Summary
In mouse and man Y chromosome deletions are frequently associated with spermatogenic defects. Mice with extensive deletions of non-pairing Y chromosome long arm (NPYq) are infertile and produce sperm with grossly misshapen heads, abnormal chromatin packaging and DNA damage. The NPYq-encoded multi-copy gene Sly controls the expression of sex chromosome genes after meiosis and Sly deficiency results in a remarkable upregulation of sex chromosome genes. Sly deficiency has been shown to be the underlying cause of the sperm head anomalies and infertility associated with NPYq gene loss, but it was not known whether it recapitulates sperm DNA damage phenotype. We produced and examined mice with transgenically (RNAi) silenced Sly and demonstrated that these mice have increased incidence of sperm with DNA damage and poorly condensed and insufficiently protaminated chromatin. We also investigated the contribution of each of the two Sly-encoded transcript variants and noted that the phenotype was only observed when both variants were knocked down, and that the phenotype was intermediate in severity compared with mice with severe NPYq deficiency. Our data demonstrate that Sly deficiency is responsible for the sperm DNA damage/chromatin packaging defects observed in mice with NPYq deletions and point to SLY proteins involvement in chromatin reprogramming during spermiogenesis, probably through their effect on the post-meiotic expression of spermiogenic genes. Considering the importance of the sperm epigenome for embryonic and fetal development and the possibility of its inter-generational transmission, our results are important for future investigations of the molecular mechanisms of this biologically and clinically important process.
Key words: Sperm DNA damage, Chromatin remodeling, Y chromosome, Male infertility
Introduction
Deletions of the Y chromosome are frequently associated with spermatogenic defects both in mice and in humans. In mice, the male specific, non-pairing Y chromosome long arm (NPYq) encompasses ∼90% of the Y-specific DNA content and comprises mostly repetitive sequences including multiple copies of at least four distinct genes that are expressed in spermatids: Ssty1/2, Sly, Asty, Orly (Touré et al., 2005). These genes show a progressive reduction in transcript levels with increasing NPYq deficiency and are candidates for contributing to the sperm defects associated with NPYq deletions. Mice with severe NPYq deletions are infertile in vivo and in vitro and all the sperm have head shape defects (Burgoyne et al., 1992). Live offspring were obtained from the infertile males when intracytoplasmic sperm injection (ICSI) was used (Ward and Burgoyne, 2006; Yamauchi et al., 2009), but low efficiency of assisted reproduction suggested that sperm impairment reached beyond their inability to transmit the paternal genome to the oocyte in vivo, and might have involved DNA changes.
In support of this notion we have recently shown that sperm from mice with severe NPYq deficiencies had DNA damage and abnormal chromatin packaging (Yamauchi et al., 2010). Epididymal sperm from mice with severe NPYq deficiency (i.e. deletion of nine-tenths or the entire NPYq gene complement) were impaired in oocyte activation ability following ICSI, and there was an increased incidence of oocyte arrest and paternal chromosome breaks. Comet assays revealed higher DNA damage in both epididymal and testicular sperm from these mice relative to controls, with epididymal sperm the more severely affected. Moreover, epididymal sperm from mutant mice also suffered from impaired membrane integrity, abnormal chromatin condensation and suboptimal chromatin protamination, making it likely that the increased DNA damage associated with NPYq deficiency is a consequence of disturbed chromatin remodeling during spermiogenesis.
Sly (Sycp3-like Y-linked) is present on NPYq in at least 100 copies, 70 of which retain an open reading frame, and encode a protein that is highly expressed in round spermatids, with nuclear localization at spermiogenesis steps 2/3 to 9 (Cocquet et al., 2009). The SLY protein contains a conserved COR1 domain implicated in chromatin binding (Ellis et al., 2005). The characterization of ‘shSLY mice’, in which Sly expression has been knocked down by transgenically-delivered short hairpin RNAs, showed that Sly deficiency is the major underlying cause of the sperm head anomalies and infertility associated with NPYq deficiency (Cocquet et al., 2009). This study preceded our finding of chromatin packaging defects and increased DNA damage in NPYq-deficient mice (Yamauchi et al., 2010), and it is therefore not known if shSLY mice recapitulated this aspect of NPYq-deficient phenotype.
In the present study we tested the hypothesis that Sly deficiency is responsible for the sperm DNA damage/chromatin packaging defects observed in mice with NPYq deletions, and established Sly transcript requirements for this phenotype.
Results
Characterization of a new shSLY line in which Sly1 but not Sly2 expression is knocked down
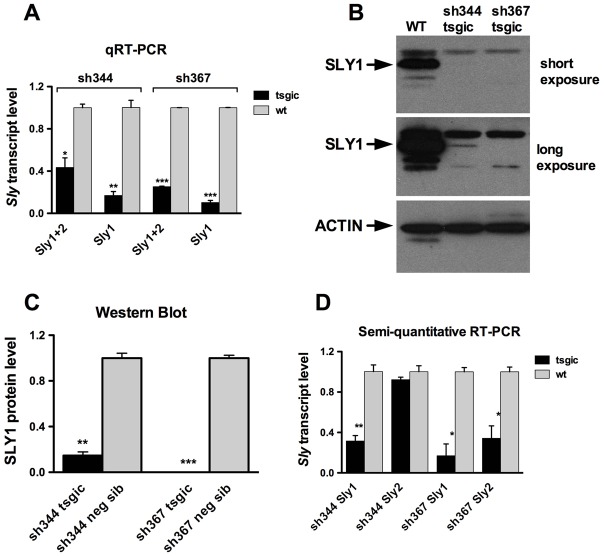
The NPYq specific gene Sly encodes two transcript variants, Sly_v1 and Sly_v2 (Reynard et al., 2009), hereafter called Sly1 and Sly2 (Fig. 1A). Sly1 is a full-length isoform and encodes a ∼40 kDa protein (referred to as SLY1), detected by our anti-SLY1 antibody (Reynard et al., 2009). Exons 5–6, which arose from a duplication of exons 3–4 (Ellis et al., 2007), are spliced out in the Sly2 isoform. Our anti-SLY1 antibody does not detect SLY2 protein but Sly2 can be translated: a transgene-derived SLY2 protein fused to a FLAG tag was detectable by an anti-FLAG antibody (supplementary material Fig. S1). We previously reported that, on an MF1 background, transgenic delivery of Sly-specific short hairpin sh367 RNAs led to significant decrease of Sly RNA and protein levels, with both splice variants being affected (Cocquet et al., 2009). These Sly-deficient males presented severe sperm head abnormalities and reduced fertilizing abilities. Here, we characterized a new line, sh344 (Fig. 1A,B), which was not reported on earlier. As in line sh367, expression of sh344 RNA was associated with a decrease of global Sly transcript level and of SLY1 protein level in the testes: sh367 mice showed 75% reduction of global Sly (Sly1+2) and 90% reduction of Sly1 transcripts while in sh344 the reduction was ∼50% of Sly1+2 and ∼80% of Sly1 transcripts (Fig. 2A; supplementary material Fig. S2A). No SLY1 protein was detected in sh367 testes by western blotting, even with prolonged exposure, whereas in sh344 SLY1 the protein level was decreased by 85% (Fig. 2B,C).
Fig. 1.
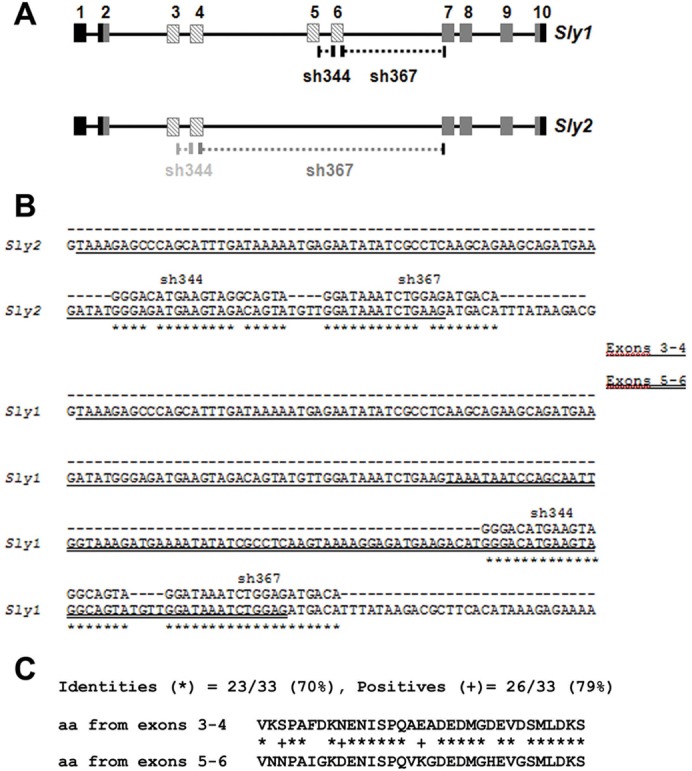
Structure of Sly gene. (A) Schematic diagram of the structure of the Sly gene (GeneID: 100040308) and of its alternative splice variants: Sly1 and Sly2. The boxes numbered 1 to 10 represent exons, with the coding region in gray and alternatively spliced exons in hatched gray. Short hairpin RNAs (sh344 and sh367) were designed to target sequences located in exons 5 and 6 (indicated in black beneath the schematic of the Sly gene). Because exons 3–4 share ∼86% identity with exons 5–6, sh344 and sh367 can partially recognize sequences in exons 3–4 (indicated in grey beneath the schematic of the Sly gene). (B) Alignment of sh344 and sh367 sequences with Sly1 and with Sly2 cDNA sequence. (C) Alignment of the amino acid sequence derived from exons 3–4 and exons 5–6 of Sly gene.
