Abstract
Systems approaches have long been used in pharmacology to understand drug action at the organ and organismal levels. The application of computational and experimental systems biology approaches to pharmacology allows us to expand the definition of systems pharmacology to include network analyses at multiple scales of biological organization and to explain both therapeutic and adverse effects of drugs. Systems pharmacology analyses rely on experimental “omics” technologies that are capable of measuring changes in large numbers of variables, often at a genome-wide level, to build networks for analyzing drug action. A major use of omics technologies is to relate the genomic status of an individual to the therapeutic efficacy of a drug of interest. Combining pathway and network analyses, pharmacokinetic and pharmacodynamic models, and a knowledge of polymorphisms in the genome will enable the development of predictive models of therapeutic efficacy. Network analyses based on publicly available databases such as the U.S. Food and Drug Administration’s Adverse Event Reporting System allow us to develop an initial understanding of the context within which molecular-level drug-target interactions can lead to distal effectors in a process that results in adverse phenotypes at the organ and organismal levels. The current state of systems pharmacology allows us to formulate a set of questions that could drive future research in the field. The long-term goal of such research is to develop polypharmacology for complex diseases and predict therapeutic efficacy and adverse event risk for individuals prior to commencement of therapy.
Keywords: enhanced pharmacodynamics, genomics and personalized therapy, drug discovery, adverse event predictions
INTRODUCTION
Over the past 60 years, drug therapy for many complex noncommunicable diseases has been quite successful. Drugs are now routinely used to control hypertension and treat peptic ulcers, asthma, and many types of cancers. In spite of these successes over past decades, it has become clear that the drug discovery process has slowed down as the costs of bringing a drug to market have gone up tremendously (1). New targets are most often identified by linking individual cellular components to an organismal- or tissue/organ-level phenotype. Such distal correlations, although a good starting point, often do not work for drug development because the cellular and tissue/organ-level systems are treated as black boxes. This leads to a lack of mechanistic understanding of how drug interactions at the molecular level manifest themselves as alterations in tissue/organ-level function. This lack of understanding, in turn, leads to confounding situations at various stages during the drug discovery process. Drugs that are promising in cell-based assays often do not work in vivo, and even when they do, they show variable efficacy. Most new drugs often fail in Phase II and Phase III trials. Another limitation of the black-box approach is the inability to predict adverse events when the drug is brought to market and used by the population at large. The occurrence of serious and sometimes fatal adverse events has led to the withdrawal of or tight restrictions on the use of drugs that are beneficial to the majority of the population. Such regulatory caution is warranted because of our lack of ability to predict who among the target users of a drug will have a serious adverse event such that the risk of adverse events outweighs the therapeutic advantage. These problems have led to calls for different approaches to drug discovery and therapeutics (2).
The advances in mammalian genomics, biochemistry, molecular and cell biology, and physiology have allowed us initial glimpses of the complexity of human and mammalian biology at multiple scales that involve organization at the atomic/molecular, cellular and tissue, organ, and organismal levels. Although this understanding is still far from complete, the emerging picture indicates that network analysis, wherein we study the organization (i.e., topology) of interactions among components of a system, can provide a useful approach for multiscale understanding. Network analyses allow us to define the relationship between emergent functions and topology at each level of organization (atomic/molecular, cellular/tissue, organ, and organismal) and connections between levels that give rise to organ- and organismal-level functions. Understanding the explicit relationships between scales (i.e., levels) of organization allows us to appreciate how drugs that interact with molecular components and have their first effects at the cellular level are able to produce organ- and organismal-level effects, both therapeutic and adverse. In this approach, which focuses on what we term multiscale mechanisms, black-box assumptions are purposefully avoided and qualitative relationships or quantitative parameters are directly related to molecular interactions or cellular functions.
The term systems pharmacology now describes a field of study that uses experimental and computational approaches to provide us with a broad view of drug action rooted in molecular interactions between the drug and its targets in the context of such targets interacting with and regulating other cellular components. This newer definition expands the older usage, in which systems pharmacology was used to describe drug action in a specific organ system such as ocular pharmacology or reproductive pharmacology (3). Here we review the current status of this new field and describe how, in our view, integrating network analysis with dynamical quantitative approaches in pharmacokinetic and pharmacodynamic models can help us develop a mechanistic understanding of drug action in the context of an individual’s genomic status and environmental exposure.
TYPES OF NETWORKS USED IN ANALYZING DRUG ACTION
A network is defined as a series of entities connected to one another on the basis of a defined criterion. The entities in a network are named nodes, which represent different types of objects such as genes (4), proteins (5), drugs (6), and disease (7, 8). Nodes in a network can also be used to specify the state of a system. Such specifications can be computed using Boolean dynamics (9, 10), in which each node has a chance to exist in two states (inactive or active), or using concentrations of the nodes with dynamical models based on ordinary differential equations (9, 11). The latter approach is most commonly used in pharmacokinetic-pharmacodynamic models.
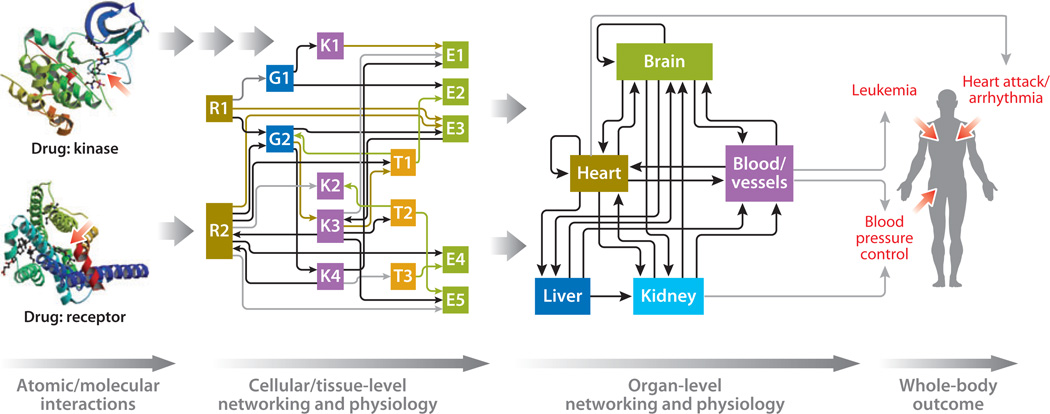
The connections between the nodes are termed edges and can be specified using criteria of interest. In the context of studying drug action, edges may represent protein-protein interactions (5), drug-target interactions (12), or transcriptional regulation (13). Edges can also be defined on the basis of similarities between two nodes such as the chemical or therapeutic similarity (6), the similarities between proteins based on the shared number of diseases (14), or the similarities of diseases based on the shared number of genes (14).These types of complex definitions for edges allow the networks to transcend multiple scales of interactions from the atomic- and molecular-level drug-target interactions to the coordinated functional outputs of multiple organs, i.e., phenotype. Such multiscale organization is featured in Figure 1, which shows networks at two levels: the cellular/ tissue level and the organ level. There is a possibility that tissue-level networks are distinct from cellular-level networks. This distinction will have to be specified on a case-by-case basis. Nevertheless, developing networks at various scales allows us to explicitly track drug effects from atomic-level interactions to organismal physiology.
Figure 1.
A schematic representation of the multiscale networks needed to understand and predict drug action. Atomic interactions between drug and target lead to alterations in the function of cellular regulatory networks, which lead to changes in cellular- and tissue-level physiology, which, in turn, lead to alterations in organ-level networking, which lead to changes in whole-body functions. Networks at both the cellular/tissue level and organ level are needed to understand the mechanism of drug action and to predict therapeutic efficacy and adverse event probability. The drug-protein structures are taken from structures deposited in the Protein Data Bank (http://www.pdb.org) with PDB IDs of 3QC4 and 2Y03 (88, 89), with the authors’ permission.
