Abstract
There is a genetic contribution to fetal alcohol spectrum disorders (FASD), but the identification of candidate genes has been elusive. Ethanol may cause FASD in part by decreasing the adhesion of the developmentally critical L1 cell adhesion molecule through interactions with an alcohol binding pocket on the extracellular domain. Pharmacologic inhibition or genetic knockdown of ERK2 did not alter L1 adhesion, but markedly decreased ethanol inhibition of L1 adhesion in NIH/3T3 cells and NG108-15 cells. Likewise, leucine replacement of S1248, an ERK2 substrate on the L1 cytoplasmic domain, did not decrease L1 adhesion, but abolished ethanol inhibition of L1 adhesion. Stable transfection of NIH/3T3 cells with human L1 resulted in clonal cell lines in which L1 adhesion was consistently sensitive or insensitive to ethanol for more than a decade. ERK2 activity and S1248 phosphorylation were greater in ethanol-sensitive NIH/3T3 clonal cell lines than in their ethanol-insensitive counterparts. Ethanol-insensitive cells became ethanol sensitive after increasing ERK2 activity by transfection with a constitutively active MAP kinase kinase 1. Finally, embryos from two substrains of C57BL mice that differ in susceptibility to ethanol teratogenesis showed corresponding differences in MAPK activity. Our data suggest that ERK2 phosphorylation of S1248 modulates ethanol inhibition of L1 adhesion by inside-out signaling and that differential regulation of ERK2 signaling might contribute to genetic susceptibility to FASD. Moreover, identification of a specific locus that regulates ethanol sensitivity, but not L1 function, might facilitate the rational design of drugs that block ethanol neurotoxicity.
Prenatal alcohol exposure causes fetal alcohol spectrum disorders (FASD) in up to 2–5% of school-age children and is the leading preventable cause of mental retardation in the Western world (1, 2). The prevalence and presentation of FASD are influenced by the quantity, frequency, and timing of drinking and are modified by a variety of environmental, nutritional, epigenetic, and genetic factors (3–7). The observation that there is greater concordance for fetal alcohol syndrome (FAS) in monozygotic twins than in dizygotic twins suggests that there are susceptibility genes for FASD (8); however, their identification remains elusive. The identification of molecular pathways that regulate sensitivity to ethanol teratogenesis would be helpful in the search for FASD susceptibility genes.
One potentially important target of ethanol in the pathogenesis of FASD is the developmentally critical immunoglobulin neural cell adhesion molecule, L1. The homophilic binding of L1 molecules on adjacent cells mediates neuronal migration, axon guidance, and axon fasciculation (9)—developmental events that are disrupted in FASD (10-12). Mutations in the human L1 gene cause brain lesions and neurological abnormalities. Some of these mutations also disrupt L1 homophilic binding (13–16). We noted that brain lesions in children with FASD resemble those of children with mutations in the gene for L1 and demonstrated that concentrations of ethanol attained after just one or two drinks inhibit L1 adhesion in cerebellar granule neurons, neural cell lines, and NIH/3T3 fibroblasts (17-19). Importantly, drugs that block ethanol inhibition of L1 adhesion also prevent ethanol teratogenesis in mice (20–25).
L1 adhesion is not universally inhibited by ethanol. For example, ethanol does not inhibit the adhesion of human L1 (hL1) when expressed in myeloma cells or Drosophila S2 cells (26, 27). Even clonal NIH/3T3 cell lines derived from a single transfection with hL1 have shown either an ethanol-sensitive or ethanol-insensitive phenotype over multiple passages and many years (19). These findings suggest that cell-specific factors regulate the sensitivity of L1 to ethanol. The characterization of these factors might prove valuable in identifying candidate genes that govern susceptibility to ethanol teratogenesis.
L1 homophilic binding is mediated by its extracellular domain (ECD), which comprises six Ig and five fibronectin type III (Fn) repeats (9). Homophilic binding is potentiated by the folding of the L1-ECD into a horseshoe structure in which the Ig1 and Ig4 domains lie opposed to each other (28–30). Using photolabeling, we demonstrated the interaction of alcohols with a binding pocket at this functionally important Ig1–Ig4 domain interface (31). Mutation of a single alcohol binding residue, Glu-33 on Ig1, did not reduce L1 adhesion, but markedly altered the pharmacology of alcohol inhibition of L1 adhesion (31). These findings suggest that subtle changes in the conformation of an alcohol binding pocket can significantly alter alcohol inhibition of L1 adhesion.
If ethanol inhibits L1 adhesion by interacting with an extracellular binding pocket, how, then, might intracellular events regulate these extracellular interactions? The L1 cytoplasmic domain (L1-CD) is highly conserved across species and contains numerous sites for phosphorylation by casein kinase II, p90 ribosomal S6 kinase, extracellular signal-regulated kinase 2 (ERK2) [a member of the mitogen-activated protein kinase (MAPK) family], pp60c-src, chicken embryonic kinase 5, and potentially other kinases (32–35). Phosphorylation of the L1-CD regulates the conformation and function of the extracellular domain (ECD) (36, 37), a phenomenon known as “inside-out” signaling and could conceivably modify the conformation of the alcohol binding pocket in the L1-ECD. Here, we show that L1 sensitivity to ethanol is regulated by phosphorylation of S1248, an ERK2 substrate on the L1-CD. Furthermore, differences in MAPK activity and S1248 phosphorylation determine the ethanol-sensitive or ethanol-insensitive phenotype of clonal L1-expressing NIH/3T3 cells. Finally, two substrains of C57BL mice that differ in susceptibility to ethanol teratogenesis show corresponding differences in MAPK activity.
Results
Ethanol Does Not Inhibit Adhesion of the Purified L1-ECD.
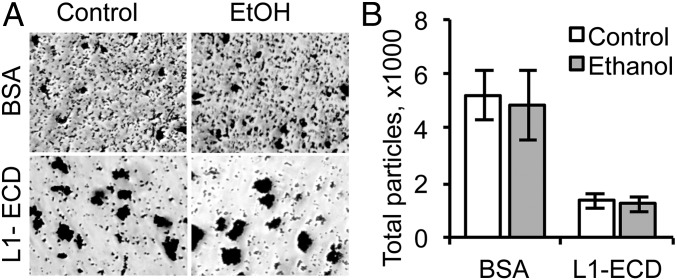
To evaluate the intrinsic sensitivity of the L1-ECD to ethanol, we studied the effects of ethanol on the adhesive properties of the purified L1-ECD attached to microspheres. Microspheres coated with mouse L1 (mL1)-ECD showed significantly greater aggregation than microspheres coated with BSA (Fig. 1). High concentrations of ethanol (100 mM) had no effect on the aggregation of microspheres coated with either BSA or mL1. These findings imply that cellular presentation is necessary for the L1-ECD to assume an ethanol-sensitive conformation.
