Abstract
Reprogramming of mouse fibroblasts toward a myocardial cell fate by forced expression of cardiac transcription factors or microRNAs has recently been demonstrated. The potential clinical applicability of these findings is based on the minimal regenerative potential of the adult human heart and the limited availability of human heart tissue. An initial but mandatory step toward clinical application of this approach is to establish conditions for conversion of adult human fibroblasts to a cardiac phenotype. Toward this goal, we sought to determine the optimal combination of factors necessary and sufficient for direct myocardial reprogramming of human fibroblasts. Here we show that four human cardiac transcription factors, including GATA binding protein 4, Hand2, T-box5, and myocardin, and two microRNAs, miR-1 and miR-133, activated cardiac marker expression in neonatal and adult human fibroblasts. After maintenance in culture for 4–11 wk, human fibroblasts reprogrammed with these proteins and microRNAs displayed sarcomere-like structures and calcium transients, and a small subset of such cells exhibited spontaneous contractility. These phenotypic changes were accompanied by expression of a broad range of cardiac genes and suppression of nonmyocyte genes. These findings indicate that human fibroblasts can be reprogrammed to cardiac-like myocytes by forced expression of cardiac transcription factors with muscle-specific microRNAs and represent a step toward possible therapeutic application of this reprogramming approach.
Keywords: cardiogenesis, cell fate specification, phenotypic switching, regeneration
The ability to convert one cell type to another by forced expression of transcription factors has been known for decades (1, 2). For example, overexpression of the skeletal muscle basic helix–loop–helix transcription factor, MyoD or related factors in fibroblasts, converts these cells into skeletal muscle (3, 4). Similarly, forced expression of the cardiovascular coactivator myocardin is sufficient to convert fibroblasts into smooth muscle cells (5–9). However, no single factor has yet been shown to possess the ability to activate the complete cardiac muscle gene program. In contrast, ectopic expression of three cardiac transcription factors, GATA binding protein 4 (GATA4), myocyte enhancer factor 2C (Mef2c), and T-box5 (Tbx5) (referred to as GMT), was reported to initiate cardiac gene expression in mouse cardiac and tail-tip fibroblasts and to allow for maturation of reprogrammed cells to a spontaneously contractile state at low efficiency (10). Inclusion of the basic helix–loop–helix transcription factor Hand2 with GMT (in a four-factor combination called GHMT) increased reprogramming efficiency (11). Moreover, introduction of GMT or GHMT into cardiac fibroblasts of mice following myocardial infarction (MI) resulted in the formation of cardiomyocytes that enhanced cardiac function and diminished fibrosis, suggesting a strategy for cardiac repair (11, 12).
Despite successful lineage conversions of mouse fibroblasts into a variety of clinically relevant cell types (10, 11, 13–15), only neuronal direct reprogramming has been demonstrated in human cells (16, 17). However, neuronal lineage conversion of human cells generates only functionally immature cells with lower efficiency than mouse cells and requires longer maturation time (16, 17). Because human cells are more resistant to the reprogramming process, it is reasonable to speculate that additional regulatory events are required to propel human cells toward alternative cell fates. Although the majority of reprogramming factors identified to date are transcription factors, microRNAs (miRNAs) have recently been shown to exert synergistic or independent phenotypic reprogramming activities. Two miRNAs, miR-9/9* and miR-124, in combination with neuronal transcription factors, were reported to efficiently convert human fibroblasts into neuron-like cells (18). More recently, it was reported that a combination of muscle-specific miRNAs, including miR-1, -133, -208, and -499, without any exogenous transcription factor, reprogrammed mouse fibroblasts into cardiomyocyte-like cells in vitro and in vivo (19).
In the present study, we investigated whether human adult fibroblasts can be reprogrammed into cardiac-like myocytes by forced expression of cardiac transcription factors and muscle-specific miRNAs. We show that specific combinations of factors, which are different from the defined combinations in mouse fibroblasts, are able to induce a cardiac-like phenotype in human neonatal foreskin fibroblasts, adult cardiac fibroblasts, and adult dermal fibroblasts. These reprogrammed cells expressed multiple cardiac markers, showed suppression of nonmyocyte genes, and developed sarcomere-like structures. In addition, a small subset of these reprogrammed cells exhibited spontaneous contractility. These findings represent an important step toward potential therapeutic application of this reprogramming technology.
Results
Reprogramming Human Foreskin Fibroblasts with Human Cardiac Transcription Factors.
We first examined whether the same four cardiac transcription factors, GHMT, shown previously to direct the reprogramming of mouse fibroblasts to induced cardiac-like myocytes (iCLMs) (11), were able to reprogram neonatal human foreskin fibroblasts (HFFs) toward a cardiac phenotype. Unlike mouse fibroblasts, retroviral transduction of GHMT in HFFs failed to efficiently activate cardiac marker expression after 2 wk (Fig. S1). Thus, we selected 14 additional transcription factors that are known to be important in heart development and 3 muscle-specific miRNAs in an effort to identify optimal combinations of factors for human cardiac reprogramming (Fig. S2 A and B).
