Abstract
Circadian clocks exhibit ‘temperature compensation’, meaning that they show only small changes in period over a broad temperature range. Several clock genes have been implicated in the temperature-dependent control of period in Arabidopsis. We show that blue light is essential for this, suggesting that the effects of light and temperature interact or converge upon common targets in the circadian clock. Our data demonstrate that two cryptochrome photoreceptors differentially control circadian period and sustain rhythmicity across the physiological temperature range. In order to test the hypothesis that the targets of light regulation are sufficient to mediate temperature compensation, we constructed a temperature-compensated clock model by adding passive temperature effects into only the light-sensitive processes in the model. Remarkably, this model was not only capable of full temperature compensation and consistent with mRNA profiles across a temperature range, but also predicted the temperature-dependent change in the level of LATE ELONGATED HYPOCOTYL, a key clock protein. Our analysis provides a systems-level understanding of period control in the plant circadian oscillator.
Keywords: circadian rhythm, genetic network, mathematical model, systems biology
Introduction
The circadian clock is an endogenous 24 h timer, found throughout nature(McWatters and Devlin, 2011). It allows an organism to temporally orchestrate metabolic, physiological, biochemical and developmental processes. A defining feature of circadian clocks is the remarkable control of the clock’s pace (the circadian period) over a physiological range of constant temperatures, which is termed temperature compensation (Dunlap et al, 2003). It is unclear whether, in general, temperature compensation is owing to specific molecular mechanisms that have evolved in order to provide unusually temperature-dependent or temperature-insensitive regulation. The obvious alternative is that there are no special molecular mechanisms but that the clock gene network has evolved so that the many, milder effects of temperature upon its components produce an overall balance. This alternative hypothesis is akin to the idea of Hastings and Sweeney (1957), where they describe a balance of period-lengthening and period-shortening effects.
A mathematically coherent characterisation of these hypotheses is provided by Ruoff’s balance equation (Ruoff et al, 1997). This applies to any differential equation model of the clock. It asserts that the overall effect of temperature T on period is a sum of terms, one for each of the temperature-dependent parameters kj, and that each of these terms is a product of two quantities: a measure of the way in which the period depends upon the parameter (the parameter’s control coefficient 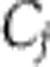 ) and a measure dj of the way the parameter depends upon T. We can write this
) and a measure dj of the way the parameter depends upon T. We can write this 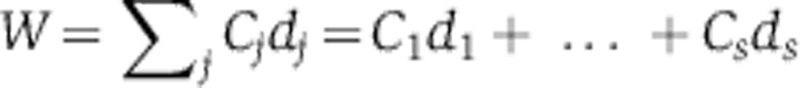 where W measures the change in period for a unit change in temperature. Thus, for temperature compensation a necessary and sufficient condition is
where W measures the change in period for a unit change in temperature. Thus, for temperature compensation a necessary and sufficient condition is 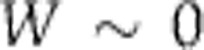 . In the first hypothesis above, specific molecular structures have evolved so that some subset of the
. In the first hypothesis above, specific molecular structures have evolved so that some subset of the 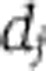 s have a significant functional dependence upon T, which makes the principal contribution to ensure that
s have a significant functional dependence upon T, which makes the principal contribution to ensure that 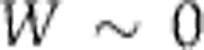 (for a more extensive discussion see the Supplementary Material). The ‘isoform switch’ mechanism that has been proposed for the Neurospora clock (Akman et al, 2008) is an example of this. The relative amounts of two isoforms of FRQ protein are highly temperature dependent (Liu et al, 1997; Diernfellner et al, 2007) and in the model (Akman et al, 2008) this provides special terms
(for a more extensive discussion see the Supplementary Material). The ‘isoform switch’ mechanism that has been proposed for the Neurospora clock (Akman et al, 2008) is an example of this. The relative amounts of two isoforms of FRQ protein are highly temperature dependent (Liu et al, 1997; Diernfellner et al, 2007) and in the model (Akman et al, 2008) this provides special terms 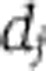 for the parameters associated with the production of the two isoforms. In the alternative hypothesis, rather than evolving the special temperature dependencies
for the parameters associated with the production of the two isoforms. In the alternative hypothesis, rather than evolving the special temperature dependencies 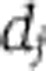 , it is the network and its control coefficients
, it is the network and its control coefficients 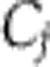 that have evolved so that
that have evolved so that 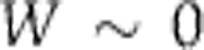 . In this scenario, the
. In this scenario, the 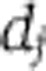 would either come from standard thermodynamic principles for the way in which rate constants vary with temperature or would arise from simple molecular processes. We would expect them to be simple functions of temperature, as opposed to the highly nonlinear dependencies seen in the isoform switching example. We will say that systems that satisfy the latter hypothesis are network balanced. An alternative approach to temperature compensation in Neurospora proposes that it is owing to a balance between FRQ degradation and FRQ synthesis. This an example of such network balancing (Ruoff et al, 2005). Positions intermediate between these two hypotheses where both effects have occurred are obviously possible. In the cyanobacterial KaiABC oscillator, the period is strongly controlled (large Cj) by an unusually temperature-insensitive process (small dj). A recent model argues that other processes with significant Cjalso contribute, and that their effects are network-balanced (Brettschneider et al, 2010). It is currently unclear how the temperature compensation of the plant clock works and it would clearly be interesting to provide strong evidence for or against the above hypotheses.
would either come from standard thermodynamic principles for the way in which rate constants vary with temperature or would arise from simple molecular processes. We would expect them to be simple functions of temperature, as opposed to the highly nonlinear dependencies seen in the isoform switching example. We will say that systems that satisfy the latter hypothesis are network balanced. An alternative approach to temperature compensation in Neurospora proposes that it is owing to a balance between FRQ degradation and FRQ synthesis. This an example of such network balancing (Ruoff et al, 2005). Positions intermediate between these two hypotheses where both effects have occurred are obviously possible. In the cyanobacterial KaiABC oscillator, the period is strongly controlled (large Cj) by an unusually temperature-insensitive process (small dj). A recent model argues that other processes with significant Cjalso contribute, and that their effects are network-balanced (Brettschneider et al, 2010). It is currently unclear how the temperature compensation of the plant clock works and it would clearly be interesting to provide strong evidence for or against the above hypotheses.
The genetic circuit of the Arabidopsis circadian clock, in common with other organisms, comprises several interlocking feedback loops (Harmer, 2009). A recent mathematical model proposes a three-loop structure as a framework for analysis (Pokhilko et al, 2010), though several components have still to be located within the circuit. One loop, the morning loop, comprises the morning expressed genes LATE ELONGATED HYPOCOTYL (LHY) and CIRCADIAN CLOCK ASSOCIATED 1 (CCA1), the PSEUDO-RESPONSE REGULATORs PRR9 and PRR7 and night inhibitor (NI) (Pokhilko et al, 2010). LHY/CCA1 promote PRR7/9 expression, while PRR7 and PRR9 are transcriptional repressors of CCA1/LHY (Ito et al, 2003; Farré et al, 2005; Salome and McClung, 2005; Nakamichi et al, 2010). TOC1 is thought to form a second loop, the evening loop, with GIGANTEA (GI) and other components that are expressed in the evening (Locke et al, 2006; Martin-Tryon et al, 2007). A third loop is formed between morning and evening loops. Several of these genes have been proposed as having important roles in temperature compensation (Gould et al, 2006; Salomé et al, 2010). This model has been recently modified to add the evening complex (EC), comprised of LUX (LUX ARRHYTHMO), ELF3 and ELF4 (Pokhilko et al, 2012). The binding of the EC to the promoters of the target genes PRR9 and LUX suppresses their transcription (Nusinow et al, 2011). In addition, TOC1 has been changed from an activator to a repressor (Gendron et al, 2012; Huang et al, 2012; Pokhilko et al, 2012). Both of the above mentioned mathematical models are based solely on data from plants grown in standard laboratory conditions close to 22°C. They are able to match much of the experimental data including altered circadian phenotypes of mutated components.
In classical studies of temperature compensation, the focus is on the period of the free-running oscillator, under constant conditions. In most physiologically relevant situations, the clock is entrained by daily environmental cycles, comprising both light and temperature fluctuations. In this work, we hypothesised that daily cycles of light and temperature should not disrupt each other’s entrainment effects, and that this requirement might be most simply satisfied if the two signals acted through the same machinery. The same mechanisms for temperature input likely function also in temperature compensation, suggesting that the mechanisms of temperature compensation might in principle be linked to the mechanisms of light input. Entraining light signals impact the plant clock mechanism at multiple points. It has previously been shown that cycles of either red light (RL) or blue light (BL) can entrain the clock; reducing the fluence rate of either light quality or mutating the phy or cry photoreceptors lengthens the circadian period (Somers et al, 1998). At least three classes of photoreceptor proteins are involved (reviewed in Millar, 2004); the four RL photoreceptors, phytochromes (PHY) A, B, D and E, the two BL photoreceptors, cryptochromes (CRY) 1 and 2, and the three LOV-domain, F-box proteins in the ZEITLUPE (ZTL) family (Somers et al, 1998; Kim et al, 2007; Baudry et al, 2010). The photoreceptors affect multiple molecular mechanisms, including transcriptional activation of LHY, CCA1, PRR9 and GI by the phy and cry pathways, and degradation of PRR proteins by the ZTL family (reviewed in Harmer (2009)).
