Summary
Microtubules (MTs) are the major constituent of the mitotic apparatus. Deregulation of MT dynamics leads to chromosome missegregation, cytokinesis failure and improper inheritance of genetic materials. Here, we describe the identification and characterization of KIAA1383/MTR120 (microtubule regulator 120 kDa) as a novel MT-associated protein. We found that MTR120 localizes to stabilized MTs during interphase and to the mitotic apparatus during mitosis. MTR120 overexpression results in MT bundling and acetylation. In vitro, purified MTR120 protein binds to and bundles preassembled MTs. Moreover, depletion of MTR120 by RNA interference leads to cytokinesis failure and polyploidy. These phenotypes can be rescued by wild-type MTR120 but not by the MT non-binding mutant of MTR120. Together, these data suggest that MTR120 is a novel MT-associated protein that directly stabilizes MTs and hence ensures the fidelity of cell division.
Key words: Microtubule, Cell cycle, Cytokinesis
Introduction
A microtubule (MT) is a polarized cylindrical structure composed of α/β-tubulin dimers. Its plus ends are dynamic and subjected to frequent growing and shrinking, whereas its minus ends are inert and capped by a γ-tubulin ring complex that also provides a template for MT nucleation (Kollman et al., 2011; Lüders and Stearns, 2007). MTs play a pivotal role in coordinating numerous biological processes, including chromosome segregation and cell motility. MT-associated proteins (MAPs) can bind to MTs in various manners, either on the ends or along the lattice (Jiang and Akhmanova, 2011). MAPs are believed to play indispensable roles in modulating many MT-based cellular functions.
MAPs regulate the functions of the MT-based mitotic spindle structure via several different mechanisms. For example, MT plus-end-directed motor Eg-5 establishes spindle bipolarity by crosslinking MTs outward from the spindle poles (Kapitein et al., 2005). Minus-end-directed motor Dynein transports MTs and spindle pole components toward the MT minus-end, contributing to spindle pole organization (Merdes et al., 2000). Non-motor MAPs such as NuMA are required for spindle pole focusing (Silk et al., 2009). TPX2 activates and mediates Aurora A kinase localization onto spindle MTs (Kufer et al., 2002). MT plus-end master EB1 stabilizes the plus ends of spindle MT and therefore influences chromosome alignment (Green et al., 2005).
Distinct MAPs regulate midzone MT to facilitate cytokinesis. For instance, PRC1, an evolutionarily conserved MAP, plays an important role in midzone assembly, an early polarization step (Mollinari et al., 2002; Mollinari et al., 2005). Kinesin family member MKLP1, together with CYK4, forms the central spindling complex that promotes midzone MT bundling and recruits regulators for late abscission (Pavicic-Kaltenbrunner et al., 2007). In the chromosome passenger complex, INCENP binds to and activates another chromosome passenger complex component, Aurora B kinase, which in turn phosphorylates central spindlin complex, facilitating midbody formation (Guse et al., 2005; Sessa et al., 2005).
Because MAPs play significant roles in various cellular processes, including mitosis, a more complete inventory may contribute to a better understanding of spindle assembly, chromosome segregation, and more importantly, genome stability maintenance. In a study of subcellular localization of human ORFs, we found that KIAA1383 had a robust MT localization pattern when overexpressed. In this study, we found that KIAA1383 is a novel MAP that promotes MT stability and participates in cytokinesis. Given its major function in stabilizing MT, we named this protein MTR120 (microtubule regulator 120 kDa).
Results
Identification of new MT-associated proteins using ORFeome screening
To determine the subcellular localization of individual ORFs in the human ORFeome library, we subcloned human ORFeomes into a gateway-compatible destination vector that contains a HA-FLAG tag (Fig. 1A). Constructs containing these ORFs were then transfected into HeLa cells and subjected to immunostaining. The fluorescence images were captured using an automated imaging system (Fig. 1A), and the subcellular localization of these ORFs was determined by visual inspection. During the course of this study, we identified 39 ORFs with an MT-like pattern. To validate our results, we performed a literature search and found that 22 of these 39 ORFs (56.4%) had been previously reported to localize to MTs (Fig. 1B), suggesting that our localization screen is capable of identifying tubulins and MAPs. Several uncharacterized proteins were discovered that target the MT network (Fig. 1C).
Fig. 1.
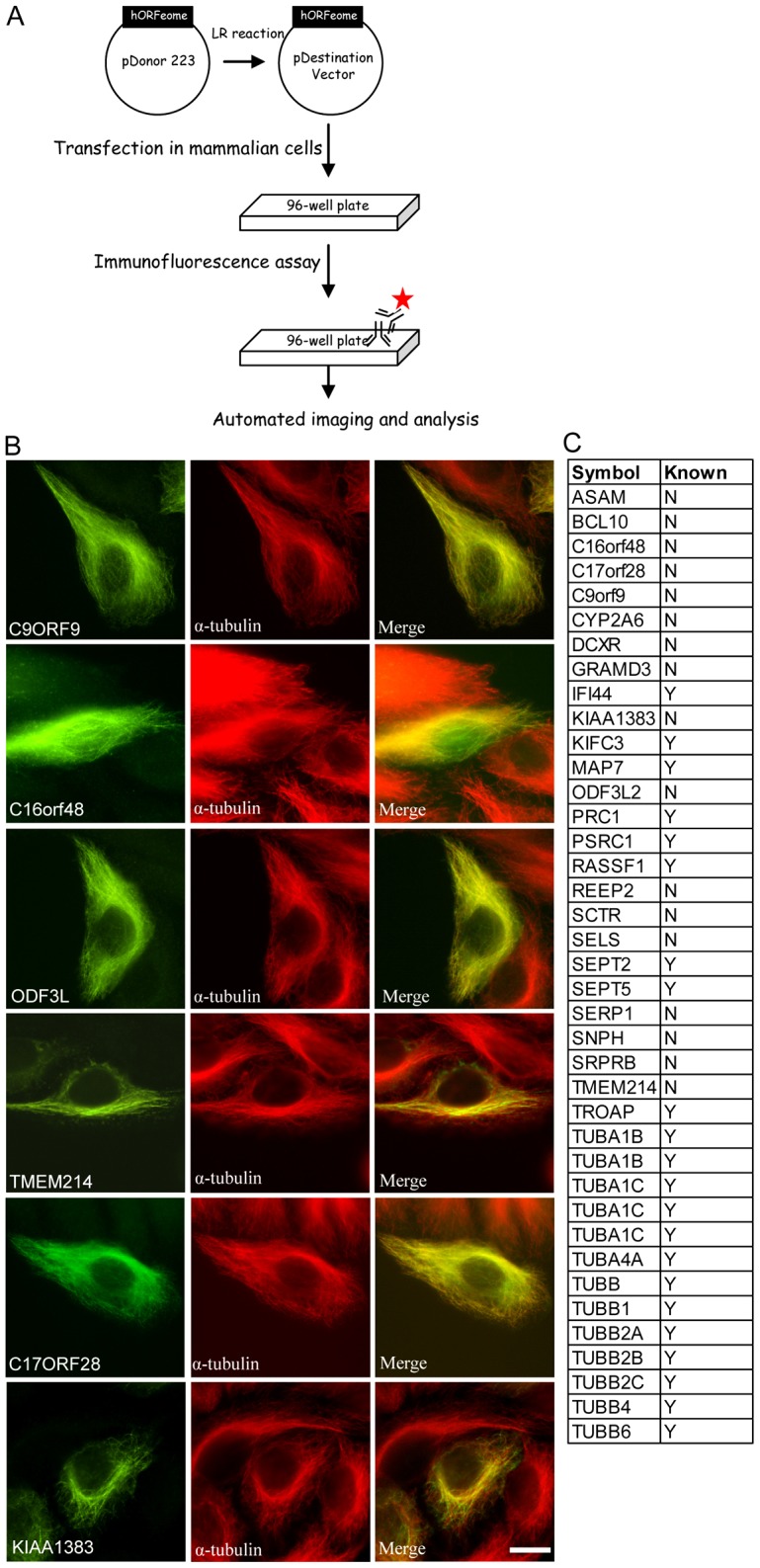
High-throughput imaging of human ORFeome to identify proteins that localize to MT. (A) Schematic flowchart of construction of the ORFeome library and large-scale screening for protein localization. (B) Representative images show several uncharacterized proteins that target the MT network. Scale bar: 10 µm. (C) List of ORFs scored as potential proteins with MT localization (N, no; Y, yes).
