Abstract
Major histocompatibility complex (Mhc) genes are believed to play a key role in the genetic basis of disease control. Although numerous studies have sought links between Mhc and disease prevalence, many have ignored the ecological and epidemiological aspects of the host–parasite interaction. Consequently, interpreting associations between prevalence and Mhc has been difficult, whereas discriminating alleles for qualitative resistance, quantitative resistance and susceptibility remains challenging. Moreover, most studies to date have quantified associations between genotypes and disease status, overlooking the complex relationship between genotype and the properties of the Mhc molecule that interacts with parasites. Here, we address these problems and demonstrate avian malaria (Plasmodium) parasite species-specific associations with functional properties of Mhc molecules (Mhc supertypes) in a wild great tit (Parus major) population. We further show that correctly interpreting these associations depends crucially on understanding the spatial variation in risk of infection and the fitness effects of infection. We report that a single Mhc supertype confers qualitative resistance to Plasmodium relictum, whereas a different Mhc supertype confers quantitative resistance to Plasmodium circumflexum infections. Furthermore, we demonstrate common functional properties of Plasmodium-resistance alleles in passerine birds, suggesting this is a model system for parasite–Mhc associations in the wild.
Keywords: major histocompatibility complex, avian malaria, supertype, resistance, Plasmodium, great tit (Parus major)
1. Introduction
Infectious disease is a major driver of ecological and evolutionary processes within wild populations, and characterizing the genetic component of host immunity is crucial to understand the genetic basis of variation in infection and to assess the evolutionary impacts of diseases [1]. Despite recent advances in genetic technologies, detecting novel resistance loci remains difficult and our understanding of immunogenetics in wild populations is still based on a small number of genes, for example, major histocompatibility complex (Mhc) genes [2] and interleukin 2 genes [3] (though see [4–6] for new markers being developed in wildlife disease studies). Hence, further work on these candidate genes has the potential to provide valuable information to explain the variation in disease susceptibility in natural populations [7]. In particular, the critical role of Mhc genes in triggering an immune response makes them excellent candidate markers for disease resistance studies, and Mhc-dependent disease resistance has been reported across taxa in natural populations of non-model species [8–10].
Early empirical evidence of a link between specific Mhc alleles and disease resistance was provided by the classic paper of Hill et al. [11], which suggested that specific Mhc alleles conferred protection from severe malaria infections (Plasmodium spp.) among African children. Avian malaria parasites, like their human-borne counterparts, are intracellular pathogens that invade host erythrocytes in the bloodstream. Thus, it is highly likely that the Mhc class I molecules of avian hosts recognize the peptides derived from malaria parasites and initiate cell destruction. Avian malaria parasites have been shown to compromise host fitness in both immunologically naive populations [12], as well as in species with which they are assumed to have shared a long evolutionary history [13–15], and are thus likely to exert strong selection on their hosts. These findings suggest that Mhc-linked malaria resistance should also exist in wild avian hosts, a premise supported by the results of several studies in passerine birds [16–20].
However, although significant associations between Mhc alleles and avian malaria resistance have been reported, the interpretation of these associations varied considerably in all these studies because both positive and negative associations were observed. Alleles that are negatively correlated with host infection are easily interpreted as disease-resistance alleles that provide qualitative protection against the parasite [17,19]. Positive correlations between Mhc alleles and host infection are more difficult to explain, because selection against susceptibility alleles should eliminate them from populations. Potentially, such positive associations may arise as a consequence of pleiotropy, whereby an allele that confers susceptibility to infection simultaneously confers protection to other pathogens [18]. Alternatively, they may arise as a consequence of quantitative disease-resistance alleles that limit the deleterious effects of disease without eradicating it completely. For example, in great reed warbler (Acrocephalus arundinaceus) populations, a positive relationship between a specific Mhc allele and Plasmodium infection was interpreted as quantitative resistance because parasitaemia (parasite load) was lower in individuals carrying the allele [21]. Selection might favour such alleles if individuals carrying them were better able to suppress acute infection and hence experienced improved survival relative to those without it, generating a positive relationship between presence of this allele and host infection. Another mechanism that may result in a link between Mhc genotype, and avian malaria infection is heterozygote advantage [22,23]: possessing more Mhc alleles may allow more effective parasite presentation [2,9,24]. Alternatively, maximal Mhc diversity may lead to excessive T-cell elimination during negative selection in the thymus [25,26]; hence, individuals possessing an intermediate, optimal number of Mhc alleles may have higher fitness [8,27].
Determining whether specific alleles confer susceptibility or quantitative resistance for the host is crucial for understanding the dynamics of complex host–parasite relationships. Using measures of parasitaemia as a surrogate measure of tolerance to initial infection makes a number of untested assumptions regarding the relationship of this variable to host immunocompetence. Exploring the presence of pleiotropic effects, on the other hand, is very difficult, given the need to test for and diagnose many different potential pathogens. An alternative approach to discriminate advantageous alleles from disadvantageous alleles is to investigate the fitness consequences of the parasite strain (e.g. malaria) in terms of host survival rates [28–30]. If a mortality cost for infection during the acute stage can be shown in the population, then a positive association between prevalence of infection and the immuno-allele would be interpreted as quantitative disease resistance. Investigating the fitness consequences of Mhc alleles in terms of host survival rates will also discriminate advantageous Mhc alleles from disadvantageous alleles [31,32]. However, determining such patterns is a major challenge in wild population studies, and none of the studies cited earlier were able to assess survival of hosts in relation to their Mhc or disease status (see [28,31,33]).
Difficulties in understanding the mechanisms by which Mhc alleles confer susceptibility or resistance may also stem from other factors. First, risk of infection, particularly for vector-borne diseases, varies extensively at both large and small spatial scales [34–36]. Such factors, if not controlled for, may confound or conceal relevant patterns between Mhc and disease status [7]. Moreover, the phenotypic effects of Mhc alleles are rarely taken into account [37], despite the fact that it is the properties of the antigen-binding sites (ABS) that determine the interactions between Mhc molecules and parasites. Indeed, a growing body of evidence across taxa highlights the importance of grouping Mhc alleles into supertypes (defined based on functional properties of the ABS) in order to classify functionally distinct Mhc types [38,39].