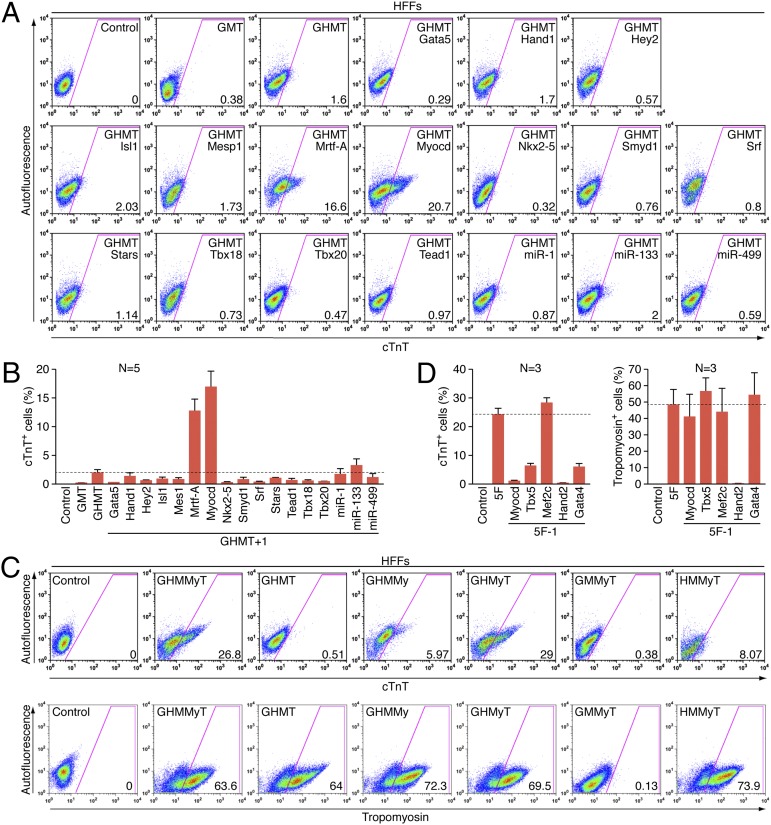
Two weeks after transducing HFFs with retroviruses expressing GMT, GHMT, or GHMT with the addition of an extra factor, we quantified endogenous cardiac marker expression using flow cytometry. GMT and GHMT activated cardiac Troponin T (cTnT) expression in ∼0.2% and ∼2% of cells, respectively. In contrast, the addition of myocardin (Myocd or My) or myocardin-related transcription factor-A (Mrtf-A) to GHMT, significantly increased the number of cTnT+ cells to ∼17% and ∼13%, respectively (Fig. 1 A and B). The ability of each combination of factors (GHMT plus each of the 14 transcription factors and 3 miRNAs) to activate cardiac gene expression was also analyzed by the percentage of tropomyosin+ cells using flow cytometry. Although the percentage of tropomyosin+ cells induced by GMT was only ∼1%, GHMT was able to induce ∼24% of cells to adopt the tropomyosin+ phenotype, indicating the critical role of Hand2 in cardiac reprogramming of human fibroblasts. Consistent with cTnT expression, addition of either Myocd or Mrtf-A significantly enhanced the percentage of tropomyosin+ cells compared with GHMT alone (∼39% and ∼45%, respectively; Fig. S3 A and B). However, neither Myocd nor Mrtf-A alone was able to activate cTnT or tropomyosin expression in HFFs (Fig. S3C).
Fig. 1.
Screening for additional factors able to activate cardiac gene expression. (A) Representative flow cytometry plot for analyses of cTnT+ cells 2 wk after infection of HFFs with retroviruses expressing indicated combinations of factors. Cells infected with empty vector retrovirus were used as a control. The numbers in each plot indicate the percentage of cTnT+ cells. (B) Summary of flow cytometry analyses. Percentage of cTnT+ cells following infection of HFFs with empty vector retrovirus (control) or retroviruses expressing GHMT with an indicated individual factor as shown. Data from five independent experiments are presented as mean ± SD. Dotted line indicates the percentage of cTnT+ cells induced by retroviruses expressing GHMT alone. (C) Representative flow cytometry plot for subtractive analyses to determine the requirement of each individual factor in 5F (GHMMyT). Two weeks after infection of HFFs with retroviruses expressing indicated combinations of factors, cTnT+ (Upper) and tropomyosin+ (Lower) cells were quantified by flow cytometry. Cells infected with empty vector retrovirus were used as a control. The numbers in each plot indicate the percentage of cTnT+ or tropomyosin+ cells. (D) Summary of flow cytometry analyses. Percentage of cTnT+ (Left) or tropomyosin+ (Right) cells following infection of HFFs with empty vector retrovirus (control), or retroviruses expressing 5F (GHMMyT) or 5F minus an indicated individual factor as shown. Data from three independent experiments are presented as mean ± SD. Dotted line indicates the percentage of cTnT+ or tropomyosin+ cells induced by retroviruses expressing 5F.
Because Myocd can activate smooth muscle gene expression in fibroblasts (6–9), we analyzed the expression of smooth muscle markers including smooth muscle myosin heavy chain and smooth muscle protein 22 alpha that are not expressed in adult cardiomyocytes (20–22). These smooth muscle markers were up-regulated in adult human cardiac fibroblasts (AHCFs) transduced with GHMT plus Myocd [referred to as the five factors (5F) or GHMMyT] (Fig. S4A). However, the level of expression was insignificant compared with that of human aorta. These findings suggested that myocardin in combination with other cardiac transcription factors activates the cardiac gene program more prominently than the smooth muscle gene program.
We next determined whether all five factors, GHMMyT, are necessary for optimal activation of cardiac marker expression in HFFs by withdrawing each factor individually from the 5F pool. Removing Gata4, Hand2, Tbx5, or Myocd markedly decreased the percentage of cells expressing cTnT (Fig. 1 C and D). In contrast, removal of Mef2c did not diminish the expression of cTnT in response to the other four factors. On the other hand, the activation of tropomyosin expression was ablated when Hand2 was withdrawn, but was not significantly altered by withdrawal of any other factor from the 5F pool (Fig. 1 C and D). Indeed, Hand2 when combined with any three of the four GMMyT factors resulted in activation of tropomyosin expression in 60–70% of HFFs. These findings suggest that the GMMyT factors act redundantly to complement the reprogramming activity of Hand2 in activation of tropomyosin expression in HFFs. Surprisingly, the absence of Hand2 almost eliminated both cTnT and tropomyosin expression in HFFs, demonstrating the irreplaceable activity of this factor for activation of cardiac contractile gene programs in human fibroblasts, unlike its synergistic role in mouse myocardial reprogramming (11).
Because withdrawing any of the five factors (GHMMyT) failed to further increase cardiac marker expression, we searched for an additional sixth factor that could cooperatively enhance cardiac gene expression in HFFs by adding each factor into the 5F pool. No other factor added to the 5F pool allowed for further optimization of the reprogramming process. On the contrary, most other factors demonstrated inhibitory effects (Fig. S5 and Fig. 2A).
