Abstract
The 83-residue conopeptide (p21a) and its corresponding non-hydroxylated analog were isolated from the injected venom of Conus purpurascens. The complete conopeptide sequences were derived from Edman degradation sequencing of fragments from tryptic, chymotryptic and cyanogen bromide digestions. p21a has a unique, ten-cystine/5-disulfide 7-loop framework with extended 10-residue N-terminus and a 5-residue C-terminus tails, respectively. p21a has a 48% sequence homology with a recently described 13-cystine conopeptide, con-ikot-ikot, isolated from the dissected venom of the fish-hunting snail Conus striatus. However, unlike con-ikot-ikot, p21a does not form a dimer of dimers. MALDI-TOF mass spectrometry suggests that p21a may form a non-covalent dimer. p21a is an unusually large conotoxin and insofar is the largest isolated from injected venom. p21a provides evidence that the Conus venom arsenal includes larger molecules that are directly injected into the prey. Therefore, cone snails can utilized toxins that are comparable in size to the ones commonly found in other venomous animals.
Keywords: Cone snails, venom, Conus purpurascens, conotoxins, injected venom, framework 21, p21a, posttranslational modifications, long toxins
Introduction
Cone snails (genus Conus) are versatile marine predators that utilize venom to paralyze and capture their prey. Conus venom is a complex mixture of modified peptides (conopeptides), which are directed with great specificity towards a wide range of physiological targets. The venom components are produced in a tubular duct that contains secretory cells and are discharged into prey through a radular tooth or harpoon1. Most of the cone snail venom components are small peptides (7-30 amino acids in length) that are constrained by disulfide bonds and contain other post-translational modifications2.
Conus species are classified into three groups, depending on their preferential prey: worm hunters (vermivorous), mollusk hunters (molluscivorous), and fish hunters (piscivorous). Piscivorous species use two main strategies to capture fish: 1) hook-and-pull and 2) netting. Hook-and-pull requires rapid immobilization of the prey upon venom delivery, with subsequent pulling of the immobilized prey, which is then engulfed and consumed by the snail3. Conus purpurascens is the only fish-hunting cone snail species found in the tropical Eastern Pacific region, which ranges from the Gulf of California to Perú. C. purpurascens uses the hook-and-pull strategy to capture fish (Figure 1). The injected venom of Conus purpurascens has been shown to elicit an excitotoxic shock effect followed by neuromuscular blocking that produces instant immobilization of the prey4.
Fig. 1.
Extraction of injected venom from C. purpurascens. (A) A fish is placed in front of the snail for stimulation. The stimulated snail extends its proboscis seeking to strike prey (B) The snail strikes the vial with the trap, where the injected venom is collected.
Most conopeptides have been isolated from venom obtained from dissected venom ducts. However, the components of dissected venom do not necessarily correspond to those that are actually injected into prey. The venom collected through the radular tooth during feeding (injected venom) is not identical to the dissected venom of the same animal as the profile of peptides in the injected venom is far less complex than the one observed in dissected venom5,6.
The majority of Conus venom components isolated so far are peptides with less than 30 residues in length. Larger components, including proteins, are seldom described; particularly, those found in injected venom. Just as the smaller conopeptides, the larger polypeptides in Conus venom (either from the ducts or the delivered concoction to the prey) can perform important functions such as acting as toxins, carrier proteins, degradative enzymes (proteases and lipases) or toxin processing7.
Here we report the isolation and characterization of a 9.3 kDa 83-residue component present in the injected venom of Conus purpurascens that defines a new 10-cystine/5-disulfide 7-loop conotoxin framework (framework 21). p21a is also expressed as non-hydroxylated products, at two of its Hyp residues. p21a has a similar cystine arrangement to the recently reported con-ikot-ikot conopeptide. However, p21a does not form a covalent intermolecular disulfide dimer like con-ikot-ikot. p21a is the largest component reported so far from the injected venom of a cone snail, demonstrating that larger components comparable in size to the toxins of other animals, are part of the neurochemical strategy used by cone snails to capture prey.
Results
Peptide purification
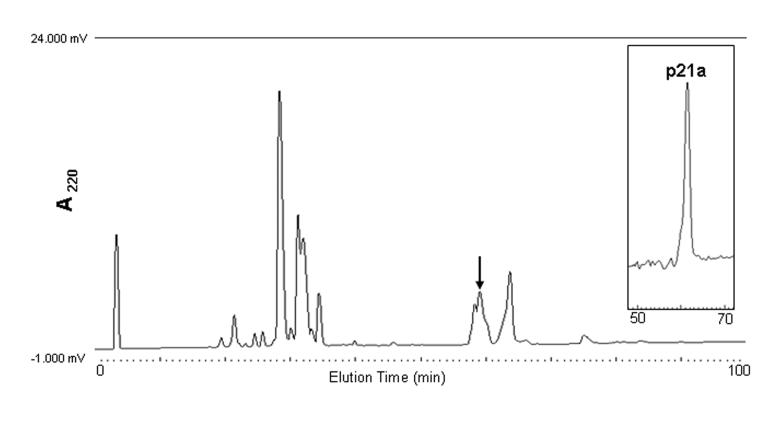
The RP-HPLC of the injected venom of C. purpurascens is shown in figure 2. The peak of interest eluted at 59 min. This peak was re-chromatographed under same conditions yielding a purified p21a (Fig. 2 inset).
