Abstract
MicroRNAs (miRNAs) are known to regulate most biological processes and have been found dysregulated in a variety of diseases, including multiple sclerosis (MS). In this study, we characterized miRNAs that associate with susceptibility to develop experimental autoimmune encephalomyelitis (EAE) in rats, a well-established animal model of MS. Using Illumina next-generation sequencing, we detected 544 miRNAs in the lymph nodes of EAE-susceptible Dark Agouti and EAE-resistant Piebald Virol Glaxo rats during immune activation. Forty-three miRNAs were found differentially expressed between the two strains, with 81% (35 out of 43) showing higher expression in the susceptible strain. Only 33% of tested miRNAs displayed differential expression in naive lymph nodes, suggesting that a majority of regulated miRNAs are EAE dependent. Further investigation of a selected six miRNAs indicates differences in cellular source and kinetics of expression. Several of the miRNAs, including miR-146a, miR-21, miR-181a, miR-223, and let-7, have previously been implicated in immune system regulation. Moreover, 77% (33 out of 43) of the miRNAs were associated with MS and other autoimmune diseases. Target genes likely regulated by the miRNAs were identified using computational predictions combined with whole-genome expression data. Differentially expressed miRNAs and their targets involve functions important for MS and EAE, such as immune cell migration through targeting genes like Cxcr3 and cellular maintenance and signaling by regulation of Prkcd and Stat1. In addition, we demonstrated that these three genes are direct targets of miR-181a. Our study highlights the impact of multiple miRNAs, displaying diverse kinetics and cellular sources, on development of pathogenic autoimmune inflammation.
Introduction
Twenty years after the discovery of the first microRNA (miRNA) (1), they are now acknowledged as important regulators of most biological functions. Mature miRNAs are evolutionary conserved, small, ∼22 nt, noncoding RNAs (2). They act by binding through partial complementarity of the seed (nt 2–8) to sequences in the 3′ untranslated regions (UTRs) of mRNAs (3). This binding results in negative regulation of gene transcription in a posttranscriptional manner, either by RNA degradation or translational inhibition (4). It is estimated that a single miRNA has the potential to regulate hundreds of target genes, and therefore, >90% of all human genes have the potential to be under regulation by miRNAs (5).
miRNAs are important regulators of immune cell development and immune response (6), and they have been found dysregulated in a variety of inflammatory diseases such as rheumatoid arthritis (RA) (7, 8), psoriasis (9), systemic lupus erythematosus (10, 11), and multiple sclerosis (MS) (12). MS is a chronic inflammatory disease characterized by infiltration of autoreactive immune cells into the CNS, leading to demyelination and axonal loss. Both genetic (13) and environmental factors (14) contribute to MS susceptibility and progression. Nevertheless, despite current advancement in MS research, many questions on disease etiology remain unexplained. In order to find relevant and stable biomarkers in MS, miRNAs have been profiled in patient PBMCs (15–17), whole blood (18, 19), plasma (20), lymphocytes (21, 22), regulatory T cells (23), cerebrospinal fluid (24), as well as in active and inactive MS lesions (25). Together, these studies demonstrate involvement of miRNAs in MS. They also illustrate the complexity of miRNA expression, in which tissue and time point of sampling influence the miRNA expression profile.
Experimental autoimmune encephalomyelitis (EAE) is a well-established and reproducible experimental model extensively used to study MS-like neuroinflammation. Myelin oligodendrocyte glycoprotein (MOG)–induced EAE in Dark Agouti (DA) rats is characterized by inflammation and demyelination in the CNS, resulting in a disease progression mimicking the relapsing-remitting form of MS (26). Importance of miRNAs in EAE has emerged with the discovery of Th17 regulation by miR-326 and macrophage/microglia regulation by miR-124 (27, 28) in murine EAE. In addition, EAE in mice can be ameliorated through silencing of miR-155 (29). To date, no comprehensive miRNA studies have been reported in rodent EAE, and no studies have been reported in rat EAE.
In this study, we aimed to establish an miRNA profile that distinguishes pathogenic immune activation, leading to EAE development, from immune activation that resolves and does not cause disease. We studied two inbred rat strains, the EAE-susceptible DA and the EAE-resistant Piebald Virol Glaxo (PVG) strain, with respect to their miRNA expression profile during induction of EAE. Using Illumina next-generation sequencing (NGS) (Illumina, San Diego, CA), we discovered 43 mature miRNAs (from 41 precursors) to be differentially expressed between the two strains. By integrating target gene predictions with gene expression data, we identified an enrichment of genes involved in homeostasis and immune-response mechanisms that are likely regulated by miRNA. Furthermore, consistent with our prediction analysis, we demonstrated that three predicted targets, Cxcr3, Prkcd, and Stat1, are directly regulated by miR-181a, a differentially expressed and lymphocyte-specific miRNA.
Materials and Methods
Animals and EAE induction
Inbred DA rats were originally obtained from the Zentralinstitut für Versuchstierzucht (Hanover, Germany) and MHC-identical PVG rats from Harlan UK Limited (Blackthorn, U.K.). Animals were bred in the animal facility at Karolinska University Hospital (Stockholm, Sweden) in a pathogen-free and climate-controlled environment in polystyrene cages containing aspen wood shavings with free access to standard rodent chow and water with regulated 12-h light/dark cycles. MOG, aa 1–125 from the N terminus, was expressed in Escherichia coli and purified to homogeneity by chelate chromatography (30). The purified protein, dissolved in 6 M urea, was dialyzed against PBS to obtain a physiological preparation that was stored at −70°C. Age-matched female rats were anesthetized with isoflurane (Forene; Abbott Laboratories, Chicago, IL) and injected s.c. in the tail base with a 200 μl inoculum containing 25 μg recombinant MOG in PBS, emulsified 1:1 with IFA (Sigma-Aldrich, St. Louis, MO). All experiments in this study were performed in accordance with the ethical permit approved by Stockholms norra djurförsöksetiska nämnd (North Stockholm animal ethics committee).
Sample collection for NGS and RNA isolation
Animals were sacrificed using CO2 7 d post–EAE induction, before signs of clinical disease. Draining inguinal lymph nodes were collected and immediately frozen in liquid nitrogen. Tissue was homogenized using Matrix D tubes (MP Biomedicals, Irvine, CA) on a FastPrep homogenizer (MP Biomedicals), and total RNA was isolated using standard TRIzol protocol (Invitrogen). RNA concentration and purity were determined by measurement of A260/A280 ratios with a NanoDrop ND-1000 Spectrophotometer (NanoDrop Technologies, Wilmington, DE). RNA samples were immediately frozen and stored at −70°C.
NGS of miRNA and data analysis
Small RNA library preparation, including quality control and NGS, was performed by FASTERIS SA (Plan-les-Ouates, Switzerland). The library preparation was performed on 10 μg total RNA per sample using the alternative v1.5 protocol from Illumina and standard adapters. The samples were indexed, using multiplex tags, to facilitate sequencing of all samples on one flow cell channel. Library preparation quality control showed <5% adapter–adapter contamination and 85% of the sequences being miRNAs. The samples were subsequently sequenced on the Illumina HiSeq2000 instrument (Illumina) using 1 × 50 + 7 (index) sequencing cycles. A total of 93.6% of bases had a base quality ≥Q30, and the real-time error rate of spiked PhiX was 0.25%.
After sequencing, the data were obtained in Illumina FASTQ format (Illumina). Data filtering and sequence annotation can be visualized in Fig. 1B. In short, the sequence reads were sorted on sample type based on matches to the multiplex tags (six bases), and the 3′ adapter was trimmed from the read. In the cases in which no perfect adapter match was found, the last base of the adapter sequence was removed in successive steps, and the sequence was searched at the end of the reads. The minimum adapter size of 8 bases permit identifying reads of up to 42 bases.
FIGURE 1.
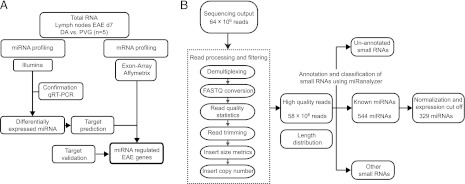
Study design and data filtering. (A) Schematic illustration of study design. DA (n = 5) and PVG (n = 5) rats were immunized with MOG to induce EAE. Seven days after immunization, lymph nodes were dissected and small RNAs subjected to NGS (Illumina). Sequencing results were confirmed using TaqMan miRNA assays in the same as well as an independent lymph node material. Targets of differentially expressed miRNAs were predicted using TargetScan and miRanda algorithms and combined with expression data generated using Affymetrix exon arrays (Affymetrix). Direct interaction of miRNA and target 3′ UTR was validated using a luciferase reporter assay. (B) Schematic illustration of the data filtering and analysis following NGS. NGS generated ∼64 million reads, which were processed and filtered to remove multiplexing tags and sequence adapters. The remaining 58 million high-quality reads were analyzed using miRanalyzer, a web-based tool that can identify miRNAs and other small RNAs and predict novel miRNAs. After normalization and expression cutoff, 329 unique miRNAs were quantified in the DA and PVG samples.