The edges of a network can be directed, in which the source node causes an effect on the target node, and the relationship is valid in only one direction. An example of directed edges is the protein kinase activation of a transcription factor and the regulation of a target gene by that transcription factor (13). Alternatively, the edges can be undirected, in which interactions can occur in both directions. Examples of undirected edges include the interactions between a protein and its scaffold. The edges can also be given weight based on the strength of their association. These weights can be derived from numerous criteria, ranging from statistical correlations for distal relationships (such as gene-disease relationships) to kinetic rate constants for direct physical interactions (such as hormone or drug binding to receptors).
In analyzing drug actions, one can use a variety of networks based on different types of nodes and edges. The simplest network has a directed edge connecting a drug node to its target protein node (12).The target protein node is then connected to other proteins that physically interact with the drug-target protein, and these proteins can be linked to additional proteins using the same criterion (15). In this network, all edges have the same weight, implying that they have the same extent of connectivity. This simplifying assumption is not always true, and we need to be careful in ascertaining when the network as depicted is a reasonable representation of the system. These types of networks are known as interaction networks. Interaction networks allow us to quickly determine the potential downstream and upstream interactors of a particular node (16), which can be useful in identifying pathways for signal flow and regulatory motifs such as feed-forward and feedback loops that have information processing capability.
Interaction networks are also the foundation of more highly specified networks that are typically used to study biological systems such as dynamical, Boolean, and stochastic networks. However, the interaction networks require the least amount of knowledge regarding nodes and edges, allowing them to be easily constructed and applied to a variety of problems (6, 17–19). The simplicity of interaction networks allows networks at different scales of organization to be combined on the basis of the notion that they “interact”; i.e., they are related. A protein-protein interaction network can be expanded by connecting proteins to their physiological function (20), and, in doing so, one can analyze drug-to-physiological function through combining edges that represent drug-protein, protein-protein, and protein-physiology interactions. The types of network used to analyze drug action depend on the type of action of interest. These networks can be constructed, taken apart, and reorganized from different data sets that define methodologies for identifying edges that connect the nodes. Performing the analyses correctly requires knowledge about the edges’ limitations and meanings.
EXPERIMENTAL APPROACHES IN SYSTEMS PHARMACOLOGY
The data sets needed to build networks require simultaneous measurements on a large number of variables in response to a perturbation, such as a pathophysiological state or drug treatment. Systems biology uses “omics” experiments, in which a large number of output variables is measured in response to one or more perturbations. Typically, such experiments fall into one of three categories: genomics, proteomics, or metabolomics.
Genomics Analyses
Genomics analyses involve the sequencing or characterization of many genes, typically the whole genome simultaneously. At the DNA level, genomics involves sequencing of the genome to identify variations and to determine transcriptional binding sites and epigenetic status. At the mRNA level, genome-wide profiling is largely focused on characterizing gene expression patterns in a disease state or before and after drug treatment. This type of profiling was accomplished mostly through the use of microarrays, but in the past few years, direct sequencing, termed mRNA seq, has become more widely used.
As the sequences of many organisms were determined, it became apparent that positional variations exist within the DNA sequences of the individuals in a single species and even between the sequences of pairs of chromosomes. These variations are named single-nucleotide polymorphisms (SNPs). Sometimes SNPs fall within the coding regions of genes, leading to changes in protein primary sequences and activity (21). Although coding-region SNPs are infrequent, they are important in drug action because genes for several drug-metabolizing enzymes have coding-region SNPs, such as rs1799853 in cytochrome P450–2C9 (CYP2C9*2) for warfarin dosing (22). Consequently, genomics analysis that is focused on DNA-sequencing methodologies has become important in understanding drug action. Sequencing technologies are rapidly changing, and the cost of sequencing is decreasing (23). These developments indicate that whole-genome sequencing is likely to play an important role in systems pharmacology.
An interesting approach for characterizing drug action has been the use of gene expression patterns as a means to connect drugs with disease states. Combining profiling experiments with pattern-matching software, Lamb et al. (24) have created a library of gene expression signatures from adding drugs—both U.S. Food and Drug Administration (FDA)-approved drugs and non-therapeutic small molecules used in laboratory research—to human cell lines in vitro to obtain whole-genome gene expression patterns. These studies indicate that structurally different compounds that converge on common targets can yield the same gene expression signature. As the field progresses from cell-based to tissue-based analyses of drug action, such gene expression signatures can be useful in understanding drug action in multiple tissues and organs.
Proteomics Analyses
Proteomics involves the study of changes in the levels or states of large numbers of proteins in a sample of interest such as a cell extract, the plasma, or a tissue sample. Typically, the measurement of proteins is by mass spectrometry, although sometimes protein arrays are also used. In contrast to genomics approaches, the use of proteomics in drug discovery and study of drug action has been limited. A major issue is the difficulty in obtaining tissue biopsies sufficient for proteomics analyses to correlate changes in target tissues and organs with drug action in humans. Most proteomics studies have focused on human cancer cell lines and can be used for target profiling (25) and mechanism-based classification of potential drugs (26).
Metabolomics Analyses
Metabolomics focuses on measuring changes in a large number of metabolites simultaneously (27). The method of choice for identification of metabolites is mass spectrometry, generally preceded by chromatographic resolution. The most readily available source for metabolic profiling in humans is plasma. Several studies have shown identifiable metabolic signatures associated with drug treatment. A study on 50 patients with schizophrenia being treated with antidepressants showed identifiable changes in lipid patterns after treatment (28). These observations raise the possibility that metabolic signatures of drug treatment could be an additional tool in assessing drug therapy in patients. A recent study (29) on patients with major depressive disorders has shown an interesting relationship between genomics and metabolomics in predicting drug action. Metabolomics was used to characterize levels of amino acids in plasma. Patients who were nonresponsive to therapy with the serotonin reuptake inhibitor citalopram showed higher baseline levels of glycine, which remained unaltered after treatment. Genomics analyses indicated that in nonresponders, the SNP rs10975641 in the glycine dehydrogenase gene was associated with treatment outcome. The authors of the study (29) did not elucidate the mechanisms by which the SNP in the glycine dehydrogenase gene can result in elevated plasma levels of glycine, but the study is important because it provides an approach for developing mechanistic multiscale studies wherein genomic changes can be correlated with the biochemical profile of the plasma, which, in turn, can be correlated with treatment outcomes. Such multilevel correlation can be used to specify mechanisms that operate within cells and tissues.
PHARMACOGENOMICS AS A SUCCESSFUL MODEL FOR SYSTEMS PHARMACOLOGY
A success story in explicitly connecting genomic status and drug action has come from the field of pharmacogenomics. This type of linkage is important for understanding drug action and effects at an individual level. Genomics is thought to account for a significant part of interindividual variability (30) in the drug effect, while eliciting consistent intraindividual responses (31). Many FDA-approved drugs now contain pharmacogenomic information within their labels. Pharmacogenomic specification is used for a wide range of drugs from antiasthmatics (e.g., montelukast) (32) to cancer therapeutics (e.g., cetuximab) (33). The study of genomics variations in altering drug responses can be divided into three major areas: pharmacokinetics, pharmacodynamics, and responsiveness to therapy. A list of select pharmacogenomic biomarkers in the various areas for FDA-approved drugs is given in Table 1.
Table 1.
