Abstract
To understand the structure and function of large molecular machines, accurate knowledge of their stoichiometry is essential. In this study, we developed an integrated targeted proteomics and super-resolution microscopy approach to determine the absolute stoichiometry of the human nuclear pore complex (NPC), possibly the largest eukaryotic protein complex. We show that the human NPC has a previously unanticipated stoichiometry that varies across cancer cell types, tissues and in disease. Using large-scale proteomics, we provide evidence that more than one third of the known, well-defined nuclear protein complexes display a similar cell type-specific variation of their subunit stoichiometry. Our data point to compositional rearrangement as a widespread mechanism for adapting the functions of molecular machines toward cell type-specific constraints and context-dependent needs, and highlight the need of deeper investigation of such structural variants.
Keywords: fluorophore counting, nucleoporin, protein complex-based analysis, super-resolution microscopy, targeted proteomics
Introduction
Little is known about remodeling and composition of protein complexes across different cell types in higher eukaryotes. Previous studies have shown that protein complex composition can be subjected to temporal variation in yeast, for example, throughout the cell cycle (de Lichtenberg et al, 2005). To the best of our knowledge, only a handful of protein complexes have been shown to vary across different cell types so far (Noda et al, 2000; Murata et al, 2007; Liu et al, 2008; Wu et al, 2009), but whether protein complexes are rearranged as a function of cell type has never been addressed on a global scale. Here, we systematically study the exact compositions of nuclear pore complexes (NPCs) from several human tissue cultures and also quantify cell type-dependent variations of well-characterized nuclear protein complexes in general.
The NPC is one of the most intricate molecular machines of eukaryotic cells and conducts the transport of molecules in and out of the nucleus. It is built from ∼30 nucleoporins (Nups) that assemble in multiple copies to form an eight-fold rotationally symmetric complex (Hoelz et al, 2011). NPCs possess different hierarchical levels of structural organization: Nups first assemble into hetero-oligomeric subcomplexes, which in turn, serve as modular building blocks to form larger structures. The best currently characterized modules are the so-called Nup107 and Nup93 subcomplexes, which are essential architectural elements of the NPC scaffold, and the Nup62 subcomplex that constitutes a major transporter module (Brohawn et al, 2009). Accurate knowledge of Nup copy numbers per NPC is a crucial prerequisite toward the generation of structural models that explain how subcomplexes are arranged in the higher order assembly and for a detailed understanding of the transport mechanism. Based on semi-quantitative investigations of Nup stoichiometry (Rout et al, 2000; Cronshaw et al, 2002), a head-to-tail arrangement of the yNup84 subcomplex (human homolog Nup107) has been proposed, which relies on 16 copies of this scaffold motif per NPC (2 copies per unit cell) (Alber et al, 2007b; Kampmann and Blobel, 2009). Alternatively, the fence pole (Debler et al, 2008) and the lattice model (Brohawn and Schwartz, 2009) have assumed four copies per unit cell. Since there is no consensus on the relative as well as absolute stoichiometry, those models disagree even on fundamental aspects, such as the overall size, the orientation of known subcomplexes and the total molecular weight. In this study, we used targeted proteomics and super-resolution microscopy to establish the abundance of all human Nups within the NPC on an absolute scale. In contrast to previous studies, our data imply a higher baseline of copy numbers and the existence of multiple distinct structural species of subcomplexes in situ. Furthermore, we found that the abundance of a large subset of peripheral Nups is rearranged as a function of the cell type, while the NPC scaffold structure remains steady. To investigate whether the compositional changes of the NPC are the rule or an exception, we used shotgun proteomics to analyze a set of well-defined nuclear protein complexes across five human cell lines. With only a few cell types at hand, we already found rearrangements for 38% of the studied complexes, implying that most molecular machines are likely to be fine-tuned at the quaternary structure level to comply with the particularities of different cell types.
Results
Absolute abundance of NPC components
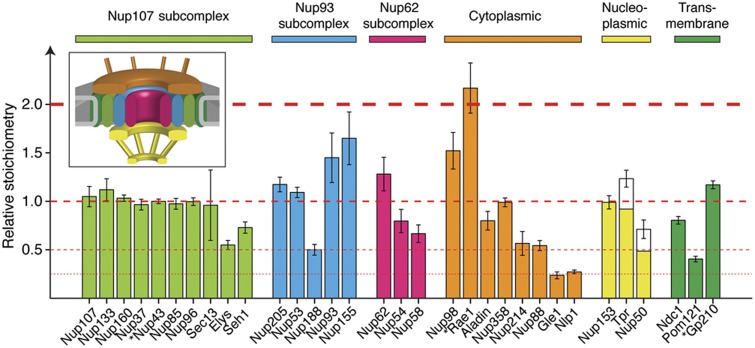
To determine the exact stoichiometry of all human Nups when subcomplexes are assembled into NPCs, we used targeted mass spectrometry (multiple reaction monitoring, MRM) in combination with absolutely quantified (AQUA) internal standard peptides, as previously described (Gerber et al, 2003; Picotti et al, 2009). We quantified all Nups within nuclear envelopes (NEs) that were purified to homogeneity from HeLa cells using an improved sub-cellular fractionation procedure (Supplementary Materials and Methods). We performed a series of quality control experiments for our preparations to rule out leakage of NPC components during the isolation and ensure efficient removal of nucleoplasmic material following chromatin digestion (Supplementary Figure S1). For 29 out of 32 Nups, the absolute abundance was determined with high confidence, namely with two or more independent peptide measurements per protein (Figure 1; Supplementary Table S1). More than 80% of the selected assays displayed <20% variation across four biological replicates (Supplementary Figure S1F), corroborating the high quality of the data. Nup abundances span about one order of magnitude and, in most cases, occur as multiples of each other (Figure 1). Remarkably, our data reveal an unexpected compositional architecture of the NPC. Within subcomplexes that were biochemically defined in vitro (Siniossoglou et al, 2000; Kampmann and Blobel, 2009; Amlacher et al, 2011), Nups often do not occur iso-stoichiometrically in situ. These data indicate either the co-existence of the same subunit in different assemblies or the presence of hetero-oligomers that are not iso-stoichiometric. For example, two members of the Nup93 subcomplex, Nup205 and Nup53, have the same abundance as the majority of members of the Nup107 subcomplex. However, Nup188 occurs at half of this abundance and Nup93 as well as Nup155 are 1.5 times as abundant. Interestingly, the abundance of Nup93 equals the sum of Nup188 and Nup205 abundances supporting the existence of two independent structural species (Kosova et al, 1999; Theerthagiri et al, 2010). Here, we provide highly detailed stoichiometric information of all Nups, which is a key prerequisite for further structural investigations, in particular studies relying on the reconstitution of subcomplexes and structural modeling.
Figure 1.
Stoichiometry of the human nuclear pore complex (NPC). Nuclear envelopes (NEs) were isolated from HeLa cells and the abundances of all Nups were determined by targeted proteomics using intrinsic heavy labeled standards (AQUA peptides). Relative stoichiometries are displayed as median light to heavy ratio±median absolute deviation (across four biological replicates and multiple independent measurements). Nups are grouped in subcomplexes or according to their predicted localization in the NPC (inset). Stars indicate cases that were quantified using a single PTP, and therefore represent lower confidence quantifications. The abundances of Tpr and Nup50 were corrected for the estimated nucleoplasmic pool that remained associated to the NEs. All abundance levels are related to the Nup107 subcomplex and the major abundance steps (multiples of Nup107) are indicated with dashed red lines. See also Supplementary Figure S1 and Supplementary Table S1.