MTR120 is a cell-cycle-regulated protein with distinct mitotic localizations
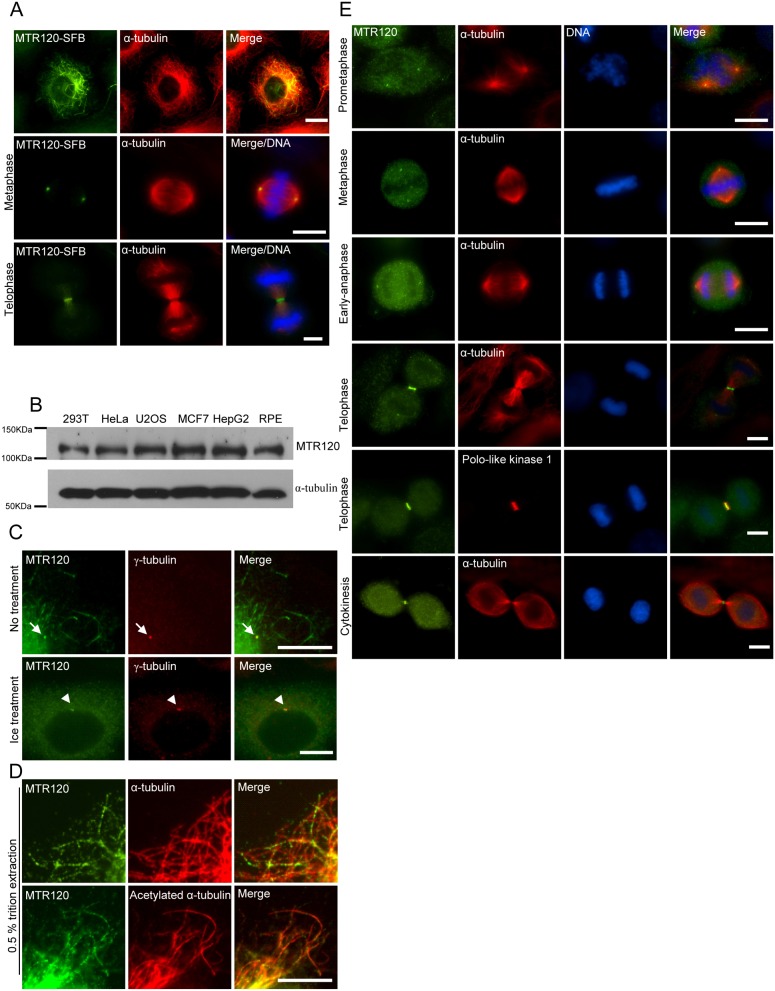
We found that one ORF, MTR120/KIAA1383, robustly localizes to the MT-like structure. MTR120 is an uncharacterized protein of 1047 residues with a predicted molecular mass of 120.4 kDa and a predicted isoelectric point of 8.16. In addition, its orthologs can be found in many vertebrates (supplementary material Fig. S1). A DNA topoisomerase I conserved domain with unknown significance was identified at the carboxyl region of its mouse homolog. We generated stable clones that expressed SFB (S-protein tag, FLAG epitope tag, and streptavidin-binding peptide tag)-tagged MTR120 and found that tagged MTR120 localized to filamentous structures that partially overlapped with MTs during interphase. Moreover, the transgene was found to be at spindle poles in metaphase and spindle midzone during telophase (Fig. 2A).
Fig. 2.
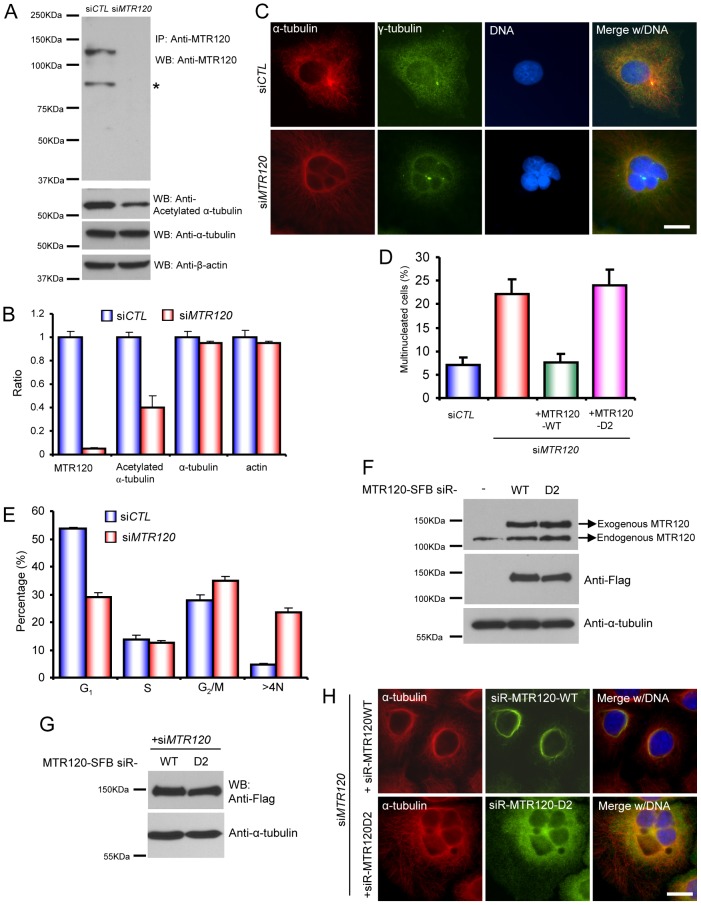
MTR120 is targeted to MTs and centrosomes. (A) Cells that stably express MTR120-SFB were fixed and immunostained for MTR120-SFB (anti-FLAG), MTs (anti-α-tubulin antibodies) and DNA (DAPI). (B) Various mammalian cell extracts were subjected to western blot analysis using anti-MTR120 and anti-α-tubulin antibodies. (C) Cells on the coverslip were untreated (upper row) or placed in ice water for 1 hour (lower row) before being fixed and immunostained for endogenous MTR120 and centrosomes (anti-γ-tubulin). Arrows and arrowheads indicate the centrosomes. (D) Cells on the coverslip were pre-extracted with 0.5% Triton X-100 before being fixed and immunostained for endogenous MTR120 and MTs (α-tubulin) (upper row) or acetylated MTs (anti-acetylated-α-tubulin) (lower row). (E) Mitotic cells were co-stained with anti-MTR120 and anti-α-tubulin antibodies (a marker for spindle MTs). Anti-Polo-like kinase 1 was used to stain the spindle midzone at telophase. DAPI staining was used to stain the congressed chromosomes. Scale bars: 10 µm.
To confirm our immunostaining results using exogenously expressed proteins, we generated rabbit polyclonal antibodies, raised against purified GST-tagged MTR120 carboxyl terminus (residues 697–1047). This antibody specifically recognized a 120-kDa band from HeLa cell lysate but not in the cells treated with MTR120-specific siRNA (see later). Moreover, this antibody recognized MTR120 protein in different cell lines (Fig. 2B), indicating that MTR120 is ubiquitously expressed. Similar to the results obtained using tagged protein, we observed the enrichment of MTR120 at centrosomes when cells were subjected to cold treatment to remove cytoplasmic MTs (Fig. 2C, bottom panels), whereas the centrosome signal of MTR120 was not easily detected without treatment (Fig. 2C, upper panels). After detergent pre-extraction, MTR120 was found along a population of MTs (Fig. 2D, upper panels). Because MTR120 does not co-localize perfectly with α-tubulin, we speculated that it preferentially localizes to a subset of MTs, such as stabilized MTs. To test this possibility, we co-stained endogenous MTR120 with acetylated α-tubulin, which is a marker for MT stabilization, and observed substantial co-localization of these proteins (Fig. 2D, bottom panels). We also noticed distinct localization of MTR120 at various stages of mitosis (Fig. 2E). During prometaphase and metaphase, MTR120 accumulated at spindle poles. At late anaphase, MTR120 was targeted to the center of spindle midzone, as shown by its co-localization with Polo-like kinase 1 (Fig. 2E). During cytokinesis, MTR120 localized at the midbody. To validate the specificity of these immunostaining results, we used MTR120-specific siRNA to downregulate the expression of endogenous MTR120. MTR120s fluorescence signals normalized to internal reference signals (γ-tubulin) at various cell cycle stages and were dramatically reduced in siRNA-treated cells compared with in control cells (supplementary material Fig. S2), confirming that MTR120 localizes to MTs and mitotic apparatuses that contain MTs.
