Abstract
Seasonal environments present fundamental physiological challenges to a wide range of insects. Many temperate insects surmount the exigencies of winter by undergoing photoperiodic diapause, in which photoperiod provides a token cue that initiates an alternative developmental programme leading to dormancy. Pre-diapause is a crucial preparatory phase of this process, preceding developmental arrest. However, the regulatory and physiological mechanisms of diapause preparation are largely unknown. Using high-throughput gene expression profiling in the Asian tiger mosquito, Aedes albopictus, we reveal major shifts in endocrine signalling, cell proliferation, metabolism, energy production and cellular structure across pre-diapause development. While some hallmarks of diapause, such as insulin signalling and stress response, were not important at the transcriptional level, two genes, Pepck and PCNA, appear to show diapause-induced transcriptional changes across insect taxa. These processes demonstrate physiological commonalities between Ae. albopictus pre-diapause and diapause strategies across insects, and support the idea of a genetic ‘toolkit’ for diapause. Observations of gene expression trends from a comparative developmental perspective suggest that individual physiological processes are delayed against a background of a fixed morphological ontogeny. Our results demonstrate how deep sequencing can provide new insights into elusive molecular bases of complex ecological adaptations.
Keywords: Aedes albopictus, RNA-Seq, diapause preparation, embryonic development, molecular physiology, invasive species
1. Introduction
Many organisms living in temperate habitats must coordinate their life cycles to accomplish growth, development and reproduction during favourable seasons and appropriately initiate dormancy or migration to avoid seasons that are unfavourable. Insects in seasonally variable environments can exhibit adaptive polyphenisms, in which discrete, alternative phenotypes develop in response to threshold environmental cues [1]. Photoperiodic diapause, in particular, provides a crucial adaptive mechanism whereby a token cue (photoperiod) stimulates pre-programmed physiological changes, leading to the onset of dormancy in advance of unfavourable winter conditions [2]. Studies in a diverse range of insects have identified a number of common physiological themes underpinning the diapause response. These results suggest the possibility of a ‘genetic toolkit’ for diapause, in which sets of genes have a conserved role in mediating the diapause response across insect taxa by regulating common physiological processes at the molecular level [3]. However, the complexity of the diapause programme, and its diverse manifestations in different insect species, has complicated the effort to find such common molecular mechanisms underlying diapause [4]. High-throughput gene expression profiling (‘RNA-Seq’) provides a new method to generate a comprehensive view of global transcriptional changes in ‘non-model’ organisms with a well-understood diapause programme, in order to identify shared or unique patterns of gene expression across insect taxa.
Diapause-destined insects undergo a series of preparatory events before the onset of developmental arrest; the stage comprising these events is called diapause preparation [5], or, here, ‘pre-diapause’. During pre-diapause, insects must complete developmental processes while at the same time retaining the pre-programmed information specifying developmental arrest later in ontogeny [5]. Insects in pre-diapause often show an extended developmental period and undergo physiological changes such as additional provisioning with lipid reserves, cuticular hydrocarbons and nutrient storage proteins (reviewed in [6]). The pre-diapause phase is thus critical for survival of the insect during adverse conditions, and therefore is also of potential relevance for management of pest species. However, comparatively little is known about the specific trajectory, timing and molecular basis of physiological changes that occur during the preparatory stage. One way to approach this question is to compare gene expression changes over time between pre-diapause and non-diapause insects.
The Asian tiger mosquito, Aedes albopictus, is an outstanding model system for the evolution and molecular physiology of insect diapause. Temperate populations of Ae. albopictus undergo a photoperiodic diapause in which exposure of the pupa and adult female to short day-lengths induces the production of diapausing offspring that enter developmental arrest as pharate larvae inside the chorion of the egg (see the electronic supplementary material, figure S1) [7,8]. Thus, the diapause induction, preparation and initiation phases are clearly separated by distinct life-history stages, which greatly facilitates the study of pre-diapause in this species. Urbanski et al. [9] have recently demonstrated rapid adaptive evolution of the diapause response in invasive populations of Ae. albopictus, underscoring the crucial ecological significance of diapause during the rapid global spread of this mosquito. Previous work has elucidated the molecular underpinnings of increased desiccation resistance during diapause in Ae. albopictus [10] and also molecular mechanisms related to increased lipid content of diapause versus non-diapause eggs [11]. Both of these properties are likely to be related to the ability of Ae. albopictus eggs to survive long-distance transport, and thus may contribute to the invasive success of this mosquito, one of the fastest-spreading animals on Earth [12]. Finally, information from related model species such as Aedes aegypti and Drosophila melanogaster has allowed us to develop extensive transcript me resources [13].
Here, we use RNA-Seq and a novel Ae. albopictus transcriptome to identify mechanisms of diapause preparation in Ae. albopictus. We analyse gene expression differences between pre-diapause and non-diapause embryos from two separate time points, encompassing major developmental transitions. We examine global gene expression trends, and analyse pathways and functional groups of biochemically related genes to infer major physiological shifts during pre-diapause development. Within this framework, we additionally ask whether genes and pathways previously identified as important during diapause in other organisms are also relevant in Ae. albopictus pre-diapause, to provide evidence for a ‘genetic toolkit’ for diapause. Finally, we perform comparisons with time-series data from D. melanogaster [14,15] to investigate the trajectory of gene expression in pre-diapause Ae. albopictus embryos relative to gene expression during D. melanogaster embryonic development. We argue that our approach not only illuminates mechanisms of diapause preparation, but also demonstrates a strategy for the analysis of seasonal polyphenisms in general.