Calibration of stoichiometries to copies per NPC
In order to translate the Nup stoichiometries (protein ratios) into copy numbers per NPC, previous studies relied on the assumption that, due to the eight fold rotational symmetry, the lowest abundant Nups occur in eight copies per NPC (1 copy per asymmetric unit) (Cronshaw et al, 2002). It has been argued that Nups occur at three discrete abundance levels corresponding to 8, 16 and 32 copies per NPC (Alber et al, 2007a). As a consequence, 16 copies per NPC were assumed for the majority of scaffold Nups, which would correspond to 2 copies per asymmetric unit distributed pseudo-symmetrically across the NE plane. In contrast to the aforementioned studies, we observed two discrete populations of Nups that are less abundant than the scaffold level (two- and four-fold less abundant; Figure 1). If the lowest abundant Nups occur in eight copies per NPC as previously assumed, then this finding implies that at least 12 Nups (including, e.g., Nup107) occur in 32 copies per NPC, while at least 5 others (including, e.g., Elys) occur in 16 copies. Finally, Gle1 and Nlp1 are only a quarter as abundant, which would correspond to 8 copies per NPC.
To directly measure the copy number of Nup107 per NPC, we used an integrated approach. First, we accurately counted nuclei isolated from HeLa cells by FACS using an internal fluorescent bead standard for calibration and cross-validated this method by an independent counting scheme (Supplementary Materials and Methods). Subsequently, we quantified the total copies of Nup107 contained in a defined number of nuclei by targeted MS. We estimated Nup107 to occur at ∼125±6 thousand copies per nucleus. Combined with the average number of NPCs per NE, these data should ultimately reveal the copies of Nup107 per NPC. Next, we acquired a large set of cryo electron tomograms NEs and determined the density of NPCs (Supplementary Figure S2A). To extrapolate the NPC density to the number of NPCs per nucleus, we measured the nucleus surface area using fluorescent dyes and confocal microscopy (Supplementary Figure S2B). By combining these measurements, we estimated an average number of 3376±1304 NPC per nucleus, in agreement with previous reports (Maul et al, 1972; Ribbeck and Gorlich, 2001). Taken together with the number of molecules per nucleus, these data result in an average of 37±14 Nup107 copies per NPC.
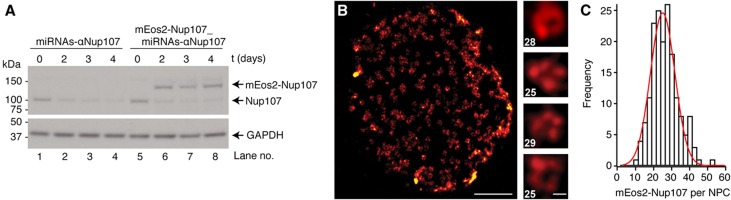
As an independent measurement, we used super-resolution microscopy to count the Nup107 protein copies per NPC. Fluorophore counting of the mEos2 protein by iterative photo-conversion with subsequent bleaching was previously established (Annibale et al, 2011; McKinney et al, 2009). To efficiently integrate mEos2 into NPCs, we engineered a stable cell line derived from human embryonic kidney cells (HEK293-Flp-In-T-REx) that co-expresses two microRNAs silencing endogenous Nup107 and a replacement gene encoding Nup107 N-terminally fused to mEos2. We observed efficient silencing of the endogenous Nup107 and co-occurring expression of the mEos2-tagged protein with a replacement efficiency of about 80–90% (Figure 2A). The use of genetically encoded fluorescent proteins suffers from one major disadvantage, that is, these proteins do not necessarily mature into functional fluorophores (Ulbrich and Isacoff, 2007). Since this maturation efficiency cannot be assessed easily, fluorophore counting will only permit to determine a minimal number for mEos2-Nup107 fusion protein copies in the NPC. In other words, if the experimentally determined copy number is significantly larger than 16, then the NPC could contain 32, 64 or more, but not 8 or 16 copies of mEos2-Nup107 (Supplementary Figure S3; Supplementary Materials and Methods). In order to observe signals with high optical sectioning near the coverslip, we performed imaging of purified nuclei that were allowed to settle on a coverslip and observed using total internal reflection fluorescence (TIRF) microscopy. We acquired time-lapse movies of sequential photo-conversion/photo-bleaching of mEos2 molecules until complete bleaching was reached, as previously described (Betzig et al, 2006; Supplementary Movie S1). A clustering algorithm based on density (DBSCAN) (Ester et al, 1996) allowed us to select photo-conversion events corresponding to isolated, individual NPCs. The reconstructed images show individual NPCs as circular patterns (Figure 2B; Supplementary Figures S4A–D), which is to our knowledge the first time that the ring-like arrangement of the major scaffold component Nup107 was directly visualized by super-resolution microscopy. The number of photo-conversion events detected in each NPC was corrected for fluorophore blinking using the method developed by Annibale et al (2011). Although the approach employed here is likely to systematically underestimate the copy number of mEos2-Nup107 (as explained above), the majority of all measured events (90%) account for >16 copies per NPC (Figure 2C).
Figure 2.
Super-resolution microscopy in combination with fluorophore counting provides a lower limit estimate of the copies of Nup107 per NPC. (A) Endogenous Nup107 was efficiently replaced with Nup107 N-terminally fused to mEos2. HEK293-Flp-In-T-REx cells were stably transfected with two microRNAs targeting both UTRs of the Nup107 gene (miRNAs-αNup107, lanes 1–4) or with a vector containing the same miRNA cassette together with the human mEos2-Nup107 fusion (mEos2-Nup107_miRNAs-αNup107, lanes 5–8), both under an inducible promoter. Nuclear extracts were analyzed by western blot and probed with antibodies against Nup107 (top panel) or GAPDH (bottom panel), as loading control. (B) Representative super-resolution image of a nucleus isolated from transfected HEK293 cells shows the characteristic circular NPC staining. Zoomed images of selected nuclear pores are shown together with the respective number of counted fluorophores (after correction for labeling efficiency; scale bars: 1 μm and 50 nm for zoomed images). (C) Distribution of the measured copies of mEos2-Nup107 per NPC obtained from super-resolution-based fluorophore counting (after correction for the labeling efficiency). The majority of all measured NPCs contain >16 copies of mEos2-Nup107. The final copy number frequency distribution was obtained from 172 clusters imaged from 5 nuclei and corrected for labeling efficiency. See also Supplementary Figures S3 and S4, and Supplementary Movie S1. Source data for this figure is available on the online supplementary information page.
To validate our workflow and test if we could, in principle, resolve an overlay of multiple structural species of different copy numbers, we performed Monte-Carlo simulations considering three alternative scenarios, 16, 32 and 64 copies of Nup107 per NPC. These simulations take into account a stochastic NPC position within the NE, a stochastic labeling density across individual NPCs, a rotational distribution of fluorophores around the central channel as well as the photo-conversion, blinking and bleaching of mEos2. All input parameters are based on experimentally determined values (see also Supplementary Materials and Methods and Supplementary Figure S4E). The synthetic data recapitulate the experimental results very well (Supplementary Figures S4F–H), that is, they are most consistent with a scenario of 32 copies and argue for a monodisperse distribution of structural species.