Conserved MTR120 region is required for its MT localization
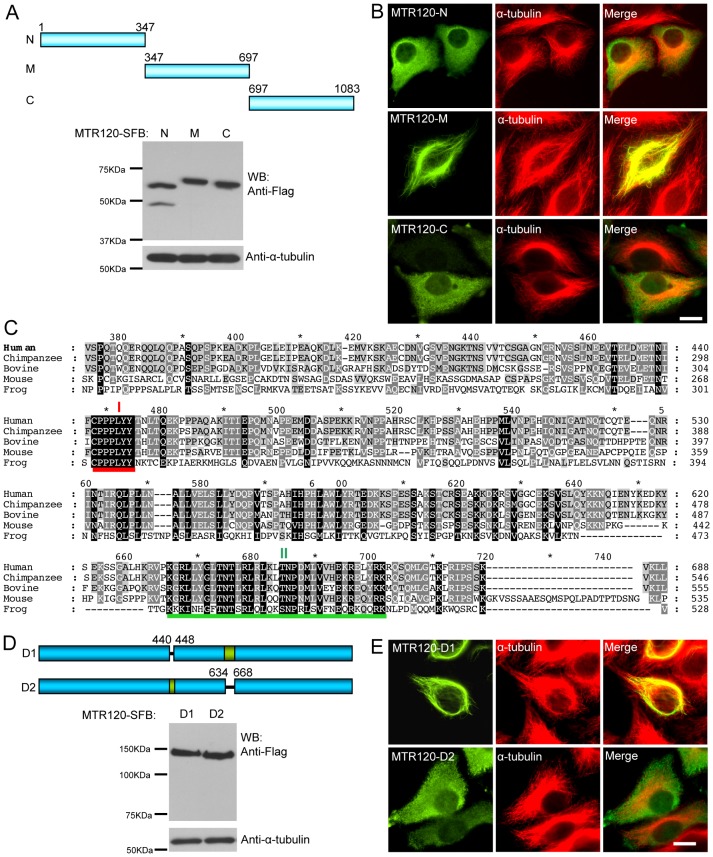
We further delineated the MT-targeting domain on MTR120. Several SFB-tagged MTR120 fragments were generated and expressed in cells (Fig. 3A). Eighty-five percent of cells (n = 100) that expressed MTR120s middle region (M; residues 347–697) showed MT localization (Fig. 3B), whereas 95% or 90% of cells that expressed the NH2-terminal (N) (n = 60) or COOH-terminal (C) fragment (n = 70) demonstrated diffuse cytoplasm localization. To further define the MT-targeting domain, we performed sequence alignment of MTR120 from various species and found two highly conserved regions (I: residues 440–448 and II: residues 634–668) in the middle region of MTR120 (Fig. 3C). Moreover, region II harbors several conserved positive-charge residues, which is a general feature of known MT-binding domains. The internal deletion mutants of these conserved regions were constructed and expressed in cells (Fig. 3D). As expected, the D2 mutant, which lacks the conserved region II, failed to co-localize with MTs (94%, n = 50), whereas D1 mutant localized (80%, n = 40) in the same manner as full-length protein (Fig. 3E). These data indicate that MTR120 uses a conserved region to localize to MTs.
Fig. 3.
A conserved MT-targeting domain is present in the central region of MTR120. (A) Schematic representation and expression of MTR120 fragments. (B) HeLa cells were stained for ectopically expressed MTR120 fragments and MTs using anti-FLAG and anti-α-tubulin antibodies, respectively. (C) Sequence alignment of MTR120 homologs identified in different species. The GenBank accession numbers are as follows: human, NP_061963.2; chimpanzee, XP_525093.2; bovine, NP_001091617.1; mouse, NP_083184.1; and frog, XP_002935866.1. The red and green lines indicate the first and second conserved regions, respectively. (D) Schematic representation and expression of internal deletion mutants of MTR120. (E) HeLa cells were transiently transfected with constructs encoding the indicated MTR120 mutants and stained with anti-FLAG and anti-α-tubulin antibodies to detect MTR120 proteins and MTs, respectively. Scale bars: 10 µm.
MTR120 promotes MT stability in vivo and binds MT in vitro
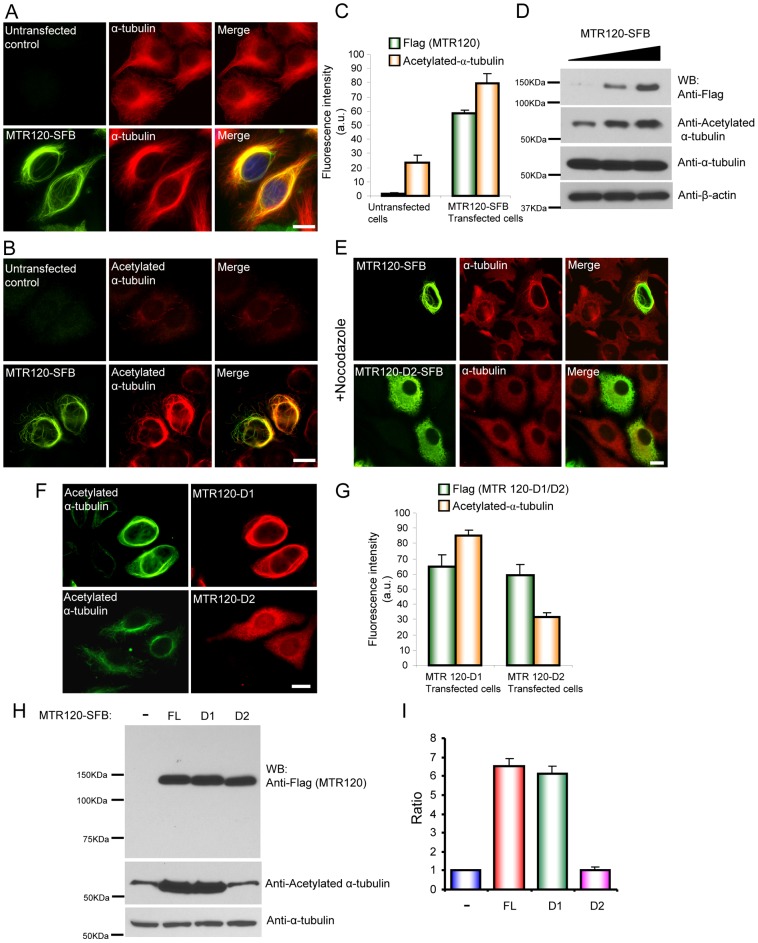
Because MTR120 selectively localizes to stabilized MTs (Fig. 2D), we determined whether it promoted MT stabilization in cells. Interestingly, we found that 70% of cells with MTR120 overexpression had perinuclear rings of MT bundles, which was not observed in control cells (Fig. 4A). Consistent with MTR120s not co-localizing perfectly with MT network, we found that the MT bundle induced by MTR120 co-localized well with acetylated α-tubulin (Fig. 4B, lower panels). The results of a quantification indicated that the fluorescence intensity of acetylated α-tubulin in MTR120-overexpressed cells was 4-fold higher than that in untransfected controls (Fig. 4C). Moreover, a western blotting analysis revealed that the amount of acetylated-α-tubulin increased when MTR120 was overexpressed, whereas the total amount of α-tubulin did not change (Fig. 4D).
Fig. 4.
MTR120 promotes MT stability in vivo. (A) HeLa cells were untransfected or transfected with construct encoding SFB-tagged, full-length MTR120. Immunostaining was conducted with anti-FLAG antibody and anti-α-tubulin antibodies. (B) HeLa cells were untreated or transfected with SFB-tagged, full-length MTR120. Immunostaining was conducted using anti-FLAG antibody and anti-acetylated α-tubulin antibodies. (C) Quantification revealed the fluorescence intensities of anti-FLAG and anti-acetylated α-tubulin in untreated or MTR120-overexpressing cells; 50 cells were surveyed in three independent experiments. (D) HEK293T cells were transfected with increasing amounts of plasmids that encode MTR120-SFB (0–2 µg). Whole-cell extracts were prepared and analyzed by western blotting using the indicated antibodies. (E) At 24 hours after transfection, cells that expressed full-length MTR120 or D2 mutant were treated with 5 µM nocodazole for 1 hour and then fixed and immunostained using the indicated antibodies. (F) HeLa cells expressing internal deletion mutants of MTR120 were stained using anti-FLAG antibody and anti-acetylated α-tubulin.(G) Quantification revealed the fluorescence intensities of anti-FLAG and anti-acetylated α-tubulin in D1- or D2-positive cells; 50 cells were surveyed in three independent experiments. (H) HeLa cells were mock treated (−) or transfected with constructs that encoded full-length (FL), D1 or D2 mutant MTR120; they were then harvested and subjected to western blot analysis using the indicated antibodies. (I) Bar graphs show the ratio of acetylated α-tubulin levels in treated cells relative to those in mock-treated cells (n = 3 independent experiments). Error bars represent s.d. Scale bars: 10 µm.