Fig. 1.
Ethanol does not inhibit the adhesion of beads coated with the purified L1-ECD. (A) Representative phase contrast photomicrographs of 1-µm polystyrene beads coated with purified L1-ECD or BSA after agitation for 30 min in the absence or presence of 100 mM ethanol. (B) Mean ± SEM number of particles ≥1 µm per image from three independent experiments.
Inhibition of ERK2 Blocks Ethanol Inhibition of L1 Adhesion.
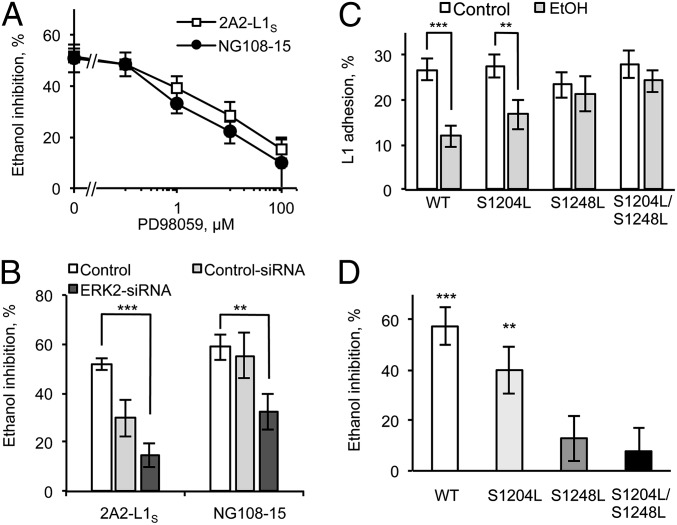
The use of computational selection and candidate gene prioritization identified MAPK among leading candidate genes for susceptibility to FAS (3). Therefore, we studied ERK1/2 modulation of ethanol sensitivity in two model systems in which ethanol always inhibits L1 adhesion: 2A2-L1s, an NIH/3T3 clonal cell line that stably expresses hL1, and NG108-15, a neural cell line that expresses mL1 after treatment with bone morphogenetic proteins (17, 19, 23, 38, 39). Pretreatment for 1 h with PD98059, a noncompetitive inhibitor of MAP kinase kinase 1 (MEK1), reduced ERK1/2 activity in 2A2-L1s (Fig. S1). PD98059 caused a dose-dependent reduction in ethanol inhibition of L1 adhesion in 2A2-L1s and NG108-15 cells (Fig. 2A and Table S1). In contrast, PD98059 did not alter L1 adhesion, except for a small decrease at the highest dose in NG108-15 cells. Similarly, transfection with an ERK2 siRNA had no effect on L1 adhesion but significantly reduced ethanol inhibition of L1 adhesion in both 2A2-L1s and NG108-15 cells (Fig. 2B). These findings indicate that inhibition of ERK2 activity can block ethanol inhibition of hL1 and mL1 adhesion in fibroblast and neural cell lines, respectively, without modulating L1 adhesion.
Fig. 2.
ERK2 phosphorylation of S1248 is necessary for ethanol inhibition of L1 adhesion. (A) PD98059 dose dependently blocks ethanol inhibition of L1 adhesion in 2A2-L1s cells and BMP-7–treated NG108-15 cells. (B) Effect of ERK2-siRNA transfection on ethanol inhibition of L1 adhesion in BMP-7–treated NG108-15 and 2A2-L1s cells. Bars represent results of treatment with Lipofectamine, the transfection reagent (Control), control-siRNA, or ERK2-siRNA. **P < 0.01, n = 4; ***P < 0.001, n = 8. (C and D). Effect of ethanol on cell adhesion in WT and mutant-L1–expressing NIH/3T3 cells. Constructs containing L1-WT, L1-S1204S, L1-S1248S, or L1-S1204S/L1248S were transiently transfected in NIH/3T3 cells as shown in Fig. S2. (D) Ethanol inhibition of cell adhesion calculated from paired experiments shown in C. Data shown are the mean ± SEM for percent cell adhesion (C) or percent ethanol inhibition of cell adhesion (D) in 9–14 independent experiments; **P = 0.0020, ***P < 0.0001.
ERK2 Phosphorylation of S1248 on the L1 Cytoplasmic Domain Is Necessary for Ethanol Sensitivity.
There are two known ERK2 phosphorylation sites at S1204 and S1248 on the hL1-CD (40). To determine whether phosphorylation of these sites is necessary for L1 sensitivity to ethanol, we transiently transfected NIH/3T3 cells with plasmids containing hL1 constructs in which S1204, S1248, or both were mutated to leucine to prevent phosphorylation (32). Cells expressing L1-S1204L showed normal L1 expression and adhesion (Fig. S2) and a statistically insignificant reduction in sensitivity to ethanol (Fig. 2 C and D). Cells expressing L1-S1248L or L1-S1204L/S1248L also showed normal L1 expression and adhesion but were insensitive to high concentrations of ethanol (100 mM). These data provide compelling evidence that ERK2 phosphorylation of the L1-CD is necessary for ethanol inhibition of L1 adhesion.
ERK2 Activity Differs in Ethanol-Sensitive and Insensitive Cell Lines.
We next asked whether differences in ERK2 activity could account for the ethanol-sensitive or ethanol-insensitive phenotype of stably transfected hL1-expressing NIH/3T3 clonal cell lines (19). The normalized ratio of phosphorylated ERK1/2 (pERK1/2) to total ERK1/2, as measured by ELISA, was significantly higher in two ethanol-sensitive clonal cell lines (0.94 ± 0.01) than in three ethanol-insensitive clonal cell lines (0.70 ± 0.03, n = 18, P < 0.0001) (Fig. 3A). Furthermore, ERK1/2 activity, as measured by the phosphorylation of myelin basic protein, was significantly higher in the ethanol-sensitive 2A2-L1s cell line (8.2 ± 0.4 OD units per mg of protein, n = 4) than in the ethanol-insensitive 3C3-L1i cell line (5.7 ± 0.4 OD units per mg of protein, n = 4, P = 0.0057) (Fig. S3B). Of note, treatment of 2A2-L1s cells with 100 µM PD98059, which blocked ethanol sensitivity, also reduced ERK1/2 activity to levels observed in 3C3-L1i cells (Figs. S1A and S3B). Hence, constitutive differences in ethanol sensitivity in L1-expressing clonal cell lines are associated with differences in ERK1/2 activity.
Fig. 3.