In this study, we investigate whether Mhc class I supertypes are associated with Plasmodium infection and host survival in a population of great tits infected with two divergent Plasmodium parasite species, Plasmodium relictum and Plasmodium circumflexum [40], while controlling for confounding factors, such as local risk of infection and host-related effects. Previous studies in this population on the closely related sympatric host species, blue tits (Cyanistes caeruleus), have shown that the two Plasmodium species differ substantially in their spatial distributions and impacts [15,35]. While P. circumflexum infections are associated with reduced survival, particularly during the acute stage of infection [15], P. relictum infections are linked with reproductive costs [14]. Here, we aimed to understand the role that Mhc play in determining host resistance and susceptibility to Plasmodium infections; and tested the three hypotheses that relate Mhc diversity to disease resistance: (i) the maximal diversity hypothesis; (ii) the optimal diversity hypothesis and (iii) effect of specific Mhc supertypes. Moreover, we aimed to determine whether specific Mhc supertypes confer susceptibility or quantitative resistance to Plasmodium infections.
2. Material and methods
(a). Study population and sampling
We studied the relationship between avian malaria, Mhc supertypes and fitness in 576 adult great tits from a nest-box breeding population sampled over 2 years (2008 and 2009 breeding seasons, April–June), and monitored for survival to 2011. One hundred and seventeen individuals were sampled in both years; hence, 693 samples were screened for Plasmodium infections. This population has been monitored continuously since the early 1960s and is located in Wytham Woods near Oxford, UK (51°46′ N, 1°20′ W). The great tit is a small, year-round resident, short-lived passerine bird species. Between 250 and 450 great tit pairs breed in the study population annually and display high breeding site fidelity following postnatal dispersal. Breeding birds were captured during the nestling phase, either within the nest-box by hand or using traps, or with mist nets in front of the nest entrance. All adults and nestlings are processed and ringed with aluminium bands for individual recognition. Blood was collected by wing or jugular venipuncture and stored in ‘SET’ lysis buffer (0.015M NaCL, 0.05M Tris, 0.001M EDTA, pH 8.0) at −80°C until DNA extraction.
(b). Screening of Plasmodium infections
Total DNA was measured using a Picogreen assay (Quant-iT Picogreen dsDNA assay kit, Invitrogen, Grand Island, NY, USA) and diluted to a concentration of 2 ng µl−1 for Plasmodium quantification [35]. A quantitative-polymerase chain reaction (qPCR)-based assay was used for measuring parasitaemia. The primers L9 (5′-AAACAATTCCTAACAAAACAGC-3′) and new R (5′-ACATCCAATCCATAATAAAGCA-3′) were chosen to ensure Plasmodium-specific amplification and to differentiate the two Plasmodium species [14]. The two Plasmodium species are a clade of lineages based on sequence data from approximately 450 bp of the mitochondrial cytochrome b gene fragment [34]. To calculate the parasitaemia in each sample, standard curves were created using Plasmodium pSGS1 lineage. qPCR amplifications were run in a final volume of 25 µl. Each DNA sample was analysed three times, and Plasmodium parasitaemia was scored as the mean value. Twenty-five samples were randomly chosen, re-quantified and re-amplified to estimate the repeatability of the method. Full details regarding qPCR screening are provided in Knowles et al. [14].
(c). 454 pyrosequencing of Mhc class I genes
The Mhc characterizations and genotyping methodology used in this study has been described in Sepil et al. [41], and complete details of all molecular protocols can be found therein; brief details are presented here. Bidirectional 454 pyrosequencing was performed on 1536 great tit samples, using the 16 lanes of a Picotiter plate gasket. We used 10 forward and 10 reverse fusion primers to amplify a 212–221 base pair fragment of great tit Mhc class I exon 3. Amplifications were run in a final volume of 25 µl and purified as described in Sepil et al. [41]. Samples with different primer combinations were pooled together in approximate equimolar quantities, a single pool was prepared for each lane and the pools were sent for bidirectional 454 pyrosequencing.
High numbers of artefacts can be generated during PCR and 454 pyrosequencing; therefore, we adopted a five-step variant validation procedure to distinguish real alleles from PCR/sequencing errors. From the 638 501 reads that the experiments generated, final genotypes were based on 214 357 reads. Eight hundred and fifty-seven individuals passed our reliability criteria, of which 576 were captured in an extensive sampling of the breeding population in 2008–2009 and screened for avian malaria infection and hence were used in this study. A total of 862 Mhc class I alleles (GenBank nos JQ034624–JQ035485) were detected; 755 alleles were classified as functional, and the presence of at least 16 functional loci was shown. There was clear evidence that the functional alleles were under strong balancing selection based on the detection of positively selected sites using a likelihood ratio test [41]. Lastly, functional alleles were translated into a matrix based on the physico-chemical amino acid properties of their positively selected sites [42], and the matrix was subjected to a k-means clustering algorithm [43]; the optimal number of supertypes was identified as 17. The association between specific Mhc supertypes and disease prevalence was assessed for 13 supertypes; the remaining four supertypes identified (supertypes 1, 2, 9 and 11) were removed from analyses as they were nearly fixed in the population with frequencies higher than 95 per cent [41].
(d). Measures of local infection risk
Spatial analysis was based on the geographical information system (GIS)-derived measures of tit nest-box locations [44]. Breeding tits are territorial and forage in the vicinity of their nests [45]; hence, the nest-box coordinates give an accurate representation of an individual's location during and either side of the breeding season, when transmission is most likely [46]. Previous work on the Wytham Woods tit populations has shown that there is pronounced spatial variation in the distribution of the two Plasmodium species [34,35,47]. The local risk of infection with either P. relictum or P. circumflexum was obtained for each Mhc-typed great tit in 2008 and 2009 and calculated as the prevalence of infected great tits within a 500 m buffer (based on disease cluster distances obtained in Wood et al. [34] and Lachish et al. [47]) of the focal individuals' nest-box. The analysis was carried out using GIS software (MapInfo Professional v. 7.8, Stamford, CT, USA).
(e). Statistical analyses
Statistical associations between Mhc supertypes and the probability of infection with either P. relictum or P. circumflexum were tested using generalized linear models with binomial errors and a logit link. If 2 years' data were available for an individual (which was the case for 20% of individuals), one was randomly excluded from the analysis, although inclusion of all the samples in the dataset did not qualitatively change the results. The probability of being infected was assessed in relation to the following covariates: the presence of each of the 13 Mhc supertypes described earlier, the total number of supertypes and alleles an individual possessed, the square of the total number of supertypes and alleles, local infection risk, year, sex, age class (categorized as 1 year of age, 2 years of age and 3 years or older) and a quadratic age class term (colinearity among explanatory variables was low; see the electronic supplementary material, table S1). Supertype number, its square, allele number, its square, age class and its quadratic function were mean-centred before inclusion in the model, by subtracting the mean from each data point, to reduce the covariance between the terms.