Various types of pharmacogenomic effects in drug action
| Drug | Gene | Effect | ||
|---|---|---|---|---|
| Pharmacokinetics | ||||
| Codeine | CYP2D6 (34) | Increase in the amount of active drug by variants | ||
| Clopidogrel | CYP2C19 (80) | Increase in the amount of active drug by variants | ||
| Warfarin | CYP2C9 (81) | Changes in drug levels in blood by variants | ||
| Pharmacodynamics | ||||
| Warfarin | VKORC1 (21) | Increase or decrease of effectiveness of drug | ||
| Capecitabine | DPD (82) | Decrease in breakdown of 5-FU metabolite | ||
| Responsiveness | ||||
| Panitumumab | k-RAS (83) | Requirement of wild-type k-RAS for drug efficacy | ||
| Imatinib | c-KIT (84) | Requirement of wild-type c-KIT for drug efficacy | ||
| Tretinoin | PML/RARα translocation (85) | Increased drug responsiveness | ||
| Unknown mechanisms | ||||
| Carbamazepine | HLA-B*1502 (86) | Increased risk of Stevens-Johnson syndrome and toxic epidermal necrolysis | ||
| Abacavir | HLA-B*5701 (87) | Multiorgan systemic hypersensitivity, which may lead to death | ||
Abbreviations: 5-FU, fluorouracil; CYP, cytochrome P450; DPD, dihydropyrimidine dehydrogenase; HLA, human leukocyte antigen; PML, promyelocytic leukemia; RAR, retinoic acid receptor; VKOR, vitamin K epoxide reductase.
A major use of genomic information is in relating genomic status to drug dosage and metabolism (pharmacokinetics) because drug-metabolizing enzymes play a large role in converting prodrugs into active metabolites (e.g., codeine) (34) or active drugs into inactive drugs or toxic metabolites (e.g., nortriptyline) (35). Sometimes, the same metabolizing enzyme can do both: Both codeine and nortriptyline are metabolized by the cytochrome P450 isoform CYP2D6 (35). Codeine is converted from the prodrug to the active drug, whereas nortriptyline is converted from the active form to the inactive form. In both cases, the relationship between the intake dose of the drug and the active drug in the plasma is related to the activity of CYP2D6. These findings have led to tests for drug-metabolizing proteins in individuals (36, 37) so that intake dosage can be set at a safe level for each patient. Other drug-binding proteins with polymorphisms include drug transporters such as the ATP-binding cassette family and solute carrier organic anion transporter family of membrane transporters. These transporters affect plasma concentrations of non-nucleoside reverse transcriptase inhibitors sunitinib (a tyrosine kinase inhibitor) and methotrexate (a dihydrofolate reductase enzyme inhibitor) (29, 38, 39).
The cytochrome P450 protein CYP2C9 regulates the metabolism of warfarin, which is used as an anticoagulant. CYP2C9 has two polymorphisms that reduce the level of enzyme activity; consequently, increased levels of warfarin in the blood result in an increased risk of bleeding (22). Thus, knowing if a patient has CYP2C9 polymorphisms allows the physician to titrate the dosage of warfarin to optimize therapy while reducing the risk of bleeding as a serious adverse event. Testing for warfarin metabolism has become a common approach to titrating warfarin dosage in clinical practice.
The relationship between genomic status and pharmacodynamics can also be important. An example is the polymorphisms in vitamin K epoxide reductase (VKOR) that alter an individual’s sensitivity to warfarin (21). VKOR activity is required for the γ-carboxylation of multiple clotting factors, and warfarin exerts its action by inhibiting VKOR and thus reducing clotting. Polymorphisms in the VKOR genes result in variants that have lower or higher sensitivity to warfarin; thus, the dosage of warfarin needs to be adjusted in patients with different polymorphisms (21). In the case of warfarin, both pharmacokinetic and pharmacodynamic properties are regulated by SNPs, so both aspects need to be considered in the dosing of individual patients.
Genomic status can also predict responsiveness in therapy. This type of relationship has been used mainly in cancer therapy—the presence of certain mutant oncogenes is an indicator for lack of responsiveness to targeted therapy. KIT oncogene mutations reduce the responsiveness of gastrointestinal stromal tumors to imatinib (40); k-RAS oncogene mutations in colorectal cancer reduce responsiveness to cetuximab (33); and epidermal growth factor receptor mutations in non-small-cell lung cancer alter responsiveness to gefitinib or erlotinib (41, 42). Other examples in which a genotype–drug response phenotype is known but the underlying mechanism is not fully understood include patients with variants in major histocompatibility complex class IB (HLA-B*5701) who show hypersensitivity to the antiviral drug abacavir (43) and musculoskeletal toxicity from aromatase inhibitors used to treat breast and ovarian cancers (44).
Several tests are commercially available for identifying polymorphisms associated with drug responses. Those approved by the FDA include the microarray-based Roche AmpliChip® for cytochrome P450 genotyping. Because warfarin effects are regulated by polymorphisms in both cytochrome P450 enzymes and VKOR, the FDA recommends testing both CYP2C9 and VKORC1 polymorphisms for warfarin (45). The FDA has also approved the Invader® UGT1A1 Molecular Assay for the polymorphisms that increase the risk of neutropenia associated with the colon cancer drug irinotecan, which inhibits DNA topoisomerase. The FDA recommends the testing of HLA-B*5701 for abacavir; low-density lipoprotein receptor variants for atorvastatin; thiopurine S-methyltransferase for azathioprine; HLA-B*1502 for carbamazepine; epidermal growth factor receptor for cetuximab; cytochrome P450 protein CYP2C19 for clopidogrel; breakpoint cluster region–Abl tyrosine kinase translocation for dasatinib and imatinib; UDP glucuronosyltransferase 1 family, polypeptide A1 for irinotecan; k-RAS oncogene for panitumumab; glucose-6-phosphate dehydrogenase for rasburicase; erythroblastic leukemia viral oncogene homolog 2 for trastuzumab; and carbamoyl-phosphate synthase 1 for valproic acid. However, no specific tests for these polymorphisms have been approved by the FDA.
NEED FOR DETAILED PHARMACODYNAMIC MODELS THAT INCLUDE CELLULAR AND TISSUE MECHANISMS
Although in the case of warfarin one can explicitly relate drug interactions with metabolizing enzymes and target proteins to physiological events such as thrombosis and bleeding at the organismal level, explicitly identifying such multiscale relationships for most drugs is difficult. A major challenge for systems pharmacology is the development of a mechanistic understanding of how different cellular- and tissue-level regulatory networks control variability in drug response at the organismal level. For this, we need enhanced pharmacodynamic models that couple detailed models of cellular regulatory networks with measurable pharmacokinetic and pharmacodynamic parameters. A recent pharmacodynamic modeling study (46) on antagonists of the calcium-sensing receptor and its relationship to parathyroid hormone (PTH) secretion has shown how negative allostericmodulators of the calcium-sensing receptor, coupled with negative feedback, can explain the observed homeostatic relationship among plasma calcium, PTH, and calcium absorption. The mechanisms of the PTH-driven negative feedback are not known; this demonstrates the need to build detailed models of intra- and intercellular signal networks so that the molecular and cellular bases of homeostasis can be explicitly described and understood. It has long been known that intracellular signaling networks form regulatory motifs such as positive feedback loops that can function as bistable switches (47) that are involved in cellular state change such as synaptic plasticity (48). Large signaling networks are full of regulatory motifs such as feedback and feed-forward loops that can process information as signal flows through (16). Characterizing the topology of cellular regulatory networks and understanding the dynamic capability of the topology can help explain both therapeutic and adverse drug action. In the calcium-sensing receptor and PTH, identifying the molecular components that participate in the negative feedback loop and how they can be modulated can help us design better antagonists at the calcium-sensing receptor or develop polypharmacology for treatment of osteoporosis.
Detailed studies of intracellular pathways, such as metabolic or signaling pathways, can be useful for understanding the efficacy of drug action in humans. Panetta et al. (49) built models of methotrexate metabolism and action for the treatment of childhood acute lymphoblastic leukemia. The model incorporated the production of methotrexate polyglutamate metabolites and the regulation of the folate pathway enzymes by both methotrexate and its metabolites in T and B cells. The development of the detailed model allowed for simulations that were able to explain how changing dosage and duration of infusion affected efficacy of treatment. The simulations also took into account how SNPs in folate metabolism genes affect drug responses. This combination of pathway simulations and pharmacokinetic and pharmacodynamic patient data with knowledge of genomic status can be useful in predicting drug efficacy in individual patients.
