Abstract
We evaluated the presence/absence of proteins encoded by 14 077 genes in adipocytes obtained from different tissue samples using immunohistochemistry. By combining this with previously published adipocyte-specific proteome data, we identified proteins associated with 7340 genes in human adipocytes. This information was used to reconstruct a comprehensive and functional genome-scale metabolic model of adipocyte metabolism. The resulting metabolic model, iAdipocytes1809, enables mechanistic insights into adipocyte metabolism on a genome-wide level, and can serve as a scaffold for integration of omics data to understand the genotype–phenotype relationship in obese subjects. By integrating human transcriptome and fluxome data, we found an increase in the metabolic activity around androsterone, ganglioside GM2 and degradation products of heparan sulfate and keratan sulfate, and a decrease in mitochondrial metabolic activities in obese subjects compared with lean subjects. Our study hereby shows a path to identify new therapeutic targets for treating obesity through combination of high throughput patient data and metabolic modeling.
Keywords: adipocyte, flux balance analysis, genome-scale metabolic model, obesity, proteome
Introduction
White adipose tissue (WAT) serves as an important buffer for handling daily lipid flux by uptake of fatty acids (FAs) in the post-prandial state and release in the post-absorptive state (Frayn, 2002). Furthermore, studies in recent years have revealed that WAT is a major endocrine organ sending out hormones and signaling molecules that regulate and coordinate energy homeostasis, insulin sensitivity, lipid metabolism, substrate selection, satiety and appetite (Cristancho and Lazar, 2011). WAT also cooperates with other tissues including liver, muscle, pancreas, heart and brain through the release of FAs (lipokines), secretory factors (adipokines) and pro-inflammatory cytokines. WAT dysfunction or overload of its lipid storage capacity can lead to wide range of diseases (e.g., immunological and inflammatory diseases), including metabolic diseases such as obesity and its adverse outcomes (Lago et al, 2007; Auffray et al, 2009). Obesity can lead to preventable cause of death and increases the likelihood of coronary heart disease, diabetes and several forms of cancer (Cao, 2010) and ∼33% of the adults older than 20 years living in the United States and 20% of European adult population are obese. Obesity is therefore considered to be one of the greatest threats to global human health (Caveney et al, 2011). However, obesity is not directly related with particular etiological factors and it is therefore essential to enable stratification and prediction of disease risks among obese subjects to ensure early treatment either through diet intervention, exercise or surgery.
An increased understanding of the mechanisms behind obesity and related diseases will provide valuable insights into their etiology, pathogenesis and may lead to new treatment strategies. It is inherently difficult to find the exact disease onset, since such systemic diseases are caused by a combination of different genetic and environmental factors and often result in similar disease phenotypes. Therefore, it remains challenging to identify the cellular and molecular mechanisms associated with obesity, in particular as the metabolism of the single cell involves thousands of metabolites and interconnected chemical reactions that occur simultaneously. Proper understanding of such a complex system requires a holistic approach. For this purpose, the so-called genome-scale metabolic models (GEMs) are suitable as they allow for analysis of metabolism at the genome scale but at the same time ensures identification of specific pathways, enzymes and metabolites associated with specific phenotypes (Nielsen, 2009; Thiele and Palsson, 2010).
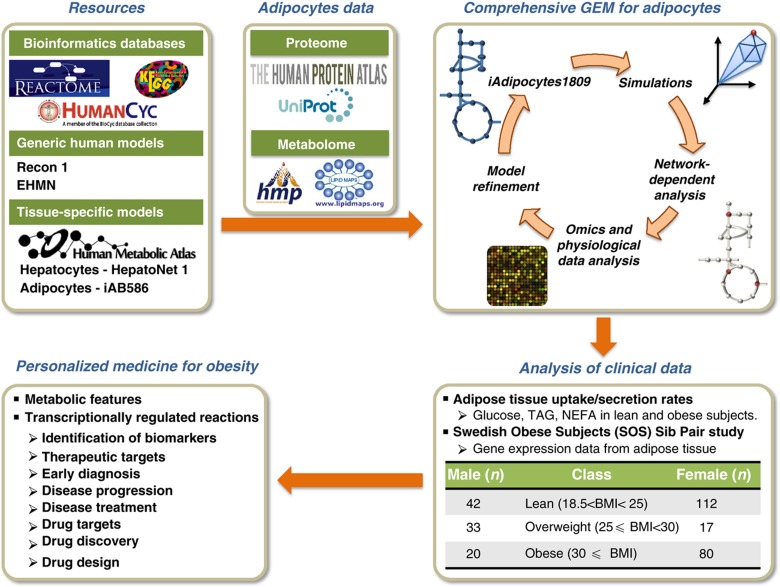
GEMs integrate biochemical and physiological data on genes and enzymatic reactions and allow the study of relationships between networks, functions, diseases and patients at a systems level. Reconstructed large-scale GEMs can serve as scaffolds for data analysis and identify biologically meaningful correlations and mechanistic relationships between components of different sub-systems (Patil and Nielsen, 2005). Hereby, GEMs can be employed for understanding the underlying mechanisms of complex diseases, and hence be used for identification of novel therapeutic and drug targets and discovery of new biomarkers. The diseases that we are discussing are not difficult to diagnose (e.g., obesity and diabetes) but efficient biomarkers would be of interest to predict prognosis and outcome of the disease and thereby enabled patient stratification which will be the basis for developing personalized medicine (Mardinoglu and Nielsen, 2012; Nielsen, 2012; Figure 1).
Figure 1.
Genome-scale metabolic models (GEMs) provide a scaffold for integrative analysis of clinical data. Schematic illustration of how a comprehensive and functional GEM for adipocytes may provide links between specific molecular processes and subject phenotypes and hereby contribute to the development of personalized medicine for obesity. Here, the GEM iAdipocytes1809 was reconstructed to bridge the gap between genotype and phenotype through the use of proteome, metabolome, lipidome and transcriptome data, literature-based models (Recon 1, Edinburg Human Metabolic Network (EHMN) and HepatoNet1) and public resources (Reactome, HumanCyc, KEGG and the Human Metabolic Atlas). We first performed global protein profiling of adipocytes encoded by 14 077 genes to study adipocyte biology at the genome-wide level using antibodies generated within the Human Protein Atlas (HPA). We further used information on metabolome and lipidome data from the Human Metabolome Database (HMDB) and LIPID MAPS Lipidomics Gateway, respectively. Model driven simulations, network-dependent analysis and condition-specific transcriptome data allowed for model refinement. iAdipocytes1809 was used for the analysis of gene expression data obtained from subjects with different body mass indexes in the Swedish Obese Subjects (SOS) Sib Pair study and other adipose tissue relevant clinical data such as uptake/secretion rates in lean and obese subjects. Employing iAdipocytes1809 for integration of transcriptome and fluxome enables understanding of adipocyte metabolism in obese subjects compared with lean subjects. Furthermore, the results of the study lead to identification of molecular mechanisms underlying obesity and its adverse outcomes. This can be useful for identification of new therapeutic and drug targets and discovery of new biomarkers for predicting prognosis and outcome of the disease, developing a system-oriented drug design strategy, obtaining novel diagnostic and therapeutic techniques and eventually determining effective personalized medicines for treatment of obesity-related diseases.
Two generic literature-based GEMs of human metabolism, Recon 1 (Duarte et al, 2007) and the Edinburgh human metabolic network (EHMN; Ma et al, 2007) have been reconstructed previously. Although these first reconstructions represent a major advancement, human metabolism is specialized in different cell types, and hence there is a need for reconstruction of cell type or tissue-specific GEMs. In this context, tissue-specific GEMs have been developed for liver (hepatocytes) (Gille et al, 2010; Jerby et al, 2010), cardiomyocytes (Karlstaedt et al, 2012), kidney (Chang et al, 2010), brain (Lewis et al, 2010) and alveolar macrophage (Bordbar et al, 2010) and recently, three small cell type-specific GEMs of hepatocytes, myocytes and adipocytes (iAB586) were developed (Bordbar et al, 2011). Furthermore, using the Integrative Network Inference for Tissues (INIT) algorithm, we previously reconstructed cell type-specific draft GEMs for 69 different cell types and 16 cancer types (Agren et al, 2012).
With the objective of gaining new insight into adipocyte metabolism at the genome level, we first used human antibodies to evaluate the presence/absence of 14 077 proteins in adipocytes found in breast and two different soft tissue samples and checked their presence call with previously published adipocyte proteome data. Second, we manually reconstructed a high-quality, simulation ready GEM for adipocytes by using all adipocyte-specific proteome data. The model is based on previously published GEMs but also on publicly available databases on metabolism. Third, we employed the functional GEM for the analysis of microarrays that profile the gene expression from subcutaneous adipose tissue (SAT) of subjects from the Swedish Obese Subjects (SOS) Sib Pair Study, which includes nuclear families with body mass index (BMI)–discordant sibling pairs (BMI difference⩾10 kg/m2). The male and female participants of the study were divided into three different groups based on their BMIs: lean, overweight and obese. Analysis of transcription data from this study recently demonstrated that there are differences in mitochondrial function between men and women (Nookaew et al, 2012), but there was not performed any analysis on the effect of obesity. We incorporated differentially expressed genes between obese and lean subjects into the GEM. Besides the gene expression data from the SOS Sib Pair Study, additional clinical data (e.g., plasma and WAT lipid concentrations) were also incorporated into the model. By integrating gene expression data and adipose tissue uptake/secretion rates with the reconstructed GEM, we identified metabolic differences between individuals with different BMIs by using the concept of Reporter Metabolites (Patil and Nielsen, 2005) and transcriptionally controlled reaction fluxes (Bordel et al, 2010; Figure 1).
Results
Immunohistochemistry-based proteomics of human adipocytes
Several studies have reported proteomics data of human WAT that include not only adipocytes but also the connective tissue matrix, nerve tissue, stromal vascular cells and immune cells (Peinado et al, 2012). Recently, Xie et al (2010) characterized the proteome of human adipocytes and reported existence of proteins encoded by 1574 genes in subcutaneous abdominal adipocytes taken from three healthy lean subjects. Although this first adipocyte-specific proteome study is promising to understand the unique characteristics of the adipocytes, it is still necessary to expand the protein coverage to be able to study adipocyte biology at the genome-wide level.
The Human Protein Atlas (HPA) is a knowledge-based portal that cover the annotated protein expression feature for protein targets analyzed with one or more antibodies and the main subcellular localization of protein targets in all major human cell types and cell lines (Uhlen et al, 2010). Here, we expanded the coverage of the HPA so as to define the protein profiles of adipocytes found in breast and two different soft tissues and examined the spatial distribution and the relative abundance of proteins encoded by 14 077 genes in adipocytes (Supplementary Dataset 1a). Soft tissue cores with 1 mm diameter were sampled from different locations of the human body and included in tissue microarrays (TMAs) together with breast tissue core as previously described (Kampf et al, 2012). A total of 17 296 high-throughput generated affinity-purified antibodies (Supplementary Dataset 1a) against 14 077 proteins were arranged according to abundance of the corresponding protein target. Immunohistochemically stained sections from TMAs blocks were scanned in high-resolution scanners and separated to individual spot images to represent each core. Here, proteins with strong, moderate and weak relative expression were included in the reconstruction process of the GEM for adipocytes. It is important to point out that the absolute levels of each protein have not been determined and could vary by orders of magnitude. The high-resolution images together with annotation of the presence or absence of a particular protein target in adipocytes are publically available through the HPA (http://www.proteinatlas.org).