Fig. 2.
Purification of p21a. The fraction that corresponds to p21a is marked with an arrow. Three squirts of injected venom were applied to an analytical Vydac C18 column and eluted with a linear gradient of 1 % B/min over 100 min at a flow rate of 1 ml/min. The buffers were 0.1 % TFA (Buffer A) and 0.1% TFA in 60% acetonitrile (Buffer B). The fraction that corresponded to p21a was re-purified under the same conditions (inset).
Mass Spectrometry of p21a
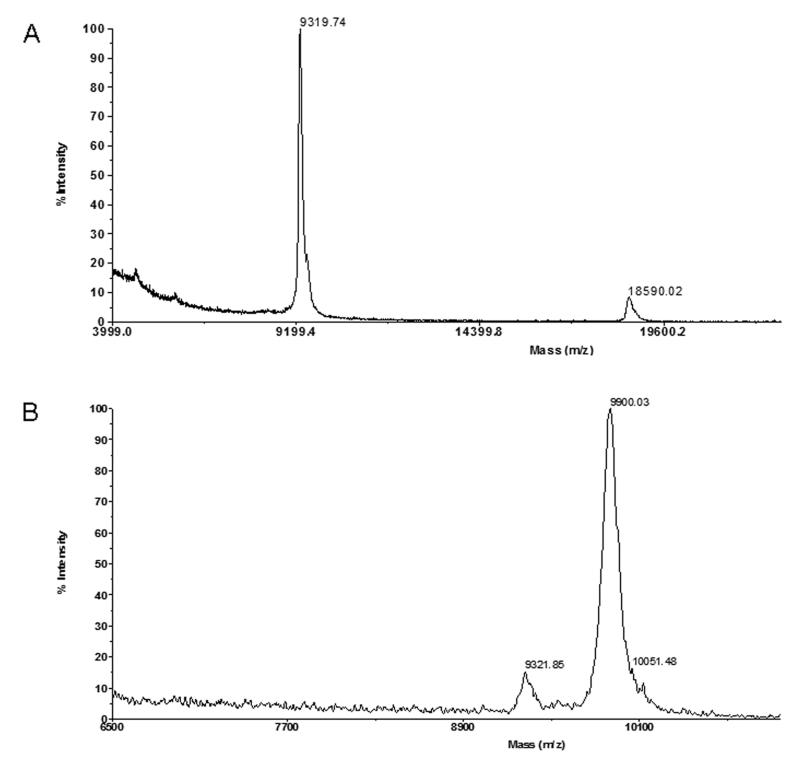
Mass spectrometry of the purified RP-HPLC fraction carried out using MALDI-TOF in linear mode yielded an average molecular mass of 9320 Da (Fig.3A). Mass analysis between the reduced/alkylated peptide and the native peptide showed a mass difference of 554 Da, which is consistent with the presence of ten cystine residues (Fig. 3B).
Fig. 3.
MALDI-TOF mass spectra (linear mode) of: (A) fraction corresponded to p21a. (B) p21a after reduction/alkylation with DTT/Iodoacetamide, indicating the presence of ten cysteines.
Sequence of p21a
Direct N-terminal sequencing of p21a was obtained until Gln-35 (Table 1). Subsequently, p21a was digested using trypsin, chymotrypsin and CNBr respectively. The fragments from these digestions were separated by RP-HPLC and sequenced by Edman degradation (Table 1). The sequence of p21a contained five Pro residues; however, at positions 24 and 43 γ-hydroxyprolines (Hyp) residues were predominant over their Pro counterparts. Upon deduction of the total sequence of p21a from overlapping fragments, the MS-determined molecular weight for the peptide was in agreement with its calculated molecular weight, which included the two Hyp residues (Mwcalc. = 9322 Da vs Mwexp. = 9320 Da).
Table 1.
Sequences of the fragments of p21a obtained by the cleavage with Trypsin, Chymotrypsin and CNBr. O = Hydroxyproline (blue). P = partial sequence. Cystine residues were highlighted in red and hydroxyproline residues were highlighted in blue.