Fig. 2.
shRNA disrupts Sly expression in sh367 and sh344 differently. (A) Sly transcripts levels in shSLY transgenic mice (tsgic, n = 3 for each sh344 and sh367) and controls (neg sib, n = 2 for each sh344 and sh367) obtained by real-time RT-PCR with actin as a loading control. (B) Western blots of SLY1 protein in testis extract from shSLY transgenic mice and controls with actin as loading control. (C) Levels of SLY1 protein expression in shSLY transgenic mice (tsgic, n = 4 for sh344 and n = 3 for sh367) and controls (n = 2 for sh344 and n = 3 for sh367) quantified with ImageJ software and normalized to actin signal. (D) Sly1 and Sly2 transcript levels in shSLY transgenic mice (n = 3 for each sh344 and sh367) and controls (n = 2 for each sh344 and sh367) obtained by semi-quantitative RT-PCR, quantified with ImageJ software and normalized to actin. In A, C and D values are means ± s.e.m. Statistical significance with respect to corresponding control: *P<0.05, **P<0.01, ***P<0.001 (t-test). Similar results were obtained for mice on C57BL/6 and MF1 backgrounds. Neg sib, negative siblings. Primer sequences are shown in supplementary material Table S3.
Because exons 3–4 are very similar to exons 5–6 (∼86% identity, Fig. 1C) we failed to design real-time PCR primers that were specific to Sly2 transcripts. We therefore quantified the expression of Sly1 and Sly2 transcripts independently with semi-quantitative RT-PCR, using primers, which span the alternatively spliced exons 5–6. These experiments demonstrated that, in line sh344, Sly1 but not Sly2 transcripts were knocked down, while both Sly1 and Sly2 transcripts were knocked down in line sh367 (Fig. 2D; supplementary material Fig. S2C). Alignment of Sly2 transcript sequence with sh344 and sh367 RNA sequences shows the presence of two mismatches with sh344 sequence while there is only one with sh367 sequence (Fig. 1B). Owing to the lack of an SLY2-specific antibody, we could not measure/observe SLY2 protein level, but one can assume that SLY2 level mimics that of Sly2 transcript and, as a consequence, SLY2 protein level is expected to be unchanged in sh344 testes but decreased in sh367 testes.
These expression analyses demonstrate that, on a predominantly C57BL/6 background, sh367 is deficient for both Sly1 and Sly2 (i.e. Sly-deficient males) while sh344 line is only deficient for Sly1 (i.e. Sly1-only-deficient males). To determine the impact of each Sly variant on sperm differentiation, we analyzed and compared the testicular phenotypes of these two lines.
Sly-deficient but not Sly1-only-deficient males have increased sperm head shape defects and impaired sperm function in vitro
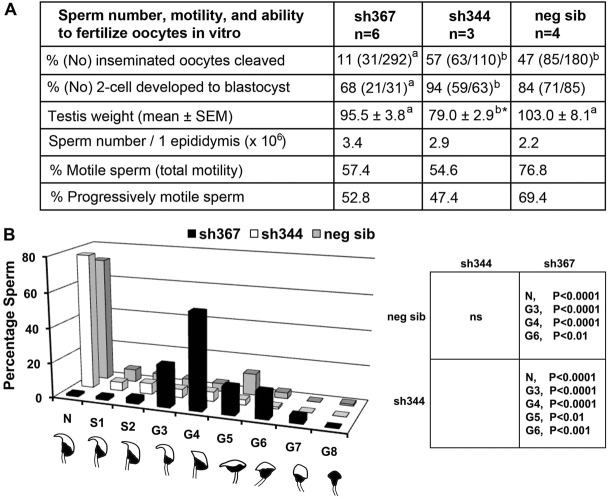
Sly-deficient sh367 mice were previously shown to have impaired sperm function on MF1 genetic background (Cocquet et al., 2009). We sought to determine if C57BL/6 sh367 mice and Sly1-only-deficient mice (i.e. sh344) also presented impaired sperm function. When we assessed fertility in vitro, sperm from sh367, but not from sh344 mice, had significantly impaired ability to fertilize oocytes and those oocytes that were successfully fertilized exhibited poorer development to blastocyst (Fig. 3A). This sperm dysfunction was not related to decrease in testis size, decrease in cauda epididymal sperm number or insufficient sperm motility, which were all normal and comparable between both Sly- and Sly1-only-deficient lines and controls. Detailed cauda epididymal sperm head shape analysis differentiating between sperm that were normal, slightly deformed, and grossly deformed demonstrated that in Sly-deficient sh367 mice virtually all sperm were morphologically abnormal, and had gross head shape defects. However, Sly1-only-deficient mice were not significantly different from non-transgenic siblings, both having >85% of morphologically normal sperm (Fig. 3B).
Fig. 3.
Sperm analyses in Sly-deficient mice. The analysis was performed for two transgenic lines with Sly deficiency (sh344 and sh367) and non-transgenic siblings controls (neg sib). (A) Analysis of sperm number, motility and ability to fertilize oocytes in vitro. Numbers with different superscripts within rows are statistically different (P<0.05); *Testes from only two males were weighted. (B) Sperm morphology analysis. Data shown in the y-axis are the percentage of sperm analyzed. Three hundred sperm were scored per male, with number of males n = 3, n = 3 and n = 5 for sh344, sh367, and neg sib, respectively. The x-axis represents individual categories of sperm head shapes: N, normal; S1–S2, slight defects; G3–G8, gross defects. Examples of each head shape are shown under each sperm morphology designation. The table on the right shows significant differences between the genotypes in the proportion of specific morphology defects (two-way ANOVA). ns, non statistically significant.
To summarize, sperm analyses point to severe sperm head shape defects and impairment in the ability of sperm to fertilize oocytes in vitro in Sly-deficient, but not in Sly1-only-deficient mice.
Sly-deficient but not Sly1-only-deficient males have increased sperm DNA damage
We have previously shown that testicular sperm from normal wild-type mice have more comet assay detectable sperm DNA fragmentation than cauda epididymal sperm, and that freezing increases DNA damage in sperm from both sources; in mice with severe NPYq deficiencies this sperm DNA fragmentation is markedly increased with epididymal sperm more affected than testicular sperm, and with both types of sperm more susceptible to freezing (Yamauchi et al., 2010). Here, we tested fresh and frozen cauda epididymal and fresh and frozen testicular sperm from Sly (sh367)- and Sly1-only (sh344)-deficient transgenic lines, with their negative siblings as controls.
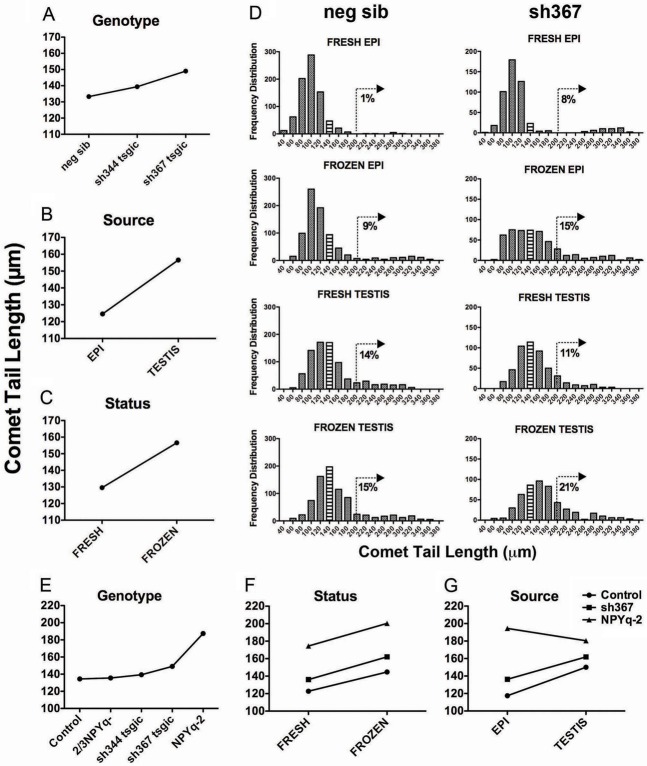
As in our previous study, because of the non-normality of the underlying data, we have used male means for the analysis. A preliminary ANOVA analysis revealed no significant differences between sh344 and sh367 negative siblings, so the control data were pooled. We then carried out ANOVA analysis of sh344, sh367, and pooled controls, with genotype, sperm source (testis or epididymis) and sperm status (fresh or frozen) as factors (Fig. 4A–C). As in controls described in our previous study, in all genotypes there was a significant (P<0.000001) effect of sperm source (testicular sperm more affected than epididymal; Fig. 4B) and sperm status (frozen sperm more affected than fresh sperm, Fig. 4C). There was also a significant effect of genotype (Fig. 4A, P = 0.0004) with sh367 mice, but not sh344 mice, having significantly (P = 0.00009) longer comet tail lengths than controls. The histograms of the raw data for each sperm type shed light on the nature of sperm DNA damage revealed by the ANOVA analysis (Fig. 4D). In the controls the baseline is provided by fresh epididymal sperm, in which 99% (792/800) of sperm fall in a normally distributed population of sperm with comet tails in the range 41–188 µm, and the remaining 1% (8/800) with comet tails ranging from 210 to 308 µm, indicating severe sperm DNA fragmentation. Therefore, as a yardstick for increases in sperm DNA damage for the remaining distributions we have calculated the percentage of sperm with comet tail lengths exceeding 200 µm. Freezing the epididymal sperm shifts the entire distribution a little to the right, and increases the proportion of sperm with severe DNA damage; as a consequence 9% (69/800) of sperm now have comet tail lengths exceeding 200 µm. Comparison of distributions of fresh testicular sperm with fresh epididymal sperm (14%, 113/800 versus 1%, 8/800 >200 µm) indicates a more marked shift of the distribution to the right but a less marked increase in sperm with the highest DNA damage. In agreement with the ANOVA analysis, sperm in sh367 mice are more affected than in controls resulting in further increases in the proportion of sperm with comet tails >200 µm in all categories except fresh testis; nevertheless, for fresh testis it is apparent that the ‘normal’ part of the distribution has moved to the right which is consistent with the ANOVA result (Fig. 4D).
Fig. 4.
