Abstract
Major urinary proteins (Mups) are important for rodent scent communication and sexual behaviour. Recent evidence suggests that Mup1 may be regulated by fasting and re-feeding (RF). However, other Mup isoforms are poorly investigated, and data on the impact of long-term dietary restriction (DR) and ad libitum RF on Mup expression are missing. We investigated the effects of long-term 25 per cent DR and subsequent RF on Mup expression in male C57BL6 mice. DR significantly decreased Mup gene expression, hepatic and urinary protein levels compared with ad libitum (AL) fed control mice, with the greatest downregulation found for Mup5 expression. The decline in Mup expression was inverted by six months of RF. Because of inhibitory glucocorticoid response elements in the genomic sequence of the Mup5 gene, the observed inverse correlation of nuclear glucocorticoid receptor levels with Mup expression in response to DR and subsequent RF is a possible regulatory mechanism. Additionally, gene-expression-inhibiting histone deacetylation (H3K9) occurred in the region of the Mup5 gene in response to DR. We assume that Mup may act as a molecular switch linking nutritional status to sexual behaviour of mice, and thereby regulating male fertility and reproduction in response to food supply.
Keywords: Mup, glucocorticoid receptor, histone modification, leptin, nutritional status
1. Introduction
Prolongation of lifespan by dietary restriction (DR) has been repeatedly observed in different model organisms, including fishes, flies and mice [1].
However, in primates, it remains unclear whether DR promotes longevity, as the latest study on DR in rhesus monkeys showed no beneficial effect on lifespan [2,3]. Primates and other mammals differ greatly regarding their reproductive behaviour, and Shanley & Kirkwood [4] stated that in periods of food shortage mice reduce their investment in reproduction to maintain individual survival, resulting in an increased lifespan merely to permit sufficient amounts of progeny when food supply increases again [5,6]. However, in studies investigating DR and reproduction, the focus has been on female mice, thereby disregarding the effect of DR on reproduction in males.
Despite intensive research, the underlying molecular mechanisms of how DR and subsequent re-feeding (RF) influence reproduction in mice still remain to be elucidated.
An important aspect in reproduction as a part of murine behaviour is at least to some extent influenced by pheromone-triggered scent communication [7–16]. The volatile pheromones are hydrophobic ligands that are bound, transported and released by major urinary proteins (Mups) [17].
Mups comprise at least 21 highly polymorphic isoforms that belong to the multi gene family of lipocalins located at the Mup a locus on chromosome 4 [18–20]. Because all Mup genes derive from the same ancestral gene, they exhibit high sequence homology. After hepatic synthesis, Mup proteins are secreted into the circulation, and owing to their small size, they are rapidly excreted via urine [17,21–24]. Mups account for 99 per cent of the protein content in mouse urine and provide persistent, highly diverse signals in mouse urinary scent marks [25,26].
Males express fivefold higher hepatic Mup mRNA levels compared with females [19,27]. Mup expression patterns are mouse strain-specific [28] and provide information on the genetic and individual identity to avoid events such as inbreeding [15,29–31]. Mup gene expression is thought to be subjected to hormonal control, including testosterone, thereby explaining the higher Mup levels found in males [32].
Mups promote the onset of puberty and induce the oestrus in females, thus accelerating their reproductive potential [33,34]. Sensing of Mups occurs at the vomeronasal organ, where they also promote regeneration, survival and proliferation of neurons [35,36]. It has been shown that Mups stimulate the phosphorylation of ERK, Akt and CREB through G protein-coupled pheromone receptor neuron (V2R) signalling [35,37].
Apart from their role in chemical communication, Mups also seem to exert metabolic functions. Most insights come from studies on Mup1 that appears to be involved in increasing energy expenditure, core body temperature, glucose tolerance and insulin sensitivity in mice [38]. In mice with genetic- or diet-induced diabetes, serum and urine concentrations of Mup1 were markedly decreased and contrarily elevated Mup1 levels exerted beneficial metabolic effects. By decreasing the expression of important gluconeogenic and lipogenic genes, Mup1 was shown to suppress gluconeogenesis and lipogenesis in liver [38,39]. These findings suggest a function of Mup1 in the signalling pathways that regulate glucose and lipid metabolism [16].
Furthermore, it has been shown that the amount of dietary intake can affect Mup1 expression and modulate its secretion [40]. Interestingly, overnight fasting for 18 h reduced Mup1 gene expression in the liver of mice. This was abolished after RF for 1 h [38]. However, to date, there are no systematic studies investigating the effect of DR on Mup expression.
In this study, we have explored the impact of a six-month DR followed by a six-month RF phase on hepatic Mup expression in male C57BL6 mice. Levels of total Mup were determined in liver and urine. Mup5 transcription was the most affected upon DR and thus of special interest in the following analysis of possible regulatory mechanisms, including histone acetylation and DNA binding of the glucocorticoid receptor (GR). This is the first study, to the best of our knowledge, to provide evidence of DR and subsequent RF directly influencing male Mup expression (possibly via glucocorticoid signalling), a finding that could help us to explain how sexual behaviour and reproduction in mice are influenced by food supply. Moreover, we suspect that Mups may, at least, partly mediate the lifespan prolonging effects of DR. A putative role of Mup in lifespan extension could explain the different outcomes of DR and lifespan studies in primates and give hints as to what effect DR might have on human lifespan. As humans and primates, in contrast to mice, do not express various Mup homologues, it is possible that DR does not improve longevity in these species.
2. Material and methods
(a). Mice and diet
Male C57BL6 wild-type mice (six- to eight-weeks old, n = 33) were purchased from Taconic Europe (Ry, Denmark). Mice were housed individually in macrolon cages under controlled environmental conditions (55% relative humidity, 21–25°C and 12 L : 12 D cycle). Mice were maintained on a commercial pelleted standard chow diet (Ssniff special diets, Soest, Germany) consisting, by energy, of 58 per cent carbohydrates, 33 per cent protein and 9 per cent fat from grain and soya bean, and they had free access to tap water. Mice were divided into two groups with 14 animals in the ad libitum fed group (AL) and 19 animals in the DR group. DR mice were fed dietarily restricted for six months with 75 per cent of the amount consumed by the AL control group; however, all micronutrients, including vitamins and minerals were provided at an adequate level. After this period, the DR animals were re-fed by granting ad libitum access to standard chow diet for another six months. Over the entire study period, food intake was controlled daily, and body weight was determined weekly. After six months of DR and after six months of RF, animals of each group (DR: AL n = 7, DR n = 9; RF: AL n = 7, DR n = 10) were fasted overnight, anaesthetized and killed by cervical dislocation. Liver and visceral adipose tissues were removed from the animal's post-mortem. Adipose tissue was weighed, and the liver was stored at −80°C until analysis. The blood was collected in tubes containing EDTA, and the plasma was separated by centrifugation (3000g, 10 min, 4°C) and also stored at 80°C. One week before termination of each feeding period, mice from the respective groups were kept in metabolic cages (Tecniplast, Seeheim/Ober-Beerbach, Germany) for 48 h to collect urine samples.