|
The monoisotopic molecular masses of the digestion products, obtained in the reflector mode, were identical to their corresponding calculated counterparts. The fragment corresponding to the carboxyterminal region of p21a is amidated on the C-terminal as we found a difference of 1 Da between the monoisotopic molecular mass and its calculated molecular mass.
p21a is a 83-residue conopeptide with an arrangement of 10-Cys/5-disulfide and 7-loop framework. Additionally, p21a has a 10-residue N-terminus tail and a 5-residue C-terminus tail. A BLAST search8 of the databases (Swissprot/EMBL) revealed 32% sequence homology (query p21a 9-81: Identities = 24/75 (32%), Positives = 36/75 (48%), Gaps = 2/75 (2%) ) with a recently reported 13-cystine conopeptide from Conus striatus (con-ikot-ikot) that forms dimers of dimers (Table 2)9.
Table 2.
Sequence homology between conotoxins con-ikot-ikot and p21a. O indicates hydroxyproline residues, grey highlighted residues indicates identical amino acids and underlined amino acids are molecularly similar residues,

|
indicates carboxyterminus amidation.
Analysis of p21a by SDS-PAGE
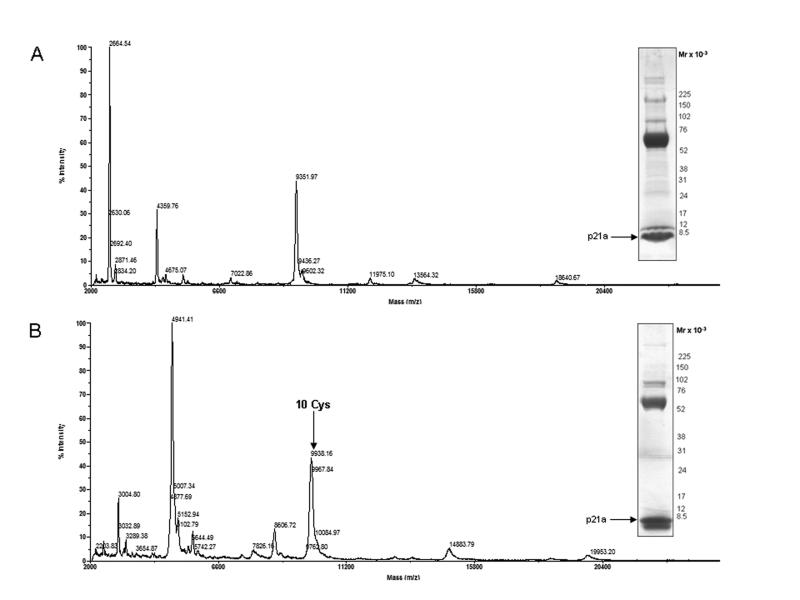
In order to assess whether p21a has a multimeric arrangement, the injected venom was analyzed by gel electrophoresis under native (Fig 4A) and reducing conditions (Fig. 4B). In addition to the p21a band, a major band was observed at 60 kDa along with other larger proteins. However, we did not observe a band at 38 kDa in either gel, which would correspond to the p21a peptide forming dimer of dimers, or around 19 kDa that would correspond a dimer of p21a (Fig. 4). The electrophoresis profile obtained under reducing conditions was similar to the profile obtained under native conditions, with the exception of the bands migrating at molecular masses above 70 kDa (Fig. 4B) and the band of 12 kDa, which disappeared under reducing conditions and generated a new band with a molecular mass lower than the p21a band.
Fig. 4.
SDS-PAGE and MALDI-TOF MS of the injected venom of C. purpurascens (A) native (B) reducing/alkylating conditions.
We used MALDI-TOF mass spectrometry (linear mode) to analyze the large components of the injected venom of Conus purpurascens under native (Fig. 4A) and reducing/alkylating conditions (DTT + IAM), respectively (Fig. 4B). A molecular mass peak corresponding to 18.6 kDa was observed on the MALDI-TOF MS of the injected venom under native conditions (Fig.4A) suggesting possible dimerization of p21a. This peak was also observed on the MALDI-TOF MS of purified p21a (Fig. 3A). Likewise, a molecular mass peak of 19953 Da was observed under reducing/alkylating conditions (Fig. 4B), which corresponds to the dimer of the reduced/alkylated form of p21a (Mw = 9.9 kDa). This result indicates that the tendency of p21a to form dimers does not disappear when the disulfide bonds are reduced.
The results obtained by SDS-PAGE and mass spectrometry suggest that p21a does not form a covalently disulfide-linked dimer as con-ikot-ikot does. The con-ikot-ikot monomer has thirteen cystines; presumably, twelve of these residues are involved in intramolecular disulfide bonding and the remaining cystine is responsible for covalent dimerization. By way of contrast, p21a has an even number of cystines (ten) that are forming five intramolecular disulfide bonds.