Finally, the sum of the molecular weight of all copies of all Nups in our study is 110±10 MDa when 32 copies of Nup107 are assumed (Supplementary Table S1). This is in excellent agreement with molecular weight measurements of the vertebrate NPC by scanning transmission electron microscopy (112±11 MDa) (Reichelt et al, 1990). We furthermore assessed the structural plausibility of the molecular mass by volumetric matching into the cryo electron microscopy map of the human NPC (Maimon et al, 2012; Supplementary Figure S2C). Although the calibration experiments described above are on their own less accurate than the stoichiometric measurements by MS, taken together, several independent lines of evidence suggest that 32 copies per NPC have to be alternatively considered for the major scaffold module.
The composition of the NPC varies across different human cell lines
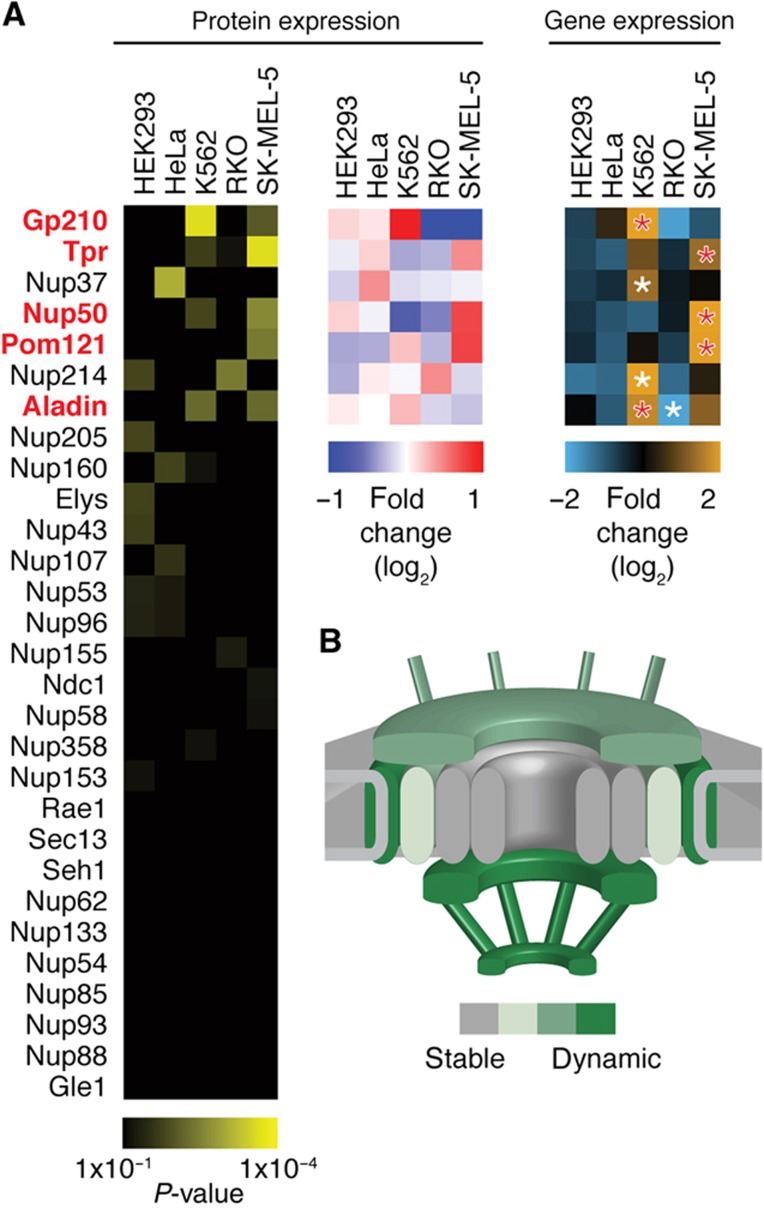
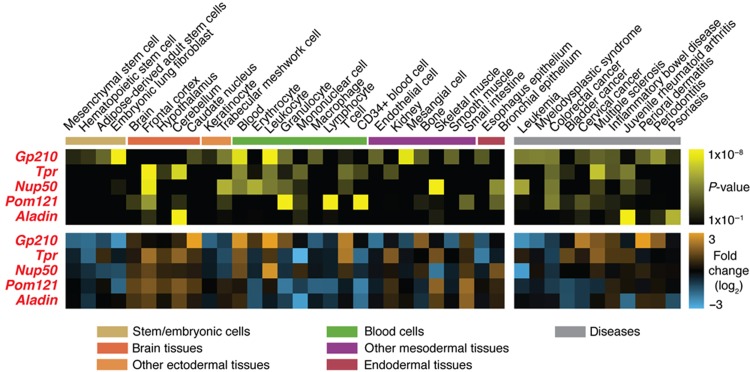
To ensure that the established stoichiometry serves as a general principle for all human cells, but also to test for the opportunity of cell type-specific adjustments that might have a functional impact, we used our targeted proteomic assays to quantify NPC composition across various human tissue culture cells. First, we used gene expression data to screen for cell lines that displayed very pronounced variations in Nup mRNA expression levels and identified three human cancer-derived cell lines: K562, an erythroleukemic cell line; RKO, a colon carcinoma-derived cell line; and SK-MEL-5, a malignant melanoma-derived cell line (Supplementary Figure S5). To investigate whether the different mRNA profiles are reflected by Nup protein abundance and ultimately differential NPC composition, we compared nuclei isolated from these three cell lines as well as HeLa and non-tumor derived HEK293 cells by targeted proteomics. We consistently analyzed the profile of 29 Nups across the 5 cell lines in three biological replicates and identified significant changes in the abundance of 7 Nups across at least two cell types (Figure 3A). We observed cell-specific changes for two nucleoplasmic Nups (Nup50 and Tpr), two cytoplasmic Nups (Nup214 and Aladin), two out of the three transmembrane Nups (Gp210 and Pom121) and one scaffold Nup (Nup37). Our analysis thus implies dynamic rearrangements of NPC stoichiometry across cell types. This is primarily true for peripheral Nups, while the NPC scaffold structure remains static (Figure 3B). The observed differences prompted us to study Nups of varying abundance in a variety of human tissues. For five out of these seven Nups (Gp210, Pom121, Aladin, Nup50 and Tpr), the variation at the protein level could be correctly inferred from gene expression data (Figure 3A). Therefore, we analyzed mRNA expression profiles of these 5 Nups across 44 tissue types and 30 disease states (Figure 4). We detected significant tissue-specific expression in distinct cell types such as stem cells, brain-derived tissues and certain classes of immune cells as well as in several diseases including various cancers and inflammatory disorders (Figure 4).
Figure 3.
NPC stoichiometry varies across human cell lines. Relative Nup protein abundances were measured by targeted proteomics in nuclei isolated from five different cell lines. (A) Three biological replicates each were analyzed using linear mixed-effect models (Supplementary Materials and Methods). In significant cases (adjusted P-value<0.01, corresponding to the top seven Nups), the mean protein fold change is shown (blue-red heat map) and compared with the mean fold change from gene expression data (light blue-orange heat map). Differential gene expression per cell line was assessed by unpaired, two-sided t-test and starred boxes indicate highly significant cases (adjusted P-value<1 × 10−10). For five of the seven significantly regulated Nups (highlighted in red), protein and mRNA expression data gave consistent results. (B) Schematic representation highlighting the peripheral and transmembrane localization of the dynamic NPC subunits, while the majority of all scaffold components remains unchanged. The color code is based on the fraction of dynamic subunits identified in each group. See also Supplementary Figure S5.
