Significance
Ubiquitin is a 76 residue protein that is attached to target proteins as a posttranslational modification. This modification is dependent on the successive activity of three enzymes, designated E1, E2, and E3. We developed a high-throughput mutagenesis strategy to probe the mechanism of E3-catalyzed transfer of ubiquitin from the E2 to the target protein. By scoring the effect of nearly 100,000 mutations in an E3, we identified mutations that affect direct and allosteric interactions between the E3 and the E2. These results highlight the general utility of high-throughput mutagenesis in delineating the molecular basis of enzyme activity.
Keywords: NMR, phage display, ubiquitin E3 ligase, protein stability, deep mutational scanning
Abstract
Although ubiquitination plays a critical role in virtually all cellular processes, mechanistic details of ubiquitin (Ub) transfer are still being defined. To identify the molecular determinants within E3 ligases that modulate activity, we scored each member of a library of nearly 100,000 protein variants of the murine ubiquitination factor E4B (Ube4b) U-box domain for auto-ubiquitination activity in the presence of the E2 UbcH5c. This assay identified mutations that enhance activity both in vitro and in cellular p53 degradation assays. The activity-enhancing mutations fall into two distinct mechanistic classes: One increases the U-box:E2-binding affinity, and the other allosterically stimulates the formation of catalytically active conformations of the E2∼Ub conjugate. The same mutations enhance E3 activity in the presence of another E2, Ube2w, implying a common allosteric mechanism, and therefore the general applicability of our observations to other E3s. A comparison of the E3 activity with the two different E2s identified an additional variant that exhibits E3:E2 specificity. Our results highlight the general utility of high-throughput mutagenesis in delineating the molecular basis of enzyme activity.
Covalent modification of proteins by ubiquitin (Ub) has an impact on nearly all eukaryotic cell biology. The attachment of Ub is accomplished by three enzymes: an E1 Ub-activating enzyme, an E2 Ub-conjugating enzyme, and an E3 Ub ligase (1). In the final stage of the pathway, the E3 ligase facilitates transfer from a Ub-loaded E2 (termed an E2∼Ub conjugate) onto a substrate lysine. Minimally, functional E3s contain an E2-binding domain and a substrate-recognition domain, enabling them to bind an E2∼Ub and a substrate simultaneously. The majority of E3s harbor either a RING (really interesting new gene) domain or a related U-box domain to bind cognate E2s. RING-type E3s enhance transfer of Ub directly from an E2’s active site to a substrate lysine without an intermediate transfer step of Ub to the E3 itself, as occurs with homology to E6AP carboxyl-terminus (HECT)-type ligases. In addition to providing proximity between the Ub attached to the active site of an E2 and a substrate amino group, RING-type E3s activate E2∼Ub conjugates allosterically (2–11). Allosteric activation relies on promotion of catalytically active “closed” conformations of an E2∼Ub conjugate that presumably arrange the E2 active site thioester for access and attack by an incoming nucleophile. Thus, two processes contribute to the rate enhancement of Ub transfer by RING-type E3s: (i) proximity (and, in some cases, orientational) effects and (ii) promotion of reactive states of the E2∼Ub. Recent studies (8–10) have shown that minimal RING-type domains that lack a substrate-binding activity (and therefore cannot provide a proximity enhancement) are able to enhance the intrinsic reactivity of E2∼Ub species, demonstrating that the two sources of rate enhancement are separable and independent. Although substrate proximity effects are likely to be idiosyncratic for each E3/substrate pair, the mechanisms that underlie allosteric activation appear to be shared among E3s. Furthermore, allosteric activation of an E2∼Ub is necessary and sufficient for RING-type E3 ligase activity. Therefore, a more thorough understanding of the allosteric mechanism and its structural bases should prove insightful for a majority of Ub E3 ligases.
Recent structural studies of RING and U-box domains with E2∼Ub moieties reveal common features: (i) a closed E2∼Ub conformation and (ii) an intermolecular hydrogen bond between a conserved E3 side chain and an E2 backbone carbonyl (2, 3, 11). Mutational analyses demonstrate that both are critical for E3 allosteric enhancement of Ub transfer (3). Despite these conserved features, E3 ligases display large differences in Ub transfer activity even when working with the same E2, suggesting that other, possibly more subtle, effects are in play. Traditional approaches for dissecting enzymatic processes involve detailed structural studies and targeted mutational analyses, but these approaches are limited by the number of mutants that can be analyzed. Typically, the mutations generated disrupt activity and are often restricted to protein/protein interfaces, thereby ignoring much of the possible sequence space. Such limitations prompted us to use a high-throughput method, known as “deep mutational scanning” (12, 13), that can assess the effects of over 105 sequence variants of a single protein simultaneously. We adapted this method to generate a sequence–function map of the U-box of the murine E3 ligase ubiquitination factor E4B (Ube4b). The U-box domain of Ube4b is an ideal candidate for this approach because it functions as a monomer; its structure has been solved both in isolation and in complex with the E2 UbcH5c; and with an extended N-terminal region, it exhibits auto-ubiquitination activity in vitro (14, 15). In vivo, the human homolog UBE4B polyubiquitinates the tumor suppressor p53 to target it for degradation by the proteasome. Increased expression of UBE4B and amplification of the genomic locus of UBE4B have been found in patient-derived medulloblastoma tumors that also have lower levels of p53 (16), suggesting that increased UBE4B activity might be oncogenic by reducing p53 abundance.
We used the E2 UbcH5c in reactions to assess the Ub ligase activity of nearly 100,000 unique protein variants of the Ube4b U-box domain, over 900 of which contained single amino acid substitutions. Although most single mutations decreased enzymatic activity, a few mutations greatly increased activity both in in vitro reactions and in promoting the degradation of p53 in a human cell line. We used NMR to characterize four activity-enhancing mutations and found two classes: One class enhances E2∼Ub catalysis by increasing E3:E2-binding affinity, and the other augments the allosteric capacity of the U-box to promote catalytically active E2∼Ub conformational states. These two classes of mutations are distinct, and mutations from both classes can be combined to enhance activity even further. Selecting for functional U-box variants in the presence of a different E2, Ube2w, resulted in a similar set of Ube4b-activating mutations, consistent with the hypothesis that mechanisms by which E3s allosterically activate intrinsic E2∼Ub reactivity are shared and that enhancing mutations will be generalizable. Additionally, we identified a Ube4b variant that has reduced activity with UbcH5c but retains activity with Ube2w, suggesting that it may be possible to use deep mutational strategies to identify and manipulate sources of E3:E2 specificity. Our deep mutational scans and structural analyses further define the molecular basis for E3-induced E2∼Ub allosteric activation and provide tools for future structural and functional studies of E3 ligases.
Results
Sequence–Function Map of the Ube4b U-Box Domain.