ERK1/2 activity and phosphorylation of S1248 determine ethanol sensitivity in L1-expressing clonal NIH/3T3 cell lines. (A) Ratio of pERK1/2 to ERK1/2, determined by ELISA, n = 13–18. Pooled values of ERK1/2 phosphorylation for the two ethanol-sensitive cell lines were significantly higher than in the three ethanol-insensitive cell lines (F = 16.0; P < 0.0001). (B and C) L1 was immunoprecipitated from 2A2-L1s and 3C3-L1i cells or NIH/3T3 cells transiently transfected with wild-type L1 (WT) or L1-S1248L using mAb 5G3. Immunoblots were prepared by using polyclonal antibody SC-1508, which recognizes the L1-CD, and antiserum dp-S1248, which preferentially recognizes the nonphosphorylated form of L1-S1248. The ratio of bands labeled by SC-1508 and dp-S1248 was normalized in each gel to values from NIH/3T3 cells transiently transfected with wild-type L1 (WT) to control for variable exposure on individual Western blots. (B) Representative gel showing immunolabeling of L1 immunoprecipitated from NIH/3T3 cells transiently transfected with wild-type L1 (WT) or L1-S1248L (S1248L) and from stably transfected 2A2-L1s (2A2) and 3C3-L1i (3C3) cells. IP indicates the immunoprecipitating antibody 5G3; IB indicates the antibodies dp-S1248 and SC-1508 used for the immunoblot. (C) Bars represent the mean ± SEM ratio of dp-S1248 (nonphosphorylated L1-S1248) to SC-1508 (total L1) immunolabeling from eight independent experiments. Lower values indicate higher levels of phosphorylation. *P = 0.02; **P = 0.003. (D) The effect of transfection of a constitutively active MEK1 (MEK1*) on L1 adhesion in the absence and presence of 100 mM ethanol in 2A2-L1s cells, *P = 0.0350, n = 4; **P = 0.0017, n = 8; ***P < 0.0001, n = 4; and in 3C3-L1i cells, ***P < 0.0001, n = 14. (E) Effect of MEK1* transfection in 2A2-L1s (PolyFect, n = 4; MEK1, n = 8) and 3C3-L1i cells (PolyFect, n = 4; MEK1, n = 14) on the percentage of ethanol inhibition of L1 adhesion calculated from the indicated number of paired experiments. ***P = 0.0005.
L1-S1248 Phosphorylation Is Greater in Ethanol-Sensitive than in Ethanol-Insensitive NIH/3T3 Cells.
If L1-S1248 phosphorylation is necessary for ethanol sensitivity of L1 and differences in ERK2 activity account for the differences in the ethanol sensitivity of 2A2-L1s and 3C3-L1i cells, then levels of S1248 phosphorylation (pS1248) should be higher in 2A2-L1s than in 3C3-L1i cells. To test this hypothesis, we developed a rabbit antiserum, dp-S1248, that preferentially recognizes an oligopeptide (L1-D1242-L1256) containing nonphosphorylated versus phosphorylated L1-S1248 (Fig. S4) and immunoprecipitates from cells transfected with L1-S1248L versus wild-type L1-S1248 (Fig. 3 B and C). Antiserum dp-S1248 immunolabeling was 46.7 ± 5.1% lower (P < 0.0001, n = 8) in L1 immunoprecipitates from 2A2-L1s cells than in those from 3C3-L1i cells. This dp-S1248 immunolabeling was preferentially inhibited when antibody was absorbed with the nonphosphorylated oligopeptide compared with the phosphorylated oligopeptide (Fig. S4 C and D). These findings suggest that S1248 phosphorylation levels are higher in ethanol-sensitive 2A2-L1s cells than in ethanol-insensitive 3C3-L1i cells.
Ethanol-Insensitive Phenotype Can Be Transformed by Increasing ERK1/2 Activity.
If constitutive ERK1/2 activity determines the sensitivity of L1 to ethanol and inhibition of ERK1/2 activity blocks ethanol inhibition of L1 adhesion, then increasing ERK1/2 activity should render ethanol-insensitive cells sensitive to ethanol. To test this hypothesis, we transiently transfected the 2A2-L1s and 3C3-L1i cell lines with constitutively active MEK1 (MEK1*) (41, 42), the kinase that phosphorylates ERK1/2. Transfection with MEK1* significantly increased ERK1/2 phosphorylation in 3C3-L1i cells to levels observed in control 2A2-L1s cells, but did not increase ERK2 phosphorylation in 2A2-L1s cells (Fig. S5). MEK1* transfection did not alter L1 adhesion in either cell line and did not enhance ethanol inhibition of L1 adhesion in 2A2-L1s cells (Fig. 3 D and E). However, after transfection with MEK1*, ethanol robustly inhibited L1 adhesion in 3C3-L1i cells (43.7 ± 5.3% inhibition; P = 0.0005, n = 4–14). These findings support the hypothesis that ERK2 phosphorylation of L1 modulates its sensitivity to ethanol.
ERK1/2 Activity Differs in Mouse Substrains That Differ in Sensitivity to Ethanol Teratogenesis.
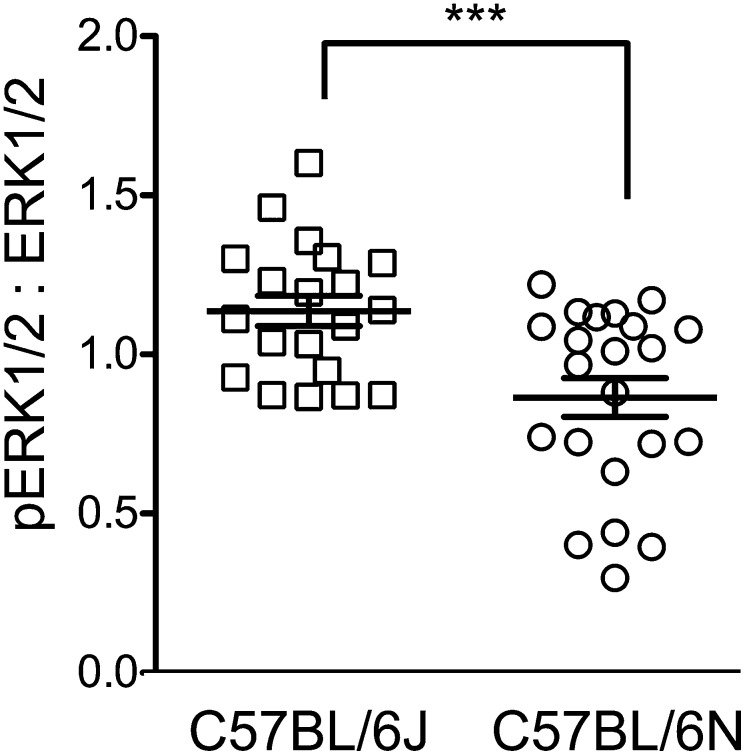
We next asked whether ERK1/2 activity correlates with sensitivity to ethanol teratogenesis in animals. C57BL/6J (6J) mice from the Jackson Laboratory and C57BL/6Nhsd (6N) mice from Harlan Laboratories are derived from the same strain but have diverged slightly after hundreds of generations of inbreeding. Both substrains were administered one dose of ethanol (2.9 g/kg) at the beginning of gestational day (GD) 7 and another dose 4 h later. This well-established ethanol exposure paradigm induces ocular defects similar to those observed in children with FAS (43). Examination of GD 14 fetuses from ethanol-exposed dams revealed right ocular defects in 50.9% of 6J mice (n = 55) and 28.2% of 6N mice (n = 71; P = 0.0104, Fisher’s exact test) and left ocular defects in 41.8% of 6J mice and 11.3% of 6N mice (P = 0.0001). Blood alcohol concentrations 30 min after the second injection did not differ significantly between the two substrains (6J: 420 ± 18 mg/dL, n = 7; 6N: 403 ± 33 n = 5; P = 0.63). These data demonstrate that 6J mice are significantly more susceptible to the teratogenic effects of early ethanol exposure than 6N mice. Of note, levels of pERK1/2, measured by ELISA, were 31% higher in GD 10 embryos from untreated, ethanol-sensitive 6J mice (114 ± 4 relative units) than in those from the comparably staged less sensitive 6N mice (87 ± 6 relative units; n = 26; P = 0.0004, ANOVA) (Fig. 4). These differences in ERK1/2 activity are comparable to those measured in ethanol-sensitive and ethanol-insensitive NIH/3T3 clonal cell lines (Fig. 3A and Fig. S3B).