For mapping of sequencing output, we used miRanalyzer version 0.2 (31, 32), a Web server tool detecting all known miRNA sequences annotated in the miRBase registry (release 16.0) (33). miRanalyzer also aligns reads to Rfam and RefSeq libraries and predicts novel miRNAs. Unique reads and their corresponding copy number were uploaded into miRanalyzer using default settings (with the exception “of number of mismatches [known miRNA],” where 0 mismatches were allowed). miRNA expression, measured as reads per million (RPM), was normalized by dividing the miRNA read counts to the total number of reads aligning to RNA sequence libraries in the specific samples. The RPM cutoff for expression analysis was set at > RPM with average expression larger than the SD. Student t test was performed using GraphPad Prism 5 (GraphPad Software, San Diego, CA). All miRNAs with RPM >1000 and p < 0.05 as well as miRNAs with RPM <1000 with p < 0.05 and fold change >1.5 were considered differentially expressed. miRNAs with low expression (<1000 RPM) and low fold change (<1.5) were not considered for further analysis.
EAE kinetics and FACS
Draining inguinal lymph nodes were collected from naive state and days 3, 7, and 25 postimmunization (p.i.). Organs were placed in DMEM (Life Technologies-BRL, Grand Island, NY), enriched with 10% FCS, 1% L-glutamine, 1% penicillin-streptomycin, and 1% pyruvic acid (all from Life Technologies, Paisley, Scotland) before being mechanically separated by passing through a mesh screen. Cells were stained for 20 min at 4°C with CD3, CD4, CD8a, and CD45RA Abs (all from BD Biosciences, San Jose, CA) and sorted with a MoFlo cell sorter (Beckman Coulter). After sorting, total RNA was isolated from each cell subset using standard TRIzol protocol (Invitrogen, Karlsruhe, Germany).
Quantitative real-time PCR
Expression levels of selected miRNAs were confirmed by quantitative real-time PCR (qRT-PCR) using the TaqMan MicroRNA Assay Kit for specific mature miRNAs (Applied Biosystems, Barcelona, Spain) following the manufacturer's protocol. qRT-PCR of target mRNAs was performed on cDNA-converted RNA (iScript; Bio-Rad, Hercules, CA) using the following primers: rCxcr3_F: 5′-TGT ACC TTG AGG TCA GTG AAC G-3′ and rCxcr3_R: 5′-GGG AGT CAG AGA AGT CGC TTT-3′; rPrkcd_F: 5′-CTA TGA AGG CCG TGT CAT CC-3′ and rPrkcd_R: 5′-CAG GTC CAG CCA GAA CTC AG-3′; rStat1_F: 5′-GCC CAG CGA TTT AAT CAG GCC C-3′ and rStat1_R: ATG CTC TAT GCA CAT GAC TTG GTC C-3′; rHPRT_F: 5′-CTC ATG GAC TGA TTA TGG ACA-3′ and rHPRT_R: 5′-GCA GGT CAG CAA AGA ACT TAT-3′; and rβ-ACTIN_F: 5′-CGT GAA AAG ATG ACC CAG ATC A-3′ and rβ-ACTIN_R: 5′-TCC CAT TCT CAG CCT TGA CTG-3′. miRNA and mRNA expression levels were quantified by the Bio-Rad CFX384 Real-Time Detection System and analyzed using CFX manager software v2.0 (Bio-Rad, Hercules, CA). Relative quantification of miRNA was quantified using the 2−ΔΔCT method (34) and normalized against RNU6B for each sample. Relative quantification of mRNA was performed using the standard curve method and normalized against the geometrical mean of β-actin and Hprt. The Student t test was performed using GraphPad Prism 5 (GraphPad, San Diego, CA).
Target prediction and integration with mRNA expression
Targets of differentially expressed miRNAs were predicted using two different target prediction tools, TargetScan and miRanda. The predictions were performed using relaxed parameters; for TargetScan, both conserved and nonconserved targets were considered, and for miRanda, all mirsvr scores were considered regardless of conservational status. Removing the conservation filter in the individual target prediction tools is acceptable in this setting, as combining the two tools will be a conserved approach without introducing bias through individual tools' scoring systems. Targets predicted by both prediction tools were considered for further analysis. Genome-wide expression analysis previously performed in the same strains and the same tissue is reported in detail by Gillett et al. (35). In short, gene expression data were generated for samples from lymph node cells of DA and PVG inbred rat strains 7 d after EAE induction using Affymetrix Gene Chip Rat Exon 1.0 ST Arrays (Affymetrix, Santa Clara, CA). Differentially expressed transcripts (p < 0.05), predicted as miRNA targets by both TargetScan and miRanda that displayed an inverse expression to the miRNA expression were considered as potential miRNA targets and selected for pathway analysis.
Pathway and functional annotation analysis
The list of miRNA targets was analyzed with Ingenuity pathway analysis (IPA; Ingenuity Systems, http://www.ingenuity.com/) using default settings (with the exception of selection of tissues and cell lines during creation of core analysis in which “nervous system” was removed). The data were analyzed focusing on Biological Functions, only considering “Molecular and Cellular Functions.” The Functional Analysis identified the biological functions that were most significant in the data set. Right-tailed Fisher’s exact test was used to calculate p values. Top 10 functions also passed significance using Benjamini-Hochberg correction for multiple testing.
miRNA inhibition and overexpression
Owing to high miRNA/3′ UTR conservation between mammals, we used HEK293 cells to avoid technical difficulties and limitations of transfecting primary rat lymphocytes. The HEK293T cells were transfected with 100 nM miR-181a inhibitor or inhibitor control (Exiqon, Vedbaek, Denmark) or miR-181a mimic or mimic control (Ambion, Carlsbad, CA) using Lipofectamine 2000 (Invitrogen, Karlsruhe, Germany). Six to 48 h after transfection, cells were harvested and subjected to reverse transcription and real-time PCR as described above using the following primers hCXCR3_F: 5′-TCC TTG AGG GGT CCC TGG-3′ and hCXCR3_R: 5′-TCC TAT AAC TGT CCC CGC CA-3′; hPRKCD_F: 5′-GGA GCC AGG ACT AAG GAC AAG-3′; and hPRKCD_R: 5′-TGT TTT CCC ACG CTC TGT GC-3′; hSTAT1_F: AAA GGA AGC ACC AGA GCC AAT-3′ and hSTAT1_R: 5′-TCC GAG ACA CCT CGT CAA AC-3′; and hGAPDH_F: 5′-AGG GCT GCT TTT AAC TCT GGT AAA-3′ and hGAPDH_R: 5′-CAT ATT GGA ACA TGT AAA CCA TGT AGT TG-3′. GAPDH expression was used as endogenous control. The Student t test was performed using GraphPad Prism 5 (GraphPad).
Luciferase reporter system
HEK293T cells were cultured in DMEM supplemented with 10% FBS, 1% penicillin/streptomycin, 2 mM L-glutamine, and 1 mM sodium pyruvate at 37°C in 5% CO2. The full 3′ UTR sequence of Cxcr3 and Prkcd as well as 500 bp of the sequence surrounding the miR-181a binding site in the 3′ UTR of Stat1 were PCR amplified and cloned into the pmirGLO Dual-Luciferase miRNA Target Expression Vector (Promega, San Luis Obispo, CA) using SacI and XbaI restriction enzyme sites. Positive clones and insert orientation were verified by PCR and sequencing. HEK293T cells at a density of 2 × 105 cells/well in 24-well plates were cotransfected with the reporter plasmid (200 ng) and miR-181a mimic or mimic control (50 nM) using Lipofectamine 2000 (Invitrogen). Reporter assays were performed 24 h after transfection using a dual luciferase reporter assay system (Promega) according to the manufacturer’s instructions. Luciferase activities were measured on a GloMAX-Multi Detection System plate reader (Promega). The results are expressed as relative luciferase activity (Firefly luciferase/Renilla luciferase).
Results
Study design
We investigated miRNAs in the draining lymph nodes 7 d after immunization with MOG. At this stage, susceptible DA and resistant PVG rats mount a strong immune response, which subsequently leads to infiltration of cells into CNS and signs of clinical disease only in DA rats (36). To identify and quantify miRNAs, we used NGS of small RNAs (Illumina HiSeq2000; Illumina). Differentially expressed miRNAs were validated by qRT-PCR using specific TaqMan miRNA assays. Targets of differentially expressed miRNA were predicted using a combination of TargetScan and miRanda algorithms. To identify targets that are more likely to be regulated by miRNA during EAE, we integrated target prediction with genome-wide expression analysis (35). Moreover, by using a luciferase reporter system, we determine direct binding to selected target genes. The experimental design is summarized in Fig. 1A.
NGS of lymph node miRNAs in EAE-susceptible and -resistant rat strains
NGS enables sequencing of the entire small RNA transcriptome within each sample, providing the complete set of miRNAs transcribed at a given time. Sequencing generated 64 × 106 reads, ranging from 2–9 × 106 reads per sample. After read processing and filtering, a total of 58 × 106 high-quality reads were selected for further analysis (Fig. 1B). Length distribution of the reads, as expected, showed a main peak centered at 22 bases. Using the miRanalyzer web tool, we found 544 specific read sequences to be annotated as mature miRNAs in the miRBase registry (release 16) (Table I). After normalization and application of expression cutoff, we could reliably quantify 329 miRNAs (Supplemental Table I).
