Abstract
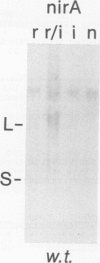
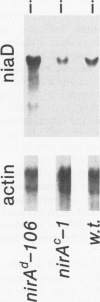
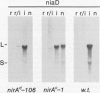
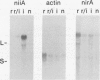
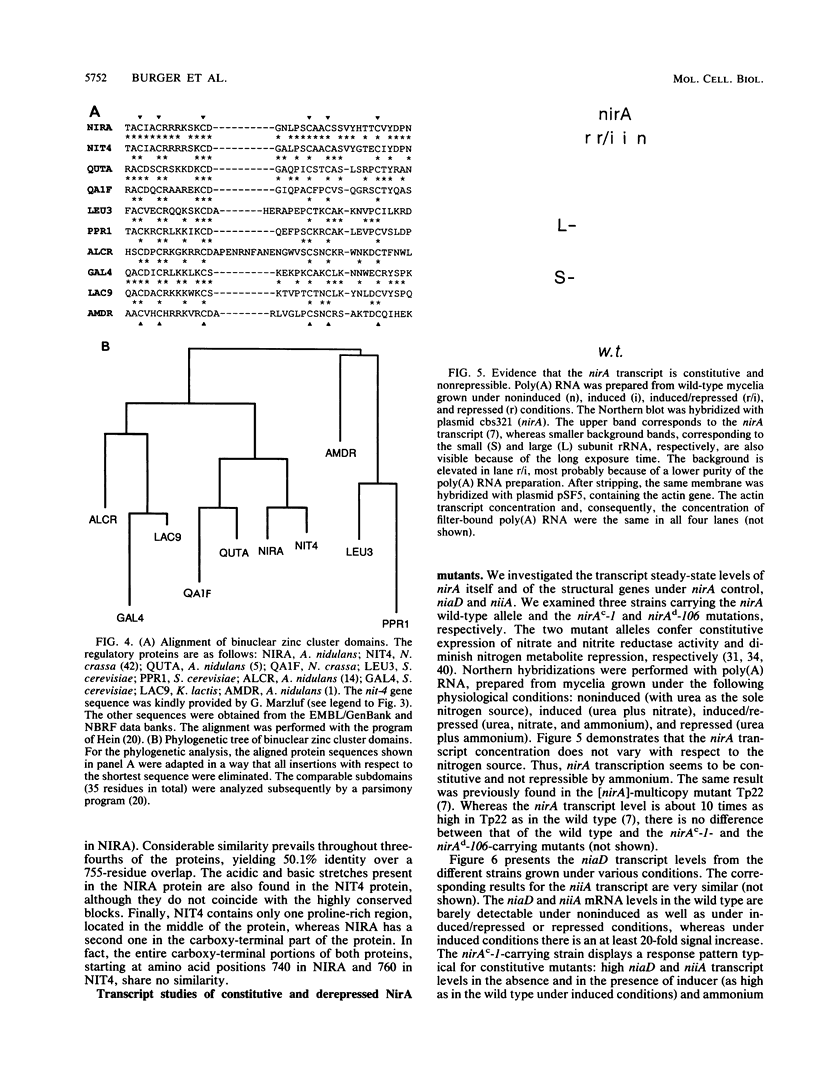
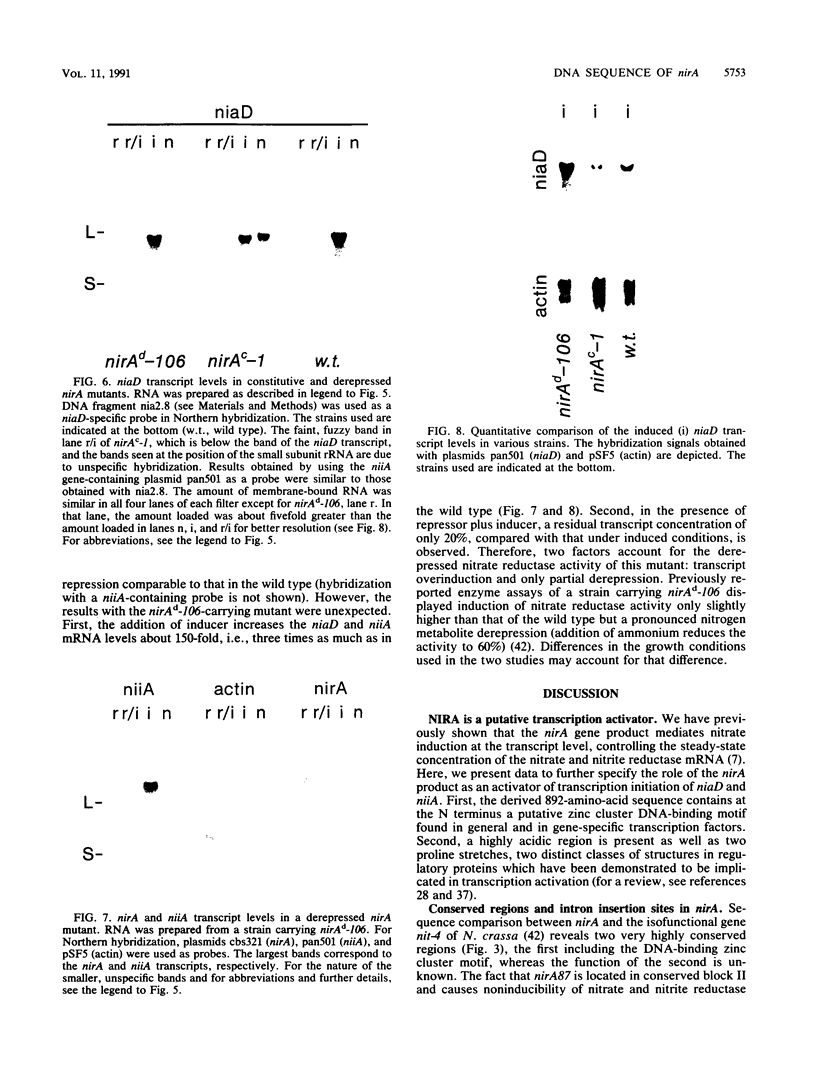
The nucleotide sequence of nirA, mediating nitrate induction in Aspergillus nidulans, has been determined. Alignment of the cDNA and the genomic DNA sequence indicates that the gene contains four introns and encodes a protein of 892 amino acids. The deduced NIRA protein displays all characteristics of a transcriptional activator. A putative double-stranded DNA-binding domain in the amino-terminal part comprises six cysteine residues, characteristic for the GAL4 family of zinc finger proteins. An amino-terminal highly acidic region and two proline-rich regions are also present. The nucleotide sequences of two mutations were determined after they were mapped by transformation with overlapping DNA fragments, amplified by the polymerase chain reaction. nirA87, a mutation conferring noninducibility by nitrate and nitrite, has a -1 frameshift at triplet 340, which eliminates 549 C-terminal amino acids from the polypeptide. Under the assumption that the truncated polypeptide is stable, it comprises the zinc finger domain and the acidic region, which seem not sufficient for transcriptional activation. nirAd-106, an allele conferring nitrogen metabolite derepression of nitrate and nitrite reductase activity, includes two transitions, changing a glutamic acid to a lysine and a valine to an alanine, situated between a basic and a proline-rich region of the protein. Northern (RNA) analysis of the wild type and of constitutive (nirAc) and derepressed (nirAd) mutants show that the nirA transcript does not vary between these strains, being in all cases constitutively expressed. On the other hand, transcript levels of structural genes (niaD and niiA) do vary, being highly inducible in the wild type but constitutively expressed in the nirAc mutant. The nirAd mutant appears phenotypically derepressed, because the niaD and niiA transcript levels are overinduced in the presence of nitrate but are still partially repressed in the presence of ammonium.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andrianopoulos A., Hynes M. J. Sequence and functional analysis of the positively acting regulatory gene amdR from Aspergillus nidulans. Mol Cell Biol. 1990 Jun;10(6):3194–3203. doi: 10.1128/mcb.10.6.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballance D. J. Sequences important for gene expression in filamentous fungi. Yeast. 1986 Dec;2(4):229–236. doi: 10.1002/yea.320020404. [DOI] [PubMed] [Google Scholar]