(b). RNA isolation and quantitative real-time polymerase chain reaction
Total RNA of mouse liver tissue was isolated, using the RNeasy mini kit according to the manufacturer's protocol (Qiagen, Hilden, Germany). RT-PCR primers were designed, using Primer v. 3 input software (v. 0.4.0) (β-actin: F: GACAGGATGCAGAAGAGATTACT, R: TGATCCACATCTGCTGGAAGGT; Mup5: F: ATGGAGCTCTTTGGTCGA, R: TGTATGGAAGGGAAGGGATG). The one-step quantitative reverse transcriptase PCR was carried out, using the SensiMix SYBR No-ROX one step kit (Bioline, Luckenwalde, Germany) and with SybrGreen detection on a Rotorgene 6000 cycler (Corbett Life Science, Sydney, Australia).
(c). RNA-microarray
One-colour labelling of four individual liver mRNA samples per treatment group was performed, using the low input quick amp labelling kit (Agilent Technologies, Böeblingen, Germany) according to the manufacturer's protocol. The labelled samples were purified with the Qiacube, hybridized to the RNA-microarray (Sure Print G3 mouse gene expression 8 × 60 K arrays (design: 28005; Agilent) and scanned with the microarray scanner (Agilent). The arrays were analysed using Feature Extractions software (v. 10.5.1.1.; Agilent).
(d). Western blot analysis
Liver nuclear and cytoplasmatic extracts were prepared as described in Wagner et al. [41]. Owing to the limited excretion of protein in the urine and the resulting lack of a loading control, protein concentrations in urinary samples were determined with the Coomassie plus protein assay kit (Thermo Scientific, Schwerte, Germany) according to the manufacturer's instructions to ensure equal loading amounts of protein onto the gel. Proteins were separated by SDS–PAGE and transferred onto a PVDF membrane. Target proteins were identified, using respective primary (anti-Mup (1 : 500), anti-GR (1 : 200), anti-α-tubulin (1 : 200) and anti-LaminB1 (1 : 200); all Santa Cruz Biotechnology Inc., Heidelberg, Germany) and secondary antibodies. Protein bands were visualized with ECL reagent (Fisher Scientific, Schwerte, Germany) in a ChemiDoc XRS system (BioRad, Munich, Germany).
(e). Determination of plasma leptin levels
Leptin levels were determined in plasma diluted 1 : 2, using the Quantikine mouse leptin ELISA kit (R&D Systems, Abingdon, UK) according to the manufacturer's instructions.
(f). Chromatin-immunoprecipitation–ChIP assay
After cross-linking of liver tissue, ChIP experiments were performed with the shearing optimization kit and OneDay ChIP kit (Diagenode, Liège, Belgium) according to the manufacturer's protocol with slight modifications. Chromatin was sheared for 5 s, using a MICCRA D-1 disperser (speed: 26 000 rpm; ART Prozess & Labortechnik, Müllheim-Hügelheim, Germany) and incubated on ice for 30 min with anti-histone 3, anti-histone 3 acetyl K9 (both from Abcam, Cambridge, UK) or the provided anti-rabbit IgG in an ultrasonic bath (Sonorex; Bandelin Electronic, Berlin, Germany). Reversion of the formaldehyde cross-linking and DNA purification was performed, using the IPure kit (Diagenode) according to the manufacturer's instructions. Chromatin was amplified, using the WGA2 kit according to the manufacturer's instructions (Sigma-Aldrich Chemie GmbH, Steinheim, Germany). DNA samples were labelled Cy3 (input) and Cy5 (IP) (dual-colour DNA labelling kit, Roche, Mannheim, Germany) and hybridized with the mouse microarray (ChIP-chip 385K RefSeq promoter arrays 100718_MM9_RefSeq_Prom_ChIP_HX3, Roche). Arrays were scanned on a NimbleGen scanner MS200 (Roche), and the data were analysed with NimbleScan software v. 2.6. A ChIP find-peaks analysis was performed to detect differences between IP and input samples, for further information see http://www.nimblegen.com/products/lit/NimbleScan_v2p6_usersguide.pdf.
(g). Statistical analysis
The statistical analysis was performed, using SPSS v. 15.0 (SPSS GmbH Software, Munich, Germany). Kolmogorov–Smirnov and Shapiro–Wilk tests were used to test the data for normal distribution. Data following a Gaussian distribution were analysed by Student's t-test. In the case of a non-Gaussian distribution, a Mann–Whitney U-test was performed. Results are presented as means and standard error of mean. p-values < 0.05 were considered statistically significant.
3. Results
(a). Initial difference in body weight between ad libitum and dietary restriction-fed mice is abolished through re-feeding
Mouse body weight did not differ between groups at the beginning of the experiment (figure 1). During the following six months, the body weight in the AL group increased, and DR mice approximately maintained their initial weight so that after six months, a significant difference in body weight was observed between groups. During the following period of ad libitum feeding, body weight in DR mice increased so that the difference in body weight between groups was abolished after six months of RF.
Figure 1.
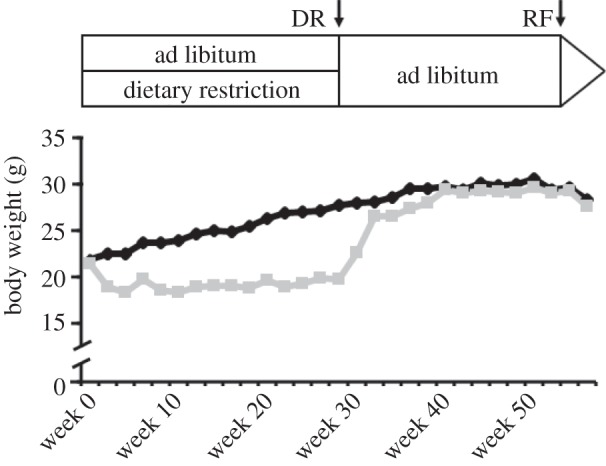
Body weight development of male C57BL6 mice fed ad libitum (AL; black line) or dietarily restricted (DR; grey line). After reducing the dietary intake successively (5% per week), the DR mice were fed 75% of the AL mice feed intake for six months. Following DR, mice were re-fed by granting ad libitum access to diet for additional six months (starting at week 29). Data are shown as means (n = 7 for AL; n = 10 for DR).