We used Akaike's information criterion (AIC) to determine the combination of variables that best explained the data with minimal parameters. Model selection was performed by backward stepwise elimination, and the fit of each new model was assessed by comparison of AIC values. Terms were eliminated from the model when their removal reduced AIC by at least 2 and were retained if their removal increased AIC by at least 2. Where the removal of a term resulted in a model with an approximately equal fit (i.e. a change in AIC of less than 2), the model with fewer terms was considered the most parsimonious model [48]. To confirm the validity of the minimum model, removed variables were added individually to assess any potential improvement in the model fit. Estimates of effect sizes (odds ratios) were calculated for each Mhc supertype to confirm the strength of association between Mhc and probability of infection. All analyses were conducted using R v. 2.12.1 [49].
The earlier-mentioned analyses revealed a positive relationship between the presence of one Mhc supertype (supertype 6) and infection with P. circumflexum (§3), suggesting that this supertype confers either susceptibility or quantitative resistance to P. circumflexum infection. To differentiate between these two possibilities, we conducted further analyses to investigate whether the parasite has a mortality cost during acute stage infection and whether the supertype conferred a survival advantage to its carriers (which would favour the idea that this supertype confers quantitative resistance to infection).
First, as proposed by Westerdahl et al. [21], we assessed whether P. circumflexum parasitaemia varied according to the presence of the Mhc supertype, using a linear mixed effects model. Again, if 2 years data were available for an individual, one was randomly excluded from the analysis. Parasitaemia was log-transformed [ln(x + 1)] to meet assumptions of constant variance and normal errors. Because parasitaemia values were calculated through comparison with standard curve values (see details above), the particular standard used was treated as a random factor to eliminate any bias arising from the use of different standards. Second, we assessed whether host survival probability varied between individuals that either possessed or did not possess supertype 6, as a function of individual infection status and the local risk of P. circumflexum infection. We reasoned that, if supertype 6 conferred quantitative resistance to P. circumflexum infection, then uninfected hosts that lack the supertype would have lower survival in high-risk areas (where they are at greater risk of becoming infected) than in low-risk areas. This should not be the case for infected birds, as the majority of these individuals would already have survived the brief acute stage of infection and thus harbour only chronic infections [50]. Moreover, we wished to assess whether host survival varied as a function of the local risk of P. circumflexum infection, because such a relationship would support a mortality cost for this parasite in this species. We predicted that uninfected hosts would have lower survival in high-risk areas than in low-risk areas, if P. circumflexum infection has a mortality cost during the acute stage. The absence of a pattern for uninfected birds would either indicate no or weak survival cost of P. circumflexum infection or imply that some of the uninfected birds have already had and cleared infection, concealing the identification of any relevant pattern. We determined host survival by scoring whether each individual was recaptured in the subsequent breeding seasons (using data on recaptures, including 2011) or not. As annual recapture rates of great tits in the breeding season are high in Wytham Woods (higher than 0.80 [51]), this provides a reasonably robust index of survival rates. Analyses were performed using a generalized linear model with binomial errors and a logit link, and the starting model included the three variables (presence of supertype 6, infection status and local risk of P. circumflexum infection) and their interaction. Third, we assessed whether the frequency of the Mhc supertype varied between first year and older individuals as a function of the local risk of P. circumflexum infection. We predicted that the frequency of supertype 6 should be higher in older individuals, particularly in areas with elevated risk of infection, as a result of the selective disappearance of individuals that lack this supertype. We separated high- and low-infection-risk areas by scoring whether an individual s nest-box was located within 500 m of the River Thames or not; P. circumflexum infection risk is substantially elevated in areas within 500 m of this large water body [34,35,47]. We assessed whether the frequency of the Mhc supertype varied as a function of host age (categorized as first year and older), and P. circumflexum infection risk, using a generalized linear model with binomial errors and a logit link, with both terms and their interaction included in the starting model. Calculating local infection risk as the percentage of infected great tits within a 500 m buffer, rather than using an arbitrary-cut of point, did not quantitatively change the results of the analysis (results not shown). Lastly, we explored the effects of supertype/allelic diversity, Mhc supertype 6 and their interaction on P. circumflexum prevalence. We predicted that if Mhc supertype 6 conferred susceptibility, individuals with lower supertype or allelic diversity would be more prone to P. circumflexum infection as a result of homozygosity (as in Worley et al. [30]). Great tit Mhc class I, Plasmodium infection and survival data files are included in the electronic supplementary material.
In a final separate analysis, we explored whether the associations we found between the two Mhc supertypes and the probability of Plasmodium infections (§3) were comparable to the findings of earlier studies. Of five studies investigating a link between passerine Mhc class I and Plasmodium infection, only two studies, both on house sparrows (Passer domesticus), presented sequence information for Mhc alleles identified as being linked to P. relictum infection (GenBank nos EU715815–EU715817 and EF429132; [18,19]). The similarity of these house sparrow alleles to the Mhc supertypes examined in this study at their putative ABS (to infer functional similarity) was assessed by combining these additional four alleles with our 755 functional class I alleles and re-running the k-means clustering algorithm [41]. Hence, in this analysis, we aimed to determine whether Mhc alleles that are associated with avian malaria in house sparrows cluster, in terms of their functional properties, with alleles of the two Mhc supertypes linked with Plasmodium infection in great tits.
3. Results
Overall 55 per cent of individuals were infected with Plasmodium; P. circumflexum prevalence (37.8%) was significantly higher than P. relictum prevalence (19.8%; 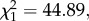 p < 0.0001). Fifteen individuals (2.7%) were co-infected with the two Plasmodium species and were included in both sets of analyses. Parasitaemia of infected individuals did not differ between the two Plasmodium species (
p < 0.0001). Fifteen individuals (2.7%) were co-infected with the two Plasmodium species and were included in both sets of analyses. Parasitaemia of infected individuals did not differ between the two Plasmodium species ( p = 0.184, ΔAIC =−0.3); the repeatability of the parasitaemia data was 0.779 (Pearson's product–moment correlation, p < 0.001), similar to previous estimates of repeatability from blue tits (r = 0.71) [14].
p = 0.184, ΔAIC =−0.3); the repeatability of the parasitaemia data was 0.779 (Pearson's product–moment correlation, p < 0.001), similar to previous estimates of repeatability from blue tits (r = 0.71) [14].
Our analyses revealed there to be significant associations between two different Mhc supertypes and the probability of infection with P. relictum and P. circumflexum. The probability of P. relictum infection was negatively associated with the presence of supertype 17 (see figure 1a and the electronic supplementary material, table S2), such that individuals lacking the supertype were twice as likely as individuals carrying it to become infected with P. relictum (odds ratio: 0.51, 95% CI: 0.39–0.66, p = 0.009; electronic supplementary material, figure S1), indicative of qualitative resistance. No other supertype was significantly associated with probability of P. relictum infection; nor was P. relictum infection related to the total number of supertypes or total number of alleles an individual possessed (either linearly or as a quadratic function; electronic supplementary material, table S2 and figure S1). The probability of P. relictum infection increased significantly with age and local infection risk, but did not vary between the sexes or the year of the study (see the electronic supplementary material, table S2).