Discussion
Here we described the isolation and characterization of the conopeptide p21a from the injected venom of Conus purpurascens. This 83-residue conopeptide is the longest sequence reported so far from the injected venom of a cone snail. This finding in injected venom is significant as it indicates that p21a is part of the actual arsenal used by C. purpurascens to immobilize its prey. p21a has ten cystines that are forming five intramolecular disulfide bonds. There are two pairs of vicinal cystines that define a total of seven loops in the sequence. This particular arrangement of cystines is unique among disulfide-constrained peptides/proteins and defines a new conotoxin framework (framework 21). p21a features a 10-residue N-terminal and a 5-residue C-terminal tails respectively. These tails are unsual among conotoxin scaffolds; however, they are common among the long neurotoxins of scorpions10, spiders11 and snakes12. These tails can have significant influence in the functionality of the toxins, as they affect targeting10, stability by incorporation of D-amino acids11, and overall shape of the toxin as they can (non-covalently) participate in the overall folding of the toxin12 .
The cystine pattern found in p21a is clearly related to a recently reported 13-cystine conotoxin that forms a 36 kDa dimer of dimers (con-ikot-ikot)9. Con-ikot-ikot disrupts the desensitization of AMPA receptors (AMPARs). The dimer of dimers arrangement suggests that con-ikot-ikot acts as a molecular four-legged clamp that keeps AMPARs in the open state. AMPARs are a subfamily of ionotropic glutamate receptors mediating most of the excitatory transmission in the central nervous system (CNS). Thus, disruptors of AMPARs desensitization will have an excitatory effect, which in conjunction with other excitatory toxins in the venom will produce a synergistic effect (excitatory cabal) that will lead to efficacious prey immobilization. Whether p21a is an AMPAR modulator similar to con-ikot-ikot is currently under investigation; however, extensive screening of p21a remains a challenge due to limited supply of the native toxin and its current synthetic inaccessibility.
p21a and con-ikot-ikot are related large toxins found in fish-hunting cone snails that use the hook-and-pull strategy. They are likely to be complementary to other excitatory toxins such as the δ-PVIA and δ-SVIE found in Conus purpurascens and Conus striatus, respectively. These δ-conotoxins inhibit the rapid inactivation of voltage-gated sodium channels causing a state of hyperexcitation, which leads muscle contraction and rigid paralysis. Thus, large conotoxins that modulate AMPARs, such as con-ikot-ikot and possibly p21a, can be considered the ligand-gated components of the excitatory arsenal in the venom.
It is likely that p21a belongs to the same gene superfamily as con-ikot-ikot. However, there might be features in the sequence of the precursors of these conopeptides that encode for the formation of dimer-of dimers such as con-ikot-ikot as opposed to monomers such as p21a. In spite of its large size, the precursor of con-ikot-ikot has similar features to those of other conotoxins: it has a unique signal sequence (that defines it within a new gene superfamily), a pro region, and a mature region that encodes the toxin. Formation of covalent dimers within known frameworks occurs in framework V conotoxins (T-superfamily). A 5-cystine conotoxin (TxXIIIA) was found to form a covalent homodimer of a framework V conotoxin13. Similarly, con-ikot-ikot can be the dimeric equivalent of p21a. While p21a does not form covalent dimers such as con-ikot-ikot, αD-VxXXs or TxXIIIA, we found by mass spectrometry that p21a may form a non-covalent dimer. The connotation of this finding will be addressed at a later time.
The overall sequence of p21a was determined by direct sequencing of the mature toxin, which represented a challenging proposition given the size of this conopeptide. While conopeptide discovery can be performed by deducing putative mature sequences from cDNA libraries, the precise sites of cleavage of the precursor can be difficult to determine. Likewise, it is difficult to predict sites that have undergone posttranslational modifications. In fact, it would be difficult to predict from the cDNA of p21a the existence of the N-terminal tail or the precise sites of hydroxylation found in the isolated toxin (see below). Therefore, direct primary structure determination of conopeptides, even for large components, remains as a definite tool in conopeptide discovery.
p21a is selectively hydroxylated at Hyp-24 and Hyp-43; the other three Pro residues are not hydroxylated. However, a non-hydroxylated version of p21a at positions 24 and 43 coexist in the venom. This differential and selective hydroxylation has been observed for the α-PIVA conotoxin also found in the venom of C. purpurascens14. The presence of hydroxyprolines is frequent in conopeptides15. The reason for Pro hydroxylation in conopeptides is still unclear16. One possibility could be to increase the capacity to form hydrogen bonds and also facilitate the binding strength and selectivity toward a target17. It is possible that the hydroxyprolines along with other residues are involved in the non-covalent dimerization of p21a
As with other components of the venom, p21a is differentially expressed by Conus purpurascens. For some species of cone snails, there are significant intraspecies differences in the composition of venom5,6, which suggests the existence of a regulatory mechanism to produce specific venom peptides for injection into prey. These intraspecies differences can be a result of a combination of genetic and environmental factors. The differential expression of venom components, such as in the case p21a, represents a neurochemical paradigm that warrants further investigation.