Table I. miRanalyzer output.
| Read Counts/Sample | DA 1 | DA 2 | DA 3 | DA 4 | DA 5 | PVG 1 | PVG 2 | PVG 3 | PVG 4 | PVG 5 |
|---|---|---|---|---|---|---|---|---|---|---|
| miRNAs (unique miRNAs) | 3,014,491 (427) | 1,455,446 (391) | 4,492,515 (439) | 3,406,906 (447) | 4,997,298 (468) | 6,131,684 (451) | 3,293,980 (438) | 5,060,088 (457) | 4,373,368 (467) | 4,491,832 (419) |
| mRNA/Rfam | 1,392 | 884 | 2,250 | 3,317 | 3,368 | 3,650 | 1,970 | 3,456 | 2,694 | 2,128 |
| Mapping to the genome | 488,156 | 236,915 | 712,816 | 859,31 | 1,031,468 | 1,754,037 | 852,844 | 1,420,302 | 1,471,645 | 1,012,062 |
| Predicted novel miRNAs | 24,762 | 11,854 | 35,711 | 38,196 | 42,513 | 24,4519 | 34,864 | 69,334 | 52,566 | 38,889 |
| Not mapping | 402,763 | 188,863 | 602,435 | 749,711 | 830,668 | 513,726 | 770,571 | 1,245,324 | 1,503,135 | 901,413 |
| Total | 3,906,802 | 1,882,108 | 5,810,016 | 5,019,465 | 6,862,802 | 8,403,097 | 4,919,365 | 7,729,170 | 7,350,842 | 6,407,435 |
Mapping to the genome, number of reads mapping to other genomic regions; miRNAs, number of reads mapping to miRBase; mRNA/Rfam, number of reads mapping to other RNA libraries; Not mapping, reads not mapping to known RNA sequences; Predicted novel miRNAs, number of reads predicted to be previously unknown miRNA; unique miRNAs, number of unique mature miRNAs detected in the sample.
miRNAs are differentially expressed between DA and PVG
We next determined the miRNAs that differed between DA and PVG at induction of EAE. In total, 64 miRNAs displayed differential expression between the two strains (at nominal p < 0.05 and without fold change cutoff). Based on: 1) the ability to confirm differentially expressed miRNAs with an independent method, 2) the fold change differences, and 3) the fact that lymph node tissue has a heterogeneous cell composition, we chose to subdivide differentially expressed miRNAs into high- and low-abundance miRNAs (Table II). We decided to also report low-abundant miRNAs, as they may only be expressed and play a role in cell types that are underrepresented within the lymph node, but also due to bias of different methods in estimating abundance (37). For these miRNAs, we applied a fold-change requirement, as small fluctuations in expression levels will greatly influence the fold change. The targets and pathways regulated by these low-abundant miRNAs will not be addressed in this article.
Table II.
Differentially expressed miRNAs between DA and PVG
| miRNAs with Higher Expression in the Susceptible Strain (DA) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| High-Abundance miRNAs |
Low-Abundance miRNAs |
||||||||
| Average RPM |
Average RPM |
||||||||
| miRNA | DA | PVG | Fold Change | p Value | miRNA | DA | PVG | Fold Change | p Value |
| rno-miR-146b | 1990.3 | 1055.0 | 1.9 | 4.2E-04 | rno-miR-92b | 14.2 | 7.2 | 1.96 | 8.8E-05 |
| (rno-miR-146a | 9889.5 | 8010.5 | 1.2 | 9.1E-02) | rno-miR-210 | 61.4 | 28.5 | 2.16 | 2.2E-04 |
| rno-miR-128 | 2111.3 | 1671.2 | 1.3 | 1.0E-03 | rno-miR-466b | 9.4 | 5.0 | 1.87 | 3.6E-04 |
| rno-miR-7a | 12386.6 | 8929.5 | 1.4 | 1.2E-03 | rno-miR-132* | 23.8 | 13.7 | 1.73 | 5.8E-04 |
| (rno-miR-7b | 1.4 | 0.9 | 1.5 | 9.6E-02) | rno-miR-1 | 21.1 | 9.4 | 2.23 | 2.5E-03 |
| rno-miR-125b-5p | 5192.6 | 4034.3 | 1.3 | 2.3E-03 | rno-miR-196c | 4.4 | 2.4 | 1.82 | 1.0E-02 |
| rno-miR-872 | 1815.1 | 1532.7 | 1.2 | 2.3E-03 | rno-miR-434 | 5.4 | 2.5 | 2.14 | 1.4E-02 |
| rno-miR-143 | 34563.4 | 27667.8 | 1.2 | 2.8E-03 | rno-miR-31 | 27.9 | 14.5 | 1.92 | 1.5E-02 |
| rno-miR-92a | 3424.3 | 3061.6 | 1.1 | 5.8E-03 | rno-miR-541 | 10.4 | 5.0 | 2.08 | 1.7E-02 |
| rno-miR-25 | 2852.1 | 2504.3 | 1.1 | 9.9E-03 | rno-miR-3545-3p | 6.8 | 2.5 | 2.73 | 1.9E-02 |
| (rno-miR-92b | 14.2 | 7.2 | 2.0 | 8.8E-05) | rno-miR-132 | 15.1 | 9.8 | 1.54 | 2.1E-02 |
| rno-let-7i | 22792.6 | 18823.1 | 1.2 | 6.2E-03 | rno-miR-369-3p | 2.7 | 1.2 | 2.33 | 2.1E-02 |
| rno-let-7e | 1092.4 | 812.3 | 1.3 | 4.4E-02 | rno-miR-203 | 63.1 | 39.8 | 1.59 | 2.3E-02 |
| rno-let-7d | 3641.6 | 2884.7 | 1.3 | 5.0E-02 | rno-miR-218 | 109.4 | 72.7 | 1.51 | 3.2E-02 |
| (rno-let-7b | 5402.1 | 4234.8 | 1.3 | 1.1E-01) | rno-miR-224 | 3.9 | 1.0 | 4.08 | 4.1E-02 |
| rno-miR-181b | 1770.3 | 1424.5 | 1.2 | 1.3E-02 | rno-miR-133a | 2.1 | 1.2 | 1.70 | 4.4E-02 |
| rno-miR-181a | 9389.8 | 8201.1 | 1.1 | 3.3E-02 | rno-miR-127 | 10.3 | 4.7 | 2.19 | 4.7E-02 |
| rno-miR-21 | 86834.9 | 54672.0 | 1.6 | 2.1E-02 | |||||
| rno-miR-199a-3p | 5138.5 | 3777.8 | 1.4 | 2.5E-02 | |||||
| rno-miR-223 | 3632.8 | 2432.8 | 1.5 | 3.6E-02 | |||||
| rno-miR-140* | 3873.0 | 3372.4 | 1.1 | 4.9E-02 | |||||
| rno-miR-93 | 3008.3 | 2711.1 | 1.1 | 4.9E-02 | |||||
| miRNAs with Higher Expression in the Resistant Strain (PVG) | |||||||||
| rno-miR-150 | 7269.4 | 8907.0 | 1.23 | 7.2E-03 | rno-miR-3559-3p | 20.5 | 44.4 | 2.17 | 2.0E-06 |
| rno-miR-29c | 3422.2 | 4205.1 | 1.23 | 2.4E-02 | rno-miR-3559-5p | 218.6 | 440.6 | 2.01 | 5.4E-05 |
| rno-miR-16 | 6187.6 | 7479.3 | 1.21 | 2.9E-02 | rno-miR-190b | 14.8 | 31.6 | 2.14 | 3.9E-04 |
| 8907.0 | 1.23 | 7.2E-03 | rno-miR-551b | 1.9 | 4.4 | 2.32 | 3.3E-02 | ||
| rno-miR-653 | 0.6 | 1.7 | 3.03 | 4.6E-02 | |||||
miRNAs in parentheses and italics refer to miRNA family members with p > 0.05.
In total, 35 miRNAs displayed higher expression in the susceptible DA strain (18 high- and 17 low-abundance miRNAs, respectively), and 8 showed higher expression in the resistant PVG (3 high- and 5 low-abundance miRNAs, respectively) (Table II). Also included in the high-abundance group are miRNA family members displaying expression with similar patterns, although with reduced significance. These were not counted as differentially expressed. The genetic location and disease association of the miRNAs and their family members is summarized in Supplemental Table II. More than 92% (37 out of 40) of the miRNAs with human homologs show sequence identity of the mature sequence to their human equivalent, and, strikingly, 77% (33 out of 43) of differentially expressed miRNAs have been previously associated to MS and other autoimmune diseases.
We next validated the results of the sequencing with an independent method, the TaqMan miRNA assays. We found that robust significant differences in expression could be confirmed for 76% (13 out of 17) miRNAs tested on the total RNA used for NGS (Fig. 2). Out of these miRNAs, we could confirm 80% (12 out of 15) in an independent material (data not shown). Furthermore, tested miRNA expression at naive state showed that a majority of the miRNAs, 67% (10 out of 15), were not differentially expressed between DA and PVG (data not shown).
FIGURE 2.
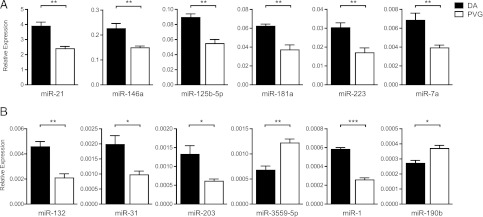
Validation of differentially expressed miRNAs. Relative expression of mature miRNAs was performed using specific TaqMan miRNA assays for selected high- (A) and low-abundance (B) miRNAs. miRNA expression was measured in total RNA from the samples subjected to NGS, DA (n = 5) and PVG (n = 5). Relative expression was calculated using the ΔΔ threshold cycle method and normalized against RNU6B. Error bars represent SEM. *p < 0.05, **p < 0.01, ***p < 0.001.