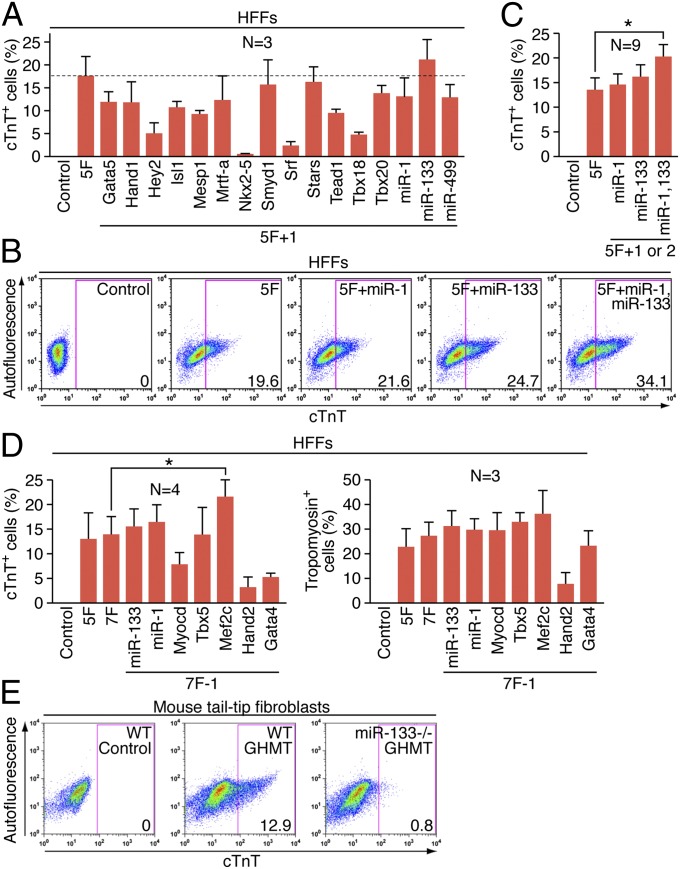
Fig. 2.
Determining the optimal combination of factors to activate cardiac gene expression. (A) Summary of flow cytometry analyses of cTnT+ cells 2 wk after infection of HFFs with retroviruses expressing indicated combinations of factors. Percentage of cTnT+ cells following infection of HFFs with empty vector retrovirus, or retroviruses expressing 5F (GHMMyT) or 5F plus an indicated individual factor as shown. Data from three independent experiments are presented as mean ± SD. Dotted line indicates the percentage of cTnT+ cells induced by retroviruses expressing 5F. (B) Effect of miR-1 and miR-133 on activation of cardiac markers. Representative flow cytometry plot for analyses of cTnT+ cells 2 wk after infection of HFFs with retroviruses expressing indicated combinations of factors. Cells infected with empty vector retrovirus were used as a control. The numbers in each plot indicate the percentage of cTnT+ cells. (C) Summary of flow cytometry analyses. Percentage of cTnT+ cells following infection of HFFs with empty vector retrovirus, or retroviruses expressing 5F (GHMMyT) or 5F plus miR-1, miR-133, or both miR-1 and miR-133 as shown. Data from nine independent experiments are presented as mean ± SD. *P = 0.0062 (D) Summary of flow cytometry analyses to determine the necessity of individual 7F (GHMMyT, miR-1, miR-133). Percentage of cTnT+ (Left) or tropomyosin+ (Right) cells following infection of HFFs with empty vector retrovirus (control), or retroviruses expressing 7F (GHMMyT miR-1 miR-133) or 7F minus an indicated individual factor as shown. Data from three or four independent experiments are presented as mean ± SD. *P = 0.041. (E) Inefficient cardiac gene activation in the absence of miR-133. Representative flow cytometry plot for analyses of cTnT+ cells 1 wk after infection of retroviruses expressing GHMT into tail-tip fibroblasts isolated from either wild-type (WT) or miR-133 knockout (miR-133−/−) mice. Tail-tip fibroblasts isolated from wild type mice infected with empty vector were used as a control.
Influence of miRNAs on Cardiac Reprogramming.
In an effort to further optimize the reprogramming process, we tested whether various muscle-specific miRNAs could augment the activity of GHMT or GHMMyT. Prior studies reported that miR-1 alone or in combination with miR-133, -208, and -499 allowed for efficient reprogramming of mouse cardiac fibroblasts to a cardiomyocyte-like phenotype (19). However, in our hands, retroviral expression of these miRNAs failed to activate expression of cardiac markers in mouse fibroblasts (Fig. S6A). We transduced HFFs with 5F and either miR-1 or miR-133 or both. The expression of endogenous cardiac markers, including cTnT and tropomyosin, was quantified by flow cytometry 2 wk after infection. GHMMyT with both miRNAs (termed 7F) increased the percentage of cTnT+ cells (Fig. 2 B and C), whereas there was no significant effect of these miRNAs on tropomyosin expression (Fig. S6 B and C). The effect on cardiac marker expression by both miRNAs was relatively mild, but highly reproducible. Next, we tested whether these miRNAs were able to replace any of the 5Fs or whether all 7Fs were necessary for optimal activation of cardiac gene expression by withdrawing each factor from the 7F pool. Unexpectedly, the deletion of Mef2c significantly increased the percentage of cells expressing cardiac markers, as quantified by flow cytometry (Fig. S7 and Fig. 2D). Although neither miR-1 nor miR-133 was required, the six factors including these two muscle-specific miRNAs (termed 6F; Gata4, Hand2, Tbx5, Myocd, miR-1, and miR-133) represented the most optimal combination of factors for efficient initiation of cardiac gene expression in human fibroblasts among the combinations of factors we tested. We henceforth refer to 5F-, 6F-, or 7F-transduced human fibroblasts expressing endogenous cardiac markers as human-induced cardiac-like myocytes (hiCLMs).
We also examined whether miR-133 was necessary for the activation of endogenous cardiac-specific gene expression using tail-tip fibroblasts isolated from mice lacking miR-133 (23). We found that genetic deletion of miR-133 markedly decreased the percentage of cells expressing cTnT (Fig. 2E).