NETWORK ANALYSIS FOR DISCOVERY OF NEW DRUG TARGETS
Drug discovery for complex diseases requires the identification of therapeutic targets that can be used to achieve the desired therapeutic effect while reducing the risk of adverse events. Network analysis methods provide computational tools for pharmacologists and physiologists to identify and rank potential targets, which can then be used to develop drugs. The tasks of simultaneously identifying the appropriate target, determining efficacy for the therapeutic effect, and predicting adverse events constitute a problem that cannot be solved through the use of high-throughput experimental techniques alone owing to the high dimensional size of the problem. Determination of efficacy requires detailed dynamical models based on the biochemical kinetic parameters of the target and other proteins involved in the phenotypic responses. Network analysis can be used, in an unbiased way, to define the system that needs to be dynamically modeled and identify targets that, in theory, would enable one to have a higher specificity for the selected drug targets. Such combined network and dynamical analysis should both increase therapeutic efficacy and decrease the adverse events (15).
The mammalian signaling and regulatory network is complex, and even perturbations on the intended targets may lead to adverse events owing to propagation of signal to distal effectors in multiple cell types or tissues. Certain proteins, such as GRB2 or MYC, may directly interact with several hundred different proteins, according to a PubMed search in March 2011. Most protein kinases have 10 or more substrates. Such multiplicity of connections poses serious challenges in designing drugs that affect single pathways that lead to the desired therapeutic effects. To identify new drug targets, it is important to know how specificity of signal flow is achieved within pathways. Network analysis can be used (a) to identify how many different proteins will be affected by the targeting of a particular protein or (b) to identify if a protein participates in a motif (16). This approach involves the use of what is known as a path discovery. The simplest and oldest is Dijkstra’s (50) shortest-path algorithm: When given a seed node, the algorithm finds the shortest path between that node and potentially every other node. By looking for a path—that is, a series of edges from a starting node to an ending node—one can identify relationships and topology for the network. This algorithm can be expanded by setting requirements for the path; for example, one can require a path to start at a receptor, go through specific types of intracellular proteins, and eventually reach a known transcription factor. However, imposing such requirements would require a longer run time that increases with their complexity. A protein that has a high connectivity is commonly lethal, whereas diseased genes with lower connectivity are nonlethal but lead to a diseased phenotype (7). This type of analysis can produce initial ranked lists of potential targets that can be further vetted by additional criteria.
Both positive and negative effects can be predicted for a potential target on the basis of network analysis methods. For example, a network algorithm utilizing mean first passage time (MFPT) can, on the basis of a set of known genes that cause a prolonged long QT interval, identify drugs that may lead to a long QT interval event (15). In this method, Berger et al. (15) used the MFPT as a distance measure to assess how “close” a protein is to another protein that is known to be related to long QT intervals. Other methods of measuring functional distance can also be used. The nearest neighbor method assesses distance by measuring the shortest length of a path between a protein of interest and any of the known long QT–causing proteins. It is important to know the scoring metric used in the algorithm; for example, two methods looking at protein-protein interactions may measure them differently according to the problem of interest (6, 51). Network analysis methods can be used to determine the association of a protein with a physiological phenotype of interest on the basis of a defined set of proteins related to the phenotype. Therefore, proteins can be selected as potential targets on the basis of their high proximity to the phenotype of interest and their distance from proteins involved in adverse events. The potential of finding one target that can be optimized for a variety of criteria simultaneously is likely to be low. Computational network analysis allows us to explore combinatorial targets and thus increases our chances of finding useful therapeutic agents for complex diseases.
Network Analysis to Define the Context of Targets Involved in Therapeutic and Adverse Action
Network analysis can be used to identify physiologically relevant targets and the neighborhood within which these targets have their action. The network-building techniques require the selection of a seed list of proteins that are related to the physiology or pathophysiology of interest. A seed list is a set of related nodes in a network based on some predefined characterization; for example, the list of gene mutations related to the congenital prolonged QT interval used by Berger et al. (15) is a typical seed list. Other examples include lists of genes identified through microarray experiments or genes associated with a particular disease phenotype in the Online Mendelian Inheritance in Man® catalog (OMIM®). Characterization of the seed list can be based on key phrases that describe an organismal-level physiological event such as water retention or clotting. The list of relevant genes/proteins involved in these processes can be identified through the use of databases such as the Kyoto Encyclopedia of Genes and Genomes (KEGG) (52), the Gene Ontology project (53), and Reactome (54), or the list may be manually curated on the basis of the physiology of interest (16, 55). These seed lists serve as inputs to network-building algorithms (e.g., MFPT, nearest-neighbor, or other network distance metrics) that identify proteins within the neighborhood associated with the phenotype of interest. To identify candidates that are more easily druggable, researchers can further filter identified proteins for ontological classifications such as receptors, plasma membrane proteins, or cytoplasmic protein kinases. Thus, both potential drug targets and their functional neighborhood can be identified.
A similar approach can be used with a known drug: One can explore functional distances that are downstream of known drug targets to understand their therapeutic and adverse effects. For example, starting with rosiglitazone as the drug and peroxisome proliferator-activated receptor γ (PPARγ) as the target, we can identify a series of potentially important PPARγ-regulated effectors such as PTGS2(prostaglandin 2 synthase), SERPINE1 (plasminogen activator inhibitor), VEGFA (vascular endothelial growth factor A), APOB (apolipoprotein B), TSPO (mitochondrial translocator protein), MMP9 (metalloproteinase 9), IL-6 (interleukin-6), CASP3 (caspase 3), and CA2/4 (carbonic anhydrase 2/4). Several of these proteins are associated with cardiac function and myocardial infarction (56–63) and may in part be responsible for the observed association between rosiglitazone and myocardial infarction (64). Thus, using information from known drug targeting and building networks of related proteins can allow us to understand how drugs can have varied effects, some beneficial and others detrimental.
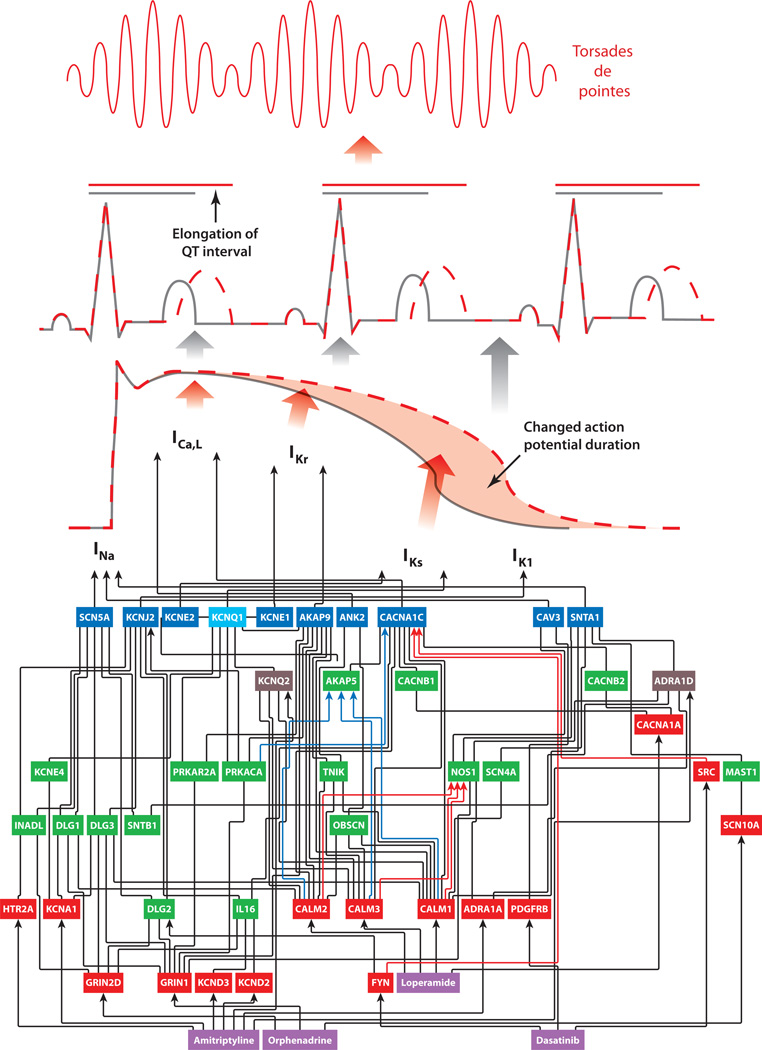
Such network building can also be used to understand how drug-induced effects percolate through multiple layers of organization. This is shown in Figure 2 for a few drugs that induce long QT syndrome as an adverse event. Targets of these drugs are part of the cellular networks that directly regulate the ion channels whose activity shapes the cardiac myocyte action potential. Changes in this cell physiological phenotype (myocyte action potential) lead to the observed organ-level phenotype: the prolonged QT interval in the electrocardiogram. Such prolongation can lead to arrhythmias that can result in life-threatening events such as torsades de pointes. Although this diagram implies that we understand the multiscale mechanism by which these drugs cause long QT syndrome and fatal arrhythmias, the intracellular network does not explain why such adverse events are observed in only a few patients. The answer may lie in building tissue-level networks to explain how changes in myocyte action potential may or may not lead to changes in electrocardiogram profiles. Building such multicellular networks that are anchored in molecular interaction networks will be the required next step to address multiscale biological problems of this kind.