Here, we tested the hypothesis that light and temperature share common input mechanisms to the clock. We do this by producing a large data set in which temperature and light quality were varied independently. Outputs included the scoring of circadian period in wild-type (WT) and Arabidopsis photoreceptor mutants. We hypothesised that the dominant temperature effects in the balance equation  discussed above are in the terms
discussed above are in the terms 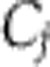 corresponding to parameters of the light input pathways with the djs just having passive thermodynamic variation. We applied linear regression and mixed-effects models to period estimates and identified significant variation in light, genotype, temperature and marker in our data. Using statistical modelling, we identified a strong interaction between temperature and BL in the control of circadian period. In view of this, we decided to test the hypothesis that temperature control is not delivered through dedicated molecular mechanisms as in the initial hypothesis, but is instead by network balancing through the BL input pathway. This led to the development of a fully temperature-compensating model of the Arabidopsis circadian oscillator simply by adding passive temperature effects through the known BL input pathways within the context of the Pokhilko et al, 2010 model of the clock. Remarkably, this model was not only capable of matching multiple non-trivial features of our data, but could also predict an unexpected and previously unobserved change in the abundance of a key clock protein with temperature. This success suggests that our model captures the salient features of the plant circadian system, which control temperature compensation, and that our systems-level approach that considers multiple temperature-dependent processes in parallel proves a powerful method to understand the control of period by temperature.
corresponding to parameters of the light input pathways with the djs just having passive thermodynamic variation. We applied linear regression and mixed-effects models to period estimates and identified significant variation in light, genotype, temperature and marker in our data. Using statistical modelling, we identified a strong interaction between temperature and BL in the control of circadian period. In view of this, we decided to test the hypothesis that temperature control is not delivered through dedicated molecular mechanisms as in the initial hypothesis, but is instead by network balancing through the BL input pathway. This led to the development of a fully temperature-compensating model of the Arabidopsis circadian oscillator simply by adding passive temperature effects through the known BL input pathways within the context of the Pokhilko et al, 2010 model of the clock. Remarkably, this model was not only capable of matching multiple non-trivial features of our data, but could also predict an unexpected and previously unobserved change in the abundance of a key clock protein with temperature. This success suggests that our model captures the salient features of the plant circadian system, which control temperature compensation, and that our systems-level approach that considers multiple temperature-dependent processes in parallel proves a powerful method to understand the control of period by temperature.
Results
Temperature compensation requires blue light and cryptochromes
Previous experiments tested light inputs to the circadian clock close to 22°C. We conducted a large experiment to measure circadian period under multiple conditions of temperature, photoreceptor complement and light quality, in order to test for interactions among these regulators. Multiple, independent transgenic Arabidopsis lines were prepared in the Columbia (Col-0) genetic background and in the photoreceptor mutants, cry1-304 (Mockler et al, 1999), cry2-1 (Guo et al, 1998), phyA-211 (Nagatani, et al, 1993), phyB-9 (Reed et al, 1994) and the cry1-304; cry2-1 double mutant. Each contained a LUCIFERASE (LUC) reporter gene fused to a well-characterised, circadian-controlled promoter, either from COLD-CIRCADIAN RHYTHM-RNA BINDING 2 (CCR2, also known as AtGRP7) or CHLOROPHYL A/B-BINDING PROTEIN 2 (CAB2, also known as LHCB1*1). After seedlings were grown in light–dark cycles, circadian rhythms of LUC reporter gene activity were measured by in vivo imaging under constant light and temperature conditions. Two seedling groups were measured over 96 h, for at least three independent transgenic lines in six genotypes and two reporter combinations, in two to three biologically independent experiments, at three temperatures (12, 17 and 27°C) and three light conditions (RL, BL, and mixed RL and BL, termed R/BL (Figure 1 and Supplementary Figure 1, 2, 3 and 4).
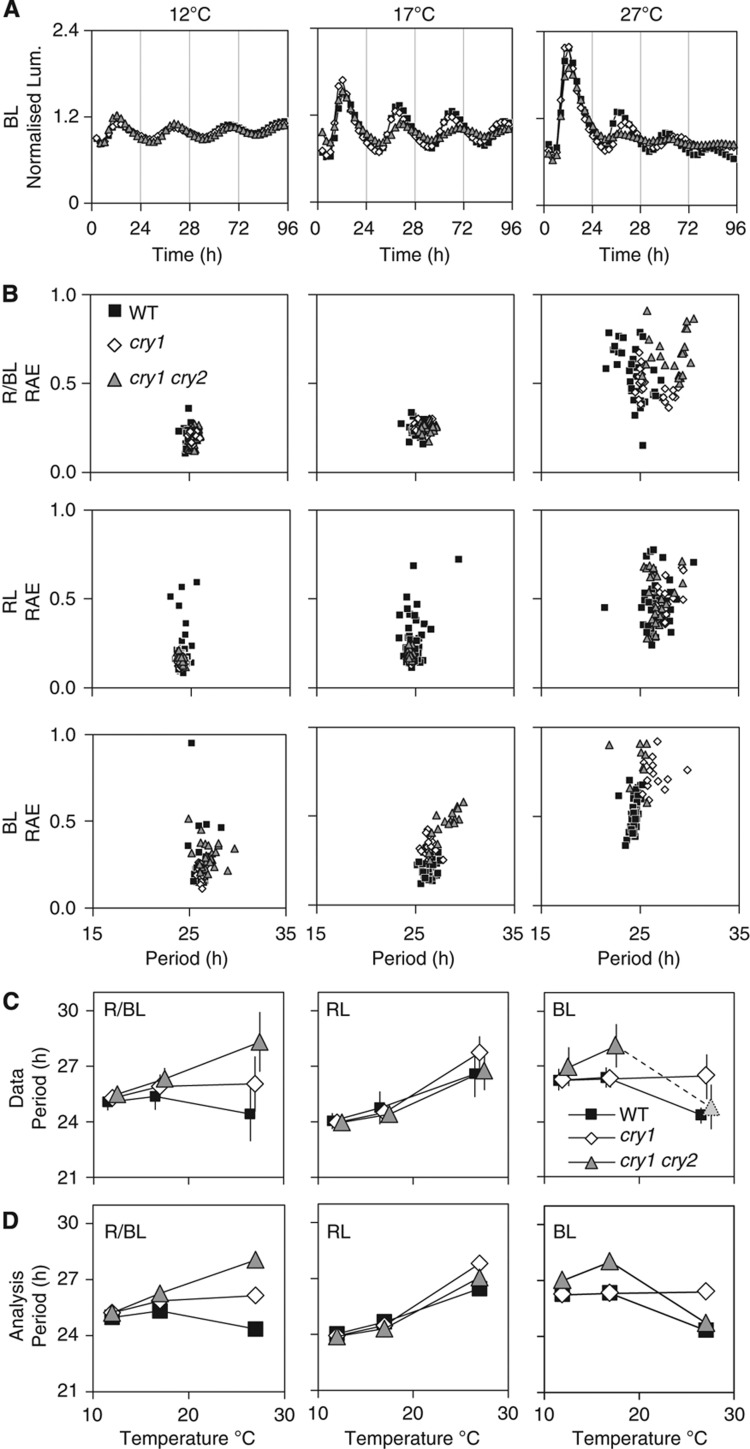
Figure 1.
Light and temperature responses interact strongly to control circadian period. Transgenic Col-0 WT (black squares), cry1 mutant (white diamonds) and cry1 cry2 double mutant (grey triangles) seedlings carrying the CCR2:LUC reporter gene were entrained under 12L:12D cycles for 7 days, transferred at ZT 0 to 12, 17 or 27°C and imaged under constant BL, RL or R/BL. (A) Representative luminescence profiles for plants under BL at 12, 17 and 27°C. Each trace is normalised to the mean expression level over the entire time course, error bars indicate s.e. (B) Plot of the circadian period of luminescence in each seedling sample against its RAE, where low RAE values indicate robust rhythms. Each condition includes 16–50 samples, comprising of three independently transformed transgenic lines per genotype, tested in two to three independent experiments. (C) Arithmetic mean periods of the data in (B). The cry1 cry2 double mutant at 27°C is largely arrhythmic, denoted by a shaded symbol, though the average period of the weak remaining rhythms is close to WT. Markers are offset to show error bars (1 s.d.). (D) Mean periods from only the fixed effects of the mixed-effect statistical model (Supplementary Table 1), which capture the variation in the data.
The rhythmic period of each sample was calculated from the luminescence time-series data, along with a measure of rhythm robustness (the relative amplitude error, RAE), using the Fast Fourier Transform—Non Linear Least Squares (FFT–NLLS) algorithm (Plautz et al, 1997) (Figure 1B). Under R/BL at 17°C, rhythms were robust with a period of 24–25 h. The period in WT plants was shorter at 27°C and rhythmic amplitude fell during the experiments (Figure 1A). Their circadian rhythms at 27°C were less robust than at 12 and 17°C, and this was reflected in higher RAE values (Figure 1B), consistent with earlier results (Gould et al, 2006). The rhythms of WT in BL appeared more robust than in RL, with lower variation in period values (Figure 1B and C).
To identify the sources of variation within the 2748 period values, we used two statistical models (Pinheiro and Bates, 2000), as described in the Supplementary Methods online. The whole data set was first analysed using the linear mixed-effects model (Bates and Maechler, 2011) (Supplementary Table 1), weighting each period value by its associated uncertainty. Variation between replicated experiments or between independent transgenic lines was almost negligible (Supplementary Methods online). The statistical model including only the experimentally controlled manipulations (fixed effects, Figure 1D) accounted for almost all variation in the data, closely replicating the arithmetic means (Figure 1C). The CAB2 reporter (Supplementary Figure 1) had shorter period in general than the CCR2 reporter (Figure 1), again consistent with earlier reports (for example, Eriksson et al, 2003). The periods measured by both reporters showed a strong interaction between light quality and temperature, which was analysed separately in the RL and BL data using standard linear regression (Supplementary Tables 1 and 2). The largest interaction was between RL and 27°C, because the period lengthened in these conditions whereas in BL and R/BL the period shortened at this temperature, leaving a 2 h period difference among the light conditions (Figure 1C and D, Supplementary Tables 1 and 2). The long period at 27°C is consistent with a reduced effect of RL on period length, because it tends towards the long period of plants in constant darkness (DD). The shortening of period in R/BL at 27°C might thus reflect mainly the effect of BL on period (Figure 1C and D).