Reprogramming Human Adult Cardiac and Dermal Fibroblasts.
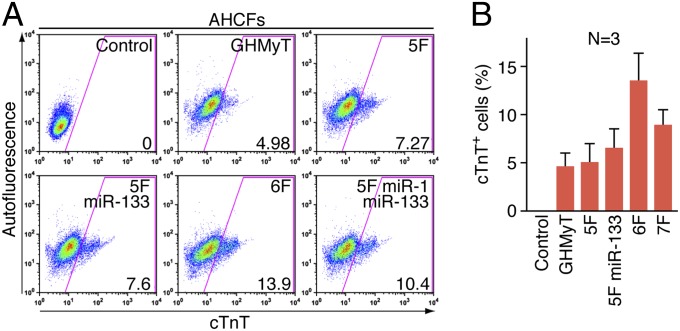
Cardiac fibroblasts represent the most prevalent cell type in the adult human heart and are the principal mediators of cardiac fibrosis and scar formation post-MI (24, 25). Thus, AHCFs are an ultimate target cell type for cardiac reprogramming. To examine whether combinations of factors could activate cardiac gene expression in this population of cells, we isolated AHCFs from human hearts provided by heart transplantation recipients or disqualified organ donors using an explant culture in which AHCFs migrated from minced heart tissue in fibroblast growth medium. This method avoids contamination of adult cardiomyocytes, which are unable to migrate or survive in this medium. We transduced into AHCFs multiple combinations of factors including the 6F combination and expression of cardiac markers was analyzed 2 wk later. The 6F combination induced ∼13% of AHCFs to become cTnT+, whereas other combinations we tested showed lower efficiency of generating cTnT+ cells (Fig. 3 A and B). Overall, the reprogramming efficiency of AHCFs was much lower than that of HFFs. This difference results, at least in part, from relatively inefficient retroviral transduction of AHCFs compared with HFFs because of the slower proliferation rate of AHCFs. Moreover, AHCFs from humans of at least 20 y of age have likely established more stable epigenetic programs, which are more refractory to reprogramming than HFFs isolated from newborns.
Fig. 3.
Induction of cardiac markers in AHCFs. (A) Representative flow cytometry plot for analyses of cTnT+ cells 2 wk after infection of AHCFs with retroviruses expressing indicated combinations of factors. Cells infected with empty vector retrovirus were used as a control. The numbers in each plot indicate the percentage of cTnT+. (B) Summary of flow cytometry analyses in AHCFs. Percentage of cTnT+ cells following infection of AHCFs with empty vector retrovirus (control) or retroviruses expressing indicated combinations of factors as shown. Data from three independent experiments are presented as mean ± SD.
We also tested whether adult human dermal fibroblasts (AHDFs) could be reprogrammed into hiCLMs. Two weeks after transduction of AHDFs with retroviruses expressing 6F or 7F, cardiac marker expression was quantified by flow cytometry. Consistent with the reprogramming of HFFs and AHCFs, the presence of 6F activated a higher percentage of AHDFs to express cTnT than 7F (9.5% vs. 4.4%), whereas there was no significant difference in tropomyosin expression with each combination (Fig. S8).
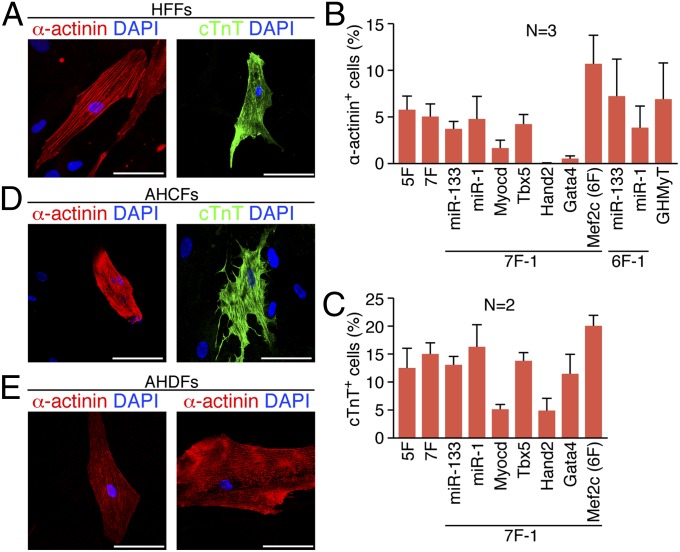
Analysis of the time course of cardiac marker expression in HFFs transduced with 6F showed that the percentage of cells expressing cTnT and tropomyosin reached 2.3% and 22.4%, respectively, at 1 wk and continuously increased up to 35% and 42.5% at 4 wk after transduction, respectively (Fig. S9 A and B). Compared with the relatively rapid activation of cardiac gene expression seen in mouse tail-tip fibroblasts (TTFs), which showed a peak at 1 wk after viral transduction of GHMT (11), the initiation as well as full maturation of cardiac gene expression in HFFs was much slower. After extended culture periods of 4–5 wk postinfection, HFFs infected with various combinations of factors, including at least GHMyT, showed strong immunostaining of the sarcomeric proteins α-actinin and cTnT with cellular striations resembling sarcomere structures (Fig. 4A). With viral transduction of 6F, ∼12% or ∼19% of HFFs became α-actinin+ or cTnT+, respectively, by immunostaining, which was greater than with any other combinations we tested (Fig. 4 B and C). However, 7F including Mrtf-A instead of Myocd were unable to activate α-actinin expression in HFFs, indicating that Mrtf-A is not an alternative factor to Myocd despite its comparable ability to induce cTnT and tropomyosin expression. Similarly, we also observed strong α-actinin staining in AHCFs and AHDFs transduced with 6F (Fig. 4 D and E).
Fig. 4.