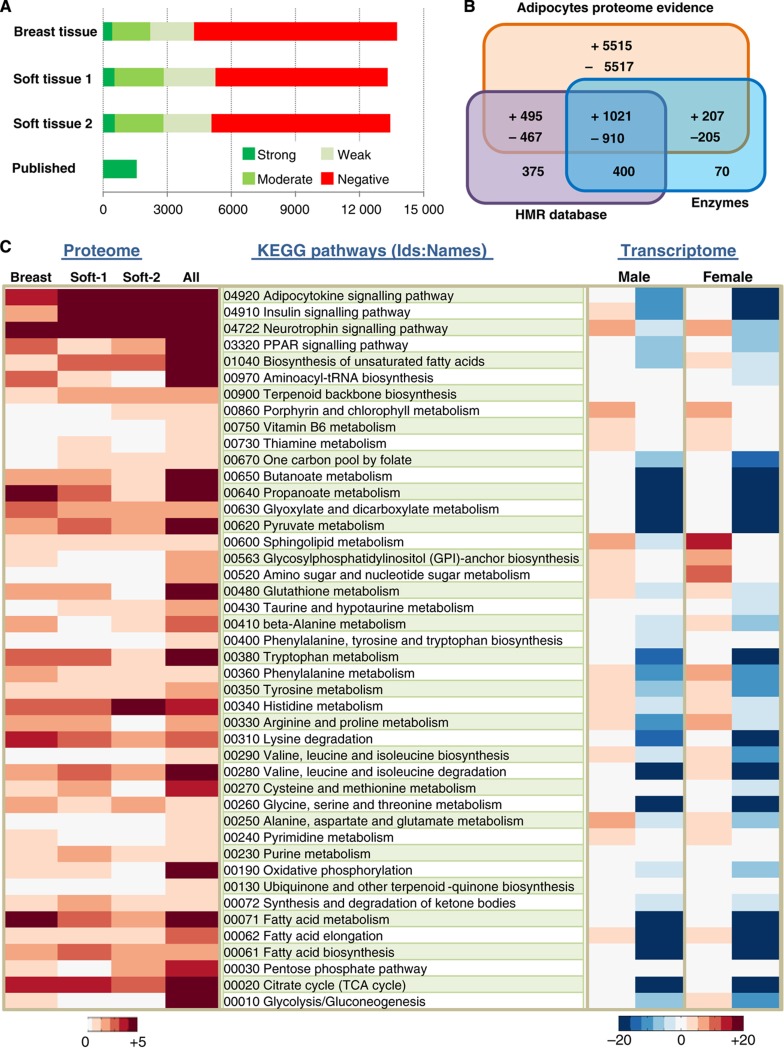
The proteome data generated for adipocytes found in breast and two different soft tissues were merged with previously published proteome data as presented in Figure 2A (Supplementary Dataset 1b). In total, we have proteome evidence for the presence/absence of proteins associated with 14 337 genes in adipocytes and together with the genes in our Human Metabolic Reaction (HMR) database (see later), we cover 98% of the enzymes (Supplementary Dataset 1c) reported in the HPA database (Figure 2B). Our proteomics analysis resulted in provision of evidence for presence of proteins associated with 7340 genes in adipocytes that could be used for our GEM reconstruction. Each protein that has evidence for presence/absence in adipocytes was annotated with UniProt id (Apweiler et al, 2011) and the corresponding encoding genes were annotated with Ensembl (Flicek et al, 2011) id for standardization.
Figure 2.
Adipocytes proteome data and its correlation with transcriptome data. (A) Proteins encoded by 14 077 genes were annotated for their presence or absence in adipocytes found in breast and two different soft tissues sampled from different locations within the body using antibodies generated within the Human Protein Atlas (HPA) project. The proteome evidence for adipocytes in each tissue sample and previously published adipocytes proteome data were merged and there are proteome evidence for the presence/absence of proteins associated with 14 337 genes in adipocytes. (B) The here generated proteome data combined with published adipocytes specific proteome data encodes for a total of 7340 genes that were used to reconstruct a comprehensive genome-scale metabolic model (GEM) for adipocytes, iAdipocytes1809. The Venn diagram shows how these genes overlap with genes in an updated version of the Human Metabolic Reaction (HMR) database and enzymes in the HPA. (+) indicates the presence of protein encoded genes whereas (−) means the absence of protein encoded genes in adipocytes based on the here generated HPA data. (C) Enrichment of KEGG pathways is presented (P-value from hypergeometric distribution is used) to check the coverage of here generated adipocyte-specific proteome data found in breast, soft tissue 1 (Soft-1), soft tissue 2 (Soft-2) and all adipocyte-specific proteome data. The enrichment of differentially expressed genes in subcutaneous adipose tissue (SAT) of male and female obese subjects compared with lean subjects was identified in the enriched KEGG pathways in all adipocyte-specific proteome data.
Since adipocytes obtained from breast and two different soft tissues were used for the protein profiling, there was some variation between the samples. To estimate the effect of these variations on the functionality of the adipocytes, we used the functional annotation tool DAVID to calculate the enrichment in KEGG pathways (Huang et al, 2009). The results are presented in Figure 2C for breast, the two types of soft tissues, as well as for all proteomics data used for the GEM reconstruction. The analysis demonstrated that for all the adipocyte-specific proteome data, there is enrichment in terms of metabolic pathways including FA metabolism, elongation and biosynthesis as well as major signaling pathways including adipocytokine, insulin, neurotrophin and PPAR signaling pathways. The figure shows that the samples from different tissues exhibit some differences, but that the overall pattern is similar. We therefore decided to reconstruct a general GEM for adipocytes by incorporating all proteins that were expressed in any of the three tissues.
Subcellular localization of the proteins and associated reactions
Metabolism is distributed across subcellular compartments of the cell and precise coordination is important for overall cellular function and maintenance of metabolic homeostasis. For instance, β-oxidation of very long chain FAs (VLCFAs) is a process that starts in the peroxisome and ends in the mitochondria. The subcellular localization of reactions has large implications on the functionality of GEMs, as only a portion of metabolites can be transported between compartments. Furthermore, compartments can be individually redox and/or energy balanced. It is therefore important to understand the locations of individual enzymes as well as their associated reactions when metabolism is going to be studied at the genome scale. The HPA includes subcellular profiling data using immunofluorescence-based confocal microscopy in three human cancer cell lines of different origin to be used for further in-depth functional studies.
Here, proteins were classified into eight different compartments following our HMR database standard (Agren et al, 2012): cytosol, nucleus, endoplasmic reticulum (ER), Golgi apparatus (GA), peroxisome, lysosome, mitochondria and extracellular space. Reactions were assigned to compartments through their association with proteins in these different compartments. We assigned a confidence score ranging from one to three for each protein due the availability of knowledge in HPA and Uniprot (Supplementary Dataset 2a). Proteins localized in the aggresome, centrosome and cytoskeletons were assigned to the cytoplasm, whereas proteins in cell junctions and focal adhesions were assigned to the extracellular compartment (Supplementary Dataset 2b). Lysosome and peroxisome were merged into vesicles in the HPA data, but proteins associated with vesicles in our model were separated based on Uniprot data. In the case a protein has different locations in the two resources, we chose to assign the protein in the location reported based on the HPA data. In cases where the subcellular localization information was absent in HPA, Uniprot and literature (indirect physiological evidence), proteins and their associated reactions were assigned to be present in the cytosol.
Transcriptome data for SAT and its correlation with proteome data
Gene expression in SAT of 304 subjects involved in the SOS Sib Pair Study was analyzed by using Affymetrix U133 Plus 2.0 microarrays (Affymetrix, Santa Clara, CA, USA). The participants of the study, 209 female and 95 male subjects, were divided into three different groups according to their BMI: lean (18.5<BMI <25), overweight (25⩽ BMI<30) and obese (30⩽BMI) (Supplementary Table S1). Our study considers the differences in male and female obese subjects compared with lean male and female subjects independently. Differentially expressed genes between obese, overweight and lean subjects for male and female subjects were identified and differentially expressed genes on male and female obese subjects were incorporated during the reconstruction process of GEM for adipocytes to analyze gene expression data from the SOS Sib Pair Study.
Although the generated proteome data for adipocytes cover the entire set of cellular processes (e.g., signaling, metabolism and cell cycle), GEMs are applicable only for the study of metabolism. To get a general overview of the global changes between obese, overweight and lean subjects, the enrichment of differentially expressed genes was calculated for KEGG pathways (Supplementary Figures S1 and S2) and for biological process Gene Ontology (BP:GO) terms (Supplementary Figures S3 and S4). This was done using DAVID and for male and female subjects (Huang et al, 2009). To check the correlation of the genome-wide transcription data of SAT with the proteome data, the enrichment of differentially expressed genes in male and female obese subjects was calculated for the most significant KEGG pathways from the analysis of the proteome data (Figure 2C). This was done as the transcriptome data for SAT represent not only adipocytes but also other cell types; including immune cells and preadipocytes linked with different BMIs.
The analysis demonstrates that some of the metabolic and signaling pathways found to be enriched in adipocytes based on the proteome data also show significant changes in gene expression between lean and obese subjects, both in males and in females. Similarly, we find that BP:GO terms that are enriched based on the proteome data (assessed with DAVID; Huang et al, 2009) also show enrichment based on the transcriptome data (Supplementary Dataset 3). Thus, enriched BP:GO terms such as post-translational protein modification, cellular protein metabolic process, lipid metabolic process, cellular lipid metabolic process and FA metabolic process are found both from the adipocyte-specific proteome data and from comparison of expression data for lean and obese subjects.
Reconstruction of the adipocytes GEM iAdipocytes1809
GEMs are reconstructed based on high-throughput data such as genome, transcriptome, proteome, metabolome and fluxome and they provide an excellent scaffold for integrative analysis of this kind of data (Patil and Nielsen, 2005; Cakir et al, 2006). To reconstruct a large-scale comprehensive GEM for adipocytes, biochemical and genetic evidence were combined with data on protein expression and localization. Besides the proteins associated with 7340 genes identified from our proteomics analysis, we further used differentially expressed genes in SAT of obese male and female subjects for our reconstruction process. However, SAT contains not only adipocytes but also other cell types and it is therefore necessary to check the functionality of the genes before including into the GEM.