Fig. 4.
ERK1/2 phosphorylation is higher in C57BL/6J than in C57BL/6N mouse substrains. Total ERK1/2 and pERK1/2 were measured by ELISA in GD10 embryos from the ethanol-sensitive 6J mice (n = 26) and less-sensitive 6N mice (n = 22). Each point represents the pERK1/2 to ERK1/2 ratio for an individual embryo normalized to values from a pooled sample of GD10 embryos from both substrains. Also shown is the mean ± SEM for the pERK1/2 to ERK1/2 ratios for each group, ***P = 0.0004.
Discussion
A major finding of this work is that ethanol inhibition of L1 adhesion has two requirements: L1 must be expressed in cells and must be phosphorylated by ERK2 at S1248 in the L1-CD. Ethanol had no effect on adhesion mediated by beads coated with the purified L1-ECD. It appears likely that presentation of purified L1 outside of a cellular context disrupts the specific conformation of the alcohol binding pocket that is required for ethanol inhibition of L1 adhesion. This observation is consistent with our recent report that an alcohol binding pocket at the Ig1–Ig4 interface of L1 is susceptible to minor structural modifications (31, 44).
Ethanol inhibition of L1 adhesion in cells was strongly inhibited by 1 µM PD98059, a concentration that selectively inhibits MEK1 (45), the kinase that phosphorylates and activates ERK1/2. The effect of PD98059 was evident at similar concentrations in fibroblasts expressing hL1 and neural cells expressing mL1, indicating that MEK1 inhibition affects L1 sensitivity to ethanol across multiple cell lineages. Although kinase inhibitors are never perfectly selective for individual kinases, targeted knockdown of ERK2 also reduced L1 sensitivity to ethanol. Taken together, these experiments indicate that ERK2 activation is necessary for ethanol inhibition of L1 adhesion, but do not establish whether ERK2 acts by directly phosphorylating the L1-CD or by phosphorylating other proteins that interact with L1 (9). To address this question, we studied NIH/3T3 cells transiently transfected with L1 constructs in which S1204, S1248, or both were mutated to leucine residues, thereby preventing phosphorylation of the two known ERK2 substrates on the L1-CD. The S1204L mutation did not significantly reduce ethanol inhibition of L1 adhesion, whereas the S1248L or the S1204L/S1248L mutations abolished ethanol inhibition of L1 adhesion. These findings indicate that ERK2 regulates L1 sensitivity to ethanol by directly phosphorylating S1248 in the L1-CD.
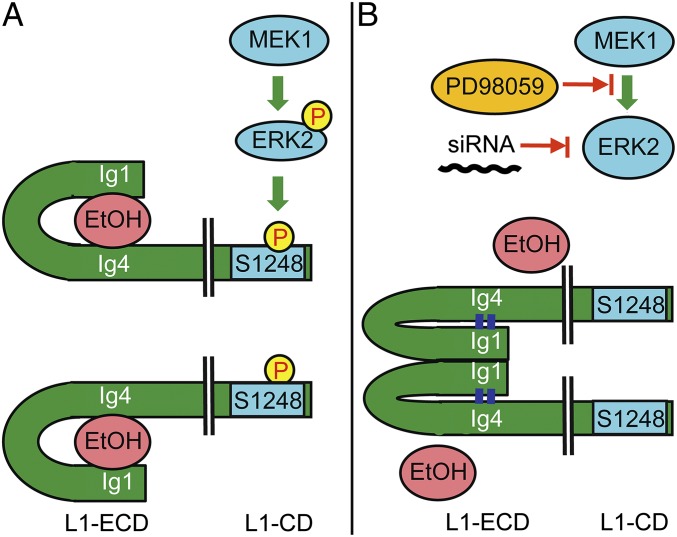
The cytoplasmic domain is not necessary for the adhesive properties of neuroglian, the Drosophila L1 homolog (46); however, modifications to the neuroglian-CD can influence its adhesion through inside-out signaling (36). Phosphorylation of the L1-CD has also been shown to modify the structure and function of the L1-ECD. Chen et al. (35) reported that phosphorylation of T1172 in the L1-CD altered L1 binding to integrins and masked or unmasked specific epitopes in the L1-ECD. Of note, L1 adhesion in the absence of ethanol was not altered by pharmacological inhibition of MEK1, ERK2 knockdown, or mutation of S1248 to leucine, although each of these interventions reduced or abolished ethanol inhibition of L1 adhesion. Presumably, dephosphophorylation of S1248 alters the conformation of an alcohol binding pocket on the L1-ECD sufficiently to prevent the interaction of ethanol with L1, while preserving structural features of the L1-ECD that are necessary for L1 homophilic adhesion (Fig. 5).
Fig. 5.
ERK2 phosphorylation of S1248 in the L1-CD is necessary for ethanol inhibition of L1 adhesion. (A) The L1 extracellular domain (L1-ECD) folds into a horseshoe structure that favors L1 adhesion. An alcohol binding pocket has been identified at the interface between Ig1 and Ig4, and ethanol appears to inhibit L1 adhesion by disrupting this horseshoe structure (31, 44). Phosphorylation of S1248 by ERK2 is proposed to induce a conformational change in the L1-ECD (inside-out signaling) that favors ethanol binding within this pocket. (B) PD98059, an inhibitor of MEK1/2, or knockdown of ERK2 using siRNA decrease ERK2 phosphorylation of S1248 and reduce ethanol inhibition of L1 adhesion. Likewise, mutation of S1248 to L1248 prevents phosphorylation at this site and abolishes ethanol inhibition of L1 adhesion.