Figure 1.

Probability of (a) Plasmodium relictum infection in great tit hosts (±s.e.) as a function of the presence of supertype 17; (b) Plasmodium circumflexum infection in great tit hosts (±s.e.) as a function of the presence of supertype 6.
By contrast, results of the generalized linear model for P. circumflexum infections in hosts revealed, there was a significant positive association between the probability of P. circumflexum infection and the presence of supertype 6 (see figure 1b and the electronic supplementary material, table S3). Individuals carrying the supertype were 58 per cent more likely to be infected with P. circumflexum (odds ratio: 1.58, 95% CI: 1.11–2.27, p = 0.009; electronic supplementary material, figure S2) than were the individuals that did not possess this Mhc supertype. No other supertype was significantly associated with P. circumflexum infection, nor was there any significant relationship between the total number of supertypes or total number of alleles an individual possessed (either linearly or as a quadratic function) and its infection status (see the electronic supplementary material, table S3 and figure S2). As in the P. relictum analysis mentioned earlier, the probability of P. circumflexum infection did not vary between the sexes, nor between the years of the study, but did vary significantly with P. circumflexum infection risk and host age, with older individuals more likely to be infected (see the electronic supplementary material, table S3).
The earlier-mentioned analyses showed that individuals carrying Mhc supertype 6 were more likely to be infected with P. circumflexum, suggesting that supertype 6 may confer either susceptibility or quantitative resistance to P. circumflexum infection. Contrary to our predictions, P. circumflexum parasitaemia did not vary between individuals that did or did not possess supertype 6 (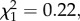 p = 0.641, ΔAIC =−1.78), nor did survival probabilities vary between individuals with and without supertype 6 (see the electronic supplementary material, table S4). Moreover, we found no significant interaction between the presence of supertype 6 and supertype/allelic diversity in terms of P. circumflexum infection (supertype 6 × supertype diversity,
p = 0.641, ΔAIC =−1.78), nor did survival probabilities vary between individuals with and without supertype 6 (see the electronic supplementary material, table S4). Moreover, we found no significant interaction between the presence of supertype 6 and supertype/allelic diversity in terms of P. circumflexum infection (supertype 6 × supertype diversity, 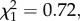 p = 0.4, ΔAIC =−1.28; supertype 6 × allelic diversity,
p = 0.4, ΔAIC =−1.28; supertype 6 × allelic diversity, 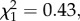 p = 0.51, ΔAIC =−1.57). However, our results showed that uninfected, but not infected, hosts experienced lower survival rates in high-infection-risk areas than in low-risk areas, indicating that this pathogen entails a mortality cost for hosts during the acute stage of infection (see figure 2 and the electronic supplementary material, table S4). In addition, as predicted, our analyses revealed that the frequency of supertype 6 varied between first year and older individuals as a function of P. circumflexum infection risk (local infection risk × host age,
p = 0.51, ΔAIC =−1.57). However, our results showed that uninfected, but not infected, hosts experienced lower survival rates in high-infection-risk areas than in low-risk areas, indicating that this pathogen entails a mortality cost for hosts during the acute stage of infection (see figure 2 and the electronic supplementary material, table S4). In addition, as predicted, our analyses revealed that the frequency of supertype 6 varied between first year and older individuals as a function of P. circumflexum infection risk (local infection risk × host age,  p = 0.033, ΔAIC =+2.55). The frequency of supertype 6 increased with age in high-risk areas: suggesting that individuals possessing this supertype are better able survive the lethal acute stage of infection, and that selection progressively increases the frequency of this supertype (figure 3). As none of these findings lends support for the susceptibility hypothesis, the evidence favours quantitative resistance.
p = 0.033, ΔAIC =+2.55). The frequency of supertype 6 increased with age in high-risk areas: suggesting that individuals possessing this supertype are better able survive the lethal acute stage of infection, and that selection progressively increases the frequency of this supertype (figure 3). As none of these findings lends support for the susceptibility hypothesis, the evidence favours quantitative resistance.
Figure 2.

Mean survival rates of (a) uninfected great tits (±s.e.) as a function of infection risk; (b) Plasmodium circumflexum infected great tits (±s.e.) as a function of infection risk.
Figure 3.
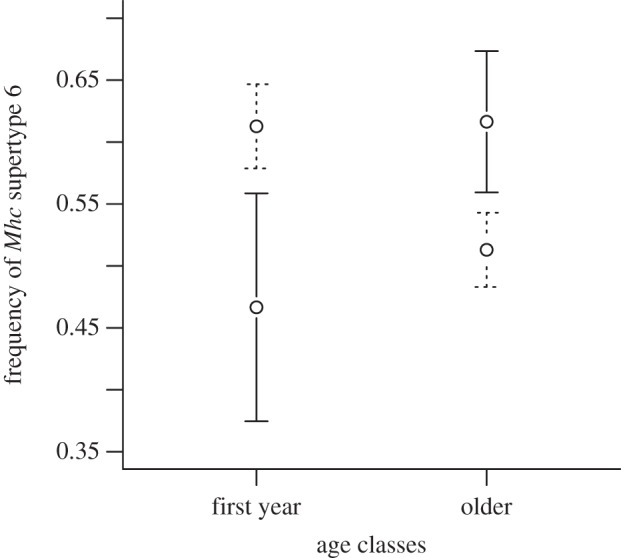
Frequency of Mhc supertype 6 in great tit hosts (±s.e.) in high P. circumflexum infection-risk areas (solid lines) and low-P. circumflexum infection-risk areas (dotted lines) as a function of host age.
Finally, we assessed the functional similarity of the Mhc class I alleles associated with Plasmodium prevalence in passerines by running k-means clustering algorithm on the combination of four house sparrow Mhc alleles and 755 great tit Mhc alleles. Two of the house sparrow alleles (EU715815 and EF429132) clustered with 18 great tit alleles, of which 14 were originally designated supertype 17, whereas a third house sparrow allele (EU715816) clustered with 31 great tit alleles, of which 18 were supertype 6 in our original clustering (see the electronic supplementary material, figure S3). The fourth allele (EU715817) clustered with four great tit alleles that were originally designated supertype 4 (see the electronic supplementary material, figure S3). Hence, three of the four alleles identified as being associated with malaria in another bird species cluster statistically, in terms of their functional properties, with those identified as being malaria-associated in this study.