Most conotoxins characterized to date fall in a size range of 10 to 30 amino acids; with 2 or 3 disulfide bonds (Table 3). It has been shown that dissected Conus venom is abundant in proteins32. In addition to p21a and con-ikot-ikot, other large size conopeptides have been reported: Conophysin-R31 (84 residues), Conkunitzin-S123 (60 residues) and the dimeric conotoxins, αD-VxXXA-F33,34. However, unlike the aforementioned conopeptides, p21a was isolated from injected venom. Clearly, cone snails inject in their prey larger components than the typical 6 - 30 residue conotoxins. Larger components will also have important physiological effects on their prey. It is evident from Figure 4 that even larger proteins (60 kDa and above) than p21a are injected in the prey.
Table 3.
Disulfide-rich conopeptides. Cystine patterns are shown for each superfamily or conopeptide.
| Number of Disulphide Bonds |
Conopeptide or Superfamily |
Cys pattern | Reference |
|---|---|---|---|
| 2 | A-Superfamily | CC-C-C | 18 |
| J-Superfamily | C-C-CXC | 19 | |
| F-superfamily | C-C-C-C | 20 | |
| T-Superfamily | CC-CC CC-CXOC |
21
22 |
|
| Conkunitzin-S1 | C-C-C-C | 23 | |
| 3 | A-Superfamily | CC-C-C-C-C | 24 |
| M-Superfamily | CC-C-C-CC | 25 | |
| O-Superfamily | C-C-CC-C-C | 26 | |
| P-Superfamily | C-C-C-CXC-C | 27 | |
| 4 | I-Superfamily | C-C-CC-CC-C-C | 28 |
| 5 | S-Superfamily | C-C-C-CXCXC-C-CXCXC | 29 |
| D-Superfamily | C-CC-C-CC-CXC-CXC | 33 | |
| Di16a | C-C-C-CCC-C-CXC-C | 30 | |
| p21a | CC-C-C-C-CC-C-C-C | This work | |
| 6 | Con-ikot-ikot | CC-CC-C-C-C-CC-C-C-C-C | 9 |
| 7 | Conophysin-R | C-C-C-CC-C-C-C-C-C-CC-C-C | 31 |
Toxins from the venom of spiders, snakes and scorpions are usually larger molecules than most reported conopeptides. Discovery and characterization of larger conotoxins represents a new facet of Conus venom studies that correlates well with the biochemical strategies used by other venomous animals. These understudied larger components in Conus venom may have effects such as tissue degradation, necrosis, edema among others, as well as possibly acting as a target for receptors or ion channels.
Material and Methods
Materials
Octadecyl (C18) reversed-phase high performance liquid chromatography (RP-HPLC) analytical column was obtained from Vydac (238TP54, 4.6 mm × 250 mm; 5 μm particle diameter; 300 Å pore size, Illinois, United States). HPLC-grade acetonitrile (ACN), Trifluoroacetic acid (TFA), Dithiothreitol (DTT) and water were purchased from Fisher Scientific (Pennsylvania, United States). Iodoacetamide (IAM) was from Sigma-Aldrich (Missouri, United States). Zip Tip (C18, size P10) was purchase from Millipore (Massachusetts, United States). Calibration mix for MALDI, Calmix 1 and Calmix 2, were acquired from Applied Biosystems (California, United States). Reagents for N-terminal sequencing were acquired from Applied Biosystems (California, United States). 4-Hydroxy-3,5-dimethoxy-cinnamic acid and α-cyano-4-hydroxycinnamic acid matrix were obtained from Sigma-Aldrich (Missouri, United States). Trypsin was acquired from Promega (Wisconsin, United States), Chymotrypsin was purchased from Sigma-Aldrich (Missouri, United States), Cyanogen bromide (CNBr) was acquired from Acros Organics (Geel, Belgium).
Specimen collection
Five specimens of Conus purpurascens (20-50 mm) were collected from intertidal areas at the shores of the Republic of Ecuador and kept alive in aquaria for over four years for venom extraction. Only one specimen expresses sufficient amounts of the p21a conopeptide. Conus purpurascens specimens from other Panamic locations that we currently keep in our laboratory (total of 25) do not express significant amounts of p21a.