2. Material and methods
(a). Mosquito rearing and tissue generation
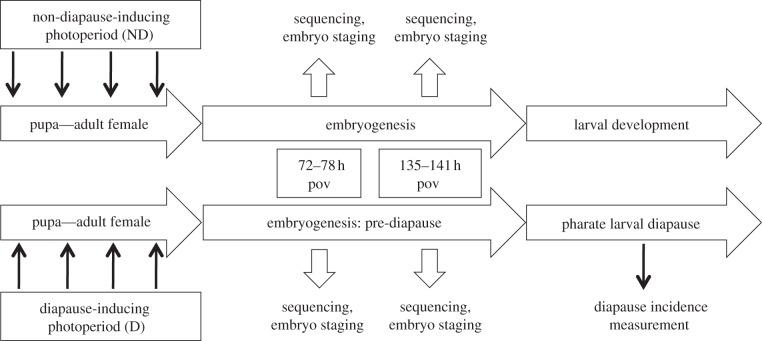
The experimental design (figure 1), tissue and RNA collection, transcriptome sequencing, assembly, and annotation are described in detail in the electronic supplementary material (tables S1 and S2, and figures S1–S3). Briefly, six cohorts of a laboratory F9 Ae. albopictus strain were reared at 16 L : 8 D, 21°C and approximately 80 per cent relative humidity until the pupal stage [16,17]. Upon pupation, we established three replicate cohorts (biological replicates) of approximately 500 individuals each under a diapause-inducing photoperiod (D; 8 L : 16 D), and three cohorts under a non-diapause-inducing photoperiod (ND; 16 L : 8 D). Eggs were collected over a 6 h oviposition (pov) period, and were divided to provide 3-day-old (3d pov; 72–78 h) and 6-day-old (6d pov; 135–141 h) embryos for RNA extraction. In addition, embryos were reserved to assess developmental stage and diapause incidence (see below). The 3d and 6d developmental stages were chosen to represent clear morphological landmarks in embryonic development, and were classified based on comparisons of cleared Ae. albopictus embryos with published images of Ae. aegypti embryos [18–20]. The 3d period encompasses the developmental landmarks of germ band retraction and dorsal closure; the 6d period comprises the end of embryonic development, and is characterized by complete segmentation and clear separation of the head from the thorax. Illumina paired-end mRNA-Seq library construction and sequencing were performed on each replicate at the University of Maryland Genomics Institute. Two flow-cell lanes on an Illumina HiSeq high-throughput sequencer were used to sequence the libraries: all 3d libraries were sequenced on one lane, and all 6d libraries on the second lane. Raw reads are accessible in NCBI's short read archive (SRA) under submission numbers SRA044835 and SRA051478. The assembly is available at http://www.albopictusexpression.org/?q=data.
Figure 1.
Schematic of the experimental design. Individual elements of the experimental design are depicted in the context of the diapause programme within the Ae. albopictus life cycle. 72–78 h pov and 135–141 h pov refer to the embryo collection dates of 3 days (72–78 h) post-oviposition and 6 days (135–141 h) post-oviposition, respectively.
(b). Assessment of developmental stage and diapause incidence
Subsets of eggs from each treatment and replicate (50–388 per replicate) retained for morphological staging were bleached to clear the chorion following methods outlined by Trpis [21]. Morphology was scored using dissecting scopes to quantify the percentage of embryos in the appropriate developmental stage for each sample. Eggs retained for diapause incidence calculations (50–273 per replicate) were treated following Poelchau et al. [13], based on methods of Hawley et al. [22]; the D photoperiod treatment resulted in a high diapause incidence (98.89%).
(c). Gene expression quantification and analysis
We describe gene expression quantification in detail in the electronic supplementary material. Briefly, we quantified expression for each unigene using RSEM [23], followed by TMM-normalization (trimmed mean of M values) in edgeR [24]. Normalized read counts are available in the electronic supplementary material (table S2). We independently verified the gene expression results with qRT-PCR results from Reynolds et al. [11] (see the electronic supplementary material, figure S4).
We used the limma library [25,26] in BioConductor to identify differentially expressed genes between D and ND conditions at either 3d or 6d pov. Genes with a Benjamini–Hochberg corrected p-value smaller than 0.05 and greater than 0.5 absolute log2-fold change were considered differentially expressed (DE).
(d). Pathway and gene ontology category enrichment analyses
Pre-diapause and non-diapause developing embryos are expected to show major physiological differences, which should be manifest in the differential expression of suites of genes that underlie or regulate that physiological trait. Enrichment analyses identify whether these a priori defined suites of genes with related functions contain more members that are differentially expressed than expected by chance. We used DAVID [27,28] to ask which Kegg pathways [29,30] or functional groups of genes (Gene Ontology [31]; the DAVID ‘GO FAT’ lists were used) were enriched in genes DE between diapause and non-diapause treatments at 3d or 6d pov. While transcript length can bias results from enrichment analyses in RNA-Seq datasets [32], we determined in a separate analysis that there was no substantial length bias in our dataset, and therefore a DAVID analysis would give valid results (described in detail in the electronic supplementary material). Analyses were performed using D. melanogaster orthologues for each gene model (one-to-one and apparent one-to-one, retrieved from BioMart [33,34]). Because different functional groups can exhibit substantial overlap in gene composition, we used DAVID to cluster functional groups with similar gene compositions into ‘annotation clusters’. We performed downstream analyses using the functional group in each annotation cluster with the most comprehensive gene set to avoid redundant calculations. Groups with a false discovery rate (FDR) < 0.05 were considered significantly enriched.
We additionally used GeneMerge v. 1.2 [35] to determine whether gene lists unavailable in DAVID were over-represented. Based on Ragland et al. [4] and Wu & Brown [36], we reconstructed the insulin signalling pathway, which has been implicated in diapause [4]. We also compiled a list of heat-shock proteins from text-based searches within gene descriptions and the gene ontology category ‘response to stress’ (GO:0006950). For these gene sets, we identified genes from Ae. aegypti annotations. Finally, we composed a list of ecdysone-signalling genes from the ‘molting’ category of the Interactive Fly database [37].
(e). Developmental comparisons with Drosophila melanogaster
Embryonic development is characterized by strong changes in gene expression that coordinate dramatic physiological changes [38]. Furthermore, in Ae. albopictus, gene expression changes in preparation for diapause must occur within the context of regular embryonic development. To correctly interpret expression changes during pre-diapause, it is useful to consider these changes within the context of the non-diapause developmental trajectory. Preliminary observations of expression differences between D and ND photoperiods in Ae. albopictus cell-cycle regulators suggested that expression in D embryos was delayed relative to ND embryos, given the expected trend of downregulation of cell-cycle regulators over the course of embryonic development in other insects [38]. This observation raised the question of whether this phenomenon could be observed in other DE genes. To ask this question in a quantitative framework would require a detailed comparison of the time series of gene expression across development in non-diapause and pre-diapause embryos. In the absence of such a dataset, we generated qualitative comparisons with a time series of embryonic gene expression from another dipteran species: a comprehensive RNA-Seq time series from D. melanogaster, where embryo RNA was sampled every 2 h post-oviposition [14,15] (downloaded as RPKM from http://flybase.org/reports/FBrf0212041.html). We used this time series to visualize a representation of the course of gene expression during regular development in a dipteran embryo. Differences in gene expression between D and ND embryos can be compared against this background to assess whether fine-scale temporal shifts in gene expression occur during diapause preparation.