Kinetics of miRNA expression during the course of EAE was evaluated for a selected number of high-abundance miRNAs, namely miR-181a, miR-128, miR-146a, miR-125b-5p miR-199a-5p, and miR-223 (Fig. 3A). For half of the miRNAs tested (miR-181a, miR-128, and miR-146a), the differential expression was evident at all time points, as opposed to other miRNAs (miR-223 and miR-125b-5p) in which difference in expression was only observed at day 7 p.i. In this material, we were unable to reproduce differential expression for miR-199a-3p at day 7, but it was instead observed at day 25.
FIGURE 3.
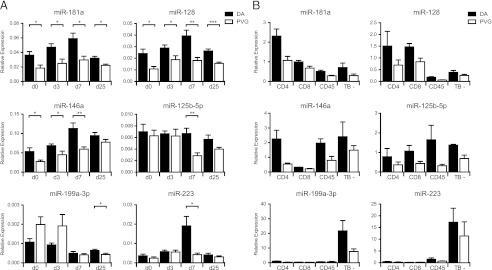
EAE kinetics and cell-type origin of miRNA expression. Relative expression of selected high-abundance miRNAs was performed using specific TaqMan miRNA assays for different time points of EAE (i.e., days 0 [naive], 3, 7, and 25 p.i. [n = 5]) (A) and for different cell types from lymph nodes at day 7 p.i. (i.e., Th [CD3+CD4+], cytotoxic T cells [CD3+CD8+], B cells [CD3−CD45RA+], and non-TB cells [TB−; CD3−CD4−CD8−CD45−]) (n = 3) (B). Relative expression was calculated using the ΔΔ threshold cycle method and normalized against RNU6B. Error bars represent SEM. *p < 0.05, **p < 0.01, ***p < 0.001.
We also investigated the cell-type origin of expression for the six miRNAs on four cellular fractions sorted from lymph nodes at day 7 p.i. (i.e., Th cells, cytotoxic T cells, B cells, and nonlymphocyte cells) (Fig. 3B). The result displayed different patterns of origin of miRNA expression in which miR-181a and miR-128 were primarily expressed in T cells, whereas miR-199a-3p and miR-223 were predominantly expressed in the nonlymphocyte fraction. miR-146a and miR-125b-5p were more ubiquitously expressed. We could observe differences in expression between DA and PVG mostly in lymphocytes, representing what we observed in the whole lymph node.
Target prediction and integration with genome-wide mRNA expression
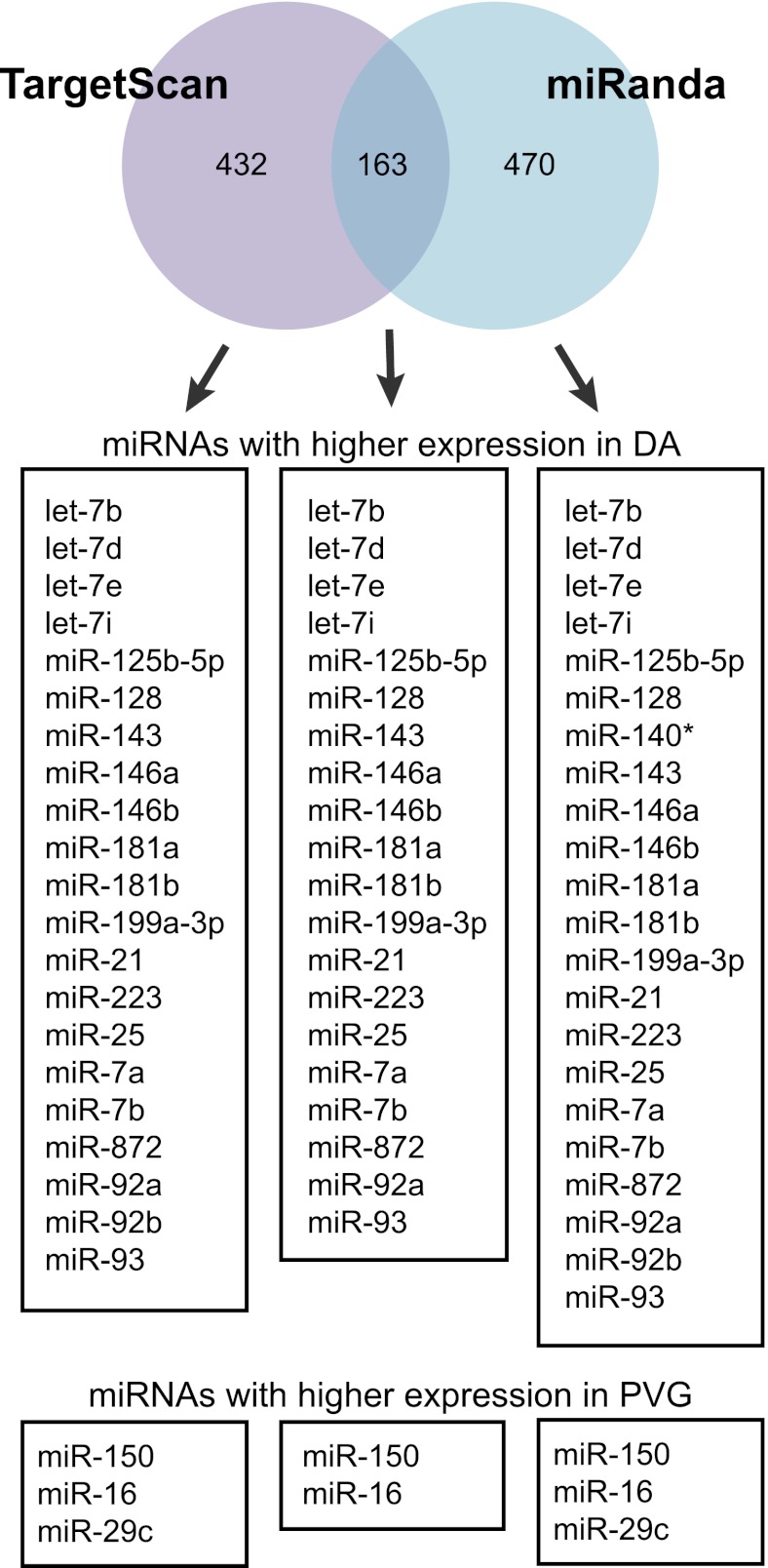
As only a handful of rat miRNA targets have been experimentally validated, we used target prediction tools to predict target mRNAs of the miRNAs. In general, only a fraction of predicted targets can be confirmed to be regulated by a given miRNA in a specific tissue (38). Therefore, to increase likelihood of predicting genuine targets of the miRNAs, we considered only targets that: 1) were predicted by two well-established prediction tools, TargetScan and miRanda; and 2) displayed according downregulation in lymph nodes between the strains. In total, 432 and 470 of the targets predicted by TargetScan and miRanda, respectively, were differentially expressed in the opposite direction in the target tissue. Of these, 38% (163 out of 432) of the targets were shared between the two prediction tools (109 and 54 targets of miRNAs with higher expression in DA and PVG, respectively) and considered for future functional annotation analysis (Fig. 4 and Supplemental Table III).
FIGURE 4.
Target prediction of the differentially expressed high-abundance miRNAs. mRNA targets were predicted using TargetScan and miRanda algorithms. Predictions were made with relaxed parameters (including nonconserved targets for TargetScan and all mirSVR scores for miRanda) and then combined with gene-expression information (Affymetrix). Numbers indicate unique targets showing differential expression in opposite direction to the miRNA in the gene-expression data set. TargetScan predicted 432 target genes (269 targets of DA miRNAs and 163 of PVG miRNAs, and miRanda predicted 470 target genes (218 targets of DA miRNAs and 252 of PVG miRNAs). Out of these, 163 targets were shared between the two algorithms (111 targets of DA miRNAs and 54 targets of PVG miRNAs). Predictions by both algorithms could be made for all miRNAs accept for miR-140*, which could only be predicted by miRanda. miR-29c and miR-92b had no targets predicted by both TargetScan and miRanda. None of the targets for miR-92a, let-d, and let-7d were found to be differentially expressed.
Functional annotations
We next sought to identify functions that associate with predicted targets of the high abundance miRNAs using IPA (Ingenuity). Targets of miRNAs that displayed higher expression in susceptible DA rats were enriched for functions such as cellular movement, amino acid metabolism, cellular function and maintenance, cellular development, and cell-to-cell signaling and interaction (Table III). The representative subgroups revealed more specific functions such as cell movement of leukocytes, accumulation of amino acids, and influx of Ca2+. Different functions emerged for the targets of miRNAs with higher expression in resistant PVG rats (Table III). More specific functions involved differentiation of cells, transcription of RNA, and generation of B-1a lymphocytes. This implicated functions that are likely regulated by miRNAs and suggests regulation of different pathways during immune activation in susceptible and resistant rats.