To substantiate MTR120s role in promoting MT stability, we treated MTR120-overexpressing cells with nocodazole, a drug that induces MT depolymerization. MT bundles persisted in 75% of cells (Fig. 4E, upper panels). We then determined whether MT's association with MTR120 is required for its ability to promote MT stabilization. For this, we took advantage of the D2 deletion mutant, which lacks the MT-binding domain. Ninety-five percent of D2-positive cells did not maintain the filamentous MT network after nocodazole treatment (Fig. 4E, lower panels). In another experiment, the D2 mutant, but not the D1 mutant, of MTR120 did not increase the fluorescence intensity of acetylated α-tubulin, as demonstrated by immunofluorescence staining (Fig. 4F,G). Similarly, quantification of the western blotting results revealed that both the full-length and D1 mutant of MTR120 significantly increased the level of acetylated MT by 6-fold compared with the untransfected control or D2-positive cells (Fig. 4H,I). Together, these results suggest that MTR120 associates with MT and enhances its stability in vivo.
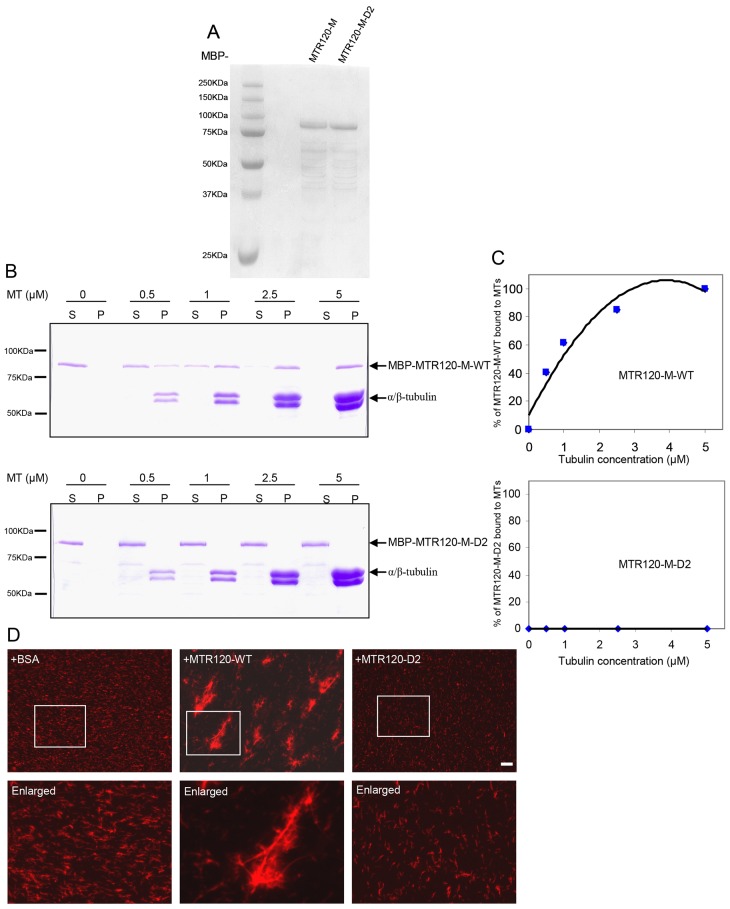
To provide biochemical evidence of the MT-stabilizing activity of MTR120, we expressed and purified recombinant MBP-tagged MTR120-M that harbors its MT-targeting domain and MBP-tagged MTR120-M-D2 in which the conserved region (residues 634–668) was deleted in Escherichia coli (Fig. 5A). In an MT co-sedimentation assay, MTR120-M-WT, but not MTR120-M-D2, bound directly to polymerized MTs. A quantification revealed that the dissociation constant (Kd) (concentration of tubulin needed to pellet 50% of MTR120-M) was ∼0.8±0.24 µM (Fig. 5C). The co-pelleted MTR120-M was not simply precipitated due to protein aggregation because the protein stayed in supernatant when polymerized MT was not added (Fig. 5B).
Fig. 5.
MTR120 binds to and stabilizes MTs in vitro. (A) MBP-tagged fusion proteins were purified from E. coli, separated by SDS-PAGE and stained with Coomassie Blue. (B) We incubated 200 nM of the indicated proteins with 0, 0.5, 1, 2.5 or 5 µM paclitaxel-stabilized MTs and subjected them to ultracentrifugation to pellet the polymerized MT. The supernatant and pellet fractions were run on 10% polyacrylamide gel and stained using Coomassie Blue. (C) Quantitative analysis was performed of the binding properties between MTs and MTR120-M-WT or MTR120-M-D2. The percentages of bound protein were plotted against tubulin concentrations. The disassociation constant (Kd) was determined from the best-fit curve. The data were collected from three independent experiments. (D) An MT bundling assay was performed using 4 µM rhodamine-labeled MTs. We used 0.1 µM BSA (as the negative control), wild-type (WT) or the D2 mutant MTR120-SFB. Boxed areas are enlarged. Scale bars: 10 µm.
To determine whether full-length MTR120 is capable of binding to MTs in vitro, we expressed and affinity-purified SFB-tagged, wild-type (WT) and D2 mutant MTR120 from mammalian cells. After extensive washing using 1 M NaCl and 12 mM deoxycholate, we eluted the fusion proteins from streptavidin beads and subjected the eluates to sodium dodecyl sulfate-PAGE (SDS-PAGE) (supplementary material Fig. S3). WT MTR120 co-pelleted with polymerized MTs, whereas the D2 mutant of MTR120 was barely detectable in the pellet (data not shown). We determined whether MTR120 can bundle paclitaxel-preassembled MTs. The addition of full-length, wild-type MTR120 protein bundled the preassembled MTs into long, thick fibers (Fig. 5D). In contrast, the MTs remained short and thin in samples containing the D2 mutant of MTR120 or BSA, which served as a negative control in this experiment (Fig. 5D). Together, these findings indicate that MTR120 is a MT-binding protein that can promote MT stabilization in vitro.
MTR120 downregulation causes cytokinesis defects and leads to polyploidy
To further determine MTR120s cellular functions, we used MTR120-specific siRNA to reduce its expression by more than 95% in HeLa cells (Fig. 6A,B). Of note, downregulation of MTR120 expression resulted in a reduction (∼40%) of acetylated α-tubulin (Fig. 6B), indicating that MTR120 is required for MT stabilization in vivo. In addition, the number of multinucleated cells increased significantly (∼23%) compared with those observed in cells transfected with control siRNA (∼6%) (Fig. 6C,D). We also found centrosome clusters (indicated by bright γ-tubulin staining) in multinucleated cells, indicating that polyploidy progenies inherit multiple centrosomes and that some fuse and display bright γ-tubulin staining. This phenotype was reproducible when a second siRNA was used to downregulate MTR120 expression (supplementary material Fig. S4). Consistent with results from immunofluorescence staining, a fluorescence-activated cell sorting analysis also revealed a significant increase in the >4N population in MTR120 knockdown cells (Fig. 6E; supplementary material Fig. S5), indicating that MTR120 downregulation causes defects in cell division, preventing cells fail from maintaining normal ploidy.
Fig. 6.