Figure 4.
Nucleoporin gene expression varies across multiple human tissues and diseases. Gene expression of the five dynamic Nups displaying correlated protein and mRNA expression profiles in human tissue culture cells (Figure 3A) was analyzed across a panel of tissues and diseases. The analyzed Nups displayed characteristic expression profiles that are consistent between tissue and disease groups. The significance of condition-specific differential expression was assessed using an unpaired, two-sided t-test (FDR corrected, adjusted P-value<1 × 10−2 with an additional constraint of absolute log2-fold change>1). Only conditions displaying at least one significant case are shown. Gene expression data were retrieved from the Gene Expression Atlas (Lukk et al, 2010; Kapushesky et al, 2012). For details on data source and analysis, see Supplementary Materials and Methods.
Cell type-specific remodeling of multiple nuclear protein complexes
To determine whether cell type-specific compositional changes, as observed in case of the NPC, are rare or represent a more generic principle underlying functional variations of molecular machines, we analyzed the expression profiles of the subunits of a defined set of well-characterized nuclear protein complexes by label-free shotgun proteomics. For this purpose, we used nuclear extracts from the same five cell lines that we used to investigate NPC composition. Since many protein complexes share components (Gavin et al, 2006; Krogan et al, 2006), we manually selected 34 complexes with minimal overlap that were detected with sufficient coverage across the 5 cell lines in our data set (see Materials and methods, Supplementary Figure S6 and Supplementary Table S2). As an intrinsic measure for protein complex integrity, we used coherence measurements of the abundance of their subunits as previously described (Wang et al, 2009) (Supplementary Figure S6D, inset).
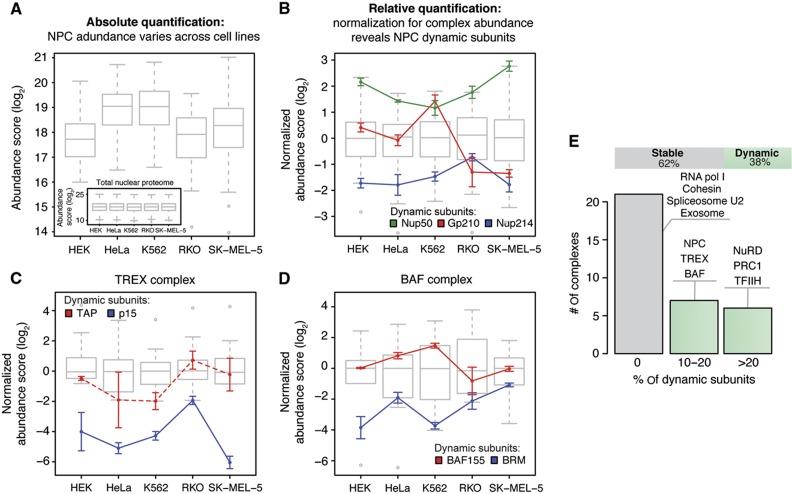
To test whether the selected subset of well-defined complexes are subject to compositional variations, we implemented a two-step analysis procedure. We first analyzed the absolute abundance of the each complex as a whole, which was often highly variable across the five cell lines (Figure 5A). Subsequently, we normalized the expression level of each subunit to the absolute abundance of the respective protein complex and used significance testing to identify subunits deviating from the expression profile of the other members of the same protein complex, which is indicative of a variable composition (Figure 5B; Supplementary Figure S6E). We validated our method using the NPC as a test case (Figures 5A and B) and found that it could to a large extent recapitulate the compositional variations that were accurately quantified by targeted proteomics beforehand (Figure 3). When we examined the data for significant, cell type-specific changes in the relative expression of other protein complex components, we found that the subunits of 21 out of 34 protein complexes (62%) displayed remarkably stable expression, indicating a conserved complex stoichiometry (Figure 5E). These cases include for example the cohesin complex and the core complexes of RNA polymerases I and II. For the remaining protein complexes (13 out of 34, 38%), the expression of specific subunits appeared to be more dynamic, that is, significantly changed in one or more of the analyzed cell lines, and decoupled from the expression of other components of the same complex. This indicates that these complexes might undergo compositional rearrangements in different cell types. This group contains not only the NPC (Figure 5B), but also the TREX complex that is involved in mRNPs packaging and export (Figure 5C), and complexes involved in chromatin remodeling and histone modification such as NuRD and BAF complexes (Figure 5D). To exclude a potential bias due to the manual selection of protein complexes, we repeated our analysis using a recently published data set of soluble protein complexes that were biochemically defined in two of the cell lines used in our study (HEK293 and HeLa) (Havugimana et al, 2012) and obtained similar results (Supplementary Figure S6F). Since we tested only five cell lines, it is likely that much more stoichiometric variations will exist when more cell types are analyzed, that is, the 38% we observe here likely represent a lower limit.
Figure 5.
Cell type-specific stoichiometries are detected for ∼38% of all investigated nuclear protein complexes. To analyze protein expression profiles in a complex-centered manner, a two-step normalization procedure was used. (A) First, the data were corrected for the variation of the total amount of nuclear protein observed in each sample using quantile normalization (inset), as commonly done for microarray data (Bolstad et al, 2003). Variable abundances of protein complexes across cell types are revealed (shown as box plot for the NPC). (B) Subsequently, the data were corrected for variable abundances of protein complexes by median subunit abundance centering. Variable stoichiometries were detected by relative subunit quantification across cell types (unpaired, two-sided t-test; significance cutoffs were set at adjusted P-value<0.01 after FDR correction and absolute log2-fold change>0.9). The NPC is shown as a test case; all the three significant dynamic subunits that were detected are in agreement with targeted proteomic measurements (Figure 3). Box plots represent the normalized distribution of NPC subunit abundances across the five cell lines; colored lines display the expression profiles of dynamic components. For each cell line, three biological replicates are grouped and displayed values are mean±standard deviation. (C, D) Same as in (B), but for selected protein complexes that displayed stoichiometric variations. In case of TREX complex (C), the profile of TAP is displayed using a dashed line, despite being below the significance threshold (adjusted P-value=0.066, log2-fold change=1.9 in RKO), since it is known to form a heterodimer with the significantly changed subunit p15 (Katahira and Yoneda, 2009). (E) Thirty-eight percent of all analyzed nuclear protein complexes displayed dynamic stoichiometric arrangements (shown as bar chart). Complexes were defined as dynamic if the expression of at least one subunit was significantly changed across the cell lines. Selected examples showing different percentages of dynamic subunits are listed. See also Supplementary Figure S6 and Supplementary Table S2.