Immunostaining of cardiac markers in 5F-, 6F-, or 7F-transduced human fibroblasts. Immunofluorescence staining for α-actinin (red) or cTnT (green) was performed 5 wk after transduction of (A) HFFs, (D) AHCFs, or (E) AHDFs with 5F, 6F, or 7F. (Scale bar, 100 μm.) (B) Quantification of α-actinin+ cells. (C) Quantification of cTnT+ cells. Percentage of α-actinin+ (B) or cTnT+ (C) cells after infection of HFFs with 5F, 7F, or 7F or 6F minus an indicated individual factor as shown. Data from two or three independent experiments are presented as mean ± SD.
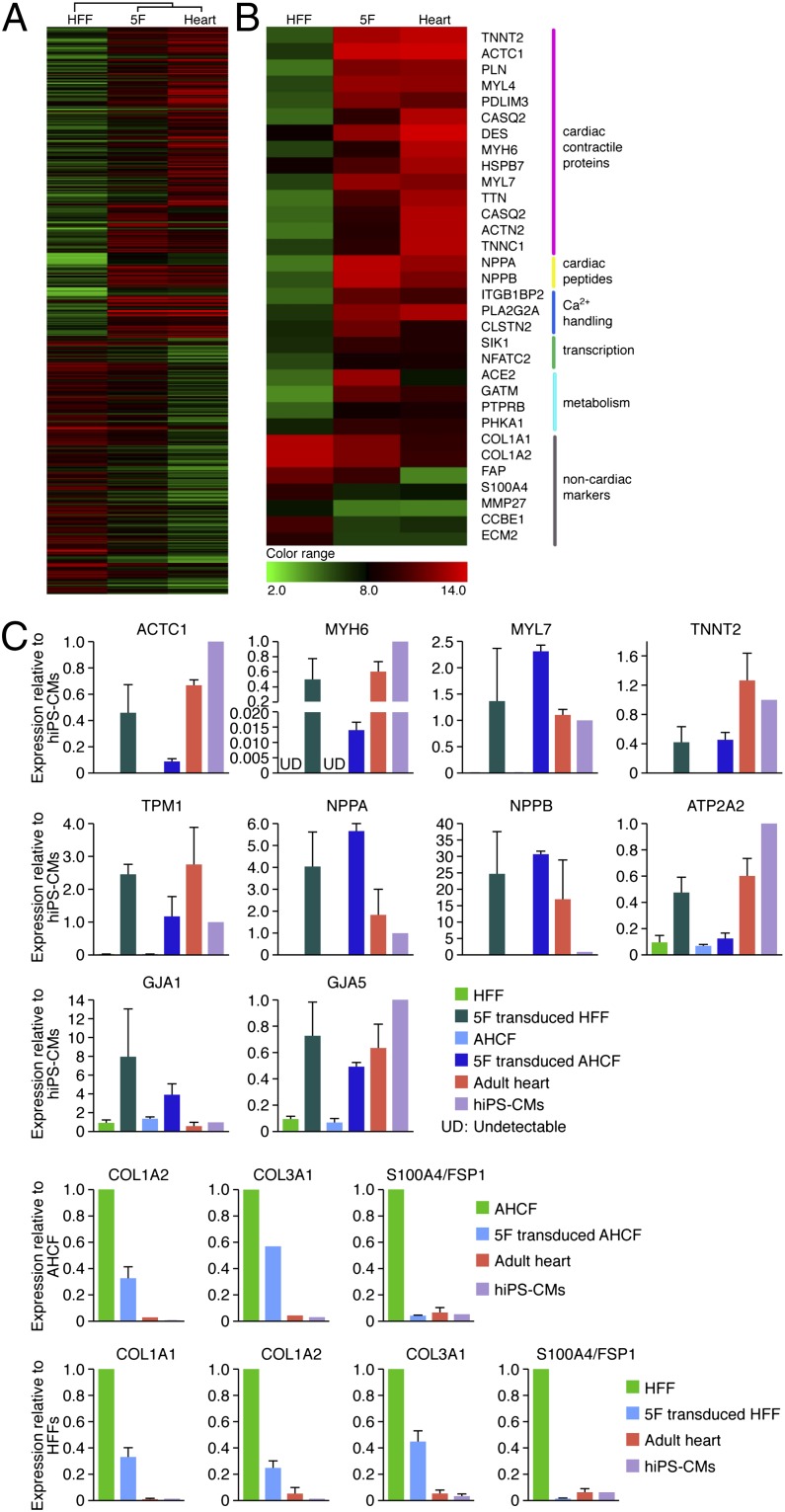
Microarray analysis of gene expression patterns showed expression of a broad range of cardiac genes, and concomitant suppression of nonmyocyte genes in HFFs transduced with 5F at 4 wk (Fig. 5 A and B). We further analyzed gene expression profiles of HFFs, AHCFs, and AHDFs transduced with 5F or 6F compared with adult human heart (Fig. 5C and Fig. S4 B and C), in addition to human induced pluripotent stem (iPS)-cell derived cardiomyocytes (Fig. 5C), using quantitative PCR. Retroviruses encoding 5F or 6F initiated expression of a broad range of cardiac genes, including actin, alpha, cardiac muscle 1 (ACTC1); troponin T type 2 (cardiac) (TNNT2); myosin, heavy chain 6, cardiac muscle, alpha (MYH6); myosin, light chain 7, regulatory (MYL7); tropomyosin 1 (alpha) (TPM1); ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 (ATP2A2); gap junction protein, alpha 1 (GJA1); GJA5; natriuretic peptide A (NPPA); and NPPB in HFFs, AHCFs (Fig. 5C), and AHDFs (Fig. S4B). Moreover, genes encoding ion channels and neurohormonal components that are important in cardiomyocyte physiology were significantly up-regulated in 5F-transduced HFFs (Fig. S4C). In contrast, expression of nonmyocyte genes, collagen, type I, alpha 2 (COL1A2); COL3A1; and fibroblast specific protein 1 (FSP1) (fibroblast-specific protein 1, or S100A4) was significantly down-regulated. We observed variations in the levels of expression of different cardiac markers between the different cardiac samples. For example, cardiac natriuretic peptides (NPPA and NPPB), as well as ACE2, were expressed at higher levels in 5F-transduced HFFs, whereas the cardiac sodium channel SCN5A was expressed at reduced levels in 5F-transduced HFFs compared with human iPS cell–derived cardiomyocytes or adult heart (Fig. 5C and Fig. S4C). These results further substantiate the cardiac-like phenotype evoked by the 5F combination in human fibroblasts. However, there were also cardiac genes that were not up-regulated in response to 5F or 6F, indicating that the cardiac-like phenotype is incomplete and likely variable between cells in the population.