To identify the photoreceptor pathways responsible for this temperature-dependent effect, we analysed the phenotypes of cry1 and cry2 mutants under BL. The cry1 mutation singularly had no detectable effect on period at 12 or 17°C under BL relative to WT (Figure 1D) but prevented the shortening of period at 27 °C, resulting in a long period phenotype (cry1 26.3 h; WT 24.3 h) and a strong statistical interaction (Table I). This suggests that cry1 is a major photoreceptor involved in shortening the period at 27°C in WT under BL. Analysis of the phyA and phyB mutants revealed a series of subtle temperature-dependent phenotypes (Supplementary Figure 2, Table 1, Supplementary Tables 1, 2). For the phyA mutant, under both R/BL and RL there was a subtle shortening of period at low temperatures and lengthening at high temperatures. In BL, period in phyA was shorter than WT at 12 and 17°C, and identified as having a statistical interaction at all temperatures (Supplementary Figure 2). For phyB, a specific lengthening in RL was observed at 27°C (phyB 27.5 h; WT 26.6 h). This is consistent with the lengthening observed in Devlin and Kay (2000) and the temperature dependence of the phyB flowering phenotype (Halliday et al, 2003). An interaction with phyB and BL was also identified at 17 and 27 °C causing a period shortening, again matching observations in Devlin and Kay (2000). The cry2 single mutant had little detectable phenotype in BL (Supplementary Figures 3 and 4, Table I and Supplementary Tables 1, 2). Both statistical analyses indicated smaller effects of the cry mutations under RL (Supplementary Tables 1 and 2), in line with previous data (Devlin and Kay, 2000).
Table 1. Statistical analysis of the circadian period under BL, using linear regression.
| Factors | Estimate (h) | s.e. | P-value |
|---|---|---|---|
| Intercept | 25.06 | 0.16 | <2e–16 |
| cry1cry2 | 0.85 | 0.25 | <<0.05 |
| phya | −0.88 | 0.26 | <<0.05 |
| 27C | −1.9 | 0.22 | <2e–16 |
| CCR2 | 1.12 | 0.1 | <2e–16 |
| phyb:17°C | −0.78 | 0.39 | 0.05 |
| cry1:27°C | 1.66 | 0.39 | <<0.05 |
| phyb:27°C | −1.01 | 0.38 | <0.05 |
Significant main effects and interactions are presented together with corresponding P-values as retrieved by the function lm in R. The intercept term corresponds to the reference mean period length, that is, the mean period for the Col-0 WT measured at 12°C, using the CAB marker. The estimated effect of each significant factor (second column) can be interpreted as the change, in hours, in period length relative to the reference mean period.
The cry1 cry2 double mutant had a long period at all temperatures under BL (Figure 1D) and, most remarkably, failed to sustain robust rhythms at 27°C. Rhythmic amplitude collapsed by the second cycle (Figure 1A), only 38% of cry1 cry2 mutants (9/24 samples) returned a circadian period estimate (compared with 42/42 in WT) and those with estimates had high RAE values (mean 0.8±s.e.m. 0.04, cf. WT RAE mean 0.5±0.01), indicative of weak rhythms (Figure 1B). The CAB2 marker gave a very similar result (7/16 cry1 cry2 samples rhythmic, 44%; Supplementary Figure 1). The fact that not all samples returned a period would be consistent with 27°C being at or close to the temperature at which the clock fails in the cry1 cry2 double mutant under BL. Thus the cry1 cry2 double mutant showed more severe phenotypes than either single mutant at all temperatures under BL. The addition of RL in R/BL conditions supported rhythms at 27°C in all the double-mutant samples, but with a long, 28 h period (Figure 1B and D). To summarise, our data showed that BL controlled circadian period via cryptochromes most strongly at 27°C, suggesting that temperature effects on the clock converged with light input pathways. While temperature-dependent phenotypes were observed for the phy mutants, the effects were small.
Temperature does not affect CRY abundance in Arabidopsis
The temperature dependence of cry-regulated circadian period could simply reflect temperature-specific variation in cryptochrome expression or abundance. The expression profiles of CRY1 and CRY2 mRNA and their cognate proteins were therefore tested under the conditions described above, using qRT-PCR and comparative western blotting with native antibodies against CRY1 and CRY2 (Supplementary Figure 5). The profiles showed evidence of weak circadian rhythmicity in mRNA levels (Toth et al, 2001) but not in protein levels. The slight tendency towards lower mRNA levels at 12°C contrasts with a tendency for the proteins to be more abundant at 12°C. Neither level of molecular data explained the marked requirement for cry photoreceptor function at 27°C, suggesting that temperature signals converge with light inputs downstream of the CRY protein accumulation.
The expression of clock genes change little with temperature
To extend this analysis, we tested the expression of the circadian clock-related genes, which are the ultimate targets of both light and temperature input, using qRT-PCR assays. Seedlings of WT and the cry1 cry2 double mutant were grown in the conditions used above for period assays, under constant BL at 12, 17 and 27°C. WT plants showed rhythmic expression of all the genes across the temperature range (Figure 2). A phase advance in the mRNA rhythms at 27°C relative to lower temperatures was consistent with the period shortening observed using reporter genes (Figure 1). Changes in the mean levels and rhythmic amplitudes of clock gene expression were remarkably limited. Previous results showed that LHY transcript levels decreased with increasing temperatures (Gould et al, 2006). These data were acquired from plants of the Wassilewskija accession, grown under white light, using a qRT-PCR primer set that detected only a subset of LHY gene models, missing the gene model At1g1060.4, that was absent from earlier genome annotations. Our current data are from the Col-0 accession and assayed under BL conditions, using qRT-PCR primers carefully designed to allow the amplification of all the gene models of LHY currently in TAIR 10. A recent publication illustrates that there are temperature-dependent changes at least within a subset of splice variation events in LHY, (James et al, 2012) with spliced transcripts decreasing with increasing temperature, at least between 20 and 4°C. This temperature-induced alteration in splice variant accumulation is the most likely explanation for the differences between the data presented here and the Gould et al, (2006) published data set.
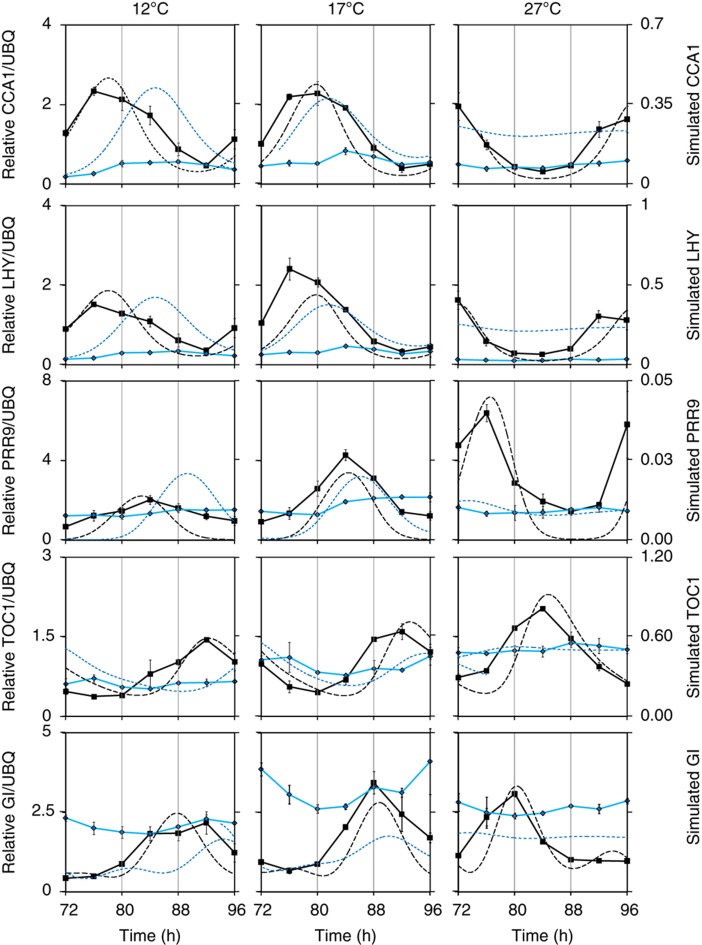
Figure 2.
Temperature-specific effects of cry photoreceptors on the expression profiles of clock genes are matched in the temperature-dependent model. Measured (solid lines, left axis) and simulated (dashed lines, right axis) mRNA expression profiles of clock genes in WT (black lines) and cry1 cry2 double-mutant plants (blue lines) under constant BL. Col-0 and cry1 cry2 seedlings were grown in 12L:12D entrainment conditions for 7 days before transferring at ZT0 to constant BL and 12, 17 or 27°C. Groups of seedlings were harvested every 4 h from 72–96 h after transfer. From each tissue, sample total RNA was extracted and assayed by qRT–PCR for the accumulation of CCA1, LHY, TOC1, GI and PRR9 relative to an internal UBIQUITIN10 (UBQ10) control. Plots represent average relative expression in each genotype for two biological replicates, error bars indicate s.e. Simulated mRNA expression profiles are shown for clock model components LHY/CCA1 (top two rows), PRR9, GI and TOC1 in the WT and cry1 cry2 double-mutant models under the same conditions. Simulated time series at 27°C were normalised to match the average level of the data at 27°C, because component levels in the model are arbitrary; the levels of simulated RNAs at 12 and 17°C follow from the model. As phase in LL is not well-defined, the time of peak simulated LHY/CCA1 was normalised to match the LHY data at 27°C; the timing of other simulated profiles follows from the model.
The mean PRR9 mRNA level changed most with temperature, by a threefold increase from 12 to 27°C, consistent with regulated PRR9 levels having an active role in shortening the period of WT plants. A smaller, temperature-dependent increase in TOC1 mRNA and little change in CCA1 or GI mRNA levels were consistent with earlier results (Gould et al, 2006).