We selected ‘one-to-one’ and ‘apparent one-to-one’ orthologues between Ae. albopictus gene models and D. melanogaster [33], and created heat maps of z-standardized expression scores of Ae. albopictus genes from enriched categories at 3d or 6d pov. These were contrasted with D. melanogaster orthologues from morphologically similar developmental stages (the ranges of morphological characters observed within Ae. albopictus at 3d and 6d pov correspond roughly with those observed 8–12 h and 18–22 h pov in D. melanogaster, respectively [37]). This allowed us to qualitatively, yet systematically compare expression trends of D versus ND embryos in the context of expression change over embryonic development in D. melanogaster.
3. Results and discussion
(a). Assessment of developmental stage
Cleared subsets of staged eggs verified that there were no distinguishable differences at the morphological level between chronologically matched D and ND embryos, as 87–96 per cent of embryos were morphologically similar (see the electronic supplementary material, table S3 and figure S5).
(b). Gene expression quantification
Differential gene expression relevant to diapause preparation increased between 3d and 6d pov. Many more genes were significantly DE between D and ND photoperiods at the 6d stage than at 3d (figure 2; 6d: 4337; 3d: 2506). The range of log-fold change was also much wider at 6d versus 3d (figure 2). Gene expression profile clustering additionally revealed strong ontogenetic changes in transcriptional profiles (see the electronic supplementary material, figure S6).
Figure 2.
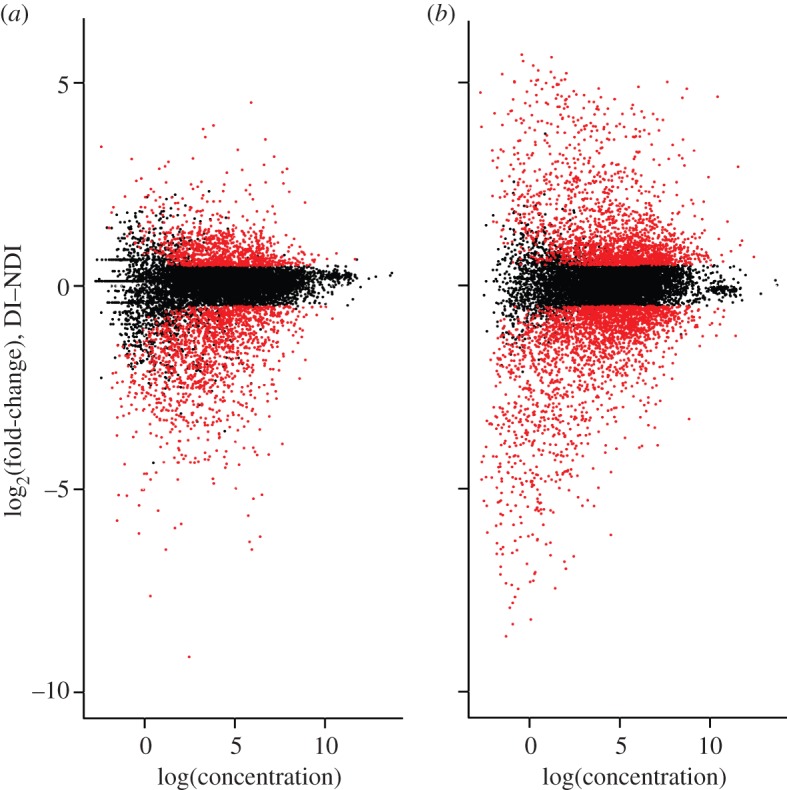
Log-fold-change expression (M) versus log abundance (A) of gene expression at (a) 3 days and (b) 6 days post-oviposition. Gene expression data are TMM-normalized. Genes with higher expression under diapausing (D) conditions have positive M values, and genes with higher expression under non-diapausing (ND) conditions have negative M values. Genes that qualified as significantly differentially expressed (DE; corrected p < 0.05; absolute log2-fold change greater than 0.5) are in red.
(c). Identification of diapause-relevant genes and pathways
Enriched functional groups shifted dramatically from intermediate to late embryological development (table 1). There was no overlap in enriched functional groups between the two time points (see the electronic supplementary material, table S4), indicating clear changes over time in the processes that contribute to pre-diapause development. While several pathways with known relevance for diapause were identified as enriched, lists of insulin-signalling genes and stress-related genes were not enriched during pre-diapause (all e-scores = 1). Below, we describe the implications of each enriched functional category at the 3d and 6d pov developmental stages.
Table 1.
Representative functional groups that were significantly enriched for DE genes at 3 days (3d) and 6 days (6d) post-oviposition. Functional groups are derived from the gene ontology (GO) biological processes, molecular function and cellular component clusters (BP, MF and CC, respectively). Only representative functional groups with FDR < 0.05 are reported; representative groups from each annotation cluster were designated as the group with the largest number (and therefore most representative set) of genes, in order to avoid redundant calculations.
