10% FBS, 100 units/mL penicillin, and 100 µg/mL streptomycin at 37 °C and 5% CO2.
Inhibition of [3H]-Gabapentin Uptake.
Uptake studies were performed as described (55). Briefly, HEK-LAT1 cells were seeded at a density of 2 × 105 cells per well in poly-D-lysine–coated 24-well (BD Falcon) plates and grown to 80–90% confluence. Cells were rinsed with prewarmed, sodium-free choline buffer at pH 7.4 (140 mM choline chloride, 2 mM KCl, 1 MgCl2 mM, 1 CaCl2 mM, 1 M Tris) and then incubated in 0.3 mL of prewarmed choline buffer containing 1 μM unlabeled gabapentin and 10 nM [3H]-gabapentin (American Radiolabeled Chemicals) for 3 min at 37 °C in the presence of 10 and 100 μM test compound (Sigma-Aldrich). The reaction was terminated by washing cells twice with 1.0 mL of ice-cold choline buffer, followed by addition of 700 μL of lysis buffer (0.1% SDS vol/vol, 0.1 N NaOH). Intracellular radioactivity was determined by scintillation counting and normalized per well of protein content as measured by bicinchoninic acid protein assay (Pierce). Concentration-dependent inhibition was measured under the same conditions as for the single-point measurements. Cells were incubated with 0.5, 1, 10.0, 50.0, 100.0, and 200.0 μM 3,5 diiodo-L-tyrosine or 10.0, 50.0, 100.0, 400.0, 800.0, and 1,600.0 μM acivicin. The concentration at which 50% of [3H]-gabapentin accumulation was inhibited (IC50) was computed by fitting the data using GraphPad Prism (Version 5.0).
Trans-stimulation of [3H]-L-Leucine Efflux.
Trans-stimulation studies were performed by monitoring intracellular L-leucine efflux from HEK-LAT1 cells stimulated by extracellular addition of known or putative LAT-1 substrates. HEK-LAT1 cells were seeded under the same conditions described for inhibition experiments. Cells were rinsed with prewarmed choline buffer and then preloaded with [3H]-L-Leucine (Perkin-Elmer) by incubating cells in 0.3 mL of prewarmed choline buffer containing 1 μM unlabeled and 10 nM radiolabeled substrate for 5 min at 37 °C. Uptake was terminated by washing cells twice with 1.0 mL of ice-cold choline buffer, and [3H]-L-Leucine efflux was then induced by addition of 1 mM test compound (Sigma-Aldrich) in prewarmed choline buffer for 1 min at 37 °C. Trans-stimulation was terminated by washing cells twice with 1.0 mL of ice-cold choline buffer, followed by addition of 700 μL of lysis buffer (0.1% SDS vol/vol, 0.1 N NaOH). Intracellular radioactivity was determined as described above.
Cell Proliferation Assay.
T98G-KD and -EV cells were seeded at 2.5 × 103 cells per well in 96-well plates (Corning Life Sciences), and on the following day cells were exposed to growth medium containing either drug or vehicle (0.85% saline solution) for 48 h. Cell density was measured on the treatment day and 48 h after treatment by using the CellTiter-Glo cell viability kit (Promega) according to the manufacturer’s instructions. Cell lysates were transferred to white opaque 96-well plates (Corning Life Sciences), and bioluminescence was measured on a Glomax luminometer (Promega). Proliferation of each cell line after 48 h was first normalized to the density measured on treatment day (0 h), followed by normalization of drug to vehicle treatment.
Statistical Analysis.
Data were analyzed by one-way ANOVA followed by Dunnett’s multiple comparison test, two-way ANOVA followed by Bonferroni correction for multiple testing, or two-tailed unpaired t test. Probability values of <0.05 were considered statistically significant.
Supplementary Material
Acknowledgments
We thank Ursula Pieper, Ben Webb, and Elina Tjioe for technical assistance and maintenance of the computational resources required for this study; Jonathan Sockolosky for technical assistance in cloning LAT-1; and University of California, San Francisco ViraCore for the production of custom lentivirus. The project was supported by National Institutes of Health Grants R01 GM54762 (to A. Sali), U54 GM074929 and U01 GM61390 (to A. Sali and K.M.G.), P01 GM71790 (to A. Sali), F32 GM088991 (to A. Schlessinger), and T32 GM007175 (to E.G.G.). We also received funding for computing hardware from Hewlett Packard, IBM, NetApps, Intel, Ron Conway, and Mike Homer.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1218165110/-/DCSupplemental.