(b). Most dietary restriction-regulated genes return to a normal state after re-feeding
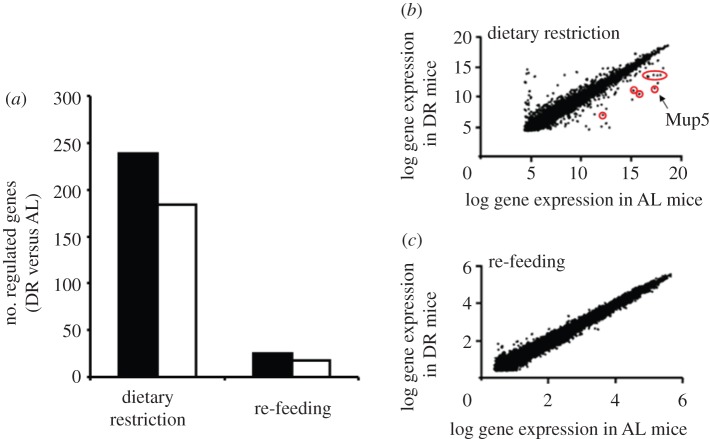
Global gene expression in the DR group was related to global gene expression in the AL group. Values for individual genes are given as ratio (DR/AL; AL = 1). In response to DR, more than 200 genes were significantly upregulated, and approximately 180 significantly downregulated. Most DR-regulated genes return to normal state after RF (figure 2a–c). There are more genes upregulated than downregulated in DR compared with AL mice. However, the group of downregulated genes includes a remarkably high number of Mup gene isoforms (figure 2b).
Figure 2.
DR leads to regulation of numerous genes that is abolished by re-feeding (RF) in most cases. The number of regulated genes in response to DR and subsequent RF identified by microarray analysis (change at least two-fold, p ≤ 0.05) is shown in (a). Global gene expression of DR mice was related to global gene expression of AL mice after six months of 25 per cent DR and six months of subsequent RF. In the DR group, the total number of upregulated genes is higher than that of downregulated genes at all time points. Black bars denote increased gene expression; white bars denote decreased gene expression. A comparison of log gene expression in AL and DR mice illustrates the diminishment of previously DR-induced differences in the gene expression pattern by RF (b,c). (b) Circled data points indicate Mup isoforms. (Online version in colour.)
(c). Mup expression decreases upon dietary restriction
RNA-microarray analysis identified numerous genes with twofold increased or decreased expression in DR when compared with AL mice. For our purpose, the microarray data were scanned only for expression of Mup isoforms. Following DR, several Mup isoforms showed significantly decreased mRNA levels compared with age-matched AL controls (table 1). After six months of RF, no statistical differences between AL and former DR mice could be found for the expression levels of the measured Mup isoforms (data not shown).
Table 1.
Mup isoforms that are significantly downregulated in response to DR as determined by microarray analysis. Global gene expression of DR mice was related to global gene expression of AL mice after six months of dietary restriction. Data are means of four mice in the AL group and five mice in the DR group, p-values are given for compared DR with AL.
| Mup isoform | ratio DR/AL ± s.e.m. | p-value |
|---|---|---|
| Mup2 | 0.029 ± 0.008 | 0.001 |
| Mup3 | 0.097 ± 0.022 | 0.001 |
| Mup4 | 0.112 ± 0.029 | 0.001 |
| Mup5 | 0.019 ± 0.006 | 0.001 |
| Mup6 | 0.046 ± 0.029 | 0.018 |
| Mup9 | 0.073 ± 0.020 | 0.001 |
| Mup20 | 0.059 ± 0.013 | 0.001 |
| Mup21 | 0.061 ± 0.008 | 0.001 |
(d). Mup transcription is decreased in dietary restriction mice and overcompensated by subsequent re-feeding
RT-PCR confirmed the results of the microarray analysis and indicated a significant decrease in hepatic mRNA expression of Mup5 in DR mice compared with AL mice after six months of DR (figure 3a). After six months of RF, the Mup5 gene expression in the DR mice was markedly increased compared with AL mice (figure 3b). However, probably due to a high s.e. of mean, this effect is not significant.
Figure 3.
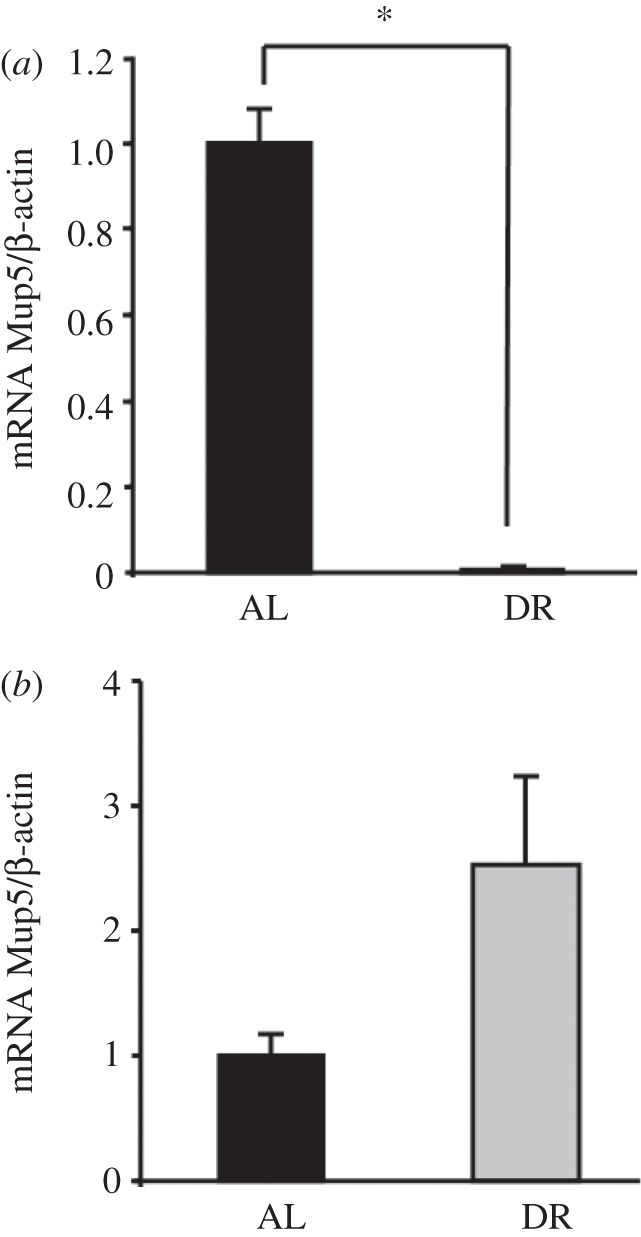
(a) After six months of dietary restriction hepatic Mup5 mRNA levels in male C57BL6 mice decrease compared with ad libitum fed mice. (b) Re-feeding for six months increased Mup5 gene expression compared with AL mice. The Mup5 mRNA to β-actin mRNA ratios for AL- and DR-fed mice as determined by real-time PCR are shown. The ratio for the AL control was set to 1. Data are shown as means +s.e.m, n ≥ 7 per group. *p ≤ 0.05 compared DR with AL.