Fig. 5.
Gene expression profile in human fibroblasts transduced with 5F. Gene expression profile was analyzed by microarray or quantitative PCR (qPCR) in HFFs or AHCFs 4 wk after transduction with 5F. (A) Heat map of microarray data illustrating differentially expressed 2,436 genes in HFFs, 5F-transduced HFFs, and adult human heart. Red indicates up-regulated genes; green indicates down-regulated genes. (B) Heat map of selected genes. Genes that encode cardiac contractile proteins, cardiac peptides, calcium handling genes, cardiac transcription factors, and genes involved in cardiac metabolism were up-regulated. In contrast, genes encoding nonmyocyte markers were down-regulated. (C) Gene expression analyses by qPCR in HFF and AHCFs transduced with 5F. ACTC1, TNNT2, MYL7, MYH6, and TPM1 are sarcomere genes; ATP2A2, GJA1, and GJA5 are cardiac channel genes; NPPA and NPPB are cardiac peptide genes; COL1A2, COL3A1, and S100A4 are nonmyocyte genes. Expression of cardiac and nonmyocyte genes was quantified by qPCR. UD, undetectable.
Contractility of hiCLMs.
Upon maintaining hiCLMs in culture for 4 wk, we observed calcium transients in ∼10% of HFFs (Fig. 6A) and ∼15% of AHCFs (Fig. 6B) in response to potassium chloride stimulation. Using coinfection with a cTnT-green fluorescent protein (GFP) CaMP5 lentiviral reporter, in which expression of GCaMP5, a fluorescent calcium sensor, is controlled by the cardiac-specific cTnT promoter, we were able to identify a low percentage of hiCLMs generating spontaneous calcium transients at 8 wk after infection (Movie S1). By extending the culture period for ∼11 wk, a small subset of hiCLMs derived from AHCFs exhibited spontaneous contractions (Movies S2 and S3). We did not observe spontaneous contractions in hiCLMs derived from HFFs or AHDFs. These findings indicate that human fibroblasts can be converted into functional hiCLMs at lower efficiency and need a longer maturation time compared with mouse fibroblasts. Expression of the GCaMP5 reporter is specific for cardiomyocytes because of the cardiac specificity of the cTnT promoter, ruling out the possibility that GCaMP5-positive cells arise from a small population of fibroblasts reprogrammed to smooth muscle-like cells by Myocd. Moreover, smooth muscle cells do not display the type of spontaneous, rhythmic contractility observed in 5F-transduced fibroblast cultures.
Fig. 6.
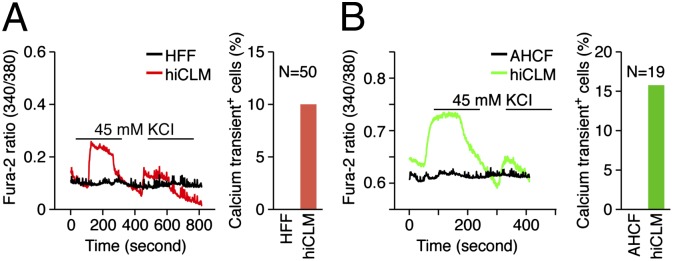
Measurement of calcium transient in hiCLMs. Calcium transient of a single cell was traced upon KCL stimulation in hiCLMs derived from (A) HFFs and (B) AHCFs. HFFs or AHCFs were used as a negative control accordingly. The graph bar represents the percentage of cells displaying calcium transients. The total number of cells recorded for calcium transients was 50 for HFFs and 19 for AHCFs.
Discussion
An attractive strategy in regenerative medicine is to generate therapeutically useful cells from an adult patient’s own ordinary tissue and replenish dying or diseased cells with these new cells (26). This autologous therapeutic approach can produce immunologically matched tissues and circumvent immunogenicity. In this context, we and others recently showed that mouse fibroblasts can be converted into functional cardiac-like myocytes by forced expression of three or four core cardiac transcription factors or muscle-specific miRNAs (10–12, 19, 27). Furthermore, direct delivery of these transcription factors into the myocardium of mice following MI reduced scar formation and blunted worsening of cardiac function, which appears to be at least partially attributable to reprogramming of nonmyocytes into cardiac-like myocytes (11, 12). To advance this strategy toward clinical application, reprogramming human fibroblasts is a prerequisite. In the present study, we identified a combination of factors capable of initiating cardiac gene expression in human fibroblasts. Whereas we previously found GHMT to be the optimal combination of factors for cardiac reprogramming of mouse fibroblasts (11), these factors alone were ineffective in activating cardiac gene expression in human fibroblasts and required the addition of Myocd for human cardiac gene expression. Two muscle-specific miRNAs, miR-1 and miR-133, further improved myocardial conversion of human fibroblasts and eliminated the requirement of Mef2c. We showed previously that miR-1 and miR-133 are regulated by Mef2c (23), which likely contributes to their ability to replace this transcription factor in the reprogramming mixture. The different requirements for reprogramming of mouse and human fibroblasts likely reflect differences in the mouse and human fibroblast populations and the susceptibility of cardiac genes to activation in these different cells.