| development stage | functional category | type | ID | term description |
|---|---|---|---|---|
| 3d | cell cycle | GO:BP | GO:0007088 | regulation of mitosis |
| 3d | cell cycle | GO:BP | GO:0022402 | cell-cycle process |
| 3d | cell cycle | GO:CC | GO:0005694 | chromosome |
| 3d | cell cycle | GO:BP | GO:0006260 | DNA replication |
| 3d | cell cycle | GO:BP | GO:0007059 | chromosome segregation |
| 3d | cell cycle | GO:BP | GO:0065004 | protein–DNA complex assembly |
| 3d | cell cycle | GO:CC | GO:0030894 | replisome |
| 3d | cell cycle/DNA repair | GO:BP | GO:0006259 | DNA metabolic process |
| 3d | cytoskeleton | GO:CC | GO:0044430 | cytoskeletal part |
| 3d | DNA binding | GO:MF | GO:0003677 | DNA binding |
| 3d | DNA repair | Kegg | Dme 03430 | mismatch repair |
| 6d | cuticle | GO:MF | GO:0005214 | structural constituent of chitin-based cuticle |
| 6d | cuticle | GO:BP | GO:0005976 | polysaccharide metabolic process |
| 6d | energy production | GO:BP | GO:0055114 | oxidation reduction |
| 6d | energy production | GO:CC | GO:0005739 | mitochondrion |
| 6d | extracellular region | GO:CC | GO:0005576 | extracellular region |
| 6d | metabolism | GO:BP | GO:0044271 | nitrogen compound biosynthetic process |
| 6d | metabolism/ protein synthesis | GO:BP | GO:0034660 | ncRNA metabolic process |
| 6d | transport | GO:MF | GO:0022890 | inorganic cation transmembrane transporter activity |
(d). Enriched functional categories at 3-day post-oviposition
Cell-cycle arrest contributes to developmental arrest in diapausing insects, but its manifestation and regulation during diapause are not well understood [39]. Previous gene expression studies of cell-cycle regulators are consistent with a decrease or halt of cell cycle and DNA replication processes in diapause, which resume once diapause is terminated [39–43]. In our study of the pre-diapause stage, the majority of DE genes involved in the cell cycle and DNA repair were positive cell-cycle regulators, and had higher expression in D embryos relative to ND embryos at 3d pov (see the electronic supplementary material, table S6). This increase in expression should reflect increases, rather than decreases, in cellular proliferation. However, later in development, downregulation of these genes occurred irrespective of the diapause status of the resulting pharate larva: most genes implicated in these categories had significantly lower expression at 6d than 3d pov in both pre-diapause and non-diapause embryos (D: 93%; ND: 83%; data not shown; e.g. electronic supplementary material, figure S7). The expression level of cell-cycle regulators such as cyclin-D can affect both cell size and cell number in growing tissue [44], and thus the higher expression of cell-cycle-related genes in pre-diapause 3d embryos may reflect increased size of diapause eggs, consistent with the observation that diapausing Ae. albopictus eggs are larger than non-diapause eggs [11] (but see §3g).
The category ‘DNA binding’ contained numerous genes related to ecdysone signalling (see the electronic supplementary material, tables S5 and S6). Endocrine signalling plays a central role governing developmental transitions in insects, as well as regulating transitions into and out of diapause, primarily via changing titres of ecdysone or juvenile hormone (reviewed in [6,45]). A post hoc enrichment analysis of 1 : 1 orthologues of ecdysone-signalling genes confirmed that this pathway was significantly enriched in DE genes at 3d pov (e-score = 0.011), which suggests an intriguing role for ecdysone signalling in promoting physiological changes at an intermediate stage of pre-diapause.
The enriched group ‘cytoskeletal part’ contained genes that were mainly structural in nature, or related to cell motility (see the electronic supplementary material, table S6). Expression differences of cell structure genes could result from observed expression differences in ecdysone-related transcription factors, as embryonic ecdysone is important for cuticle deposition and structural development [46].
(e). Enriched functional categories at 6-day post-oviposition
Reduced metabolism, especially reduced oxidative processes, and the redirection of energy production are among the defining characteristics of diapause [47,48]. Consistent with this pattern, the functional groups metabolism, energy production and transport were enriched in DE genes at 6d pov. Many transcripts in these groups had reduced expression in pre-diapause embryos (table 1; electronic supplementary material, table S6), including several NADH-dehydrogenases, as well as several members of the TCA cycle. This pattern implies that overall metabolic and energy production rates are lower in late pre-diapause embryos, similar to the lower metabolism consistently observed in other insects during diapause.
Further enriched categories at 6d pov included ‘extracellular region’, which contained a diverse assemblage of genes with varied expression patterns; and ‘structural constituent of chitin-based cuticle’ and ‘polysaccharide metabolism’, dominated by genes encoding insect cuticle proteins and chitin-binding domains (Interpro ID IPR002557; electronic supplementary material, table S6). The identified chitin-binding proteins could contribute to Ae. albopictus serosal cuticle formation in eggs, which are more desiccation-resistant under diapause conditions [10,49]. The majority of genes in these two categories are over-expressed in D embryos. Therefore, it is possible that the enrichment of these groups reflects greater provisioning of larval cuticular structures in pre-diapause embryos.
(f). Genes with potential for universal roles in diapause
Physiological and gene expression studies of insect diapause have uncovered processes commonly involved in the maintenance (reviewed in [6]) or termination [43,50] of diapause. These hallmarks include shifts in metabolic pathways [4,51–53], upregulation of stress response genes [45,54], changes in cell-cycle arrest [40,41,55] and changes in insulin signalling [52,56–58]. Despite the identification of some common physiological processes during diapause, it has been difficult to identify genes involved in regulating these processes across species, at least at a broad taxonomic scale [4]. Here, we identify two genes that may share common regulatory roles in diapause across insects and diapause stages, and thus may contribute to a ‘genetic toolkit’ of diapause.
We identified striking expression patterns in Pepck (phosphoenolpyruvate carboxykinase), a rate-limiting enzyme in gluconeogenesis. Pepck is upregulated in diapause-related gene expression scans in a diversity of organisms (pupae of Sarcophaga crassipalpis and Rhagoletis pomonella, larvae of Wyeomyia smithii, pre-diapause oocytes of Ae. albopictus, and in dauer larvae of Caenorhabditis elegans) [4,13,43,50,59]. In the present study, two of the three Pepck homologues (Ae. aegypti IDs AAEL000006, AAEL000025, AAEL000080) had significantly higher expression during pre-diapause at both 3d and 6d pov (see the electronic supplementary material, table S2). An increase in Pepck expression probably reflects an increase in gluconeogenesis and glucose production, and, in a diapause context, could reflect the transition between oxygen-rich and oxygen-poor environments [50,52]. Pepck may be a promising component of a universal diapause toolkit, and as such represents a candidate for attempts to disrupt the gluconeogenic component of the diapause response as a tool for pest management.