- Berg J. M. DNA binding specificity of steroid receptors. Cell. 1989 Jun 30;57(7):1065–1068. doi: 10.1016/0092-8674(89)90042-1. [DOI] [PubMed] [Google Scholar]
- Berg J. M. Zinc fingers and other metal-binding domains. Elements for interactions between macromolecules. J Biol Chem. 1990 Apr 25;265(12):6513–6516. [PubMed] [Google Scholar]
- Beri R. K., Whittington H., Roberts C. F., Hawkins A. R. Isolation and characterization of the positively acting regulatory gene QUTA from Aspergillus nidulans. Nucleic Acids Res. 1987 Oct 12;15(19):7991–8001. doi: 10.1093/nar/15.19.7991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger G., Tilburn J., Scazzocchio C. Molecular cloning and functional characterization of the pathway-specific regulatory gene nirA, which controls nitrate assimilation in Aspergillus nidulans. Mol Cell Biol. 1991 Feb;11(2):795–802. doi: 10.1128/mcb.11.2.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke J. M., RajBhandary U. L. Intron within the large rRNA gene of N. crassa mitochondria: a long open reading frame and a consensus sequence possibly important in splicing. Cell. 1982 Dec;31(3 Pt 2):509–520. doi: 10.1016/0092-8674(82)90307-5. [DOI] [PubMed] [Google Scholar]
- Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988 Nov 25;16(22):10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cove D. J. Genetic studies of nitrate assimilation in Aspergillus nidulans. Biol Rev Camb Philos Soc. 1979 Aug;54(3):291–327. doi: 10.1111/j.1469-185x.1979.tb01014.x. [DOI] [PubMed] [Google Scholar]
- Cove D. J. The induction and repression of nitrate reductase in the fungus Aspergillus nidulans. Biochim Biophys Acta. 1966 Jan 11;113(1):51–56. doi: 10.1016/s0926-6593(66)80120-0. [DOI] [PubMed] [Google Scholar]
- Davis M. A., Hynes M. J. Complementation of areA- regulatory gene mutations of Aspergillus nidulans by the heterologous regulatory gene nit-2 of Neurospora crassa. Proc Natl Acad Sci U S A. 1987 Jun;84(11):3753–3757. doi: 10.1073/pnas.84.11.3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeley R. G., Gordon J. I., Burns A. T., Mullinix K. P., Binastein M., Goldberg R. F. Primary activation of the vitellogenin gene in the rooster. J Biol Chem. 1977 Nov 25;252(22):8310–8319. [PubMed] [Google Scholar]
- Felenbok B., Sequeval D., Mathieu M., Sibley S., Gwynne D. I., Davies R. W. The ethanol regulon in Aspergillus nidulans: characterization and sequence of the positive regulatory gene alcR. Gene. 1988 Dec 20;73(2):385–396. doi: 10.1016/0378-1119(88)90503-3. [DOI] [PubMed] [Google Scholar]
- Fidel S., Doonan J. H., Morris N. R. Aspergillus nidulans contains a single actin gene which has unique intron locations and encodes a gamma-actin. Gene. 1988 Oct 30;70(2):283–293. doi: 10.1016/0378-1119(88)90200-4. [DOI] [PubMed] [Google Scholar]