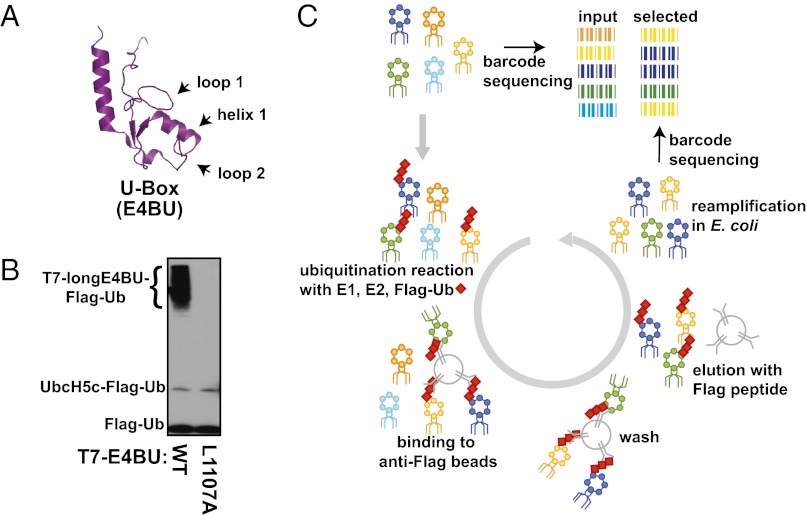
Deep mutational scanning uses a protein display format in which a large library of protein variants is placed under selection for the activity of interest, with their identity and frequency in the population before and after selection determined by high-throughput DNA sequencing. A critical requirement for such a selection is the linkage between genotype (i.e., the encoded variant) and phenotype (i.e., the activity of interest). In the case of some E3 ligases, auto-ubiquitination, in which the E3 catalyzes ubiquitination of its own lysine residues distant from the E2-binding domain, can be used to generate this genotype-phenotype linkage. Furthermore, use of auto-ubiquitination, as opposed to substrate ubiquitination, avoids the identification of substrate-specific mutations that simply enhance the recruitment of a specific substrate and, instead, focuses the selection on E3 mutations that enhance Ub transfer per se. To perform a deep mutational scan of an E3 ligase domain, we fused the extended U-box domain (the carboxyl-terminal 102 amino acids) of Ube4b, termed longE4BU, to the T7 bacteriophage coat protein for display. This region of Ube4b was chosen because it is sufficient in the presence of an E1, E2, and Ub for auto-ubiquitination activity; in addition, the U-box domain (the carboxyl-terminal 82 amino acids) of Ube4b (termed E4BU) folds independently and its structure has been solved (Fig. 1A). In vitro auto-ubiquitination assays confirmed that the phage-displayed U-box domain is an active Ub ligase (Fig. 1B, lane 1 and Fig. S1 A and B) and that mutation of a residue (L1107A) critical for E3 function (17) abrogates activity with UbcH5c (Fig. 1B, lane 2). As proof of principle, we found that WT T7-longE4BU phages were preferentially selected over their L1107A counterparts when immunoprecipitated with an antibody against tagged Ub and that this selection was E2-dependent (Fig. S1C). This control experiment demonstrates two essential requirements: (i) auto-ubiquitination in the context of the phage display system requires a functional E3:E2 interaction for catalysis; and (ii) auto-ubiquitination in this context occurs in cis, establishing a direct link between the E3 variant genotype and auto-ubiquitination phenotype.
Fig. 1.
Highly parallel method for examining Ub ligase activity. (A) Cartoon representation of the 82-aa E4BU solution structure (Protein Data Bank ID code 2kr4). Residues within loop 1, loop 2, and helix 1 form the E3:E2 interface. The longE4BU domain contains an extra 20 amino-terminal segment required for auto-ubiquitination activity. (B) Western blot for Flag-Ub shows Ub ligase activity of longE4BU expressed on the phage surface. LongE4BU-WT (lane 1) or longE4BU-L1107A (lane 2) was fused to the coat protein of T7 bacteriophage. Amplified phage lysate was incubated with recombinant E1, UbcH5c, ATP/Mg, and Flag-Ub. The reaction was analyzed by Western blot with anti-Flag to follow Flag-Ub. (C) Library of longE4BU sequence variants was generated from doped oligonucleotides, with a degenerate 18-base barcode inserted 3′ of the stop codon. The variable region and barcode were cloned into the phage genome and displayed as a carboxyl-terminal fusion with the T7 coat protein. The library of T7-longE4BU sequence variants was subjected to multiple rounds of selection for functional Ub ligase activity by incubation with recombinant 6× His-E1, UbcH5c, ATP/Mg, and Flag-Ub. The ubiquitinated phages were selected on anti-Flag agarose, and unbound phages were washed away. Bound Flag-ubiquitinated T7-longE4BU variants were eluted by competition with 3× Flag peptide. Eluted phages were reamplified and subjected to additional rounds of selection. DNA was purified from the input and selected T7-longE4BU populations, Illumina libraries were constructed, and the barcodes were sequenced by 36-base single end reads.
We created and sequenced (18) a library of phages that encode longE4BU variants containing, on average, two nucleotide mutations per variant (Fig. S1 D and E and Table S1). Because longE4BU contains more amino acids than previously used in the deep mutational scanning approach, we advanced the method through the use of a barcode-directed subassembly approach designed to sequence larger contiguous segments of DNA by short-read Illumina technology (19) (SI Materials and Methods). Using 163,829 unique protein variants identified from high-throughput sequencing and subassembly, we performed successive rounds of selection for Ub ligase activity. Each round of selection consisted of in vitro phage auto-ubiquitination reactions with an E1, the E2 UbcH5c, and Flag-Ub, followed by enrichment of the phage on anti-Flag agarose beads and reamplification of the phage in Escherichia coli (Fig. 1C). We determined the frequency at which each variant was present in the starting population and in the population after three rounds of selection (Fig. S2 A and B and Table S2). The ratio of the selected frequency of each variant vs. its starting frequency, called here the “enrichment ratio” or E, provides a measure of the performance of each variant during the selection for E3 function. All E scores in this study are normalized to the performance of the WT U-box (E = 1.06).
We were able to calculate E scores for 98,289 unique protein variants after three rounds of selection (Dataset S1). These included 932 variants with one amino acid mutation, 54,507 with two, and 42,850 with three or more. Because the performance of the double- and triple-mutation variants is confounded by the contributions from each mutation, we first focused on variants that contain only one amino acid change. The log2-transformed E score for each single variant is represented in a sequence–function map (Fig. 2A). Mutations in loops 1 and 2 and in helix 1 were more likely to be deleterious for Ub transfer than mutations in other portions of the U-box domain, whereas residues near either the amino- or carboxy-terminus were more tolerant to mutation (Mann–Whitney U test, P = 2.2 × 10−16). Ube4b positions L1107 (buried in the E3:E2 interface) and R1143 (the hydrogen bond donor necessary for activation of the E2) were highly sensitive to mutation (Fig. 2A, yellow boxes), demonstrating that the selection was capable of identifying positions involved in the E3’s binding and allosteric mechanisms.
Fig. 2.
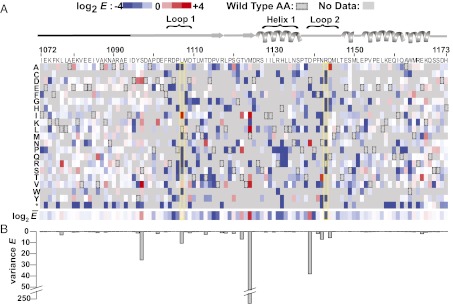
Effects of known inactivating mutations and >900 unique mutations on E3 function are uncovered by deep mutational scanning of longE4BU. (A) Sequence–function map of log2 E scores for 932 T7-longE4BU variants with a single amino acid change. Blue, white, and red boxes represent T7-longE4BU variants that were depleted, neutral, or enriched, respectively, during the selection process; gray represents no data that passed quality filters; and boxed rectangles represent the WT residue. A schematic of the E4BU secondary structure is shown above. The amino-terminal 20 amino acids that were not included in either deposited structure are represented by a black line. Loop 1, loop 2, and helix 1 are indicated. The longE4BU sequence is represented on the x axis, and the possible amino acid substitutions are represented on the y axis. Below are the position-averaged E scores for each position in T7-longE4BU. (B) Variance of E scores for each position represented in a bar graph is shown.
Positions with high variance in E result from both enriching and depleting mutations occurring at the same site (Fig. 2B). These are the same positions at which mutations with the highest E scores occurred. This correlation suggests that only specific amino acid changes are capable of activating this E3 ligase. The highest variance occurred at M1124, followed by S1097, L1107, T1122, D1139, and N1142. The functional importance of some of these residues was further illustrated by the ability of enriched mutations at these positions (M1124L, M1124I, M1124V, D1139N, and N1142T plus F1141Y) to rescue detrimental mutations when paired in the same double-mutation variant (Fig. S2C).
Highly Enriched Mutants Are More Active E3 Ub Ligases in Vitro and in Cells.