Proliferating cell nuclear antigen (PCNA) is central to cell cycle progression [60], and is of particular interest in diapause cell-cycle arrest owing to its relevance in other insect species. PCNA expression decreases during larval diapause in the fly Chymomyza costata [39], and increases following pupal diapause termination in flesh flies [40] and the apple maggot fly, R. pomonella [43]. In Ae. albopictus, PCNA has higher expression in 3d pre-diapause embryos relative to non-diapause embryos, whereas at 6d pov, PCNA expression is only modestly increased (see the electronic supplementary material, table S2). Expression decreases in both pre-diapause and non-diapause embryos over time (log2(3d/6d), D: 2.31; log2(3d/6d), ND: 1.58; p < 0.001). This gene warrants further attention as a potential regulator of diapause-associated changes in the cell cycle, both in preparation for and during termination of diapause.
(g). Developmental comparisons with Drosophila melanogaster
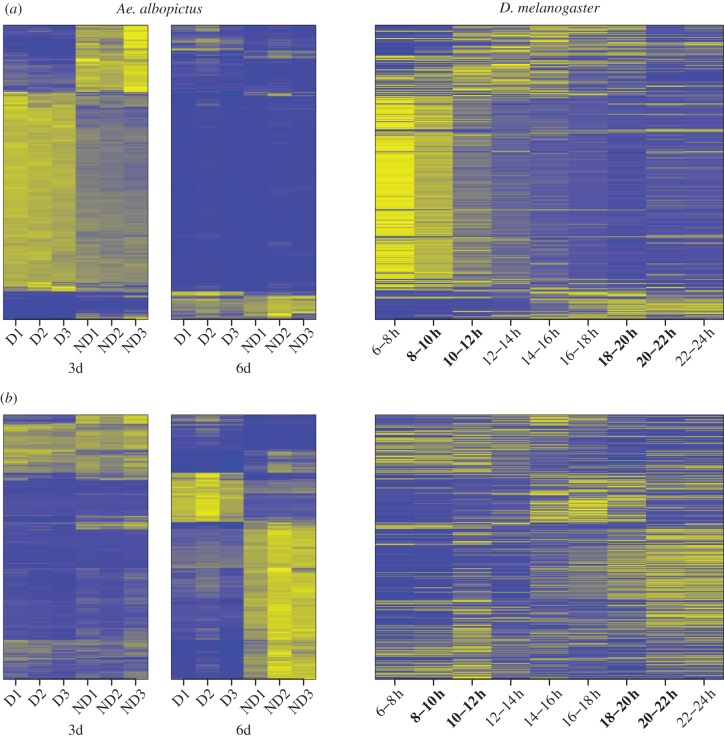
Qualitative comparisons of Ae. albopictus gene expression with a D. melanogaster time series show striking patterns at both 3d and 6d pov. At 3d pov, most genes in enriched categories that are upregulated under D conditions also have orthologues with higher expression early relative to late embryonic development in D. melanogaster (figure 3a). Conversely, many genes with lower D expression show lower expression early in D. melanogaster embryonic development. While the displayed genes were not DE at 6d pov, their orthologues also appear to show little change late in D. melanogaster embryonic development. Similar patterns are observed for genes DE at 6d pov in enriched categories, although these patterns are not apparent for as many genes (figure 3b).
Figure 3.
Heat maps of Ae. albopictus and D. melanogaster 1 : 1 orthologues for all DE genes in enriched categories at (a) 3d and (b) 6d pov. Expression values are depicted as z-standardized scores, where blue represents low expression and yellow are high. Expression values from all twelve Ae. albopictus libraries are shown, and a full time course of D. melanogaster embryonic development; times on the x-axes indicate the day or hour post-oviposition that RNA was collected. Time periods in D. melanogaster that correspond to Ae. albopictus 3d and 6d post-oviposition, based on correspondence of morphological characters, are emboldened.
One explanation for these patterns could be a delay of diapause expression relative to non-diapause expression in some physiological processes. We propose that such a delay could occur on the background of a fixed morphological ontogenetic programme under both conditions, because D and ND embryos were morphologically indistinguishable at both 3d pov and 6d pov. These data suggest that Ae. albopictus pre-diapause could in part involve prolonged or delayed gene expression in certain developmental processes to generate the diapause phenotype. Our interpretation assumes that the regular embryonic trajectory of these genes is conserved between D. melanogaster and Ae. albopictus. While this assumption may not be true for some genes, expression patterns of most genes during embryonic development are similar between D. melanogaster and a different mosquito species, Anopheles gambiae [61], implying that these gene expression trajectories are likely to be similar to those occurring in regular Ae. albopictus development. We could not quantitatively confirm our proposal of a developmental delay in pre-diapause gene expression owing to the lack of a developmental time series with more than two time points under each condition. However, this proposal could be tested in any organism with a well-defined pre-diapause phenotype, using comprehensive gene expression time series from both pre-diapause and non-diapause stages, to ask whether expression at time t in non-diapause was more consistent with expression at time t, or at time t−1, in pre-diapause.
4. Conclusions
Diapause is a widespread seasonal adaptation critical for survival of many insect species in seasonally changing environments. Like many polyphenic adaptations, the molecular regulation of this complex alternative developmental programme has proved elusive: while isolated pieces of information from a variety of insects exist, there are few comprehensive examples of gene expression changes during diapause development from a single organism. Here, we provide one of the first analyses of global gene expression change during insect pre-diapause development. Our results show strong shifts in gene expression patterns throughout diapause preparation in Ae. albopictus. Increasing divergence of global gene expression between D and ND embryos over time, combined with differences in the expression of ecdysone-signalling genes at 3d pov, may reflect cascading effects of early differences in regulatory pathways, leading to stronger gene expression differences at 6d pov. Pathways and key groups of genes shift in their importance to pre-diapause throughout embryonic development. Many of the identified changes show commonalities with processes important for diapause in other insect species. However, other components of the molecular regulation of diapause identified in other species do not appear to be regulated at the transcriptional level as part of the diapause preparation programme of Ae. albopictus, perhaps because these particular components are relevant further upstream of diapause preparation (i.e. insulin signalling), come into play after the developmental arrest has been initiated (i.e. stress response proteins) or may be less important for diapause at the embryonic stage.