Discussion
NPC stoichiometry and cell type-specific remodeling
The newly established stoichiometry of the human NPC calls for a new generation of structural models that take this into account. The determined Nup abundances provide a direct insight into the modularity of the NPC by revealing specific stoichiometries of different subcomplexes within assembled pores. The stoichiometric abundances of the majority of the components of the Nup107 subcomplex are consistent with biochemical and structural data obtained for the homolog Nup84 subcomplex from yeast (Rout et al, 2000; Alber et al, 2007b; Kampmann and Blobel, 2009). However, two Nups, namely Seh1 and Elys, are sub-stoichiometric relative to the other components. Interestingly, Seh1 was located to the periphery of the yeast Nup84 subcomplex and does not directly contribute to the structural core of the complex (Fernandez-Martinez et al, 2012), as previously thought (Nagy et al, 2009). In case of the chromatin binding protein Elys (Rasala et al, 2006), the lower copy number might be explained by its potential asymmetric occurrence on the nuclear face of the NPC. Moreover, both human Seh1 and Elys have been shown to be more loosely associated with the Nup107 subcomplex than other components (Loiodice et al, 2004; Franz et al, 2007). Taken together, these data suggest that a second, distinct structural instance of the Nup107 subcomplex might exist in humans, which has not yet been biochemically characterized. The two subspecies might be associated with an alternative subset of peripheral components, a possibility that has been neglected by all previous structural models. Our comprehensive stoichiometry data provide not only a general framework for understanding Nup interactions but also hint at some heterogeneity of certain Nup populations, in particular peripheral ones. This information might also facilitate the in vitro reconstruction of subcomplexes that account for distinct native structural species. The presented experimental strategy is universal and paves the way toward elucidating the detailed architecture of other molecular machines.
Recently, it has been reported that temporal regulation of the transmembrane nucleoporin Gp210 occurs in myoblast and stem cells and that varying Gp210 expression is required for differentiation to myotubes and neural cells (D'Angelo et al, 2012). Here, we show that Gp210 expression also varies spatially between different human cancer cell lines, at both the transcript and protein level (Figure 3A). In addition, we identified six other, mostly peripheral, Nups with a similar dynamic behavior, while the expression of the majority of the scaffold Nups remains stable (Figure 3B), presumably due to their essential structural role as seen in gene silencing experiments (Boehmer et al, 2003; Walther et al, 2003; Hawryluk-Gara et al, 2008; Mitchell et al, 2010), or due to deleterious subunit dosage effects (Papp et al, 2003). At least five of the seven significantly regulated nucleoporins have been shown to be non-essential NPC components (Aladin, Nup214, Nup50, Tpr and Gp210) (Smitherman et al, 2000; Cronshaw and Matunis, 2003; Hase and Cordes, 2003; Antonin et al, 2005; Hutten and Kehlenbach, 2006), and six out of seven have a predicted peripheral localization (Figure 3B). Since protein and gene expression strongly indicate copy number variation across a number of cell types as well as healthy and pathological tissues, rearrangements of the NPC stoichiometry could influence several mechanisms that link Nups to the regulation of cell function. Stable integration of the relevant Nups into NPCs at the NE has been well established in terms of both localization and mean residence times (Rabut et al, 2004). Therefore, it is highly unlikely that these subunits are part of other protein assemblies and thus differentially expressed, except for Nup50 and Tpr for which we also detected significant nucleoplasmic pools of protein (see also Supplementary Materials and Methods). Different arrangements of peripheral Nups could affect nucleocytoplasmic transport by varying the docking sites available for transport factors, thus having an impact on processes such as translocation of signaling molecules (Xylourgidis et al, 2006) and mRNA export (Forler et al, 2004). In addition, changes in the expression of nucleoplasmic Nups could affect the spatial organization of chromatin in the proximity of the NE (Krull et al, 2010), regulate the activity of histone modifying enzymes (Kehat et al, 2011), and have an impact on transcription regulation via direct interaction with chromatin (Capelson et al, 2010; Kalverda et al, 2010). Fine-tuning of the function of the NPC, the machinery that indirectly influences gene expression by controlling the composition of the nuclear compartment and the export of regulatory and messenger RNAs, might thus be a more general mechanism for reprogramming the cell machinery according to the needs of individual cell types or even cell states, for example, during development or disease.
Remodeling of nuclear protein complexes
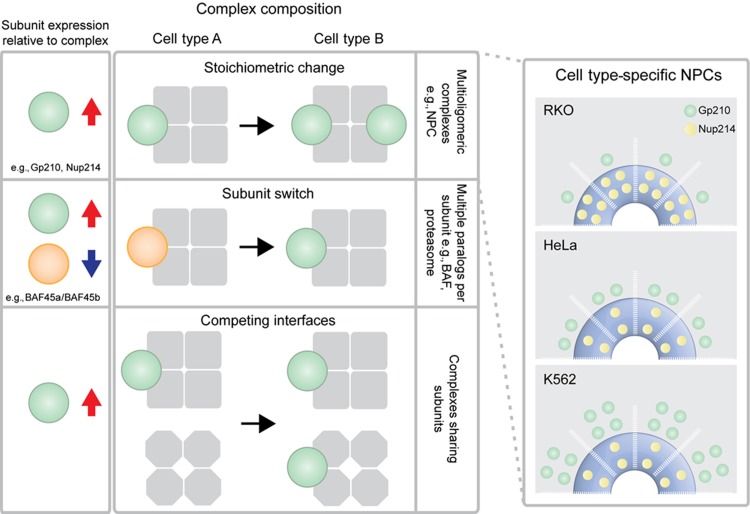
Shotgun proteomics confirmed the cell type-specific variation of NPCs and revealed that more than one third of the well-characterized molecular machines studied show a dynamic expression of at least one subunit across the five cell lines in which we studied nuclear pore composition. This finding indicates that these complexes might similarly undergo compositional rearrangements as a function of the cell type (Figure 5), although this remains to be ultimately proven by their biochemical isolation from the different cell types in the future. The characteristic changes in expression of components of the same protein complex might occur through one of at least three different mechanistic scenarios (Figure 6): (i) a change in the stoichiometry of the protein complex rendering the function, as in case of the NPC; (ii) a switch in subunit composition that is driven by the downregulation of one or more components coupled to a balancing expression of homologous proteins, as observed for the chromatin remodeling complex BAF (Lessard et al, 2007; Ho et al, 2009) and the proteasome (Noda et al, 2000; Murata et al, 2007); or (iii) an adjustment in expression as a result of the association of the subunit with another complex that varies in abundance (in case of overlapping complexes). Since we minimized subunit overlap, the majority of dynamic protein complexes studied would appear to follow one of first two scenarios. Our data are generated from the combined bulk of nuclei/NEs and they comprise therefore an average over multiple single cell species. It is thus possible that for a significant fraction of protein complexes, multiple distinct compositional states co-exist in a single cell with certain stoichiometries being more abundant than others. However, the observed differences are highly robust and reproducible and thus also cell-type specific. The compositional variations that we describe co-occur with remarkable changes in total complex abundance between different cells (as shown for the NPC in Figure 5). Both these mechanisms might therefore contribute to adapt the activity of protein complexes toward cell type-specific needs.
Figure 6.
Three different scenarios for the fine-tuning of protein complexes can potentially underlie the observed cell type-specific variations. For each scenario, the left panel shows the expected change in subunit expression after normalization for complex abundance. Red arrow indicates upregulation and blue arrow downregulation. In case of stoichiometric change, the NPC is presented as an example. Cell type-specific NPC stoichiometries imply a variable copy number of dynamic Nups, e.g., Gp210 or Nup214 and are shown in the right panel according to experimentally determined stoichiometries. The number of copies is shown for half a spoke, assuming an asymmetric distribution of Nup214 across the nuclear envelope plane. Alternatively, subunit switches as previously described for BAF (Lessard et al, 2007; Ho et al, 2009) and the proteasome (Noda et al, 2000; Murata et al, 2007) might occur. This scenario generally requires subunits to exist in multiple paralogs in order to be switched. Finally, subunits might compete for interfaces across multiple, overlapping protein complexes and re-distribute according to the variable abundance of the given complexes/subunits.