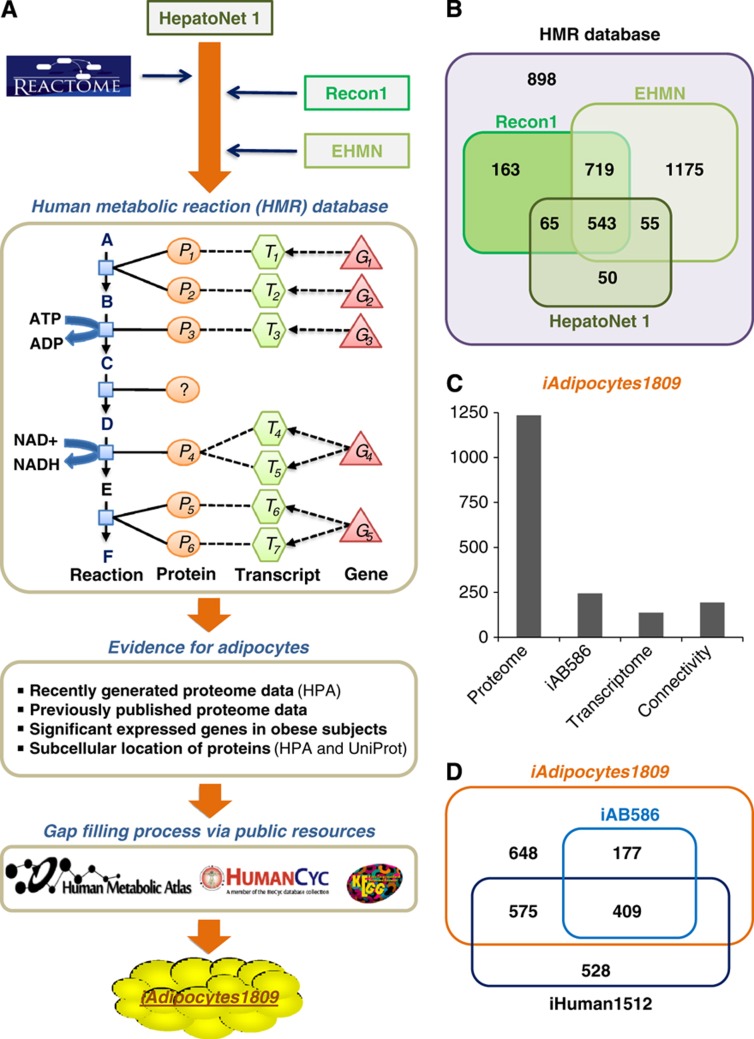
HepatoNet1, a GEM for hepatocytes which is reconstructed based on the manual evaluation of the original scientific literature (Gille et al, 2010), was used as a starting point for our reconstruction process and used to generate an initial candidate list of network components (Figure 3A). First, metabolism of lipids and lipoproteins in Reactome, a manually curated and peer-reviewed pathway database (Croft et al, 2011), was merged into HepatoNet1. Second, the resulting network was combined with the evidence-based generic human models Recon1 (Duarte et al, 2007) and the compartmentalized EHMN (Hao et al, 2010). This combined reaction list resulted in an updated version of our HMR database that is available at http://www.metabolicatlas.org. The HMR database contains 6000 metabolites in 8 different compartments (3160 unique metabolites), 8100 reactions and 3668 genes associated with those reactions. HMR includes all of the genes and gene-associated reactions in HepatoNet1, Recon1 and EHMN and other isolated gene-associated reactions, and represents the most comprehensive human reaction database for genome-scale modeling (Figure 3B). Third, the existence of each protein coding genes associated with a reaction in HMR was assessed for the presence or absence in adipocytes using previously published and the here generated adipocyte-specific proteome data. Differentially expressed genes and associated reactions in obese subjects compared with lean subjects were also included in the model by checking their functionality. This process provided us with a list of reactions that occur in adipocytes. Gaps in the resulting network were filled using our recently developed iHuman1512 metabolic network (Agren et al, 2012), public databases such as KEGG (Kanehisa et al, 2010) and HumanCyc (Romero et al, 2005) and manual evaluation of the literature about adipocyte metabolism. This gap filling resulted in generation of iAdipocytes1809 that is a functional and fully connected GEM for adipocytes (Figure 3A). Reactions were included in the GEM depending on evidence from previously published models and databases (Supplementary Table S2) or on the availability of specific experimental evidence, for example, enzyme assay or protein identification, for the occurrence of the corresponding reaction in adipocytes. Reaction directionality in the model was treated as in the original resource and the directionality was only changed if it was necessary to perform successful simulations of known biological functions of adipocytes. iAdipocytes1809 contains 6160 reactions and 4550 metabolites in 8 different compartments (2497 unique metabolites) (Supplementary Table S3) and it is available in the Systems Biology Mark-up Language (SBML) format at http://www.metabolicatlas.org. There are 1809 genes in the model and 80% of the reactions are associated with one or more genes. In iAdipocytes1809, individual metabolites rather than generic pool metabolites for 59 FAs (Supplementary Table S4) have been used and this allowed us to incorporate measured concentrations of different FAs in human plasma and adipocytes into the model.
Figure 3.
Illustration of the reconstruction process for iAdipocytes1809. (A) Human Metabolic Reaction (HMR) database was updated through the use of literature and previously published GEMs like HepatoNet1, Recon1 and EHMN and the Reactome database, with focus on the extensive lipid metabolism in adipocytes. HMR provides gene-transcript protein reaction (GTPR) links and GTPR represents functional relationships between genes/proteins and the corresponding reactions that they catalyze or control. Genes are first mapped to their transcripts, accounting for alternative splicing, the transcripts are mapped to the proteins and the proteins are then mapped to reactions based on the current knowledge of their effects on the reactions. In HMR, 75% of the reactions have GTPR and this allow us to incorporate proteome and transcriptome evidence in the adipocyte-specific network. The resulting gaps in the adipocyte-specific network were identified using the RAVEN Toolbox and filled manually using publicly available resources like KEGG, HumanCyc and Human Metabolic Atlas. Based on this, the comprehensive functional genome-scale metabolic model (GEM) for adipocytes, iAdipocytes1809, was reconstructed. (B) The Venn diagram illustrating the comparison of genes associated with reactions in the HMR and previously published literature-based models such as Recon 1, EHMN and HepatoNet 1. HMR includes all of the genes and associated reactions in previous published models. (C) Protein encoding genes and associated reactions with those genes included in iAdipocytes1809 based on proteome (1235 genes) and transcriptome (137 genes) data and evidence from previously published small-scale adipocytes model, iAB586 (244 genes). Another 193 genes and their associated reactions were included in the model due to connectivity constraints and known function of adipocytes. (D) iAdipocytes1809 is compared with iAB586 and fully connected generic human network, iHuman1512. iAdipocytes1809 contains all of the genes and associated reaction in iAB586 and all of the adipocyte-specific genes and associated reactions in iHuman1512.
Adipocyte-specific metabolome data from the Human Metabolome Database (HMDB) (Wishart et al, 2009) and a comprehensive database for lipid biology, Lipidomics Gateway (Harkewicz and Dennis, 2010), were used in the reconstruction process. To ensure standardization, each metabolite in the GEM was assigned at least one of the following: HMDB ids, Lipidomics Gateway ids, KEGG ids (Kanehisa et al, 2010), Chemical Entities of Biological Interest (ChEBI) (Degtyarenko et al, 2008), or International Chemical Identifiers (InChI). In the model, new genes were assigned to the reactions in iAdipocytes1809 using EC numbers from UniProt (Apweiler et al, 2011) and the Lipid Map proteome database (Cotter et al, 2006).
The reconstruction of iAdipocytes1809 involved a comprehensive review of lipid metabolism and gene products related to the lipid metabolism in publicly available databases were closely examined for their existence in adipocytes. In all, 1235 protein encoding genes with proteome evidence in adipocytes and 244 genes in iAB586 were included in iAdipocytes1809 (Figure 3C). Another 137 genes were included in the model due to positive evidence for the presence of these proteins from the transcriptome data. These 137 genes were significantly higher expressed both in male and in female obese subjects compared with lean subjects, and are hence relevant for the analysis of gene expression data from the SOS Sib Pair Study. Another 193 genes and their associated reactions were included in the model due to connectivity constraints and known function of adipocytes and these genes are mainly associated with transport across membranes. In all, 73 of these genes do not have any negative evidence whereas 120 of these genes have negative HPA scores. These 120 genes are likely to be false negatives in the HPA, and they can be explained by either poor antibody hybridization to adipocytes or condition-dependent protein expression. We also compared iAdipocytes1809 with iAB586 and the previously published generic human network, iHuman1512, and we found that iAdipocytes1809 contains all of the genes and associated reactions in iAB586 and adipocyte-specific reactions and associated genes of iHuman1512 (Figure 3D).
In iAdipocytes1809, 59 different common long and very long chain FAs (Supplementary Table S4) in human plasma can be taken up as NEFAs and lipoproteins (Supplementary Figures S5–S7). Cholesterol and its 59 different CEs can also be taken up from low-density lipoproteins (LDLs) and high-density lipoproteins (HDLs). In the post-prandial state, FAs and CEs can be incorporated into LD structures and in the post-absorptive state LDs can be broken down to FAs and CEs. The comprehensive iAdipocytes1809 covers all metabolic pathways known to exist in adipocytes as well as extensive knowledge of lipid metabolism in human cells and adipocytes in particular. Lipid metabolism in adipocytes involve uptake of FAs from two potential sources: non-esterified FAs (NEFAs) and lipoproteins (Supplementary Table S5) including chylomicrons, very low-density lipoprotein (VLDL), LDL and HDL through LPL. iAdipocytes1809 also cover the transport of FAs, cholesterol and cholesterol esters (CEs) across the plasma membrane, small amount of de novo FA and cholesterol synthesis, FA transport into mitochondria and peroxisomes for β-oxidation, FA esterification into triacylglycerols (TAGs), lipolysis and lipid droplet (LD) formation and maintenance (see Supplementary information).
Model validation and formation of lipid droplets
Due to the large size of GEMs (typically thousands of metabolic reactions), proper validation is essential to ensure that a reconstructed model has predictive ability (Mardinoglu and Nielsen, 2012). A common problem is lack of mass balancing of the individual reactions, and this typically involves metabolites with ill-defined formulas, such as polymers or metabolite pools, but different protonation states can also be problematic. This type of problem results in a situation where the model can take up or excrete metabolites in an unbalanced manner. The model was tested so that all reactions except pool reactions were mass balanced. A second issue is that reactions can violate thermodynamic constraints by being written in the wrong direction or where irreversible reactions are written as reversible. This was tested by making sure that the model could not generate high-energy compounds from low-energy compounds (such as ATP and water from ADP and phosphate or any organic compound from CO2 and water). A third issue is that reactions may be dead-ends, meaning that they cannot carry flux, either because the model is not able to produce/take up some of the substrates or because it is not able to consume/excrete some of the products. Dead-end reactions tend to propagate as one unconnected reaction can give rise to many more. Therefore, many GEMs contain a large proportion of dead-end reactions. Considerable efforts were spent in making sure that the number of dead-end reactions was kept to a minimum. Furthermore, with the simulation ready iAdipocytes1809, the production of all metabolites in the model was checked with minimum input to the model (Supplementary Dataset 4) to ensure the connectivity. Lastly, even a well-connected, thermodynamically correct and balanced model may not be able to perform all relevant metabolic functions, or it may be able to perform functions that it should not do (such as synthesis of essential amino acids or FAs). The model was therefore validated for 250 known metabolic functions of adipocytes, adapted from the definitions provided in connection with setting up HepatoNet1 (Gille et al, 2010; Supplementary Dataset 5). All validation steps and simulations were performed using the RAVEN Toolbox (Agren et al, 2013).