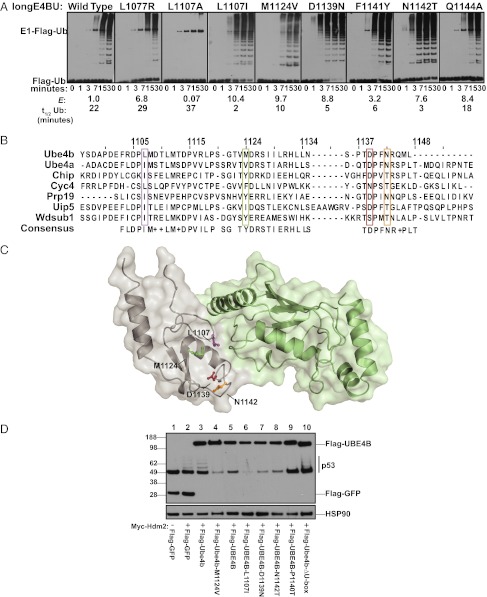
We expressed and purified highly enriched variant longE4BUs to examine their ability to catalyze auto-ubiquitination in vitro. Equal concentrations of WT and mutant U-boxes (Fig. S3A) were incubated with E1, UbcH5c, ATP/Mg, and Flag-tagged Ub over a time course from 1 to 30 min. For WT longE4BU, high-molecular-weight proteins bearing Flag-tagged Ub were visible after 7 min (Fig. 3A). The L1107A mutant, which was depleted in the phage assay, had no observable auto-ubiquitination activity (Fig. 3A). In contrast, almost all the variants that were enriched relative to WT in the phage assay also exhibited enhanced ubiquitination activity in vitro. The longE4BU variants L1107I, M1124V, D1139N, F1141Y, and N1142T produced high-molecular-weight ubiquitinated species within 3 min (Fig. 3A). The disappearance of Ub during the course of the assay was detected by Coomassie staining and quantified by densitometry to calculate approximate half-lives for each of the ubiquitination reactions (Fig. S3 B and C). The half-lives trended with E values determined from the phage assay, with the exception of the L1077R and Q1144A variants, which enriched in the phage assay but showed no increase in ligase activity (Fig. 3A). The L1077R and Q1144A mutations occur at positions with low variance compared with their activating counterparts (Fig. 2B), and the L1077R mutation could not rescue deleterious mutations (Fig. S2C). Thus, although the high E scores for the single mutants were generally predictive of high in vitro activity (correct for five of seven variants tested), the combined information gleaned from E values, variance, and rescue scores more accurately predicted activity. The strong correlation between the most highly enriched T7-longE4BU variants and the most active longE4BU ligases in vitro demonstrates that the phage competition provided a good selection for bona fide enhanced Ub transfer activity.
Fig. 3.
Enriched longE4BU variants are more active Ub ligases than the WT protein. (A) Ubiquitination assays. Recombinant WT longE4BU and the indicated variants were incubated with recombinant E1, UbcH5c, ATP/Mg, and Flag-Ub at 37 °C for the indicated time. Ubiquitination products were monitored by Western blot to follow Flag-Ub. E scores and the approximate t1/2 of unmodified Ub calculated from densitometry of Coomassie-stained reactions are indicated below (see also Fig. S3 B and C). (B) Multiple sequence alignment of the U-box domains for the seven murine U-box–containing proteins. Positions corresponding to E4BU L1107 (purple), M1124 (green), D1139 (red), and N1142 (orange) are indicated. (C) Sites of mutation in highly enriched T7-longE4BU variants mapped to the E4BU:UbcH5c crystal structure (Protein Data Bank ID code 3L1Z). L1107 (purple), M1124 (green), D1139 (red), and N1142 (orange) are depicted on gray E4BU. UbcH5c is pale green. (D) H1299 cells were transfected with pCMV-neo-p53, and cells in lanes 2–8 were transfected with pCMV-Myc-Hdm2. Flag-tagged, full-length mouse Ube4b or human UBE4B constructs were transfected as indicated. Blots were probed for p53 and the Flag epitope. Endogenous heat shock protein 90 (HSP90) was used as a protein loading control (see also Fig. S4).
The activity-enhancing variants identified in the selection represent conservative amino acid changes. Analysis of Ube4b sequences across species shows that substitutions equivalent to L1107I, M1124V, D1139N, and N1142T exist in metazoans (20). Furthermore, alignment of the seven murine U-box–containing proteins revealed that the amino acids resulting in increased Ube4b activity appear in other U-box contexts. Ubiquitination factor E4A (Ube4a) contains the equivalent of the L1107I and M1124V mutations; cyclophilin-like protein Cyp-60 (Cyc4) contains the equivalent of the D1139N mutation; and carboxy terminus of Hsp70-interacting protein (CHIP), Cyc4, ubiquitin-conjugating enzyme 7-interacting protein 5 (Uip5), and WD repeat, SAM and U-box domain-containing protein 1 (Wdsub1) contain the equivalent of the N1142T mutation (Fig. 3B). Thus, the positions of mutations that enhance the activity of Ube4b may represent sites at which E3 activity can be fine-tuned. The positions of the L1107I, M1124V, D1139N, and N1142T mutations are highlighted on the structure of E4BU bound to UbcH5c (Fig. 3C): L1107 is in loop 1 and directly contacts the E2; M1124 is buried behind loops 1 and 2 and helix 1; and D1139 and N1142 are located in loop 2, slightly away from the E2 interface.
We expected that Ube4b variants with enhanced Ub transfer in an auto-ubiquitination reaction would also exhibit enhanced Ub transfer to substrates. We tested this hypothesis against a relevant cellular target, p53. Mouse Ube4b and human UBE4B have both been shown to possess E4 activity for the tumor suppressor p53 (16). Members of the E4 group of E3 ligases polymerize Ub chains on existing mono-Ub groups; in this case, Ube4b extends Ub chains on p53 initiated by mouse double minute 2 (Mdm2) or human HDM2. The chain extension on p53 results in degradation of p53 by the proteasome, with implications for tumorigenesis.
We hypothesized that a hyperactive UBE4B would promote increased ubiquitination and, therefore, proteasome-dependent degradation of p53 in cell culture. Cotransfection of p53 and Myc-Hdm2 into the H1299 human cell line led to discrete laddering of p53 bands, likely representing multiple modifications of p53 with single or short Ub chains, because there was no loss of steady-state protein levels (Fig. 3D, lanes 1 and 2). As previously reported (16), transfection of the WT UBE4B expression plasmid led to a decrease in p53 protein levels (Fig. 3D, lane 5), in a U-box–, HDM2-, and proteasome-dependent fashion (Fig. 3D, lane 10 and Fig. S4 A and B). Transfection of a plasmid expressing UBE4B with any of the M1124V, L1107I, D1139N, or N1142T mutations led to more dramatic loss of steady-state p53 protein levels (Fig. 3D, lanes 4 and 6–8). As with the in vitro auto-ubiquitination assays, the relative levels of p53 degradation on transfection of each UBE4B mutant trended with E values from the phage selection.
A single human polymorphism, P1140T, has been identified in the U-box domain of UBE4B (dsSNP identifier rs15191). This mutation is apparently deleterious for E4BU function because it disappeared from the round three population of the deep mutational scan (Dataset S1). Transfection of a plasmid expressing UBE4B harboring a P1140T mutation resulted in no detectable change in p53 protein level, validating this variant as a loss-of-function allele (Fig. 3D, lane 9). Together, the results from these cell-based assays confirm that activity-enhancing and activity-lowering mutations identified in the high-throughput assay retain their phenotype in the context of the full-length protein on a relevant substrate in human cells. Additionally, the sequence–function map will provide data on the effect of other polymorphisms of UBE4B as future genome or tumor sequencing efforts identify them.
Activating Mutations Enhance the General Reactivity of an E2∼Ub Conjugate.