Figure 2.
An intracellular network to explain how drug-induced adverse events can propagate across scales of organization. Drug interaction with the target leads through the network to an alteration of channel activity, which leads to a change in duration of the myocyte action potential, which leads to prolongation of the QT interval as seen in the electrocardiogram. This can result in fatal arrhythmias such as torsades de pointes. This network explains how drugs used to treat very different pathophysiologies such as diarrhea (loperamide) and cancer (dasatanib) can lead to long QT syndrome as an adverse event. However, this network does not explain why only some people show drug-induced long QT syndrome and why only some patients with long QT syndrome develop fatal arrhythmias. Additional information on genomic status and tissue- and multiorgan-level networks may be needed to explain individual susceptibility. The drugs are shown in purple boxes. Red boxes are drug targets, green boxes are intermediate nodes, and blue boxes are channels responsible for the various phases of the myocyte action potential. The light blue box represents a node that is an intermediate and also a channel involved in myocyte action potential, and each brown box represents a node that is both an intermediate and also a drug target. Black arrows indicate edges that are either undirected or directed with an unknown effect type (inhibition or activation), red arrows indicate edges that are activating, and blue arrows indicate edges that are inhibitory. Abbreviation: I, current that arises from the functioning of the channel protein. Adapted from Berger et al. (15) with permission.
Network Analyses of the FDA Adverse Events Reporting System Database
Building organ-level and organismal-level networks to identify concurrence of therapeutic and adverse phenotypes requires that one define the loci identifying relationships between drug-target interactions and phenotypes in humans. The FDA Adverse Events Reporting System (AERS) is a publicly available database that records drug-induced adverse events in people using one or more drugs. The FDA AERS database, in combination with other databases such as DrugBank, enables one to examine drug target relationships to phenotypes that are adverse events in an unbiased way in the context of multitherapeutic systems without control subjects. This allows us to associate drugs with possible adverse effects without having to conduct specialized trials. AERS can be used to determine relative adverse event profiles. For example, using AERS to select for patients who are being treated for schizophrenia and who experienced tardive dyskinesia as an adverse event, we identify haloperidol, promazine, risperidone, quetiapine, ziprasidone, and clozapine. All these drugs are associated with this adverse event in a clinical trial (65, 66).
Network analysis can be used to associate distal drug targets with particular adverse events. Using these drug targets as seed nodes, we can use methods such as MFPT to look for genes that are closely connected to them. This approach would help us understand and identify potentially useful targets for combination therapies that might mitigate the adverse events or help us predict drug targets that should be avoided because of their potential to lead to adverse events. A major limitation of using the FDA AERS database, however, is that it does not have data on the total number of people using a drug of interest. This information is critical in determining the prevalence of incidents and reporting bias. These problems can be ameliorated in the future through the use of electronic medical records, as they become more commonplace in hospitals and clinics. Mining data from electronic medical records to identify unknown drug interactions and adverse events is likely to be useful.
RELATING TARGETS OF A DRUG TO ITS STRUCTURE
The network analyses described above focus on drug targets as macromolecules that function within the context of cellular regulatory systems. Such an analysis does not take into account atomic-level interactions between the drug and its targets. As we understand more about cellular-level and tissue/organ-level networks, potential drug targets will have to be filtered by structural criteria to determine the ability of the target to be regulated by drugs. Interactions between a drug and its target depend on structural determinants both in the drug and in the target. This is a well-studied area in medicinal chemistry and structural pharmacology. The binding pockets of targets are often analyzed to understand how the drugs fit and the types of conformational changes they induce. Agonists and antagonists interact with the same binding pocket, and the differences in detailed interactions lead to either activation or inhibition of the receptor. The structure of a drug can be used to determine the targeting of a drug through various computational algorithms (67–72) and for ADMET (absorption, distribution, metabolism, excretion, and toxicity) predictions (73). The identification of targets that bind or metabolize structural variants of a drug differently can serve as the starting point for network analysis in the identification of off-target physiological events. However, there are no tools to conduct such scalable computation easily. To build such integrated networks, we require knowledge of the structures of both the drug and the targets of the drug. Often, target structures are not readily available. This lack of structural knowledge can lead to false-positive drug-target interactions. The potential of false positives may be decreased through the application of orthogonal experimental techniques such as high-content screening (74), in vitro ADMET (75), medicinal chemistry techniques (76, 77), binding screens (78), and gene regulation (79). Methods based on binding screens and gene regulation may also be used to develop drug-specific target interactions as this information is made available through the differential binding interactions and gene regulation with and without the drug.
PERSPECTIVE
Although in its infancy, the field of systems pharmacology has enormous potential to impact both drug development and drug usage in the future. Currently, drug development is largely focused on noncommunicable diseases such as cancers, metabolic diseases, type 2 diabetes, psychiatric disorders, and immune disorders, as well as communicable diseases wherein the pathology arises from complex host-pathogen interactions and is not susceptible to simple treatment approaches such as the use of antibiotics. Developing drugs for these diseases through classical empirical methods has not proven to be productive. Many molecules that show good therapeutic effects in cellular or animal models fail to be efficacious in humans. These failures arise from our lack of understanding of human biology as defined by the multiscale mechanisms that underlie the propagation of effects from molecular-level drug-target interactions to organismal-level phenotypes. In addition to understanding and predicting efficacy of therapy, it is becoming increasingly imperative that treatments are personalized so that the risks associated with the drug therapy are understood before treatment commences. These considerations lead to a set of key questions (see sidebar, Questions in Systems Pharmacology) that need to be addressed by research in the field of systems pharmacology. As such research progresses, the pace of drug discovery and therapy should become more proportional to the pace of discovery in basic biomedical sciences.
QUESTIONS IN SYSTEMS PHARMACOLOGY.
What are the characteristics of diseases for which drugs at a single target may not be therapeutically efficacious?
How does intracellular and intercellular networking give rise to adverse events?
How do we relate the efficacy of (poly)pharmacology to the genomic status of the individual, and how does genomic status interact with environment and behavior to control (poly)pharmacology efficacy?
How do we determine what combinations of targets are most likely to be effective for polypharmacology of complex diseases?
Can we use the human interactome and the genomic status of the individual to predict therapeutic efficacy and adverse event probability prior to commencement of therapy?