Our findings integrate with previous studies in the burgeoning field of diapause molecular regulation by confirming the importance of several major genes and pathways for diapause across insects. This study supports a model of a genetic toolkit of diapause that is initiated and detectable in the diapause preparatory stage. Furthermore, our data propose that developmental delays in the expression of some genes, occurring on the background of a fixed morphological ontogeny, contribute to the physiological shifts during diapause preparation. This observation warrants further investigation in the diapause programme of Ae. albopictus and other insects. If developmental delays in gene expression are validated, this suggests that the mechanisms by which the alternative developmental programme of diapause is mediated are firmly integrated into the developmental trajectory of the pre-diapause insect. This emphasizes the importance of a developmental perspective not only for the identification of relevant pathways for diapause preparation, but also for polyphenic adaptations, in general.
Acknowledgements
We would like to thank the Armbruster, Elsik and Denlinger laboratories, and R. Scott Cornman for helpful comments and suggestions on this work. Comments from the Associate Editor and two anonymous referees greatly improved the quality of this manuscript. Christopher Childers was instrumental in setting up the website http://AlbopictusExpression.org. This work was supported by the National Institutes of Health (grant no. 5R21AI081041-02 to P.A.A., C.G.E. and D.L.D.) and Georgetown University.
References
- 1.Nijhout HF. 1999. Control mechanisms of polyphenic development in insects . Bioscience 49, 181–192 10.2307/1313508 (doi:10.2307/1313508) [DOI] [Google Scholar]
- 2.Tauber MJ, Tauber CA, Masaki S. 1986. Seasonal adaptations of insects. New York, NY: Oxford University Press [Google Scholar]
- 3.Carroll SB, Grenier JK, Weatherbee SD. 2001. From DNA to diversity: molecular genetics and the evolution of animal design. Malden, MA: Blackwell Science [Google Scholar]
- 4.Ragland GJ, Denlinger DL, Hahn DA. 2010. Mechanisms of suspended animation are revealed by transcript profiling of diapause in the flesh fly . Proc. Natl Acad. Sci. USA 107, 14 909–14 914 10.1073/pnas.1007075107 (doi:10.1073/pnas.1007075107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kostal V. 2006. Eco-physiological phases of insect diapause . J. Insect Physiol. 52, 113–127 10.1016/j.jinsphys.2005.09.008 (doi:10.1016/j.jinsphys.2005.09.008) [DOI] [PubMed] [Google Scholar]
- 6.Denlinger DL. 2002. Regulation of diapause . Annu. Rev. Entomol. 47, 93–122 10.1146/annurev.ento.47.091201.145137 (doi:10.1146/annurev.ento.47.091201.145137) [DOI] [PubMed] [Google Scholar]
- 7.Wang RL. 1966. Observations on the influence of photoperiod on egg diapause in Aedes albopictus Skuse . Acta Entomol. Sin. 15, 75–77 [Google Scholar]
- 8.Mori A, Oda T, Wada Y. 1981. Studies on the egg diapause and overwintering of Aedes albopictus in Nagasaki . Trop. Med. 23, 79–90 [Google Scholar]
- 9.Urbanski JM, Mogi M, O'Donnell DL, DeCotiis M, Toma T, Armbruster PA. 2012. Rapid adaptive evolution of photoperiodic response during invasion and range expansion across a climatic gradient . Am. Nat. 179, 490–500 10.1086/664709 (doi:10.1086/664709) [DOI] [PubMed] [Google Scholar]
- 10.Urbanski JM, Benoit JB, Michaud MR, Denlinger DL, Armbruster PA. 2010. The molecular physiology of increased egg desiccation resistance during diapause in the invasive mosquito, Aedes albopictus . Proc. R. Soc. B 277, 2683–2692 10.1098/rspb.2010.0362 (doi:10.1098/rspb.2010.0362) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reynolds JA, Poelchau MF, Rahman Z, Armbruster PA, Denlinger DL. 2012. Transcript profiling reveals mechanisms for lipid conservation during diapause in the mosquito, Aedes albopictus . J. Insect Physiol. 58, 966–973 10.1016/j.bbr.2011.03.031 (doi:10.1016/j.bbr.2011.03.031) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benedict MQ, Levine RS, Hawley WA, Lounibos LP. 2007. Spread of the tiger: global risk of invasion by the mosquito Aedes albopictus . Vector Borne Zoonotic Dis. 7, 76–85 10.1089/vbz.2006.0562 (doi:10.1089/vbz.2006.0562) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poelchau MF, Reynolds JA, Denlinger DL, Elsik CG, Armbruster PA. 2011. A de novo transcriptome of the Asian tiger mosquito, Aedes albopictus, to identify candidate transcripts for diapause preparation . BMC Genomics 12 10.1186/1471-2164-12-619 (doi:10.1186/1471-2164-12-619) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roy S, et al. 2010. Identification of functional elements and regulatory circuits by Drosophila modENCODE . Science 330, 1787–1797 10.1126/science.1198374 (doi:10.1126/science.1198374) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graveley BR, et al. 2011. The developmental transcriptome of Drosophila melanogaster . Nature 471, 473–479 10.1038/nature09715 (doi:10.1038/nature09715) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Armbruster PA, Conn JE. 2006. Geographic variation of larval growth in North American Aedes albopictus (Diptera: Culicidae) . Ann. Entomol. Soc. Am. 99, 1234–1243 10.1603/0013-8746(2006)99[1234:GVOLGI]2.0.CO;2 (doi:10.1603/0013-8746(2006)99[1234:GVOLGI]2.0.CO;2) [DOI] [Google Scholar]
- 17.Armbruster PA, Hutchinson RA. 2002. Pupal mass and wing length as indicators of fecundity in Aedes albopictus and Aedes geniculatus (Diptera: Culicidae) . J. Med. Entomol. 39, 699–704 10.1603/0022-2585-39.4.699 (doi:10.1603/0022-2585-39.4.699) [DOI] [PubMed] [Google Scholar]