Table III. Top five unique IPA functions enriched in targets of differentially expressed high-abundance miRNAs between DA and PVG.
| Category | p Value | No. of Molecules* | Representative Subgroups | p Value | No. of Molecules |
|---|---|---|---|---|---|
| DA | |||||
| Cellular movement | 8.16E-06–1.78E-02 | 28 (CXCR3, STAT1) | Migration of cells | 2.90E-05 | 13 |
| Cell movement of leukocytes | 5.13E-04 | 25 | |||
| Amino acid metabolism | 1.05E-04–1.64E-02 | 12 (PRKCD) | Accumulation of neutral amino acid | 1.05E-05 | 2 |
| Transport of D-serine | 2.09E-04 | 2 | |||
| Cellular function and maintenance | 1.31E-04–1.78E-02 | 18 (CXCR3, PRKCD, STAT1) | Compartmentalization of cells | 1.31E-04 | 3 |
| Influx of Ca2+ | 1.97E-03 | 5 | |||
| Cellular development | 1.37E-04–1.78E-02 | 31 (PRKCD, STAT1) | Differentiation of cells | 3.35E-04 | 24 |
| Growth of tumor cells | 1.07E-03 | 6 | |||
| Cell-to-cell signaling and interaction | 2.14E-04–1.78E-02 | 26 (CXCR3, STAT1) | adhesion of immune cells | 2.14E-04 | 9 |
| Chemoattraction of mononuclear leukocytes | 4.29E-04 | 3 | |||
|
p Value |
No. of Molecules |
Function Annotation |
p Value |
No. of Molecules |
|
| PVG | |||||
| Cellular development | 7.64E-05–4.21E-02 | 19 | Differentiation of cells | 5.75E-05 | 16 |
| Developmental process of B lymphocytes | 7.00E-03 | 4 | |||
| Gene expression | 5.24E-05–3.96E-02 | 19 | Transcription of RNA | 5.24E-05 | 17 |
| Expression of RNA | 5.48E-05 | 18 | |||
| Cell death | 6.83E-05–4.21E-02 | 18 | Cell death of organ | 6.83E-05 | 11 |
| Apoptosis of tumor cell lines | 1.78E-03 | 10 | |||
| Cellular growth and proliferation | 7.99E-05–3.6E6-02 | 9 | Inhibition of cancer cells | 7.99E-05 | 2 |
| Generation of B-1a lymphocytes | 5.72E-03 | 1 | |||
| Lipid metabolism | 4.35E-04–3.66E-02 | 4 | Esterification of fatty acid | 4.35E-04 | 2 |
| Biosynthesis of phosphatidylglycerol | 2.86E-03 | 1 |
miRNA target genes in parentheses were used in in vitro studies.
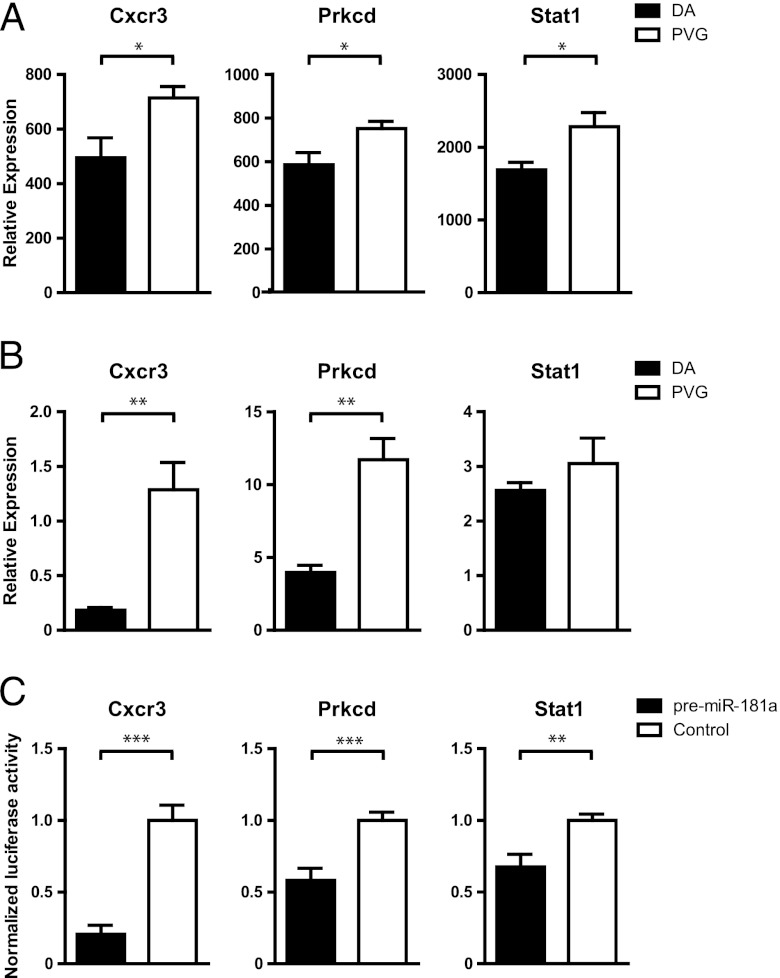
Validation of targets for miR-181a
Highly abundant lymphocyte miRNA miR-181a is predicted, following our criteria, to target 12 differentially expressed genes in EAE. Frequently represented in the top IPA functions (Table III) were Cxcr3, Prkcd, and Stat1 (Fig. 5A), which showed similar expression differences in independent material (Fig. 5B). In order to establish that miR-181a can directly interact with the 3′ UTR of the target genes, we used a luciferase reporter system. We cloned the 3′ UTRs of Cxcr3, Prkcd, and Stat1 into a vector downstream of the luciferase gene. By cotransfecting the vector with pre–miR-181a, we could confirm direct binding of miR-181a with the cloned 3′ UTR sequence by the subsequent translational block of luciferase protein and activity for all three targets (Fig. 5C). In addition, we studied impact of either miR-181a inhibitor or mimic (pre–miR-181) on expression of the target genes in vitro. Inhibition of miR-181a resulted in increased expression of all three target genes (Supplemental Fig. 1A) compared with the scrambled control. Transfection with miR-181a mimic reduced the expression of CXCR3 and STAT1 compared with the control (Supplemental Fig. 1B). For PRKCD, there was only a tendency for downregulation, which is likely due to the high intrinsic expression of the gene. Together, these data indicate that these genes are likely direct targets of miR-181a and that the combination of in silico target prediction with whole genome expression data improves the discovery of true miRNA targets.
FIGURE 5.
miR-181a target validation. (A) Relative expression from Affymetrix whole-genome expression data from lymph nodes of DA (n = 4) and PVG (n = 4) at day 7 p.i. (B) Relative mRNA expression compared with β-actin measured by qRT-PCR in lymph node cells of DA (n = 5) and PVG (n = 7) at day 7 p.i. (C) Normalized luciferase activity in lysates from HEK293 cells cotransfected with either pre–mir-181a mimic or negative scrambled control together with pmirGLO Dual-Luciferase miRNA Target Expression Vector cloned with 3′ UTR sequence of Cxcr3, Prkcd, or Stat1. Data represent at least three independent experiments in which values are normalized to the mean of the control for each experiment. Error bars represent SEM. *p < 0.05, **p < 0.01, ***p < 0.001.
Discussion
miRNA profiling has proven extremely valuable in unraveling regulation of biological functions (6, 39) and establishing biomarkers of disease states (40–42). Studies of miRNAs in mouse EAE focused on a few key miRNAs as regulators of pathogenic Th1 and Th17 cell subsets (27, 29, 43) as well as macrophages and microglia (28). To date, 12 profiling studies have been published assessing the role of miRNAs in MS (15–25, 44), albeit with little overlap between identified miRNAs. This can be attributed to different cellular compartments and disease conditions that the different studies have investigated. Moreover, high genetic heterogeneity in human populations could have contributed to the inconsistent findings of miRNA differences in MS. Therefore, our approach was to establish miRNAs that discriminate the pathogenic immune response from the resolving, using two distinct inbred rat strains. Following immunization with MOG, EAE-susceptible DA and EAE-resistant PVG rats both mount a potent immune response in the draining lymph nodes. In DA rats, a pathogenic immune response leads to subsequent cell infiltration and damage to the CNS (45). In PVG rats, the initial immune response resolves, and the rats remain unaffected. The outcome of this activation does not associate with the changes in the major cell subsets but rather with the quality of immune response (45). This separates our study from many others in which miRNAs have been compared between affected and healthy subjects, resulting in a large number of miRNAs that differ between inflamed and naive states as opposed to those associating with susceptibility to develop disease.
Interestingly, a majority of the miRNAs displayed higher expression in the susceptible strain. These miRNAs are likely either promoting disease or they are expressed in an attempt to downregulate immune activation in the susceptible rats (46). A well-known example of the latter is miR-146, which is upregulated upon TLR activation by LPS or by proinflammatory cytokines such as IL-1 and TNF. Both IL-1 and TNF are elevated during MS and EAE (45, 47, 48). miR-146 acts by negatively regulating IL-1R–associated kinase 1 and TNFR-associated factor 6, thereby producing a negative-feedback loop to fine-tune the immune response (49). Due to its important role in regulating the magnitude of the immune response, it is easy to see how dysregulation of miR-146 can contribute to disease development. This is further highlighted by miR-146 association with most autoimmune diseases, including RA (8), systemic lupus erythematosus (11), and MS (15).
Many of the miRNAs are also known to have disease-promoting functions in MS and other autoimmune diseases. Two examples are miR-128 and miR-203, which showed higher expression in the susceptible DA rats. miR-128 is upregulated in naive T cells of MS patients compared with healthy controls (50). We also found a predominant expression of miR-128 in T cells with higher expression in susceptible DA rats in the naive state and after EAE induction. It was shown that upregulation of miR-128 results in a shift from Th2 to Th1 cytokine production. A similar shift toward preferential Th1/Th17 response has been reported in susceptible DA rats in several autoimmune conditions (45) and could in part be attributed to miR-128. The other, miR-203, has previously been associated with psoriasis (9) and RA (51). In the latter, the authors found a miR-203 associated upregulation of IL-6. Higher miR-203 in DA may also contribute to elevated levels of IL-6 observed in DA compared with PVG rats (45). Collectively, these findings strongly suggest that the miRNAs identified in our study are of relevance in immune responses and development of autoimmunity. It is therefore likely that identified miRNAs, which have not been previously reported to associate with autoimmune diseases, may comprise novel mechanisms in autoimmunity.