Comet assay analyses of sperm from Sly-deficient mice. (A–C) Sly-deficient mice, sh344 tsgic and sh367 tsgic, and their non-transgenic siblings (neg sib) were compared for the effects of genotype (A), sperm source (B; epididymal and testicular), and sperm status (C; fresh and frozen). (D) The frequency distribution of all comet tail lengths for each sperm type from neg sib and sh367. (E–G) Sly-deficient mouse lines were compared with mice with moderate (2/3NPYq-) and severe (NPYq-2) NPYq deficiency. The controls here were negative siblings of shSLY mice and wild-type controls of mice deficient for NPYq. In F and G, sh344 and 2/3NPYq- data are omitted due to lack of significant difference from controls. The number of males was n = 8, n = 10, n = 3, n = 5, n = 2, and n = 2 for neg, sib, controls, sh344 tsgic, sh367 tsgic, 2/3NPYq- and NPYq-2, respectively; 100 sperm were scored per sperm type (fresh epididymal, frozen epididymal, fresh testicular, frozen testicular) per male. Bin size: 20.
Next, we compared Sly-deficient mice with mice with moderate (2/3NPYq-) and severe (NPYq-2) NPYq deficiency (Fig. 4E–G). The controls in this comparison were negative siblings of sh344 and sh367 mice and wild-type controls of NPYq-deficient mice; the controls did not differ from each other and were therefore pooled. ANOVA analysis demonstrated a significant (P<0.000001) effect of genotype (Fig. 4E) with sh367 and NPYq-2, but not 2/3NPYq- and sh344, mice significantly different from controls. In both Sly and NPYq-deficient mice, there was a significant effect of sperm status (Fig. 4F; P<0.000001) with frozen sperm having significantly longer comet tail lengths than fresh; the same effect was seen in controls. However, while sh344, sh367 and control mice had overall longer comet tail lengths in testicular sperm than in epididymal sperm, this was not true for NPYq-2 mice, in which epididymal sperm were more affected (Fig. 4G; genotype/source interaction P = 0.0007).
Overall, comet assay analysis points to Sly-deficient but not Sly1-only-deficient mice having an increased susceptibility to sperm DNA damage. However, this DNA damage is less severe than in mice with complete NPYq deficiency, and is not preferentially affecting epididymal sperm as was observed in severely NPYq-deficient mice.
Sly deficiency is not associated with impairment of early fertilization steps after ICSI and does not lead to increased incidence of paternal chromosome breaks in the zygotes
We have previously shown that when cauda epididymal sperm from mice with severe NPYq deficiencies were injected into the oocytes by ICSI, there was an impairment in oocyte activation, increased incidence of oocyte arrest at the pronuclei stage, and presence of paternal chromosome breaks in the zygote (Yamauchi et al., 2010). Here, following the design of the previous study, we injected fresh and frozen cauda epididymal and fresh and frozen testicular sperm from Sly (sh367)- and Sly1-only (sh344)-deficient mice (and transgene-negative siblings serving as controls), into the oocytes and examined early post-fertilization steps.
No significant differences between the same sperm types from Sly- and Sly1-only-deficient mouse lines and controls, and between epididymal and testicular sperm regardless of whether sperm were fresh or frozen (‘within genotype’ comparison) were observed in incidence of oocyte activation, oocyte arrest, and oocytes with normal paternal chromosomes (supplementary material Table S1). We also carried out a single analysis of Sly-deficient mice and controls by ANOVA with genotype, sperm source (testis or epididymis) and sperm status (fresh or frozen) as factors (data not shown). In all measured end points there was no effect of genotype.
We then placed the data obtained with sh367 and sh344 mice in perspective of data obtained earlier with NPYq-deficient mice (Yamauchi et al., 2010) (supplementary material Fig. S3), pooling the data obtained after ICSI with four types of sperm, and using as a control pooled controls from both studies. Single ANOVA analysis demonstrated a significant (P<0.001) effect of genotype, with sh367, sh344 and 2/3NPYq mice being no different from controls and significantly less affected than mice with severe NPYq- deficiency (9/10NPYq- and NPYq-2) in incidence of arrested oocytes (supplementary material Fig. S3A), oocytes with normal paternal chromosomes (supplementary material Fig. S3B), and aberration rate (supplementary material Fig. S3C).
To assess whether lack of ‘ICSI phenotype’ in Sly-deficient mice was due to milder sperm head shape defects and unintentional preselection during injection, we compared sh367 frozen cauda epididymal sperm purposely preselected according to their head shape into two groups: sperm with slight (categories S1–2 shown in Fig. 3) and gross (G4–6 shown in Fig. 3) head shape defects (supplementary material Fig. S4). No differences in the incidence of oocytes activated, arrested, and with normal paternal chromosomes were noted after injection of sperm with slight versus gross head shape defects (Fisher's exact test).
Overall, ICSI data imply that contrary to what is seen in mice with severe NPYq deficiency, Sly knockdown does not impair sperm function in assisted fertilization. Sperm from Sly-deficient mice activate oocytes normally yielding no chromosome damage in the ICSI-generated zygotes.
Sly-deficient but not Sly1-only-deficient mice have poor sperm chromatin condensation
Transmission electron microscopy was used to examine chromatin compaction in cauda epididymal sperm from sh367 Sly-deficient mice, sh344 Sly1-only-deficient mice, and their negative siblings as controls (Fig. 5). Sperm were categorized into three categories: those with properly condensed, slightly decondensed, and severely decondensed chromatin (Fig. 5A–C). In controls and sh344 mice the vast majority of sperm had properly condensed chromatin (87% and 77%); less than 1% of sperm had severely decondensed chromatin. In sh367 males, sperm with properly condensed chromatin were significantly less frequent (50%) and the incidence of sperm severely affected increased to 3.4%. Single ANOVA analysis comparing the incidence of sperm with properly condensed chromatin in these three genotypes revealed that sh367 mice were significantly different from control (P = 0.00002) and from sh344 (P = 0.003; Fig. 5D).
Fig. 5.
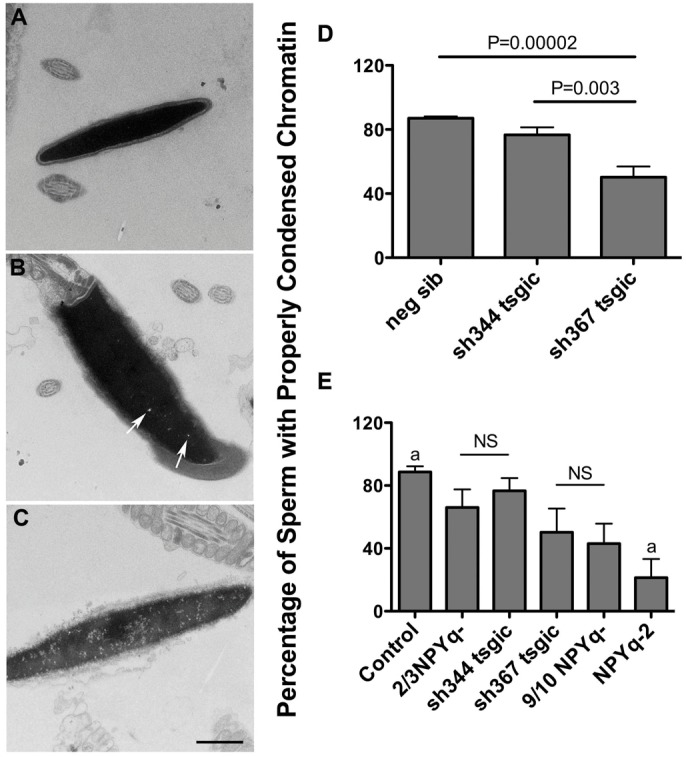
Transmission electron microscopy analysis of chromatin condensation in sperm from Sly-deficient mice. (A–C) shows examples of levels of chromatin condensation: (A) properly condensed chromatin; (B,C) slightly (arrows) and severely decondensed chromatin, respectively. (D,E) Two transgenic mouse lines with Sly deficiency (sh367 and sh344) and their negative siblings serving as controls were compared in single ANOVA analysis, revealing that sh367 had significantly less sperm with properly condensed chromatin than controls and sh344 (D). When Sly-deficient mice were put in the context of mice with moderate (2/3NPYq-) and severe (9/10NPYq- and NPYq-2), single ANOVA analysis showed a significant effect of genotype (P<0.000001), with decrease in proportion of sperm with properly condensed chromatin in order: control>2/3NPYq- = sh344>sh367 = 9/10NPYq->NPYq-2 (E). Statistical significance: a = different (P<0.05) from all other genotypes; NS = non significant. Values are means ± s.e.m. Number of males examined: n = 7, n = 3, n = 5, n = 7, n = 13, n = 3, n = 2, n = 3 for neg sib, sh344, sh367, control, 2/3NPYq-, 9/10NPYq-, and NPYq-2, respectively; 100 sperm heads were scored per male. Scale bar: 1 µm.
When the data obtained with Sly- and Sly1-only-deficient mice were put in the context of data obtained before with sperm from mice with NPYq deficiencies (Yamauchi et al., 2010), single ANOVA analysis showed a significant effect of genotype (P<0.000001; Fig. 5E). All tested genotypes were significantly different than controls, which in this analysis were pooled controls from both studies. However, a clear gradation in chromatin condensation deficiency could be observed, with sh344 mice being similar to mice with a moderate NPYq gene loss (2/3NPYq-), sh367 mice being similar to those with extensive NPYq deletion (9/10NPYq-), and mice lacking the entire NPYq gene complement being most affected.
Overall, the data point to Sly-deficient but not Sly1-only-deficient mice exhibiting impairment in sperm chromatin condensation.