- 18.Farnesi LC, Martins AJ, Valle D, Rezende GL. 2009. Embryonic development of Aedes aegypti (Diptera: Culicidae): influence of different constant temperatures . Mem. Inst. Oswaldo Cruz 104, 124–126 10.1590/S0074-02762009000100020 (doi:10.1590/S0074-02762009000100020) [DOI] [PubMed] [Google Scholar]
- 19.Rezende GL, Martins AJ, Gentile C, Farnesi LC, Pelajo-Machado M, Peixoto AA, Vale D. 2008. Embryonic desiccation resistance in Aedes aegypti: presumptive role of the chitinized serosal cuticle . BMC Dev. Biol. 8, 182. 10.1186/1471-213X-8-82 (doi:10.1186/1471-213X-8-82) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vital W, Rezende GL, Abreu L, Moraes J, Lemos FJA, da SV, Logullo C. 2010. Germ band retraction as a landmark in glucose metabolism during Aedes aegypti embryogenesis . BMC Dev. Biol. 10, 25. 10.1186/1471-213X-10-25 (doi:10.1186/1471-213X-10-25) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trpis M. 1970. A new bleaching and decalcifying method for general use in zoology . Can. J. Zool. 48, 892–893 10.1139/z70-158 (doi:10.1139/z70-158) [DOI] [Google Scholar]
- 22.Hawley WA, Reiter P, Copeland RS, Pumpuni CB, Craig GB. 1987. Aedes albopictus in North America—probable introduction in used tires from northern Asia . Science 236, 1114–1116 10.1126/science.3576225 (doi:10.1126/science.3576225) [DOI] [PubMed] [Google Scholar]
- 23.Li B, Dewey C. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome . BMC Bioinformatics 12, 323. 10.1186/1471-2105-12-323 (doi:10.1186/1471-2105-12-323) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a bioconductor package for differential expression analysis of digital gene expression data . Bioinformatics 26, 139–140 10.1093/bioinformatics/btp616 (doi:10.1093/bioinformatics/btp616) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smyth GK. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments . Stat. Appl. Genet. Mol. Biol. 3, 3. 10.2202/1544-6115.1027 (doi:10.2202/1544-6115.1027) [DOI] [PubMed] [Google Scholar]
- 26.Smyth GK. 2005. Limma: linear models for microarray data. In Bioinformatics and computational biology solutions using R and Bioconductor (eds Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W.), pp. 397–420 New York, NY: Springer [Google Scholar]
- 27.Huang DW, Sherman BT, Lempicki RA. 2009. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists . Nucleic Acids Res. 37, 1–13 10.1093/nar/gkn923 (doi:10.1093/nar/gkn923) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang DW, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources . Nat. Protoc. 4, 44–57 10.1038/nprot.2008.211 (doi:10.1038/nprot.2008.211) [DOI] [PubMed] [Google Scholar]
- 29.Kanehisa M, Goto S. 2000. KEGG: Kyoto encyclopedia of genes and genomes . Nucleic Acids Res. 28, 27–30 10.1093/nar/28.1.27 (doi:10.1093/nar/28.1.27) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. 2012. KEGG for integration and interpretation of large-scale molecular data sets . Nucleic Acids Res. 40, D109–D114 10.1093/nar/gkr988 (doi:10.1093/nar/gkr988) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ashburner M, et al. 2000. Gene Ontology: tool for the unification of biology . Nat. Genet. 25, 25–29 10.1038/75556 (doi:10.1038/75556) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Young M, Wakefield M, Smyth G, Oshlack A. 2010. Gene ontology analysis for RNA-seq: accounting for selection bias . Genome Biol. 11, R14. 10.1186/gb-2010-11-2-r14 (doi:10.1186/gb-2010-11-2-r14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haider S, Ballester B, Smedley D, Zhang JJ, Rice P, Kasprzyk A. 2009. BioMart Central Portal—unified access to biological data . Nucleic Acids Res. 37, W23–W27 10.1093/nar/gkp265 (doi:10.1093/nar/gkp265) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Waterhouse RM, Zdobnov EM, Tegenfeldt F, Li J, Kriventseva EV. 2011. OrthoDB: the hierarchical catalog of eukaryotic orthologs in 2011 . Nucleic Acids Res. 39, D283–D288 10.1093/nar/gkq930 (doi:10.1093/nar/gkq930) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Castillo-Davis C, Hartl DL. 2003. GeneMerge—post-genomic analysis, data mining, and hypothesis testing . Bioinformatics 19, 891–892 10.1093/bioinformatics/btg114 (doi:10.1093/bioinformatics/btg114) [DOI] [PubMed] [Google Scholar]
- 36.Wu Q, Brown MR. 2006. Signaling and function of insulin-like peptides in insects . Annu. Rev. Entomol. 51, 1–24 10.1146/annurev.ento.51.110104.151011 (doi:10.1146/annurev.ento.51.110104.151011) [DOI] [PubMed] [Google Scholar]
- 37.Brody T. 1999. The interactive fly: gene networks, development and the Internet . Trends Genet. 15, 333–334 10.1016/S0168-9525(99)01775-8 (doi:10.1016/S0168-9525(99)01775-8) [DOI] [PubMed] [Google Scholar]
- 38.Arbeitman MN, et al. 2002. Gene expression during the life cycle of Drosophila melanogaster . Science 297, 2270–2275 10.1126/science.1072152 (doi:10.1126/science.1072152) [DOI] [PubMed] [Google Scholar]
- 39.Kostal V, Simunkova P, Kobelkova A, Shimada K. 2009. Cell cycle arrest as a hallmark of insect diapause: changes in gene transcription during diapause induction in the drosophilid fly, Chymomyza costata . Insect Biochem. Mol. Biol. 39, 875–883 10.1016/j.ibmb.2009.10.004 (doi:10.1016/j.ibmb.2009.10.004) [DOI] [PubMed] [Google Scholar]
- 40.Tammariello SP, Denlinger DL. 1998. G0/G1 cell cycle arrest in the brain of Sarcophaga crassipalpis during pupal diapause and the expression pattern of the cell cycle regulator, proliferating cell nuclear antigen . Insect Biochem. Mol. Biol. 28, 83–89 10.1016/S0965-1748(97)00082-9 (doi:10.1016/S0965-1748(97)00082-9) [DOI] [PubMed] [Google Scholar]