References
- 1.Kanai Y, et al. Expression cloning and characterization of a transporter for large neutral amino acids activated by the heavy chain of 4F2 antigen (CD98) J Biol Chem. 1998;273(37):23629–23632. doi: 10.1074/jbc.273.37.23629. [DOI] [PubMed] [Google Scholar]
- 2.Roberts LM, et al. Subcellular localization of transporters along the rat blood-brain barrier and blood-cerebral-spinal fluid barrier by in vivo biotinylation. Neuroscience. 2008;155(2):423–438. doi: 10.1016/j.neuroscience.2008.06.015. [DOI] [PubMed] [Google Scholar]
- 3.Alexander GM, Schwartzman RJ, Grothusen JR, Gordon SW. Effect of plasma levels of large neutral amino acids and degree of parkinsonism on the blood-to-brain transport of levodopa in naive and MPTP parkinsonian monkeys. Neurology. 1994;44(8):1491–1499. doi: 10.1212/wnl.44.8.1491. [DOI] [PubMed] [Google Scholar]
- 4.Wang Y, Welty DF. The simultaneous estimation of the influx and efflux blood-brain barrier permeabilities of gabapentin using a microdialysis-pharmacokinetic approach. Pharm Res. 1996;13(3):398–403. doi: 10.1023/a:1016092525901. [DOI] [PubMed] [Google Scholar]
- 5.Killian DM, Hermeling S, Chikhale PJ. Targeting the cerebrovascular large neutral amino acid transporter (LAT1) isoform using a novel disulfide-based brain drug delivery system. Drug Deliv. 2007;14(1):25–31. doi: 10.1080/10717540600559510. [DOI] [PubMed] [Google Scholar]
- 6.Gynther M, et al. Brain uptake of ketoprofen-lysine prodrug in rats. Int J Pharm. 2010;399(1-2):121–128. doi: 10.1016/j.ijpharm.2010.08.019. [DOI] [PubMed] [Google Scholar]
- 7.Kaira K, et al. Prognostic significance of L-type amino acid transporter 1 expression in resectable stage I-III nonsmall cell lung cancer. Br J Cancer. 2008;98(4):742–748. doi: 10.1038/sj.bjc.6604235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. doi: 10.1227/01.neu.0000316018.51292.19. Kobayashi K, et al. (2008) Enhanced tumor growth elicited by L-type amino acid transporter 1 in human malignant glioma cells. Neurosurgery 62(2):493–503, discussion 503–504. [DOI] [PubMed] [Google Scholar]
- 9.Kaira K, et al. l-type amino acid transporter 1 and CD98 expression in primary and metastatic sites of human neoplasms. Cancer Sci. 2008;99(12):2380–2386. doi: 10.1111/j.1349-7006.2008.00969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nicklin P, et al. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2009;136(3):521–534. doi: 10.1016/j.cell.2008.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oda K, et al. L-type amino acid transporter 1 inhibitors inhibit tumor cell growth. Cancer Sci. 2010;101(1):173–179. doi: 10.1111/j.1349-7006.2009.01386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohkawa M, et al. Oncogenicity of L-type amino-acid transporter 1 (LAT1) revealed by targeted gene disruption in chicken DT40 cells: LAT1 is a promising molecular target for human cancer therapy. Biochem Biophys Res Commun. 2011;406(4):649–655. doi: 10.1016/j.bbrc.2011.02.135. [DOI] [PubMed] [Google Scholar]
- 13.Shennan DB, Thomson J. Inhibition of system L (LAT1/CD98hc) reduces the growth of cultured human breast cancer cells. Oncol Rep. 2008;20(4):885–889. [PubMed] [Google Scholar]
- 14.Verrey F, et al. CATs and HATs: the SLC7 family of amino acid transporters. Pflugers Arch. 2004;447(5):532–542. doi: 10.1007/s00424-003-1086-z. [DOI] [PubMed] [Google Scholar]
- 15.Gao X, et al. Structure and mechanism of an amino acid antiporter. Science. 2009;324(5934):1565–1568. doi: 10.1126/science.1173654. [DOI] [PubMed] [Google Scholar]
- 16.Shaffer PL, Goehring A, Shankaranarayanan A, Gouaux E. Structure and mechanism of a Na+-independent amino acid transporter. Science. 2009;325(5943):1010–1014. doi: 10.1126/science.1176088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao X, et al. Mechanism of substrate recognition and transport by an amino acid antiporter. Nature. 2010;463(7282):828–832. doi: 10.1038/nature08741. [DOI] [PubMed] [Google Scholar]