Phosphorylation of S1248 is necessary for ethanol inhibition of L1 adhesion; however, our data do not exclude a role for phosphorylation of other residues in the L1-CD in regulating ethanol sensitivity. Both T1247 and S1248 are required for L1-mediated ERK activation and cell motility (47), and conceivably a combination of phosphorylation and dephosphorylation of a variety of cytoplasmic residues is required for ethanol inhibition of L1 adhesion. Hence, a variety of kinases or phosphatases could separately influence the ethanol sensitivity of L1 by modulating the phosphorylation state of key residues in the L1-CD. The identification of loci that regulate ethanol sensitivity, but not L1 function, might facilitate the rational design of drugs that block ethanol neurotoxicity.
Shortly after reporting that ethanol inhibits L1 adhesion, we learned that not all L1-expressing cells are sensitive to ethanol. A series of cell lines that had been clonally selected after the transfection of a single NIH/3T3 stock with hL1 (19) consistently expressed either an ethanol-sensitive or ethanol-insensitive phenotype over more than a decade. L1 expression and L1 adhesion did not differ between ethanol-sensitive and ethanol-insensitive cells; however, purified Fab fragments from a polyclonal antibody blocked L1 adhesion in the ethanol-sensitive cells, but not in the ethanol-insensitive cells (19). We speculated that the L1-ECD required a particular conformation for ethanol sensitivity and that the conformation of the L1-ECD in ethanol-insensitive cells masked epitopes recognized by this polyclonal antibody (19).
Our current data suggest that constitutive differences in ERK2 activity contribute to the ethanol-sensitive or ethanol-insensitive phenotype of these clonal cell lines. ERK1/2 activity was approximately one-third higher in two ethanol-sensitive cell lines than in three ethanol-insensitive cell lines. These findings could not be attributed to differences in cell density or cell–cell interactions before assay (48), because ethanol-sensitive and ethanol-insensitive cell lines were harvested at the same density. These modest increases in ERK1/2 activity were amplified at the level of substrate phosphorylation; S1248 phosphorylation in ethanol-sensitive 2A2-L1s cells was approximately double that of ethanol-insensitive 3C3-L1i cells. Finally, the long-standing ethanol-insensitive phenotype of 3C3-L1i cells was transformed to an ethanol-sensitive phenotype by increasing ERK2 activity through the expression of a constitutively active MEK1* molecule. These findings indicate that the ERK1/2 signaling pathway is an important determinant of L1 sensitivity to ethanol in clonal cell lines. Differential phosphorylation of S1248 and other cytoplasmic residues might also explain the inability of ethanol to inhibit L1 adhesion in myeloma and insect cells (26, 27).
To demonstrate the relevance of our findings to ethanol teratogenesis, we also identified differences in susceptibility to ethanol teratogenesis in two highly related mouse substrains. Like our clonal cell lines, the two substrains of C57BL/6 mice were derived from a common stock and likely differed only minimally in their genetic and epigenetic profiles. The C56BL/6J mice proved more susceptible to ethanol-induced ocular defects than their C56BL/6N counterparts. Blood alcohol levels were comparable in the two strains, indicating that their differential susceptibility to ethanol teratogenesis was not related to differences in ethanol pharmacokinetics. In contrast, ERK1/2 activity was significantly higher in the more sensitive C56BL/6J mice than in the less sensitive C57BL/6N mice. Thus, increased ERK1/2 activity is associated with increased ethanol inhibition of L1 adhesion in clonal cell lines and increased susceptibility to ethanol teratogenesis in mice. Taken together, these data suggest that genes for ERK2 and the signaling molecules that regulate ERK2 activity might influence genetic susceptibility to FASD. The identification of additional regulatory pathways for L1 sensitivity to ethanol might further expand this repertoire of candidate genes.
Materials and Methods
Bead Aggregation Assay for L1 Adhesion.
Mouse L1-ECD-Fc (mL1-Ig1-6-Fn3-1–5-Fc) was prepared as described (44, 49) and digested with AcTEV Protease (Invitrogen) to remove Fc. Polybead Carboxylate Blue Dyed 1-μm microspheres (Polysciences Inc.) were coated with mL1-ECD protein at a ratio of 8 × 109 beads per μg of L1 protein or 8 × 109 beads per 3 μg of BSA using the Glutaraldehyde Kit (Polysciences Inc.), according to the manual. Beads were suspended in PBS in 96-well plates (1 × 108 beads per 100 μL of PBS for each well) in the absence and presence of 100 mM ethanol and agitated at 60 rpm for 30 min at room temperature on a rotary shaker (Stovall Life Science). Plates were set for 5 min on an inverted-stage Nikon/Eclipse-MA100 microscope to allow beads to settle, and images were acquired at 200× magnification by using Openlab software (PerkinElmer). The total number of particles ≥1.0 μm in each image was analyzed with National Institutes of Health ImageJ software by using the particle analysis function (50).
Cell Culture.
We studied five stably transfected hL1-expressing NIH/3T3 clonal cell lines, including two—2A2-L1s and 2B2-L1s—in which ethanol inhibited L1 adhesion (ethanol-sensitive, denoted by L1s) and three—3C3-L1i, 50FE-L1i and 3E2-L1i—in which ethanol did not inhibit L1 adhesion (ethanol insensitive, denoted by L1i); cells were cultured as described (18, 19). In other experiments, NIH/3T3 cells were transiently transfected with plasmid DNA containing WT-L1, mutant L1, or MEK1* plasmid, (Upstate Biotech) and cultured for 24 h before use in experiments (31). The neuroblastoma × glioma hybrid cell line NG108-15 was cultured as described (38, 51). Three days before cell adhesion assays, cells were switched to serum-free medium (51) with or without 10 ng/mL BMP-7 (Creative BioMolecules) to induce the expression of mL1 (17, 39, 51). The MEK1 inhibitor PD98059 was dissolved in DMSO and applied directly into culture medium to the designated final concentration. The L1-S1204L, L1-S1248L, and L1-S1204L/S1248L mutants were kind gifts from B. Schmitz (Department of Biochemistry, University of Bonn, Bonn, Germany) (32).
siRNA Transfection of NIH/3T3 and NG108-15 Cells.
Cells were transfected with ERK2-siRNA and negative control siRNA (Thermo Scientific Dharmacon RNAi Technologies) using Lipofectamine 2000 (Invitrogen). Forty-eight hours after transfection, the cells were harvested for cell adhesion assays or to examine ERK2 expression with immunofluorescence microscopy and Western blotting by using a mouse anti-ERK2 mAb (clone 05–157, Millipore).