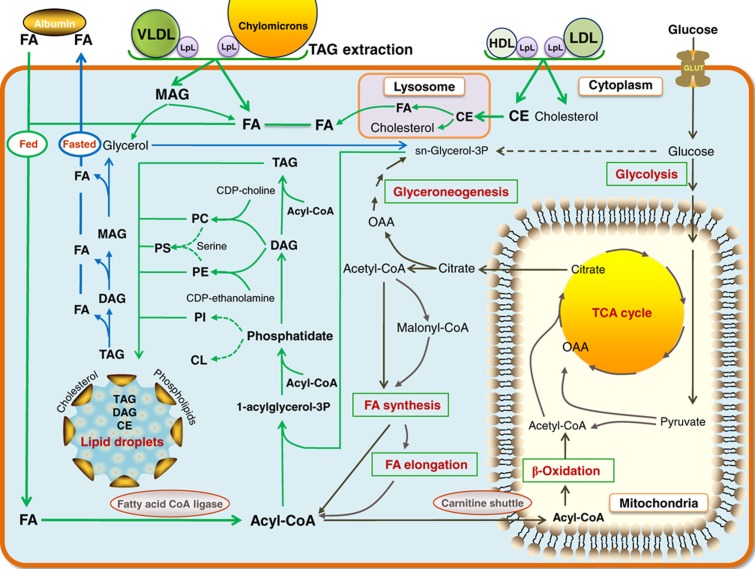
Formation of LDs (Figure 4) is included in iAdipocytes1809. LDs protect the cell from the lipotoxic effects of unesterified lipids, but they are also associated with the development of metabolic diseases, such as obesity, type 2 diabetes, atherosclerosis and liver steatosis that are characterized by the extensive accumulation of LDs. LDs represent a significant energy storage that can be mobilized by catabolism and this process is highly regulated by hormones and signaling pathways. Although LDs are recognized as an organelle called adiposomes (Farese and Walther, 2009), we represented it as a large composite metabolite in our model. LDs have polar surfaces that contain a variety of proteins, PLs and sterols and have a non-polar core that contain CEs, DAGs and TAGs (Gross et al, 2011; Supplementary Table S6). The diameter of LDs varies from 50 nm to 200 μm and LDs may grow to enormous sizes with different mechanisms including localized synthesis of lipids, transport of lipids and coalescence of LDs.
Figure 4.
Uptake of fatty acids through NEFA and lipoproteins and formation of lipid droplet in adipocytes. Adipocytes in white adipose tissue (WAT) store lipid mainly in the form of triacylglycerols (TAGs) and cholesterol esters (CEs) and form lipid droplets (LDs) in the post-prandial state (green arrows) and release them by degrading LDs in the post-absorptive state (blue arrows) to provide energy for other tissues. The released fatty acids (FAs) from adipocytes are transported to other tissues by albumin. The FAs are taken up from non-esterified FAs (NEFAs) and lipoproteins, including chylomicrons, very low-density lipoprotein (VLDL) and CEs together with cholesterol are taken up with low-density lipoproteins (LDLs) and high-density lipoproteins (HDLs) through LPL. CEs taken up from lipoproteins are degraded to cholesterol and FAs in lysosomes and transported to the endoplasmic reticulum and cytosol to be stored in LDs. Adipocytes also take up glucose to be used in the de novo synthesis of FAs (black arrows) that occurs at low level in adipocytes. LDs are rich in TAGs, CEs and an unknown neutral lipid that migrated between CEs and TAGs, ether neutral lipid monoalk(en)yl diacylglycerol (MADAG). LDs also contain small amounts of free FAs, cholesterol and phospholipids including phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylinositol (PI), ether-linked phosphatidylcholine (ePC), ether-linked phosphatidylethanolamine (ePE), lyso phosphatidylcholine (LPC), lysophosphatidylethanolamine (LPE), phosphatidylserine (PS) and sphingomyelin (SM). Formation of ePC, ePE, LPC, LPE and SM is included in the genome-scale metabolic model for adipocytes, iAdipocytes1809 and are not shown in figure.
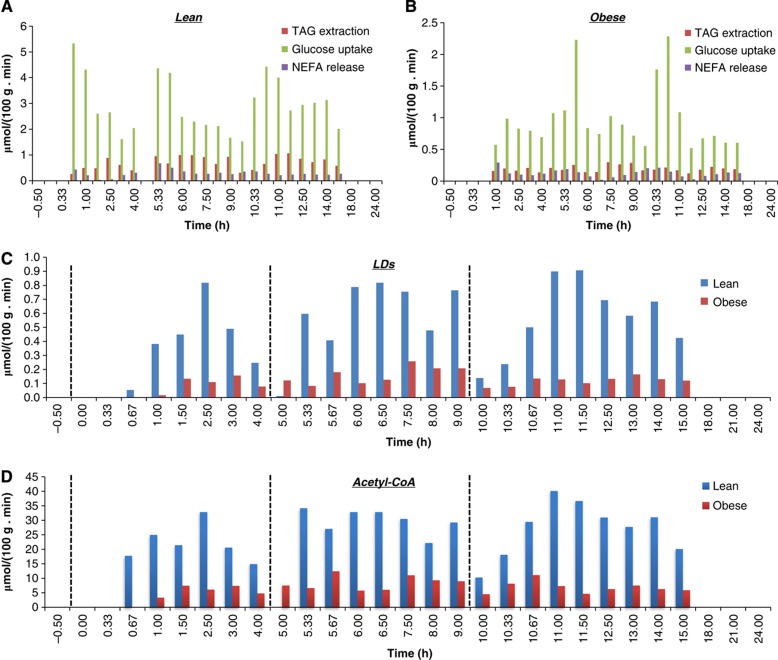
The function of iAdipocytes1809 was examined by estimating the formation of LDs and by means of available physiological, biochemical and genetic evidence in lean and obese subjects and clinically observed data. Recently, McQuaid et al (2011) measured the delivery and transport of FAs in adipose tissue using multiple and simultaneous stable-isotope FA tracers in lean and obese subject groups over 24-h period. Even though abdominally obese subjects have greater adipose tissue mass than control lean subjects, the rates of delivery of NEFAs were downregulated in obese subjects. Clinical data on Supplementary Table S1, NEFA release, TAG extraction, glucose uptake (Supplementary Dataset 6; McQuaid et al, 2011) and amino-acid uptake rates (Supplementary Dataset 7; Patterson et al, 2002) and FA concentrations in plasma and adipocytes (Supplementary Dataset 8) were incorporated into the model to predict the formation of LDs. Based on measurements of the uptake of glucose and TAG and the release of NEFAs over a 24-h period (Figure 5A and 5B), we simulated the change in LD size (Figure 5C; Supplementary Dataset 9a). We found from our simulations that lean subjects have large dynamic changes in LD formation compared with obese subjects, which is in agreement with experimental data (Arner et al, 2011). Furthermore, we predicted a lower acetyl-CoA production in obese subjects, as shown in Figure 5D (Supplementary Dataset 9b).
Figure 5.
Simulation results for lipid droplets and acetyl-CoA production. Formation of lipid droplets (LDs) and acetyl-CoA that is the central node in the mitochondria has been simulated and qualitative amounts were predicted through the use of iAdipocytes1809. Acetyl-CoA is central metabolite in the mitochondria and the reduced dynamics in LD levels in obese clearly results in lower fluxes through this key metabolite in obese subjects. The uptake rates for glucose and triacylglycerols (TAGs) and release rates for non-esterified fatty acids (NEFAs) in adipocytes for lean (A) and obese (B) subjects have been used as lower and upper bounds for input reactions (McQuaid et al, 2011) and the amount of LDs (C) and acetyl-CoA (D) is generated for 24 h. In addition, the NEFA concentrations in adipocytes and plasma and FA content of lipid structures are incorporated into the model during the generation of pool metabolites. The dashed lines at time=0, 5, 10 represent breakfast, lunch and dinner, respectively, for each participants of the study. iAdipocytes1809 cannot produce LDs and acetyl-CoA at some time points since adipocytes degrade the LDs at post-absorptive state to provide FAs to other tissues as energy sources. iAdipocytes1809 cannot produce LDs at time=5 in lean subject groups, whereas it can produce LDs in the obese subject groups as in clinical data.
Identification of obesity-specific metabolic features
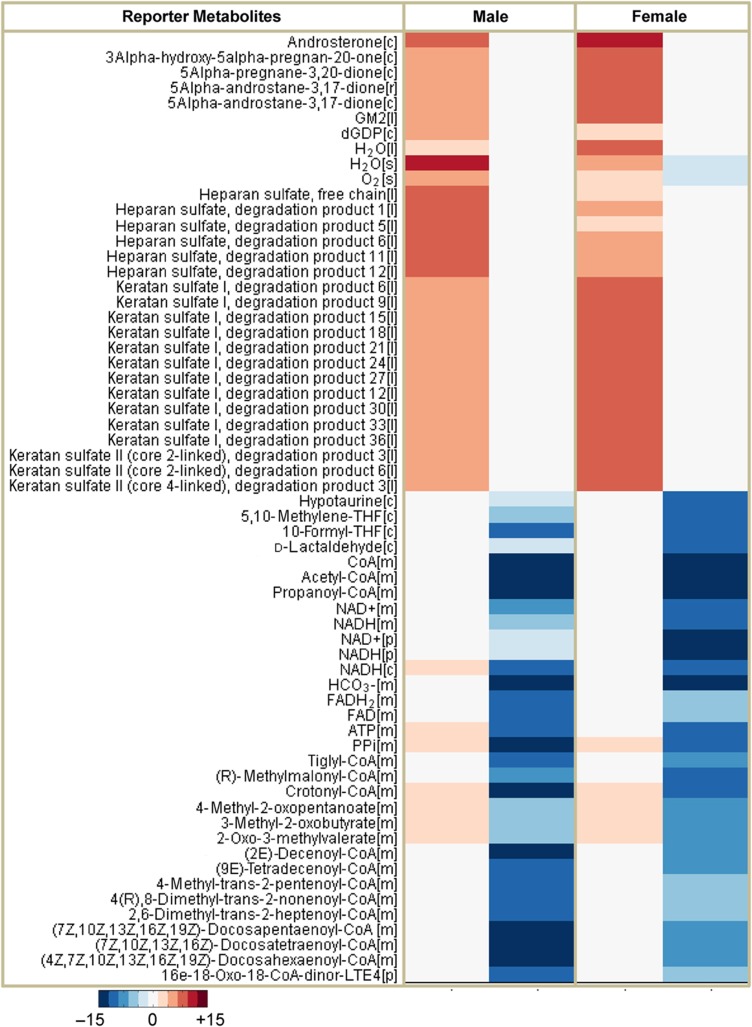
Modeling using iAdipocytes1809 can be applied to predict metabolic states under various perturbations, study regulation of the adipocytes, identify potential therapeutic targets and discover novel biomarkers for the development of more effective therapies. Here, we used the model to identify Reporter Metabolites (Patil and Nielsen, 2005) of male and female obese subjects compared with lean subjects using gene expression data obtained from the SOS Sib Pair Study. Reporter Metabolites are metabolite nodes in the metabolic network around which there are significant transcriptional changes. Here, 20 statistically significant Reporter Metabolites are presented for upregulated and downregulated genes in male and female obese subjects through the employment of iAdipocytes1809 (Figure 6). The most significant results from our Reporter Metabolites analysis for upregulated and downregulated genes are correlated with the KEGG pathways enrichment results of significantly expressed genes in the obese subject groups (Supplementary Figures S1 and S2).