Since human tissue gene expression data also point to considerable spatial variation and many other functional states of cells are conceivable in which protein complex composition might be different (e.g., development or disease state), we believe that only a comparative study of homogeneous cell populations on a temporal and spatial scale will reveal the true extent of compositional rearrangements of molecular machines in multicellular organisms. The repertoire of context-dependent complex variants should be the starting point to uncover both the mechanisms leading to differential regulation and the functional consequences of varying quarternary structures.
Materials and methods
Isolation of nuclei and NEs from human cell lines
Nuclei were isolated by hypotonic cell lysis using a dounce homogeneizer followed by centrifugation through a sucrose cushion. NEs were obtained by a combined DNase and RNase treatment of nuclei followed by an additional sucrose cushion step. Detailed procedures for the different cell lines and additional experiments performed to ensure the integrity of the obtained organelles are described in Supplementary Materials and Methods and Supplementary Figure S1.
Quantification of Nups by targeted proteomics
Nups were quantified in nuclear or NE extracts using targeted proteomics in combination with the spike-in of isotopically labeled peptides used as internal standard. For absolute quantification, the abundance of each Nup was calculated from the median ratio between the intensities of its endogenous proteotypic peptides (PTPs, light) and the corresponding spiked-in AQUA peptides (heavy). A panel of 90 MRM assays for 76 PTPs was used for absolute quantification. For relative quantification of Nup abundances across cell lines, crude synthetic peptides were used as internal standard instead of AQUA peptides. Therefore, a larger panel of 142 MRM assays for 119 PTPs was used. A detailed description of MRM assays development, data acquisition and processing is available in Supplementary Materials and Methods and Supplementary Figures S1E–G. A list of the MRM assays employed is available in Supplementary Table S1. Raw MRM data and method files are available at http://www.peptideatlas.org/PASS/PASS00189 for absolute quantification, and http://www.peptideatlas.org/PASS/PASS00188 for relative quantification across cell lines.
Measurement of Nup107 copies per NPC by super-resolution microscopy
Acquisition of PALM measurement of purified nuclei with native Nup107 replaced with mEos2-Nup107 was acquired following Betzig et al (2006). A clustering algorithm based on density (DBSCAN) (Ester et al, 1996) was employed to select photo-conversion events corresponding to isolated NPCs. The counted number of photo-conversion events detected in each pore was then corrected for fluorophore blinking with the method proposed by Annibale et al (2011) in order to retrieve the number of fluorophores in each pore. More details can be found in Supplementary Materials and Methods and Supplementary Figures S3 and S4.
Measurement of Nup107 copies per NPC by targeted MS, electron and light microscopy
The average copy number per nucleus of Nup107 was measured using targeted proteomics on HeLa nuclei that were FACS sorted (because nuclei are more reliably sorted then NEs; a nuclear protein pool of Nup107 was not detected). After sorting, the concentration of isolated nuclei was assessed by two independent methods using CountBright Absolute Counting Beads (Life Technologies), according to manufacturer instructions, and, in parallel, using a Neubauer chamber. In both cases, the measurements were performed in triplicate. We observed a high correlation between the two methods (data not shown). Next, for each biological replicate between 5 and 7 × 105 counted nuclei were spiked with AQUA peptides at a concentration of 1 pmol per peptide/1 × 106 nuclei, digested as described in Supplementary Materials and Methods and measured in scheduled MRM mode. The Nup107 concentration was derived directly from the light to heavy ratios of two distinct PTPs and transformed into protein copy numbers using Avogadro’s number. In order to estimate the average number of NPC per nucleus, we measured NPC density using cryo electron microscopy and estimated nuclear surface area by membrane staining and fluorescence microscopy (Supplementary Figures S2A and B). Isolated NEs were deposited on the EM grid (Copper R2/1, Quantifoil Micro Tools GmbH, Jena, Germany), blotted and immediately plunge frozen. Tilt series of intact cryo-fixed NEs were collected in the range of ±60 degree with 3 degrees increment on a Polara TEM (300 kV) (FEI, equipped with Gatan Camera 2k x 2k and energy filter). Tomograms were reconstructed from the tilt series using IMOD (Kremer et al, 1996). The number of NPCs per tomogram was counted manually. The surface area was estimated in each tomogram by fitting a surface onto the membrane curvature. For membrane staining, nuclei were incubated for 1 min on ice with 3 μg/μl of FM1-43FX (Life Technologies) in ice-cold PBS immediately before mounting the sample on glass coverslips in Mowiol. Confocal z-stacks were acquired on a Zeiss LSM780 using the 458 nm laser line for excitation. The surface was obtained by fitting the acquired z-stacks using Imaris software (Bitplane, Zürich, Switzerland) with automatic thresholding.
Acquisition of shotgun proteomics data
We acquired shotgun MS data from nuclei extracts of the five cell lines that we selected to investigate NPC composition. For each cell line, three biological replicates were analyzed and protein abundance scores based on PTP intensities were calculated as described in Supplementary Materials and Methods. In total, we quantified 1159 proteins using at least two PTPs consistently identified in at least two out of three replicates for each cell line. We assessed the consistency of our measurements using hierarchical clustering (Spearman correlation with average linkage) and found that biological replicates of cell lines clustered together, indicating high reproducibility (data not shown). Raw shotgun MS data are available at http://www.peptideatlas.org/PASS/PASS00190.
Analysis of human nuclear protein complexes
We selected 57 large protein complexes, composed of at least 5 proteins with predicted nuclear localization. A limitation of our approach lies on the analysis of subunits that are shared between different protein complexes. For these cases, a change in abundance of one out of a subset of protein complexes sharing a subunit could result in the false positive detection of dynamic stoichiometries for the others (since our second normalization step is based solely on the median abundance of the complex analyzed). In order to minimize redundancy, complexes were manually selected from minimal endogenous modules (MEMOs) described in a recent large-scale affinity-purification MS study (Malovannaya et al, 2011), entries from the CORUM database (Ruepp et al, 2010) and literature mining (Supplementary Figures S6A–D; Supplementary Table S2). Subsequently, the shotgun proteomics data were used to extract protein complexes having at least 50% of their components cross-quantified and, in any case, a minimal number of four quantified proteins was required. In total, we selected 34 protein complexes, totaling 274 quantified proteins, largely not redundant. Less than 4% of the proteins were shared between two different protein complexes, with the remaining 96% being uniquely assigned to one protein complex (Supplementary Figure S6; Supplementary Table S2). We then designed a workflow to analyze subunit expression profiles in a complex centered manner as explained in Figure 5 and Supplementary Figure S6E.
Supplementary Material
Acknowledgments
We gratefully acknowledge support from EMBL’s proteomics, flow cytometry, advanced light microscopy core facilities and mechanical workshop, and particular want to thank Toby Gibson, Holger Dinkel and Jeroen Krijgsveld. We thank Christine Köhler, Amanda DiGuilio and Lukas Reiter for technical support; Joseph Glavy, Ulrike Kutay, Alexander Schmidt, Iain Mattaj, Vera van Noort and Katja Beck for critical advice and reagents. AO was supported by postdoctoral fellowships from the Alexander von Humboldt Foundation and Marie Curie Actions. HKB was supported by postdoctoral fellowships from the Swiss National Science Foundation, the European Molecular Biology Organization and Marie Curie Actions. EAL acknowledges funding by the Emmy Noether program of the German Research Foundation. MB acknowledges funding by the European Research Council (Grant No. 309271/NPCAtlas).