In contrast to WT, the loss of cry photoreceptors profoundly altered gene expression levels. The cry1 cry2 double mutant showed LHY and CCA1 mRNA levels at 12°C peaking at trough WT expression levels, with lower fold amplitude (3.2-fold, compared with WT 4.3-fold). Thus, rhythms in CAB2 and CCR2 target genes were surprisingly robust in the double mutants at 12°C (Figure 1 and Supplementary Figure 1), considering the severe molecular effects of mutating the cryptochrome pathway (Figure 2). Clock gene expression was arrhythmic in the cry1 cry2 double mutant at 27°C, as expected, with LHY and CCA1 mRNA levels at or below the WT trough level.
A temperature-compensated model of the Arabidopsis circadian oscillator
The key observation of the statistical analysis was a strong coupling between the temperature and light input pathways of the clock. We decided to test the hypothesis that the effects of light and temperature converge to control common targets in the clock. It was impossible to test intuitively whether this hypothesis was consistent with our data on circadian period and on molecular regulation, not least because light is known to affect multiple processes in the clock mechanism. We therefore asked if there is a realistic, temperature-dependent mathematical model of the Arabidopsis circadian clock that can reproduce the data. To test the convergence hypothesis, we required that the effects of temperature on the clock model were mediated by changes in the parameters associated with light inputs. Thus, we sought to match the period and gene expression results by introducing temperature dependence into a highly constrained Arabidopsis clock model (Pokhilko et al, 2010).
Previous Arabidopsis clock models have constrained period with data produced under standard laboratory conditions, using a temperature of around 22°C (Locke et al, 2005, 2006; Pokhilko et al, 2010). The period of our recent model (Pokhilko et al, 2010) simulated in constant light was similar to the period measured at 27°C under BL in WT plants, so we used the published parameter set (Pokhilko et al, 2010) to represent this condition (Supplementary Methods). Next, we introduced temperature dependence into the model. Several parameters are associated with light inputs into the system. Sensitivity analysis revealed that only a subset of these had substantial control coefficients for period length (the quantities Ci introduced above), indicating that they could alter period in constant light. Two critical period-lengthening parameters were identified, associated with LHY transcription and translation, along with several period-shortening parameters (Supplementary Table 3). Parameters with a small control coefficient, Ci, will have little effect on period even if they are temperature-dependent, because the change in period owing to temperature effects on this parameter is of the form Cidi (see the balance equation of Ruoff (1) discussed above). Thus the temperature effects of such parameters can be safely ignored and one can legitimately avoid unnecessary complexity, by only introducing temperature dependence to those parameters associated with light inputs that have a significant control coefficient.
As a result, seven light-regulated parameters were assumed to be temperature dependent and the quantitative effect of temperature on these was assumed to satisfy an Arrhenius relationship kj=Aje−Ej/RT. These parameters control LHY/CCA1 transcription, mRNA degradation and LHY protein translation, degradation of TOC1 protein and its modified form (TOC1mod), PRR7 protein degradation and GI transcription. The prediction that the parameters associated with LHY/CCA1 (representing LHY and CCA1) were important in temperature compensation already has support in the literature (Gould et al, 2006), suggesting this approach had promise.
Attempts were then made to fit the model to the period and the mean mRNA expression levels in BL (Figures 1 and 2) for the three temperatures (12, 17 and 27°C), initially for the WT data. Matching such a complex and information-rich data set with a model is a difficult task, because the data greatly constrains the model. Such matching cannot be easily done without using abstract mathematical understanding of the temperature-dependent models, and the associated analytical tools. For example, by using the calculated balance equations, we could show that this model was capable of reproducing the observed period lengthening of the WT at lower temperatures (Supplementary Table 3). Even in the relatively large parameter set of the Pokhilko et al (2010) model, the small number of light-sensitive parameters with significant control coefficients and the Arrhenius form of the temperature dependence provided strong additional constraints on the parameterisation of our temperature-sensitive clock model. It was therefore unclear whether the model could fit the molecular data as well as the circadian periods. Importantly, because the resulting model is so highly constrained, model behaviours that were not fitted to data are also constrained, and thus can be regarded as predictions to test experimentally.
Experimental testing of the temperature-dependent clock model
Figure 2 shows that the model successfully captured the key aspects of mRNA expression in WT plants, matching the amplitude, amplitude changes and relative phases of gene expression at 12, 17 and 27°C. LHY/CCA1 levels remained nearly constant and mean PRR9 mRNA levels increased with temperature, as observed. We also observed subtle increases in TOC1 expression (Figure 2) in the simulation matching increases in experimental data and Gould et al (2006).
One of the critical features underpinning the success of the model was its ability to increase the levels of LHY protein with temperature, while mean LHY mRNA levels remained relatively constant, as in the data. An increase in LHY translation rate (parameter p1) with temperature, specified by the fitting procedure, resulted in a net increase in LHY protein (Figure 3A). This result was surprising, because parameter p1 (and the LHY transcription rate) had the strongest period-lengthening control coefficients (Supplementary Table 3). Increasing these parameters alone lengthens period in the model, yet the circadian period in the plant shortens with temperature (Figure 1).
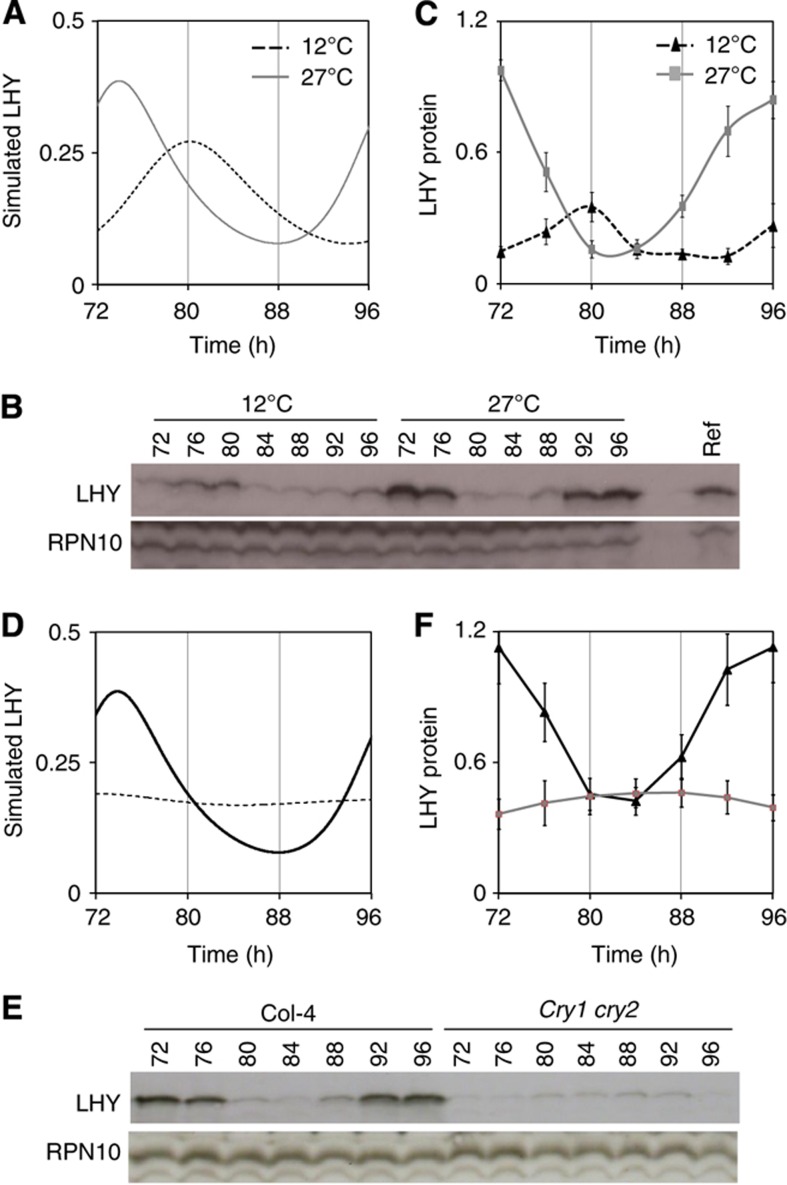
Figure 3.
Predicted and observed temperature effects on LHY protein abundance. (A) Model simulation of LHY protein expression profiles at 12°C (black dotted line) and 27°C (grey line) in WT. (B) LHY protein levels from plant extracts assayed by western blotting. Col-0 seedlings were entrained under 12L:12D cycles for 7 days, before transferring at ZT0 to 12 or 27°C in constant BL. Plant tissue was harvested every 4 h from 72–96 h after transfer. From each tissue sample, total protein was extracted and assayed by western blotting for the accumulation of LHY protein. RPN10 was used as loading control. A reference sample (Ref., containing moderate levels of LHY protein) was loaded onto each blot to allow normalisation among blots. (C) Quantification of LHY protein levels. Mean LHY protein levels were determined from biological triplicate samples; error bars, 1 s.e.m. (D) Model simulation of LHY protein expression in the WT at 27°C (black line) and the cry1 cry2 double mutant (dotted line). (E) LHY protein levels from plant extracts assayed by western blotting. Treated as described in Figure 2F, for WT at 27°C and the cry1 cry2 mutant at 27°C. (F) Quantification of LHY protein level WT at 27°C (black line) and cry1 cry2 mutant at 27°C (grey line). Protein levels were determined from triplicates, error bars, 1 s.e.m.