ACKNOWLEDGMENTS
The authors’ research is supported by grant GM54508 and Systems Biology Center New York grant P50-GM071558, both from the NIH’s National Institute of General Medical Sciences (NIGMS). S.Z. is currently supported by a predoctoral training grant in pharmacological sciences (T32-GM062754) and is a trainee in the NIGMS Medical Scientist Training Program.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Li JW-H, Vederas JC. Drug discovery and natural products: end of an era or an endless frontier? Science. 2009;325:161–165. doi: 10.1126/science.1168243. [DOI] [PubMed] [Google Scholar]
- 2.Fitzgerald G. Drug development needs a new brand of science. Nature. 2010;468:869. doi: 10.1038/468869a. [DOI] [PubMed] [Google Scholar]
- 3.Brunton LL, Chabner BA, Knollman BC. Goodman & Gilman’s The Pharmacological Basis of Therapeutics. 12th ed. New York: McGraw-Hill; 2010. p. 1808. [Google Scholar]
- 4.Davidson EH. Emerging properties of animal gene regulatory networks. Nature. 2010;468:911–920. doi: 10.1038/nature09645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stark C, Breitkreutz B-J, Chatr-Aryamontri A, Boucher L, Oughtred R, et al. The BioGRID Interaction Database: 2011 update. Nucleic Acids Res. 2011;39:D698–D704. doi: 10.1093/nar/gkq1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao S, Li S. Network-based relating pharmacological and genomic spaces for drug target identification. PLoS ONE. 2010;5:e11764. doi: 10.1371/journal.pone.0011764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barabási A-L, Gulbahce N, Loscalzo J. Network medicine: a network-based approach to human disease. Nat. Rev. Genet. 2011;12:56–68. doi: 10.1038/nrg2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vidal M, Cusick ME, Barabási A-L. Interactome networks and human disease. Cell. 2011;144:986–998. doi: 10.1016/j.cell.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferrell JE, Tsai TY-C, Yang Q. Modeling the cell cycle: Why do certain circuits oscillate? Cell. 2011;144:874–885. doi: 10.1016/j.cell.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 10.Singhania R, Sramkoski RM, Jacobberger JW, Tyson JJ. A hybrid model of mammalian cell cycle regulation. PLoS Comput. Biol. 2011;7:e1001077. doi: 10.1371/journal.pcbi.1001077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neves SR, Tsokas P, Sarkar A, Grace EA, Rangamani P, et al. Cell shape and negative links in regulatory motifs together control spatial information flow in signaling networks. Cell. 2008;133:666–680. doi: 10.1016/j.cell.2008.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wishart DS, Knox C, Guo AC, Cheng D, Shrivastava S, et al. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008;36:D901–D906. doi: 10.1093/nar/gkm958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao F, Xuan Z, Liu L, Zhang MQ. TRED: a Transcriptional Regulatory Element Database and a platform for in silico gene regulation studies. Nucleic Acids Res. 2005;33:D103–D107. doi: 10.1093/nar/gki004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goh K-I, Cusick ME, Valle D, Childs B, Vidal M, Barabási AL. The human disease network. Proc. Natl. Acad. Sci. USA. 2007;104:8685–8690. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berger SI, Ma’ayan A, Iyengar R. Systems pharmacology of arrhythmias. Sci. Signal. 2010;3:ra30. doi: 10.1126/scisignal.2000723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma’ayan A, Jenkins SL, Neves S, Hasseldine A, Grace E, et al. Formation of regulatory patterns during signal propagation in a mammalian cellular network. Science. 2005;309:1078–1083. doi: 10.1126/science.1108876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chatr-aryamontri A, Ceol A, Peluso D, Nardozza A, Panni S, et al. VirusMINT: a viral protein interaction database. Nucleic Acids Res. 2009;37:D669–D673. doi: 10.1093/nar/gkn739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chautard E, Ballut L, Thierry-Mieg N, Ricard-Blum S. MatrixDB, a database focused on extracellular protein-protein and protein-carbohydrate interactions. Bioinformatics. 2009;25:690–691. doi: 10.1093/bioinformatics/btp025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharan R, Ulitsky I, Shamir R. Network-based prediction of protein function. Mol. Syst. Biol. 2007;3:88. doi: 10.1038/msb4100129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schaefer CF, Anthony K, Krupa S, Buchoff J, Day M, et al. PID: the Pathway Interaction Database. Nucleic Acids Res. 2009;37:D674–D679. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rost S, Fregin A, Ivaskevicius V, Conzelmann E, Hörtnagel K, et al. Mutations in VKORC 1 cause warfarin resistance and multiple coagulation factor deficiency type 2. Nature. 2004;427:537–541. doi: 10.1038/nature02214. [DOI] [PubMed] [Google Scholar]
- 22.Aithal GP, Day CP, Kesteven PJ, Daly AK. Association of polymorphisms in the cytochrome P450 CYP2C9 with warfarin dose requirement and risk of bleeding complications. Lancet. 1999;353:717–719. doi: 10.1016/S0140-6736(98)04474-2. [DOI] [PubMed] [Google Scholar]
- 23.Drmanac R, Sparks AB, Callow MJ, Halpern AL, Burns NL, et al. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science. 2010;327:78–81. doi: 10.1126/science.1181498. [DOI] [PubMed] [Google Scholar]
- 24.Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 25.Rix U, Remsing Rix LL, Terker AS, Fernbach NV, Hantschel O, et al. A comprehensive target selectivity survey of the BCR-ABL kinase inhibitor INNO-406 by kinase profiling and chemical proteomics in chronic myeloid leukemia cells. Leukemia. 2010;24:44–50. doi: 10.1038/leu.2009.228. [DOI] [PubMed] [Google Scholar]
- 26.Muroi M, Kazami S, Noda K, Kondo H, Takayama H, et al. Application of proteomic profiling based on 2D-DIGE for classification of compounds according to the mechanism of action. Chem. Biol. 2010;17:460–470. doi: 10.1016/j.chembiol.2010.03.016. [DOI] [PubMed] [Google Scholar]
- 27.Kaddurah-Daouk R, Kristal BS, Weinshilboum RM. Metabolomics: a global biochemical approach to drug response and disease. Annu. Rev. Pharmacol. Toxicol. 2008;48:653–683. doi: 10.1146/annurev.pharmtox.48.113006.094715. [DOI] [PubMed] [Google Scholar]
- 28.Kaddurah-Daouk R, McEvoy J, Baillie RA, Lee D, Yao JK, et al. Metabolomic mapping of atypical antipsychotic effects in schizophrenia. Mol. Psychiatry. 2007;12:934–945. doi: 10.1038/sj.mp.4002000. [DOI] [PubMed] [Google Scholar]
- 29.Ji Y, Hebbring S, Zhu H, Jenkins GD, Biernacka J, et al. Glycine and a glycine dehydrogenase (GLDC) SNP as citalopram/escitalopram response biomarkers in depression: pharmacometabolomics-informed pharmacogenomics. Clin. Pharmacol. Ther. 2010;89:97–104. doi: 10.1038/clpt.2010.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Evans WE, McLeod HL. Pharmacogenomics—drug disposition, drug targets, and side effects. N. Engl. J. Med. 2003;348:538–549. doi: 10.1056/NEJMra020526. [DOI] [PubMed] [Google Scholar]