- 18.Kowalczyk L, et al. Molecular basis of substrate-induced permeation by an amino acid antiporter. Proc Natl Acad Sci USA. 2011;108(10):3935–3940. doi: 10.1073/pnas.1018081108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schlessinger A, et al. Comparison of human solute carriers. Protein Sci. 2010;19(3):412–428. doi: 10.1002/pro.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fang Y, et al. Structure of a prokaryotic virtual proton pump at 3.2 A resolution. Nature. 2009;460(7258):1040–1043. doi: 10.1038/nature08201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamashita A, Singh SK, Kawate T, Jin Y, Gouaux E. Crystal structure of a bacterial homologue of Na+/Cl—dependent neurotransmitter transporters. Nature. 2005;437(7056):215–223. doi: 10.1038/nature03978. [DOI] [PubMed] [Google Scholar]
- 22.Forrest LR, Krämer R, Ziegler C. The structural basis of secondary active transport mechanisms. Biochim Biophys Acta. 2011;1807(2):167–188. doi: 10.1016/j.bbabio.2010.10.014. [DOI] [PubMed] [Google Scholar]
- 23.Shi L, Weinstein H. Conformational rearrangements to the intracellular open states of the LeuT and ApcT transporters are modulated by common mechanisms. Biophys J. 2010;99(12):L103–L105. doi: 10.1016/j.bpj.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jardetzky O. Simple allosteric model for membrane pumps. Nature. 1966;211(5052):969–970. doi: 10.1038/211969a0. [DOI] [PubMed] [Google Scholar]
- 25.Guan L, Kaback HR. Lessons from lactose permease. Annu Rev Biophys Biomol Struct. 2006;35:67–91. doi: 10.1146/annurev.biophys.35.040405.102005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen MY, Sali A. Statistical potential for assessment and prediction of protein structures. Protein Sci. 2006;15(11):2507–2524. doi: 10.1110/ps.062416606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eramian D, Eswar N, Shen MY, Sali A. How well can the accuracy of comparative protein structure models be predicted? Protein Sci. 2008;17(11):1881–1893. doi: 10.1110/ps.036061.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schlessinger A, et al. Structure-based discovery of prescription drugs that interact with the norepinephrine transporter, NET. Proc Natl Acad Sci USA. 2011;108(38):15810–15815. doi: 10.1073/pnas.1106030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang N, Shoichet BK, Irwin JJ. Benchmarking sets for molecular docking. J Med Chem. 2006;49(23):6789–6801. doi: 10.1021/jm0608356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fan H, et al. Molecular docking screens using comparative models of proteins. J Chem Inf Model. 2009;49(11):2512–2527. doi: 10.1021/ci9003706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mensch J, Oyarzabal J, Mackie C, Augustijns P. In vivo, in vitro and in silico methods for small molecule transfer across the BBB. J Pharm Sci. 2009;98(12):4429–4468. doi: 10.1002/jps.21745. [DOI] [PubMed] [Google Scholar]
- 32.Smith BJ, et al. P-glycoprotein efflux at the blood-brain barrier mediates differences in brain disposition and pharmacodynamics between two structurally related neurokinin-1 receptor antagonists. J Pharmacol Exp Ther. 2001;298(3):1252–1259. [PubMed] [Google Scholar]
- 33.Park S, Sinko PJ. The blood-brain barrier sodium-dependent multivitamin transporter: a molecular functional in vitro-in situ correlation. Drug Metab Dispos. 2005;33(10):1547–1554. doi: 10.1124/dmd.105.005231. [DOI] [PubMed] [Google Scholar]
- 34.Abbott NJ, Patabendige AA, Dolman DE, Yusof SR, Begley DJ. Structure and function of the blood-brain barrier. Neurobiol Dis. 2010;37(1):13–25. doi: 10.1016/j.nbd.2009.07.030. [DOI] [PubMed] [Google Scholar]
- 35.Yamamoto A, Akanuma S, Tachikawa M, Hosoya K. Involvement of LAT1 and LAT2 in the high- and low-affinity transport of L-leucine in human retinal pigment epithelial cells (ARPE-19 cells) J Pharm Sci. 2010;99(5):2475–2482. doi: 10.1002/jps.21991. [DOI] [PubMed] [Google Scholar]