To test this prediction, we measured LHY protein levels through 24 h in BL using western blot analysis. The peak of LHY protein at both 12 and 27°C is consistent with the LHY mRNA (Figure 2) with the protein peak lagging mRNA expression peak. Our experimental data matched the prediction, as peak LHY levels were 2.7-fold higher at 27°C than at 12°C (Figure 3B and C). The rise in PRR9 mRNA expression levels in the model with increasing temperature was owing to the increase in LHY protein, rather than any temperature-dependent increase in the parameters governing PRR9 transcription. LHY has been experimentally shown to activate PRR9 transcription (Farré et al, 2005; Salome et al, 2005) and this regulation was included in the model (Pokhilko et al, 2010). Thus, our model captured the behaviour of the Arabidopsis circadian oscillator across a broad temperature range, and correctly predicted an observed rise in LHY protein levels with temperature. This analysis strongly supports our hypothesis that at a systems level, temperature compensation is primarily effected through temperature-dependent modulation of light input pathways into the morning loop of the clock.
Using the model to test the temperature-dependent effects of the cry mutants
Loss of both cryptochromes severely alters light signalling, resulting in long-period, lower-amplitude oscillations at 12° and 17°C and completely abolishing rhythms at 27°C (Figures 1 and 2). We therefore tested the extent to which these data constrain the temperature-dependent model. We made three assumptions, first considering only the same set of light-dependent parameters as above. Second, the parameter values must rise with temperature, following the Arrhenius relationships in the model. Third, the loss of photoreceptor activity in the mutants was assumed to limit the rate of light-dependent processes, reducing or increasing (depending on the parameter) their parameter values relative to WT. No single parameter change could match the data (Supplementary Information), consistent with the notion that cryptochromes affect multiple, relevant processes. The model structure was sufficient to match the 27°C mutant phenotype (Supplementary Figure 6 and Supplementary Table 4). The model also predicted that in the cry1 cry2 double-mutant LHY protein levels at 27°C would be low and arrhythmic. This raises the question of whether the low protein levels could be an outcome of low LHY mRNA levels. Interestingly, the mathematical model suggests that this is not the case as model LHY mRNA levels at 27°C, though lower than peak WT levels, are still not nearly as low as the data suggests, Figure 2.
We have analysed whether switching to the more recent clock model of Pokhilko et al (2012) would lead to improvement of the fits of the mathematical model to data. In particular, it is natural to ask whether the experimentally confirmed negative feedback of TOC1 on LHY mRNA would aid this. We conclude that switching models would lead to some improvement of the fits (e.g., that of LHY mRNA), but at the expense of others. The resulting trade-offs are presented in Table II. Most importantly, an attempt to create a WT temperature-compensated clock model from Pokhilko et al (2012) would potentially not be able to capture one of the two striking features of the WT data at lower temperature: the increased period and the decreased PRR9 mRNA at the lower temperature. A detailed description of the analysis to back up these statements (and those in Table II) is given in the Supplementary Information (Supplementary Figures 7–11 and Supplementary Table 6).
Table 2. Summary of the fit of the temperature-dependent cry1cry2 mutant model to 27°C data and the improvements that can be made by switching the underlying model structure to that of the (Pokhilko et al (2012)) model.
| Characteristics | Fit of temperature-dependent model at 27°C | Improvements by switch to Pokhilko et al (2012) model |
|---|---|---|
| LHY mRNA and protein | mRNA levels are too high compared with data while protein levels show a good match. | Can be improved at the expense of worse fit for two out of three following mRNAs: PRR9, TOC1 and GI. Choice of which two depends on choice of parameters. |
| PRR9 and TOC1 mRNA | Good | |
| GI mRNA | Shape is good but levels are too low. | Any improvement comes at the expense of the PRR9 mRNA fit. |
The modelling results suggest that CRY1 and CRY2 were regulating multiple transcription and degradation parameters, including at least a subset of the model’s seven temperature-dependent processes, and possibly others (discussed below).
Simulating the temperature-dependent effects of other clock mutants
To further our tests of the temperature-dependent model, simulations were carried out for known temperature-compensation mutants. CCA1 and LHY have previously been identified as having a role in temperature compensation with CCA1 functioning at low temperatures and LHY at high temperatures (Gould et al, 2006). In the Pokhilko model, the LHY and CCA1 are treated as a single factor called LHY (Pokhilko et al, 2010). Therefore, we simulated the lhy cca1 double mutant by reducing the translation of LHY to zero. The simulation identified that a residual short-period oscillator was still functional in the double mutant and that the mutant was still capable of temperature compensation; this seemed surprising, therefore, we tested the prediction identifying a close match with experimental data, thus further validating our model (Supplementary Figure 12A–D). It also predicted the subtle reduction in rhythm robustness at lower temperatures. Similarly the prr7 prr9 double mutant has been shown to have a temperature-dependent phenotype (Salomé et al, 2010), with period increasing and rhythm robustness decreasing with a rise in temperature. When the prr7 prr9 double mutant was simulated by reducing the translation parameters (of PRR7 and PRR9, only) close to zero, a temperature-compensation phenotype was produced that qualitatively matched that of the published mutant (Salomé et al, 2010). The simulation showed period lengthening with a rise in temperature and a dampening in rhythm robustness (Supplementary Figure 12E–G). Although temperature-dependent phenotype was correctly predicted, the model underestimated the absolute period value by ∼2.5 h across the temperature range. This is removed in Supplementary Figure 12G because this underestimate follows from the dynamics of the P2010 model that models 27°C in the WT as this also underestimated the period of the prr7 prr9 mutant.
Discussion
Light and temperature signalling control many plant physiological processes in common, such as growth, germination and flowering (Penfield, 2008). The circadian system’s unusually robust control of its rhythmic period is thought to maintain accurate timing under a range of environmental conditions, matching the constant period of the Earth’s rotation. Likewise, circadian period is only modestly affected by the fluence rates of constant white light used in laboratory studies, until very dim light levels are reached (Somers et al, 1998). Either red or blue photoreceptor pathways alone are sufficient to support similar rhythms (Figure 1), and previously it had been shown that plants lacking both cryptochromes (Devlin and Kay, 2000) or all phytochromes retained circadian rhythms in constant WL (Strasser et al, 2010).
Most recent work on temperature compensation focuses on the effects of specific molecular components. Here, we adopted a different systems-level approach that produced a temperature-dependent model of the Arabidopsis clock. The model was highly constrained by previous data acquired in standard conditions around 22°C and by our initial, information-rich data set on temperature effects on the circadian period and mRNA profiles of WT plants. This model produced a specific non-trivial molecular prediction that was validated. This validation provides evidence that temperature compensation in plants is based on the network-balancing model, rather than relying on unusually temperature-(in)sensitive components.
It should be noted that Arrhenius relations typical of elementary chemical processes were used to model the temperature dependence of parameters. However, the effect is to produce parameters that vary roughly linearly with temperature, that is, 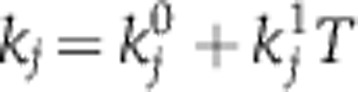 . A similar temperature dependence could be produced by complex molecular processes, for example, degradation of mRNA could be increasingly aided at higher temperatures by nonsense-mediated degradation of splice-defective mRNA. In fact, evidence at least for temperature regulation of splice-defected mRNA has been identified for a number of clock genes (James et al, 2012) and this may well contribute to the balancing of transcription rates and mRNA degradation.
. A similar temperature dependence could be produced by complex molecular processes, for example, degradation of mRNA could be increasingly aided at higher temperatures by nonsense-mediated degradation of splice-defective mRNA. In fact, evidence at least for temperature regulation of splice-defected mRNA has been identified for a number of clock genes (James et al, 2012) and this may well contribute to the balancing of transcription rates and mRNA degradation.
Our experimental results showed an unexpected interaction between light quality and temperature regulation. At warmer temperatures, we found that BL was essential for maintenance of period, and that WT plants had a 2 h longer period in RL than BL (Figure 1). Under BL and R/BL, circadian period decreased in the WT as temperature increased. The temperature independence of period in the cry1 mutant (Figure 1) suggests that ‘perfect’ compensation is achievable. Therefore, there might be an evolutionary advantage in the WT plant’s controlled change of period, possibly in globally changing the circadian phase of clock-regulated processes to occur earlier in the day when temperatures are warmer. Intriguingly, the desert succulent K. fedtschenkoi also has a clock that oscillates with a short period (Anderson and Wilkins, 1989). The long period phenotype in the cry mutants at 27°C under R/BL, therefore, suggests an important role of the cry1 and cry2 BL receptors in temperature compensation. Cryptochromes were absolutely required for robust oscillatory behaviour at 27°C, in a striking parallel with previous work in Drosophila (Kaushik et al, 2007).
One surprising observation was that clear circadian rhythms in the clock outputs CCR2 and CAB2 were observed at 12 and 17°C under BL in the cry1 cry2 double mutant (Figure 1). The molecular rhythms of clock component RNAs were more strongly affected (Figure 2), though LHY and CCA1 retained rhythmicity with only slightly lower fold amplitude than in WT. This is not without precedent, as clear oscillations with near-normal periods were observed in constant far-red light, despite major shifts in the levels of clock gene mRNAs (Wenden et al, 2011). Complex output pathways that include nonlinear feedback structures can, in general, filter and potentially amplify rhythmic signals, and these structures are present in the control of CCR2 and CAB2. The EPR1 transcription factor, for example, is rhythmically expressed, controls CAB2 expression and shows autoregulation (Kuno et al, 2003). The CCR2 RNA-binding protein, likewise, contributes to the control of CCR2 mRNA levels (Schöning et al, 2008).
This interaction between light and temperature is unlikely to be through a direct temperature regulation of photoreceptor abundance as neither CRY mRNA or protein levels was dependent on temperature (Supplementary Figure 4). Instead, our mathematical analysis showed that the gene circuit of the clock model described previously (Pokhilko et al, 2010) could support the period control observed in WT plants, with the major effects of temperature mediated through a small number of light-sensitive processes (parameters). However, not only could we show that control of the model’s period was achievable in principle, but in practice the predicted deviations of period across our experimental range closely matched those we observed in WT. The model was neither designed for temperature input nor constructed with data from varying temperature, so this result was not guaranteed. This represented the more general hypothesis that light-responsive processes were the most important temperature inputs, which was suggested by the strong effects of the photoreceptors in our experimental data. Added justification for this hypothesis comes from the fact that almost all the genes previously implicated in the control of period by temperature (LHY, CCA1, GI, PRR9 and PRR7) are light-responsive at either the transcriptional or, for PRR7, the protein level (Gould et al, 2006; Salomé et al, 2010). However, in suggesting this hypothesis, we are not ruling out that other components of the clock are temperature dependent, indeed we would expect this to be the case. We are proposing that the balance equation 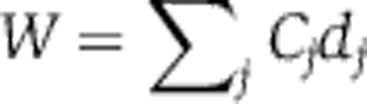 is dominated by the effect of the parameters associated with light input pathways.
is dominated by the effect of the parameters associated with light input pathways.