E3 auto-ubiquitination activity could potentially involve a contribution from substrate proximity because the substrate lysine is part of the E3 in this assay. To assess whether activating mutations somehow alter the substrate recruitment role of the E3, we tested the activating effects of the M1124V, L1107I, D1139N, and N1142T mutations in a substrate-independent assay. Monitoring Ub transfer from the E2∼Ub conjugate to the free amino acid lysine removes E3 contributions due to substrate recognition and/or presentation, leaving only the raw catalytical rate enhancement. In these assays, the shorter E4BU construct that lacks auto-ubiquitination activity was incubated with preformed UbcH5c∼Ub and the free amino acid lysine. In the absence of an E3, the UbcH5c∼Ub conjugate reacted slowly with lysine, with conjugate still observed after 15 min. Addition of WT E4BU significantly enhanced reactivity, with most of the conjugate gone by 5 min (Fig. 4). The M1124V, L1107I, D1139N, and N1142T E4BU mutants all exhibited increased activation of the UbcH5c∼Ub conjugate toward lysine compared with WT, with most of the E2∼Ub consumed by 3 min (Fig. 4). The apparent rate enhancement in reactivity toward lysine agrees with both in vitro auto-ubiquitination and cellular p53 degradation assays, confirming our hypothesis that the activating mutants identified in the phage selection enhance Ub transfer in a general, substrate-independent capacity, presumably through altered E3:E2 interactions (see below).
Fig. 4.
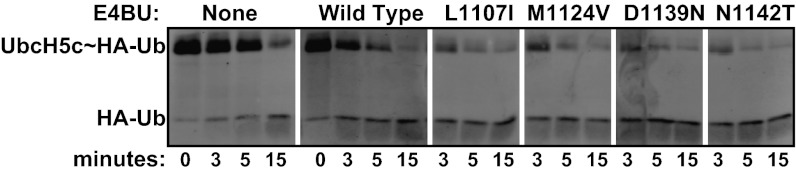
Activating mutations enhance the general reactivity of an E2∼Ub conjugate lysine reactivity assay. Recombinant minimal U-box domains of WT E4BU (1,092–1,173) and the indicated variants were incubated with purified UbcH5c∼HA-Ub and free lysine at 37 °C for the times indicated. Breakdown of the UbcH5c∼HA-Ub thioester linkage was monitored by Western blot with anti-HA antibodies.
Some E3 Activating Mutations Combine to Act Synergistically.
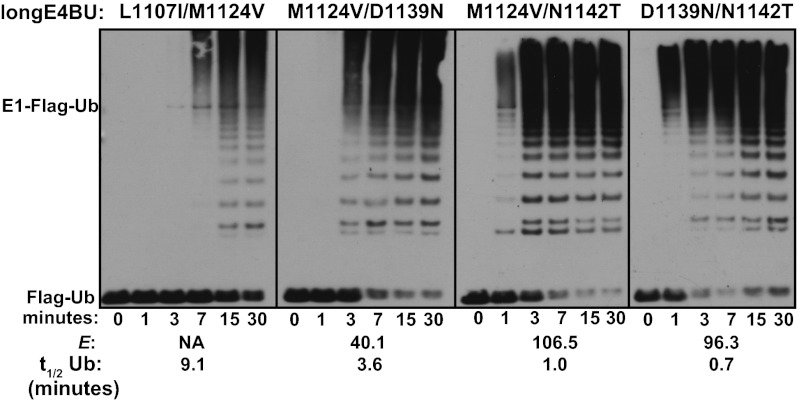
Based on their large positive E scores, we further characterized the activity of the double-mutation–containing variants M1124V/D1139N, M1124V/N1142T, and D1139N/N1142T (E = 40, E = 106, and E = 96, respectively). In vitro auto-ubiquitination assays revealed that the M1124V mutation combined with either the D1139N or N1142T mutation resulted in more rapid Ub transfer activity than any of these single mutations on their own. The combination of the D1139N and N1142T mutations also enhanced ligase activity, with high-molecular-weight poly-Ub products visible after only 1 min (Fig. 5). Quantification of the free Ub band in Coomassie-stained gels as a function of reaction time showed that Ub was consumed with a t1/2 of ∼1 min by both the M1124V/N1142T and D1139N/N1142T double mutants, which is considerably faster than the nearly 22-min Ub t1/2 catalyzed by the WT (Fig. S3 B and C). The rate enhancement observed for the M1124V/N1142T and D1139N/N1142T mutants [i.e., kobs(E4BUmutant)/kobs(E4BUwild type)] corresponds to a ∆∆G‡ of ∼2 kcal/mol. For comparison, the rate enhancement for WT E4BU relative to an otherwise uncatalyzed UbcH5c∼Ub hydrolysis reaction (3) corresponds to a ∆∆G‡ of ∼1 kcal/mol. The rate enhancements observed for double mutants incorporating M1124V, D1139N, or N1142T correspond to more than the sum of the individual mutations’ rate enhancements (∆∆G‡ values ranging from 0.5 to 1 kcal/mol), indicating that these mutations work in synergy. However, not all combinations of enhancing mutations further increase activity. The L1107I/M1124V double mutant exhibited activity below the level observed for the L1107I mutant alone (Figs. 3A and 5 and Fig. S3C). Therefore, some but not all combinations of enhancing mutations are synergistic.
Fig. 5.
Effects of some activating mutations are synergistic and create hyperactive U-box domains. Ubiquitination assays. The indicated purified variants of longE4BU were incubated with recombinant E1, UbcH5c, ATP/Mg, and Flag-Ub at 37 °C for the indicated times. Ubiquitination products were monitored by Western blot analysis to follow Flag-Ub. E scores and the approximate t1/2 of unmodified Ub calculated from densitometry of Coomassie-stained reactions are indicated below (see also Fig. S3).
Mechanistic Classification of Mutations That Enhance E3 Ligase Activity.
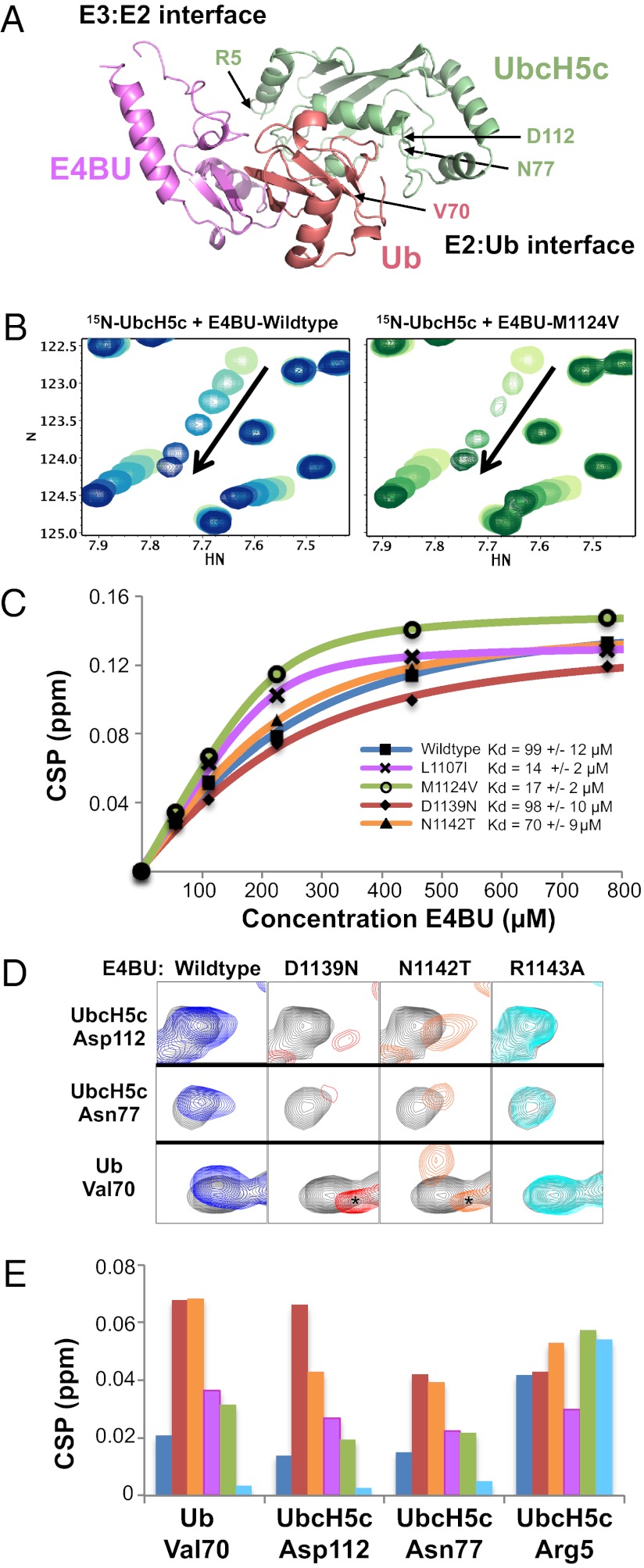
To contribute to the growing understanding of E3 ligase function (2, 3, 11), we sought to determine the structural bases for the enhancements in ligase activity due to activating mutations. CD spectra of purified WT E4BU and the four mutants showed a similar secondary structure (Fig. S5A). Furthermore, the activating mutants showed no trend in stability toward thermal denaturation, with various mutant proteins exhibiting decreased, similar, or increased thermal stability relative to the WT (Fig. S5B). L1107, D1139, and N1142 are positioned in and around the E2-binding site, raising the likely possibility that mutations at these positions would alter this interaction. An NMR-heteronuclear single quantum coherence (HSQC) spectrum of E4BU harboring the most buried mutation, M1124V, showed a number of changes centered around the site of the mutation that maps to E4BU loops 1 and 2, suggesting a potentially altered E2-binding interface (Fig. S5C). The capacity of each E4BU variant to bind UbcH5c was assessed using 1H, 15N HSQC transverse relaxation-optimized spectroscopy (TROSY) experiments in which increasing quantities of E4BU were titrated into 15N-UbcH5c. The spectra confirm that each E4BU variant binds to the same E2 surface composed of residues in helix 1, loop 4, and loop 7 of UbcH5c (Fig. 6A). However, the magnitude and character of the chemical shift perturbations (CSPs) differed among E4BU mutants. Although the D1139N and N1142T mutants exhibited fast exchange behavior similar to WT E4BU, both the L1107I and M1124V mutants had shifted toward the intermediate exchange regime, indicating a longer lifetime, and hence higher affinity of the E4BU:UbcH5c complex (Fig. 6B).