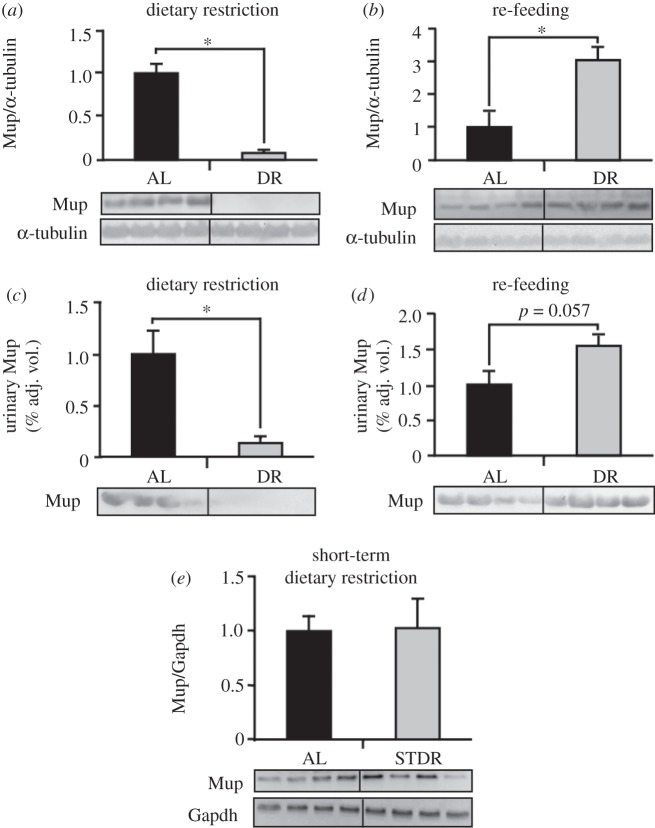
(e). A decreased Mup expression is reflected in lowered protein levels
Differences in Mup expression after six months of DR were not only observed in hepatic mRNA but also in hepatic (figure 4a,b) and urinary protein levels (figure 4c,d) as determined by Western blotting. Interestingly, decreased total Mup levels in DR mice rose again after six months of RF and even exceeded the Mup protein levels of age-matched AL mice. However, short-term DR (three weeks) had no effects on hepatic Mup expression (figure 4e).
Figure 4.
Hepatic and urinary Mup protein levels decrease upon dietary restriction and return to higher than ad libitum fed mice levels after re-feeding (RF). (a,b) Hepatic and (c,d) urinary Mup protein levels in male C57BL6 mice after six months of dietary restriction and six months of RF were detected by Western blotting. (e) A three-week dietary restriction does not affect hepatic Mup levels. Representative Western blots of four mice per group are shown and for the corresponding densitometries of all animals were used. As protein excretion in urine is limited, detecting a loading control in urinary samples was not possible.
(f). Plasma leptin levels are affected by dietary restriction and the amount of visceral adipose tissue decreases
As previously described for Mup2, Mups may be regulated via glucocorticoids [42], which are controlled by the adipose-tissue-derived hormone leptin [43]. Plasma leptin levels and visceral adipose tissue weight were significantly lower after six months of DR compared with AL mice (table 2). Plasma leptin levels and the amount of visceral adipose tissue were increased in response to RF after six months of DR, and no significant differences were evident between the groups after RF. Interestingly, similar to Mup levels, during short-term DR, plasma leptin concentration was not changed compared with AL fed mice (data not shown).
Table 2.
Effect of dietary restriction and re-feeding on plasma leptin levels and white adipose tissue mass of male C57BL6 mice compared with ad libitum fed mice. Leptin concentrations were determined using a specific ELISA kit. Data are shown as means ± s.e.m. Statistically significant differences (p ≤ 0.05) between dietary restricted (DR) and corresponding ad libitum fed (AL) animals are indicated as asterisk (n = 7 for AL; n = 9 for DR).
| dietary restriction |
re-feeding |
|||
|---|---|---|---|---|
| AL | DR | AL | DR | |
| leptin (ng ml−1) | 1.15 ± 0.44 | 0.18 ± 0.03* | 1.24 ± 0.29 | 0.55 ± 0.15 |
| white adipose tissue (mg) | 339 ± 41 | 54 ± 7* | 516 ± 45 | 478 ± 41 |
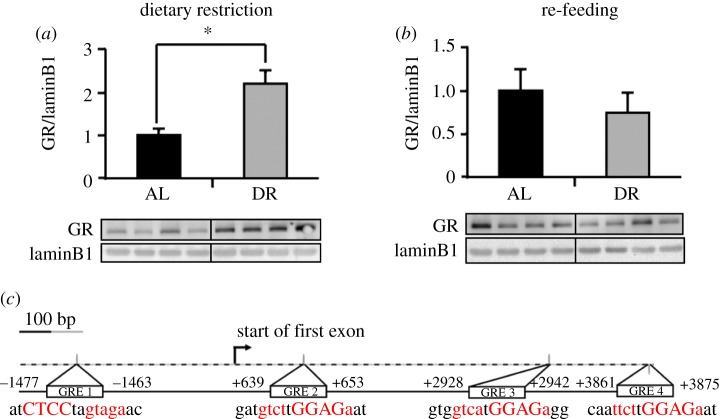
(g). Dietary restriction-fed mice show increased nuclear glucocorticoid receptor levels
As it has been discussed that Mup levels may be influenced by glucocorticoids [42], we compared the amount of GR in the nucleus of DR mice with AL-fed mice by Western blotting. DR mice exhibited higher nuclear GR levels than age-matched AL mice (figure 5a). Subsequent RF for six months, however, decreased the amount of nuclear GR in DR mice leading to lower nuclear GR levels in the former DR group (figure 5b).
Figure 5.
Dietary restriction leads to increased glucocorticoid receptor levels in the liver of DR-fed compared with AL-fed mice. The hepatic nuclear GR levels after (a) six months of DR and (b) six months of RF as determined by Western blot are shown. The corresponding densitometries are shown above the blots. (c) The genomic sequence of the Mup5 gene and its promoter contains four glucocorticoid response elements. Representative Western blots of four mice per group are shown and for the corresponding densitometries samples of all animals were used. (Online version in colour.)
A transcription factor analysis of the Mup5 gene has revealed inhibitory glucocorticoid response elements (GREs) in the Mup5 promoter and also in the Mup5 gene sequence (figure 5c). Therefore, translocation of the GR to the nucleus may be responsible for a decline in Mup5 expression as observed in the DR mice.