Injected venom extraction procedure
Extraction of injected venom from the cone snails was carried out following the procedure of Hopkins et al.14 (Fig. 1). Conus purpurascens were “milked” once a week. A micro-centrifuge tube was used in conjunction with the tail of a gold fish (Carassius auratus) and a piece of latex as a trap. The gold fish was placed in front of the snail to induce a full extension of the proboscis and then we interpose the microcentrifuge tube trap; once the cone snail strikes the tube we separated the harpoon from the latex and fed the snail with the same fish. The injected venom (about 5-30 μL) was centrifuged and saved at −80 °C.
Purification of p21a
Three injected venom samples were dissolved in 0.1% TFA and applied onto the RP-HPLC analytical column with a flow of 1 ml/min. The buffers were 0.1 % TFA (Buffer A) and 0.1% TFA in 60% ACN (Buffer B). The peptide was eluted with an incremental linear gradient of 1% B/min. All fractions were manually collected, lyophilized, and kept at −40 °C prior to further use. Each fraction was subjected to re-chromatography under the same conditions for further purification.
Mass spectrometry
Positive ion MALDI-TOF mass spectrometry was carried out on an Applied Biosystems Voyager-DE STR spectrometer. Samples were dissolved in 0.1% TFA/ 60% acetonitrile, and applied on 4-hydroxy-3,5-dimethoxy-cinnamic acid (sinapinic acid) matrix. Molecular masses of the digestion products were measured with α-cyano-4-hydroxycinnamic acid matrix (cinnamic acid). Mass spectra were obtained in the linear and reflector mode (M/ΔM resolution ~10000) using Calmix 1 and Calmix 2 (Applied Biosystems) as external calibration standards. Two separated aliquots (5 μl each) of fresh injected venom were subjected for molecular mass analysis with sinapinic acid. One aliquot of the injected venom was used to carry out the reduction and alkylation of the cystine groups as described in the next section without the Zip Tip step. The other aliquot was analyzed under native conditions. Calculated molecular masses were obtained using Protein Prospector35.
Reduction and alkylation of the purified peptide
Reduction and alkylation of cystine groups were carried out as previously described20 with slight modifications. An aliquot of the peptide (~1 pmol) was dried, re-dissolved in 0.1 M Tris-HCl (pH 6.2), 5 mM EDTA, 0.1 % sodium azide and reduced with 6 mM DTT. Following incubation at 60 °C for 30 min, peptides were alkylated in a final volume of 15 μl with 20 mM IAM and 2 μl of NH4OH (pH 10.5), at room temperature, for 1 h, in the dark. The reduced and alkylated peptides were purified using a Zip Tip.
Peptide sequencing
Reduced and alkylated peptides were adsorbed onto Biobrene-treated glass fiber filters and sequenced using Edman degradation on an Applied Biosystems Procise model 491A sequencer.
Digestion of p21a
Protease digestions
1 nmol of the lyophilized reduced/alkylated peptide was dissolved in 0.5 mM NH4HCO3, pH 8.0 and then digested using either trypsin or chymotrypsin in a protease:peptide ratio of 1:20 to a final volume of 25 μl at 37 °C for 4 h. An aliquot of 0.5 μL of the digestion mixture was analyzed by MALDI-TOF and the rest was dissolved in 300 μl of 0.1 % TFA and applied to a C18 analytical HPLC column. The resulting peptide fragments were dried for further sequence by Edman degradation.
CNBr cleavage
1 nmol of the reduced/alkylated peptide was dissolved in 20 μl of 70 % formic acid. Lysis of the peptide with CNBr was performed using a CNBr:peptide mass ratio of 2:1. The peptidic sample was incubated for 20 h, at room temperature, in the dark, and the reaction was terminated by diluting the mixture with 300 μl of 0.1 % TFA. The quenched reaction was submitted to RP-HPLC analysis. The fractions were then sequenced by Edman degradation.
Analysis of p21a by SDS-PAGE
Injected venom was subjected to SDS-PAGE analysis using pre-cast 4-20 % polyacrylamide gradient gels (Invitrogen) under native and reducing conditions. Under reducing conditions, the venom was incubated with an equal volume of sample buffer in the presence of 10 mM DTT at 60 °C for 10 min previous to the separation. The gels were fixed and stained with Novex-Colloidal Blue Stain (Invitrogen) according to the manufacturer’s protocol. Band extraction from the gel was carried out according to the protocol previously reported by Mirza36 with some minor changes. Individual bands between 8.5 and 12 kDa were excised using a scalpel and placed in 1.5 ml tubes. Each gel piece was washed with 100 μl of 50% methanol/5% acetic acid under agitation for 2 h. Then, the liquid was removed and the piece of gel was washed three times with water and incubated with 100 μl of 60% acetonitrile/0.1% TFA under agitation for a minimum of 4 h. This extraction procedure was repeated three times; the resulting solutions were pooled and dried in a speed vacuum apparatus. Samples were re-suspended in 5 μl of 60% ACN/0.1% TFA for MALDI-TOF mass spectrometry analysis.