Fig. 6.
NMR analysis reveals that E4BU activating mutations fall into two classes. (A) One member from the E4BU:UbcH5c∼Ub model (3) is shown for reference with the E3:E2 and E2:Ub interfaces annotated. (B) Representative portion of the UbcH5c HSQC spectrum shows CSPs on addition of 0.25, 0.5, 1, 2, and 3.4 mol eq of E4BU WT (Left, blue) or M1124V (Right, green) to 225 μM 15N-UbcH5c. The M1124V mutant exhibits binding phenomena that approach intermediate exchange, as illustrated by line-width broadening at early titration points. (C) CSP values fit with quadratic binding equations using NMRView. Kd values were determined for WT (blue, 99 ± 12 μM), L1107I (magenta, 14 ± 2 μM), M1124V (green, 17 ± 2 μM), D1139N (red, 98 ± 10 μM), and N1142T (orange, 70 ± 9 μM) E4BU titrated into 15N-UbcH5c. (D) Resonances corresponding to D112 in UbcH5c helix 2, N77 near the UbcH5c active site, and V70 within the Ub hydrophobic surface act as indicators for closed, active UbcH5c∼Ub conformations promoted by E4BU binding. Compared with the addition of 0.25 molar equivalences of WT E4BU (blue) to 225 μM 15N-UbcH5c-O∼15N-Ub, both the D1139N (red) and N1142T (orange) mutants induce further population of closed UbcH5c∼Ub conformations. We could not collect NMR data at higher additions of E4BU due to the catalyzed hydrolysis of the oxyester effected by the E3. Addition of the catalytically inactive E4BU mutant R1143A (cyan) is shown for reference. Signal arising from small quantities of free, hydrolyzed Ub is marked by asterisks. (E) CSPs shown in D are quantified and shown in histogram format for the addition of 0.25 mol eq of WT (blue), D1139N (red), N1142T (orange), L1107I (magenta), M1124V (green), and R1143A (cyan) E4BU to 225 μM 15N-UbcH5c-O∼15N-Ub. Perturbations that reflect E3:E2 binding directly are largely unaffected, as shown by UbcH5c residue R5.
To quantify the differences, we fit CSPs of a minimum of 10 resonances from each titration to quadratic binding equations to calculate Kd values (Fig. S6A). WT E4BU and the D1139N and N1142T mutants bound UbcH5c with similar affinity (Kd of ∼99, ∼98, and ∼70 μM, respectively), whereas the L1107I and M1124V mutants bound with higher affinity (Kd of ∼14 and ∼17 μM, respectively) (Fig. 6C). Therefore, we classify the L1107I and M1124V variants as enhanced E2-binding mutants. Combining the L1107I and M1124V mutations had no additional effect on binding (Kd of ∼21 μM) compared with the individual mutations. Similarly, the double mutant D1139N/N1142T had an E2-binding affinity (Kd of ∼100 μM) that was unchanged from either single mutant (Fig. S6A). Combining the E2-binding mutation M1124V with a mutation that did not affect binding (D1139N) yielded a double mutant that bound UbcH5c with an intermediate affinity (Kd of ∼77 μM) relative to the two single mutants, suggesting that mutation of the buried Met side chain may have a complicated effect on loops 1 and 2 of the E2-binding interface, as might be expected from the widespread changes observed in the HSQC NMR spectrum of such a mutant (Fig. S5C).
Because enhanced binding affinity for the E2 per se does not provide an explanation for the activity enhancement of some mutants, we asked whether the variants have altered interactions with the E2∼Ub conjugate. NMR experiments with the UbcH5c∼Ub conjugate can provide a direct observation of interactions involved in Ub transfer but are complicated by the reactivity of the UbcH5c∼Ub thioester bond. We used the active site cysteine-to-serine mutation to create the more stable oxyester-linked UbcH5c-O∼Ub conjugate (21). A subset of resonances in the UbcH5c-O∼Ub spectrum serve as indicators for assembly of an active E4BU:UbcH5c∼Ub ternary complex. These indicators correspond to residues within the closed UbcH5c-O∼Ub interface (Ub V70 and UbcH5c D112) and a residue within the E2 active site (UbcH5c N77), all of which undergo CSPs on addition of WT E4BU (3) (Fig. 6A). Addition of 0.25 mol equivalents of each E4BU variant (L1107I, M1124V, D1139N, and N1142T) to 15N-UbcH5c-O∼Ub increased the magnitude of CSPs in these resonances beyond those observed for addition of WT E4BU (Fig. 6 D and E). In contrast, addition of the R1143A mutant, in which the hydrogen bond required for allosteric activation of UbcH5c∼Ub is lost, failed to elicit CSPs in the indicators of an active E3:E2∼Ub conformation (Fig. 6 D and E).
Each of the four mutants promoted increased formation of closed, active E2∼Ub conformations, with the D1139N and N1142T mutants exhibiting the strongest induction of these states (Fig. 6E). Larger closed-state-specific CSPs observed on addition of substoichiometric amounts of the D1139N and N1142T mutants could arise for two reasons: (i) higher binding affinity for the E2∼Ub conjugate or (ii) more efficient induction of closed states on binding. To distinguish between these two possibilities, we performed NMR-binding assays with the most highly active variant, the D1139N/N1142T double mutant (Fig. 5). To test if the mutant U-box has an affinity for free Ub, the E4BU double mutant was added in large excess over free 15N-Ub. No interaction was detected (Fig. S6B, Left). In an NMR experiment in which the ability of the mutant to compete with WT for binding to UbcH5c-O∼Ub was assessed, the double mutant outperformed the WT U-box (Fig. S6B, Right). The enhancement is modest, with binding simulations estimating an approximately threefold enhancement in binding affinity over WT. This difference corresponds to a ∆∆G of less than 1 kcal/mol and does not account for the ∼25-fold increase in activity observed for this mutant (which corresponds to a ∆∆G for activation of ∼2 kcal/mol). Thus, it appears that the D1139N and N1142T mutations activate Ub transfer by binding the E2∼Ub conjugate more tightly and by more effectively inducing the active, closed conformation of the ternary complex on binding. Altogether, results from the NMR-binding studies delineate two classes of activating mutation: mutations that predominantly increase the binding affinity to the E2 itself and mutations that primarily increase binding specifically to the E2∼Ub conjugate and augment the ability of the U-box to activate the UbcH5c∼Ub conjugate allosterically.