Protein Analysis for Cultured Cells.
Cells were grown to 75–85% confluence, harvested in PBS plus 2 mM EDTA, and pelleted by centrifugation. Cells were resuspended in Nonidet P-40 lysis buffer [150 mM NaCl, 50 mM Tris at pH 8.0, and 1.0% (vol/vol) Nonidet P-40] supplemented with 1/100 (vol/vol) Protease Arrest and 1/100 (vol/vol) Phosphatase Inhibitor Mixture (Calbiochem), incubated on ice for 5 min, and centrifuged at 4 °C at 16,000 × g for 15 min to remove the insoluble fraction. Protein was separated and analyzed by Western blot using goat polyclonal antibody SC-1508 for L1 (31), rabbit polyclonal anti-pERK1/2 antibody (SC-23759-R), mouse monoclonal anti-ERK1/2 antibody (SC-135900) (all from Santa Cruz Biotechnology), and rabbit polyclonal ab11317 for γ-tubulin (Abcam).
Mouse Embryo Collection and Assay.
The 6J mice were obtained from The Jackson Laboratories, and the 6N were obtained from Harlan Laboratories. The mice were bred by placing one or two females with a single male of the same substrain for 1 h and then examined for the presence of a copulation plug. The beginning of the 1-h mating period in which a copulation plug was found was designated as GD 0. On GD 10, the dams were euthanized by CO2 asphyxiation, and the embryos were removed and dissected from the extraembryonic tissue. After staging each embryo by counting the number of somite pairs, embryos were quickly frozen and stored in PBS with 5 mM EDTA at −80 °C. Only embryos having 26–30 somites were used for analyses. These embryos were thawed on ice and placed directly in 200–300 µL of Nonidet P-40 lysis buffer with the protease and phosphatase inhibitors above, lysed by vigorous pipetting and vortexing, and centrifuged at 16,000 × g to collect supernatant for pERK1/2 and ERK1/2 analysis below. All animal use protocols were approved by the University of North Carolina at Chapel Hill Institutional Animal Care and Use Committee (IACUC).
Production of an Antiserum, dp-S1248, to the Nonphosphorylated Form of S1248.
A rabbit antiserum was generated by Antagene by immunization with L1-D1242-L1256 [Cys-DSSGAT(p)SPINPAVAL], a phosphopeptide derived from the L1-CD. The antiserum was passed through a phosphopeptide column [Cys-DSSGAT(p)SPINPAVAL], and the eluent was then loaded onto the corresponding nonphosphopeptide column. Specificity of the antisera for S1248 or pS1248 was tested by using dot blots impregnated with serial dilutions of the immunizing phosphorylated and nonphosphorylated peptides. Dot blots were air dried on a PVDF membrane and blocked with 3% (wt/vol) BSA in TBST, incubated with HRP-conjugated secondary antibody, and visualized by using chemiluminescence (Millipore) (31). The effluent from the nonphosphopeptide column preferentially labeled the nonphosphorylated peptide; this antiserum is referred to as dp-S1248. The flow-through from the nonphosphopeptide column did not preferentially label the phosphorylated peptide.
ERK1/2 Phosphorylation and Activity Assays.
Phospho-ERK1/2 (pERK1/2) and total ERK1/2 in whole-cell lysates from cultured cells and mouse embryos were measured with pERK1/2 (pThr185/pTyr187) and ERK1/2 ELISA kits, respectively (Sigma-Aldrich), according to the manuals. Absorbance at 450 nm was normalized to protein content of the whole-cell lysates, as determined by the Bradford method (31), and expressed as mean ± SEM optical density (O.D.) per mg of protein. Alternatively, pERK1/2 levels were measured by immunoblotting of whole-cell extracts with mouse polyclonal antibody SC23759 against pERK1/2 and mouse mAb SC135900 against total ERK1/2 (Santa Cruz). ERK1/2 activity in 2A2-L1s and 3C3-L1i cell lines was measured by using a MAPK (ERK1/2) Activity Assay Kit (Millipore), which assays the phosphorylation of myelin basic protein (MBP) by ERK1/2 in total cell lysates. ERK1/2 activity was expressed as O.D. units at 450 nm per mg of protein of total cell lysates, and the relative level of ERK1/2 activity in 2A2-L1s and 3C3-L1i cell lines was calculated as the mean ± SEM ratio of ERK1/2 activities in the two cell lines.
Cell Surface Expression of L1.
L1 immunostaining was performed in attached live cells and visualized by using confocal microscopy, as described (31), using 5G3 mAb, which recognizes an epitope overlapping the Ig1 and Ig2 domains of L1 (35). Cell surface expression of L1 was quantified by flow cytometry with mAb 5G3 (31). Data were presented as the geometric mean ± SEM.
Cell Aggregation Assay.
L1-mediated cell–cell adhesion (L1 adhesion) in NIH/3T3 cells and NG108-15 cells was determined by separating cells into single-cell suspensions, agitating, and measuring the percentage of adherent cells using phase contrast microscopy, as described (19, 31, 38). For comparisons of L1 adhesion between ethanol-sensitive and ethanol-insensitive cell lines, all cells were harvested at the same density. We used 100 mM ethanol, a concentration much higher than the half-maximal concentration of ∼5 mM (18, 31), to demonstrate the robustness of the antagonistic effects of kinase inhibitors, gene knockdown, and L1 point mutation on ethanol inhibition of L1 adhesion.
Statistical Analysis.
Data are expressed as mean ± SEM, except as noted. Statistical analysis was performed by using the ANOVA with Tukey’s correction for multiple comparisons and two-tailed paired t tests using Prism 5 (Graphpad Software). Statistical significance was defined as P < 0.05.
Supplementary Material
Acknowledgments
This work was supported by the National Institute on Alcohol Abuse and Alcoholism (NIAAA) Grant R01-AA12974 (to M.E.C.); NIAAA Grant U24-AA014811, as a component of the Collaborative Initiative on Fetal Alcohol Spectrum Disorders (CIFASD) (to M.E.C.); the Medical Research Service, Department of Veterans Affairs (M.E.C.); NIAAA Grant AA017124 (CIFASD) (to K.K.S.); NIAAA Grant AA011605 (to K.K.S.); and NIAAA Grant K99/R00-AA018697 (to S.E.P.).
Footnotes
The authors declare no conflict of interest.
†This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1221386110/-/DCSupplemental.
See Commentary on page 5285.