The model simulations predicted non-trivial changes in clock components across our experimental temperature range. The most notable of these was that LHY protein levels would increase with rising temperature. Remarkably, we confirmed this temperature-induced change (Figure 3), which was consistent with James et al (2012) where similar temperature-dependent changes where observed between 20 and 4°C (James et al, 2012). The increased LHY protein at 27°C is presumably active, as it correlated with higher levels of its target gene, PRR9 (Figure 2). This demonstrated that our model captures essential features of the in vivo temperature-compensation mechanism in WT plants. It does so without any unusual temperature-dependent steps, suggesting that the plant clock is network-balanced.
Model analysis was essential, because no simple molecular hypothesis accounts for all the data. Increasing LHY levels are expected to lengthen period: the period shortening of plants with reduced LHY or CCA1 is well established (Gould et al, 2006) and consistent with this, increasing LHY translation in the model-lengthened period (the control coefficient for period is positive, Supplementary Table 3). However, rising temperature increased LHY levels in the plants (Figure 3) but shortened period (Figure 1). This increase in LHY levels at 27°C is consistent with previous experimental evidence that the lhy mutant has a more severe effect on period at higher temperatures (Gould et al, 2006), which had seemed paradoxical. PRR9 overexpression can shorten period but also reduces LHY and CCA1 mRNA levels in constant light (Matsushika et al, 2002). The fact that LHY and CCA1 mRNA levels did not change with temperature (Figure 2) strongly suggests that another temperature-dependent process abrogated the effect of increased PRR9 expression at 27°C. Thus temperature effects on multiple processes were required to explain the data, in line with the network-balancing hypothesis, and these could only be integrated using the model analysis.
Our novel data and theoretical analysis strongly support a notion that light and temperature affect the clock by a common regulatory pathway, that impacts upon multiple points in the clock gene network. This convergence may be an important design principle for clocks to perform robustly over a wide variety of environments (Edwards et al, 2010). An obvious justification is the strong correlation between daily light and temperature cycles in the environment, which might drive such signalling convergence as an operating principle of environmental sensing. Hence, it might be no surprise that warm temperature and light pulses have similar resetting effects on circadian clocks across many organisms (Dunlap et al, 2003).
Using our temperature-dependent model, we took a systems-level approach to uncover the mechanism of temperature-dependent arrhythmia in the cry1 cry2 double mutant. This analysis confirmed that cryptochromes affect the clock by regulating multiple light/temperature-dependent parameters, involved in both transcriptional regulation and protein degradation. The notion that cryptochromes have non-transcriptional effects on the clock is consistent with the severe effects of the cry1 cry2 double mutant. In contrast, the most highly temperature-regulated of the transcriptional targets, PRR9, can be overexpressed or mutated with only mild effects (Matsushika et al, 2002; Eriksson et al, 2003). CRY proteins are not known to interact directly with plant clock proteins except for the interaction of CRY1 with ZTL reported from assays in yeast (Jarillo et al, 2001). The known inhibition of COP1 by cryptochromes (reviewed in Lin and Shalitin (2003) could be introduced as a new temperature-regulated process in a future, more complex model. For example, a clock protein(s) might be marked for degradation by COP1-mediated ubiquitination, in the manner reported for ELF3 (Yu et al, 2008), but in a cry- and temperature-dependent manner.
In view of the recent discovery that TOC1 functions as a repressor (Gendron et al, 2012; Huang et al, 2012) and the incorporation of this into a revised model (Pokhilko et al, 2012), it is natural to ask whether the new model would enable an improved fit to the expression data for the cry1 cry2 double mutant. As discussed above, we note that the analysis presented in the Supplementary Methods suggests that although some improvement of the fits can be obtained by using the later model, this is only at the expense of others.
One observation from our data is that the cry1 cry2 double mutant was not completely arrhythmic, in fact, 38% of the samples returned a rhythmic value for their expression of CCR2. The FFT-NLLS analysis method identifies a minority of near-circadian periods with high RAE in the fluctuating expression patterns of all arrhythmic mutants, such as elf3 (Hicks et al, 1996). However, the remaining rhythms could also be explained from our model. In BL, the cry1 cry2 double mutant at 12, 17 and 27°C is close to a Hopf bifurcation (see discussion in Supplementary Material). This means that by small alterations in some of the parameters one can change the damped oscillations to a sustained oscillation or vice versa. At 12°C, the data and model suggest that there is a relatively low amplitude limit cycle and therefore sustained oscillations. At 27°C, the model suggests that the system is so close to a Hopf bifurcation that one should expect to see a mixture of behaviours for the experimental system. We believe that this is why we observe that at 27°C ∼40% of cry1 cry2 mutant samples are still rhythmic and with period lengths clustered around the WT, rather than a completely arrhythmic phenotype.
Inevitably, both mathematical models suffer from limitations that arise from lack of knowledge and uncertainty about specific connections in the system (Pokhilko et al, 2012). In particular, the identity and detailed mechanism of the assumed acute light activation of LHY, PRR9 and GI transcription after dawn remains to be clarified. In addition, the modelling of the sequential activation of the transcription of the PRRs by each other and by LHY/CCA1 protein to reproduce the experimentally observed wave of PRR9, PRR7 and PRR5 (NI) inhibitors is only partially supported by data (Farré et al, 2005) and remains to be elucidated. The mechanistic details of the inhibition of LHY/CCA1 by TOC1 are currently unknown. Recent data suggest the involvement of additional proteins in the regulation of gene expression by LHY and CCA1 (Lau et al, 2011) and these are not included in these models. Although they are included in the 2012 model, the exact mechanisms of the formation of the protein complexes between ELF3 and ELF4, ELF3 and GI, and between ELF3, ELF4, LUX (EC) and their functional significance need to be further elucidated. In the 2012 model, it is assumed that post-translational regulation of ELF3 protein complexes is by different forms of the COP1 ubiquitin E3ligase. The details of this are unknown and the dynamics of COP1-containing complexes awaits further elucidation. Improvement of future mathematical models will rely on improved understanding of interactions as above and on measurements of the stoichiometry, modification, dynamics and abundance of multiprotein complexes.
The arrhythmia seen in the cry1 cry2 double mutants observed at 27°C has striking parallels in other systems. The fly cryptochrome (dCRY) mediates temperature signalling to the clock by interacting with the clock protein Timeless (Kaushik et al, 2007), leading to Tim degradation, as well as mediating light signalling via Tim and the F-box protein Jetlag (Peschel et al, 2009). Cryptochromes of Arabidopsis and Drosophila are not orthologous, because their protein sequences derive from different branches of the photolyase gene family (Cashmore et al, 1999). In mouse, a cry double mutant causes arrhythmia (van der Horst et al, 1999) but their role as circadian photoreceptors is controversial. Recently, Rhodopsin has also been implicated in temperature sensing in Drosophila (Shen et al, 2011). Thus, cryptochromes and light sensors have been co-opted across different kingdoms to have important roles in sensing temperature and sustaining circadian rhythms, although the molecular mechanisms through which crys interact with the clock architecture appear to be unique in plants and animals.
Materials and methods
Plant material
All the mutants used are in the Col-0 background. The mutant alleles used were cry1-304 (Mockler et al, 1999), cry2-1(Guo et al, 1998), phyA-211 (Nagatani et al, 1993) and phyB-9 (Reed et al, 1994). The multiple photoreceptor mutant cry1 cry2 was described previously (Mockler et al, 1999, 2003). The CCR2:LUC+ transgene was transformed into both mutant and WT plants using an Agrobacterium-mediated floral dip method (Davis et al, 2009). For each mutant, at least three independent lines were characterised.
Growth conditions and rhythm analysis
Seeds were surface-sterilised, kept at 4°C for 2–4 days, and grown for experiments on solid Murashige and Skoog (MS) medium containing 3% sucrose and 1.5% agar (for imaging) or 2% agar (for RNA samples) at 22°C in 12L:12D cycles of white light at 80 μmol m−2 s−1, as described previously (Gould et al, 2009). Seedlings were transferred to constant conditions at ZT0. Temperature during each experiment was monitored using Hobo temperature loggers (Onset Computer, Bourne, MA). The illumination within the cabinet was provided by either a RL LED array (30 μmol m−2 s−1), a BL LED array (30 μmol m−2 s−1) or a R/BL mixed array (20 μmol m−2 s−1 RL, 20 μmol m−2 s−1 BL). Luminescence levels of groups of seedlings (10–20 seedlings) were analysed by low-light imaging as described previously (Gould et al, 2006), normalising levels to the mean LUC activity over the whole time course for each group. Time-series data from imaging assays were analysed as described (Hall and Brown, 2007) using FFT–NLLS (Plautz et al, 1997) through the Biological Rhythms Analysis Software Suite v3 (Edwards et al, 2010) (available from www.amillar.org), or through the online BioDare interface (www.biodare.ed.ac.uk). To avoid light-driven responses and ensure consistency across all experiments, 96 h of data were analysed for each plant starting from ZT12, retaining any rhythm estimates with 15–35 h period. Arrhythmia was defined as a failure of FFT-NLLS to identify any period in this range. The raw data are available from the BioDare database, http://www.biodare.ed.ac.uk, for specific experimental IDs see Supplementary Table 7. The data have also been included as Supplementary Data with this manuscript.