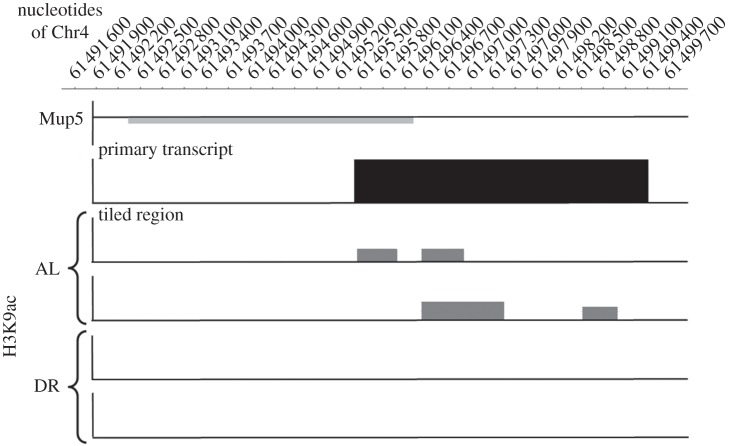
(h). Histone 3 K9 deacetylation is likely to inhibit Mup5 expression in dietary restriction mice
Deacetylation of lysine residues provides a positive charge at the histones, leading to attraction of the negatively charged DNA strand and tightly packed chromatin. In this state, the DNA is difficult to access for the RNA polymerase which results in lowered gene expression. Contrarily, gene expression in regions with acetylated histones is enhanced. Significant differences in the acetylation of lysine 9 in histone 3 (H3K9ac) were detected in close proximity to the Mup5 gene on chromosome 4 by ChIP–Chip analysis. In AL mice, acetylation of H3K9 could be detected, thereby enabling gene expression of Mup5, whereas in DR mice, H3K9 was deacetylated, thus explaining the observed inhibition of Mup5 gene expression (figure 6).
Figure 6.
Dietary restriction leads to deacetylation of the lysine 9 residue in histone 3 (H3K9) close to the Mup5 gene on chromosome 4 (light grey bar). The black bar indicates the region tiled by the microarray. Acetylation of H3K9 within this region in the liver of DR-fed compared with AL-fed mice is indicated with grey bars. Following DR, H3K9 is deacetylated, leading to inhibition of gene expression. Representative results of two mice per group are shown.
4. Discussion
In this study, we show that male Mups are downregulated in response to DR that could explain how sexual behaviour is linked to nutrient intake. Previous studies on DR and reproduction have been mostly conducted in females. Urinary excretion of Mups in male mice was shown to attract females and correlate with the onset of male sexual maturity in mice [9,17,32,44] which is why male Mups could play a dominant role in the control of reproduction. Considering the fact that the urine of male mice typically contains up to 5 mg ml−1 Mup protein [45], production of Mups represents a substantial metabolic cost [46]. Therefore, decreasing Mup expression seems to save energy during DR periods. In addition, reproduction is prevented in times of insecure survival of the offspring.
In this study, expression and secretion of a broad range of Mup isoforms was downregulated in response to long-term DR. Mice that were subsequently re-fed ad libitum, however, exhibited higher Mup expression than their age-matched controls. Data on DR and subsequent RF concerning Mup expression are limited. In the case of Mup1, it could be shown that DR-caused downregulation of the gene is quickly abrogated by RF and leads to elevated mRNA expression within 1 h [38]. This is in line with our results showing that after a DR period, RF leads to elevated levels of total Mup. Consistent with these findings, different studies showed higher reproduction rates in re-fed DR mice compared with control mice fed AL [5,6]. Therefore, elevated Mup levels in re-fed mice that had been dietarily restricted could be a mechanism to compensate for the reduced reproduction rate during the time of limited access to feed.
A variety of hormones, including testosterone, have been described as regulating Mups and as impacting on Mup expression patterns [17]. However, in a study by Knopf et al. [32], expression of Mup1 and Mup5 was regulated by administration of thyroxine, but not testosterone. We therefore assume that the decreased Mup production in our DR animals may be caused by effects related to nutritional status other than testosterone.
Interestingly, higher nuclear GR levels were evident in the livers of the DR mice. Glucocorticoids have been reported to be involved in the production of α2u-globulin in the rat which is a homologue of murine Mups [47], and activation of the GR has recently been shown to impact on Mup2 expression [42]. Furthermore, we identified inhibitory GREs within the promoter region and genomic sequence of the Mup5 gene. By binding to these inhibitory GREs, nuclear GR may inhibit Mup5 expression. The finding that Mup levels rise to higher than AL control levels after RF, whereas nuclear GR levels fall to lower than control levels under the same conditions also points to the notion that there could be an inverse relationship of glucocorticoid levels and Mup transcription. In this context, it is interesting to note that glucocorticoid release could be induced by falling leptin levels [43]. Leptin is an adipocyte-derived hormone with a key role in regulating energy metabolism [48]. As a result of reduced body fat, our DR mice exhibited significantly lower leptin levels than AL mice that could have led to the observed lowered Mup levels via increased glucocorticoid release and subsequent transcriptional inhibition because of GRE in the Mup5 promoter region. Interestingly, leptin levels as well as Mup levels did not change in response to short-term DR (three weeks) emphasizing the potential role of leptin in regulation of Mup expression.
Studies in obese mice also suggest a relation between leptin signalling and Mup expression. Genetically obese leptin-receptor-deficient mice as well as dietary obese leptin-resistant mice showed lowered Mup1 levels [38,39]. As mediators of reproduction, decreased Mup levels are probably responsible for the observed infertility of these mice [49] and in leptin-deficient obese mice (ob/ob), reproduction parameters were normalized by leptin administration [50–52].
Accordingly, leptin resistance in obese mice [53] followed by low Mup levels could explain why energy excess accelerates reproduction only after periods of energy deficit as observed in our re-fed mice by exhibiting elevated Mup levels.
We hypothesize that the RF-induced leptin increase in our former DR mice suppresses glucocorticoid secretion and therefore Mup5 inhibition which, in turn, leads to increased Mup5 in liver and urine. On the contrary, in obese mice, permanently elevated leptin levels show no effect on Mup expression because the organism has developed a resistance towards this adipocyte-derived hormone. Taken together, our data suggest that Mup5 expression is at least in part regulated by glucocorticoid signalling as a consequence of falling leptin levels during DR.
An additional mechanism that leads to lowered Mup5 expression appears to involve histone deacetylation. In liver samples from DR mice, deacetylation of lysine 9 at histone 3 leading to suppressed gene expression in the region of the Mup5 gene could be detected. An important histone deacetylase thought to be regulated by DR is sirtuin 1 (SIRT1). SIRT1 activity depends on the NAD/NADH ratio which is closely connected to the metabolic rate [54] and increases in response to DR [55]. Therefore, a DR-mediated activation of SIRT1 may have caused the H3K9 deacetylation, thus inhibiting Mup5 gene expression.