MTR120 is required for maintaining normal diploid status. (A) Cells transfected with control siRNA (siCTL) or siRNA targeting MTR120 (siMTR120) were harvested 96 hours after transfection. Cell lysates were analyzed by western blotting for MTR120, acetylated α-tubulin, α-tubulin and β-actin. Asterisks indicate a degraded or smaller form of MTR120. (B) Quantification revealed the ratios of indicated protein levels in siMTR120-treated cells relative to those in siCTL-treated cells (three independent experiments). (C) Immunofluorescence images of cells transfected with siCTL or siMTR120. γ-tubulin and α-tubulin are the respective markers for centrosomes and MTs. DAPI was used to stain DNA. (D) Quantification revealed the mean percentages of multinucleated cells in the indicated groups; 100 cells from each group were counted from three independent experiments. (E) The bar graphs reveal the percentages of cells in different cell cycle phases. We analyzed 10,000 cells from each group (three independent experiments). (F) Parental cells and cells harboring siRNA-resistant wild-type MR120 or the D2 mutant were harvested and subjected to an immunoblotting analysis using anti-MTR120, anti-FLAG and anti-α-tubulin antibodies. (G,H) Cells that stably expressed siRNA-resistant wild-type MTR120 or the D2 mutant were transfected with siMTR120. The expression of siRNA-resistant wild-type or D2 mutant was confirmed by western blotting analysis (G). Cells were fixed and subjected to immunofluorescence staining with anti-α-tubulin and anti-FLAG (MTR120) antibodies (H). Scale bars: 10 µm. Error bars represent s.d.
Because MTR120 is a MAP, we determined whether MT is required for MTR120s function. We first generated cell lines that stably expressed the siRNA-resistant form of wild-type or D2 mutant MTR120. The expression of exogenous proteins was slightly higher than that of endogenous MTR120 (Fig. 6F). Next, we depleted endogenous MTR120 using siRNA in these cell lines; thus, these cells only expressed wild-type or D2 mutant MTR120 (Fig. 6G). The expression of wild-type MTR120, but not D2 mutant, rescued the polyploidy phenotype in cells transfected with MTR120 siRNA (Fig. 6D,H). The introduction of exogenous wild-type MTR120 led to the formation of perinuclear rings, suggesting that at least some MTs are bundled in these cells. Nonetheless, these cells proliferated normally as parental cells (data not shown), indicating that the expression level of these exogenous proteins is relatively low and thus does not severely disturb cell cycle progression.
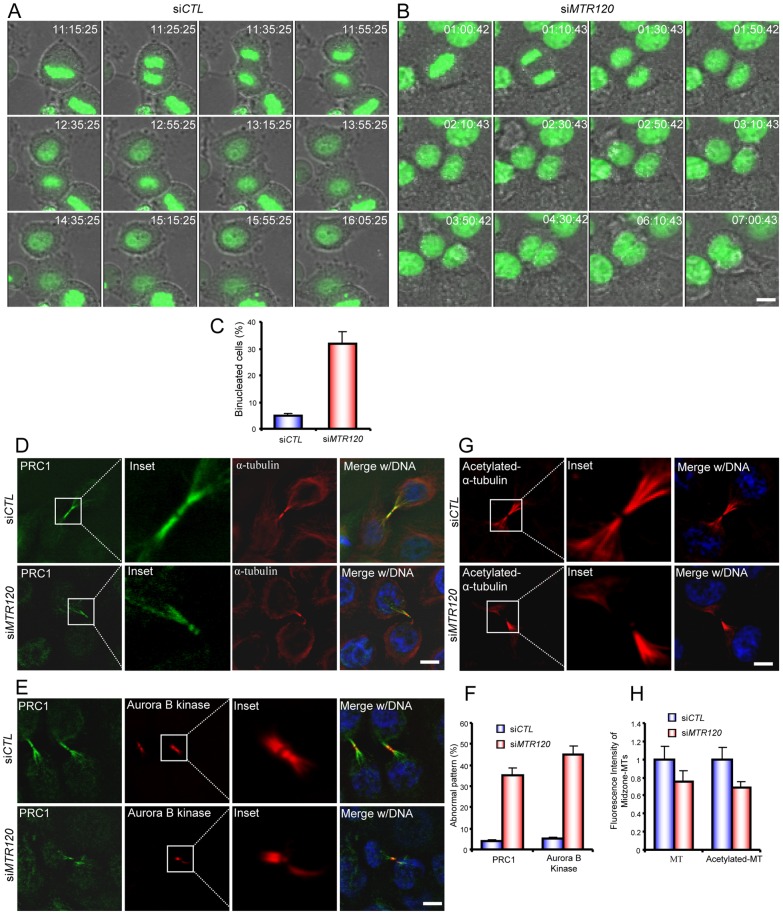
To better describe MTR120s involvement in mitosis and cytokinesis, we used time-lapse microscopy to image M-phase events in HeLa cells that stably expressed GFP-H2B. In the control population, more than 90% of cells underwent mitosis with aligned chromosomes, and the cleavage furrow successfully ingressed so that the cells eventually separated from each other (Fig. 7A,C; supplementary material Movie 1). On the contrary, we observed a cytokinesis defect in MTR120-depleted cells. Although the cells underwent chromosome segregation after anaphase onset and the segregated chromosomes migrated to the daughter cells, the furrow failed to ingress completely. As a result, cytokinesis failed to complete, and bi-nucleated cells formed (∼30%) (Fig. 7B,C; supplementary material Movie 2). Together, these results suggest that MTR120 depletion leads to a cytokinesis defect.
Fig. 7.
MTR120 downregulation leads to cytokinesis defects and mislocalization of spindle midzone components. (A,B) Time series of live images of HeLa cells transfected with control (A) or MTR120-specific siRNA. (B) Representative merged images of GFP-H2B and brightfield images. (C) Quantification reveals the mean percentages of bi-nucleated cells resulting from cytokinesis defects; 50 control and 60 siMTR120-treated cells were analyzed in three independent experiments. (D,E) Telophase cells were stained with anti-PRC1 and anti-α-tubulin antibodies (D) or anti-PRC1 and anti-Aurora B kinase antibodies (E) to mark the spindle midzone. The insets reveal the enlarged spindle midzone region. (F) The bar graph shows the mean percentages of telophase cells with an abnormal PRC1 or Aurora kinase B pattern. PRC1 or Aurora kinase B staining with no anti-parallel pattern was scored as ‘abnormal’; 50 cells from each indicated group were counted from three independent experiments. (G) Control or siMTR120-treated cells were fixed, and anti-acetylated α-tubulin was used to detect stabilized MTs during telophase. The insets reveal the enlarged spindle midzone region. (H) Quantification revealed the fluorescence intensities of α-tubulin or acetylated α-tubulin staining in the midzone region in control and siMTR120-treated cells; 50 cells were surveyed from three independent tests (see Materials and Methods). A statistical analysis was performed using Student's unpaired, two-tailed t-test. The fluorescence intensities of both midzone MTs and midzone acetylated MTs are significantly different for control and MTR120 knockdown cells (P<0.05). Error bars represent s.d. Scale bars: 10 µm.
The central spindle or spindle midzone is a highly ordered MT structure and is enriched with bundled MTs. Sequential recruitment of factors to this structure is essential for central spindle build-up and signaling for cell cleavage (Glotzer, 2009; Wheatley and Wang, 1996). We determined the organization of the spindle midzone in MTR120-knockdown cells by determining the localization pattern of central spindle proteins such as PRC1 and Aurora B kinase. We observed a compact, organized, MT structure marked by PRC1 and α-tubulin (Fig. 7D) or Aurora B kinase staining (Fig. 7E) in normal telophase cells. In MTR120 knockdown cells, PRC1 and Aurora B kinase failed to localize to the midzone MTs (Fig. 7D,E). The quantification indicated that ∼35% and 45% of MTR120-silenced cells had distorted PRC1 and Aurora B kinase patterns, respectively (Fig. 7F). We also observed that the α-tubulin and acetylated α-tubulin labeling in the midzone region was less robust: the intensity decreased by ∼24% and 30%, respectively (Fig. 7G,H). These data indicate that as a regulator of MT stability, MTR120 participates in the organization of the spindle midzone.
Discussion
In this study, we identified MTR120 as a novel MAP in a high-throughput proteomic screen and demonstrated that it promotes MT stability. Moreover, we found that the MT-binding ability is essential for MTR120s function in stabilizing MTs, which is critical for its roles in mitotic progression and cytokinesis.
To fully understand MAPs' roles in mitosis, several research groups have used various approaches to identify MAPs, mainly based on biochemical purification (Sauer et al., 2005; Torres et al., 2011). Here, we carried out a large-scale analysis of the subcellular localization of all 15,483 ORFs in the human ORFeome v5.1 collection, which is complementary to previous approaches. We identified tubulins, previously characterized MAPs, and putative new MAPs, including several uncharacterized proteins, such as C9ORF9, C16ORF48, ODF3L, TMEM214, C17ORF28 and KIAA1383/MTR120 (Fig. 1B,C). These putative MAPs provide an opportunity to understand how MAPs participate in diverse cellular processes. Our study only focuses on one, KIAA1383/MTR120.