Mice with severe NPYq deficiency and Sly-deficient mice have impaired sperm chromatin protamination
Poor chromatin condensation can result from insufficient DNA protamination. Chromomycin A3 (CMA3) is a fluorescent dye that intercalates into DNA but because of its size it can only bind to sperm DNA when it is not completely condensed and supposedly loosely bound to DNA binding proteins. Therefore, CMA3 staining has been used as an indirect measure of the level of protamination in sperm, in humans, mice and other species (Bianchi et al., 1993; Delbes et al., 2007; Meyer-Ficca et al., 2009; Noblanc et al., 2012; Simões et al., 2009). Here, we used this approach to examine cauda epididymal sperm from Sly- and Sly1-only-deficient mice and their negative siblings as controls (Fig. 6). Because CMA3 staining was not included in our previous analysis of NPYq-deficient mice (Yamauchi et al., 2010), we also tested three NPYq-deficient genotypes and their specific controls.
Fig. 6.
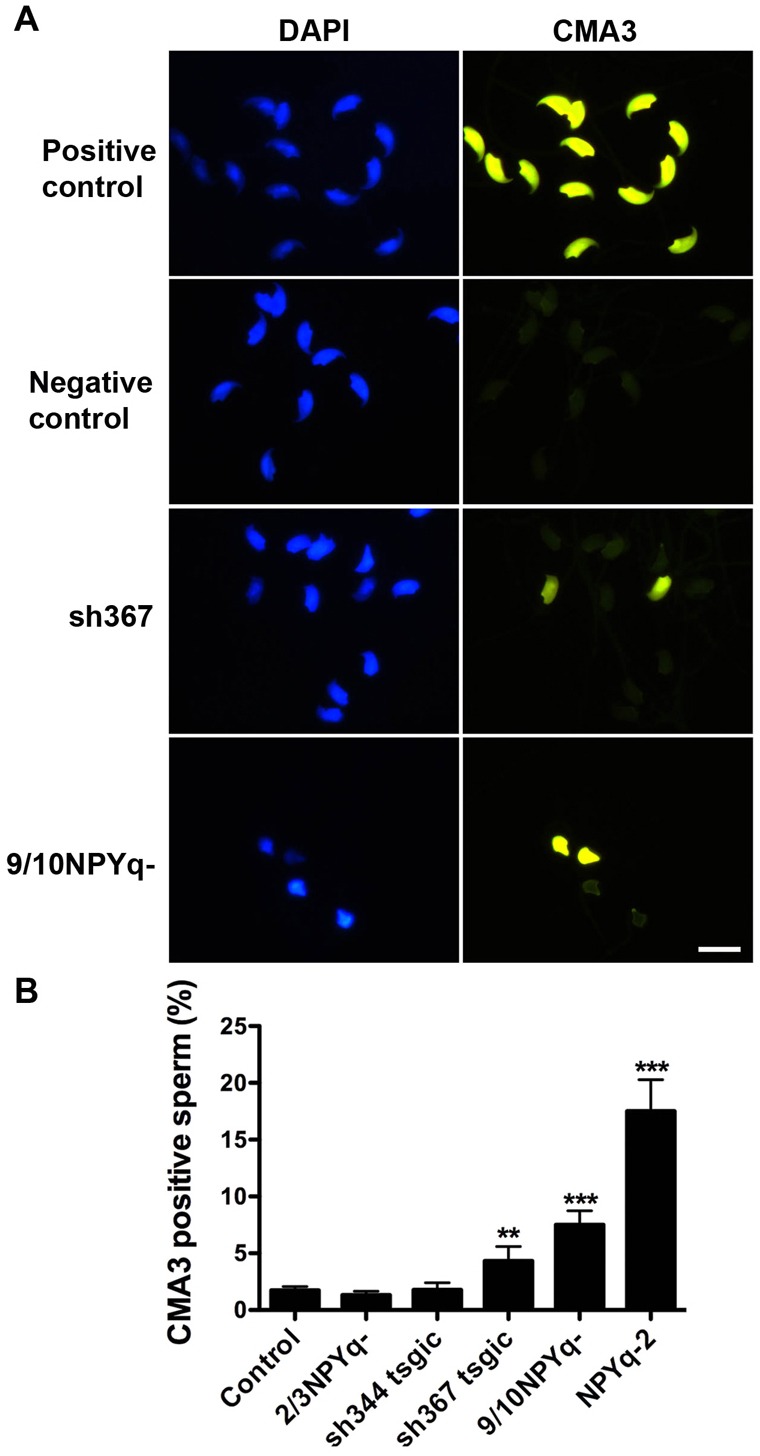
CMA3 staining of sperm from Sly-deficient and NPYq-deficient mice. (A) Examples of CMA3 staining (yellow) of epididymal sperm from Sly-deficient mice (sh367) and mice with severe NPYq deficiency (9/10NPYq-); the presence of CMA3-positive sperm indicates that these mice have impaired sperm chromatin protamination. Positive controls were sperm treated with 2 mM DTT/0.5% Triton X-100 to reduce disulfide bridges and destabilize sperm chromatin. Negative controls were sperm from negative siblings of sh367 mice. DAPI (blue) was used to stain nuclei. Scale bar: 10 µm. (B) Two Sly-deficient lines (sh344 and sh367) and mice with moderate (2/3NPYq-) and severe (9/10NPYq- and NPYq-2) NPYq deficiency were compared to controls. The control represent negative siblings of Sly-deficient males and wild-type controls of NPYq-deficient mice, which were not different from each other and were therefore pooled. 100 sperm were scored per male, and the number of males examined per genotype was: n = 27, n = 9, n = 6, n = 3, n = 6, n = 6 for controls, sh344, sh367, 2/3NPYq-, 9/10NPYq-, and NPYq-2, respectively. Statistical significance: **P<0.01; ***P<0.001 versus control. Values are means ± s.e.m.
The proportion of CMA3 positive sperm was similar among the controls justifying pooling for the next comparisons. Single ANOVA analysis on pooled controls and all Sly- and NPYq-deficient genotypes (Fig. 6B) revealed a significant effect of genotype (P<0.0001) with sh367 (P = 0.0098), 9/10NPYq- (P = 0.000013) and NPYq-2 (P<0.000001) having more CMA3 positive sperm when compared to controls. Mice with moderate NPYq gene loss (2/3NPYq-) and sh344 mice were not significantly different from controls, and were significantly less affected than the remaining three genotypes. The results from this multi-genotype ANOVA were further supported by one-to-one ANOVA comparisons of each genotype with its specific control. In this analysis sh344 and 2/3NPYq- were not different from their specific controls, but sh367 (P = 0.017), 9/10NPYq- (P = 0.0007) and NPYq-2 (P = 0.0001) were.
In support for CMA3 analysis we also examined protamine processing in cauda epididymal sperm from sh367 mice using acid-urea gel electrophoresis and western blotting (supplementary material Fig. S5), following the strategy described earlier for NPYq-deficient mice (Yamauchi et al., 2010). Separated sperm nuclear proteins corresponding to the same sperm number were assessed on Coomassie-Blue-stained gels and by immunoblot with Hub2B and PreP2 antibodies recognizing mature and immature protamine P2 forms, respectively. The analysis was performed with two-way ANOVA, with gel and genotype as factors. We did not observe significant differences between Sly-deficient and control mice in the intensities of bands representing mature protamines. No preP2 bands originating from sperm were detected on any of Coomassie-Blue-stained gels. When the gels were blotted with anti-preP2 antibody, bands were detected in four out of five Sly-deficient and two out of five control males. Although the difference in average preP2 band intensity did not reach significance, it was more than twice higher in sh367 mice.
Overall, mice with severe NPYq deficiency had significantly increased incidence of CMA3 positive sperm suggesting impaired protamination; Sly-deficient mice, but not Sly1-only-deficient mice, exhibited the same phenotype but were less severely affected. Impaired protamination in sperm from Sly-deficient mice was also shown directly using nuclear protein analysis but, as in CMA3 analysis, it was much less severe than that noted before for mice with severe NPYq deficiency (Yamauchi et al., 2010).
Sly1-only-deficient males have a minor de-repression of X-and Y-encoded spermiogenic genes compared to global de-repression in Sly-deficient males
Sly-deficient males display a remarkable upregulation of sex chromosome genes after meiosis and we have proposed that the multiple defects associated with Sly deficiency (head shape malformation, impairment of sperm function, etc.) may likely be a consequence of the massive and global upregulation of spermiogenic genes (Cocquet et al., 2009). Since Sly1-only-deficient males do not display any of the above-mentioned defects we have analyzed the level of expression of several X and Y spermiogenic genes. Results presented in Fig. 7 show a de-repression of some of the X and Y encoded genes analyzed in Sly1-only-deficient males but this de-repression is significantly less pronounced than in Sly-deficient males. Microarray analysis (supplementary material Table S2) validated the qRT-PCR data, with sh344 males showing the same widespread overexpression of X-linked spermatid genes previously seen in sh367 (Cocquet et al., 2009), but to a much lesser extent.
Fig. 7.
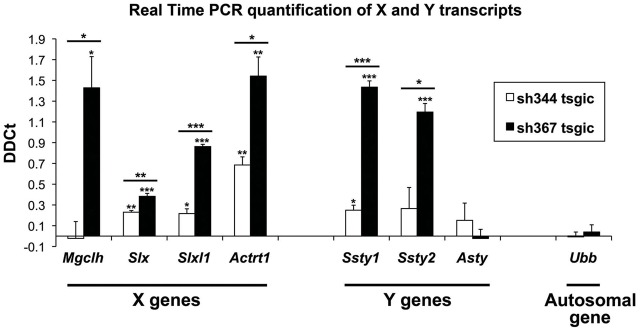
X-Y gene upregulation in Sly-deficient mice. Gene expression in testis from two Sly-deficient lines (sh344 and sh367) was analyzed by real-time PCR. Values plotted are ΔΔCt ± standard errors. The difference in PCR cycles with respect to the spermatid-specific gene Acrv1 (ΔCt), the expression of which is not regulated by Sly, for a given experimental sample was subtracted from the mean ΔCt of the reference samples (negative siblings of each transgenic line) (ΔΔCt). Three to five animals were analyzed per genotype. Asterisks indicate significant difference between sh344 and sh367 or between Sly-deficient mice and control values (t-test) with *P<0.05, **P<0.01 and ***P<0.001. Primer sequences are shown in supplementary material Table S3.