In the light of DR prolonging lifespan in mice (reviewed by Fontana et al. [1]), it is intriguing to speculate how Mups may relate to this phenomenon. In mice that are subjected to fluctuating feed supply and therefore periods with lowered fertility, it makes sense that lifespan is prolonged by DR until sufficient nutrients for successful reproduction are available [56]. On the one hand, abundant feed is likely to promote reproduction and onset of puberty in male mice, but on the other hand work against longevity [9,17,44]. It is generally believed that downregulation of metabolic activity favours lifespan extension. Administration of Mup1 increased energy expenditure, locomotor activity and core body temperature in mice [38], indicating an activating function of Mups on energy/glucose and lipid metabolism. Thus, DR-mediated Mup suppression may directly favour a type of energy-preserving, life-prolonging state in the mouse. However, administration of Mup1 protein also exerted beneficial effects on hyperglycaemia, glucose intolerance and insulin sensitivity in an obese mouse model while suppressing hepatic gluconeogenetic as well as lipogenetic genes encoding proteins such as PEPCK, glucose-6-phosphatase, fatty acid synthase, carbohydrate response element binding protein, stearoyl-CoA desaturase-1 and peroxisome proliferator-activated receptor-γ [38,39]. In our study, the observed decrease in Mup levels in response to DR is accompanied by significantly increased expressions of hepatic gluconeogenetic genes such as glucose-6-phosphatase, fructose-1,6-bisphosphatase and PEPCK (see the electronic supplementary material, table S1) which could mean that there is an inverse bidirectional relation between Mup signalling and glucose signalling. Therefore, some type of nutritional status-dependent Mup interaction with energy metabolism seems likely, and it has been suggested that Mups serve as both chemical and metabolic signals that coordinate sexual behaviour as well as nutrient metabolism [16].
As observed in our study, subsequent RF and energy excess led to Mup over-production. While it is known that intermittent fasting is beneficial for mouse health [57], longer periods of energy excess were shown to be detrimental for health as, for example, in diet-induced obese mice that exhibit low levels of Mup [39].
Even though DR repeatedly induced lifespan prolongation in rodents, DR does not increase lifespan in all mouse strains [58]. In wild mice, DR even increased mortality in early life [59]. In this context, it has to be kept in mind that laboratory animals generally show a higher dietary intake than wild mice [60]. Excessive energy intake in laboratory animals may lead to accelerated growth and earlier reproduction [58]. Therefore, it is possible that the dietary intake of our DR-mice and subsequently Mup expression were only lowered to the levels of wild mice in their natural habitat. Nevertheless, our results show that varying dietary intake affects Mup expression.
It would be of great interest to observe how the changes in Mup expression caused by DR and RF influence the behaviour and reproduction of mice on the one hand and their lifespan on the other. Moreover, clarifying the role of Mups in lifespan extension through DR is especially important when using mice as a model for longevity studies because humans do not have a functional Mup homologue.
Acknowledgements
Mice were kept according to the German Regulations of Animal Care with permission from the responsible authority.
We are grateful to the BMBF (project: ‘Epifood’) and the DFG Cluster of Excellence ‘Inflammation at Interfaces’ for financial support.
References
- 1.Fontana L, Partridge L, Longo VD. 2010. Extending healthy life span: from yeast to humans. Science 328, 321–326 10.1126/science.1172539 (doi:10.1126/science.1172539) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Colman RJ, et al. 2009. Caloric restriction delays disease onset and mortality in rhesus monkeys. Science 325, 201–204 10.1126/science.1173635 (doi:10.1126/science.1173635) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mattison JA, et al. 2012. Impact of caloric restriction on health and survival in rhesus monkeys from the NIA study. Nature 489, 318–321 10.1038/nature11432 (doi:10.1038/nature11432) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shanley DP, Kirkwood TB. 2006. Caloric restriction does not enhance longevity in all species and is unlikely to do so in humans. Biogerontology 7, 165–168 10.1007/s10522-006-9006-1 (doi:10.1007/s10522-006-9006-1) [DOI] [PubMed] [Google Scholar]
- 5.Ball ZB, Barnes RH, Visscher MB. 1947. The effects of dietary caloric restriction on maturity and senescence, with particular reference to fertility and longevity. Am. J. Physiol. 150, 511–519 [DOI] [PubMed] [Google Scholar]
- 6.Selesniemi K, Lee HJ, Tilly JL. 2008. Moderate caloric restriction initiated in rodents during adulthood sustains function of the female reproductive axis into advanced chronological age. Aging Cell 7, 622–629 10.1111/j.1474-9726.2008.00409.x (doi:10.1111/j.1474-9726.2008.00409.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruce HM. 1959. An exteroceptive block to pregnancy in the mouse. Nature 184, 105. 10.1038/184105a0 (doi:10.1038/184105a0) [DOI] [PubMed] [Google Scholar]
- 8.Mugford RA, Nowell NW. 1970. Pheromones and their effect on aggression in mice. Nature 226, 967–968 10.1038/226967a0 (doi:10.1038/226967a0) [DOI] [PubMed] [Google Scholar]
- 9.Mucignat-Caretta C, Caretta A, Cavaggioni A. 1995. Acceleration of puberty onset in female mice by male urinary proteins. J. Physiol. 486, 517–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bronson FH, Caroom D. 1971. Preputial gland of the male mouse; attractant function. J. Reprod. Fertil. 25, 279–282 10.1530/jrf.0.0250279 (doi:10.1530/jrf.0.0250279) [DOI] [PubMed] [Google Scholar]
- 11.Jemiolo B, Xie TM, Novotny M. 1991. Socio-sexual olfactory preference in female mice: attractiveness of synthetic chemosignals. Physiol. Behav. 50, 1119–1122 10.1016/0031-9384(91)90570-E (doi:10.1016/0031-9384(91)90570-E) [DOI] [PubMed] [Google Scholar]