Ube4b Mutations That Enhance Activity with UbcH5c Also Enhance Activity with the E2 Ube2w.
E2 enzymes play an important role in dictating the length and specific linkages of Ub chains and the processivity of the reaction (22). Our library of T7-longE4BU variants provided the opportunity to ask whether E3 mutations differentially affect activity depending on the paired E2. We tested the auto-ubiquitination activity of the U-box domain (longE4BU) with a panel of human E2s; we found polyubiquitination activity with UbcH5a, UbcH5b, and UbcH5c and monoubiquitination activity with Ube2w (Fig. S7A). Because Ube2w is more divergent from UbcH5c, both in sequence and activity, than the other E2s tested, we sought to identify mutations in the U-box that affect its activity in the presence of Ube2w. We therefore performed phage-based selection on longE4BU with Ube2w as the E2 and obtained a sequence–function map (Fig. S7B). The range of E scores is not as wide with Ube2w (log2 E score of −5 to 2) as with UbcH5c (log2 E score of −9.5 to 5.5). A likely reason for the limited range of E scores is that the monoubiquitination activity of Ube2w limits the sensitivity of the antibody-based selection of ubiquitinated phages.
Most longE4BU variants performed similarly with Ube2w and with UbcH5c (Spearman’s ρ = 0.54 across all mutations common between the two datasets; Fig. 7A). The four UbcH5c-activating mutations L1107I, M1124V, D1139N, and N1142T (labeled in Fig. 7A) also enriched over WT in a selection using Ube2w, although only modestly for M1124V. We performed in vitro ubiquitination reactions to compare the activities of WT and mutant longE4BU proteins directly when paired with either UbcH5c or Ube2w (Fig. 7B). Persistence of the Coomassie-stained longE4BU bands in the Ube2w assay shows that the M1124V variant is not as active as the other mutants in enhancing auto-monoubiquitination, in accord with the phage experiment. The level of activation afforded by the M1124V mutation thus shows a modest dependence on the partnered E2. Because M1124V is a mutation that enhances E3:E2 binding, this finding may point to subtle differences in the E3:E2 interface containing UbcH5c compared with Ube2w.
Fig. 7.
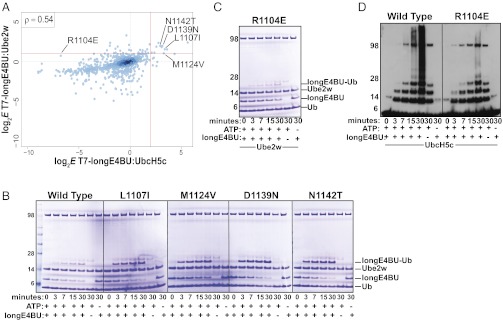
E4BU activating mutation behaves differently when paired with a different E2, Ube2w. (A) Log2-transformed E scores calculated from the deep mutational scan of T7-longE4BU performed with Ube2w as the E2 (y axis) compared with log2-transformed E scores from the deep mutational scan of T7-longE4BU performed with UbcH5c (x axis). Only E scores from variants with a single amino acid change are compared. The red lines signify a twofold increase in variant E scores over WT for Ube2w and a fourfold increase for UbcH5c. A Spearman’s rank correlation coefficient of 0.54 was calculated. (B and C) Ubiquitination assays. The indicated purified variants of longE4BU were incubated with recombinant E1, UbcH5c or Ube2w ATP/Mg, and Ub at 37 °C for the indicated time. Ubiquitination products were monitored by Coomassie staining. (D) Ubiquitination assays. The indicated purified variants of longE4BU were incubated with recombinant E1, UbcH5c or Ube2w ATP/Mg, and Ub at 37 °C for the indicated time. Ubiquitination products were monitored by Western blot analysis to follow Ub.
From the UbcH5c vs. Ube2w enrichment plot shown in Fig. 7A, we identified the R1104E variant as showing a minor enrichment in the Ube2w selection (E = 2.6) but a significant depletion in the UbcH5c selection (E = −44.6). In in vitro ubiquitination assays, the R1104E variant showed little decrease in ligase activity with Ube2w (Fig. 7C) but drastically decreased the amount of high-molecular-weight Ub adducts produced with UbcH5c (Fig. 7D). In sum, comparisons across the two high-throughput selections revealed that most mutations activate both of the diverse E2 enzymes, consistent with the notion of a common allosteric mechanism. The observation of a mutation that has a differential effect depending on the E2 present suggests that it may be possible to evolve more highly specific E3:E2 pairs.
Discussion
Our deep mutational scan of Ube4b has provided the most in-depth analysis to date of U-box sequence–function relationships. The activating mutations identified from this approach represent changes in RING/U-box E3s that broadly enhance ligase activity in a substrate-independent fashion. The selection strategy was predicated on the hypothesis that the ability of E3s to activate Ub transfer by E2∼Ubs can be separated into substrate-dependent and substrate-independent contributions, and that mutations that increase substrate-independent processes will also be enhanced for substrate ubiquitination activity. The hypothesis was borne out because mutations identified using Ube4b auto-ubiquitination as the readout also exhibited enhanced activity in cellular degradation of the substrate p53 and in biochemical assays monitoring transfer to the free amino acid lysine. Many of the most enriched variants contain a conservative mutation, suggesting that the weak interactions and catalytical functions of U-box ligases can be specifically tuned to suit their biological roles. Despite the capacity of a single point mutation to enhance ligase activity greatly, Ube4b has evolved in mammals to function with more moderate ubiquitination activity. The importance of human UBE4B ligase activity in the turnover of p53 (16) argues that precise levels of ubiquitination activity are critical for cellular function. Overactive ubiquitination by UBE4B would offer an opportunity for tumor cells to suppress the p53-dependent cell cycle arrest and/or apoptotic response. Because other U-boxes contain some of the amino acids in their native sequences that activated Ube4b, their activities may also be subject to fine-tuning.
Gain-of-function mutations that aid in the elucidation of enzymatic mechanisms are commonly sought but rarely discovered using conventional mutagenesis strategies. The rare nature of ligase-activating mutations emphasizes the power of the deep mutational scanning method: Only 25 single mutants (2.7% of the total) exhibited threefold or greater enrichment over WT after three rounds of selection. Many of the activity-enhancing mutations are chemically similar, evolutionarily conserved, and physically distant from known protein/protein interfaces, making it unlikely that they would have been predicted even with available high-resolution structural information. As mutations that significantly activate RING-type E3s, this set provides a unique tool to further the understanding of ligase function. Our NMR-based analyses illustrate that the E4BU-activating mutations fall into two classes: those that predominantly affect the binding affinity of E4BU for the E2 (L1107I and M1124V) and those that primarily affect the ability of E4BU to activate E2∼Ub conjugates (D1139N and N1142T) allosterically.
Because E4BU binds both the free and Ub-conjugated forms of the E2 with relatively similar affinities (3), it was unclear whether tighter binding would be beneficial or detrimental for Ub transfer. On the one hand, an extremely tight-binding E2 would transfer only the first Ub before acting as an inhibitor because it would be unable to be recharged by the E1 Ub-activating enzyme. On the other hand, a longer-lived E3:E2∼Ub complex (as is observed with the L1107I and M1124V variants) would allow more time for the formation of active E2∼Ub conformations and the approach of an incoming substrate lysine. We postulate that along the continuum of E3:E2-binding affinities, a point must exist at which Ub transfer reactivity is maximized before additional binding energy inhibits the reaction; at that point, the E3:E2 dissociation step becomes rate-limiting. WT Ube4b must be low enough on this binding energy vs. catalysis curve such that tighter binding can lead to enhanced Ub transfer; the hyperactive L1107I and M1124V variants showed as much as a sixfold enhancement in binding affinity.