- Frankel A. D., Berg J. M., Pabo C. O. Metal-dependent folding of a single zinc finger from transcription factor IIIA. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4841–4845. doi: 10.1073/pnas.84.14.4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohman M. A., Dush M. K., Martin G. R. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y. H., Marzluf G. A. nit-2, the major positive-acting nitrogen regulatory gene of Neurospora crassa, encodes a sequence-specific DNA-binding protein. Proc Natl Acad Sci U S A. 1990 Jul;87(14):5331–5335. doi: 10.1073/pnas.87.14.5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein J. Unified approach to alignment and phylogenies. Methods Enzymol. 1990;183:626–645. doi: 10.1016/0076-6879(90)83041-7. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Kudla B., Caddick M. X., Langdon T., Martinez-Rossi N. M., Bennett C. F., Sibley S., Davies R. W., Arst H. N., Jr The regulatory gene areA mediating nitrogen metabolite repression in Aspergillus nidulans. Mutations affecting specificity of gene activation alter a loop residue of a putative zinc finger. EMBO J. 1990 May;9(5):1355–1364. doi: 10.1002/j.1460-2075.1990.tb08250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulmburg P., Prangé T., Mathieu M., Sequeval D., Scazzocchio C., Felenbok B. Correct intron splicing generates a new type of a putative zinc-binding domain in a transcriptional activator of Aspergillus nidulans. FEBS Lett. 1991 Mar 11;280(1):11–16. doi: 10.1016/0014-5793(91)80193-7. [DOI] [PubMed] [Google Scholar]
- Lang B. F., Burger G. A collection of programs for nucleic acid and protein analysis, written in FORTRAN 77 for IBM-PC compatible microcomputers. Nucleic Acids Res. 1986 Jan 10;14(1):455–465. doi: 10.1093/nar/14.1.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang B. F., Burger G. A rapid, high resolution DNA sequencing gel system. Anal Biochem. 1990 Jul;188(1):176–180. doi: 10.1016/0003-2697(90)90548-n. [DOI] [PubMed] [Google Scholar]
- Lang B. F. The mitochondrial genome of the fission yeast Schizosaccharomyces pombe: highly homologous introns are inserted at the same position of the otherwise less conserved cox1 genes in Schizosaccharomyces pombe and Aspergillus nidulans. EMBO J. 1984 Sep;3(9):2129–2136. doi: 10.1002/j.1460-2075.1984.tb02102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermod N., O'Neill E. A., Kelly T. J., Tjian R. The proline-rich transcriptional activator of CTF/NF-I is distinct from the replication and DNA binding domain. Cell. 1989 Aug 25;58(4):741–753. doi: 10.1016/0092-8674(89)90108-6. [DOI] [PubMed] [Google Scholar]