- 31.Drazen JM, Silverman EK, Lee TH. Heterogeneity of therapeutic responses in asthma. Br. Med. Bull. 2000;56:1054–1070. doi: 10.1258/0007142001903535. [DOI] [PubMed] [Google Scholar]
- 32.Mougey EB, Feng H, Castro M, Irvin CG, Lima JJ. Absorption of montelukast is transporter mediated: a common variant of OATP2B1 is associated with reduced plasma concentrations and poor response. Pharmacogenet. Genomics. 2009;19:129–138. doi: 10.1097/FPC.0b013e32831bd98c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Roock W, Jonker DJ, Di Nicolantonio F, Sartore-Bianchi A, Tu D, et al. Association of KRAS p.G13D mutation with outcome in patients with chemotherapy-refractory metastatic colorectal cancer treated with cetuximab. JAMA. 2010;304:1812–1820. doi: 10.1001/jama.2010.1535. [DOI] [PubMed] [Google Scholar]
- 34.Ciszkowski C, Madadi P, Phillips MS, Lauwers AE, Koren G. Codeine, ultrarapid-metabolism genotype, and postoperative death. N. Engl. J. Med. 2009;361:827–828. doi: 10.1056/NEJMc0904266. [DOI] [PubMed] [Google Scholar]
- 35.Dalén P, Dahl ML, Bernal Ruiz ML, Nordin J, Bertilsson L. 10-Hydroxylation of nortriptyline in white persons with 0, 1, 2, 3, and 13 functional CYP2D6 genes. Clin. Pharmacol. Ther. 1998;63:444–452. doi: 10.1016/S0009-9236(98)90040-6. [DOI] [PubMed] [Google Scholar]
- 36.Beer B, Erb R, Pitterl F, Niederstätter H, Maroñas O, et al. CYP2D6 genotyping by liquid chromatography-electrospray ionization mass spectrometry. Anal. Bioanal. Chem. 2011;400(8):2361–2370. doi: 10.1007/s00216-010-4597-4. [DOI] [PubMed] [Google Scholar]
- 37.Guengerich FP. Cytochrome P450 enzymes in the generation of commercial products. Nat. Rev. Drug Discov. 2002;1:359–366. doi: 10.1038/nrd792. [DOI] [PubMed] [Google Scholar]
- 38.Treviño LR, Shimasaki N, Yang W, Panetta JC, Cheng C, et al. Germline genetic variation in an organic anion transporter polypeptide associated with methotrexate pharmacokinetics and clinical effects. J. Clin. Oncol. 2009;27:5972–5978. doi: 10.1200/JCO.2008.20.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Erp NP, Eechoute K, van der Veldt AA, Haanen JB, Reyners AKL, et al. Pharmacogenetic pathway analysis for determination of sunitinib-induced toxicity. J. Clin. Oncol. 2009;27:4406–4412. doi: 10.1200/JCO.2008.21.7679. [DOI] [PubMed] [Google Scholar]
- 40.Heinrich MC, Corless CL, Demetri GD, Blanke CD, von Mehren M, et al. Kinase mutations and imatinib response in patients with metastatic gastrointestinal stromal tumor. J. Clin. Oncol. 2003;21:4342–4349. doi: 10.1200/JCO.2003.04.190. [DOI] [PubMed] [Google Scholar]
- 41.Kobayashi S, Boggon TJ, Dayaram T, Jänne PA, Kocher O, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 2005;352:786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 42.Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mallal S, Phillips E, Carosi G, Molina J-M, Workman C, et al. HLA-B*5701 screening for hypersensitivity to abacavir. N. Engl. J. Med. 2008;358:568–579. doi: 10.1056/NEJMoa0706135. [DOI] [PubMed] [Google Scholar]
- 44.Ingle JN, Schaid DJ, Goss PE, Liu M, Mushiroda T, et al. Genome-wide associations and functional genomic studies of musculoskeletal adverse events in women receiving aromatase inhibitors. J. Clin. Oncol. 2010;28:4674–4682. doi: 10.1200/JCO.2010.28.5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klein TE, Chang JT, Cho MK, Easton KL, Fergerson R, et al. Integrating genotype and phenotype information: an overview of thePharmGKBproject.Pharmacogenetics Research Network and Knowledge Base. Pharmacogenomics J. 2001;1:167–170. doi: 10.1038/sj.tpj.6500035. [DOI] [PubMed] [Google Scholar]
- 46.Abraham AK, Maurer TS, Kalgutkar AS, Gao X, Li M, et al. Pharmacodynamicmodel of parathyroid hormone modulation by a negative allosteric modulator of the calcium-sensing receptor. AAPS J. 2011;13:265–273. doi: 10.1208/s12248-011-9266-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bhalla US, Iyengar R. Emergent properties of networks of biological signaling pathways. Science. 1999;283:381–387. doi: 10.1126/science.283.5400.381. [DOI] [PubMed] [Google Scholar]
- 48.Tanaka K, Augustine GJ. A positive feedback signal transduction loop determines timing of cerebellar long-term depression. Neuron. 2008;59:608–620. doi: 10.1016/j.neuron.2008.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Panetta JC, Sparreboom A, Pui C-H, Relling MV, Evans WE. Modeling mechanisms of in vivo variability in methotrexate accumulation and folate pathway inhibition in acute lymphoblastic leukemia cells. PLoS Comput. Biol. 2010;6:e1001019. doi: 10.1371/journal.pcbi.1001019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dijkstra EW. A note on two problems in connexion with graphs. Numer. Math. 1959;1:269–271. [Google Scholar]
- 51.Huang T, Wang P, Ye Z-Q, Xu H, He Z, et al. Prediction of deleterious non-synonymous SNPs based on protein interaction network and hybrid properties. PLoS ONE. 2010;5:e11900. doi: 10.1371/journal.pone.0011900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38:D355–D360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Matthews L, Gopinath G, Gillespie M, Caudy M, Croft D, et al. Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res. 2009;37:D619–D622. doi: 10.1093/nar/gkn863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kandasamy K, Mohan SS, Raju R, Keerthikumar S, Kumar GSS, et al. NetPath: a public resource of curated signal transduction pathways. Genome Biol. 2010;11:R3. doi: 10.1186/gb-2010-11-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Holly TA, Drincic A, Byun Y, Nakamura S, Harris K, et al. Caspase inhibition reduces myocyte cell death induced by myocardial ischemia and reperfusion in vivo. J. Mol. Cell. Cardiol. 1999;31:1709–1715. doi: 10.1006/jmcc.1999.1006. [DOI] [PubMed] [Google Scholar]
- 57.Schaller S, Paradis S, Ngoh GA, Assaly R, Buisson B, et al. TRO40303, a new cardioprotective compound, inhibits mitochondrial permeability transition. J. Pharmacol. Exp. Ther. 2010;333:696–706. doi: 10.1124/jpet.110.167486. [DOI] [PubMed] [Google Scholar]
- 58.Abbott JD, Huang Y, Liu D, Hickey R, Krause DS, Giordano FJ. Stromal cell-derived factor-1α plays a critical role in stem cell recruitment to the heart after myocardial infarction but is not sufficient to induce homing in the absence of injury. Circulation. 2004;110:3300–3305. doi: 10.1161/01.CIR.0000147780.30124.CF. [DOI] [PubMed] [Google Scholar]
- 59.Diedrich M, Tadic J, Mao L, Wacker MA, Nebrich G, et al. Heart protein expression related to age and sex in mice and humans. Int. J. Mol. Med. 2007;20:865–874. [PubMed] [Google Scholar]
- 60.Morange PE, Saut N, Alessi MC, Yudkin JS, Margaglione M, et al. Association of plasminogen activator inhibitor (PAI)-1 (SERPINE1) SNPs with myocardial infarction, plasma PAI-1, and metabolic parameters: the HIFMECH study. Arterioscler. Thromb. Vasc. Biol. 2007;27:2250–2257. doi: 10.1161/ATVBAHA.107.149468. [DOI] [PubMed] [Google Scholar]