- 41.Reynolds JA, Hand SC. 2009. Embryonic diapause highlighted by differential expression of mRNAs for ecdysteroidogenesis, transcription and lipid sparing in the cricket Allonemobius socius . J. Exp. Biol. 212, 2074–2083 10.1242/jeb.027367 (doi:10.1242/jeb.027367) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bao B, Xu WH. 2011. Identification of gene expression changes associated with the initiation of diapause in the brain of the cotton bollworm, Helicoverpa armigera . BMC Genomics 12, 224. 10.1186/1471-2164-12-224 (doi:10.1186/1471-2164-12-224) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ragland GJ, Egan SP, Feder JL, Berlocher SH, Hahn DA. 2011. Developmental trajectories of gene expression reveal candidates for diapause termination: a key life-history transition in the apple maggot fly Rhagoletis pomonella . J. Exp. Biol. 214, 3948–3959 10.1242/jeb.061085 (doi:10.1242/jeb.061085) [DOI] [PubMed] [Google Scholar]
- 44.Datar SA, Jacobs HW, de la Cruz AF, Lehner CF, Edgar BA. 2000. The Drosophila Cyclin D–Cdk4 complex promotes cellular growth . EMBO J. 19, 4543–4554 10.1093/emboj/19.17.4543 (doi:10.1093/emboj/19.17.4543) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Denlinger DL, Yocum GD, Rinehart JL. 2005. Hormonal control of diapause. In Comprehensive molecular insect science, vol. 3 (eds Gilbert L, Iatrou K, Gill S.), pp. 615–650 Amsterdam, The Netherlands: Elsevier Press [Google Scholar]
- 46.Kozlova T, Thummel CS. 2003. Essential roles for ecdysone signaling during Drosophila mid-embryonic development . Science 301, 1911–1914 10.1126/science.1087419 (doi:10.1126/science.1087419) [DOI] [PubMed] [Google Scholar]
- 47.MacRae TH. 2010. Gene expression, metabolic regulation and stress tolerance during diapause . Cell. Mol. Life Sci. 67, 2405–2424 10.1007/s00018-010-0311-0 (doi:10.1007/s00018-010-0311-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hand SC, Menze MA, Borcar A, Patil Y, Covi JA, Reynolds JA, Toner M. 2011. Metabolic restructuring during energy-limited states: insights from Artemia franciscana and other animals . J. Insect Physiol. 57, 584–594 10.1016/j.jinsphys.2011.02.010 (doi:10.1016/j.jinsphys.2011.02.010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sota T, Mogi M. 1992. Survival-time and resistance to desiccation of diapause and non-diapause eggs of temperate Aedes (Stegomyia) mosquitoes . Entomol. Exp. Appl. 63, 155–161 10.1111/j.1570-7458.1992.tb01570.x (doi:10.1111/j.1570-7458.1992.tb01570.x) [DOI] [Google Scholar]
- 50.Emerson KJ, Bradshaw WE, Holzapfel CM. 2010. Microarrays reveal early transcriptional events during the termination of larval diapause in natural populations of the mosquito, Wyeomyia smithii . PLoS ONE 5, e9574. 10.1371/journal.pone.0009574 (doi:10.1371/journal.pone.0009574) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kukal O, Denlinger DL, Lee RE. 1991. Developmental and metabolic changes induced by anoxia in diapausing and nondiapausing flesh fly pupae . J. Comp. Phys. B Biochem. Syst. Environ. Physiol. 160, 683–689 10.1007/BF00571268 (doi:10.1007/BF00571268) [DOI] [Google Scholar]
- 52.Hahn DA, Denlinger DL. 2011. Energetics of insect diapause . Annu. Rev. Entomol. 56, 103–121 10.1146/annurev-ento-112408-085436 (doi:10.1146/annurev-ento-112408-085436) [DOI] [PubMed] [Google Scholar]
- 53.Michaud MR, Denlinger DL. 2007. Shifts in the carbohydrate, polyol, and amino acid pools during rapid cold-hardening and diapause-associated cold-hardening in flesh flies (Sarcophaga crassipalpis): a metabolomic comparison . J. Comp. Physiol. B Biochem. Syst. Environ. Physiol. 177, 753–763 10.1007/s00360-007-0172-5 (doi:10.1007/s00360-007-0172-5) [DOI] [PubMed] [Google Scholar]
- 54.Rinehart JP, Li A, Yocum GD, Robich RM, Hayward S, Denlinger DL. 2007. Up-regulation of heat shock proteins is essential for cold survival during insect diapause . Proc. Natl Acad. Sci. USA 104, 11 130–11 137 10.1073/pnas.0703538104 (doi:10.1073/pnas.0703538104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tammariello SP. 2001. Regulation of the cell cycle during diapause. In Insect timing: circadian rhythmicity to seasonality (eds Denlinger DL, Giebultowicz J, Saunders DS.), pp. 173–183 Amsterdam, The Netherlands: Elsevier Science, B.V [Google Scholar]
- 56.Tatar M, Kopelman A, Epstein D, Tu MP, Yin CM, Garofalo RS. 2001. A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function . Science 292, 107–110 10.1126/science.1057987 (doi:10.1126/science.1057987) [DOI] [PubMed] [Google Scholar]
- 57.Williams KD, Busto M, Suster ML, So A, Ben-Shahar Y, Leevers SJ, Sokolowski MB. 2006. Natural variation in Drosophila melanogaster diapause due to the insulin-regulated PI3-kinase . Proc. Natl Acad. Sci. USA 103, 15 911–15 915 10.1073/pnas.0604592103 (doi:10.1073/pnas.0604592103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sim C, Denlinger DL. 2008. Insulin signaling and FOXO regulate the overwintering diapause of the mosquito Culex pipiens . Proc. Natl Acad. Sci. USA 105, 6777–6781 10.1073/pnas.0802067105 (doi:10.1073/pnas.0802067105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McElwee JJ, Schuster E, Blanc E, Thornton J, Gems D. 2006. Diapause-associated metabolic traits reiterated in long-lived daf-2 mutants in the nematode Caenorhabditis elegans . Mech. Ageing Dev. 127, 458–472 10.1016/j.mad.2006.01.006 (doi:10.1016/j.mad.2006.01.006) [DOI] [PubMed] [Google Scholar]
- 60.Maga G, Hubscher U. 2003. Proliferating cell nuclear antigen (PCNA): a dancer with many partners . J. Cell. Sci. 116, 3051–3060 10.1242/jcs.00653 (doi:10.1242/jcs.00653) [DOI] [PubMed] [Google Scholar]
- 61.Papatsenko D, Levine M, Goltsev Y. 2011. Clusters of temporal discordances reveal distinct embryonic patterning mechanisms in Drosophila and Anopheles . PLoS Biol. 9, e1000584. 10.1371/journal.pbio.1000584 (doi:10.1371/journal.pbio.1000584) [DOI] [PMC free article] [PubMed] [Google Scholar]