Author contributions: AO designed and performed experiments, analyzed data and wrote the manuscript; NB and AA designed and performed experiments, and analyzed data; MI designed data analysis procedures, and analyzed data; CE performed experiments; HKB and LS performed experiments, and analyzed data; VS analyzed data; OR designed experiments; PB, EAL and MB coordinated the project, designed experiments, and wrote the manuscript.
Footnotes
OR and CE are employees of Biognosys AG.
References
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, Rout MP, Sali A (2007a) Determining the architectures of macromolecular assemblies. Nature 450: 683–694 [DOI] [PubMed] [Google Scholar]
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, Sali A, Rout MP (2007b) The molecular architecture of the nuclear pore complex. Nature 450: 695–701 [DOI] [PubMed] [Google Scholar]
- Amlacher S, Sarges P, Flemming D, van Noort V, Kunze R, Devos DP, Arumugam M, Bork P, Hurt E (2011) Insight into structure and assembly of the nuclear pore complex by utilizing the genome of a eukaryotic thermophile. Cell 146: 277–289 [DOI] [PubMed] [Google Scholar]
- Annibale P, Vanni S, Scarselli M, Rothlisberger U, Radenovic A (2011) Quantitative photo activated localization microscopy: unraveling the effects of photoblinking. PLoS ONE 6: e22678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonin W, Franz C, Haselmann U, Antony C, Mattaj IW (2005) The integral membrane nucleoporin pom121 functionally links nuclear pore complex assembly and nuclear envelope formation. Mol Cell 17: 83–92 [DOI] [PubMed] [Google Scholar]
- Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF (2006) Imaging intracellular fluorescent proteins at nanometer resolution. Science 313: 1642–1645 [DOI] [PubMed] [Google Scholar]
- Boehmer T, Enninga J, Dales S, Blobel G, Zhong HL (2003) Depletion of a single nucleoporin, Nup107, prevents the assembly of a subset of nucleoporins into the nuclear pore complex. Proc Natl Acad Sci USA 100: 981–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP (2003) A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19: 185–193 [DOI] [PubMed] [Google Scholar]
- Brohawn SG, Partridge JR, Whittle JR, Schwartz TU (2009) The nuclear pore complex has entered the atomic age. Structure 17: 1156–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brohawn SG, Schwartz TU (2009) Molecular architecture of the Nup84-Nup145C-Sec13 edge element in the nuclear pore complex lattice. Nat Struct Mol Biol 16: 1173–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capelson M, Liang Y, Schulte R, Mair W, Wagner U, Hetzer MW (2010) Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell 140: 372–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronshaw JM, Krutchinsky AN, Zhang W, Chait BT, Matunis MJ (2002) Proteomic analysis of the mammalian nuclear pore complex. J Cell Biol 158: 915–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronshaw JM, Matunis MJ (2003) The nuclear pore complex protein ALADIN is mislocalized in triple A syndrome. Proc Natl Acad Sci USA 100: 5823–5827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Angelo MA, Gomez-Cavazos JS, Mei A, Lackner DH, Hetzer MW (2012) A change in nuclear pore complex composition regulates cell differentiation. Dev Cell 22: 446–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lichtenberg U, Jensen LJ, Brunak S, Bork P (2005) Dynamic complex formation during the yeast cell cycle. Science 307: 724–727 [DOI] [PubMed] [Google Scholar]
- Debler EW, Ma Y, Seo HS, Hsia KC, Noriega TR, Blobel G, Hoelz A (2008) A fence-like coat for the nuclear pore membrane. Mol Cell 32: 815–826 [DOI] [PubMed] [Google Scholar]
- Ester M, Kriegel H-P, Jörg S, Xu X (1996) A density-based algorithm for discovering clusters in large spatial databases with noise. Proceedings of the Second International Conference on Knowledge Discovery and Data Mining (KDD-96) 226–231 [Google Scholar]
- Fernandez-Martinez J, Phillips J, Sekedat MD, Diaz-Avalos R, Velazquez-Muriel J, Franke JD, Williams R, Stokes DL, Chait BT, Sali A, Rout MP (2012) Structure-function mapping of a heptameric module in the nuclear pore complex. J Cell Biol 196: 419–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forler D, Rabut G, Ciccarelli FD, Herold A, Kocher T, Niggeweg R, Bork P, Ellenberg J, Izaurralde E (2004) RanBP2/Nup358 provides a major binding site for NXF1-p15 dimers at the nuclear pore complex and functions in nuclear mRNA export. Mol Cell Biol 24: 1155–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz C, Walczak R, Yavuz S, Santarella R, Gentzel M, Askjaer P, Galy V, Hetzer M, Mattaj IW, Antonin W (2007) MEL-28/ELYS is required for the recruitment of nucleoporins to chromatin and postmitotic nuclear pore complex assembly. EMBO Rep 8: 165–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dumpelfeld B, Edelmann A, Heurtier MA, Hoffman V, Hoefert C, Klein K, Hudak M, Michon AM, Schelder M, Schirle M, Remor M et al. (2006) Proteome survey reveals modularity of the yeast cell machinery. Nature 440: 631–636 [DOI] [PubMed] [Google Scholar]
- Gerber SA, Rush J, Stemman O, Kirschner MW, Gygi SP (2003) Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc Natl Acad Sci USA 100: 6940–6945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hase ME, Cordes VC (2003) Direct interaction with nup153 mediates binding of Tpr to the periphery of the nuclear pore complex. Mol Biol Cell 14: 1923–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havugimana PC, Hart GT, Nepusz T, Yang H, Turinsky AL, Li Z, Wang PI, Boutz DR, Fong V, Phanse S, Babu M, Craig SA, Hu P, Wan C, Vlasblom J, Dar VU, Bezginov A, Clark GW, Wu GC, Wodak SJ et al. (2012) A census of human soluble protein complexes. Cell 150: 1068–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawryluk-Gara LA, Platani M, Santarella R, Wozniak RW, Mattaj IW (2008) Nup53 is required for nuclear envelope and nuclear pore complex assembly. Mol Biol Cell 19: 1753–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Ronan JL, Wu J, Staahl BT, Chen L, Kuo A, Lessard J, Nesvizhskii AI, Ranish J, Crabtree GR (2009) An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc Natl Acad Sci USA 106: 5181–5186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoelz A, Debler EW, Blobel G (2011) The structure of the nuclear pore complex. Annu Rev Biochem 80: 613–643 [DOI] [PubMed] [Google Scholar]
- Hutten S, Kehlenbach RH (2006) Nup214 is required for CRM1-dependent nuclear protein export in vivo. Mol Cell Biol 26: 6772–6785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalverda B, Pickersgill H, Shloma VV, Fornerod M (2010) Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell 140: 360–371 [DOI] [PubMed] [Google Scholar]
- Kampmann M, Blobel G (2009) Three-dimensional structure and flexibility of a membrane-coating module of the nuclear pore complex. Nat Struct Mol Biol 16: 782–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapushesky M, Adamusiak T, Burdett T, Culhane A, Farne A, Filippov A, Holloway E, Klebanov A, Kryvych N, Kurbatova N, Kurnosov P, Malone J, Melnichuk O, Petryszak R, Pultsin N, Rustici G, Tikhonov A, Travillian RS, Williams E, Zorin A et al. (2012) Gene Expression Atlas update-a value-added database of microarray and sequencing-based functional genomics experiments. Nucleic Acids Res 40: D1077–D1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katahira J, Yoneda Y (2009) Roles of the TREX complex in nuclear export of mRNA. RNA Biol 6: 149–152 [DOI] [PubMed] [Google Scholar]