- 36.Zhang L, Schaner ME, Giacomini KM. Functional characterization of an organic cation transporter (hOCT1) in a transiently transfected human cell line (HeLa) J Pharmacol Exp Ther. 1998;286(1):354–361. [PubMed] [Google Scholar]
- 37.Meier C, Ristic Z, Klauser S, Verrey F. Activation of system L heterodimeric amino acid exchangers by intracellular substrates. EMBO J. 2002;21(4):580–589. doi: 10.1093/emboj/21.4.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hidalgo M, et al. A Phase I and pharmacological study of the glutamine antagonist acivicin with the amino acid solution aminosyn in patients with advanced solid malignancies. Clin Cancer Res. 1998;4(11):2763–2770. [PubMed] [Google Scholar]
- 39.Delaville C, Navailles S, Benazzouz A. Effects of noradrenaline and serotonin depletions on the neuronal activity of globus pallidus and substantia nigra pars reticulata in experimental parkinsonism. Neuroscience. 2012;202:424–433. doi: 10.1016/j.neuroscience.2011.11.024. [DOI] [PubMed] [Google Scholar]
- 40.Kroemer G, Pouyssegur J. Tumor cell metabolism: cancer’s Achilles’ heel. Cancer Cell. 2008;13(6):472–482. doi: 10.1016/j.ccr.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 41.Lahoutte T, et al. Comparative biodistribution of iodinated amino acids in rats: selection of the optimal analog for oncologic imaging outside the brain. J Nucl Med. 2003;44(9):1489–1494. [PubMed] [Google Scholar]
- 42.O’Dwyer PJ, Alonso MT, Leyland-Jones B. Acivicin: a new glutamine antagonist in clinical trials. J Clin Oncol. 1984;2(9):1064–1071. doi: 10.1200/JCO.1984.2.9.1064. [DOI] [PubMed] [Google Scholar]
- 43.Bonomi P, Finkelstein D, Chang A. Phase II trial of acivicin versus etoposide-cisplatin in non-small cell lung cancer. An Eastern Cooperative Oncology Group study. Am J Clin Oncol. 1994;17(3):215–217. doi: 10.1097/00000421-199406000-00006. [DOI] [PubMed] [Google Scholar]
- 44.Schlessinger A, et al. High selectivity of the γ-aminobutyric acid transporter 2 (GAT-2, SLC6A13) revealed by structure-based approach. J Biol Chem. 2012;287(45):37745–37756. doi: 10.1074/jbc.M112.388157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pei J, Kim BH, Grishin NV. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 2008;36(7):2295–2300. doi: 10.1093/nar/gkn072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234(3):779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 47.Krivov GG, Shapovalov MV, Dunbrack RL., Jr Improved prediction of protein side-chain conformations with SCWRL4. Proteins. 2009;77(4):778–795. doi: 10.1002/prot.22488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carlsson J, et al. Ligand discovery from a dopamine D3 receptor homology model and crystal structure. Nat Chem Biol. 2011;7(11):769–778. doi: 10.1038/nchembio.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 50.Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38(Web Server issue):W529–W533. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mysinger MM, Shoichet BK. Rapid context-dependent ligand desolvation in molecular docking. J Chem Inf Model. 2010;50(9):1561–1573. doi: 10.1021/ci100214a. [DOI] [PubMed] [Google Scholar]
- 52.Overington J. ChEMBL. An interview with John Overington, team leader, chemogenomics at the European Bioinformatics Institute Outstation of the European Molecular Biology Laboratory (EMBL-EBI). Interview by Wendy A. Warr. J Comput Aided Mol Des. 2009;23(4):195–198. doi: 10.1007/s10822-009-9260-9. [DOI] [PubMed] [Google Scholar]
- 53.Apweiler R, et al. The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2010;38(Database issue):D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang X, McManus M. Lentivirus production. J Vis Exp. 2009;(32):e1499. doi: 10.3791/1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen Y, Zhang S, Sorani M, Giacomini KM. Transport of paraquat by human organic cation transporters and multidrug and toxic compound extrusion family. J Pharmacol Exp Ther. 2007;322(2):695–700. doi: 10.1124/jpet.107.123554. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.