- 12.Morè L. 2006. Mouse major urinary proteins trigger ovulation via the vomeronasal organ. Chem. Senses 31, 393–401 10.1093/chemse/bjj043 (doi:10.1093/chemse/bjj043) [DOI] [PubMed] [Google Scholar]
- 13.Brennan PA, Kendrick KM. 2006. Mammalian social odours: attraction and individual recognition. Phil. Trans. R. Soc. B 361, 2061–2078 10.1098/rstb.2006.1931 (doi:10.1098/rstb.2006.1931) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tirindelli R, Dibattista M, Pifferi S, Menini A. 2009. From pheromones to behaviour. Physiol. Rev. 89, 921–956 10.1152/physrev.00037.2008 (doi:10.1152/physrev.00037.2008) [DOI] [PubMed] [Google Scholar]
- 15.Hurst JL. 2009. Female recognition and assessment of males through scent. Behav. Brain Res. 200, 295–303 10.1016/j.bbr.2008.12.020 (doi:10.1016/j.bbr.2008.12.020) [DOI] [PubMed] [Google Scholar]
- 16.Zhou Y, Rui L. 2010. Major urinary protein regulation of chemical communication and nutrient metabolism. Vitam. Horm. 83, 151–164 10.1016/S0083-6729(10)83006-7 (doi:10.1016/S0083-6729(10)83006-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheetham SA, Smith AL, Armstrong SD, Beynon RJ, Hurst JL. 2008. Limited variation in the major urinary proteins of laboratory mice. Physiol. Behav. 96, 253–261 10.1016/j.physbeh.2008.10.005 (doi:10.1016/j.physbeh.2008.10.005) [DOI] [PubMed] [Google Scholar]
- 18.Finlayson JS, Hudson DM, Armstrong BL. 1969. Location of the Mup-a locus on mouse linkage group 8. Genet. Res. 14, 329–331 10.1017/S0016672300002159 (doi:10.1017/S0016672300002159) [DOI] [PubMed] [Google Scholar]
- 19.Szoka PR, Paigen K. 1978. Regulation of mouse major urinary protein production by the Mup-A gene. Genetics 90, 597–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Logan DW, Marton TF, Stowers L. 2008. Species specificity in major urinary proteins by parallel evolution. PLoS ONE 3, e3280. 10.1371/journal.pone.0003280 (doi:10.1371/journal.pone.0003280) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Finlayson JS, Potter M, Runner CR. 1963. Electrophoretic variation and sex dimorphism of the major urinary protein complex in inbred mice: a new genetic marker. J. Natl. Cancer Inst. 31, 91–107 [PubMed] [Google Scholar]
- 22.Finlayson JS, Asofsky R, Potter M, Runner CC. 1965. Major urinary protein complex of normal mice: origin. Science 149, 981–982 10.1126/science.149.3687.981 (doi:10.1126/science.149.3687.981) [DOI] [PubMed] [Google Scholar]
- 23.Finlayson JS, Mushinski JF, Hudson DM, Potter M. 1968. Components of the major urinary protein complex in inbred mice: separation and peptide mapping. Biochem. Genet. 2, 127–140 10.1007/BF01458712 (doi:10.1007/BF01458712) [DOI] [PubMed] [Google Scholar]
- 24.Kimura H, Odani S, Nishi S, Sato H, Arakawa M, Ono T. 1991. Primary structure and cellular distribution of two fatty acid-binding proteins in adult rat kidneys. J. Biol. Chem. 266, 5963–5972 [PubMed] [Google Scholar]
- 25.Shahan K, Denaro M, Gilmartin M, Shi Y, Derman E. 1987. Expression of six mouse major urinary protein genes in the mammary, parotid, sublingual, submaxillary, and lachrymal glands and in the liver. Mol. Cell Biol. 7, 1947–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beynon RJ, Hurst JL. 2004. Urinary proteins and the modulation of chemical scents in mice and rats. Peptides 25, 1553–1563 10.1016/j.peptides.2003.12.025 (doi:10.1016/j.peptides.2003.12.025) [DOI] [PubMed] [Google Scholar]
- 27.Hastie ND, Held WA, Toole JJ. 1979. Mulitple genes coding for the androgen-regulated major urinary proteins of the mouse. Cell 17, 449–457 10.1016/0092-8674(79)90171-5 (doi:10.1016/0092-8674(79)90171-5) [DOI] [PubMed] [Google Scholar]
- 28.Robertson DH, Cox KA, Gaskell SJ, Evershed RP, Beynon RJ. 1996. Molecular heterogeneity in the major urinary proteins of the house mouse Mus musculus. Biochem. J. 316, 265–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sherborne AL, Thom MD, Paterson S, Jury F, Ollier WE, Stockley P, Beynon RJ, Hurst JL. 2007. The genetic basis of inbreeding avoidance in house mice. Curr. Biol. 17, 2061–2066 10.1016/j.cub.2007.10.041 (doi:10.1016/j.cub.2007.10.041) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheetham SA, Thom MD, Jury F, Ollier WE, Beynon RJ, Hurst JL. 2007. The genetic basis of individual-recognition signals in the mouse. Curr. Biol. 17, 1771–1777 10.1016/j.cub.2007.10.007 (doi:10.1016/j.cub.2007.10.007) [DOI] [PubMed] [Google Scholar]
- 31.Hurst JL, Payne CE, Nevison CM, Marie AD, Humphries RE, Robertson DH, Cavaggioni A, Beynon RJ. 2001. Individual recognition in mice mediated by major urinary proteins. Nature 414, 631–634 10.1038/414631a (doi:10.1038/414631a) [DOI] [PubMed] [Google Scholar]
- 32.Knopf JL, Gallagher JF, Held WA. 1983. Differential, multihormonal regulation of the mouse major urinary protein gene family in the liver. Mol. Cell Biol. 3, 2232–2240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reynolds J, Keverne EB. 1979. The accessory olfactory system and its role in the pheromonally mediated suppression of oestrus in grouped mice. J. Reprod. Fertil. 57, 31–35 10.1530/jrf.0.0570031 (doi:10.1530/jrf.0.0570031) [DOI] [PubMed] [Google Scholar]
- 34.Keverne EB. 1999. The vomeronasal organ . Science 286, 716–720 10.1126/science.286.5440.716 (doi:10.1126/science.286.5440.716) [DOI] [PubMed] [Google Scholar]
- 35.Xia J, Sellers LA, Oxley D, Smith T, Emson P, Keverne EB. 2006. Urinary pheromones promote ERK/Akt phosphorylation, regeneration and survival of vomeronasal (V2R) neurons. Eur. J. Neurosci. 24, 3333–3342 10.1111/j.1460-9568.2006.05244.x (doi:10.1111/j.1460-9568.2006.05244.x) [DOI] [PubMed] [Google Scholar]
- 36.Chamero P, et al. 2011. G protein G(alpha)o is essential for vomeronasal function and aggressive behaviour in mice. Proc. Natl Acad. Sci. USA 108, 12 898–12 903 10.1073/pnas.1107770108 (doi:10.1073/pnas.1107770108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Broad KD, Keverne EB. 2011. The post-natal chemosensory environment induces epigenetic changes in vomeronasal receptor gene expression and a bias in olfactory preference. Behav. Genet. 42, 461–471 10.1007/s10519-011-9523-9 (doi:10.1007/s10519-011-9523-9) [DOI] [PubMed] [Google Scholar]