Discussion
Even relatively minor errors in chromatin remodeling during spermiogenesis are associated with sperm DNA damage and infertility, yet little is known about the etiology. Mice with severe NPYq deletions are infertile due to severe sperm differentiation defects (Ward and Burgoyne, 2006; Yamauchi et al., 2009). We have recently observed that sperm from these mice presented abnormal chromatin packaging and DNA damage. Moreover, when these sperm were injected into the oocytes, a significant increase of oocyte arrest at pronuclei stage and of chromosome aberrations in the fertilized eggs were noted (Yamauchi et al., 2010). Here we provide evidence that the deficiency of NPYq encoded gene Sly is associated with sperm DNA damage and poor sperm chromatin condensation, and propose that SLY plays a role in spermatid-specific chromatin remodeling.
How can Sly/SLY be involved in sperm DNA damage phenotype? SLY protein has been shown to control the postmeiotic expression of >100 genes, the majority of which are encoded on sex chromosomes. In Sly-deficient males, the repressive epigenetic marks normally associated with the sex chromosomes (i.e. CBX1 and H3K9me3) are decreased, which leads to the upregulation of many X- and Y-encoded spermiogenic genes (Cocquet et al., 2009). The multiple spermiogenesis defects associated with Sly deficiency, including the sperm DNA damage and abnormal chromatin packaging described in the present study, are likely a consequence of this massive change in spermiogenic gene expression. In Sly1-only-deficient males, there are no clear defects of the spermiogenesis function, and the upregulation of spermiogenic X and Y genes is very limited. In all probability, a certain threshold of upregulation must be reached to significantly alter sperm differentiation and function.
In theory, the sperm differentiation defects observed in Sly-deficient and NPYq-deficient mice could result from the deregulation of any of the >100 deregulated genes, or from the collective deregulation of several of them. Nevertheless, several genes appear to be promising individual candidates. For instance, alteration in the expression of H2al1 and H2al2y genes (Ferguson et al., 2009), which encode spermatid specific histone variants (Govin et al., 2007), could affect sperm chromatin protamination and therefore spermiogenesis. A few autosomal genes were also downregulated in Sly-deficient spermatids (Cocquet et al., 2009). Among them, the gene Chaf1b is a very likely candidate to explain sperm DNA damage since it encodes a component of the Chromatin Assembly Factor 1 (CAF-1), which is involved in the DNA damage and repair process (Groth et al., 2007). RNAi silencing of the CHAF1B in proliferating human cells led to accumulation of double strand DNA breaks, increased level of phosphorylated histone H2AX, and ultimately cell death (Nabatiyan and Krude, 2004).
Sly genes are estimated to be present in between 70 and 100 copies in the mouse genome; the majority of these encode two transcript splice variants, Sly1 and Sly2 (Cocquet et al., 2009; Ellis et al., 2011; Scavetta and Tautz, 2010). In the present study, we have produced and characterized a Sly1-only-deficient mouse model, and shown that, in contrast to Sly-deficient mice, in which both transcript variants are affected, Sly1-only-deficient mice do not have sperm differentiation defects. From our analyses, we can draw two hypotheses to explain the phenotypic differences between those two models. Sly2 may be the important functional variant and its knockdown would be solely responsible for the de-repression of spermiogenic genes and the various subsequent sperm defects observed in Sly-deficient mice, or it is the global reduction of both Sly variants that matters. Several arguments can be put forward against the former scenario. First, SLY1 protein co-localizes with the loci that are deregulated in Sly-deficient males (Cocquet et al., 2009) and, therefore, is likely to play a role in the regulation of these loci. Second, SLY1 protein structure is very similar to that of SLY2. The COR1 domain, which is presumed to mediate chromatin binding, is identical in SLY1 and SLY2 proteins. SLY1 only differs from SLY2 in the 34 additional amino acids encoded by exons 5–6. Since exons 5–6 arose from a duplication of exons 3–4, the protein region encoded by exons 5–6 shares 70% identity and 79% similarity with the protein region encoded by exon 3–4 (Fig. 1C). All in all, it is more plausible that Sly1 and Sly2 genes encode proteins of similar function, and that the phenotype is correlated with the global level of knockdown of Sly1 and Sly2. Our Sly/SLY level quantification on whole testes confirms this assumption, with a lower reduction of global Sly (Sly1+2) transcript level and retention of some of SLY1 protein in phenotypically normal Sly1-only-deficient males, and a higher reduction of global Sly (Sly1+2) and no detectable SLY1 in affected sh367 Sly-deficient males (Fig. 2A,C; supplementary material Fig. S2A).
With respect to sperm chromatin packaging/DNA damage Sly-deficient mice recapitulate the NPYq-deficient phenotype only very moderately and much less significantly than other phenotypic features (Table 1). Moreover, Sly deficiency is not associated with impairment of early fertilization events after ICSI and does not lead to increased incidence of paternal chromosome breaks in the zygotes that are present in severely NPYq-deficient mice (supplementary material Table S1, Fig. S4), and this is not due to preselection of sperm with milder head shape defect (supplementary material Fig. S4). The difference in Sly transcript levels between mice with severe NPYq gene loss, i.e. deletion removing nine-tenths (9/10NPYq-, 4%) or the entire NPYq (NPYq-2, 0%), and Sly-deficient sh367 mice (25%, supplementary material Fig. S2A) could explain the differences in the phenotype in these mice. However, lack of detectable SLY1 protein in all genotypes and similar Sly1 transcript levels in Sly-deficient and 9/10NPYq- mice (10% both) put the dependence of the phenotype exclusively on Sly/SLY levels in question. Mice with severe NPYq deficiency lack other genes encoded on the Y long arm, which could contribute to spermiogenic defects in these mice. It, therefore, cannot be excluded that these other NPYq genes, which are not only present but also upregulated in Sly-deficient mice (Cocquet et al., 2009; Ellis et al., 2005), mitigate the sperm chromatin packaging/DNA damage phenotype and/or are involved in early embryonic events, either directly or indirectly through the regulation of other loci. The most plausible gene candidate would be Ssty1/2 (Spermatid-specific transcripts Y-encoded), which is present on the mouse NPYq in more than 100 copies, is specifically transcribed in the testis in round spermatids, and is the only other NPYq gene that is translated. Ssty1/2 belongs to the Spindlin family, one member of which has recently been shown to bind to methylated histone H3 (Wang et al., 2011); Ssty1/2 could therefore contribute to chromatin remodeling during spermiogenesis, along with Sly. Future work utilizing Sly and/or Ssty1/2 transgene additions to severely NPYq-deficient mice should help clarifying the contribution of Sly and Ssty1/2.
Table 1.
Phenotype characteristics of NPYq- and Sly-deficient mice
| Phenotypic effects | sh344 | sh367 | 2/3NPYq- | 9/10NPYq- | NPYq-2 |
| Sex chromosome gene upregulation | * | *** | ** | *** | **** |
| In vivo fertility | = | *** | * | **** | **** |
| In vitro fertility | = | *** | ** | **** | **** |
| Sperm number | = | = | = | ** | ** |
| Sperm motility | * | * | * | ** | ** |
| Sperm head morphology | = | ** | * | *** | **** |
| Chromatin fragmentation in comet assay | = | * | = | *** | **** |
| Oocyte activation in ICSI | = | = | = | *** | **** |
| Oocyte arrest in ICSI | = | = | = | *** | **** |
| Paternal chromosome breaks in zygotes after ICSI | = | = | = | *** | **** |
| Chromatin condensation in TEM | = | ** | * | ** | *** |
| Chromatin packaging in CMA3 staining | = | ** | = | *** | **** |
ICSI, intracytoplasmic sperm injection; TEM, transmission electron microscopy.
Estimated levels of impairment: = not affected; * slightly affected; ** moderately affected; *** severely affected; **** very severely affected. The levels of phenotypic effects of NPYq/Sly-deficient mice presented in this table are based on the present study and on our unpublished data, as well as on several previously published manuscripts from our group and few others (Cocquet et al., 2009; Ellis et al., 2005; Grzmil et al., 2007; Reynard et al., 2007; Styrna et al., 2002; Styrna et al., 2003; Touré et al., 2005; Touré et al., 2004; Ward and Burgoyne, 2006; Xian et al., 1992; Yamauchi et al., 2010; Yamauchi et al., 2009) and therefore should be considered an estimate.
What are the clinical implications of our work? Infertility is a major public health problem that affects ∼10%, or 72.4 million couples worldwide (Boivin et al., 2007). Male factor infertility contributes to at least half of infertility cases, and its primary cause is aberrant spermatogenesis. Packaging of the haploid genome into the sperm head requires extensive chromatin remodeling, during which histones are disassembled and replaced with protamines. Deficiencies in protamination that leave portions of DNA poorly compacted are associated with sperm DNA damage; in humans this is a risk factor for adverse clinical outcomes including poor fertilization, impaired development of the preimplantation embryo, miscarriage, and an increased risk of morbidity in the offspring (Lewis and Aitken, 2005). Recent evidence suggests that the remodeling of sperm chromatin leaves a significant epigenetic imprint in the form of retained histones and histone modifications (Brykczynska et al., 2010; Hammoud et al., 2009), and that disturbance of this epigenetic imprint during the remodeling could have clinically important consequences for offspring (de Boer et al., 2010; Ng et al., 2010). Indeed, a recent report on inter-generational paternal transmission of an acquired trait (impaired glucose-insulin homeostasis) in rats (Ng et al., 2010), which must have been mediated by an alteration of the epigenetic blueprint in sperm (Skinner, 2010), emphasizes the importance of the sperm epigenome for subsequent embryonic development.
All in all, a growing number of reports suggest that impairment of the sperm epigenome could have an impact on male fertility and on the health of the offspring; but the factors that lead to the retention of histones and their epigenetic marks in sperm are poorly understood. Our identification of Sly and its target genes as candidate chromatin remodeling regulators opens the door for future investigations of this clinically important process.