Figure 6.
Representation of the Reporter Metabolites for male and female obese subjects. The Reporter Metabolites algorithm marks the regions in metabolism around which significant transcriptional changes occur. Reporter Metabolites are obtained using the P-values calculated from the comparison of obese subjects with lean subjects in male and female subjects separately. Top-scoring reporter metabolites androsterone, ganglioside GM2 and degradation products of heparan sulfate and keratan sulfate are associated with the upregulated differentially expressed genes and mitochondrial metabolites are associated with downregulated differentially expressed genes in obese subjects comparing with lean subjects. Top 20 metabolites associated with upregulated and downregulated genes in male and female obese subject were presented in the figure.
To illustrate the improvement of iAdipocytes1809 over the published iAB586, the Reporter Metabolites were also calculated for male and female obese subjects by using iAB586 (Supplementary Figures S8). Reporter Metabolites involved in the mitochondrial dysfunction as well as different amino acids were identified to be similar to the Reporter Metabolite analysis using iAdipocytes1809. However, iAB586 could not detect several of the most significant and in our view most interesting Reporter Metabolites identified when using iAdipocytes1809 due to the increase in number of reactions, metabolites and genes (see Discussion). Thus, the Reporter Metabolite analysis with iAdipocytes1809 and iAB586 provides an unbiased confirmation that iAdipocytes1809 represent significant advancement of the adipocyte metabolic network compared with iAB586.
Identification of obesity-specific transcriptionally regulated reactions by random sampling
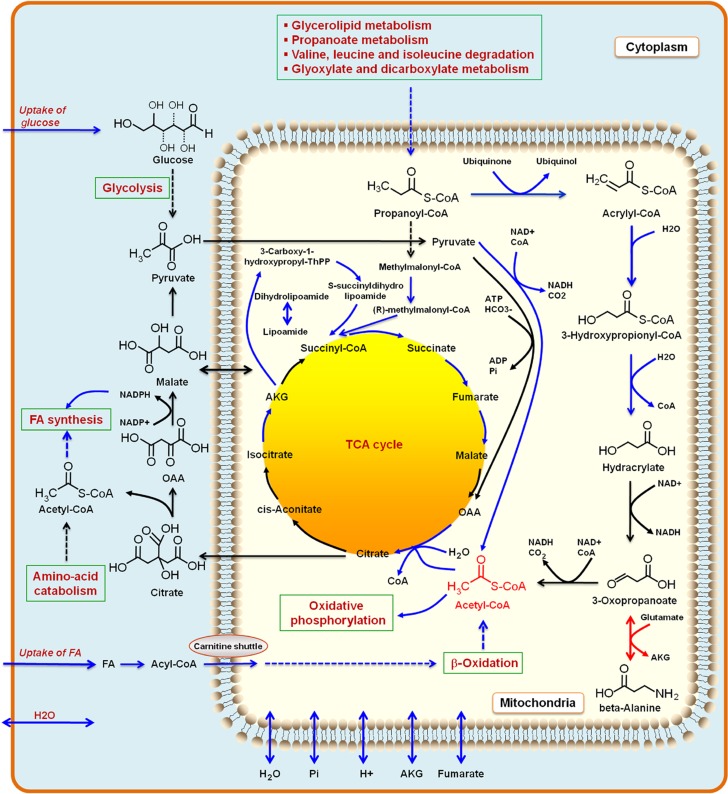
We identified changes in metabolic fluxes in response to obesity and which of these changes are likely to be associated with transcriptional changes using iAdipocytes1809. We defined a region of feasible flux distributions using uptake rates for TAGs and glucose and NEFA release rates for lean and obese subjects, and used these to calculate a set of possible flux distributions using a random sampling algorithm (Bordel et al, 2010). The average values and standard deviations for each of the fluxes in iAdipocytes1809 were calculated and the changing fluxes were compared with the significance of change in gene transcription for the corresponding enzymes. This allowed us to identify specific reactions for which flux changes are likely to be transcriptionally regulated.
The results from this analysis showed that the following pathway fluxes were transcriptionally downregulated in obese subjects: uptake of glucose, uptake of FAs, oxidative phosphorylation, mitochondrial and perixomal β-oxidation, FA metabolism and tricarboxylic acid (TCA) cycle. This analysis is consistent with the findings from the Reporter Metabolites analysis and KEGG enrichment pathway analysis (Supplementary Table S7). Furthermore, fluxes associated with beta-alanine metabolism were found to be transcriptionally downregulated in obese subjects (Figure 7). Previously, it has been reported that blood flow, glucose uptake, release of NEFA and the extraction of TAG from plasma were significantly lower in abdominally obese subjects compared with lean subjects (McQuaid et al, 2011). Our random sampling results clearly indicate that all metabolic pathways in mitochondria are downregulated (Kusminski and Scherer, 2012) similar to the mitochondrial decline that is a hallmark of different diseases associated with aging.
Figure 7.
Illustration of the reactions and metabolites obtained from random sampling. The random sampling algorithm identifies set of reactions that are transcriptionally regulated and enzymes with transcriptional regulation showed enrichment. This suggests that the regulation of metabolism in adipocytes in obese and lean subjects has evolved to contain a few flux-regulating potential transcription factors that could be the target for genetic manipulations to redirect fluxes. The random sampling algorithms indicate that uptake of glucose, fatty acids (FAs) and water, FA synthesis and mitochondrial metabolic processes such as oxidative phosphorylation, β-oxidation, TCA cycle and β-alanine metabolism is downregulated (blue arrows in figure). Increasing the metabolic activity of mitochondria by increasing the concentration of mitochondrial acetyl-CoA may provide new strategies for treatment of obesity. One way to increase the mitochondrial acetyl-CoA concentration may be through increasing the uptake of beta-alanine as found from our random sampling results. Some of the major known biological pathways of adipocytes are shown in green boxes.
Discussion
We evaluated the presence/absence of 14 077 proteins in adipocytes using human antibodies and presented a high-quality, simulation ready and functional GEM for adipocytes, iAdipocytes1809, based on proteome, metabolome and transcriptome data. iAdipocytes1809 was used to analyze clinically observed transcriptome and fluxome data to understand the mechanistic changes in adipocyte metabolism in response to obesity. Gene expression and associated metabolites in obese and lean subjects were studied using a systems biology approach, which allowed us to understand how the transcriptome data in SAT represent different metabolic functions in obesity. An increased understanding of metabolic pathways altered in complex diseases may contribute to the identification of novel therapeutic targets (Cheong et al, 2012) and here potential therapeutic targets for obesity were discovered through Reporter Metabolite analysis and random sampling of flux states. Moreover, protein coding genes related to the therapeutic targets may be used as potential drug targets.
Through our analysis, it was observed that expression of genes involved in metabolic pathways including mitochondrial and perixomal β-oxidation, FA synthesis, amino-acid metabolism, pyruvate metabolism, oxidative phosphorylation and TCA cycle are downregulated in obese subjects. Most of these pathways are linked with the mitochondrial dysfunction and different therapeutic interventions including antioxidants and chemical uncoupler treatments have proven to improve the mitochondrial dysfunction (Kusminski and Scherer, 2012). Recently, Canto et al (2012) have reported that increasing the NAD+ levels, identified as therapeutic targets in our study, through supplements (NAD+ precursors) enhances the oxidative metabolism and protects the cells against high-fat diet-induced obesity.
Mitochondrial acetyl-CoA has a central role in different pathways in the mitochondria and it reacts with oxaloacetate to form citrate, which can be transported from the mitochondria to the cytosol where it is participating in FA synthesis (Dean et al, 2009). Acetyl-CoA derived through other principal sources, including degradation of amino-acid and ketone bodies, and FA oxidation processes are insufficient for FA synthesis. Increasing the acetyl-CoA concentration and eventually FA synthesis in adipose tissue of obese subjects results in whole body regulation of metabolism, including stimulation of muscle insulin action and suppression of hepatosteatosis, as reported by Cao et al (2008). Here, we propose to boost the metabolic activity of mitochondria in the adipocytes of obese subjects by increasing the mitochondrial acetyl-CoA which is another therapeutic target identified in our study. Acetyl-CoA formation in lean and obese subjects was predicted through use of iAdipocytes1809 in lean and obese subjects and its concentration can be increased through the uptake of beta-alanine. The effect of beta-alanine as a dietary supplement was previously examined in football players and it is reported that it has effect on lean tissue accruement and body fat composition (Hoffman et al, 2006). Furthermore, rat studies reported that beta-alanine decreases the LPL enzyme activity in adipose tissue (Prabha et al, 1988) that may help to decrease the uptake of FAs to be stored in adipocytes. Our results suggest that increasing the level of beta-alanine in obese subjects may help to decrease the fat composition in obese subjects.
Furthermore, we have identified new candidates for potential therapeutic targets for obesity and their association with the obesity has been reported in different studies. One of the most significant results from the Reporter Metabolite analysis for upregulated genes was androsterone and its precursor 5alpha-androstane-3,17-dione. In obese subjects, increased secretion of androsterone was reported in serum concentrations using various indices calculated from urinary steroid excretion rates (Vierhapper et al, 2004). The measurement of androsterone-to-etiocholanolone ratio in urine is commonly used as an indicator of activities of 5α- and 5β-reductases in both male and female subjects and the ratio increased with indexes of insulin resistance, which is an adverse outcome of obesity (Tomlinson et al, 2008). Our results indicated that metabolism of obese subjects around androsterone increases and the detection of androsterone level in plasma may represent a marker for altered metabolism in obese subjects. 5alpha-pregnane-3,20-dione and 3alpha-hydroxy-5alpha-pregnan-20-one obtained from Reporter Metabolites analysis are other metabolites involved in steroid hormone synthesis specifically in pathway of progesterone metabolism. Several studies reported that sex steroids affect the adipose tissue deposition through the regulation of preadipocyte proliferation and/or differentiation as well as lipogenesis and/or lipolysis of differentiated adipocytes (Anderson et al, 2001). Zhang et al (2009) previously studied the comparison of progesterone metabolite formation in preadipocytes and lipid-storing adipocytes and they reported that the production of progesterone metabolites was significantly increased. Our study signified that metabolism around these two metabolites increase in obese subjects relative to metabolism in lean subjects.
Another high-ranking target for upregulated genes is ganglioside GM2 (GM2). Gangliosides, one of the major glycosphingolipids in mammals, have major roles as mediators for cell to cell or cell to matrix recognition and regulate the transmembrane signal transducers and cell proliferation. Gangliosides in adipose tissues are also associated with insulin signaling mechanisms and it is reported that series of gangliosides GM2, GM1 and GD1a are dramatically increased in adipose tissues of obese mice (Tanabe et al, 2009). The action of GM2 synthase also directs the metabolic flow of a-series gangliosides toward GD1a in mouse studies.