Acknowledgments
We thank Dr. Herminsul Cano and Nicole Vanderweit for their help in extracting the venom samples. This work was partially funded by the Florida Seagrant Program (Grant R/LR-MB-28) and the NIH (Grant NS066371-01).
References
- 1.Olivera BM. Mol Biol Cell. 1997;8:2101–2109. doi: 10.1091/mbc.8.11.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buczek O, Bulaj G, Olivera BM. Cell Mol Life Sci. 2005;62:3067–3079. doi: 10.1007/s00018-005-5283-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Olivera BM, Imperial JS, Bulaj G. Perspectives in Molecular Toxinology. 2002:143–158. [Google Scholar]
- 4.Terlau H, Shon K-J, Grilley M, Stocker M, Stuehmer W, Olivera BM. Nature. 1996;381:148–151. doi: 10.1038/381148a0. [DOI] [PubMed] [Google Scholar]
- 5.Bingham J-P, Jones A, Lewis RJ, Andrews PR, Alewood PF. Biochemical Aspects of Marine Pharmacology. 1996:13–27. [Google Scholar]
- 6.Jakubowski JA, Kelley WP, Sweedler JV, Gilly WF, Schulz JR. J Exp Biol. 2005;208:2873–2883. doi: 10.1242/jeb.01713. [DOI] [PubMed] [Google Scholar]
- 7.Terlau H, Olivera BM. Physiol Rev. 2004;84:41–68. doi: 10.1152/physrev.00020.2003. [DOI] [PubMed] [Google Scholar]
- 8.Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walker CS, Jensen S, Ellison M, Matta JA, Lee WY, Imperial JS, Duclos N, Brockie PJ, Madsen DM, Isaac JTR, Olivera B, Maricq AV. Curr Biol. 2009;19:900–908. doi: 10.1016/j.cub.2009.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gurevitz M, Gordon D, Ben-Natan S, Turkov M, Froy O. FASEB J. 2001;15:1201–1205. doi: 10.1096/fj.00-0571hyp. [DOI] [PubMed] [Google Scholar]
- 11.Heck SD, Siok CJ, Krapcho KJ, Kelbaugh PR, Thadeio PF, Welch MJ, Williams RD, Ganong AH, Kelly ME, Lanzetti AJ, Gray WR, Phillips D, Parks TN, Jackson H, Ahlijanian MK, Saccomano NA, Volkmann RA. Science. 1994;266:1065–1068. doi: 10.1126/science.7973665. [DOI] [PubMed] [Google Scholar]
- 12.Osipov AV, Kasheverov IE, Makarova YV, Starkov VG, Vorontsova OV, Ziganshin RK, Andreeva TV, Serebryakova MV, Benoit A, Hogg RC, Bertrand D, Tsetlin VI, Utkin YN. J Biol Chem. 2008;283:14571–14580. doi: 10.1074/jbc.M802085200. [DOI] [PubMed] [Google Scholar]
- 13.Quinton L, Gilles N, De Pauw EJ. Proteomics. 2009;72:219–226. doi: 10.1016/j.jprot.2009.01.021. [DOI] [PubMed] [Google Scholar]
- 14.Hopkins C, Grilley M, Miller C, Shon K-J, Cruz LJ, Gray WR, Dykert J, Rivier J, Yoshikami D, Olivera BM. J Biol Chem. 1995;270:22361–22367. doi: 10.1074/jbc.270.38.22361. [DOI] [PubMed] [Google Scholar]
- 15.Olivera BM, Gray WR, Zeikus R, McIntosh JM, Varga J, Rivier J, de Santos V, Cruz LJ. Science. 1985;230:1338–1343. doi: 10.1126/science.4071055. [DOI] [PubMed] [Google Scholar]
- 16.Lopez-Vera E, Walewska A, Skalicky JJ, Olivera BM, Bulaj G. Biochemistry. 2008;47:1741–1751. doi: 10.1021/bi701934m. [DOI] [PubMed] [Google Scholar]
- 17.Franco A, Pisarewicz K, Moller C, Mora D, Fields GB, Mari F. Prog Mol Subcell Biol. 2006;43:83–103. doi: 10.1007/978-3-540-30880-5_4. [DOI] [PubMed] [Google Scholar]
- 18.Santos AD, McIntosh JM, Hillyard DR, Cruz LJ, Olivera BM. J Biol Chem. 2004;279:17596–17606. doi: 10.1074/jbc.M309654200. [DOI] [PubMed] [Google Scholar]