Although the D1139N and N1142T variants showed no appreciable effect on E2 binding compared with WT, they demonstrated an enhanced ability to promote the active, closed conformations of the E2∼Ub conjugate allosterically. Neither the D1139N mutation nor the N1142T mutation could rescue an R1143 mutation in the phage assay (Dataset S1), consistent with the identification of R1143 as the “linchpin” for the allosteric activation of Ub transfer (3). D1139 and N1142 contribute significantly to the interactions that stabilize E4BU loop 2. We propose that the D1139N and N1142T mutations alter the structure and/or dynamics of loop 2, thereby enhancing the probability and/or longevity of the E4BU:UbcH5c intermolecular hydrogen bond and augmenting the allosteric role of the E3. Additionally, D1139 and N1142 are analogous to conserved zinc-binding cysteines in loop 2 of RING domains [e.g., C61 and C64 in breast cancer type 1 susceptibility protein (BRCA1)], suggesting that U-boxes may have evolved as a class of E3 ligase distinct from the RING cross-brace structure to enable additional modes of modulating their ligase activities.
The phage selection revealed thousands of sequence–function relationships for Ube4b:UbcH5c activity. Although this is likely the most relevant E3:E2 pair for the degradation of p53 (16), little is known regarding other roles Ube4b may play in the cell that potentially use different E2s. We showed that Ube4b is also active with Ube2w, an E2 linked to Fanconi anemia due to its ubiquitination of Fanconi anemia complementation group D2 (FANCD2) (23), and that hyperactivating mutations in Ube4b identified using this E2 are similar to those obtained with UbcH5c. However, the L1107I, D1139N, and N1142T mutations enhanced Ub transfer activity with Ube2w more than the M1124V mutation did. Based on the critical E3:E2 intermolecular hydrogen bond conserved in all structures solved to date, we predict that allosteric mutations will be more universal among E2s, whereas binding mutations may be more E2-specific. By comparing the enrichment values from the UbcH5c and Ube2w selections, we identified the R1104E mutation as highly detrimental for UbcH5c-dependent activity but with little effect on Ube2w-dependent activity. Therefore, future application of the deep mutational scanning technique might be used to design nonnative E3:E2 pairs or to examine biological roles of E3:E2 combinations through the engineering of variants specific for a given E2.
Beyond the practical applications for future studies of E3 ligases, the identification and characterization of Ube4b ligase-activating mutations have added molecular detail to the separable binding and allosteric roles of E3 ligases in facilitating Ub transfer. The insights into E3 ligase function provided by the deep mutational scans of Ube4b suggest that the mutational analysis of enzymes on a high-throughput basis has promise as a general means to identify key features of enzymatic mechanisms.
Materials and Methods
Full experimental and analytical details can be found in SI Materials and Methods.
Bacteriophage Library Preparation.
Oligonucleotides for cloning the E4BU library were synthesized with a 2% error rate by TriLink Biotechnologies, and all other oligonucleotides were from IDT. Stop codons were inserted in all three frames 3′ of the coding region, followed by a degenerate 18-base barcode. Molecular cloning of the library is outlined in Fig. S1D, with a detailed description and oligonucleotide sequences (Table S3) provided in SI Materials and Methods.
Ubiquitination Reactions and Ub Immunoprecipitations.
T7-longE4BU ubiquitination reactions contained 30 nM 6× His-E1, 100 nM UbcH5c, 1 mM Flag-Ub (Sigma), 2 mM ATP, 5 mM MgCl2, and 1/2 total volume bacteriophage lysate (∼5 × 108 plaque-forming units) in 50 μL of total reaction volume. Reactions were incubated for 45 min at 37 °C and stopped by addition of 2 mM DTT.
Enrichment of Flag-Ubiquitinated Bacteriophage and Reamplification.
M2-agarose beads (Sigma) were equilibrated into Flag dilution buffer [20 mM Tris (pH 7.5), 200 mM NaCl, 0.2 mM EDTA, 0.1% Nonidet P-40 (Sigma)] and added to ubiquitination reactions. After a 1-h incubation at room temperature, the beads were washed five times with Flag wash buffer [20 mM Tris (pH 7.5), 500 mM NaCl, 0.2 mM EDTA, 0.05% Nonidet P-40, 1 mM DTT] and eluted twice at 37 °C by addition of 2.5 mg/mL 3× Flag peptide (Sigma) in Flag wash buffer. Phages eluted from the M2-agarose beads were reamplified and titered according to the manufacturer’s instructions to avoid bottlenecks of the population. Individual clones were monitored by plaque PCR, followed by Sanger sequencing for population convergence.
Protein Purification.
E4BU and its variants were purified based on the method of Nordquist et al. (15); a more detailed description is available in SI Materials and Methods. The human 6× His-E1, wheat E1, UbcH5c, and Ube2w were purified as previously described (24, 25).
Ubiquitination Reactions with Purified LongE4BU.
One micromolar E1, 8 μM Flag-Ub, 4 μM E2, 2 mM ATP, 5 mM MgCl2, and 4 μM longE4BU were mixed in 10 mM Hepes, 60 mM NaCl, 5% (vol/vol) glycerol, and 0.5 mM EDTA (pH 7.9) buffer. The reactions were incubated at 37 °C for indicated times. Reactions were stopped with 2× LDS buffer (Invitrogen) containing 2.5% β-mercaptoethanol. Samples were separated on a 4–12% NuPAGE gel (Invitrogen) and either stained with Coomassie blue or transferred to a PVDF membrane. Membranes were probed with M2 (Sigma) or Ub antibody and anti–mouse-HRP (GE Healthcare).
Library Preparation for Tag-Directed Subassembly and Barcode Counting.
Phage DNA was purified by phenol chloroform extraction and ethanol precipitation. Illumina sequencing libraries were constructed by PCR amplification to sequence the 5′ end of longE4BU and the barcode. To sequence internal regions of longE4BU, nested libraries were prepared by PCR. A detailed description of the library construction can be found in Fig. S1D and SI Materials and Methods.
E Scores.
The Enrich software package (26) was used to determine the locations and identity of mutations in the assemblies, as well as the frequency at which that each variant appeared in the population. A nonspecific carryover correction factor was applied to the tally of each variant in the selected population (27). The corrected frequencies were used to calculate E scores and WT normalized E scores for variants found in both the input and selected populations.
Structure Visualization.
Structures were generated using PyMOL visualization software.
Multiple Sequence Alignment.
The U-box domains of the seven U-box domain-containing proteins found in the mouse genome were aligned using T-Coffee (28) and visualized with Jalview (29). The U-box consensus sequence was determined by aligning sequences of the 17 U-box domain Pfam protein family seed sequences (Pfam identifier PF04564) in Jalview using the default parameters.
NMR Spectroscopy.
NMR spectra were recorded on a 500-MHz Bruker Avance II at the University of Washington at 25 °C in 25 mM sodium phosphate (pH 7.0), 150 mM NaCl, and 10% D2O. The UbcH5c-O∼Ub conjugate was prepared as previously described and incorporated the C85S mutation to improve conjugate stability and the S22R mutation to prevent “self-assembly” through noncovalent Ub:UbcH5c interactions (30). Spectra were recorded on 225 μM 15N-labeled samples using 1H, 15N-HSQC-TROSY experiments, processed with NMRPipe/NMRDraw (31), and visualized with NMRView (32). CSPs were calculated using the formula Δδj = [(Δδj15N/5)2 + (Δδj1H)2]1/2. NMRView was used to fit the E4BU:UbcH5c-binding data with the quadratic equation f = A + (C − A) * (((p + x + 10B) − ((p + x + 10B)2–4 * p * x)0.5))/(2 * p), where p is the concentration of UbcH5c. A minimum of 10 representative titrating peaks were used to determine a Kd, and the error is reported as the average SD from the quadratic fits.