A third prominent group among the Reporter Metabolites for upregulated genes is the degradation products of heparan sulfate proteoglycans (HSPG) and keratan sulfate. These compounds are classified as glycosaminoglycans and attach to cell surface or extracellular matrix proteins. It has been reported that catalytically active adipose tissue LPL attaches to HSPG at the luminal surface of vascular endothelium (Olivecrona and Beisiegel, 1997; Lafontan, 2008) and hydrolyze the TAGs for uptake of FAs into the cell. The LPL moves between individual HSPG chains within the layer and this creates a high concentration of LPL along the surface layer of HSPG chains (Lookene et al, 1996). In the presence of heparin, more LPL is secreted and increased secretion was balanced by decreased degradation of LPL. There are special mechanisms that inhibit LPL and one mechanism is that LPL forms complexes with FAs (Bengtsson and Olivecrona, 1980). During the LPL hydrolysis and accumulation of FAs in the cells, the LPL is sequestered into enzyme FA complexes, lipolysis is reduced and eventually the binding of LPL to heparan sulfate is broken. If a high-affinity ligand (e.g., FAs, heparin and apoCII) is available, then the LPL detaches from the cell surface to heparan sulfate chains and without ligand in the medium, the LPL recycles into the cells where it is degraded. Furthermore, several studies have reported that more sulfated polysaccharide chains increase the affinity for binding of LPL (Olivecrona and Olivecrona, 2009). Keratan sulfate, a biomarker of proteoglycan degradation, can be expressed from stem cells in human SAT and its relevance with obesity has been reported earlier. Messier et al (2000) studied the effect of exercise and diet in weight loss in older obese adults with knee osteoarthritis and analyze the levels of total proteoglycan, keratan sulfate and interleukin-1 beta in their synovial fluid. It is reported that all participants of the study lost weight by changing their diet and exercise regimens and the level of keratan sulfate in synovial fluid decreased.
We envisage that iAdipocytes1809 is a key step to better enable links between molecular processes and patient phenotypes and hereby enable patient stratification through identification of specific molecular mechanisms in adipocyte metabolism. Here, we demonstrated this by predicting differences in the formation of LDs and acetyl-CoA in lean and obese subjects. Besides enabling patient stratification, this may also lead to identification of novel therapeutic targets for obesity through combination of our model with high throughput patient data. iAdipocytes1809 can also be used as a scaffold for a comprehensive whole adipocyte model that accounts for all of the annotated gene functions identified in adipocytes (Karr et al, 2012). Compared with the previously described adipose model, the here presented model is significantly larger in terms of metabolites/genes and reactions and this allows for identification of metabolic biomarkers that cannot be identified with iAB586. Furthermore, our model has undergone a thorough validation process where 250 metabolic functions were simulated and as our model also included a description of individual FAs and sterolesters it can much better simulate lipid metabolism, including simulation of lipid droplet formation. In conclusion, we demonstrated the high quality adipocyte GEM iAdipocytes1809 is very well suited for integration of omics data and hereby result in a comprehensive understanding of adipocytes biology in response to obesity.
Materials and methods
Proteome data for adipocytes
The proteomic profiling of adipocytes in breast and two different soft tissues using immunohistochemistry was performed as previously described (Uhlen et al, 2005). Representative formalin paraffin-embedded material from donor blocks was punched (1 mm in diameter) and placed in a recipient block TMA that includes 46 normal tissues, 20 types of cancer and 47 cell lines (Kampf et al, 2012). Thereafter, 4-μm TMA sections were cut using a microtome and placed on super frost glass slides. Breast and two different soft tissue cores together with 708 previously prepared tissue cores were analyzed for protein expression using IHC (Kampf et al, 2004). The variability introduced by the individual experimental staining protocol, including the choice of antibody dilution and antigen retrieval methods, was addressed by the use of TMAs (Uhlen et al, 2005). Information from databases, for example, Uniprot, ENSEMBL, and published data from PubMed were used as guides to establish if immunohistochemical results represent expected protein expression of intended target protein. In addition, other validation strategies were used to ensure antibody validity, for example, protein arrays, western blot and immunofluorescence experiments. When applicable, paired antibodies raised against separate, non-overlapping epitopes on the same target protein were used for extended validation (Uhlen et al, 2010). IHC-stained tissues were scanned and digitalized at × 20 magnification. Annotation of high-resolution images was manually performed by certified pathologists. Relative expression was indicated with four different color codes ranging from strong (red), moderate (orange), weak (yellow) and no expression (white) (Kampf et al, 2004). Localization information of proteins was inferred from manually curated Uniprot data (Apweiler et al, 2011) and recently generated HPA data on intracellular localization of proteins (Lundberg and Uhlen, 2010).
Transcriptome data for SAT
Gene expression in SAT of subjects involved in SOS Sib Pair Study which includes nuclear families with BMI–discordant sibling pairs (BMI difference ⩾10 kg/m2) was measured by microarray. The study group consisted of 304 subjects (209 female and 95 male) divided into three different groups according to their BMI: lean (18.5<BMI <25), overweight (25⩽ BMI<30) and obese (30⩽BMI) (Supplementary Table S1). Total RNA was prepared as previously described (Carlsson et al, 2009) and gene expression in human SAT was measured using Affymetrix U133 Plus 2.0 microarrays (Affymetrix). Probe hybridization intensity values were summarized using to Probe Logarithmic Intensity Error (PLIER) and the robust quantile method was applied to get normalized expression values. The normalization of the microarrays was carried out using the Expression Console software from Affymetrix and the quality assessment was carried out using R Statistical and Computing language and the Bioconductor software (Gentleman et al, 2004).
Data availability
The annotation of the presence or absence of protein targets in adipocytes together with the high-resolution images is publically available through the HPA (http://www.proteinatlas.org).
GEM for adipocytes, iAdipocytes1809 and HMR database is publically available in the Systems Biology Mark-up Language (SBML) format at Human Metabolic Atlas (http://www.metabolicatlas.org).
Gene expression from SAT of subjects from the SOS Sib Pair Study is publically available in Gene Expression Omnibus (GEO) database with the accession number GSE27916 http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=phadhqigowykote&acc=GSE27916.
Supplementary Material
Supplementary Tables S1–7, Supplementary Figures S1–8
The presence/absence of proteins encoded by 14 077 genes in adipocytes is examined based on immunohistochemistry and using antibodies generated within the Human Protein Atlas (HPA) project.
Xie et al. (Xie et al., 2010) characterized the proteome of human adipocytes and reported existence of proteins encoded by 1 574 genes in subcutaneous abdominal adipocytes taken from three healthy lean subjects.
The enzymes reported in the Human Protein Atlas database (http://www.proteinatlas.org) are listed.
Subcellular localization of the proteins based on the Human Protein Atlas (HPA) and Uniprot data and their assignments in the genome-scale metabolic model for adipocytes, iAdipocytes1809.
Subcellular localization of the proteins and their confidence scores in the genome-scale metabolic model for adipocytes, iAdipocytes1809 based on the Human Protein Atlas (HPA) and Uniprot data. Confidence score 2 for HPA, 1 for Uniprot and 3 for both HPA and Uniprot were assigned for each protein.
Enrichment of biological process Gene Ontology terms for encoded genes in here generated and previously published proteome data (assessed with DAVID (Huang et al., 2009)) are compared with the biological process Gene Ontology terms enrichment results of encoded genes in transcriptome data in male and female subjects.
List of the input metabolites and inferred transport reactions in the genome-scale metabolic model for adipocytes, iAdipocytes1809, to ensure the connectivity.
Known biological function of the adipocytes and the metabolic capacity was demonstrated by the simulation of 250 metabolic functions based on the definition of functions in HepatoNet 1 (Gille et al., 2010).
McQuaid et al. (McQuaid et al., 2011) measured the delivery and transport of fatty acids in adipose tissue using multiple and simultaneous stable-isotope fatty acid tracers in lean and obese subjects groups over a 24 hour period. Even though abdominally obese subjects have greater adipose tissue mass than lean control subjects, the rates of delivery of NEFAs are down regulated in obese subjects. In their study, uptake/secretion rates for NEFAs, TAGs and glucose in obese and lean subjects are reported. In adipose tissue, blood flow, glucose uptake, release of NEFAs and the extraction of TAGs from plasma was significantly lower in the abdominally obese subjects compared to lean subjects. Values are given as means ± SD.
Paterson et. al (Patterson et al., 2002) have studied the forearm and adipose tissue amino acid metabolism in lean and obese human subjects after 22 hours of fasting. They hypothesized that greater conservation of body protein observed during fasting in obese than in lean subjects and a combination of stable isotope tracer infusion and arteriovenous balance techniques was used to quantify amino acid kinetics. In their study, local net amino acid arteriovenous differences were calculated as the amino acid concentration in arterial plasma minus the concentration in venous plasma and local net fluxes were calculated as the arteriovenous difference multiplied by local plasma flow. Plasma arterial amino acid concentrations, regional subcutaneous abdominal adipose tissue arteriovenous concentration differences are adapted from the study of Paterson et. al (Patterson et al., 2002) and local net fluxes for lean and obese subjects were calculated.
Fatty acid (FA) composition in human plasma (Quehenberger et al., 2010), adipose tissue (Hodson et al., 2008; Raclot et al., 1997) and liver tissue (Shorten and Upreti, 2005) where there is not any information for adipocytes are incorporated in to the model for generation of pool reactions. Lipidomics analysis of a pooled human plasma obtained from healthy individuals after overnight fasting has been used in the model (Quehenberger et al., 2010). In their study, over 500 distinct molecular species distributed among the main lipid categories including fatty acyls, glycerolipids, glycerophospholipids, sphingolipids, sterols, and prenols were quantified. The reported 31 FAs in human plasma used in our model and normalized values are shown. In their study, some plasma FA known to exist in the structure of adipose tissue triacylglycerols and phospholipids have not been reported. These unreported FAs were included in to the structure of pool reactions by assuming their concentrations as half of the minimum concentration reported in the measurement study. Assumptions for the concentration of non reported FAs in lipidomics analysis allow us to include them into the genome-scale metabolic model for adipocytes, iAdipocytes1809. Raclot et. al. (Raclot et al., 1997) measured the FA composition of TAG in adipose tissue and originated non-esterified FA (NEFA) concentration from adipose TAGs through lipolysis. The mobilization of 34 individual fatty acids is quantified in adipose tissue of eight healthy non-obese women and used in our model to generate the pool reactions. The reported FA content was normalized to 1 mol/mol of TAG and shown. Hodson et. al. (Hodson et al., 2008) collected the data for measurements of FA composition in adipose tissue and blood lipids and their changes with different diet. The reported FA composition of NEFA is compared with the lipidomics analysis of a pooled human plasma study (Quehenberger et al., 2010) and an agreement has been seen. The FA molar composition of plasma TAGs and CEs reported in the study is normalized to 1 and used in iAdipocytes1809.
Lipid droplet formation in adipose tissue of lean and obese subjects have been predicted with the genome-scale metabolic model for adipocytes, iAdipocytes1809 and exchange reactions have been presented. Uptake/secretion rates for NEFA, TAG and glucose in obese and lean subjects are adapted from McQuaid et al. (McQuaid et al., 2011).
Acetyl-CoA formation in the mitochondria of adipose tissue in lean and obese subjects have been predicted through the usage of our genome-scale metabolic model for adipocytes, iAdipocytes1809 and exchange reactions have been presented. Uptake/secretion rates for NEFA, TAG and glucose in obese and lean subjects are adapted from McQuaid et al. (McQuaid et al., 2011)
Acknowledgments
We acknowledge Prof Fredrik Karpe and Dr Barbara A. Fielding, Oxford Centre for Diabetes, Endocrinology and Metabolism for providing adipose tissue uptake rates for lean and obese subjects. This work was supported by grants from the Knut and Alice Wallenberg Foundation, the Chalmers Foundation, the Wellcome Trust (GR079534) and the Swedish Research Council (K2010-55X-11285-13).