The middle region (residues 347–697) of MTR120, which contains the MT-binding domain, is rich in basic residues, reminiscent of the MT-binding sequences of conventional MAPs. However, scanning this region yielded no recognizable MT-binding motif, suggesting that MTR120 contains a novel MT-binding domain. Unlike other general MAPs, MTR120 is enriched at a subset of MTs (i.e. acetylated MTs) during interphase, which is reminiscent of other MAPs, such as CAP350 and centrobin/NIP2. In particular, MT-binding of CAP350 is required for modulating MT dynamics around Golgi (Hoppeler-Lebel et al., 2007). Centrobin/NIP2-tubulin interaction is involved in centriole elongation and mitotic spindle assembly (Gudi et al., 2011; Lee et al., 2010). These MAPs and MTR120 may represent a subgroup of MAPs that has a preference for stabilized MT and participates in cellular events that are highly dependent on these MTs.
Cytokinesis failure may result from centrosome malfunction because a population of MTR120 always adheres to centrosomes. In fact, surgical removal of centrosomes abolishes cell abscission and arrests cell in G1 phase (Hinchcliffe et al., 2001; Khodjakov and Rieder, 2001). Furthermore, several centrosomal proteins, such as centrin, centrolin, CEP55, and CP110, are required for proper cytokinesis, and prolong silencing of these proteins leads to the formation of multicellular syncytia or multinucleated progenies (Gromley et al., 2003; Gromley et al., 2005; Salisbury et al., 2002; Tsang et al., 2006; Zhao et al., 2006). These proteins likely act in distinct manners, but how they cooperate in cytokinesis is still unclear. We evaluated the potential centrosome-specific defects in MTR120 knockdown cells. However, we found no weakening or fragmentation of Aurora A kinase labeling in mitotic centrosomes, indicating no significant abnormality of centrosome maturation (unpublished observation). Loss of MTR120 also had no apparent effect on centrosomal MT nucleation (supplementary material Fig. S6), indicating that MTR120 is dispensable for centrosomes to act as MT's organization center. To determine when mitotic progression is disturbed in MTR120-depleted cells, we monitored M-phase using live-cell imaging. In the absence of MTR120, cells underwent chromosome segregation, indicating that spindle-MTs can attach to the kinetochore. However, the furrow failed to ingress completely during late mitosis (telophase), leading to a cytokinesis failure. As shown here, MTR120s major function is binding and stabilizing MT. Thus, we suspect that in the absence of MTR120, MT becomes less stable (as indicated by reduced tubulin acetylation), which is at least one of the mechanisms that leads to the cytokinesis defect observed in MTR120-depleted cells.
Because MT stabilization is involved in multiple steps during mitosis, we also evaluated the central spindle organization. Although PRC1, Aurora B kinase, and Polo-like kinase 1 targeted the spindle midzone in MTR120-silenced cells, they had a distorted or discontinued pattern (Fig. 7D,E). Of note, such mislocalization of PRC1 is distinct from what has been observed in Kif-4 knockdown cells. Kif-4 is a binding partner of PRC1. Kif-4 downregulation causes dispersed or diffuse patterns of PRC1 and other midzone proteins, including Aurora B kinase (Hu et al., 2011; Kurasawa et al., 2004; Zhu and Jiang, 2005). We speculate that MTR120 is not essential for loading midzone proteins onto the central spindle. Instead, its involvement is likely to be indirect and structural. Indeed, MTR120 exhibited a robust MT-bundling effect, which is reminiscent of the MT structure induced by the central spindle protein, PRC1 (Mollinari et al., 2002). Overexpressed PRC1 was co-localized with MTR120 in vivo (unpublished observation). PRC1 and other proteins may play an initial role in crosslinking MTs to form a central spindle, whereas MTR120 may play a regulatory role in preventing these MT bundles from collapsing. Alternatively, these MAPs may act synergistically during cytokinesis.
MTR120 orthologs are present in several vertebrates (supplementary material Fig. S1). In addition, MTR120 expression can be detected in a wide range of cell lines (Fig. 2B), indicating that it plays a fundamental role in coordinating cell division. In fact, when we searched the NCBI EST profile, we found that its transcript is present in both the fetus and adult, again suggesting that MTR120 has a general function in cell cycle regulation.
Tetraploidy cells that stem from cytokinesis failure may give rise to more chromosome aberration, which promotes further aneuploidy and genomic instability and eventually leads to tumorigenesis (Holland and Cleveland, 2009). For example, overexpression of oncogene Aurora A kinase causes cytokinesis failure (Meraldi et al., 2002), which may contribute to its oncogenic effect. By searching the public resource in Oncomine (Compendia Bioscience, Ann Arbor, MI), we found that MTR120 mRNA levels in various colorectal cancer tissues are low, ranging from a −2.386-fold (P = 7.05×10−4) to a −3.162-fold (P = 3.12×10−8) change compared with normal tissues using the TCGA colorectal dataset (n = 237). These data raise the possibility that MTR120 downregulation is associated with tumorigenesis. Of course, future studies are needed to test this possibility.
Materials and Methods
Construction of the ORFeome library and large-scale screening
We transferred 15,483 human ORFs (human ORFeome v5.1) in pDONR223 vectors into gateway-compatible destination vectors that contained HA-FLAG tag by LR reaction, according to the manufacturer's protocol (Invitrogen). The products were transformed into DH5α, and the transformants were positively selected using LB medium containing ampicillin (100 µg/ml). The plasmid DNAs were purified using the Purelink HQ 96-plasmid DNA purification kit (Invitrogen).
The day before transfection, 6×103 HeLa cells were seeded on 96-well optical bottom plates (Nunc). Plasmid transfection was performed with Lipofectamine 2000 (Invitrogen). Twenty-four hours after transfection, cells were fixed with 3% paraformaldehyde, permeabilized with solution containing 0.5% Triton X-100, and blocked with 3% BSA. Cells were subjected to incubation with anti-FLAG antibodies (1∶5000 dilution) for 2 hours. They were then washed extensively with phosphate-buffered saline (PBS) and incubated with rhodamine-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories) at ambient temperature for 1 hour. Nuclei were counterstained with 4, 6-diamidino-2-phenylindole (DAPI). Finally, cells were subjected to automated imaging using ImageXpress Micro (Molecular Devices), and the captured images were analyzed by MetaMorph software (Molecular Devices).
DNA constructs
Full-length MTR120 was obtained from human ORFeome as pDONR223 entry clone and transferred to a gateway-compatible destination vector for protein expression. SFB-tag is a triple-epitope tag (S-protein, FLAG and streptavidin-binding peptide) that allows efficient detection and purification of exogenously expressed proteins. Fragments of MTR120 were constructed by polymerase chain reaction. The MTR120 mutants were generated using the QuickChange site-directed mutagenesis kit (Stratagene) and verified by sequencing.
Antibodies
To generate antibodies that specifically recognize MTR120 and MBP, we used GST-fused MTR120 fragment (residues 697-1047) and full-length MBP, expressed and purified from E. coli BL21 (DE3), as an antigen to immunize rabbits. Anti-serum was affinity-purified using the AminoLink Plus immobilization kit and purification kit (Pierce Biotechnology). The following antibodies were purchased from commercial sources: anti-α and anti-γ, acetylated α-tubulin, β-actin and FLAG (M2) antibodies from Sigma, anti-PRC1 (H-70) and anti-cyclin A (C19) antibody from Santa Cruz Biotechnology, anti-Aurora B kinase antibody from BD Bioscience, and anti-PLK1 from Zymed; anti-Aurora A kinase was used as described previously (Yu et al., 2005).
Cell culture, transfection and RNA interference
HeLa and HEK293T (American Type Culture Collection, Manassas, VA) cells were maintained in DMEM supplemented with 10% fetal bovine serum and 1% penicillin and streptomycin. Plasmid transfection was performed using polyethylenimine reagent. To generate a stable cell line that expressed MTR120-SFB, we selected HeLa cells with 2 mg/ml puromycin 24 hours after transfection. Resistant clones were picked, and the expression of the tagged proteins was confirmed by western blotting analysis and immunofluorescence microscopy. Two siRNA duplexes (ThermoScientific) against MTR120 were synthesized: siMTR120: CAATATACAAGCAAGTCTA and siMTR120-2: GAATATAGGAGCAACTAAT. The siRNA duplexes were delivered into cells by transfection using oligofectamine (Invitrogen).