4. Discussion
Investigating associations between Mhc and parasite prevalence is a common means of studying genetically determined disease resistance in wild animals. Until recently, positive associations between Mhc alleles and parasite prevalence had been taken as evidence of susceptibility to disease, while the potential for quantitative resistance (immuno-alleles that reduce the development of infection) has been largely neglected (see [21]). In this study, we incorporated a detailed investigation of avian malaria infection and analysis of Mhc class I genes in a wild great tit population, to understand the role that Mhc genes play in determining host resistance and susceptibility to Plasmodium infections. We found that the presence of two Mhc supertypes (defined based on functional properties of the ABS) was significantly associated with the probability of host infection with two congeneric Plasmodium species, but in contrasting manners. The direction of the association for one Mhc supertype was indicative of Mhc-linked qualitative resistance to P. relictum infection, with individuals lacking that supertype twice as likely to be infected. However, the functional role of the second Mhc supertype, in regard to P. circumflexum infection, was more difficult to assess. Of the four hypotheses we tested, two analyses supported quantitative resistance, whereas two analyses provided equivocal results. Therefore, our results more strongly support the idea that supertype 6 may confer quantitative resistance to P. circumflexum infection. Hence, the findings of this study imply that different Mhc supertypes can confer both qualitative and quantitative resistance to different Plasmodium species in a single host population.
Different types of associations between immuno-alleles and the probability of infection are common for different levels of infection severity [21]. Virulent parasites are likely to induce mortality during the acute stage of infection and are difficult to suppress completely [52]. Hence, alleles that confer quantitative resistance (i.e. that limit the deleterious effects of infection but do not prevent infection) would be beneficial to hosts that are primarily exposed to virulent parasites. Conversely, qualitative resistance alleles that prevent parasite establishment (i.e. prevent infection) would be beneficial to hosts that are exposed to more benign parasites, as these parasites are unlikely to cause mortality and are easier to suppress [53]. Using a multi-state modelling framework, Lachish et al. [15] showed that infection with P. circumflexum was associated with reduced survival in blue tits, compared with infection with P. relictum, suggesting that P. circumflexum is more virulent than P. relictum. In line with this finding, we found that uninfected great tits experienced lower survival rates in areas where P. circumflexum infection risk is high, suggesting a mortality cost for this parasite in great tits as well. Our results showing Mhc-linked qualitative resistance to P. relictum infection by great tits carrying supertype 17, and the possibility of Mhc-linked quantitative resistance to P. circumflexum infection for individuals carrying supertype 6, are thus in agreement with the above rationale and may be explained by the differing virulence of these two parasites.
Consequently, we investigated whether uninfected hosts lacking supertype 6 had lower survival rates in high-risk areas, but found no such relationship. Although we would expect supertype 6 to confer a survival advantage for its carriers, the highest mortality from infection is likely to occur when birds are first exposed to the parasite, as juveniles [15]. Hence, the absence of an association between supertype 6 and survival rates of adults in this study is perhaps not unexpected. Moreover, recent analyses have shown that individuals possessing supertype 6 had significantly higher lifetime reproductive success (defined as the total number of recruits produced over a lifetime) and offspring recruitment probabilities at the brood level, implying a survival advantage for juveniles carrying this Mhc supertype 6 [32]. This finding supports the suggestion that supertype 6 confers quantitative resistance to P. circumflexum infection. In this study, we showed that the frequency of supertype 6 increased with age in high-infection-risk areas, further supporting the idea that supertype 6 confers selective advantage for individuals that are likely to contract the disease. The negative association found between supertype 6 frequency and host age in low-risk areas (figure 3) might indicate that there is a cost associated with carrying a disease-resistance supertype [54]. If this is true, then the absence of supertype 6 might be more beneficial for great tits breeding in areas where the vector transmitting P. circumflexum is absent. Hence, it seems possible that the selective advantage of this supertype depends on local infection risk and there might be differential selection for the supertype at this small spatial scale.
The results of this study lend support to hypotheses suggesting a fitness advantage for carriers of specific Mhc types. Therefore, it is plausible to suggest that the extraordinary level of Mhc diversity observed in this population might be maintained through the mechanisms of negative-frequency-dependent selection [55], fluctuating selection, [56] or a combination of the two mechanisms. We found no support for hypotheses that link maximal or optimal Mhc diversity with individual fitness [20,27]; neither the number of supertypes, nor the number of alleles had an effect on Plasmodium infection. However, it is worth noting that we were not able to test heterozygote advantage at a particular locus [22], because multiple loci (at least 16 loci) were analysed simultaneously. Therefore, we cannot discount the possibility that associations between disease prevalence and heterozygosity at certain loci might have been overlooked.
In this study, we used a ‘supertyping’ approach to determine the functional properties of the Mhc alleles. To date, the majority of work has assessed links between Mhc genotype and disease resistance, however, much of selection on Mhc is likely to act via the phenotypic effects of the underlying genes. Here, we adopted the bioinformatic and statistical methods described by Doytchinova & Flower [42] and Jombart et al. [43] to define the physico-chemical properties of the positively selected sites of each allele and to cluster Mhc alleles with similar peptide specificities into supertypes. Therefore, by analysing the relationship between Mhc supertypes and Plasmodium infection, we aimed to predict the functional effects of Mhc supertypes. There is increasing support for the biological relevance of the supertyping approach [57,58]. For instance, Trachtenberg et al. [38] revealed an advantage of rare human leucocyte antigen supertypes in human immunodeficiency virus disease progression, independent of the contribution of single alleles. Therefore, we believe this approach is both appropriate and justified. Here, we used a newly proposed method for Mhc genotyping [43], which is an advance on previous methods, as it does not require arbitrary clustering decisions, but uses k-means clustering algorithm and model selection approach to compute-associated summary statistics [41]. The supertype clusters changed significantly following the addition of the four house sparrow alleles. The house sparrow alleles differed substantially from the great tit alleles and included amino acids not found among the great tit variants. Hence, the apparent instability of the clusters is probably an outcome of this difference. Nevertheless, the methodology can be further developed to improve the precision of identifying distinct clusters; experimental approaches can be used instead of bioinformatics approaches to determine the peptide specificities of Mhc molecules [59]. Moreover, it is worth noting that our amplifications did not cover a polymorphic section of Mhc exon 3 that affects the antigen-binding capabilities of the alleles. Amplifying the entire exon would be beneficial for future studies [60].