- Kehat I, Accornero F, Aronow BJ, Molkentin JD (2011) Modulation of chromatin position and gene expression by HDAC4 interaction with nucleoporins. J Cell Biol 193: 21–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosova B, Pante N, Rollenhagen C, Hurt E (1999) Nup192p is a conserved nucleoporin with a preferential location at the inner site of the nuclear membrane. J Biol Chem 274: 22646–22651 [DOI] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR (1996) Computer visualization of three-dimensional image data using IMOD. J Struct Biol 116: 71–76 [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, Punna T, Peregrin-Alvarez JM, Shales M, Zhang X, Davey M, Robinson MD, Paccanaro A, Bray JE, Sheung A, Beattie B et al. (2006) Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440: 637–643 [DOI] [PubMed] [Google Scholar]
- Krull S, Dorries J, Boysen B, Reidenbach S, Magnius L, Norder H, Thyberg J, Cordes VC (2010) Protein Tpr is required for establishing nuclear pore-associated zones of heterochromatin exclusion. EMBO J 29: 1659–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard J, Wu JI, Ranish JA, Wan M, Winslow MM, Staahl BT, Wu H, Aebersold R, Graef IA, Crabtree GR (2007) An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron 55: 201–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WL, Coleman RA, Grob P, King DS, Florens L, Washburn MP, Geles KG, Yang JL, Ramey V, Nogales E, Tjian R (2008) Structural changes in TAF4b-TFIID correlate with promoter selectivity. Mol Cell 29: 81–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loiodice I, Alves A, Rabut G, van Overbeek M, Ellenberg J, Sibarita JB, Doye V (2004) The entire Nup107-160 complex, including three new members, is targeted as one entity to kinetochores in Mitosis. Mol Biol Cell 15: 3333–3344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukk M, Kapushesky M, Nikkila J, Parkinson H, Goncalves A, Huber W, Ukkonen E, Brazma A (2010) A global map of human gene expression. Nat Biotechnol 28: 322–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maimon T, Elad N, Dahan I, Medalia O (2012) The human nuclear pore complex as revealed by cryo-electron tomography. Structure 20: 998–1006 [DOI] [PubMed] [Google Scholar]
- Malovannaya A, Lanz RB, Jung SY, Bulynko Y, Le NT, Chan DW, Ding C, Shi Y, Yucer N, Krenciute G, Kim BJ, Li CS, Chen R, Li W, Wang Y, O'Malley BW, Qin J (2011) Analysis of the human endogenous coregulator complexome. Cell 145: 787–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maul GG, Maul HM, Scogna JE, Lieberman MW, Stein GS, Hsu BY, Borun TW (1972) Time sequence of nuclear pore formation in phytohemagglutinin-stimulated lymphocytes and in HeLa cells during the cell cycle. J Cell Biol 55: 433–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney SA, Murphy CS, Hazelwood KL, Davidson MW, Looger LL (2009) A bright and photostable photoconvertible fluorescent protein. Nat Methods 6: 131–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell JM, Mansfeld J, Capitanio J, Kutay U, Wozniak RW (2010) Pom121 links two essential subcomplexes of the nuclear pore complex core to the membrane. J Cell Biol 191: 505–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata S, Sasaki K, Kishimoto T, Niwa S, Hayashi H, Takahama Y, Tanaka K (2007) Regulation of CD8+ T cell development by thymus-specific proteasomes. Science 316: 1349–1353 [DOI] [PubMed] [Google Scholar]
- Nagy V, Hsia KC, Debler EW, Kampmann M, Davenport AM, Blobel G, Hoelz A (2009) Structure of a trimeric nucleoporin complex reveals alternate oligomerization states. Proc Natl Acad Sci USA 106: 17693–17698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda C, Tanahashi N, Shimbara N, Hendil KB, Tanaka K (2000) Tissue distribution of constitutive proteasomes, immunoproteasomes, and PA28 in rats. Biochem Biophys Res Commun 277: 348–354 [DOI] [PubMed] [Google Scholar]
- Papp B, Pal C, Hurst LD (2003) Dosage sensitivity and the evolution of gene families in yeast. Nature 424: 194–197 [DOI] [PubMed] [Google Scholar]
- Picotti P, Bodenmiller B, Mueller LN, Domon B, Aebersold R (2009) Full dynamic range proteome analysis of S. cerevisiae by targeted proteomics. Cell 138: 795–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabut G, Doye V, Ellenberg J (2004) Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat Cell Biol 6: 1114–1121 [DOI] [PubMed] [Google Scholar]
- Rasala BA, Orjalo AV, Shen ZX, Briggs S, Forbes DJ (2006) ELYS is a dual nucleoporin/kinetochore protein required for nuclear pore assembly and proper cell division. Proc Natl Acad Sci USA 103: 17801–17806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichelt R, Holzenburg A, Buhle EL, Jarnik M, Engel A, Aebi U (1990) Correlation between structure and mass-distribution of the nuclear-pore complex and of distinct pore complex components. J Cell Biol 110: 883–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribbeck K, Gorlich D (2001) Kinetic analysis of translocation through nuclear pore complexes. EMBO J 20: 1320–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT (2000) The yeast nuclear pore complex: composition, architecture, and transport mechanism. J Cell Biol 148: 635–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruepp A, Waegele B, Lechner M, Brauner B, Dunger-Kaltenbach I, Fobo G, Frishman G, Montrone C, Mewes HW (2010) CORUM: the comprehensive resource of mammalian protein complexes-2009. Nucleic Acids Res 38: D497–D501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S, Lutzmann M, Santos-Rosa H, Leonard K, Mueller S, Aebi U, Hurt E (2000) Structure and assembly of the Nup84p complex. J Cell Biol 149: 41–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smitherman M, Lee K, Swanger J, Kapur R, Clurman BE (2000) Characterization and targeted disruption of murine Nup50, a p27(Kip1)-interacting component of the nuclear pore complex. Mol Cell Biol 20: 5631–5642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theerthagiri G, Eisenhardt N, Schwarz H, Antonin W (2010) The nucleoporin Nup188 controls passage of membrane proteins across the nuclear pore complex. J Cell Biol 189: 1129–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulbrich MH, Isacoff EY (2007) Subunit counting in membrane-bound proteins. Nat Methods 4: 319–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther TC, Alves A, Pickersgill H, Loiodice I, Hetzer M, Galy V, Hulsmann BB, Kocher T, Wilm M, Allen T, Mattaj IW, Doye V (2003) The conserved Nup107-160 complex is critical for nuclear pore complex assembly. Cell 113: 195–206 [DOI] [PubMed] [Google Scholar]
- Wang HD, Kakaradov B, Collins SR, Karotki L, Fiedler D, Shales M, Shokat KM, Walther TC, Krogan NJ, Koller D (2009) A complex-based reconstruction of the Saccharomyces cerevisiae interactome. Mol Cell Proteomics 8: 1361–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JI, Lessard J, Crabtree GR (2009) Understanding the words of chromatin regulation. Cell 136: 200–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xylourgidis N, Roth P, Sabri N, Tsarouhas V, Samakovlis C (2006) The nucleoporin Nup214 sequesters CRM1 at the nuclear rim and modulates NF kappa B activation in Drosophila. J Cell Sci 119: 4409–4419 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.