Immunofluorescence staining and time-lapse microscopy
Cells grown on coverslips were fixed in methanol (−20°C for 10 minutes) or 4% paraformaldehyde in PBS (ambient temperature for 15 minutes). They were then subjected to immunostaining using the same protocol as in large-scale screening. The images were captured using a Nikon ECLIPSE E800 fluorescence microscope equipped with a Nikon Plan Fluor 40× oil objective lens (NA 1.30) and a SPOT camera (Diagnostic Instruments, Inc.) or Olympus IX-81 microscope with a disc-spinning confocal attachment. Images were captured with a Hamamatsu Orca II ER camera using a water immersion 60× 1.2 NA objective. Slidebook software from 3I was used to capture and analyze images. Deconvolved images were analyzed with Adobe Photoshop CS4. For time-lapse microscopy, 1×104 GFP-H2B-expressing cells grown on a 24-well sensor plate (VWR International) were transfected with the indicated siRNAs. Seventy-two hours after transfection, fluorescent and brightfield images of various time series were obtained using an Olympus IX-81 confocal microscope equipped with a Wafergen Smartslide Micro-Incubation System, which maintains the environment at 37°C and 5% CO2. Z-stacks of images were obtained every 10 minutes. After the time-lapse experiment, images were exported into Slidebook software from 3I to generate the projective images and time series movies.
Quantification analysis
To quantify the effect of MTR120 overexpression on acetylated α-tubulin from immunofluorescence images, we used NIS-Elements Basic Research software (Nikon). In brief, we chose an area that covered the entire cell image and acquired the fluorescence signals for anti-FLAG or acetylated α-tubulin. The fluorescence intensities were presented (± s.d.) after subtracting the same-sized background intensities. To quantify the midzone MTs or acetylated MT intensities from immunofluorescence images, we chose a rectangular region covering the spindle midzone between two daughter cells during telophase and acquired the fluorescence signals for α-tubulin or acetylated α-tubulin. After subtracting the same-sized background intensities, we normalized the florescence intensities to those of control cells and presented (± s.d.). To quantify the extent of MTR120 knockdown from immunofluorescence images, we chose an area that covered the entire cell image and acquired the fluorescence signals for anti-MTR120 or γ-tubulin. After subtracting the same-sized background intensities, we normalized the florescence intensities of MTR120 to γ-tubulin signals (± s.d.).
Fluorescence-activated cell sorter analysis
Ninety-six hours after transfection, control or siRNA-treated cells were rinsed twice with PBS and fixed by 70% ethanol at 4°C overnight. Cells were then washed twice with PBS, treated with RNase, subjected to propidium iodide (PI) staining, and finally analyzed by flow cytometry analysis (Accutech).
Protein purification
MBP-tagged MTR-M or MTR-M-D2 was induced and expressed in E. coli BL21 (DE3). The bacterial pellets were lysed with binding buffer (25 nM Tris pH 7.5, 100 mM NaCl, 0.5% NP-40, 1 mM EDTA, 1 mM DTT, and protease inhibitors) and clarified by centrifugation (14,000 r.p.m. at 4°C for 15 minutes). Clarified lysates were incubated with amylose resin (New England Biolabs) at 4°C for 2 hours. After extensive washing by binding buffer, the MBP-tagged proteins were eluted with 10 mM maltose. The purified proteins were separated by SDS-PAGE and verified by Coomassie Blue staining.
To purify the full-length MTR120 proteins, we lysed HEK293T cells that expressed WT or D2 mutant MTR120-SFB with binding buffer (25 nM Tris pH 7.5, 100 mM NaCl, 0.5% NP-40, 1 mM EDTA, 1 mM DTT, and protease inhibitors) and clarified them by centrifugation (14,000 r.p.m. at 4°C for 15 minutes). Clarified lysates were incubated with streptavidin Sepharose beads (GE Healthcare) at 4°C for 2 hours. After being extensively washed with binding buffer containing 1 M NaCl and 12 mM deoxycholate, the SFB-tagged proteins were eluted twice with buffer that contained 2 mg/ml biotin. The purified proteins were separated by SDS-PAGE and verified by Coomassie Blue staining.
MT co-sedimentation and bundling assay
Porcine brain tubulin and rhodamine-labeled tubulin were purchased from Cytoskeleton. An MT co-sedimentation assay was performed as described previously (Fong et al., 2008; Fong et al., 2009). In brief, MTs were preassembled at 37°C for 30 minutes in PEM buffer (80 mM PIPES, pH 6.8, 1 mM MgCl2, and 1 mM EGTA) supplemented with 40 µM paclitaxel and 1 mM guanosine triphosphate. Purified proteins were incubated with the indicated doses of MTs in paclitaxel-containing buffer at ambient temperature for 1 hour. The samples were centrifuged at 100,000 g for 15 minutes on a sucrose cushion (25% w/v) in PEM buffer. The resulting pellets and supernatants were run on 10% polyacrylamide gel and stained with Coomassie Blue. To measure the binding affinity, we used Quantity One basic (Bio-Rad) to quantify the intensities of the protein bands from Coomassie gel. The percentages of bound protein were plotted against tubulin concentrations. The dissociation constant (Kd) was determined from the best-fit curve. The data were collected from three independent experiments. To perform the MT bundling assay, we incubated 0.1 µM MTR120-WT, MTR120-D2, or BSA with 4 µM paclitaxel-stabilized rhodamine-labeled MTs at ambient temperature for 20 minutes. The reaction was fixed with 1% glutaraldehyde, and the MTs were sedimented on the coverslip. The MTs' morphological characteristics were evaluated by fluorescence microscopy.
Supplementary Material
Acknowledgments
We thank our colleagues in Junjie Chen's laboratory for insightful discussions and technical assistance. We thank Robert Qi for providing the GFP-H2B plasmid and Gargi Ghosal for critical reading of the manuscript.
Footnotes
Author contributions
K.W.F. and J.W.L. designed and carried out most of the experiments; Y.L., W.W., L.F. and W.M. constructed the Human ORFeome library; D.L., Z.S. and J.C. advised on the design of the experiments; K.W.F. and J.C. were responsible for the preparation of the manuscript.
Funding
This work was supported by a post-doctoral fellowship from the Canadian Louis Pasteur Foundation [grant number xxxxx to D.B.]; the European Union FP7 Priority [grant number 222887 to C.Z.]; ANR (DISCover, 2009) [grant number NEUR 00203 to X.X.]; and and the Pasteur-Weizmann Foundation (2010-2012) [grant number xxxxx to D.B.].