A recent study highlighted the importance of myocardin in the activation of cardiac gene expression in mouse fibroblasts, demonstrating more effective induction of a cardiac phenotype with myocardin in combination with Mef2c and Tbx5, compared with any other combination of three factors from 10 candidate transcription factors including GMT (28). Although myocardin plays a key role in vascular smooth muscle cell differentiation (29), it also activates cardiac gene expression through interactions with other cardiac transcription factors including serum response factor, Tbx5, and Mef2c (5, 30, 31). Our results indicate that smooth muscle genes are minimally activated by myocardin in the presence of GHMT, raising the possibility that the latter cardiac factors suppress the ability of myocardin to efficiently activate the smooth muscle gene program. In this regard, we showed previously that miR-133a null mice displayed disrupted sarcomeres and ectopic activation of smooth muscle genes in the developing heart (23). In addition, miR-1 and -133 have been shown to act through different mechanisms to influence the generation of cardiomyocytes from embryonic stem cells (32). Thus, we speculate that forced expression of miR-1 and -133 may play a role in development of sarcomere structure and suppression of smooth muscle gene expression in hiCLMs.
A concern with using freshly isolated primary fibroblasts as starting cells for reprogramming is that it is difficult to exclude contamination of other cell types, including immature or progenitor cells, which are known to be more susceptible to reprogramming (33). We found that a commercially available adult human dermal fibroblast cell line, which does not contain any contaminating cells, can convert to hiCLMs with comparable efficiency to freshly isolated adult fibroblasts, excluding potential contributions of stem or progenitor cells in this process. Human cardiomyocytes have also been differentiated from human iPS cells or cardiac progenitors (34, 35). The ability to reprogram human fibroblasts that were expanded multiple times after isolation could, with further optimization, eventually allow large-scale production of hiCLMs for possible transplantation. In this context, a recent study demonstrated that human ES cell–derived cardiomyocytes can integrate into an infarcted guinea pig heart and suppress development of post-MI arrhythmias (36). Given that human ES cells have limitations as a clinical source for cell transplantation, hiCLMs may be a viable alternative for this approach in the future.
As with other reprogrammed cells generated from human fibroblasts, including human iPSCs and human-induced neurons, human iCLMs are functionally immature, as indicated by their morphology, low-amplitude calcium transients in response to electrical stimulation, and relatively rare spontaneous contractility. In addition, the human iCLMs generated in this study were heterogeneous, containing cells with varying levels of expression of cardiac and noncardiac genes. Phenotypic conversion to a cardiac cell fate probably requires a precise stoichiometry as well as certain levels of expression of reprogramming factors, which are achieved only in a small subset of fibroblasts. Heterogeneity of hiCLMs is likely to reflect variations in the stoichiometry and levels of expression of reprogramming factors in individual cells. Moreover, variations in the percentage of cardiac marker–expressing cells from experiment to experiment are likely also attributable to heterogeneity of human fibroblasts that were isolated from human subjects of various ages and genetic backgrounds. Human fibroblasts whose epigenetic stabilities vary depending on their origins are likely to have a wide spectrum of susceptibility to reprogramming. In addition, the differences that exist in each viral preparation also contribute to variability in reprogramming efficiency. For similar reasons, we observed variable reprogramming efficiency from experiment to experiment even in mouse fibroblasts (11). Nevertheless, the results of this study indicate that diverse types of human fibroblasts can be reprogrammed toward a cardiac fate and establish a foundation for further optimization of this process and the eventual generation of more mature and homogeneous populations of hiCLMs. It will also be of particular interest to modify this process using pharmacologic agents and to generate specialized cells involved in cardiac conduction as a strategy for modulating cardiac contractility. Such studies are under way.
Materials and Methods
All deidentified human heart and foreskin tissues were obtained and banked with proper informed consent under approval of the University of Texas Southwestern Institutional Review Board (IRB CR00001649/STU 032011-174 and IRB 092010-193). Normal human foreskin was obtained from neonates (weighing less than 10 lb) undergoing routine circumcision. Adult human hearts were obtained from heart transplantation recipients, lung transplantation donors, or disqualified organ donors. Isolation of human fibroblasts, production of retroviruses, induction of reprogramming, flow cytometry, immunocytochemistry, real-time PCR and DNA microarray, and calcium transient measurements are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. J. Gearhart (University of Pennsylvania) for the cTnT-GCaMP5 FUdeltaGW-rtTA construct; J. Cabrera for graphics and the University of Texas Southwestern Microarray Core Facility for collecting gene expression data; Dr. Megan Kong for analyzing the microarray data; and Ankit Garg, Thomas Haden, Dr. Ning Liu, and Dr. Ji-Hoon Lee for technical advice. E.N.O. is supported by grants from the National Institutes of Health (NIH), the Donald W. Reynolds Center for Clinical Cardiovascular Research, the Robert A. Welch Foundation (Grant I-0025), the Leducq Foundation-Transatlantic Network of Excellence in Cardiovascular Research Program, the American Heart Association–Jon Holden DeHaan Foundation, and the Cancer Prevention and Research Institute of Texas; L.A.B. is supported by the Seay Endowment and a grant from the NIH; and Y.-J.N. was supported by NIH Grant K08 HL111420-02.
Footnotes
Conflict of interest statement: E.N.O., Y.-J.N., and K.S. have all filed a patent relating to reprogramming of human fibroblasts to human cardiomyocytes. This patent has been licensed by LoneStar Heart, Inc. E.N.O. is a cofounder of this company and holds equity.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1301019110/-/DCSupplemental.