References
- 1.Abel EL, Sokol RJ. Fetal alcohol syndrome is now leading cause of mental retardation. Lancet. 1986;2(8517):1222. doi: 10.1016/s0140-6736(86)92234-8. [DOI] [PubMed] [Google Scholar]
- 2.May PA, et al. Prevalence and epidemiologic characteristics of FASD from various research methods with an emphasis on recent in-school studies. Dev Disabil Res Rev. 2009;15(3):176–192. doi: 10.1002/ddrr.68. [DOI] [PubMed] [Google Scholar]
- 3.Lombard Z, et al. Computational selection and prioritization of candidate genes for fetal alcohol syndrome. BMC Genomics. 2007;8:389. doi: 10.1186/1471-2164-8-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramsay M. Genetic and epigenetic insights into fetal alcohol spectrum disorders. Genome Med. 2010;2(4):27. doi: 10.1186/gm148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Valenzuela CF, Morton RA, Diaz MR, Topper L. Does moderate drinking harm the fetal brain? Insights from animal models. Trends Neurosci. 2012;35(5):284–292. doi: 10.1016/j.tins.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Y, Ozturk NC, Ni L, Goodlett C, Zhou FC. Strain differences in developmental vulnerability to alcohol exposure via embryo culture in mice. Alcohol Clin Exp Res. 2011;35(7):1293–1304. doi: 10.1111/j.1530-0277.2011.01465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Warren KR, Li TK. Genetic polymorphisms: Impact on the risk of fetal alcohol spectrum disorders. Birth Defects Res A Clin Mol Teratol. 2005;73(4):195–203. doi: 10.1002/bdra.20125. [DOI] [PubMed] [Google Scholar]
- 8.Streissguth AP, Dehaene P. Fetal alcohol syndrome in twins of alcoholic mothers: Concordance of diagnosis and IQ. Am J Med Genet. 1993;47(6):857–861. doi: 10.1002/ajmg.1320470612. [DOI] [PubMed] [Google Scholar]
- 9.Maness PF, Schachner M. Neural recognition molecules of the immunoglobulin superfamily: Signaling transducers of axon guidance and neuronal migration. Nat Neurosci. 2007;10(1):19–26. doi: 10.1038/nn1827. [DOI] [PubMed] [Google Scholar]
- 10.Clarren SK, Smith DW. The fetal alcohol syndrome. N Engl J Med. 1978;298(19):1063–1067. doi: 10.1056/NEJM197805112981906. [DOI] [PubMed] [Google Scholar]
- 11.Wozniak JR, Muetzel RL. What does diffusion tensor imaging reveal about the brain and cognition in fetal alcohol spectrum disorders? Neuropsychol Rev. 2011;21(2):133–147. doi: 10.1007/s11065-011-9162-1. [DOI] [PubMed] [Google Scholar]
- 12.O’Leary-Moore SK, Parnell SE, Lipinski RJ, Sulik KK. Magnetic resonance-based imaging in animal models of fetal alcohol spectrum disorder. Neuropsychol Rev. 2011;21(2):167–185. doi: 10.1007/s11065-011-9164-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fransen E, et al. CRASH syndrome: Clinical spectrum of corpus callosum hypoplasia, retardation, adducted thumbs, spastic paraparesis and hydrocephalus due to mutations in one single gene, L1. Eur J Hum Genet. 1995;3(5):273–284. doi: 10.1159/000472311. [DOI] [PubMed] [Google Scholar]
- 14.Schäfer MK, et al. L1 syndrome mutations impair neuronal L1 function at different levels by divergent mechanisms. Neurobiol Dis. 2010;40(1):222–237. doi: 10.1016/j.nbd.2010.05.029. [DOI] [PubMed] [Google Scholar]
- 15.De Angelis E, Watkins A, Schäfer M, Brümmendorf T, Kenwrick S. Disease-associated mutations in L1 CAM interfere with ligand interactions and cell-surface expression. Hum Mol Genet. 2002;11(1):1–12. doi: 10.1093/hmg/11.1.1. [DOI] [PubMed] [Google Scholar]
- 16.Kenwrick S, Watkins A, De Angelis E. Neural cell recognition molecule L1: Relating biological complexity to human disease mutations. Hum Mol Genet. 2000;9(6):879–886. doi: 10.1093/hmg/9.6.879. [DOI] [PubMed] [Google Scholar]
- 17.Charness ME, Safran RM, Perides G. Ethanol inhibits neural cell-cell adhesion. J Biol Chem. 1994;269(12):9304–9309. [PubMed] [Google Scholar]
- 18.Ramanathan R, Wilkemeyer MF, Mittal B, Perides G, Charness ME. Alcohol inhibits cell-cell adhesion mediated by human L1. J Cell Biol. 1996;133(2):381–390. doi: 10.1083/jcb.133.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilkemeyer MF, Charness ME. Characterization of ethanol-sensitive and insensitive fibroblast cell lines expressing human L1. J Neurochem. 1998;71(6):2382–2391. doi: 10.1046/j.1471-4159.1998.71062382.x. [DOI] [PubMed] [Google Scholar]
- 20.Chen S-Y, Wilkemeyer MF, Sulik KK, Charness ME. Octanol antagonism of ethanol teratogenesis. FASEB J. 2001;15(9):1649–1651. doi: 10.1096/fj.00-0862fje. [DOI] [PubMed] [Google Scholar]
- 21.Chen SY, Charness ME, Wilkemeyer MF, Sulik KK. Peptide-mediated protection from ethanol-induced neural tube defects. Dev Neurosci. 2005;27(1):13–19. doi: 10.1159/000084528. [DOI] [PubMed] [Google Scholar]
- 22.Wilkemeyer MF, et al. Differential effects of ethanol antagonism and neuroprotection in peptide fragment NAPVSIPQ prevention of ethanol-induced developmental toxicity. Proc Natl Acad Sci USA. 2003;100(14):8543–8548. doi: 10.1073/pnas.1331636100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilkemeyer MF, Sebastian AB, Smith SA, Charness ME. Antagonists of alcohol inhibition of cell adhesion. Proc Natl Acad Sci USA. 2000;97(7):3690–3695. doi: 10.1073/pnas.050545697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spong CY, Abebe DT, Gozes I, Brenneman DE, Hill JM. Prevention of fetal demise and growth restriction in a mouse model of fetal alcohol syndrome. J Pharmacol Exp Ther. 2001;297(2):774–779. [PubMed] [Google Scholar]
- 25.Zhou FC, Sari Y, Powrozek TA, Spong CY. A neuroprotective peptide antagonizes fetal alcohol exposure-compromised brain growth. J Mol Neurosci. 2004;24(2):189–199. doi: 10.1385/JMN:24:2:189. [DOI] [PubMed] [Google Scholar]
- 26.Vallejo Y, Hortsch M, Dubreuil RR. Ethanol does not inhibit the adhesive activity of Drosophila neuroglian or human L1 in Drosophila S2 tissue culture cells. J Biol Chem. 1997;272(18):12244–12247. doi: 10.1074/jbc.272.18.12244. [DOI] [PubMed] [Google Scholar]