- 61.Shintani S, Murohara T, Ikeda H, Ueno T, Honma T, et al. Mobilization of endothelial progenitor cells in patients with acute myocardial infarction. Circulation. 2001;103:2776–2779. doi: 10.1161/hc2301.092122. [DOI] [PubMed] [Google Scholar]
- 62.Mukherjee D, Nissen SE, Topol EJ. Risk of cardiovascular events associated with selective COX-2 inhibitors. JAMA. 2011;286:954–959. doi: 10.1001/jama.286.8.954. [DOI] [PubMed] [Google Scholar]
- 63.Yusuf S, Hawken S, Ounpuu S, Dans T, Avezum A, et al. Effect of potentially modifiable risk factors associated with myocardial infarction in 52 countries (the INTERHEART study): case-control study. Lancet. 2004;364:937–952. doi: 10.1016/S0140-6736(04)17018-9. [DOI] [PubMed] [Google Scholar]
- 64.Nissen SE, Wolski K. Effect of rosiglitazone on the risk of myocardial infarction and death from cardiovascular causes. N. Engl. J. Med. 2007;356:2457–2471. doi: 10.1056/NEJMoa072761. [DOI] [PubMed] [Google Scholar]
- 65.Caroff SN, Hurford I, Lybrand J, Campbell EC. Movement disorders induced by antipsychotic drugs: implications of the CATIE schizophrenia trial. Neurol. Clin. 2011;29:127–148. doi: 10.1016/j.ncl.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Saltz BL, Woerner MG, Kane JM, Lieberman JA, Alvir JM, et al. Prospective study of tardive dyskinesia incidence in the elderly. JAMA. 1991;266:2402–2406. [PubMed] [Google Scholar]
- 67.Schmidtke P, Le Guilloux V, Maupetit J, Tufféry P. Fpocket: online tools for protein ensemble pocket detection and tracking. Nucleic Acids Res. 2010;38(Suppl):W582–W589. doi: 10.1093/nar/gkq383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ballester PJ, Mitchell JBO. A machine learning approach to predicting protein-ligand binding affinity with applications to molecular docking. Bioinformatics. 2010;26:1169–1175. doi: 10.1093/bioinformatics/btq112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thangudu RR, Tyagi M, Shoemaker BA, Bryant SH, Panchenko AR, Madej T. Knowledge-based annotation of small molecule binding sites in proteins. BMC Bioinformatics. 2010;11:365. doi: 10.1186/1471-2105-11-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chang RL, Xie L, Xie L, Bourne PE, Palsson BØ. Drug off-target effects predicted using structural analysis in the context of a metabolic network model. PLoS Comput. Biol. 2010;6:e1000938. doi: 10.1371/journal.pcbi.1000938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kitchen DB, Decornez H, Furr JR, Bajorath J. Docking and scoring in virtual screening for drug discovery: methods and applications. Nat. Rev. Drug Discov. 2004;3:935–949. doi: 10.1038/nrd1549. [DOI] [PubMed] [Google Scholar]
- 72.Keiser MJ, Roth BL, Armbruster BN, Ernsberger P, Irwin JJ, Shoichet BK. Relating protein pharmacology by ligand chemistry. Nat. Biotechnol. 2007;25:197–206. doi: 10.1038/nbt1284. [DOI] [PubMed] [Google Scholar]
- 73.Schneider G, Fechner U. Computer-based de novo design of drug-like molecules. Nat. Rev. Drug Discov. 2005;4:649–663. doi: 10.1038/nrd1799. [DOI] [PubMed] [Google Scholar]
- 74.Macarron R, Banks MN, Bojanic D, Burns DJ, Cirovic DA, et al. Impact of high-throughput screening in biomedical research. Nat. Rev. Drug Discov. 2011;10:188–195. doi: 10.1038/nrd3368. [DOI] [PubMed] [Google Scholar]
- 75.Gleeson MP, Hersey A, Montanari D, Overington J. Probing the links between in vitro potency, ADMET and physicochemical parameters. Nat. Rev. Drug Discov. 2011;10:197–208. doi: 10.1038/nrd3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wilson ID. High-performance liquid chromatography-mass spectrometry (HPLC-MS)-based drug metabolite profiling. Methods Mol. Biol. 2011;708:173–190. doi: 10.1007/978-1-61737-985-7_10. [DOI] [PubMed] [Google Scholar]
- 77.Lenz EM. Metabolic profiling. Methods Mol. Biol. 2011;708:299–319. doi: 10.1007/978-1-61737-985-7_18. [DOI] [PubMed] [Google Scholar]
- 78.Clemons PA, Bodycombe NE, Carrinski HA, Wilson JA, Shamji AF, et al. Small molecules of different origins have distinct distributions of structural complexity that correlate with protein-binding profiles. Proc. Natl. Acad. Sci. USA. 2010;107:18787–18792. doi: 10.1073/pnas.1012741107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Iskar M, Campillos M, Kuhn M, Jensen LJ, vanNoort V, Bork P. Drug-induced regulation of target expression. PLoS Comput. Biol. 2010;6:e1000925. doi: 10.1371/journal.pcbi.1000925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kim KA, Park PW, Hong SJ, Park J-Y. The effect of CYP2C19 polymorphism on the pharmacokinetics and pharmacodynamics of clopidogrel: a possible mechanism for clopidogrel resistance. Clin. Pharmacol. Ther. 2008;84:236–242. doi: 10.1038/clpt.2008.20. [DOI] [PubMed] [Google Scholar]
- 81.Wadelius M, Sörlin K, Wallerman O, Karlsson J, Yue Q-Y, et al. Warfarin sensitivity related to CYP2C9, CYP3A5, ABCB1 (MDR1) and other factors. Pharmacogenomics J. 2004;4:40–48. doi: 10.1038/sj.tpj.6500220. [DOI] [PubMed] [Google Scholar]
- 82.Ishikawa T, Sekiguchi F, Fukase Y, Sawada N, Ishitsuka H. Positive correlation between the efficacy of capecitabine and doxifluridine and the ratio of thymidine phosphorylase to dihydropyrimidine dehydrogenase activities in tumors in human cancer xenografts. Cancer Res. 1998;58:685–690. [PubMed] [Google Scholar]
- 83.Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J. Clin. Oncol. 2008;26:1626–1634. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 84.Debiec-Rychter M, Dumez H, Judson I, Wasag B, Verweij J, et al. Use of c-KIT/PDGFRA mutational analysis to predict the clinical response to imatinib in patients with advanced gastrointestinal stromal tumours entered on phase I and II studies of the EORTC Soft Tissue and Bone Sarcoma Group. EurJ. Cancer. 2004;40:689–695. doi: 10.1016/j.ejca.2003.11.025. [DOI] [PubMed] [Google Scholar]
- 85.Kakizuka A, Miller WH, Umesono K, Warrell RP, Frankel SR, et al. Chromosomal translocation t(15;17) in human acute promyelocytic leukemia fuses RARα with a novel putative transcription factor, PML. Cell. 1991;66:663–674. doi: 10.1016/0092-8674(91)90112-c. [DOI] [PubMed] [Google Scholar]
- 86.Chung W-H, Hung S-I, Hong H-S, Hsih M-S, Yang L-C, et al. Medical genetics: a marker for Stevens-Johnson syndrome. Nature. 2004;428:486. doi: 10.1038/428486a. [DOI] [PubMed] [Google Scholar]
- 87.Mallal S, Nolan D, Witt C, Masel G, Martin A, et al. HLA-B *5701, HLA-DR7, and HLA-DQ3 and hypersensitivity to HIV-1 reverse-transcriptase inhibitor abacavir. Lancet. 2002;359:727–732. doi: 10.1016/s0140-6736(02)07873-x. [DOI] [PubMed] [Google Scholar]
- 88.Warne T, Moukhametzianov R, Baker JG, Nehmé R, Edwards PC, et al. The structural basis for agonist and partial agonist action on a β1-adrenergic receptor. Nature. 2011;469:241–244. doi: 10.1038/nature09746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Erlanson DA, Arndt JW, Cancilla MT, Cao K, Elling RA, et al. Discovery of a potent and highly selective PDK1 inhibitor via fragment-based drug discovery. Bioorg. Med. Chem. Lett. 2011;21 doi: 10.1016/j.bmcl.2011.03.032. 3083–78. [DOI] [PubMed] [Google Scholar]