References
- 1.Graf T, Enver T. Forcing cells to change lineages. Nature. 2009;462(7273):587–594. doi: 10.1038/nature08533. [DOI] [PubMed] [Google Scholar]
- 2.Vierbuchen T, Wernig M. Direct lineage conversions: Unnatural but useful? Nat Biotechnol. 2011;29(10):892–907. doi: 10.1038/nbt.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51(6):987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- 4.Olson EN. MyoD family: A paradigm for development? Genes Dev. 1990;4(9):1454–1461. doi: 10.1101/gad.4.9.1454. [DOI] [PubMed] [Google Scholar]
- 5.Wang D, et al. Activation of cardiac gene expression by myocardin, a transcriptional cofactor for serum response factor. Cell. 2001;105(7):851–862. doi: 10.1016/s0092-8674(01)00404-4. [DOI] [PubMed] [Google Scholar]
- 6.Wang Z, Wang DZ, Pipes GC, Olson EN. Myocardin is a master regulator of smooth muscle gene expression. Proc Natl Acad Sci USA. 2003;100(12):7129–7134. doi: 10.1073/pnas.1232341100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen J, Kitchen CM, Streb JW, Miano JM. Myocardin: A component of a molecular switch for smooth muscle differentiation. J Mol Cell Cardiol. 2002;34(10):1345–1356. doi: 10.1006/jmcc.2002.2086. [DOI] [PubMed] [Google Scholar]
- 8.Du KL, et al. Myocardin is a critical serum response factor cofactor in the transcriptional program regulating smooth muscle cell differentiation. Mol Cell Biol. 2003;23(7):2425–2437. doi: 10.1128/MCB.23.7.2425-2437.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoshida T, et al. Myocardin is a key regulator of CArG-dependent transcription of multiple smooth muscle marker genes. Circ Res. 2003;92(8):856–864. doi: 10.1161/01.RES.0000068405.49081.09. [DOI] [PubMed] [Google Scholar]
- 10.Ieda M, et al. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142(3):375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song K, et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature. 2012;485(7400):599–604. doi: 10.1038/nature11139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qian L, et al. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature. 2012;485(7400):593–598. doi: 10.1038/nature11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou Q, Brown J, Kanarek A, Rajagopal J, Melton DA. In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature. 2008;455(7213):627–632. doi: 10.1038/nature07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang P, et al. Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature. 2011;475(7356):386–389. doi: 10.1038/nature10116. [DOI] [PubMed] [Google Scholar]
- 15.Sekiya S, Suzuki A. Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature. 2011;475(7356):390–393. doi: 10.1038/nature10263. [DOI] [PubMed] [Google Scholar]
- 16.Pang ZP, et al. Induction of human neuronal cells by defined transcription factors. Nature. 2011;476(7359):220–223. doi: 10.1038/nature10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qiang L, et al. Directed conversion of Alzheimer’s disease patient skin fibroblasts into functional neurons. Cell. 2011;146(3):359–371. doi: 10.1016/j.cell.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Yoo AS, et al. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature. 2011;476(7359):228–231. doi: 10.1038/nature10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jayawardena TM, et al. MicroRNA-mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circ Res. 2012;110(11):1465–1473. doi: 10.1161/CIRCRESAHA.112.269035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuro-o M, et al. Developmentally regulated expression of vascular smooth muscle myosin heavy chain isoforms. J Biol Chem. 1989;264(31):18272–18275. [PubMed] [Google Scholar]
- 21.Rovner AS, Thompson MM, Murphy RA. Two different heavy chains are found in smooth muscle myosin. Am J Physiol. 1986;250(6 Pt 1):C861–C870. doi: 10.1152/ajpcell.1986.250.6.C861. [DOI] [PubMed] [Google Scholar]
- 22.Owens GK. Regulation of differentiation of vascular smooth muscle cells. Physiol Rev. 1995;75(3):487–517. doi: 10.1152/physrev.1995.75.3.487. [DOI] [PubMed] [Google Scholar]
- 23.Liu N, et al. microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev. 2008;22(23):3242–3254. doi: 10.1101/gad.1738708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brown RD, Ambler SK, Mitchell MD, Long CS. The cardiac fibroblast: Therapeutic target in myocardial remodeling and failure. Annu Rev Pharmacol Toxicol. 2005;45:657–687. doi: 10.1146/annurev.pharmtox.45.120403.095802. [DOI] [PubMed] [Google Scholar]
- 25.Zeisberg EM, Kalluri R. Origins of cardiac fibroblasts. Circ Res. 2010;107(11):1304–1312. doi: 10.1161/CIRCRESAHA.110.231910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dimmeler S, Zeiher AM, Schneider MD. Unchain my heart: The scientific foundations of cardiac repair. J Clin Invest. 2005;115(3):572–583. doi: 10.1172/JCI24283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Srivastava D, Ieda M. Critical factors for cardiac reprogramming. Circ Res. 2012;111(1):5–8. doi: 10.1161/CIRCRESAHA.112.271452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Protze S, et al. A new approach to transcription factor screening for reprogramming of fibroblasts to cardiomyocyte-like cells. J Mol Cell Cardiol. 2012;53(3):323–332. doi: 10.1016/j.yjmcc.2012.04.010. [DOI] [PubMed] [Google Scholar]
- 29.Li S, Wang DZ, Wang Z, Richardson JA, Olson EN. The serum response factor coactivator myocardin is required for vascular smooth muscle development. Proc Natl Acad Sci USA. 2003;100(16):9366–9370. doi: 10.1073/pnas.1233635100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ghosh TK, et al. Physical interaction between TBX5 and MEF2C is required for early heart development. Mol Cell Biol. 2009;29(8):2205–2218. doi: 10.1128/MCB.01923-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang C, Cao D, Wang Q, Wang DZ. Synergistic activation of cardiac genes by myocardin and Tbx5. PLoS ONE. 2011;6(8):e24242. doi: 10.1371/journal.pone.0024242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ivey KN, et al. MicroRNA regulation of cell lineages in mouse and human embryonic stem cells. Cell Stem Cell. 2008;2(3):219–229. doi: 10.1016/j.stem.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou Q, Melton DA. Extreme makeover: Converting one cell into another. Cell Stem Cell. 2008;3(4):382–388. doi: 10.1016/j.stem.2008.09.015. [DOI] [PubMed] [Google Scholar]
- 34.Islas JF, et al. Transcription factors ETS2 and MESP1 transdifferentiate human dermal fibroblasts into cardiac progenitors. Proc Natl Acad Sci USA. 2012;109(32):13016–13021. doi: 10.1073/pnas.1120299109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zwi L, et al. Cardiomyocyte differentiation of human induced pluripotent stem cells. Circulation. 2009;120(15):1513–1523. doi: 10.1161/CIRCULATIONAHA.109.868885. [DOI] [PubMed] [Google Scholar]
- 36.Shiba Y, et al. Human ES-cell-derived cardiomyocytes electrically couple and suppress arrhythmias in injured hearts. Nature. 2012;489(7415):322–325. doi: 10.1038/nature11317. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.