An important and novel finding from this study is that the alleles that are linked with Plasmodium infections in two passerine species (great tits and house sparrows) have similar antigen-binding affinities. Although population-specific associations between Mhc alleles and Plasmodium species have been reported in house sparrow populations [17,19], the functional differences between these alleles were not assessed, and our results imply that different alleles linked with disease may be similar at their ABS even in unrelated host species. The functional similarity across species suggests that Mhc-linked malaria resistance can be a valuable area for further research to understand the genetic basis of variation in infection in wild systems. However, as this study shows, such work must be embedded in an understanding of the ecological and epidemiological processes affecting the host–parasite interaction.
Acknowledgements
Blood was collected under UK Home Office licence (PPL 30/2409).
We are very grateful to the Wytham fieldworkers who collected the data for this study. We thank Alicia Davies and Simon Lee for laboratory assistance, Tobias Uller, Colin Garroway, Kaan Oz and two anonymous reviewers for helpful discussion and advice. This study was partially funded by NERC grant nos NER/A/S/2002/00877 and NE/F005725/1 to B.C.S.
References
- 1.Acevedo-Whitehouse K, Cunningham AA. 2006. Is MHC enough for understanding wildlife immunogenetics? Trends Ecol. Evol. 21, 433–438 10.1016/j.tree.2006.05.010 (doi:10.1016/j.tree.2006.05.010) [DOI] [PubMed] [Google Scholar]
- 2.Oliver MK, Telfer S, Piertney SB. 2009. Major histocompatibility complex (MHC) heterozygote superiority to natural multi-parasite infections in the water vole (Arvicola terrestris). Proc. R. Soc. B 276, 1119–1128 10.1098/rspb.2008.1525 (doi:10.1098/rspb.2008.1525) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turner AK, Begon M, Jackson JA, Bradley JE, Paterson S. 2011. Genetic diversity in cytokines associated with immune variation and resistance to multiple pathogens in a natural rodent population. PLoS Genet. 7, e1002343. 10.1371/journal.pgen.1002343 (doi:10.1371/journal.pgen.1002343) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hellgren O, Sheldon BC. 2011. Locus-specific protocol for nine different innate immune genes (antimicrobial peptides: β-defensins) across passerine bird species reveals within-species coding variation and a case of trans-species polymorphisms. Mol. Ecol. Resource 11, 686–692 10.1111/j.1755-0998.2011.02995.x (doi:10.1111/j.1755-0998.2011.02995.x) [DOI] [PubMed] [Google Scholar]
- 5.Tschirren B, Råberg L, Westerdahl H. 2011. Signatures of selection acting on the innate immunity gene toll-like receptor 2 (TLR2) during the evolutionary history of rodents. J. Evol. Biol. 24, 1232–1240 10.1111/j.1420-9101.2011.02254.x (doi:10.1111/j.1420-9101.2011.02254.x) [DOI] [PubMed] [Google Scholar]
- 6.Grueber CE, Wallis GP, King TM, Jamieson IG. 2012. Variation at innate immunity toll-like receptor genes in a bottlenecked population of a New Zealand robin. PLoS ONE 7, e45011. 10.1371/journal.pone.0045011 (doi:10.1371/journal.pone.0045011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amos W, Driscoll E, Hoffman JI. 2011. Candidate genes versus genome-wide associations: which are better for detecting genetic susceptibility to infectious disease? Proc. R. Soc. B 278, 1183–1188 10.1098/rspb.2010.1920 (doi:10.1098/rspb.2010.1920) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Madsen T, Ujvari B. 2006. MHC class I variation associates with parasite resistance and longevity in tropical pythons. J. Evol. Biol. 19, 1973–1978 10.1111/j.1420-9101.2006.01158.x (doi:10.1111/j.1420-9101.2006.01158.x) [DOI] [PubMed] [Google Scholar]
- 9.Kekäläinen J, Vallunen JA, Primmer CR, Rättyä J, Taskinen J. 2009. Signals of major histocompatibility complex overdominance in a wild salmonid population. Proc. R. Soc. B 276, 3133–3140 10.1098/rspb.2009.0727 (doi:10.1098/rspb.2009.0727) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kloch A, Babik W, Bajer A, Siński E, Radwan J. 2010. Effects of an MHC-DRB genotype and allele number on the load of gut parasites in the bank vole Myodes glareolus. Mol. Ecol. 19(Suppl. 1), 255–265 10.1111/j.1365-294X.2009.04476.x (doi:10.1111/j.1365-294X.2009.04476.x) [DOI] [PubMed] [Google Scholar]
- 11.Hill AVS, et al. 1991. Common West African HLA antigens are associated with protection from severe malaria. Nature 352, 595–600 10.1038/352595a0 (doi:10.1038/352595a0) [DOI] [PubMed] [Google Scholar]
- 12.Benning TL, LaPointe D, Atkinson CT, Vitousek PM. 2002. Interactions of climate change with biological invasions and land use in the Hawaiian Islands: modeling the fate of endemic birds using a geographic information system. Proc. Natl Acad. Sci. USA 99, 14 246–14 249 10.1073/pnas.162372399 (doi:10.1073/pnas.162372399) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marzal A, de Lope F, Navarro C, Møller AP. 2005. Malarial parasites decrease reproductive success: an experimental study in a passerine bird. Oecologia 142, 541–545 10.1007/s00442-004-1757-2 (doi:10.1007/s00442-004-1757-2) [DOI] [PubMed] [Google Scholar]
- 14.Knowles SCL, Palinauskas V, Sheldon BC. 2010. Chronic malaria infections increase family inequalities and reduce parental fitness: experimental evidence from a wild bird population. J. Evol. Biol. 23, 557–569 10.1111/j.1420-9101.2009.01920.x (doi:10.1111/j.1420-9101.2009.01920.x) [DOI] [PubMed] [Google Scholar]
- 15.Lachish S, Knowles SCL, Alves R, Wood MJ, Sheldon BC. 2011. Fitness effects of endemic malaria infections in a wild bird population: the importance of ecological structure. J. Anim. Ecol. 80, 1196–1206 10.1111/j.1365-2656.2011.01836.x (doi:10.1111/j.1365-2656.2011.01836.x) [DOI] [PubMed] [Google Scholar]