Funding
This work was supported in part by a grant from the National Institutes of Health [grant numbers CA113381 to J.C., CA133249, GM081627 and GM095599 to Z.S.]; Department of Defense (DOD) Era of Hope Research awards to J.C. [grant numbers W81XWH-09-1-0409, W81XWH-05-1-0470]; the National Basic Research Program [grant numbers 2010CB945400 and 2012CB911201]; the National Natural Science Foundation of China [grant number NSFC 91019020]; and the Welch Foundation [grant number Q-1673]. We also acknowledge the support of the Dan L. Duncan Cancer Center [grant number P30CA125123] and the Administrative and Genome-wide RNAi Screen Cores [grant number IDDRC P30HD024064]. J.C. is a member of MD Anderson Cancer Center [grant number CA016672]. Deposited in PMC for release after 12 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.116137/-/DC1
References
- Fong K. W., Choi Y. K., Rattner J. B., Qi R. Z. (2008). CDK5RAP2 is a pericentriolar protein that functions in centrosomal attachment of the gamma-tubulin ring complex. Mol. Biol. Cell 19, 115–125 10.1091/mbc.E07-04-0371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong K. W., Hau S. Y., Kho Y. S., Jia Y., He L., Qi R. Z. (2009). Interaction of CDK5RAP2 with EB1 to track growing microtubule tips and to regulate microtubule dynamics. Mol. Biol. Cell 20, 3660–3670 10.1091/mbc.E09-01-0009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer M. (2009). The 3Ms of central spindle assembly: microtubules, motors and MAPs. Nat. Rev. Mol. Cell Biol. 10, 9–20 10.1038/nrm2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green R. A., Wollman R., Kaplan K. B. (2005). APC and EB1 function together in mitosis to regulate spindle dynamics and chromosome alignment. Mol. Biol. Cell 16, 4609–4622 10.1091/mbc.E05-03-0259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromley A., Jurczyk A., Sillibourne J., Halilovic E., Mogensen M., Groisman I., Blomberg M., Doxsey S. (2003). A novel human protein of the maternal centriole is required for the final stages of cytokinesis and entry into S phase. J. Cell Biol. 161, 535–545 10.1083/jcb.200301105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromley A., Yeaman C., Rosa J., Redick S., Chen C. T., Mirabelle S., Guha M., Sillibourne J., Doxsey S. J. (2005). Centriolin anchoring of exocyst and SNARE complexes at the midbody is required for secretory-vesicle-mediated abscission. Cell 123, 75–87 10.1016/j.cell.2005.07.027 [DOI] [PubMed] [Google Scholar]
- Gudi R., Zou C., Li J., Gao Q. (2011). Centrobin-tubulin interaction is required for centriole elongation and stability. J. Cell Biol. 193, 711–725 10.1083/jcb.201006135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guse A., Mishima M., Glotzer M. (2005). Phosphorylation of ZEN-4/MKLP1 by aurora B regulates completion of cytokinesis. Curr. Biol. 15, 778–786 10.1016/j.cub.2005.03.041 [DOI] [PubMed] [Google Scholar]
- Hinchcliffe E. H., Miller F. J., Cham M., Khodjakov A., Sluder G. (2001). Requirement of a centrosomal activity for cell cycle progression through G1 into S phase. Science 291, 1547–1550 10.1126/science.1056866 [DOI] [PubMed] [Google Scholar]
- Holland A. J., Cleveland D. W. (2009). Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat. Rev. Mol. Cell Biol. 10, 478–487 10.1038/nrm2718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppeler–Lebel A., Celati C., Bellett G., Mogensen M. M., Klein–Hitpass L., Bornens M., Tassin A. M. (2007). Centrosomal CAP350 protein stabilises microtubules associated with the Golgi complex. J. Cell Sci. 120, 3299–3308 10.1242/jcs.013102 [DOI] [PubMed] [Google Scholar]
- Hu C. K., Coughlin M., Field C. M., Mitchison T. J. (2011). KIF4 regulates midzone length during cytokinesis. Curr. Biol. 21, 815–824 10.1016/j.cub.2011.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang K., Akhmanova A. (2011). Microtubule tip-interacting proteins: a view from both ends. Curr. Opin. Cell Biol. 23, 94–101 10.1016/j.ceb.2010.08.008 [DOI] [PubMed] [Google Scholar]
- Kapitein L. C., Peterman E. J., Kwok B. H., Kim J. H., Kapoor T. M., Schmidt C. F. (2005). The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature 435, 114–118 10.1038/nature03503 [DOI] [PubMed] [Google Scholar]
- Khodjakov A., Rieder C. L. (2001). Centrosomes enhance the fidelity of cytokinesis in vertebrates and are required for cell cycle progression. J. Cell Biol. 153, 237–242 10.1083/jcb.153.1.237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollman J. M., Merdes A., Mourey L., Agard D. A. (2011). Microtubule nucleation by γ-tubulin complexes. Nat. Rev. Mol. Cell Biol. 12, 709–721 10.1038/nrm3209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kufer T. A., Silljé H. H., Körner R., Gruss O. J., Meraldi P., Nigg E. A. (2002). Human TPX2 is required for targeting Aurora-A kinase to the spindle. J. Cell Biol. 158, 617–623 10.1083/jcb.200204155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurasawa Y., Earnshaw W. C., Mochizuki Y., Dohmae N., Todokoro K. (2004). Essential roles of KIF4 and its binding partner PRC1 in organized central spindle midzone formation. EMBO J. 23, 3237–3248 10.1038/sj.emboj.7600347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Jeong Y., Jeong S., Rhee K. (2010). Centrobin/NIP2 is a microtubule stabilizer whose activity is enhanced by PLK1 phosphorylation during mitosis. J. Biol. Chem. 285, 25476–25484 10.1074/jbc.M109.099127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüders J., Stearns T. (2007). Microtubule-organizing centres: a re-evaluation. Nat. Rev. Mol. Cell Biol. 8, 161–167 10.1038/nrm2100 [DOI] [PubMed] [Google Scholar]
- Meraldi P., Honda R., Nigg E. A. (2002). Aurora-A overexpression reveals tetraploidization as a major route to centrosome amplification in p53-/- cells. EMBO J. 21, 483–492 10.1093/emboj/21.4.483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merdes A., Heald R., Samejima K., Earnshaw W. C., Cleveland D. W. (2000). Formation of spindle poles by dynein/dynactin-dependent transport of NuMA. J. Cell Biol. 149, 851–862 10.1083/jcb.149.4.851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollinari C., Kleman J. P., Jiang W., Schoehn G., Hunter T., Margolis R. L. (2002). PRC1 is a microtubule binding and bundling protein essential to maintain the mitotic spindle midzone. J. Cell Biol. 157, 1175–1186 10.1083/jcb.200111052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollinari C., Kleman J. P., Saoudi Y., Jablonski S. A., Perard J., Yen T. J., Margolis R. L. (2005). Ablation of PRC1 by small interfering RNA demonstrates that cytokinetic abscission requires a central spindle bundle in mammalian cells, whereas completion of furrowing does not. Mol. Biol. Cell 16, 1043–1055 10.1091/mbc.E04-04-0346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavicic–Kaltenbrunner V., Mishima M., Glotzer M. (2007). Cooperative assembly of CYK-4/MgcRacGAP and ZEN-4/MKLP1 to form the centralspindlin complex. Mol. Biol. Cell 18, 4992–5003 10.1091/mbc.E07-05-0468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salisbury J. L., Suino K. M., Busby R., Springett M. (2002). Centrin-2 is required for centriole duplication in mammalian cells. Curr. Biol. 12, 1287–1292 10.1016/S0960-9822(02)01019-9 [DOI] [PubMed] [Google Scholar]
- Sauer G., Körner R., Hanisch A., Ries A., Nigg E. A., Silljé H. H. (2005). Proteome analysis of the human mitotic spindle. Mol. Cell. Proteomics 4, 35–43 10.1074/mcp.M400158-MCP200 [DOI] [PubMed] [Google Scholar]
- Sessa F., Mapelli M., Ciferri C., Tarricone C., Areces L. B., Schneider T. R., Stukenberg P. T., Musacchio A. (2005). Mechanism of Aurora B activation by INCENP and inhibition by hesperadin. Mol. Cell 18, 379–391 10.1016/j.molcel.2005.03.031 [DOI] [PubMed] [Google Scholar]
- Silk A. D., Holland A. J., Cleveland D. W. (2009). Requirements for NuMA in maintenance and establishment of mammalian spindle poles. J. Cell Biol. 184, 677–690 10.1083/jcb.200810091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres J. Z., Summers M. K., Peterson D., Brauer M. J., Lee J., Senese S., Gholkar A. A., Lo Y. C., Lei X., Jung K.et al. (2011). The STARD9/Kif16a kinesin associates with mitotic microtubules and regulates spindle pole assembly. Cell 147, 1309–1323 10.1016/j.cell.2011.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang W. Y., Spektor A., Luciano D. J., Indjeian V. B., Chen Z., Salisbury J. L., Sánchez I., Dynlacht B. D. (2006). CP110 cooperates with two calcium-binding proteins to regulate cytokinesis and genome stability. Mol. Biol. Cell 17, 3423–3434 10.1091/mbc.E06-04-0371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheatley S. P., Wang Y. (1996). Midzone microtubule bundles are continuously required for cytokinesis in cultured epithelial cells. J. Cell Biol. 135, 981–989 10.1083/jcb.135.4.981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Minter–Dykhouse K., Malureanu L., Zhao W. M., Zhang D., Merkle C. J., Ward I. M., Saya H., Fang G., van Deursen J.et al. (2005). Chfr is required for tumor suppression and Aurora A regulation. Nat. Genet. 37, 401–406 10.1038/ng1538 [DOI] [PubMed] [Google Scholar]
- Zhao W. M., Seki A., Fang G. (2006). Cep55, a microtubule-bundling protein, associates with centralspindlin to control the midbody integrity and cell abscission during cytokinesis. Mol. Biol. Cell 17, 3881–3896 10.1091/mbc.E06-01-0015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C., Jiang W. (2005). Cell cycle-dependent translocation of PRC1 on the spindle by Kif4 is essential for midzone formation and cytokinesis. Proc. Natl. Acad. Sci. USA 102, 343–348 10.1073/pnas.0408438102 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.