- 19.Imperial JS, Bansal PS, Alewood PF, Daly NL, Craik DJ, Sporning A, Terlau H, Lopez-Vera E, Bandyopadhyay PK, Olivera BM. Biochemistry. 2006;45:8331–8340. doi: 10.1021/bi060263r. [DOI] [PubMed] [Google Scholar]
- 20.Moller C, Rahmankhah S, Lauer-Fields J, Bubis J, Fields Gregg B, Mari F. Biochemistry. 2005;44:15986–15996. doi: 10.1021/bi0511181. [DOI] [PubMed] [Google Scholar]
- 21.McIntosh JM, Corpuz GO, Layer RT, Garrett JE, Wagstaff JD, Bulaj G, Vyazovkina A, Yoshikami D, Cruz LJ, Olivera BM. J Biol Chem. 2000;275:32391–32397. doi: 10.1074/jbc.M003619200. [DOI] [PubMed] [Google Scholar]
- 22.Walker CS, Steel D, Jacobsen RB, Lirazan MB, Cruz LJ, Hooper D, Shetty R, DelaCruz RC, Nielsen JS, Zhou LM, Bandyopadhyay P, Craig AG, Olivera BM. J Biol Chem. 1999;274:30664–30671. doi: 10.1074/jbc.274.43.30664. [DOI] [PubMed] [Google Scholar]
- 23.Bayrhuber M, Vijayan V, Ferber M, Graf R, Korukottu J, Imperial J, Garrett JE, Olivera BM, Terlau H, Zweckstetter M, Becker S. J Biol Chem. 2005;280:23766–23770. doi: 10.1074/jbc.C500064200. [DOI] [PubMed] [Google Scholar]
- 24.Teichert RW, Rivier J, Dykert J, Cervini L, Gulyas J, Bulaj G, Ellison M, Olivera BM. Toxicon. 2004;44:207–214. doi: 10.1016/j.toxicon.2004.05.026. [DOI] [PubMed] [Google Scholar]
- 25.Gray WR, Luque A, Olivera BM, Barrett J, Cruz LJ. J Biol Chem. 1981;256:4734–4740. [PubMed] [Google Scholar]
- 26.Olivera BM, McIntosh JM, Curz LJ, Luque FA, Gray WR. Biochemistry. 1984;23:5087–5090. doi: 10.1021/bi00317a001. [DOI] [PubMed] [Google Scholar]
- 27.Lirazan MB, Hooper D, Corpuz GP, Ramilo CA, Bandyopadhyay P, Cruz LJ, Olivera BM. Biochemistry. 2000;39:1583–1588. doi: 10.1021/bi9923712. [DOI] [PubMed] [Google Scholar]
- 28.Jimenez EC, Shetty RP, Lirazan M, Rivier J, Walker C, Abogadie FC, Yoshikami D, Cruz LJ, Olivera BM. J Neurochem. 2003;85:610–621. doi: 10.1046/j.1471-4159.2003.01685.x. [DOI] [PubMed] [Google Scholar]
- 29.England LJ, Imperial J, Jacobsen R, Craig AG, Gulyas J, Akhtar M, Rivier J, Julius D, Olivera BM. Science. 1998;281:575–578. doi: 10.1126/science.281.5376.575. [DOI] [PubMed] [Google Scholar]
- 30.Chen P, Garrett JE, Watkins M, Olivera BM. Toxicon. 2008;52:139–145. doi: 10.1016/j.toxicon.2008.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lirazan M, Jimenez EC, Grey Craig A, Olivera BM, Cruz LJ. Toxicon. 2002;40:901–908. doi: 10.1016/s0041-0101(02)00079-x. [DOI] [PubMed] [Google Scholar]
- 32.Newcomb R, Miljanich G. Handbook of Neurotoxicology. 2002;1:617–651. [Google Scholar]
- 33.Loughnan M, Nicke A, Jones A, Schroeder CI, Nevin ST, Adams DJ, Alewood PF, Lewis RJ. J Biol Chem. 2006;281:24745–24755. doi: 10.1074/jbc.M603703200. [DOI] [PubMed] [Google Scholar]
- 34.Loughnan ML, Nicke A, Lawrence N, Lewis RJ. Biochemistry. 2009;48:3717–3729. doi: 10.1021/bi9000326. [DOI] [PubMed] [Google Scholar]
- 35.Clauser KR, Baker P, Burlingame AL. Anal. Chem. 1999;71:2871–2882. doi: 10.1021/ac9810516. [DOI] [PubMed] [Google Scholar]
- 36.Mirza UA, Liu YH, Tang JT, Porter F, Bondoc L, Chen GD, Pramanik BN, Nagabhushan TL. J Am Soc Mass Spectrom. 2000;11:356–361. doi: 10.1016/s1044-0305(00)00101-x. [DOI] [PubMed] [Google Scholar]