- Mitchell P. J., Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989 Jul 28;245(4916):371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- Pan T., Coleman J. E. GAL4 transcription factor is not a "zinc finger" but forms a Zn(II)2Cys6 binuclear cluster. Proc Natl Acad Sci U S A. 1990 Mar;87(6):2077–2081. doi: 10.1073/pnas.87.6.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan T., Halvorsen Y. D., Dickson R. C., Coleman J. E. The transcription factor LAC9 from Kluyveromyces lactis-like GAL4 from Saccharomyces cerevisiae forms a Zn(II)2Cys6 binuclear cluster. J Biol Chem. 1990 Dec 15;265(35):21427–21429. [PubMed] [Google Scholar]
- Pateman J. A., Cove D. J. Regulation of nitrate reduction in Aspergillus nidulans. Nature. 1967 Sep 16;215(5107):1234–1237. doi: 10.1038/2151234a0. [DOI] [PubMed] [Google Scholar]
- Pearson W. R. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 1990;183:63–98. doi: 10.1016/0076-6879(90)83007-v. [DOI] [PubMed] [Google Scholar]
- Pfeifer K., Kim K. S., Kogan S., Guarente L. Functional dissection and sequence of yeast HAP1 activator. Cell. 1989 Jan 27;56(2):291–301. doi: 10.1016/0092-8674(89)90903-3. [DOI] [PubMed] [Google Scholar]
- Rand K. N., Arst H. N., Jr Mutations in nirA gene of Aspergillus nidulans and nitrogen metabolism. Nature. 1978 Apr 20;272(5655):732–734. doi: 10.1038/272732a0. [DOI] [PubMed] [Google Scholar]
- Salmeron J. M., Jr, Johnston S. A. Analysis of the Kluyveromyces lactis positive regulatory gene LAC9 reveals functional homology to, but sequence divergence from, the Saccharomyces cerevisiae GAL4 gene. Nucleic Acids Res. 1986 Oct 10;14(19):7767–7781. doi: 10.1093/nar/14.19.7767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Helix-turn-helix, zinc-finger, and leucine-zipper motifs for eukaryotic transcriptional regulatory proteins. Trends Biochem Sci. 1989 Apr;14(4):137–140. doi: 10.1016/0968-0004(89)90145-X. [DOI] [PubMed] [Google Scholar]
- Suzuki M. SPXX, a frequent sequence motif in gene regulatory proteins. J Mol Biol. 1989 May 5;207(1):61–84. doi: 10.1016/0022-2836(89)90441-5. [DOI] [PubMed] [Google Scholar]
- Tilburn J., Scazzocchio C., Taylor G. G., Zabicky-Zissman J. H., Lockington R. A., Davies R. W. Transformation by integration in Aspergillus nidulans. Gene. 1983 Dec;26(2-3):205–221. doi: 10.1016/0378-1119(83)90191-9. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Yuan G. F., Fu Y. H., Marzluf G. A. nit-4, a pathway-specific regulatory gene of Neurospora crassa, encodes a protein with a putative binuclear zinc DNA-binding domain. Mol Cell Biol. 1991 Nov;11(11):5735–5745. doi: 10.1128/mcb.11.11.5735. [DOI] [PMC free article] [PubMed] [Google Scholar]