- 16.Westerdahl H, Waldenström J, Hansson B, Hasselquist D, von Schantz T, Bensch S. 2005. Associations between malaria and MHC genes in a migratory songbird. Proc. R. Soc. B 272, 1511–1518 10.1098/rspb.2005.3113 (doi:10.1098/rspb.2005.3113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bonneaud C, Pérez-Tris J, Federici P, Chastel O, Sorci G. 2006. Major histocompatibility alleles associated with local resistance to malaria in a passerine. Evolution 60, 383–389 10.1111/j.0014-3820.2006.tb01114.x (doi:10.1111/j.0014-3820.2006.tb01114.x) [DOI] [PubMed] [Google Scholar]
- 18.Loiseau C, Zoorob R, Garnier S, Birard J, Federici P, Julliard R, Sorci G. 2008. Antagonistic effects of a Mhc class I allele on malaria-infected house sparrows. Ecol. Lett. 11, 258–265 10.1111/j.1461-0248.2007.01141.x (doi:10.1111/j.1461-0248.2007.01141.x) [DOI] [PubMed] [Google Scholar]
- 19.Loiseau C, Zoorob R, Robert A, Chastel O, Julliard R, Sorci G. 2011. Plasmodium relictum infection and MHC diversity in the house sparrow (Passer domesticus). Proc. R. Soc. B 278, 1264–1272 10.1098/rspb.2010.1968 (doi:10.1098/rspb.2010.1968) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Radwan J, Zagalska-Neubauer M, Cichoń M, Sendecka J, Kulma K, Gustafsson L, Babik W. 2012. MHC diversity, malaria and lifetime reproductive success in collared flycatchers. Mol. Ecol. 21, 2469–2479 10.1111/j.1365-294X.2012.05547.x (doi:10.1111/j.1365-294X.2012.05547.x) [DOI] [PubMed] [Google Scholar]
- 21.Westerdahl H, Asghar M, Hasselquist D, Bensch S. 2011. Quantitative disease resistance: to better understand parasite-mediated selection on major histocompatibility complex. Proc. R. Soc. B 279, 577–584 10.1098/rspb.2011.0917 (doi:10.1098/rspb.2011.0917) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doherty P, Zinkernagel R. 1975. Enhanced immunological surveillance in mice heterozygous at the H-2 gene complex. Nature 256, 50–52 10.1038/256050a0 (doi:10.1038/256050a0) [DOI] [PubMed] [Google Scholar]
- 23.Takahata N, Nei M. 1990. Allelic genealogy under overdominant and frequency-dependent selection and polymorphism of major histocompatibility complex loci. Genetics 124, 967–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Froeschke G, Sommer S. 2005. MHC class II DRB variability and parasite load in the striped mouse (Rhabdomys pumilio) in the Southern Kalahari. Mol. Biol. Evol. 22, 1254–1259 10.1093/molbev/msi112 (doi:10.1093/molbev/msi112) [DOI] [PubMed] [Google Scholar]
- 25.Nowak MA, Tarczy-Hornoch K, Austyn JM. 1992. The optimal number of major histocompatibility complex molecules in an individual. Proc. Natl Acad. Sci. USA 89, 10 896–10 899 10.1073/pnas.89.22.10896 (doi:10.1073/pnas.89.22.10896) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woelfing B, Traulsen A, Milinski M, Boehm T. 2009. Does intra-individual major histocompatibility complex diversity keep a golden mean? Phil. Trans. R. Soc. B 364, 117–128 10.1098/rstb.2008.0174 (doi:10.1098/rstb.2008.0174) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kalbe M, Eizaguirre C, Dankert I, Reusch TBH, Sommerfeld RD, Wegner KM, Milinski M. 2009. Lifetime reproductive success is maximized with optimal major histocompatibility complex diversity. Proc. R. Soc. B 276, 925–934 10.1098/rspb.2008.1466 (doi:10.1098/rspb.2008.1466) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paterson S, Wilson K, Pemberton J. 1998. Major histocompatibility complex variation associated with juvenile survival and parasite resistance in a large unmanaged ungulate population (Ovis aries L.). Proc. Natl Acad. Sci. USA 95, 3714–3719 10.1073/pnas.95.7.3714 (doi:10.1073/pnas.95.7.3714) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lohm J, Grahn M, Langefors A, Andersen Ø, Storset A, Von Schantz T. 2002. Experimental evidence for major histocompatibility complex-allele-specific resistance to a bacterial infection. Proc. R. Soc. Lond. B 269, 2029–2033 10.1098/rspb.2002.2114 (doi:10.1098/rspb.2002.2114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Worley K, Collet J, Spurgin LG, Cornwallis C, Pizzari T, Richardson DS. 2010. MHC heterozygosity and survival in red junglefowl. Mol. Ecol. 19, 3064–3075 10.1111/j.1365-294X.2010.04724.x (doi:10.1111/j.1365-294X.2010.04724.x) [DOI] [PubMed] [Google Scholar]
- 31.Brouwer L, Barr I, Van de Pol M, Burke T, Komdeur J, Richardson DS. 2010. MHC-dependent survival in a wild population: evidence for hidden genetic benefits gained through extra-pair fertilizations. Mol. Ecol. 19, 3444–3455 10.1111/j.1365-294X.2010.04750.x (doi:10.1111/j.1365-294X.2010.04750.x) [DOI] [PubMed] [Google Scholar]
- 32.Sepil I, Lachish S, Sheldon BC. 2013. Mhc-linked survival and lifetime reproductive success in a wild population of great tits. Mol. Ecol. 22, 384–396 10.1111/mec.12123 (doi:10.1111/mec.12123) [DOI] [PubMed] [Google Scholar]
- 33.De Assunção-Franco M, Hoffman JI, Harwood J, Amos W. 2012. MHC genotype and near-deterministic mortality in grey seals. Sci. Rep. 2, 659. 10.1038/srep00659 (doi:10.1038/srep00659) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wood MJ, Cosgrove CL, Wilkin TA, Knowles SCL, Day KP, Sheldon BC. 2007. Within-population variation in prevalence and lineage distribution of avian malaria in blue tits, Cyanistes caeruleus. Mol. Ecol. 16, 3263–3273 10.1111/j.1365-294X.2007.03362.x (doi:10.1111/j.1365-294X.2007.03362.x) [DOI] [PubMed] [Google Scholar]
- 35.Knowles SCL, Wood MJ, Alves R, Wilkin TA, Bensch S, Sheldon BC. 2011. Molecular epidemiology of malaria prevalence and parasitaemia in a wild bird population. Mol. Ecol. 20, 1062–1076 10.1111/j.1365-294X.2010.04909.x (doi:10.1111/j.1365-294X.2010.04909.x) [DOI] [PubMed] [Google Scholar]