RNA analysis
Plants entrained in 12L:12D for 7 days at 22°C were transferred to constant BL at 12, 17 and 27°C. After 72 h in constant conditions, ∼100 seedlings were harvested every 4 h for the next 24 h and frozen immediately in liquid nitrogen. Total RNA was extracted using RNeasy plant mini kit (Qiagen, Crawley, UK) following the manufacturer’s instructions. RNA was extracted from two biological repeats. A 1 μg portion of total RNA was reverse transcribed using the QuantiTect Reverse Transcription Kit (Qiagen) according to the manufacturer’s instructions. The qRT-PCR reactions were set up using a liquid handling robot (Freedom Evo; Tecan, Reading, UK) and assayed in a Lightcycler 480 system (Roche, Burgess Hill, UK). Each 10 μl reaction contained 1 μl cDNA (1/5 dilution), 5 μl LightCycler 480 SYBR green master mix (Roche) and 0.3 μM of each gene-specific primer (Supplementary Table 5). New primers were designed using Perlprimer (Marshall, 2004). Data were analysed with Roche Lightcycler 480SW 1.5 software, using the 2nd derivative maximum method. Each cDNA sample was assayed in technical triplicate assays. Relative mRNA levels were obtained from a standard curve during the qRT–PCR. The relative RNA level for each sample was calculated:
Relative mRNA=relative measure of candidate mRNA/relative measure of UBQ mRNA.
Protein analysis
CRY and LHY proteins were extracted from plants grown as described above. The extraction and separation methods used are described in Devlin et al (1992). The CRY proteins were separated and detected with the native antibodies against CRY1 and CRY2 previously described (Zeugner et al, 2005), as detailed in the Supplementary Methods online. More detailed information of LHY protein detection can be found in the Supplementary Methods online.
Computational methods
Sensitivity analysis and period estimation for simulations were performed in the Time Series Sensitivity Analysis MATLAB package, described in Rand (2008) and freely available from http://www2.warwick.ac.uk/fac/sci/systemsbiology/software/. Starting parameter values and model equations were taken from Pokhilko model (Pokhilko et al, 2010), available from the BioModels database as BIOMD000273, and from the Plant Systems Modelling repository, www.plasmo.ed.ac.uk. Some parameter values and model equations were modified to replicate experimental data, as described in the Supplementary Methods online. The models presented here will be available from the same locations upon publication.
Identifying sources of variation
To determine the possible sources of variation in the present data, we used both standard linear regression models and linear mixed-effects models (Pinheiro and Bates, 2000) as described in the Supplementary Methods online. The estimated period lengths were treated as the response variable and the different factors studied as the covariates. The latter were divided into fixed effects (light, temperature, marker and genotype) and random effects (experimental replicates and lines). Inference was performed using the functions lm (R Development Core Team, 2011) and lmer (Bates and Maechler, 2011) implemented in the open source statistical programming environment R, freely available from http://www.R-project.org/.
Accession numbers
LHY, At1g01060; CCA1, At2g46830; PRR9, At2g46790; PRR7, At5g02810; GI, At1g22770; TOC1, At5g61380; CRY1, At4g08920; CRY2, At1g04400.
Supplementary Material
Supplementary Figures S1–12, Supplementary Tables S1–7
Luciferase time course data sets for
Acknowledgments
We gratefully acknowledge expert technical assistance of Jean Wood, Jack Young, Susannah Bird and Kelly Stewart, support for high-throughput qRT-PCR from Lorraine Kerr and Steven Kane at the Kinetic Parameter Facility, LHY antibodies were supplied by Dr Isabelle Carré, and CRY antibodies and mutant seed were provided by Profs Margaret Ahmad and Chentao Lin, respectively. NU was funded by University of Liverpool strategic PhD studentship. This work was supported by the BBSRC/EPSRC-funded ROBuST SABR project (BB/F005237/1, BB/F005318/1, BB/F005261/1 and BB/F005296/1) led by Karen Halliday. Work at the Kinetic Parameter Facility was supported by SynthSys, a Centre for Integrative and Systems Biology supported by BBSRC and EPSRC award D019621.
Author contributions: Period assays and analysis were initiated by NU and AJWH, and pursued by PDG and AJWH, with advice from the ROBuST project team. The statistical analysis was performed by MC and BF. Mathematical simulation and analysis was designed by MD and DAR, and performed by MD. Protein assays were performed by DM and SP. RNA assays were performed by PDG and JF. PDG, MD, AJWH, AJM, SP, DAR and KJH wrote the paper with input from all authors.
Footnotes
The authors declare that they have no conflict of interest.
References
- Akman OE, Locke JCW, Tang S, Carré I, Millar AJ, Rand DA (2008) Isoform switching facilitates period control in the Neurospora crassa circadian clock. Mol Syst Biol 4: 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson C, Wilkins M (1989) Control of the circadian-rhythm of carbon-dioxide assimilation in Bryophyllum leaves by exposure to darkness and high-carbon dioxide concentrations. Planta 177: 401–408 [DOI] [PubMed] [Google Scholar]
- Bates DM, Maechler M (2011) lme4: Linear Mixed-effects Models Using S4 Classes. R package version 0 999375–37/r1160 [Google Scholar]
- Baudry A, Ito S, Song YH, Strait AA, Kiba T, Lu S, Henriques R, Pruneda-Paz JL, Chua NH, Tobin EM, Kay SA, Imaizumi T (2010) F-box proteins FKF1 and LKP2 act in concert with ZEITLUPE to control Arabidopsis clock progression. Plant Cell 22: 606–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brettschneider C, Rose RJ, Hertel S, Axmann IM, Heck AJR, Kollmann M (2010) A sequestration feedback determines dynamics and temperature entrainment of the KaiABC circadian clock. Mol Syst Biol 6: 389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashmore AR, Jarillo JA, Wu YJ, Liu DM (1999) Cryptochromes: blue light receptors for plants and animals. Science 284: 760–765 [DOI] [PubMed] [Google Scholar]
- Davis AM, Hall A, Millar AJ, Darrah C, Davis SJ (2009) Protocol: streamlined sub-protocols for floral-dip transformation and selection of transformants in Arabidopsis thaliana. Plant Methods 5: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin P, Rood S, Somers D, Quail P, Whitelam G (1992) Photophysiology of the elongated internode (Ein) mutant of Brassica-Rapa—Ein mutant lacks a detectable phytochrome B-like polypeptide. Plant Physiol 100: 1442–1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin PF, Kay SA (2000) Cryptochromes are required for phytochrome signaling to the circadian clock but not for rhythmicity. Plant Cell 12: 2499–2509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diernfellner A, Colot HV, Dintsis O, Loros JJ, Dunlap JC, Brunner M (2007) Long and short isoforms of Neurospora clock protein FRQ support temperature-compensated circadian rhythms. FEBS Lett 581: 5759–5764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap JC, Loros JJ, DeCoursey PJ (2003) Chronobiology: Biological Timekeeping. Sunderland, USA: Sinauer Associates Inc. [Google Scholar]
- Edwards KD, Akman OE, Knox K, Lumsden PJ, Thomson AW, Brown PE, Pokhilko A, Kozma-Bognár L, Nagy F, Rand DA, Millar AJ (2010) Quantitative analysis of regulatory flexibility under changing environmental conditions. Mol Syst Biol 6: 424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson ME, Hanano S, Southern MM, Hall A, Millar AJ (2003) Response regulator homologues have complementary, light-dependent functions in the Arabidopsis circadian clock. Planta 218: 159–162 [DOI] [PubMed] [Google Scholar]
- Farré EM, Harmer SL, Harmon FG, Yanovsky MJ, Kay SA (2005) Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr Biol 15: 47–54 [DOI] [PubMed] [Google Scholar]
- Gendron J, Pruneda-Paz J, Doherty CJ, Gross AM, Kang SE, Kay SA (2012) Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc Natl Acad Sci USA 109: 3167–3172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould PD, Diaz P, Hogben C, Kusakina J, Salem R, Hartwell J, Hall A (2009) Delayed fluorescence as a universal tool for the measurement of circadian rhythms in higher plants. Plant J 58: 893–901 [DOI] [PubMed] [Google Scholar]
- Gould PD, Locke JC, Larue C, Southern MM, Davis SJ, Hanano S, Moyle R, Milich R, Putterill J, Millar AJ, Hall A (2006) The molecular basis of temperature compensation in the Arabidopsis circadian clock. Plant Cell 18: 1177–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo HW, Yang WY, Mockler TC, Lin CT (1998) Regulations of flowering time by Arabidopsis photoreceptors. Science 279: 1360–1363 [DOI] [PubMed] [Google Scholar]
- Hall A, Brown P (2007) Monitoring circadian rhythms in Arabidopsis thaliana using luciferase reporter genes. Methods Mol Biol 362: 143–152 [DOI] [PubMed] [Google Scholar]
- Halliday K, Salter M, Thingnaes E, Whitelam G (2003) Phytochrome control of flowering is temperature sensitive and correlates with expression of the floral integrator FT. Plant J 33: 875–885 [DOI] [PubMed] [Google Scholar]
- Harmer SL (2009) The circadian system in higher plants. Annu Rev Plant Biol 60: 357–377 [DOI] [PubMed] [Google Scholar]
- Hastings JW, Sweeney BM (1957) On the mechanism of temperature independence in a biological clock. Proc Natl Acad Sci USA 43: 804–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks KA, Millar AJ, Carre IA, Somers DE, Straume M, Meeks-Wagner DR, Kay SA (1996) Conditional circadian dysfunction of the Arabidopsis early-flowering 3 mutant. Science 274: 790–792 [DOI] [PubMed] [Google Scholar]