In order to understand the functional outcome of miRNA dysregulation, we used a combination of two well-established target prediction tools to elucidate potential targets (52). As miRNA binding to target 3′ UTR generally results in mRNA destabilization and degradation (53), we chose to narrow down potential targets to those showing differential expression in the opposite direction as the miRNA. This approach increases the strength to discover true target genes and functions affected by miRNA dysregulation. However, one should bear in mind that a few miRNA–mRNA interactions will be overlooked as miRNA binding also could result in translational repression with no change in mRNA levels (3).
In order to interpret the biological relevance of the high-abundance miRNAs, we investigated the pathways and functions enriched in the target genes. For the genes targeted by DA miRNAs, two of the most enriched functions involved the migration and homing of immune cells. Migration and infiltration of autoreactive cells into the CNS is critical for both MS and EAE (54, 55). Regulated by the miRNAs are several important chemokines and their receptors responsible for chemotaxis, CCL3L1/CCL3L3, CXCL13, and CXCR3, which have previously been reported in studies of MS and EAE (56–58). Other top functions like cellular function and maintenance and cell-to-cell signaling and interaction comprised genes of signaling pathways, such as STAT1 and STAT2 of the JAK/STAT pathway. As this pathway is commonly used by many cytokines (59), it is not surprising that the molecules of the JAK/STAT signaling pathway are implicated in both MS (60) and EAE (61).
As for the functions associated with targets of miRNAs found elevated in the resistant PVG strain, these include cell differentiation and apoptosis. Apoptosis plays a significant role in EAE development and pathogenesis, in which apoptosis of autoreactive cells limits the immune response and ultimately leads to disease amelioration (62, 63). The genes involved in these functions are mainly targets of miR-150, proposing this miRNA to be an important contributor to the control of autoimmunity through regulation of cell turn over. miR-150 is previously known for its role in T- and B-lymphocyte development (64). Together, this demonstrates that miRNAs can contribute to dysregulation of important immune functions as a key factor determining susceptibility to EAE.
Another miRNA known for its role in the immune system is miR-181a. In this study, we show that miR-181a is predominantly expressed in the lymphocyte population during the induction of EAE, with the biggest difference in expression between DA and PVG in CD4+ T cells. miR-181a is mainly known for its role in T cell development (65) but has also been shown to be important for TCR sensitivity (66). A reduced TCR sensitivity can cause breakdown of self-tolerance and could subsequently lead to development of autoimmunity (67). The high levels of miR-181a in the susceptible DA rats could therefore contribute to the lower activation threshold of autoreactive T cells, as higher T cell response to autoantigens has been reported in DA compared with PVG rats (45). In addition to known functions of miR-181a, we confirmed in this study that miR-181a can directly target several genes which expression is regulated during MOG-induced EAE in rats. Three of the miR-181a targets, namely Cxcr3, Prkcd, and Stat1, are essential for all of the top miRNA-regulated pathways in DA. In this study, we showed using luciferase assays, mimic, and inhibitor that they are direct targets of miR-181a. The chemokine receptor CXCR3 has been previously associated with MS and EAE. CXCR3 is important for the differentiation of naive CD4 cells to activated Th1 cells within the lymph node (68, 69) and has been found upregulated in the CNS during EAE (70). EAE pathogenesis is generally considered to begin with activation and differentiation of T cells, which then leave the lymph nodes and migrate to the target organ. The flexible expression of miR-181a may play a role in reducing CXCR3 levels on MOG-reactive T cells in the periphery, thereby facilitating their homing toward the CNS. PRKCD is a proapoptotic protein kinase previously shown to be directly targeted by miR-181a in humans (71). Besides its involvement in cancer, PRKCD was shown to play a role in B cell proliferation, in which depletion of PRKCD results in expansion of B cells and an increased expression of IL-6 (72). The importance of B cells in MS and EAE is demonstrated by the presence of Abs against myelin Ags (73) and by the success of the B cell–depleting drugs in EAE and in patients with relapsing remitting MS (74). STAT1 is a transcription factor that acts upon signaling by IFNs (types I, II, and III), inducing the transcription of a variety of IFN-regulated genes, several of which associated with MS (75). Regulation of STAT1 by miR-181a may therefore be critical for a vast number of homeostatic and pathogenic mechanisms. Most importantly, IFN-β, a type I IFN, is used as first-line treatment for MS, further enhancing the importance of IFNs and their downstream signaling molecules in neuroinflammation. Thus, with this, we demonstrate how the dysregulation of a single miRNA can impact several genes involved in crucial cellular mechanisms for the development of autoimmunity and EAE.
To add to the complexity of miRNA regulation during EAE, we show that some of the miRNAs are differentially expressed already at naive state, whereas others display a difference in expression only in the course of EAE. This indicates that miRNA dysregulation could be either inherent in the strains and/or dependent on EAE. In addition, cellular sources of miRNA are likely diverse, with some miRNAs displaying predominant expression in lymphocytes, whereas others are more specific for other cell types or equally abundant in all cell types. Together with a variety of implicated functions enriched for differentially expressed targets, this suggests that miRNA regulation of EAE depends both on the timing of expression and the cellular source. Although we can confirm that certain predicted targets can indeed be directly regulated by miRNA, the individual contribution of each miRNA and their net effect require further investigation.
In this study, we used NGS to establish miRNAs that differ between pathogenic and resolving immune activation. Combining target prediction with target expression, we defined immune functions that are at least in part regulated by miRNAs. The majority of detected miRNAs associate with MS and other autoimmune diseases, implicating their important roles in autoimmunity. Future investigation, focusing on strongly dysregulated miRNA and their target genes, will provide insights into pathogenesis of autoimmune inflammation and novel targets for therapeutic interventions.
This work was supported by grants from the Swedish Research Council, the Harald and Greta Jeanssons Foundation, the Swedish Association for Persons with Neurological Disabilities, the Åke Wibergs Foundation, the Åke Löwnertz Foundation, the Swedish Brain Foundation, Socialstyrelsen, and by Karolinska Institutet funds. A.G. was supported by a grant from the Multiple Sclerosis Society of Canada.
The online version of this article contains supplemental material.
- DA
- Dark Agouti
- EAE
- experimental autoimmune encephalomyelitis
- IPA
- Ingenuity pathway analysis
- miRNA
- microRNA
- MOG
- myelin oligodendrocyte glycoprotein
- MS
- multiple sclerosis
- NGS
- next-generation sequencing
- p.i.