Cell Culture and Transfections.
H1299 cells were purchased from American Type Culture Collection and cultured according to the instructions. The cells were transfected at ∼90% confluency in 500 μL of antibiotic-free media with 100 ng of phosphorylated CMV (pCMV)-neo-p53, 100 ng of pCMV-Myc3-Hdm2, 900 ng of Flag-GFP or pcDNA3_Flag-Ube4b, and 2.2 μL of Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. The cells were split 6 h after addition of transfection reagents. Twenty-four hours after transfection, cells were washed with cold PBS and lysed with 200 μL of lysis buffer [50 mM Tris (pH 8.0), 150 mM NaCl, 1% Nonidet P-40, 1 mM EDTA, and protease inhibitors (Roche)] on ice. Lysates were clarified by centrifugation and quantified using Bradford reagent. Twenty micrograms of protein was electrophoresed on 4–12% NuPAGE gels and transferred to a PVDF membrane. The blots were probed with anti-Flag antibody (M2; Sigma), anti-p53 (DO-1; Santa Cruz Biotechnology) and anti-heat shock protein 90 (16F1, Abcam).
Supplementary Material
Acknowledgments
We thank W. Chazin and N. Zheng for critical reading of the manuscript. We also thank W. Chazin, R. Leng, and R. James for plasmids and R. Gardner for the Ub antibody. Additionally, we acknowledge C. Lee for Illumina sequencing and A. Borst for protein purification work. This work was supported by National Institute of General Medical Sciences Grants R01 GM088055 (to R.E.K.) and P41 GM103533 (to S.F.) and by Public Health Service National Research Service Award 2T32 GM007270 (to J.N.P.). S.F. is an investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1303309110/-/DCSupplemental.
References
- 1.Pickart CM, Eddins MJ. Ubiquitin: Structures, functions, mechanisms. Biochim Biophys Acta. 2004;1695(1-3):55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 2.Dou H, Buetow L, Sibbet GJ, Cameron K, Huang DT. BIRC7-E2 ubiquitin conjugate structure reveals the mechanism of ubiquitin transfer by a RING dimer. Nat Struct Mol Biol. 2012;19(9):876–883. doi: 10.1038/nsmb.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pruneda JN, et al. Structure of an E2:E3∼Ub complex reveals an allosteric mechanism shared among RING/U-box ligases. Mol Cell. 2012;47(6):933–942. doi: 10.1016/j.molcel.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ozkan E, Yu H, Deisenhofer J. Mechanistic insight into the allosteric activation of a ubiquitin-conjugating enzyme by RING-type ubiquitin ligases. Proc Natl Acad Sci USA. 2005;102(52):18890–18895. doi: 10.1073/pnas.0509418102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reverter D, Lima CD. Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature. 2005;435(7042):687–692. doi: 10.1038/nature03588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saha A, Lewis S, Kleiger G, Kuhlman B, Deshaies RJ. Essential role for ubiquitin-ubiquitin-conjugating enzyme interaction in ubiquitin discharge from Cdc34 to substrate. Mol Cell. 2011;42(1):75–83. doi: 10.1016/j.molcel.2011.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Song J, et al. Stability of thioester intermediates in ubiquitin-like modifications. Protein Sci. 2009;18(12):2492–2499. doi: 10.1002/pro.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wenzel DM, Lissounov A, Brzovic PS, Klevit RE. UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids. Nature. 2011;474(7349):105–108. doi: 10.1038/nature09966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yin Q, et al. E2 interaction and dimerization in the crystal structure of TRAF6. Nat Struct Mol Biol. 2009;16(6):658–666. doi: 10.1038/nsmb.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang M, et al. Chaperoned ubiquitylation—Crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP-Ubc13-Uev1a complex. Mol Cell. 2005;20(4):525–538. doi: 10.1016/j.molcel.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 11.Plechanovová A, Jaffray EG, Tatham MH, Naismith JH, Hay RT. Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature. 2012;489(7414):115–120. doi: 10.1038/nature11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Araya CL, Fowler DM. Deep mutational scanning: Assessing protein function on a massive scale. Trends Biotechnol. 2011;29(9):435–442. doi: 10.1016/j.tibtech.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fowler DM, et al. High-resolution mapping of protein sequence-function relationships. Nat Methods. 2010;7(9):741–746. doi: 10.1038/nmeth.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benirschke RC, et al. Molecular basis for the association of human E4B U box ubiquitin ligase with E2-conjugating enzymes UbcH5c and Ubc4. Structure. 2010;18(8):955–965. doi: 10.1016/j.str.2010.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nordquist KA, et al. Structural and functional characterization of the monomeric U-box domain from E4B. Biochemistry. 2010;49(2):347–355. doi: 10.1021/bi901620v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu H, et al. UBE4B promotes Hdm2-mediated degradation of the tumor suppressor p53. Nat Med. 2011;17(3):347–355. doi: 10.1038/nm.2283. [DOI] [PubMed] [Google Scholar]
- 17.Brzovic PS, et al. Binding and recognition in the assembly of an active BRCA1/BARD1 ubiquitin-ligase complex. Proc Natl Acad Sci USA. 2003;100(10):5646–5651. doi: 10.1073/pnas.0836054100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patwardhan RP, et al. Massively parallel functional dissection of mammalian enhancers in vivo. Nat Biotechnol. 2012;30(3):265–270. doi: 10.1038/nbt.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hiatt JB, Patwardhan RP, Turner EH, Lee C, Shendure J. Parallel, tag-directed assembly of locally derived short sequence reads. Nat Methods. 2010;7(2):119–122. doi: 10.1038/nmeth.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marín I. Ancient origin of animal U-box ubiquitin ligases. BMC Evol Biol. 2010;10:331. doi: 10.1186/1471-2148-10-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levin I, et al. Identification of an unconventional E3 binding surface on the UbcH5∼Ub conjugate recognized by a pathogenic bacterial E3 ligase. Proc Natl Acad Sci USA. 2010;107(7):2848–2853. doi: 10.1073/pnas.0914821107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ye Y, Rape M. Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol. 2009;10(11):755–764. doi: 10.1038/nrm2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alpi AF, Pace PE, Babu MM, Patel KJ. Mechanistic insight into site-restricted monoubiquitination of FANCD2 by Ube2t, FANCL, and FANCI. Mol Cell. 2008;32(6):767–777. doi: 10.1016/j.molcel.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 24.Starita LM, et al. BRCA1-dependent ubiquitination of gamma-tubulin regulates centrosome number. Mol Cell Biol. 2004;24(19):8457–8466. doi: 10.1128/MCB.24.19.8457-8466.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Christensen DE, Brzovic PS, Klevit RE. E2-BRCA1 RING interactions dictate synthesis of mono- or specific polyubiquitin chain linkages. Nat Struct Mol Biol. 2007;14(10):941–948. doi: 10.1038/nsmb1295. [DOI] [PubMed] [Google Scholar]
- 26.Fowler DM, Araya CL, Gerard W, Fields S. Enrich: Software for analysis of protein function by enrichment and depletion of variants. Bioinformatics. 2011;27(24):3430–3431. doi: 10.1093/bioinformatics/btr577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jolma A, et al. Multiplexed massively parallel SELEX for characterization of human transcription factor binding specificities. Genome Res. 2010;20(6):861–873. doi: 10.1101/gr.100552.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Di Tommaso P, et al. T-Coffee: A web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res. 2011;39(Web Server issue):W13–W17. doi: 10.1093/nar/gkr245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25(9):1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brzovic PS, Lissounov A, Christensen DE, Hoyt DW, Klevit RE. A UbcH5/ubiquitin noncovalent complex is required for processive BRCA1-directed ubiquitination. Mol Cell. 2006;21(6):873–880. doi: 10.1016/j.molcel.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 31.Delaglio F, et al. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6(3):277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 32.Johnson BA, Blevins RA. NMR View: A computer program for the visualization and analysis of NMR data. J Biomol NMR. 1994;4(5):603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.