Materials and Methods
Chemicals
Mineral oil was purchased from Squibb and Sons (Princeton, NJ, USA) and equine (eCG) and human (hCG) chorionic gonadotrophin from Calbiochem (San Diego, CA, USA). All other chemicals were obtained from Sigma Chemical Co. (St Louis, MO, USA) unless otherwise stated.
Plasmid generation and breeding of transgenic mice
Mice shSLY carry a transgene expressing short hairpin RNAs targeting the specific degradation of Sly transcripts. Two shSLY lines were analyzed in the present study: shSLY367 (or sh367), which was previously characterized on an MF1 genetic background (Cocquet et al., 2009) and shSLY344 (or sh344; Fig. 1). A U6sh344 construct was produced using a PCR-based approach similar to that described in Harper et al. (Harper et al., 2005) with the primers shown in supplementary material Table S3. The PCR product was cloned into the pCR2.1 vector and sequenced (TOPO TA Cloning, Invitrogen, USA). The U6sh344 cassette was then subcloned into pEGFP-N1 plasmid (Clontech, Mountain View, CA, USA) using AflII restriction sites. The efficiency and specificity of the construct was checked by co-transfection as previously described for other U6shSLY constructs (Cocquet et al., 2009). The construct was then linearized by ApaL1 and BspH1, on-column purified, and microinjected into fertilized eggs from CBA/Ca6×C57BL/10. Transgenic animals carrying the U6sh344 construct were identified by PCR (supplementary material Table S3). Both shSLY lines were backcrossed to C57BL/6, to make these mice comparable with NPYq-deficient models used by us before for analysis of sperm DNA damage and chromatin remodeling (Yamauchi et al., 2010). shSLY males on a predominantly C57BL/6 (75–87.5%) genetic background were examined at 2–4 months of age. Non-transgenic siblings served as controls. Female B6D2F1 oocyte donors for in vitro fertilization (IVF) and ICSI (National Cancer Institute, Raleigh, NC) were used at 8- to 10-weeks old. The mice were fed ad libitum with a standard diet and maintained in a temperature- and light-controlled room (22°C, 14 h light/10 h dark), in accordance with the guidelines of the Laboratory Animal Services at the University of Hawaii and the National Research Council's (NCR) ‘Guide for Care and Use of Laboratory Animals’. The protocol for animal handling and treatment procedures was reviewed and approved by the Animal Care and Use Committee at the University of Hawaii.
Western blotting
Western blot analyses were performed as described previously (Reynard et al., 2009). Briefly, 10 to 15 µg of testis lysates were run on a 12% SDS/polyacrylamide gel. Following transfer and blocking, membranes were incubated overnight with a primary antibody (anti-SLY1 (Reynard et al., 2009) at 1:3,000 and anti-β actin at 1:50,000). Incubation with the corresponding secondary antibody coupled to peroxidase, and detection by chemiluminescence, were carried out as described by the manufacturer (SuperSignal West Pico, Pierce Biotechnology, Rockford, IL, USA). The analysis of sperm nuclear proteins using acid urea polyacrylamide electrophoresis (AU-PAGE) followed by western blotting were performed exactly as described by us earlier (Yamauchi et al., 2010).
Real-time RT-PCR
For real-time reverse transcriptase polymerase chain reaction (RT-PCR), total testis RNA or RNA from round spermatids purified with STAPUT method (Bellvé, 1993) was extracted using Trizol and DNaseI treatment (Ambion, Austin, TX, USA), and purified using an RNeasy kit (Qiagen, Valencia, CA, USA). Reverse transcription of polyadenylated RNA was performed with Superscript Reverse Transcriptase III, according to the manufacturer's guidelines (Invitrogen, Carlsbad, CA, USA). Real-time PCR was performed using SYBR Green PCR Master mix on an ABI StepOnePlus machine (Applied Biosystems, Carlsbad, CA, USA). PCR reactions were incubated at 95°C for 10 min followed by 35 PCR cycles (10 s at 95°C and 60 s at 60°C). For analysis of Sly expression, two types of PCR reactions were performed: (1) ‘Sly1+2’ amplifying both Sly_v1 and Sly_v2 transcripts (Reynard et al., 2009) (primers Sly Global) and (2) ‘Sly1’ amplifying only Sly_v1 (primers Sly Long). All reactions were carried out in triplicate per assay, and β-actin or Acrv1 (spermatid specific gene not regulated by Sly) was included on every plate as a loading control. The difference in PCR cycles with respect to β-actin or Acrv1 (ΔCt) for a given experimental sample was subtracted from the mean ΔCt of the reference samples (non-transgenic siblings) (ΔΔCt). For the quantification of Sly knockdown, values were further normalized to ΔΔCt values of the β-actin or Acrv1. Expression analysis was also done for several X and Y encoded transcripts to test for their upregulation in shSLY mice. Primer sequences are shown in supplementary material Table S3.
Semi-quantitative RT-PCR
RT-PCR was performed as described for real-time RT-PCR except that Sly and β-actin were run separately and with cycle number decreased (26 for Sly and 24 for β-actin) to stop the reaction prior to saturation. The products were electrophoresed, ImageJ software (Rasband, 2007) was used to measure band intensity, and Sly1 and Sly2 values were normalized to β-actin. Primer sequences are listed in supplementary material Table S3.
Gamete collection and embryo culture
Female mice were induced to superovulate with injection of 5 IU eCG and 5 IU hCG given 48 h apart. Oocyte collection and subsequent oocyte manipulation, including microinjections, were done in HEPES-buffered CZB medium [HEPES-CZB (Kimura and Yanagimachi, 1995)], with subsequent culture in CZB medium (Chatot et al., 1989) in an atmosphere of 5% CO2 in air. Sperm capacitation for IVF was done in T6 medium (Quinn et al., 1982). To obtain testicular sperm a portion of testis was cut off and minced in ETBS buffer (Kusakabe et al., 2001) or HEPES-CZB to release spermatogenic cells. To obtain epididymal sperm, caudae epididymides were dissected and sperm were expressed with needles into HEPES-CZB, ETBS, T6 or PBS. Spermatozoa were allowed to disperse for 2–3 min at room temperature. The samples of testicular or epididymal cell suspension were used for in vitro fertilization (IVF), intracytoplasmic sperm injection (ICSI), comet assay, transmission electron microscopy (TEM) analysis, chromomycin A3 (CMA3) staining, and for isolation of protein for AU-PAGE, or were frozen.
Epididymal and testicular sperm freezing
Aliquots of 10 µl of cauda epididymal sperm or testicular cell suspension in ETBS were loaded in 0.25 ml straws (Edwards Innovation, Spring Valley, VA, USA). Each straw was sealed with Critoseal (Oxford Labware, St Louis, MO, USA) and placed in a plastic holder floating on the surface of the liquid nitrogen for 10 min before immersion. For thawing, the straws were removed from the storage container and immersed in a water bath at 37°C for 10 min and the contents were expressed into a Petri dish. Spermatozoa were used immediately for ICSI or other analyses.
In vitro fertilization and embryo culture
Epididymal sperm were capacitated in T6 medium for 1.5 h at 37°C in a humidified atmosphere of 5% CO2. IVF was performed as previously described (Ajduk et al., 2006). Gametes were co-incubated for 4 h. After co-incubation, the oocytes were washed several times with HEPES-CZB, followed by at least one wash with CZB. Embryos were cultured in CZB and observed at different time points for proper development; 24 h (2-cell stage), 48 h (4- or 8-cell stage), 72 h (morula or early blastocyst), 96 h (blastocyst).
Intracytoplasmic sperm injection
Intracytoplamic sperm injection was carried out as previously described (Szczygiel and Yanagimachi, 2003), within 1–2 h from oocyte and sperm collection. Sperm-injected oocytes were transferred in CZB and cultured at 37°C. The survival and activation of injected oocytes was scored 1–2 h and 6 h after the commencement of culture, respectively. The oocytes with two well-developed pronuclei and extruded second polar body were considered activated.
Sperm chromosome analysis
Chromosome preparation and analysis were performed as described by us before (Yamauchi et al., 2010). Activated oocytes were transferred into CZB containing 0.006 mg/ml vinblastine, which was added to inhibit spindle formation and syngamy. Between 19 and 21 h after ICSI, oocytes were treated with 1% pronase (1000 tyrosine units/mg; Kaken Pharmaceuticals, Tokyo, Japan) for 5 min at room temperature to soften the zonae pellucidae. Oocytes were then treated with hypotonic solution (1∶1 mixture of 1% sodium citrate: 30% fetal bovine serum) for 5 min at 37°C or 10 min at 25°C. Chromosomes were spread on glass slides by the gradual fixation/air-drying method (Mikamo and Kamiguchi, 1983). The preparations were stained with 2% Giemsa (Merck, Darmstadt, Germany) in PBS (pH 6.8) for 10 min for conventional chromosome analysis. The percentages of zygotes with normal and abnormal chromosomes were determined. In addition to scoring normal versus abnormal karyoplates, we also calculated the incidence of chromosome aberrations, i.e. aberration rate, which represents the total number of aberrations divided by the number of oocytes examined. This aberration rate reflects the intensity of DNA damage. As chromosome aberrations we counted breaks, fragments, ring chromosomes, obvious translocations, and other structural chromosome defects detectable with conventional Giemsa staining.
Comet assay
Comet assay was performed using the Trevigen Kit (Trevigen, Gaithersburg, MD, USA, cat. no. 4250-050-K) under neutral conditions as described by us before (Yamauchi et al., 2007). For each sample tested, 100 DNA tails were photographed and analyzed per slide. The length of each tail was measured from the center of the comet to the end of the tail using ImageJ software (Rasband, 2007), and each tail was categorized into one of four tail types reflecting the severity of DNA damage. The severity of DNA damage increases proportionately with tail length and with tail type, from 1 to 4.