- postimmunization
- PVG
- Piebald Virol Glaxo
- qRT-PCR
- quantitative real-time PCR
- RA
- rheumatoid arthritis
- UTR
- untranslated region.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Lee R. C., Feinbaum R. L., Ambros V. 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75: 843–854 [DOI] [PubMed] [Google Scholar]
- 2.Bartel D. P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297 [DOI] [PubMed] [Google Scholar]
- 3.Bartel D. P. 2009. MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Engels B. M., Hutvagner G. 2006. Principles and effects of microRNA-mediated post-transcriptional gene regulation. Oncogene 25: 6163–6169 [DOI] [PubMed] [Google Scholar]
- 5.Miranda K. C., Huynh T., Tay Y., Ang Y. S., Tam W. L., Thomson A. M., Lim B., Rigoutsos I. 2006. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell 126: 1203–1217 [DOI] [PubMed] [Google Scholar]
- 6.Baltimore D., Boldin M. P., O’Connell R. M., Rao D. S., Taganov K. D. 2008. MicroRNAs: new regulators of immune cell development and function. Nat. Immunol. 9: 839–845 [DOI] [PubMed] [Google Scholar]
- 7.Kurowska-Stolarska M., Alivernini S., Ballantine L. E., Asquith D. L., Millar N. L., Gilchrist D. S., Reilly J., Ierna M., Fraser A. R., Stolarski B., et al. 2011. MicroRNA-155 as a proinflammatory regulator in clinical and experimental arthritis. Proc. Natl. Acad. Sci. USA 108: 11193–11198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pauley K. M., Satoh M., Chan A. L., Bubb M. R., Reeves W. H., Chan E. K. 2008. Upregulated miR-146a expression in peripheral blood mononuclear cells from rheumatoid arthritis patients. Arthritis Res. Ther. 10: R101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sonkoly E., Wei T., Janson P. C., Sääf A., Lundeberg L., Tengvall-Linder M., Norstedt G., Alenius H., Homey B., Scheynius A., et al. 2007. MicroRNAs: novel regulators involved in the pathogenesis of psoriasis? PLoS ONE 2: e610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dai Y., Huang Y. S., Tang M., Lv T. Y., Hu C. X., Tan Y. H., Xu Z. M., Yin Y. B. 2007. Microarray analysis of microRNA expression in peripheral blood cells of systemic lupus erythematosus patients. Lupus 16: 939–946 [DOI] [PubMed] [Google Scholar]
- 11.Tang Y., Luo X., Cui H., Ni X., Yuan M., Guo Y., Huang X., Zhou H., de Vries N., Tak P. P., et al. 2009. MicroRNA-146A contributes to abnormal activation of the type I interferon pathway in human lupus by targeting the key signaling proteins. Arthritis Rheum. 60: 1065–1075 [DOI] [PubMed] [Google Scholar]
- 12.Junker A., Hohlfeld R., Meinl E. 2011. The emerging role of microRNAs in multiple sclerosis. Nat Rev Neurol 7: 56–59 [DOI] [PubMed] [Google Scholar]
- 13.Sawcer S., Hellenthal G., Pirinen M., Spencer C. C., Patsopoulos N. A., Moutsianas L., Dilthey A., Su Z., Freeman C., Hunt S. E., et al. International Multiple Sclerosis Genetics Consortium. Wellcome Trust Case Control Consortium 2 2011. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature 476: 214–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ebers G. C. 2008. Environmental factors and multiple sclerosis. Lancet Neurol. 7: 268–277 [DOI] [PubMed] [Google Scholar]
- 15.Fenoglio C., Cantoni C., De Riz M., Ridolfi E., Cortini F., Serpente M., Villa C., Comi C., Monaco F., Mellesi L., et al. 2011. Expression and genetic analysis of miRNAs involved in CD4+ cell activation in patients with multiple sclerosis. Neurosci. Lett. 504: 9–12 [DOI] [PubMed] [Google Scholar]
- 16.Otaegui D., Baranzini S. E., Armañanzas R., Calvo B., Muñoz-Culla M., Khankhanian P., Inza I., Lozano J. A., Castillo-Triviño T., Asensio A., et al. 2009. Differential micro RNA expression in PBMC from multiple sclerosis patients. PLoS ONE 4: e6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martinelli-Boneschi F., Fenoglio C., Brambilla P., Sorosina M., Giacalone G., Esposito F., Serpente M., Cantoni C., Ridolfi E., Rodegher M., et al. 2012. MicroRNA and mRNA expression profile screening in multiple sclerosis patients to unravel novel pathogenic steps and identify potential biomarkers. Neurosci. Lett. 508: 4–8 [DOI] [PubMed] [Google Scholar]
- 18.Cox M. B., Cairns M. J., Gandhi K. S., Carroll A. P., Moscovis S., Stewart G. J., Broadley S., Scott R. J., Booth D. R., Lechner-Scott J., ANZgene Multiple Sclerosis Genetics Consortium 2010. MicroRNAs miR-17 and miR-20a inhibit T cell activation genes and are under-expressed in MS whole blood. PLoS ONE 5: e12132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keller A., Leidinger P., Lange J., Borries A., Schroers H., Scheffler M., Lenhof H. P., Ruprecht K., Meese E. 2009. Multiple sclerosis: microRNA expression profiles accurately differentiate patients with relapsing-remitting disease from healthy controls. PLoS ONE 4: e7440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Siegel S. R., Mackenzie J., Chaplin G., Jablonski N. G., Griffiths L. 2012. Circulating microRNAs involved in multiple sclerosis. Mol. Biol. Rep. 39: 6219–6225 [DOI] [PubMed] [Google Scholar]
- 21.Lindberg R. L., Hoffmann F., Mehling M., Kuhle J., Kappos L. 2010. Altered expression of miR-17-5p in CD4+ lymphocytes of relapsing-remitting multiple sclerosis patients. Eur. J. Immunol. 40: 888–898 [DOI] [PubMed] [Google Scholar]
- 22.Sievers C., Meira M., Hoffmann F., Fontoura P., Kappos L., Lindberg R. L. 2012. Altered microRNA expression in B lymphocytes in multiple sclerosis: towards a better understanding of treatment effects. Clin. Immunol. 144: 70–79 [DOI] [PubMed] [Google Scholar]
- 23.De Santis G., Ferracin M., Biondani A., Caniatti L., Rosaria Tola M., Castellazzi M., Zagatti B., Battistini L., Borsellino G., Fainardi E., et al. 2010. Altered miRNA expression in T regulatory cells in course of multiple sclerosis. J. Neuroimmunol. 226: 165–171 [DOI] [PubMed] [Google Scholar]
- 24.Haghikia A., Haghikia A., Hellwig K., Baraniskin A., Holzmann A., Décard B. F., Thum T., Gold R. 2012. Regulated microRNAs in the CSF of patients with multiple sclerosis: a case-control study. Neurology 79: 2166–2170 [DOI] [PubMed] [Google Scholar]
- 25.Junker A., Krumbholz M., Eisele S., Mohan H., Augstein F., Bittner R., Lassmann H., Wekerle H., Hohlfeld R., Meinl E. 2009. MicroRNA profiling of multiple sclerosis lesions identifies modulators of the regulatory protein CD47. Brain 132: 3342–3352 [DOI] [PubMed] [Google Scholar]
- 26.Storch M. K., Stefferl A., Brehm U., Weissert R., Wallström E., Kerschensteiner M., Olsson T., Linington C., Lassmann H. 1998. Autoimmunity to myelin oligodendrocyte glycoprotein in rats mimics the spectrum of multiple sclerosis pathology. Brain Pathol. 8: 681–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Du C., Liu C., Kang J., Zhao G., Ye Z., Huang S., Li Z., Wu Z., Pei G. 2009. MicroRNA miR-326 regulates TH-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nat. Immunol. 10: 1252–1259 [DOI] [PubMed] [Google Scholar]
- 28.Ponomarev E. D., Veremeyko T., Barteneva N., Krichevsky A. M., Weiner H. L. 2011. MicroRNA-124 promotes microglia quiescence and suppresses EAE by deactivating macrophages via the C/EBP-α-PU.1 pathway. Nat. Med. 17: 64–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murugaiyan G., Beynon V., Mittal A., Joller N., Weiner H. L. 2011. Silencing microRNA-155 ameliorates experimental autoimmune encephalomyelitis. J. Immunol. 187: 2213–2221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amor S., Groome N., Linington C., Morris M. M., Dornmair K., Gardinier M. V., Matthieu J. M., Baker D. 1994. Identification of epitopes of myelin oligodendrocyte glycoprotein for the induction of experimental allergic encephalomyelitis in SJL and Biozzi AB/H mice. J. Immunol. 153: 4349–4356 [PubMed] [Google Scholar]
- 31.Hackenberg M., Sturm M., Langenberger D., Falcón-Pérez J. M., Aransay A. M. 2009. miRanalyzer: a microRNA detection and analysis tool for next-generation sequencing experiments. Nucleic Acids Res. 37(Web Server issue): W68–W76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hackenberg M., Rodríguez-Ezpeleta N., Aransay A. M. 2011. miRanalyzer: an update on the detection and analysis of microRNAs in high-throughput sequencing experiments. Nucleic Acids Res. 39(Web Server issue): W132–W138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kozomara A., Griffiths-Jones S. 2011. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 39(Database issue): D152–D157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmittgen T. D., Livak K. J. 2008. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3: 1101–1108 [DOI] [PubMed] [Google Scholar]
- 35.Gillett A., Maratou K., Fewings C., Harris R. A., Jagodic M., Aitman T., Olsson T. 2009. Alternative splicing and transcriptome profiling of experimental autoimmune encephalomyelitis using genome-wide exon arrays. PLoS ONE 4: e7773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Graaf K. L., Barth S., Herrmann M. M., Storch M. K., Wiesmüller K. H., Weissert R. 2008. Characterization of the encephalitogenic immune response in a model of multiple sclerosis. Eur. J. Immunol. 38: 299–308 [DOI] [PubMed] [Google Scholar]
- 37.Linsen S. E., de Wit E., Janssens G., Heater S., Chapman L., Parkin R. K., Fritz B., Wyman S. K., de Bruijn E., Voest E. E., et al. 2009. Limitations and possibilities of small RNA digital gene expression profiling. Nat. Methods 6: 474–476 [DOI] [PubMed] [Google Scholar]
- 38.Martin G., Schouest K., Kovvuru P., Spillane C. 2007. Prediction and validation of microRNA targets in animal genomes. J. Biosci. 32: 1049–1052 [DOI] [PubMed] [Google Scholar]
- 39.Ambros V. 2004. The functions of animal microRNAs. Nature 431: 350–355 [DOI] [PubMed] [Google Scholar]