Author contributions: AM reconstructed iAdipocytes1809 and validated the model together with RA. AM and RA performed the analysis of clinical data. CK and AA generated the proteome data for adipocytes. PJ, AJW, PF and LMC generated gene expression data for adipose tissue and IN normalized the gene expression data. JN and MU conceived the project. AM, RA and JN wrote the paper and all authors were involved in editing the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Agren R, Bordel S, Mardinoglu A, Pornputtapong N, Nookaew I, Nielsen J (2012) Reconstruction of genome-scale active metabolic networks for 69 human cell types and 16 cancer types using INIT. PLoS Comput Biol 8: e1002518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agren R, Liu L, Shoaie S, Vongsangnak W, Nookaew I, Nielsen J (2013) The RAVEN toolbox and its use for generating a genome-scale metabolic model for Penicillium chrysogenum. PLoS Comput Biol doi:; DOI: 10.1371/journal.pcbi.1002980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson LA, McTernan PG, Barnett AH, Kumar S (2001) The effects of androgens and estrogens on preadipocyte proliferation in human adipose tissue: influence of gender and site. J Clin Endocr Metab 86: 5045–5051 [DOI] [PubMed] [Google Scholar]
- Apweiler R, Martin MJ, O'Donovan C, Magrane M, Alam-Faruque Y, Antunes R, Barrell D, Bely B, Bingley M, Binns D, Bower L, Browne P, Chan WM, Dimmer E, Eberhardt R, Fazzini F, Fedotov A, Foulger R, Garavelli J, Castro LG et al. (2011) Ongoing and future developments at the Universal Protein Resource. Nucleic Acids Res 39: D214–D219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arner P, Bernard S, Salehpour M, Possnert G, Liebl J, Steier P, Buchholz BA, Eriksson M, Arner E, Hauner H, Skurk T, Ryden M, Frayn KN, Spalding KL (2011) Dynamics of human adipose lipid turnover in health and metabolic disease. Nature 478: 110–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auffray C, Chen Z, Hood L (2009) Systems medicine: the future of medical genomics and healthcare. Genome Med 1: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtsson G, Olivecrona T (1980) Lipoprotein-lipase - mechanism of product inhibition. Eur J Biochem 106: 557–562 [DOI] [PubMed] [Google Scholar]
- Bordbar A, Feist AM, Usaite-Black R, Woodcock J, Palsson BO, Famili I (2011) A multi-tissue type genome-scale metabolic network for analysis of whole-body systems physiology. BMC Syst Biol 5: 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordbar A, Lewis NE, Schellenberger J, Palsson BO, Jamshidi N (2010) Insight into human alveolar macrophage and M. tuberculosis interactions via metabolic reconstructions. Mol Syst Biol 6: 422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordel S, Agren R, Nielsen J (2010) Sampling the solution space in genome-scale metabolic networks reveals transcriptional regulation in key enzymes. PLoS Comput Biol 6: e1000859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cakir T, Patil KR, Onsan Z, Ulgen KO, Kirdar B, Nielsen J (2006) Integration of metabolome data with metabolic networks reveals reporter reactions. Mol Syst Biol 2: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto C, Houtkooper RH, Pirinen E, Youn DY, Oosterveer MH, Cen Y, Fernandez-Marcos PJ, Yamamoto H, Andreux PA, Cettour-Rose P, Gademann K, Rinsch C, Schoonjans K, Sauve AA, Auwerx J (2012) The NAD(+) precursor nicotinamide riboside enhances oxidative metabolism and protects against high-fat diet-induced obesity. Cell Metab 15: 838–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao HM, Gerhold K, Mayers JR, Wiest MM, Watkins SM, Hotamisligil GS (2008) Identification of a lipokine, a lipid hormone linking adipose tissue to systemic metabolism. Cell 134: 933–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao YH (2010) Adipose tissue angiogenesis as a therapeutic target for obesity and metabolic diseases. Nat Rev Drug Discov 9: 107–115 [DOI] [PubMed] [Google Scholar]
- Carlsson LM, Jacobson P, Walley A, Froguel P, Sjostrom L, Svensson PA, Sjoholm K (2009) ALK7 expression is specific for adipose tissue, reduced in obesity and correlates to factors implicated in metabolic disease. Biochem Biophys Res Commun 382: 309–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caveney E, Caveney BJ, Somaratne R, Turner JR, Gourgiotis L (2011) Pharmaceutical interventions for obesity: a public health perspective. Diabetes Obes Metab 13: 490–497 [DOI] [PubMed] [Google Scholar]
- Chang RL, Xie L, Xie L, Bourne PE, Palsson BO (2010) Drug off-target effects predicted using structural analysis in the context of a metabolic network model. PLoS Comput Biol 6: e1000938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong H, Lu C, Lindsten T, Thompson CB (2012) Therapeutic targets in cancer cell metabolism and autophagy. Nat Biotechnol 30: 671–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter D, Maer A, Guda C, Saunders B, Subramaniam S (2006) Lmpd: Lipid Maps Proteome Database. Nucleic Acids Res 34: D507–D510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristancho AG, Lazar MA (2011) Forming functional fat: a growing understanding of adipocyte differentiation. Nat Rev Mol Cell Bio 12: 722–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft D, O'Kelly G, Wu GM, Haw R, Gillespie M, Matthews L, Caudy M, Garapati P, Gopinath G, Jassal B, Jupe S, Kalatskaya I, Mahajan S, May B, Ndegwa N, Schmidt E, Shamovsky V, Yung C, Birney E, Hermjakob H et al. (2011) Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Res 39: D691–D697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean JT, Tran L, Beaven S, Tontonoz P, Reue K, Dipple KM, Liao JC (2009) Resistance to diet-induced obesity in mice with synthetic glyoxylate shunt. Cell Metab 9: 525–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degtyarenko K, de Matos P, Ennis M, Hastings J, Zbinden M, McNaught A, Alcantara R, Darsow M, Guedj M, Ashburner M (2008) ChEBI: a database and ontology for chemical entities of biological interest. Nucleic Acids Res 36: D344–D350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte NC, Becker SA, Jamshidi N, Thiele I, Mo ML, Vo TD, Srivas R, Palsson BO (2007) Global reconstruction of the human metabolic network based on genomic and bibliomic data. Proc Natl Acad Sci USA 104: 1777–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farese RV, Walther TC (2009) Lipid droplets finally get a little R-E-S-P-E-C-T. Cell 139: 855–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P, Amode MR, Barrell D, Beal K, Brent S, Chen Y, Clapham P, Coates G, Fairley S, Fitzgerald S, Gordon L, Hendrix M, Hourlier T, Johnson N, Kahari A, Keefe D, Keenan S, Kinsella R, Kokocinski F, Kulesha E et al. (2011) Ensembl 2011. Nucleic Acids Res 39: D800–D806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frayn KN (2002) Adipose tissue as a buffer for daily lipid flux. Diabetologia 45: 1201–1210 [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge YC, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gille C, Bolling C, Hoppe A, Bulik S, Hoffmann S, Hubner K, Karlstadt A, Ganeshan R, Konig M, Rother K, Weidlich M, Behre J, Holzhutter HG (2010) HepatoNet1: a comprehensive metabolic reconstruction of the human hepatocyte for the analysis of liver physiology. Mol Syst Biol 6: 411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross DA, Zhan C, Silver DL (2011) Direct binding of triglyceride to fat storage-inducing transmembrane proteins 1 and 2 is important for lipid droplet formation. Proc Natl Acad Sci USA 108: 19581–19586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao T, Ma HW, Zhao XM, Goryanin I (2010) Compartmentalization of the Edinburgh Human Metabolic Network. Bmc Bioinformatics 11: 393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harkewicz R, Dennis EA (2010) Applications of mass spectrometry to lipids and membranes. Annu Rev Biochem 80: 301–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman J, Ratamess N, Kang J, Mangine G, Faigenbaum A, Stout J (2006) Effect of creatine and beta-alanine supplementation on performance and endocrine responses in strength/power athletes. Int J Sport Nutr Exerc Metab 16: 430–446 [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Jerby L, Shlomi T, Ruppin E (2010) Computational reconstruction of tissue-specific metabolic models: application to human liver metabolism. Mol Syst Biol 6: 401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampf C, Andersson AC, Wester K, Björling E, Uhlén M, Pontén F (2004) Antibody-based tissue profiling as a tool for clinical proteomics. Clin Proteomics 1: 285–299 [Google Scholar]
- Kampf C, Olsson I, Ryberg U, Sjöstedt E, Pontén F (2012) Production of tissue microarrays, immunohistochemistry staining and digitalization within the human protein atlas. J Vis Exp 63: e3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M (2010) KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res 38: D355–D360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlstaedt A, Fliegner D, Kararigas G, Ruderisch HS, Regitz-Zagrosek V, Holzhutter HG (2012) CardioNet: a human metabolic network suited for the study of cardiomyocyte metabolism. BMC Syst Biol 6: 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karr JR, Sanghvi JC, Macklin DN, Gutschow MV, Jacobs JM, Bolival B Jr, Assad-Garcia N, Glass JI, Covert MW (2012) A whole-cell computational model predicts phenotype from genotype. Cell 150: 389–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusminski CM, Scherer PE (2012) Mitochondrial dysfunction in white adipose tissue. Trends Endocrinol Metab 23: 435–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontan M (2008) Advances in adipose tissue metabolism. Int J Obes 32: S39–S51 [DOI] [PubMed] [Google Scholar]
- Lago F, Dieguez C, Gomez-Reino J, Gualillo O (2007) Adipokines as emerging mediators of immune response and inflammation. Nat Clin Pract Rheumatol 3: 716–724 [DOI] [PubMed] [Google Scholar]
- Lewis NE, Schramm G, Bordbar A, Schellenberger J, Andersen MP, Cheng JK, Patel N, Yee A, Lewis RA, Eils R, Konig R, Palsson BO (2010) Large-scale in silico modeling of metabolic interactions between cell types in the human brain. Nat Biotechnol 28: 1279–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lookene A, Chevreuil O, Ostergaard P, Olivecrona G (1996) Interaction of lipoprotein lipase with heparin fragments and with heparan sulfate: stoichiometry, stabilization, and kinetics. Biochemistry 35: 12155–12163 [DOI] [PubMed] [Google Scholar]
- Lundberg E, Uhlen M (2010) Creation of an antibody-based subcellular protein atlas. Proteomics 10: 3984–3996 [DOI] [PubMed] [Google Scholar]
- Ma H, Sorokin A, Mazein A, Selkov A, Selkov E, Demin O, Goryanin I (2007) The Edinburgh human metabolic network reconstruction and its functional analysis. Mol Syst Biol 3: 135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardinoglu A, Nielsen J (2012) Systems medicine and metabolic modelling. J Intern Med 271: 142–154 [DOI] [PubMed] [Google Scholar]
- McQuaid SE, Hodson L, Neville MJ, Dennis AL, Cheeseman J, Humphreys SM, Ruge T, Gilbert M, Fielding BA, Frayn KN, Karpe F (2011) Downregulation of adipose tissue fatty acid trafficking in obesity a driver for ectopic fat deposition? Diabetes 60: 47–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messier SP, Loeser RF, Mitchell MN, Valle G, Morgan TP, Rejeski WJ, Ettinger WH (2000) Exercise and weight loss in obese older adults with knee osteoarthritis: A preliminary study. J Am Geriatr Soc 48: 1062–1072 [DOI] [PubMed] [Google Scholar]