Sperm analyses
To analyze sperm number, motility and morphology, cauda epididymal sperm were released into HEPES-CZB, and incubated for at least 10 min at 37°C immediately before analysis. Sperm counts using a hemocytometer were the mean of three independent scorings per sample. For analysis of sperm morphology epididymal sperm were stained with silver nitrate as previously described (Mahadevaiah et al., 1998). Briefly, the sperm suspension (diluted as necessary with 0.9% NaCl) was smeared on three slides, allowed to dry, fixed in methanol and acetic acid (3∶1), and stained with silver nitrate. The slides were coded, and 100 sperm heads per slide were viewed at 1000× magnification and scored. Categorization of sperm head morphology was performed as previously described (Yamauchi et al., 2009).
Transmission electron microscope analysis
Cauda epididymal sperm were prepared for transmission electron microscopy (TEM) analysis as previously described (Yamauchi et al., 2010). Briefly, samples were fixed, dehydrated, and embedded in LX-112 epoxy resin (Ladd Research Industries, Willinston, VT, USA). Ultra-thin sections (60–80 nm) were collected on Formvar-coated copper grids, double stained with uranyl acetate and lead citrate, and viewed with a LEO 912 (Zeiss, Thornwood, NY, USA) TEM. For each sample tested, 100 sperm heads were photographed at 8000× magnification.
CMA3 staining
To indirectly visualize protamine-deficient DNA, CMA3 staining was applied to cryopreserved cauda epididymal sperm as previously described (Simões et al., 2009), with modifications. Frozen-thawed sperm were washed with PBS, fixed in methanol and acetic acid (3∶1, 5 min, on ice), smeared on microscope slides, and air-dried. The slides were then stained with CMA3 (0.25 mg/ml in McIlvaine buffer, pH 7.0, supplemented with 10 mM MgCl2) for 20 min. The slides were rinsed with buffer and mounted with polyvinyl alcohol mounting medium with DABCO (Sigma, 10981) supplemented with 1.5 µg/ml DAPI. As positive controls we used sperm subjected to deprotamination. To achieve deprotamination sperm were incubated in 5 mM DTT, 0.5% Triton X-100 in PBS for 15 min on ice, then layered on 0.5 ml of 1 M sucrose in 25 mM Tris-HCl, pH 7.4, centrifuged at 3000 g for 10 min at 4°C, resuspended with methanol–glacial acetic acid (3∶1) and fixed for 5 min on ice. Air-dried smears were incubated in 2 M NaCl, 2 mM DTT in H2O for 15 min at room temperature, washed thoroughly with distilled water, air-dried, and stained with CMA3, as described earlier. Microscopy images of the slides were captured using fluorescence Olympus IX81 microscope (Tokyo, Japan) with appropriate filter (460–470 nM), at 400× magnification. For analysis, at least 100 sperm heads of each sample were scored as either positive or negative, followed by measures of fluorescence intensity of each head using MetaMorph Digital Imaging Software (Olympus).
Experimental design and statistical analyses
In the analysis of oocyte activation, oocyte arrest, and incidence of abnormal karyoplates, Mantel-Haenszel χ2-test, which allows for inter-male variability by keeping the individual male data separate was used for ‘within genotype’ comparisons and Fisher's exact test was used to assess differences between the genotypes. These two tests were also used to analyze the TEM data. Expression data were analyzed with t-test. ANOVA (generalized linear model as provided by NCSS statistical analysis software) with genotypes, sperm source (epididymis or testis) and sperm status (fresh or frozen) as factors was used for the analysis of comet tail lengths, oocyte activation, arrest and chromosomal integrity, and for chromosome aberration rates. The linear model ANOVA (NCSS) was also used to analyze CMA3 data in epididymal sperm. For comparison of sperm head abnormalities and of sperm motility differences between genotypes were assessed by two-way ANOVA using the GraphPad Prism version 5.0 software. For ANOVA analyses, all percentages were transformed to angles.
The mice of interest in this study were two transgenic Sly-deficient genotypes (sh344 and sh367) and their negative siblings as controls. In all experiments except expression analyses the negative siblings of each transgenic line were compared, and if not different were pooled. In order to assess the comet, ICSI and TEM data obtained with Sly mice in the context of our earlier data from mice with NPYq deficiencies (Yamauchi et al., 2010), we first carried out ANOVA comparisons of the controls from the two studies to see if they were comparable. If this was the case, we then carried out an ANOVA analysis of the NPYq-deficient and Sly-deficient genotypes compared with the pooled controls.
Supplementary Material
Acknowledgments
The authors are grateful to Paul Burgoyne who provided invaluable advice during the entire duration of the study, helped with statistical analyses, contributed to data analysis, interpretation, and drafting the manuscript. The authors also thank members of M. Ward's lab, Hisami Oda and Hieu Nguyen, for help with mouse genotyping, the NIMR procedural service and O. A. Ojarikre for pronuclear injections and help with mouse breeding.
Footnotes
Funding
This work was supported by the National Institutes of Health [grant numbers HD058059, RR 024206 (Project 2), and HD072380 to M.A.W.]; a UK Medical Research Council Career Development Fellowship [grant number U117532009 to J.C.]; a Marie Curie Fellowship [grant number FP7-PEOPLE-2010-IEF-273143 to J.C.]; and the Biotechnology and Biological Sciences Research Council [grant number BB/J00877X/1 to P.J.I.E.]. Deposited in PMC for release after 6 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.114488/-/DC1
References
- Ajduk A., Yamauchi Y., Ward M. A. (2006). Sperm chromatin remodeling after intracytoplasmic sperm injection differs from that of in vitro fertilization. Biol. Reprod. 75, 442–451 10.1095/biolreprod.106.053223 [DOI] [PubMed] [Google Scholar]
- Akerfelt M., Henriksson E., Laiho A., Vihervaara A., Rautoma K., Kotaja N., Sistonen L. (2008). Promoter ChIP-chip analysis in mouse testis reveals Y chromosome occupancy by HSF2. Proc. Natl. Acad. Sci. USA 105, 11224–11229 10.1073/pnas.0800620105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellvé A. R. (1993). Purification, culture, and fractionation of spermatogenic cells. Methods Enzymol. 225, 84–113 10.1016/0076-6879(93)25009-Q [DOI] [PubMed] [Google Scholar]
- Bianchi P. G., Manicardi G. C., Bizzaro D., Bianchi U., Sakkas D. (1993). Effect of deoxyribonucleic acid protamination on fluorochrome staining and in situ nick-translation of murine and human mature spermatozoa. Biol. Reprod. 49, 1083–1088 10.1095/biolreprod49.5.1083 [DOI] [PubMed] [Google Scholar]
- Boivin J., Bunting L., Collins J. A., Nygren K. G. (2007). International estimates of infertility prevalence and treatment-seeking: potential need and demand for infertility medical care. Hum. Reprod. 22, 1506–1512 10.1093/humrep/dem046 [DOI] [PubMed] [Google Scholar]
- Brykczynska U., Hisano M., Erkek S., Ramos L., Oakeley E. J., Roloff T. C., Beisel C., Schübeler D., Stadler M. B., Peters A. H. (2010). Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nat. Struct. Mol. Biol. 17, 679–687 10.1038/nsmb.1821 [DOI] [PubMed] [Google Scholar]
- Burgoyne P. S., Mahadevaiah S. K., Sutcliffe M. J., Palmer S. J. (1992). Fertility in mice requires X-Y pairing and a Y-chromosomal “spermiogenesis” gene mapping to the long arm. Cell 71, 391–398 10.1016/0092-8674(92)90509-B [DOI] [PubMed] [Google Scholar]
- Chatot C. L., Ziomek C. A., Bavister B. D., Lewis J. L., Torres I. (1989). An improved culture medium supports development of random-bred 1-cell mouse embryos in vitro. J. Reprod. Fertil. 86, 679–688 10.1530/jrf.0.0860679 [DOI] [PubMed] [Google Scholar]
- Cocquet J., Ellis P. J., Yamauchi Y., Mahadevaiah S. K., Affara N. A., Ward M. A., Burgoyne P. S. (2009). The multicopy gene Sly represses the sex chromosomes in the male mouse germline after meiosis. PLoS Biol. 7, e1000244 10.1371/journal.pbio.1000244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer P., Ramos L., de Vries M., Gochhait S. (2010). Memoirs of an insult: sperm as a possible source of transgenerational epimutations and genetic instability. Mol. Hum. Reprod. 16, 48–56 10.1093/molehr/gap098 [DOI] [PubMed] [Google Scholar]
- Delbes G., Hales B. F., Robaire B. (2007). Effects of the chemotherapy cocktail used to treat testicular cancer on sperm chromatin integrity. J. Androl. 28, 241–249 10.2164/jandrol.106.001487 [DOI] [PubMed] [Google Scholar]
- Ellis P. J., Clemente E. J., Ball P., Touré A., Ferguson L., Turner J. M., Loveland K. L., Affara N. A., Burgoyne P. S. (2005). Deletions on mouse Yq lead to upregulation of multiple X- and Y-linked transcripts in spermatids. Hum. Mol. Genet. 14, 2705–2715 10.1093/hmg/ddi304 [DOI] [PubMed] [Google Scholar]
- Ellis P. J., Ferguson L., Clemente E. J., Affara N. A. (2007). Bidirectional transcription of a novel chimeric gene mapping to mouse chromosome Yq. BMC Evol. Biol. 7, 171 10.1186/1471-2148-7-171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis P. J., Bacon J., Affara N. A. (2011). Association of Sly with sex-linked gene amplification during mouse evolution: a side effect of genomic conflict in spermatids? Hum. Mol. Genet. 20, 3010–3021 10.1093/hmg/ddr204 [DOI] [PubMed] [Google Scholar]