- 40.Wang G. K., Zhu J. Q., Zhang J. T., Li Q., Li Y., He J., Qin Y. W., Jing Q. 2010. Circulating microRNA: a novel potential biomarker for early diagnosis of acute myocardial infarction in humans. Eur. Heart J. 31: 659–666 [DOI] [PubMed] [Google Scholar]
- 41.Mitchell P. S., Parkin R. K., Kroh E. M., Fritz B. R., Wyman S. K., Pogosova-Agadjanyan E. L., Peterson A., Noteboom J., O’Briant K. C., Allen A., et al. 2008. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 105: 10513–10518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alevizos I., Illei G. G. 2010. MicroRNAs as biomarkers in rheumatic diseases. Nat Rev Rheumatol 6: 391–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guan H., Fan D., Mrelashvili D., Hao H., Singh N. P., Singh U. P., Nagarkatti P. S., Nagarkatti M. 2013. MicroRNA let-7e is associated with the pathogenesis of experimental autoimmune encephalomyelitis. Eur. J. Immunol. 43: 104–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Waschbisch A., Atiya M., Linker R. A., Potapov S., Schwab S., Derfuss T. 2011. Glatiramer acetate treatment normalizes deregulated microRNA expression in relapsing remitting multiple sclerosis. PLoS ONE 6: e24604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thessen Hedreul M., Gillett A., Olsson T., Jagodic M., Harris R. A. 2009. Characterization of Multiple Sclerosis candidate gene expression kinetics in rat experimental autoimmune encephalomyelitis. J. Neuroimmunol. 210: 30–39 [DOI] [PubMed] [Google Scholar]
- 46.Bazzoni F., Rossato M., Fabbri M., Gaudiosi D., Mirolo M., Mori L., Tamassia N., Mantovani A., Cassatella M. A., Locati M. 2009. Induction and regulatory function of miR-9 in human monocytes and neutrophils exposed to proinflammatory signals. Proc. Natl. Acad. Sci. USA 106: 5282–5287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Okuda Y., Sakoda S., Bernard C. C., Yanagihara T. 1998. The development of autoimmune encephalomyelitis provoked by myelin oligodendrocyte glycoprotein is associated with an upregulation of both proinflammatory and immunoregulatory cytokines in the central nervous system. J. Interferon Cytokine Res. 18: 415–421 [DOI] [PubMed] [Google Scholar]
- 48.Hauser S. L., Doolittle T. H., Lincoln R., Brown R. H., Dinarello C. A. 1990. Cytokine accumulations in CSF of multiple sclerosis patients: frequent detection of interleukin-1 and tumor necrosis factor but not interleukin-6. Neurology 40: 1735–1739 [DOI] [PubMed] [Google Scholar]
- 49.Taganov K. D., Boldin M. P., Chang K. J., Baltimore D. 2006. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. USA 103: 12481–12486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guerau-de-Arellano M., Smith K. M., Godlewski J., Liu Y., Winger R., Lawler S. E., Whitacre C. C., Racke M. K., Lovett-Racke A. E. 2011. Micro-RNA dysregulation in multiple sclerosis favours pro-inflammatory T-cell-mediated autoimmunity. Brain 134: 3578–3589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stanczyk J., Ospelt C., Karouzakis E., Filer A., Raza K., Kolling C., Gay R., Buckley C. D., Tak P. P., Gay S., Kyburz D. 2011. Altered expression of microRNA-203 in rheumatoid arthritis synovial fibroblasts and its role in fibroblast activation. Arthritis Rheum. 63: 373–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sethupathy P., Megraw M., Hatzigeorgiou A. G. 2006. A guide through present computational approaches for the identification of mammalian microRNA targets. Nat. Methods 3: 881–886 [DOI] [PubMed] [Google Scholar]
- 53.Guo H. L., Ingolia N. T., Weissman J. S., Bartel D. P. 2010. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466: 835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Prat A., Biernacki K., Lavoie J. F., Poirier J., Duquette P., Antel J. P. 2002. Migration of multiple sclerosis lymphocytes through brain endothelium. Arch. Neurol. 59: 391–397 [DOI] [PubMed] [Google Scholar]
- 55.Denkinger C. M., Denkinger M., Kort J. J., Metz C., Forsthuber T. G. 2003. In vivo blockade of macrophage migration inhibitory factor ameliorates acute experimental autoimmune encephalomyelitis by impairing the homing of encephalitogenic T cells to the central nervous system. J. Immunol. 170: 1274–1282 [DOI] [PubMed] [Google Scholar]
- 56.Khademi M., Kockum I., Andersson M. L., Iacobaeus E., Brundin L., Sellebjerg F., Hillert J., Piehl F., Olsson T. 2011. Cerebrospinal fluid CXCL13 in multiple sclerosis: a suggestive prognostic marker for the disease course. Mult. Scler. 17: 335–343 [DOI] [PubMed] [Google Scholar]
- 57.O’Connor R. A., Li X., Blumerman S., Anderton S. M., Noelle R. J., Dalton D. K. 2012. Adjuvant immunotherapy of experimental autoimmune encephalomyelitis: immature myeloid cells expressing CXCL10 and CXCL16 attract CXCR3+CXCR6+ and myelin-specific T cells to the draining lymph nodes rather than the central nervous system. J. Immunol. 188: 2093–2101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vaknin-Dembinsky A., Murugaiyan G., Hafler D. A., Astier A. L., Weiner H. L. 2008. Increased IL-23 secretion and altered chemokine production by dendritic cells upon CD46 activation in patients with multiple sclerosis. J. Neuroimmunol. 195: 140–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schindler C., Levy D. E., Decker T. 2007. JAK-STAT signaling: from interferons to cytokines. J. Biol. Chem. 282: 20059–20063 [DOI] [PubMed] [Google Scholar]
- 60.Frisullo G., Angelucci F., Caggiula M., Nociti V., Iorio R., Patanella A. K., Sancricca C., Mirabella M., Tonali P. A., Batocchi A. P. 2006. pSTAT1, pSTAT3, and T-bet expression in peripheral blood mononuclear cells from relapsing-remitting multiple sclerosis patients correlates with disease activity. J. Neurosci. Res. 84: 1027–1036 [DOI] [PubMed] [Google Scholar]
- 61.Jiang Z., Li H., Fitzgerald D. C., Zhang G. X., Rostami A. 2009. MOG(35-55) i.v suppresses experimental autoimmune encephalomyelitis partially through modulation of Th17 and JAK/STAT pathways. Eur. J. Immunol. 39: 789–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schmied M., Breitschopf H., Gold R., Zischler H., Rothe G., Wekerle H., Lassmann H. 1993. Apoptosis of T lymphocytes in experimental autoimmune encephalomyelitis. Evidence for programmed cell death as a mechanism to control inflammation in the brain. Am. J. Pathol. 143: 446–452 [PMC free article] [PubMed] [Google Scholar]
- 63.Zettl U. K., Mix E., Zielasek J., Stangel M., Hartung H. P., Gold R. 1997. Apoptosis of myelin-reactive T cells induced by reactive oxygen and nitrogen intermediates in vitro. Cell. Immunol. 178: 1–8 [DOI] [PubMed] [Google Scholar]
- 64.Zhou B., Wang S., Mayr C., Bartel D. P., Lodish H. F. 2007. miR-150, a microRNA expressed in mature B and T cells, blocks early B cell development when expressed prematurely. Proc. Natl. Acad. Sci. USA 104: 7080–7085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Neilson J. R., Zheng G. X., Burge C. B., Sharp P. A. 2007. Dynamic regulation of miRNA expression in ordered stages of cellular development. Genes Dev. 21: 578–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li Q. J., Chau J., Ebert P. J., Sylvester G., Min H., Liu G., Braich R., Manoharan M., Soutschek J., Skare P., et al. 2007. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell 129: 147–161 [DOI] [PubMed] [Google Scholar]
- 67.Goodnow C. C., Sprent J., Fazekas de St Groth B., Vinuesa C. G. 2005. Cellular and genetic mechanisms of self tolerance and autoimmunity. Nature 435: 590–597 [DOI] [PubMed] [Google Scholar]
- 68.Sallusto F., Lenig D., Mackay C. R., Lanzavecchia A. 1998. Flexible programs of chemokine receptor expression on human polarized T helper 1 and 2 lymphocytes. J. Exp. Med. 187: 875–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Groom J. R., Richmond J., Murooka T. T., Sorensen E. W., Sung J. H., Bankert K., von Andrian U. H., Moon J. J., Mempel T. R., Luster A. D. 2012. CXCR3 chemokine receptor-ligand interactions in the lymph node optimize CD4+ T helper 1 cell differentiation. Immunity 37: 1091–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kohler R. E., Comerford I., Townley S., Haylock-Jacobs S., Clark-Lewis I., McColl S. R. 2008. Antagonism of the chemokine receptors CXCR3 and CXCR4 reduces the pathology of experimental autoimmune encephalomyelitis. Brain Pathol. 18: 504–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ke G., Liang L., Yang J. M., Huang X., Han D., Huang S., Zhao Y., Zha R., He X., Wu X. MiR-181a confers resistance of cervical cancer to radiation therapy through targeting the pro-apoptotic PRKCD gene. Oncogene. 2012 doi: 10.1038/onc.2012.323. DOI: 10.1038/onc.2012.323. [DOI] [PubMed] [Google Scholar]
- 72.Miyamoto A., Nakayama K., Imaki H., Hirose S., Jiang Y., Abe M., Tsukiyama T., Nagahama H., Ohno S., Hatakeyama S., Nakayama K. I. 2002. Increased proliferation of B cells and auto-immunity in mice lacking protein kinase Cdelta. Nature 416: 865–869 [DOI] [PubMed] [Google Scholar]
- 73.Cross A. H., Trotter J. L., Lyons J. 2001. B cells and antibodies in CNS demyelinating disease. J. Neuroimmunol. 112: 1–14 [DOI] [PubMed] [Google Scholar]
- 74.Hauser S. L., Waubant E., Arnold D. L., Vollmer T., Antel J., Fox R. J., Bar-Or A., Panzara M., Sarkar N., Agarwal S., et al. HERMES Trial Group 2008. B-cell depletion with rituximab in relapsing-remitting multiple sclerosis. N. Engl. J. Med. 358: 676–688 [DOI] [PubMed] [Google Scholar]
- 75.Serrano-Fernández P., Möller S., Goertsches R., Fiedler H., Koczan D., Thiesen H. J., Zettl U. K. 2010. Time course transcriptomics of IFNB1b drug therapy in multiple sclerosis. Autoimmunity 43: 172–178 [DOI] [PubMed] [Google Scholar]