- 27.Bearer CF, Swick AR, O’Riordan MA, Cheng G. Ethanol inhibits L1-mediated neurite outgrowth in postnatal rat cerebellar granule cells. J Biol Chem. 1999;274(19):13264–13270. doi: 10.1074/jbc.274.19.13264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haspel J, Grumet M. The L1CAM extracellular region: A multi-domain protein with modular and cooperative binding modes. Front Biosci. 2003;8:s1210–s1225. doi: 10.2741/1108. [DOI] [PubMed] [Google Scholar]
- 29.Gouveia RM, Gomes CM, Sousa M, Alves PM, Costa J. Kinetic analysis of L1 homophilic interaction: Role of the first four immunoglobulin domains and implications on binding mechanism. J Biol Chem. 2008;283(42):28038–28047. doi: 10.1074/jbc.M804991200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schürmann G, Haspel J, Grumet M, Erickson HP. Cell adhesion molecule L1 in folded (horseshoe) and extended conformations. Mol Biol Cell. 2001;12(6):1765–1773. doi: 10.1091/mbc.12.6.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dou X, Menkari CE, Shanmugasundararaj S, Miller KW, Charness ME. Two alcohol binding residues interact across a domain interface of the L1 neural cell adhesion molecule and regulate cell adhesion. J Biol Chem. 2011;286(18):16131–16139. doi: 10.1074/jbc.M110.209254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schultheis M, Diestel S, Schmitz B. The role of cytoplasmic serine residues of the cell adhesion molecule L1 in neurite outgrowth, endocytosis, and cell migration. Cell Mol Neurobiol. 2007;27(1):11–31. doi: 10.1007/s10571-006-9113-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nagaraj K, Hortsch M. Phosphorylation of L1-type cell-adhesion molecules—ankyrins away! Trends Biochem Sci. 2006;31(10):544–546. doi: 10.1016/j.tibs.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 34.Whittard JD, Sakurai T, Cassella MR, Gazdoiu M, Felsenfeld DP. MAP kinase pathway-dependent phosphorylation of the L1-CAM ankyrin binding site regulates neuronal growth. Mol Biol Cell. 2006;17(6):2696–2706. doi: 10.1091/mbc.E06-01-0090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen MM, et al. Inside-out regulation of L1 conformation, integrin binding, proteolysis, and concomitant cell migration. Mol Biol Cell. 2010;21(10):1671–1685. doi: 10.1091/mbc.E09-10-0900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hortsch M, et al. Structural requirements for outside-in and inside-out signaling by Drosophila neuroglian, a member of the L1 family of cell adhesion molecules. J Cell Biol. 1998;142(1):251–261. doi: 10.1083/jcb.142.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen MM, Lee CY, Leland HA, Silletti S. Modification of the L1-CAM carboxy-terminus in pancreatic adenocarcinoma cells. Tumour Biol. 2011;32(2):347–357. doi: 10.1007/s13277-010-0127-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilkemeyer MF, Pajerski M, Charness ME. Alcohol inhibition of cell adhesion in BMP-treated NG108-15 cells. Alcohol Clin Exp Res. 1999;23(11):1711–1720. [PubMed] [Google Scholar]
- 39.Perides G, Hu G, Rueger DC, Charness ME. Osteogenic protein-1 regulates L1 and neural cell adhesion molecule gene expression in neural cells. J Biol Chem. 1993;268(33):25197–25205. [PubMed] [Google Scholar]
- 40.Schaefer AW, et al. Activation of the MAPK signal cascade by the neural cell adhesion molecule L1 requires L1 internalization. J Biol Chem. 1999;274(53):37965–37973. doi: 10.1074/jbc.274.53.37965. [DOI] [PubMed] [Google Scholar]
- 41.Mansour SJ, et al. Transformation of mammalian cells by constitutively active MAP kinase kinase. Science. 1994;265(5174):966–970. doi: 10.1126/science.8052857. [DOI] [PubMed] [Google Scholar]
- 42.Catling AD, Schaeffer HJ, Reuter CW, Reddy GR, Weber MJ. A proline-rich sequence unique to MEK1 and MEK2 is required for raf binding and regulates MEK function. Mol Cell Biol. 1995;15(10):5214–5225. doi: 10.1128/mcb.15.10.5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parnell SE, et al. Magnetic resonance microscopy defines ethanol-induced brain abnormalities in prenatal mice: Effects of acute insult on gestational day 8. Alcohol Clin Exp Res. 2009;33(6):1001–1011. doi: 10.1111/j.1530-0277.2009.00921.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arevalo E, et al. An alcohol binding site on the neural cell adhesion molecule L1. Proc Natl Acad Sci USA. 2008;105(1):371–375. doi: 10.1073/pnas.0707815105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dudley DT, Pang L, Decker SJ, Bridges AJ, Saltiel AR. A synthetic inhibitor of the mitogen-activated protein kinase cascade. Proc Natl Acad Sci USA. 1995;92(17):7686–7689. doi: 10.1073/pnas.92.17.7686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hortsch M, Wang YM, Marikar Y, Bieber AJ. The cytoplasmic domain of the Drosophila cell adhesion molecule neuroglian is not essential for its homophilic adhesive properties in S2 cells. J Biol Chem. 1995;270(32):18809–18817. doi: 10.1074/jbc.270.32.18809. [DOI] [PubMed] [Google Scholar]
- 47.Gast D, et al. The cytoplasmic part of L1-CAM controls growth and gene expression in human tumors that is reversed by therapeutic antibodies. Oncogene. 2008;27(9):1281–1289. doi: 10.1038/sj.onc.1210747. [DOI] [PubMed] [Google Scholar]
- 48.Silletti S, et al. Extracellular signal-regulated kinase (ERK)-dependent gene expression contributes to L1 cell adhesion molecule-dependent motility and invasion. J Biol Chem. 2004;279(28):28880–28888. doi: 10.1074/jbc.M404075200. [DOI] [PubMed] [Google Scholar]
- 49.Haspel J, Blanco C, Jacob J, Grumet M. System for cleavable Fc fusion proteins using tobacco etch virus (TEV) protease. Biotechniques. 2001;30(1):60–61, 64–66. doi: 10.2144/01301st01. [DOI] [PubMed] [Google Scholar]
- 50.Staal J, Abràmoff MD, Niemeijer M, Viergever MA, van Ginneken B. Ridge-based vessel segmentation in color images of the retina. IEEE Trans Med Imaging. 2004;23(4):501–509. doi: 10.1109/TMI.2004.825627. [DOI] [PubMed] [Google Scholar]
- 51.Charness ME, Querimit LA, Diamond I. Ethanol induces the expression of functional delta-opioid receptors in the neuroblastoma-glioma hybrid NG108-15 cell line. J Biol Chem. 1986;261:3164–3169. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.