- Ferguson L., Ellis P. J., Affara N. A. (2009). Two novel mouse genes mapped to chromosome Yp are expressed specifically in spermatids. Mamm. Genome 20, 193–206 10.1007/s00335-009-9175-8 [DOI] [PubMed] [Google Scholar]
- García M. A., Collado M., Muñoz–Fontela C., Matheu A., Marcos–Villar L., Arroyo J., Esteban M., Serrano M., Rivas C. (2006). Antiviral action of the tumor suppressor ARF. EMBO J. 25, 4284–4292 10.1038/sj.emboj.7601302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govin J., Escoffier E., Rousseaux S., Kuhn L., Ferro M., Thévenon J., Catena R., Davidson I., Garin J., Khochbin S.et al. (2007). Pericentric heterochromatin reprogramming by new histone variants during mouse spermiogenesis. J. Cell Biol. 176, 283–294 10.1083/jcb.200604141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth A., Rocha W., Verreault A., Almouzni G. (2007). Chromatin challenges during DNA replication and repair. Cell 128, 721–733 10.1016/j.cell.2007.01.030 [DOI] [PubMed] [Google Scholar]
- Grzmil P., Golas A., Muller C., Styrna J. (2007). The influence of the deletion on the long arm of the Y chromosome on sperm motility in mice. Theriogenology 67, 760–766 10.1016/j.theriogenology.2006.10.007 [DOI] [PubMed] [Google Scholar]
- Hammoud S. S., Nix D. A., Zhang H., Purwar J., Carrell D. T., Cairns B. R. (2009). Distinctive chromatin in human sperm packages genes for embryo development. Nature 460, 473–478 10.1038/nature08162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper S. Q., Staber P. D., He X., Eliason S. L., Martins I. H., Mao Q., Yang L., Kotin R. M., Paulson H. L., Davidson B. L. (2005). RNA interference improves motor and neuropathological abnormalities in a Huntington's disease mouse model. Proc. Natl. Acad. Sci. USA 102, 5820–5825 10.1073/pnas.0501507102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura Y., Yanagimachi R. (1995). Intracytoplasmic sperm injection in the mouse. Biol. Reprod. 52, 709–720 10.1095/biolreprod52.4.709 [DOI] [PubMed] [Google Scholar]
- Kusakabe H., Szczygiel M. A., Whittingham D. G., Yanagimachi R. (2001). Maintenance of genetic integrity in frozen and freeze-dried mouse spermatozoa. Proc. Natl. Acad. Sci. USA 98, 13501–13506 10.1073/pnas.241517598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis S. E., Aitken R. J. (2005). DNA damage to spermatozoa has impacts on fertilization and pregnancy. Cell Tissue Res. 322, 33–41 10.1007/s00441-005-1097-5 [DOI] [PubMed] [Google Scholar]
- Mahadevaiah S. K., Odorisio T., Elliott D. J., Rattigan A., Szot M., Laval S. H., Washburn L. L., McCarrey J. R., Cattanach B. M., Lovell–Badge R.et al. (1998). Mouse homologues of the human AZF candidate gene RBM are expressed in spermatogonia and spermatids, and map to a Y chromosome deletion interval associated with a high incidence of sperm abnormalities. Hum. Mol. Genet. 7, 715–727 10.1093/hmg/7.4.715 [DOI] [PubMed] [Google Scholar]
- Meyer–Ficca M. L., Lonchar J., Credidio C., Ihara M., Li Y., Wang Z. Q., Meyer R. G. (2009). Disruption of poly(ADP-ribose) homeostasis affects spermiogenesis and sperm chromatin integrity in mice. Biol. Reprod. 81, 46–55 10.1095/biolreprod.108.075390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikamo K., Kamiguchi Y. (1983). Primary incidences of spontaneous chromosomal anomalies and their origins and causal mechanisms in the Chinese hamster. Mutat. Res. 108, 265–278 10.1016/0027-5107(83)90125-2 [DOI] [PubMed] [Google Scholar]
- Nabatiyan A., Krude T. (2004). Silencing of chromatin assembly factor 1 in human cells leads to cell death and loss of chromatin assembly during DNA synthesis. Mol. Cell. Biol. 24, 2853–2862 10.1128/MCB.24.7.2853-2862.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S. F., Lin R. C., Laybutt D. R., Barres R., Owens J. A., Morris M. J. (2010). Chronic high-fat diet in fathers programs β-cell dysfunction in female rat offspring. Nature 467, 963–966 10.1038/nature09491 [DOI] [PubMed] [Google Scholar]
- Noblanc A., Peltier M., Damon–Soubeyrand C., Kerchkove N., Chabory E., Vernet P., Saez F., Cadet R., Janny L., Pons–Rejraji H.et al. (2012). Epididymis response partly compensates for spermatozoa oxidative defects in snGPx4 and GPx5 double mutant mice. PLoS ONE 7, e38565 10.1371/journal.pone.0038565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn P., Barros C., Whittingham D. G. (1982). Preservation of hamster oocytes to assay the fertilizing capacity of human spermatozoa. J. Reprod. Fertil. 66, 161–168 10.1530/jrf.0.0660161 [DOI] [PubMed] [Google Scholar]
- Rasband W. S. (2007). ImageJ Software. Bethesda, MD: U.S. National Institutes of Health; 10.1530/jrf.0.0660161 [DOI] [Google Scholar]
- Reynard L. N., Turner J. M., Cocquet J., Mahadevaiah S. K., Toure A., Hoog C., Burgoyne P. S. (2007). Expression analysis of the mouse multi-copy X-linked gene Xlr-related, meiosis-regulated (Xmr), reveals that Xmr encodes a spermatid-expressed cytoplasmic protein, SLX/XMR. Biol. Reprod. 77, 329–335 10.1095/biolreprod.107.061101 [DOI] [PubMed] [Google Scholar]
- Reynard L. N., Cocquet J., Burgoyne P. S. (2009). The multi-copy mouse gene Sycp3-like Y-linked (Sly) encodes an abundant spermatid protein that interacts with a histone acetyltransferase and an acrosomal protein. Biol. Reprod. 81, 250–257 10.1095/biolreprod.108.075382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scavetta R. J., Tautz D. (2010). Copy number changes of CNV regions in intersubspecific crosses of the house mouse. Mol. Biol. Evol. 27, 1845–1856 10.1093/molbev/msq064 [DOI] [PubMed] [Google Scholar]
- Simões R., Feitosa W. B., Mendes C. M., Marques M. G., Nicacio A. C., de Barros F. R., Visintin J. A., Assumpção M. E. (2009). Use of chromomycin A3 staining in bovine sperm cells for detection of protamine deficiency. Biotech. Histochem. 84, 79–83 10.1080/10520290902843595 [DOI] [PubMed] [Google Scholar]
- Skinner M. K. (2010). Metabolic disorders: Fathers' nutritional legacy. Nature 467, 922–923 10.1038/467922a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styrna J., Bilinska B., Krzanowskaa H. (2002). The effect of a partial Y chromosome deletion in B10.BR-Ydel mice on testis morphology, sperm quality and efficiency of fertilization. Reprod. Fertil. Dev. 14, 101–108 10.1071/RD01089 [DOI] [PubMed] [Google Scholar]
- Styrna J., Kilarski W., Krzanowska H. (2003). Influence of the CBA genetic background on sperm morphology and fertilization efficiency in mice with a partial Y chromosome deletion. Reproduction 126, 579–588 10.1530/rep.0.1260579 [DOI] [PubMed] [Google Scholar]
- Szczygiel M. A., Yanagimachi R. (2003). Intracytoplasmic sperm injection. Manipulating the Mouse Embryo - A Laboratory Manual (ed Nagy A, Gertsenstein M, Vintersten K, Behringer R R.), pp. 585–597 New York, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Touré A., Szot M., Mahadevaiah S. K., Rattigan A., Ojarikre O. A., Burgoyne P. S. (2004). A new deletion of the mouse Y chromosome long arm associated with the loss of Ssty expression, abnormal sperm development and sterility. Genetics 166, 901–912 10.1534/genetics.166.2.901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touré A., Clemente E. J., Ellis P., Mahadevaiah S. K., Ojarikre O. A., Ball P. A., Reynard L., Loveland K. L., Burgoyne P. S., Affara N. A. (2005). Identification of novel Y chromosome encoded transcripts by testis transcriptome analysis of mice with deletions of the Y chromosome long arm. Genome Biol. 6, R102 10.1186/gb-2005-6-12-r102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Chen Z., Mao Z., Zhang H., Ding X., Chen S., Zhang X., Xu R., Zhu B. (2011). Nucleolar protein Spindlin1 recognizes H3K4 methylation and stimulates the expression of rRNA genes. EMBO Rep. 12, 1160–1166 10.1038/embor.2011.184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward M. A., Burgoyne P. S. (2006). The effects of deletions of the mouse Y chromosome long arm on sperm function - intracytoplasmic sperm injection (ICSI)-based analysis. Biol. Reprod. 74, 652–658 10.1095/biolreprod.105.048090 [DOI] [PubMed] [Google Scholar]
- Xian M., Azuma S., Naito K., Kunieda T., Moriwaki K., Toyoda Y. (1992). Effect of a partial deletion of Y chromosome on in vitro fertilizing ability of mouse spermatozoa. Biol. Reprod. 47, 549–553 10.1095/biolreprod47.4.549 [DOI] [PubMed] [Google Scholar]
- Yamauchi Y., Ajduk A., Riel J. M., Ward M. A. (2007). Ejaculated and epididymal mouse spermatozoa are different in their susceptibility to nuclease-dependent DNA damage and in their nuclease activity. Biol. Reprod. 77, 636–647 10.1095/biolreprod.107.062406 [DOI] [PubMed] [Google Scholar]
- Yamauchi Y., Riel J. M., Wong S. J., Ojarikre O. A., Burgoyne P. S., Ward M. A. (2009). Live offspring from mice lacking the Y chromosome long arm gene complement. Biol. Reprod. 81, 353–361 10.1095/biolreprod.109.076307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi Y., Riel J. M., Stoytcheva Z., Burgoyne P. S., Ward M. A. (2010). Deficiency in mouse Y chromosome long arm gene complement is associated with sperm DNA damage. Genome Biol. 11, R66 10.1186/gb-2010-11-6-r66 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.