- Huang W, Perez-Garcia P, Pokhilko A, Millar AJ, Antoshechkin I, Riechmann JL, Mas P (2012) Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science 336: 75–79 [DOI] [PubMed] [Google Scholar]
- Ito S, Matsushika A, Yamada H, Sato S, Kato T, Tabata S, Yamashino T, Mizuno T (2003) Characterization of the APRR9 pseudo-response regulator belonging to the APRR1/TOC1 quintet in Arabidopsis thaliana. Plant Cell Physiol 44: 1237–1245 [DOI] [PubMed] [Google Scholar]
- James AB, Syed NH, Bordage S, Marshall J, Nimmo GA, Jenkins GI, Herzyk P, Brown JWS, Nimmo HG (2012) Alternative splicing mediates responses of the Arabidopsis circadian clock to temperature changes. THE PLANT CELL ONLINE 24: 961–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarillo JA, Capel J, Tang RH, Yang HQ, Alonso JM, Ecker JR, Cashmore AR (2001) An Arabidopsis circadian clock component interacts with both CRY1 and phyB. Nature 410: 487–490 [DOI] [PubMed] [Google Scholar]
- Kaushik R, Nawathean P, Busza A, Murad A, Emery P, Rosbash M (2007) PER-TIM interactions with the photoreceptor cryptochrome mediate circadian temperature responses in Drosophila. PLoS Biol 5: e146. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kim WY, Fujiwara S, Suh SS, Kim J, Kim Y, Han L, David K, Putterill J, Nam HG, Somers DE (2007) ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature 449: 356–360 [DOI] [PubMed] [Google Scholar]
- Kuno N, Møller SG, Shinomura T, Xu X, Chua N-H, Furuya M (2003) The novel MYB protein EARLY-PHYTOCHROME-RESPONSIVE1 is a component of a slave circadian oscillator in Arabidopsis. Plant Cell 15: 2476–2488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau OS, Huang X, Charron JB, Lee JH, Li G, Deng XW (2011) Interaction of Arabidopsis DET1 with CCA1 and LHY in mediating transcriptional repression in the plant circadian clock. Mol Cell 43: 703–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Shalitin D (2003) Cryptochrome structure and signal transduction. Annu Rev Plant Biol 54: 469–496 [DOI] [PubMed] [Google Scholar]
- Liu Y, Garceau NY, Loros JJ, Dunlap JC (1997) Thermally regulated translational control of FRQ mediates aspects of temperature responses in the neurospora circadian clock. Cell 89: 477–486 [DOI] [PubMed] [Google Scholar]
- Locke JC, Kozma-Bognar L, Gould PD, Feher B, Kevei E, Nagy F, Turner MS, Hall A, Millar AJ (2006) Experimental validation of a predicted feedback loop in the multi-oscillator clock of Arabidopsis thaliana. Mol Syst Biol 2: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke JCW, Southern MM, Kozma-Bognár L, Hibberd V, Brown PE, Turner MS, Millar AJ (2005) Extension of a genetic network model by iterative experimentation and mathematical analysis. Mol Syst Biol 1(2005): 0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall OJ (2004) PerlPrimer: cross-platform, graphical primer design for standard, bisulphite and real-time PCR. Bioinformatics 20: 2471–2472 [DOI] [PubMed] [Google Scholar]
- Martin-Tryon EL, Kreps JA, Harmer SL (2007) GIGANTEA acts in blue light signaling and has biochemically separable roles in circadian clock and flowering time regulation. Plant Physiol 143: 473–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushika A, Imamura A, Yamashino T, Mizuno T (2002) Aberrant expression of the light-inducible and circadian-regulated APRR9 gene belonging to the circadian-associated APRR1/TOC1 quintet results in the phenotype of early flowering in Arabidopsis thaliana. Plant Cell Physiol 43: 833–843 [DOI] [PubMed] [Google Scholar]
- McWatters HG, Devlin PF (2011) Timing in plants--a rhythmic arrangement. FEBS Lett 585: 1474–1484 [DOI] [PubMed] [Google Scholar]
- Millar AJ (2004) Input signals to the plant circadian clock. J Exp Bot 55: 277–283 [DOI] [PubMed] [Google Scholar]
- Mockler T, Yang H, Yu X, Parikh D, Cheng YC, Dolan S, Lin C (2003) Regulation of photoperiodic flowering by Arabidopsis photoreceptors. Proc Natl Acad Sci USA 100: 2140–2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockler TC, Guo HW, Yang HY, Duong H, Lin CT (1999) Antagonistic actions of Arabidopsis cryptochromes and phytochrome B in the regulation of floral induction. Development 126: 2073–2082 [DOI] [PubMed] [Google Scholar]
- Nagatani A, Reed JW, Chory J (1993) Isolation and initial characterization of Arabidopsis mutants that are deficient in phytochrome A. Plant Physiol 102: 269–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N, Kiba T, Henriques R, Mizuno T, Chua N-H, Sakakibara H (2010) Pseudo-response regulators 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 22: 594–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusinow DA, Helfer A, Hamilton EE, King JJ, Imaizumi T, Schultz TF, Farré EM, Kay SA (2011) The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475: 398–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S (2008) Temperature perception and signal transduction in plants. New Phytol 179: 615–628 [DOI] [PubMed] [Google Scholar]
- Peschel N, Chen KF, Szabo G, Stanewsky R (2009) Light-dependent interactions between the Drosophila circadian clock factors cryptochrome, jetlag, and timeless. Curr Biol 19: 241–247 [DOI] [PubMed] [Google Scholar]
- Pinheiro JC, Bates DM (2000) Mixed-Effects Models in S and S-plus. New York, USA: Springer. [Google Scholar]
- Plautz JD, Straume M, Stanewsky R, Jamison CF, Brandes C, Dowse HB, Hall JC, Kay SA (1997) Quantitative analysis of Drosophila period gene transcription in living animals. J Biol Rhythms 12: 204–217 [DOI] [PubMed] [Google Scholar]
- Pokhilko A, Hodge SK, Stratford K, Knox K, Edwards KD, Thomson AW, Mizuno T, Millar AJ (2010) Data assimilation constrains new connections and components in a complex, eukaryotic circadian clock model. Mol Syst Biol 6: 416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokhilko A, Fernández AP, Edwards KD, Southern MM, Halliday KJ, Millar AJ (2012) The clock gene circuit in Arabidopsis includes a repressilator with additional feedback loops. Mol Sys Biol 8: 574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. (2011) R: A language and environment for statistical computing. R Foundation for Statistical Computing http://www.R-project.org/ [Google Scholar]
- Rand DA (2008) Mapping global sensitivity of cellular network dynamics: sensitivity heat maps and a global summation law. J R Soc Interface 5(Suppl 1): S59–S69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JW, Nagatani A, Elich TD, Fagan M, Chory J (1994) Phytochrome A and phytochrome B have overlapping but distinct functions in Arabidopsis development. Plant Physiol 104: 1139–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruoff P, Loros JJ, Dunlap JC (2005) The relationship between FRQ-protein stability and temperature compensation in the Neurospora circadian clock. Proc Natl Acad Sci USA 102: 17681–17686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruoff P, Rensing L, Kommedal R, Mohsenzadeh S (1997) Modeling temperature compensation in chemical and biological oscillators. Chronobiol Int 14: 499–510 [DOI] [PubMed] [Google Scholar]
- Salome PA, McClung CR (2005) Pseudo-response regulator 7 and 9 are partially redundant genes essential for the temperature responsiveness of the Arabidopsis circadian clock. Plant Cell 17: 791–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomé PA, Weigel D, McClung CR (2010) The role of the Arabidopsis morning loop components CCA1, LHY, PRR7, and PRR9 in temperature compensation. Plant Cell 22: 3650–3661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöning JC, Streitner C, Meyer IM, Gao Y, Staiger D (2008) Reciprocal regulation of glycine-rich RNA-binding proteins via an interlocked feedback loop coupling alternative splicing to nonsense-mediated decay in Arabidopsis. Nucleic Acids Res 36: 6977–6987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen WL, Kwon Y, Adegbola AA, Luo J, Chess A, Montell C (2011) Function of rhodopsin in temperature discrimination in Drosophila. Science 331: 1333–1336 [DOI] [PubMed] [Google Scholar]
- Somers DE, Devlin PF, Kay SA (1998) Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science 282: 1488–1490 [DOI] [PubMed] [Google Scholar]
- Strasser B, Sánchez-Lamas M, Yanovsky MJ, Casal JJ, Cerdán PD (2010) Arabidopsis thaliana life without phytochromes. Proc Natl Acad Sci USA 107: 4776–4781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth R, Kevei E, Hall A, Millar AJ, Nagy F, Kozma-Bognar L (2001) Circadian clock-regulated expression of phytochrome and cryptochrome genes in Arabidopsis. Plant Physiol 127: 1607–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Horst GT, Muijtjens M, Kobayashi K, Takano R, Kanno S, Takao M, de Wit J, Verkerk A, Eker AP, van Leenen D, Buijs R, Bootsma D, Hoeijmakers JH, Yasui A (1999) Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature 398: 627–630 [DOI] [PubMed] [Google Scholar]
- Wenden B, Kozma-Bognár L, Edwards KD, Hall AJW, Locke JCW, Millar AJ (2011) Light inputs shape the Arabidopsis circadian system. Plant J 66: 480–491 [DOI] [PubMed] [Google Scholar]
- Yu JW, Rubio V, Lee NY, Bai S, Lee SY, Kim SS, Liu L, Zhang Y, Irigoyen ML, Sullivan JA, Lee I, Xie Q, Paek NC, Deng XW (2008) COP1 and ELF3 control circadian function and photoperiodic flowering by regulating GI stability. Mol Cell 32: 617–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeugner A, Byrdin M, Bouly J, Bakrim N, Giovani B, Brettel K, Ahmad M (2005) Light-induced electron transfer in Arabidopsis cryptochrome-1 correlates with in vivo function. J Biol Chem 280: 19437–19440 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1–12, Supplementary Tables S1–7
Luciferase time course data sets for