- 38.Hui X, Zhu W, Wang Y, Lam KSL, Zhang J, Wu D, Kraegen EW, Li Y, Xu A. 2009. Major urinary protein-1 increases energy expenditure and improves glucose intolerance through enhancing mitochondrial function in skeletal muscle of diabetic mice. J. Biol. Chem. 284, 14 050–14 057 10.1074/jbc.M109.001107 (doi:10.1074/jbc.M109.001107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou Y, Jiang L, Rui L. 2009. Identification of MUP1 as a regulator for glucose and lipid metabolism in mice. J. Biol. Chem. 284, 11 152–11 159 10.1074/jbc.M900754200 (doi:10.1074/jbc.M900754200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dhahbi JM, Kim H-J, Mote PL, Beaver RJ, Spindler SR. 2004. Temporal linkage between the phenotypic and genomic responses to caloric restriction. Proc. Natl Acad. Sci. 101, 5524–5529 10.1073/pnas.0305300101 (doi:10.1073/pnas.0305300101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wagner AE, Ernst I, Iori R, Desel C, Rimbach G. 2010. Sulforaphane but not ascorbigen, indole-3-carbinole and ascorbic acid activates the transcription factor Nrf2 and induces phase-2 and antioxidant enzymes in human keratinocytes in culture. Exp. Dermatol. 19, 137–144 10.1111/j.1600-0625.2009.00928.x (doi:10.1111/j.1600-0625.2009.00928.x) [DOI] [PubMed] [Google Scholar]
- 42.Cho Y-H, Kim D, Choi I, Bae K. 2011. Identification of transcriptional regulatory elements required for the Mup2 expression in circadian clock mutant mice. Biochem. Biophys. Res. Commun. 410, 834–840 10.1016/j.bbrc.2011.06.074 (doi:10.1016/j.bbrc.2011.06.074) [DOI] [PubMed] [Google Scholar]
- 43.Dardeno TA, Chou SH, Moon HS, Chamberland JP, Fiorenza CG, Mantzoros CS. 2010. Leptin in human physiology and therapeutics. Front. Neuroendocrinol. 31, 377–393 10.1016/j.yfrne.2010.06.002 (doi:10.1016/j.yfrne.2010.06.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roberts SA, Simpson DM, Armstrong SD, Davidson AJ, Robertson DH, McLean L, Beynon RJ, Hurst JL. 2010. Darcin: a male pheromone that stimulates female memory and sexual attraction to an individual male's odour. BMC Biol. 8, 75. 10.1186/1741-7007-8-75 (doi:10.1186/1741-7007-8-75) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ferrari E, Lodi T, Sorbi RT, Tirindelli R, Cavaggioni A, Spisni A. 1997. Expression of a lipocalin in Pichia pastoris: secretion, purification and binding activity of a recombinant mouse major urinary protein. FEBS Lett. 401, 73–77 10.1016/S0014-5793(96)01436-6 (doi:10.1016/S0014-5793(96)01436-6) [DOI] [PubMed] [Google Scholar]
- 46.Sharrow SD, Vaughn JL, Žídek L, Novotny MV, Stone MJ. 2002. Pheromone binding by polymorphic mouse major urinary proteins. Protein Sci. 11, 2247–2256 10.1110/ps.0204202 (doi:10.1110/ps.0204202) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kurtz DT, Feigelson P. 1977. Multihormonal induction of hepatic alpha2u-globulin mRNA as measured by hybridization to complementary DNA. Proc. Natl Acad. Sci. USA 74, 4791–4795 10.1073/pnas.74.11.4791 (doi:10.1073/pnas.74.11.4791) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Friedman JM, Halaas JL. 1998. Leptin and the regulation of body weight in mammals. Nature 395, 763–770 10.1038/27376 (doi:10.1038/27376) [DOI] [PubMed] [Google Scholar]
- 49.Teerds KJ, Rooij DG, de Keijer J. 2011. Functional relationship between obesity and male reproduction: from humans to animal models. Hum. Reprod. Update 17, 667–683 10.1093/humupd/dmr017 (doi:10.1093/humupd/dmr017) [DOI] [PubMed] [Google Scholar]
- 50.Barash IA, Cheung CC, Weigle DS, Ren H, Kabigting EB, Kuijper JL, Clifton DK, Steiner RA. 1996. Leptin is a metabolic signal to the reproductive system. Endocrinology 137, 3144–3147 10.1210/en.137.7.3144 (doi:10.1210/en.137.7.3144) [DOI] [PubMed] [Google Scholar]
- 51.Cioffi JA, Shafer AW, Zupancic TJ, Smith-Gbur J, Mikhail A, Platika D, Snodgrass HR. 1996. Novel B219/OB receptor isoforms: possible role of leptin in hematopoiesis and reproduction. Nat. Med. 2, 585–589 10.1038/nm0596-585 (doi:10.1038/nm0596-585) [DOI] [PubMed] [Google Scholar]
- 52.Mounzih K, Lu R, Chehab FF. 1997. Leptin treatment rescues the sterility of genetically obese ob/ob males. Endocrinology 138, 1190–1193 10.1210/en.138.3.1190 (doi:10.1210/en.138.3.1190) [DOI] [PubMed] [Google Scholar]
- 53.Frederich RC, Hamann A, Anderson S, Löllmann B, Lowell BB, Flier JS. 1995. Leptin levels reflect body lipid content in mice: evidence for diet-induced resistance to leptin action. Nat. Med. 1, 1311–1314 10.1038/nm1295-1311 (doi:10.1038/nm1295-1311) [DOI] [PubMed] [Google Scholar]
- 54.Mathers JC. 2006. Nutritional modulation of ageing: genomic and epigenetic approaches. Mech. Ageing Dev. 127, 584–589 10.1016/j.mad.2006.01.018 (doi:10.1016/j.mad.2006.01.018) [DOI] [PubMed] [Google Scholar]
- 55.Gillum MP, Erion DM, Shulman GI. 2011. Sirtuin-1 regulation of mammalian metabolism. Trends Mol. Med. 17, 8–13 10.1016/j.molmed.2010.09.005 (doi:10.1016/j.molmed.2010.09.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Holliday R. 2006. Food, fertility and longevity. Biogerontology 7, 139–141 10.1007/s10522-006-9012-3 (doi:10.1007/s10522-006-9012-3) [DOI] [PubMed] [Google Scholar]
- 57.Anson RM, Guo Z, de Cabo R, Iyun T, Rios M, Hagepanos A, Ingram DK, Lane MA, Mattson MP. 2003. Intermittent fasting dissociates beneficial effects of dietary restriction on glucose metabolism and neuronal resistance to injury from calorie intake. Proc. Natl Acad. Sci. USA 100, 6216–6220 10.1073/pnas.1035720100 (doi:10.1073/pnas.1035720100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Swindell WR. 2012. Dietary restriction in rats and mice: a meta-analysis and review of the evidence for genotype-dependent effects on lifespan. Ageing Res. Rev. 11, 254–270 10.1016/j.arr.2011.12.006 (doi:10.1016/j.arr.2011.12.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Harper JM, Leathers CW, Austad SN. 2006. Does caloric restriction extend life in wild mice? Aging Cell 5, 441–449 10.1111/j.1474-9726.2006.00236.x (doi:10.1111/j.1474-9726.2006.00236.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Austad SN, Kristan DM. 2003. Are mice calorically restricted in nature? Aging Cell 2, 201–207 10.1046/j.1474-9728.2003.00053.x (doi:10.1046/j.1474-9728.2003.00053.x) [DOI] [PubMed] [Google Scholar]