- 36.Sehgal RNM, et al. 2011. Spatially explicit predictions of blood parasites in a widely distributed African rainforest bird. Proc. R. Soc. B 278, 1025–1033 10.1098/rspb.2010.1720 (doi:10.1098/rspb.2010.1720) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spurgin LG, Richardson DS. 2010. How pathogens drive genetic diversity: MHC, mechanisms and misunderstandings. Proc. R. Soc. B 277, 979–988 10.1098/rspb.2009.2084 (doi:10.1098/rspb.2009.2084) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trachtenberg E, et al. 2003. Advantage of rare HLA supertype in HIV disease progression. Nat. Med. 9, 928–935 10.1038/nm893 (doi:10.1038/nm893) [DOI] [PubMed] [Google Scholar]
- 39.Huchard E, Raymond M, Benavides J, Marshall H, Knapp LA, Cowlishaw G. 2010. A female signal reflects MHC genotype in a social primate. BMC Evol. Biol. 10, 96. 10.1186/1471-2148-10-96 (doi:10.1186/1471-2148-10-96) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Valkiunas G. 2005. Avian malaria parasites and other haemosporidia. Boca Raton, FL: CRC Press [Google Scholar]
- 41.Sepil I, Moghadam HK, Huchard E, Sheldon BC. 2012. Characterization and 454 pyrosequencing of major histocompatibility complex class I genes in the great tit reveal complexity in a passerine system. BMC Evol. Biol. 12, 68. 10.1186/1471-2148-12-68 (doi:10.1186/1471-2148-12-68) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Doytchinova I, Flower DR. 2005. In silico identification of supertypes for class II MHCs. J. Immunol. 174, 7085–7095 [DOI] [PubMed] [Google Scholar]
- 43.Jombart T, Devillard S, Balloux F. 2010. Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet. 11, 94. 10.1186/1471-2156-11-94 (doi:10.1186/1471-2156-11-94) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilkin TA, Perrins CM, Sheldon BC. 2007. The use of GIS in estimating spatial variation in habitat quality: a case study of lay-date in the great tit Parus major. Ibis 149, 110–118 10.1111/j.1474-919X.2007.00757.x (doi:10.1111/j.1474-919X.2007.00757.x) [DOI] [Google Scholar]
- 45.Stauss MJ, Burkhardt JF, Tomiuk J. 2005. Foraging flight distances as a measure of parental effort in blue tits Parus caeruleus differ with environmental conditions. J. Avian Biol. 36, 47–56 10.1111/j.0908-8857.2005.02855.x (doi:10.1111/j.0908-8857.2005.02855.x) [DOI] [Google Scholar]
- 46.Cosgrove CL, Wood MJ, Day KP, Sheldon BC. 2008. Seasonal variation in Plasmodium prevalence in a population of blue tits Cyanistes caeruleus. J. Anim. Ecol. 77, 540–548 10.1111/j.1365-2656.2008.01370.x (doi:10.1111/j.1365-2656.2008.01370.x) [DOI] [PubMed] [Google Scholar]
- 47.Lachish S, Knowles SCL, Alves R, Sepil I, Davies A, Lee S, Wood MJ, Sheldon BC. 2012. Spatial determinants of infection risk in a multi-species avian malaria system. Ecography 35, 1–12 10.1111/j.1600-0587.2012.07801.x (doi:10.1111/j.1600-0587.2012.07801.x) [DOI] [Google Scholar]
- 48.Burnham K, Anderson D. 2002. Model selection and multimodel inference: a practical information-theoretic approach. New York, NY: Springer [Google Scholar]
- 49.R Development Core Team 2011. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; See http://www.R-project.org. [Google Scholar]
- 50.Bensch S, Waldenström J, Jonzén N, Westerdahl H, Hansson B, Sejberg D, Hasselquist D. 2007. Temporal dynamics and diversity of avian malaria parasites in a single host species. J. Anim. Ecol. 76, 112–122 10.1111/j.1365-2656.2006.01176.x (doi:10.1111/j.1365-2656.2006.01176.x) [DOI] [PubMed] [Google Scholar]
- 51.Bouwhuis S, Choquet R, Sheldon BC, Verhulst S. 2012. The forms and fitness cost of senescence: age-specific recapture, survival, reproduction, and reproductive value in a wild bird population. Am. Nat. 179, E15–E27 10.1086/663194 (doi:10.1086/663194) [DOI] [PubMed] [Google Scholar]
- 52.Gandon S, Michalakis Y. 2000. Evolution of parasite virulence against qualitative or quantitative host resistance. Proc. R. Soc. Lond. B 267, 985–990 10.1098/rspb.2000.1100 (doi:10.1098/rspb.2000.1100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.May R, Nowak M. 1994. Superinfection, metapopulation dynamics, and the evolution of diversity . J. Theor. Biol. 170, 95–114 10.1006/jtbi.1994.1171 (doi:10.1006/jtbi.1994.1171) [DOI] [PubMed] [Google Scholar]
- 54.Sheldon BC, Verhulst S. 1996. Ecological immunology: costly parasite defences and trade-offs in evolutionary ecology. Trends Ecol. Evol. 5347, 317–321 10.1016/0169-5347(96)10039-2 (doi:10.1016/0169-5347(96)10039-2) [DOI] [PubMed] [Google Scholar]
- 55.Slade RW, McCallum HI. 1992. Overdominant vs frequency-dependent selection at MHC loci. Genetics 132, 861–862 10.2460/javma.240.8.931 (doi:10.2460/javma.240.8.931) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hedrick PW. 2002. Pathogen resistance and genetic variation at MHC loci. Evolution 56, 1902–1908 10.1554/0014-3820(2002)056[1902:PRAGVA]2.0.CO;2 (doi:10.1554/0014-3820(2002)056[1902:PRAGVA]2.0.CO;2) [DOI] [PubMed] [Google Scholar]
- 57.Bertoni R, Sidney J, Fowler P, Chesnut RW, Chisari FV, Sette A. 1997. Human histocompatibility leukocyte antigen-binding supermotifs predict broadly cross-reactive cytotoxic T lymphocyte responses in patients with acute hepatitis. J. Clin. Invest. 100, 503–513 10.1172/JCI119559 (doi:10.1172/JCI119559) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lund O, et al. 2004. Definition of supertypes for HLA molecules using clustering of specificity matrices. Immunogenetics 55, 797–810 10.1007/s00251-004-0647-4 (doi:10.1007/s00251-004-0647-4) [DOI] [PubMed] [Google Scholar]
- 59.Lenz TL. 2011. Computational prediction of MHC II-antigen binding supports divergent allele advantage and explains trans-species polymorphism. Evolution 65, 2380–2390 10.1111/j.1558-5646.2011.01288.x (doi:10.1111/j.1558-5646.2011.01288.x) [DOI] [PubMed] [Google Scholar]
- 60.Llaurens V, McMullan M, van Oosterhout C. 2012. Cryptic MHC polymorphism revealed but not explained by selection on the class IIB peptide-binding region. Mol. Biol. Evol. 29, 1631–1644 10.1093/molbev/mss012 (doi:10.1093/molbev/mss012) [DOI] [PubMed] [Google Scholar]