- Nielsen J (2009) Systems biology of lipid metabolism: from yeast to human. FEBS Lett 583: 3905–3913 [DOI] [PubMed] [Google Scholar]
- Nielsen J (2012) Translational and systems medicine introduction. J Intern Med 271: 108–110 [DOI] [PubMed] [Google Scholar]
- Nookaew I, Svensson PA, Jacobson P, Jernas M, Taube M, Larsson I, Andersson-Assarsson JC, Sjostrom L, Froguel P, Walley A, Nielsen J, Carlsson LM (2012) Adipose tissue resting energy expenditure and expression of genes involved in mitochondrial function are higher in women than in men. J Clin Endocrinol Metab 98: E370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivecrona G, Beisiegel U (1997) Lipid binding of apolipoprotein CII is required for stimulation of lipoprotein lipase activity against apolipoprotein CII-deficient chylomicrons. Arterioscl Throm Vas 17: 1545–1549 [DOI] [PubMed] [Google Scholar]
- Olivecrona T, Olivecrona G (2009) The Ins and Outs of adipose tissue. Cell Lipid Metab 315–369 [Google Scholar]
- Patil KR, Nielsen J (2005) Uncovering transcriptional regulation of metabolism by using metabolic network topology. Proc Natl Acad Sci USA 102: 2685–2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson BW, Horowitz JF, Wu GY, Watford M, Coppack SW, Klein S (2002) Regional muscle and adipose tissue amino acid metabolism in lean and obese women. Am J Physiol Endocrinol Metab 282: E931–E936 [DOI] [PubMed] [Google Scholar]
- Peinado JR, Pardo M, de la Rosa O, Malagon MM (2012) Proteomic characterization of adipose tissue constituents, a necessary step for understanding adipose tissue complexity. Proteomics 12: 607–620 [DOI] [PubMed] [Google Scholar]
- Prabha ANL, Leelamma S, Kurup PA (1988) Similar effects of beta-alanine and taurine in cholesterol-metabolism. J Biosci 13: 263–268 [Google Scholar]
- Romero P, Wagg J, Green ML, Kaiser D, Krummenacker M, Karp PD (2005) Computational prediction of human metabolic pathways from the complete human genome. Genome Biol 6: R2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe A, Matsuda M, Fukuhara A, Miyata Y, Komuro R, Shimomura I, Tojo H (2009) Obesity causes a shift in metabolic flow of gangliosides in adipose tissues. Biochem Bioph Res Co 379: 547–552 [DOI] [PubMed] [Google Scholar]
- Thiele I, Palsson BO (2010) A protocol for generating a high-quality genome-scale metabolic reconstruction. Nat Protoc 5: 93–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson JW, Finney J, Gay C, Hughes BA, Hughes SV, Stewart PM (2008) Impaired glucose tolerance and insulin resistance are associated with increased adipose 11 beta-hydroxysterold dehydrogenase type 1 expression and elevated hepatic 5 alpha-reductase activity. Diabetes 57: 2652–2660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlen M, Bjorling E, Agaton C, Szigyarto CA, Amini B, Andersen E, Andersson AC, Angelidou P, Asplund A, Asplund C, Berglund L, Bergstrom K, Brumer H, Cerjan D, Ekstrom M, Elobeid A, Eriksson C, Fagerberg L, Falk R, Fall J et al. (2005) A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol Cell Proteomics 4: 1920–1932 [DOI] [PubMed] [Google Scholar]
- Uhlen M, Oksvold P, Fagerberg L, Lundberg E, Jonasson K, Forsberg M, Zwahlen M, Kampf C, Wester K, Hober S, Wernerus H, Bjorling L, Ponten F (2010) Towards a knowledge-based Human Protein Atlas. Nat Biotechnol 28: 1248–1250 [DOI] [PubMed] [Google Scholar]
- Vierhapper H, Nowotny P, Waldhausl W (2004) Production rates of cortisol in obesity. Obes Res 12: 1421–1425 [DOI] [PubMed] [Google Scholar]
- Wishart DS, Knox C, Guo AC, Eisner R, Young N, Gautam B, Hau DD, Psychogios N, Dong E, Bouatra S, Mandal R, Sinelnikov I, Xia JG, Jia L, Cruz JA, Lim E, Sobsey CA, Shrivastava S, Huang P, Liu P et al. (2009) HMDB: a knowledgebase for the human metabolome. Nucleic Acids Res 37: D603–D610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie XT, Yi ZP, Bowen B, Wolf C, Flynn CR, Sinha S, Mandarino LJ, Meyer C (2010) Characterization of the human adipocyte proteome and reproducibility of protein abundance by one-dimensional gel electrophoresis and HPLC-ESI-MS/MS. J Proteome Res 9: 4521–4534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YH, Nadeau M, Faucher F, Lescelleur O, Biron S, Daris M, Rheaume C, Luu-The V, Tchernof A (2009) Progesterone metabolism in adipose cells. Mol Cell Endocrinol 298: 76–83 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Tables S1–7, Supplementary Figures S1–8
The presence/absence of proteins encoded by 14 077 genes in adipocytes is examined based on immunohistochemistry and using antibodies generated within the Human Protein Atlas (HPA) project.
Xie et al. (Xie et al., 2010) characterized the proteome of human adipocytes and reported existence of proteins encoded by 1 574 genes in subcutaneous abdominal adipocytes taken from three healthy lean subjects.
The enzymes reported in the Human Protein Atlas database (http://www.proteinatlas.org) are listed.
Subcellular localization of the proteins based on the Human Protein Atlas (HPA) and Uniprot data and their assignments in the genome-scale metabolic model for adipocytes, iAdipocytes1809.
Subcellular localization of the proteins and their confidence scores in the genome-scale metabolic model for adipocytes, iAdipocytes1809 based on the Human Protein Atlas (HPA) and Uniprot data. Confidence score 2 for HPA, 1 for Uniprot and 3 for both HPA and Uniprot were assigned for each protein.
Enrichment of biological process Gene Ontology terms for encoded genes in here generated and previously published proteome data (assessed with DAVID (Huang et al., 2009)) are compared with the biological process Gene Ontology terms enrichment results of encoded genes in transcriptome data in male and female subjects.
List of the input metabolites and inferred transport reactions in the genome-scale metabolic model for adipocytes, iAdipocytes1809, to ensure the connectivity.
Known biological function of the adipocytes and the metabolic capacity was demonstrated by the simulation of 250 metabolic functions based on the definition of functions in HepatoNet 1 (Gille et al., 2010).
McQuaid et al. (McQuaid et al., 2011) measured the delivery and transport of fatty acids in adipose tissue using multiple and simultaneous stable-isotope fatty acid tracers in lean and obese subjects groups over a 24 hour period. Even though abdominally obese subjects have greater adipose tissue mass than lean control subjects, the rates of delivery of NEFAs are down regulated in obese subjects. In their study, uptake/secretion rates for NEFAs, TAGs and glucose in obese and lean subjects are reported. In adipose tissue, blood flow, glucose uptake, release of NEFAs and the extraction of TAGs from plasma was significantly lower in the abdominally obese subjects compared to lean subjects. Values are given as means ± SD.
Paterson et. al (Patterson et al., 2002) have studied the forearm and adipose tissue amino acid metabolism in lean and obese human subjects after 22 hours of fasting. They hypothesized that greater conservation of body protein observed during fasting in obese than in lean subjects and a combination of stable isotope tracer infusion and arteriovenous balance techniques was used to quantify amino acid kinetics. In their study, local net amino acid arteriovenous differences were calculated as the amino acid concentration in arterial plasma minus the concentration in venous plasma and local net fluxes were calculated as the arteriovenous difference multiplied by local plasma flow. Plasma arterial amino acid concentrations, regional subcutaneous abdominal adipose tissue arteriovenous concentration differences are adapted from the study of Paterson et. al (Patterson et al., 2002) and local net fluxes for lean and obese subjects were calculated.
Fatty acid (FA) composition in human plasma (Quehenberger et al., 2010), adipose tissue (Hodson et al., 2008; Raclot et al., 1997) and liver tissue (Shorten and Upreti, 2005) where there is not any information for adipocytes are incorporated in to the model for generation of pool reactions. Lipidomics analysis of a pooled human plasma obtained from healthy individuals after overnight fasting has been used in the model (Quehenberger et al., 2010). In their study, over 500 distinct molecular species distributed among the main lipid categories including fatty acyls, glycerolipids, glycerophospholipids, sphingolipids, sterols, and prenols were quantified. The reported 31 FAs in human plasma used in our model and normalized values are shown. In their study, some plasma FA known to exist in the structure of adipose tissue triacylglycerols and phospholipids have not been reported. These unreported FAs were included in to the structure of pool reactions by assuming their concentrations as half of the minimum concentration reported in the measurement study. Assumptions for the concentration of non reported FAs in lipidomics analysis allow us to include them into the genome-scale metabolic model for adipocytes, iAdipocytes1809. Raclot et. al. (Raclot et al., 1997) measured the FA composition of TAG in adipose tissue and originated non-esterified FA (NEFA) concentration from adipose TAGs through lipolysis. The mobilization of 34 individual fatty acids is quantified in adipose tissue of eight healthy non-obese women and used in our model to generate the pool reactions. The reported FA content was normalized to 1 mol/mol of TAG and shown. Hodson et. al. (Hodson et al., 2008) collected the data for measurements of FA composition in adipose tissue and blood lipids and their changes with different diet. The reported FA composition of NEFA is compared with the lipidomics analysis of a pooled human plasma study (Quehenberger et al., 2010) and an agreement has been seen. The FA molar composition of plasma TAGs and CEs reported in the study is normalized to 1 and used in iAdipocytes1809.
Lipid droplet formation in adipose tissue of lean and obese subjects have been predicted with the genome-scale metabolic model for adipocytes, iAdipocytes1809 and exchange reactions have been presented. Uptake/secretion rates for NEFA, TAG and glucose in obese and lean subjects are adapted from McQuaid et al. (McQuaid et al., 2011).
Acetyl-CoA formation in the mitochondria of adipose tissue in lean and obese subjects have been predicted through the usage of our genome-scale metabolic model for adipocytes, iAdipocytes1809 and exchange reactions have been presented. Uptake/secretion rates for NEFA, TAG and glucose in obese and lean subjects are adapted from McQuaid et al. (McQuaid et al., 2011)
Data Availability Statement
The annotation of the presence or absence of protein targets in adipocytes together with the high-resolution images is publically available through the HPA (http://www.proteinatlas.org).
GEM for adipocytes, iAdipocytes1809 and HMR database is publically available in the Systems Biology Mark-up Language (SBML) format at Human Metabolic Atlas (http://www.metabolicatlas.org).
Gene expression from SAT of subjects from the SOS Sib Pair Study is publically available in Gene Expression Omnibus (GEO) database with the accession number GSE27916 http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=phadhqigowykote&acc=GSE27916.