Abstract
Factor XIII (FXIII) consists of catalytic A subunits (FXIII-A) and carrier B subunits. Congenital FXIII deficiency is a severe bleeding disorder. We previously identified an R260C missense mutation and an exon-IV deletion in Japanese patients’ F13A genes. To characterize the molecular basis of this disease, we expressed a wild-type and the mutant FXIII-A in yeast cells for detailed investigation, by taking advantage of yeast’s ability for mass protein production. The mutant proteins were expressed less efficiently than the wild-type and considerably aggregated; even their non-aggregated forms became aggregated with time. Ultra-centrifugation and gel-filtration analyses revealed that the mutants were of extremely high-molecular weight, and that the wild-type formed a dimer. Notably, a part of the R260C mutant was found in monomer form. This was consistent with the prediction by molecular modelling that the mutant molecule would lose the electrostatic interaction between the two monomers, leading to their inability to form a dimer. The mutants lost enzymatic activity. The mutants were only partially converted by thrombin to the cleaved form. The wild-type was fully converted and activated. These mutants might have significantly altered conformations, resulting in their aggregation in vitro, and may ultimately lead to FXIII deficiency in vivo as well.
Keywords: aggregation, dimer assembly, factor XIII deficiency, molecular basis, protein instability
Blood coagulation factor XIII (FXIII) is a pro-enzyme of plasma transglutaminase (TGase) consisting of two catalytic A (FXIII-A) and two carrier B subunits (FXIII-B) (1–3). In the final stage of the blood coagulation cascade, FXIII is activated by thrombin and promotes clot stability by forming covalent bonds between fibrin molecules; it also cross-links fibrin with other proteins including fibronectin and α2-plasmin inhibitor. Accordingly, FXIII deficiency results in defective crosslinking reactions, leading to a life-long bleeding tendency and abnormal wound healing in affected patients, and recurrent miscarriage in affected females (1, 4).
Because FXIII is composed of FXIII-A and FXIII-B, and FXIII-B protects FXIII-A in vitro (5, 6) and in vivo (7, 8), a genetic defect of either subunit leads to FXIII deficiency. Accordingly, FXIII deficiency is classified into two categories at the DNA level: FXIII-A deficiency and FXIII-B deficiency (9). This classification was approved by the scientific and standardization committee of the international society of thrombosis and haemostasis in 1999 (http://www.med.unc.edu/isth/99FXIII.html).
The gene for FXIII-A spans ∼180 kb and consists of 15 exons interrupted by 14 introns (10). The FXIII-A cDNA encodes a mature protein of 731 amino acids, including an activation peptide of 37 residues at the amino-terminus and a catalytic Cys314 residue (11). To date, ∼100 mutations in the gene for FXIII-A have been identified in patients with FXIII-A deficiency (http://www.f13-database.de) (12, 13). These mutations are highly heterogeneous and include a variety of nonsense and missense mutations, deletions and insertions with or without frame-shift/premature termination, splicing abnormalities, etc. (9). Many missense mutations of F13A have been confirmed to be causative, as they resulted in the generation of reduced amounts of proteins in mammalian cells (14–18). Most of these missense mutations have been predicted to change the conformation of their translated products, resulting in protein instability (14–17, 19); however, direct experimental evidence for this has not been demonstrated to date (no hit on Pubmed until the submission date of this manuscript), to the best of the authors’ knowledge.
We previously identified two mutations in the F13A genes of Japanese patients with severe FXIII-A deficiency: an R260C missense mutation (20) and an exon IV-deletion (Del-IV) caused by a splicing defect (21). The former mutation was also found in four other ethnic groups, i.e. Swiss, Iranian, Polish and Italian (18, 22–24), indicating this being a naturally recurring mutation. In this study, to obtain direct evidence for the molecular abnormalities of these two mutations, we expressed large quantities of the mutant proteins in yeast cells to perform detailed biochemical experiments. We demonstrate here, for the first time, that instability of the mutant recombinant FXIII-As (rFXIII-As), likely due to impaired dimer assembly, lead to their extensive aggregation as well as degeneration. We therefore propose a new molecular basis for FXIII deficiency.
Materials and Methods
Materials
A yeast strain Saccharomyces cerevisiae ZM118 and expression vectors RPOT and pRS215 were generously provided by Drs R. Seal and D. Foster (ZymoGenetics, Seattle, WA). Purified human plasma FXIII was the kind gift of Dr H. Kaetsu (The Chemo-Sero-Therapeutic Research Institute, Kumamoto, Japan). Anti-FXIII-A polyclonal antibody was obtained from Calbiochem (La Jolla, CA). An affinity-purified anti-FXIII-A antibody and a FXIII ELISA kit were kindly supplied by the late Dr P. Bishop (ZymoGenetics) and Dr K. Hirahara (Aventis, formerly Japan Hoechst, Tokyo, Japan), respectively. Bovine thrombin and bovine serum albumin (BSA) were purchased from Sigma (St Louis, MO). Fibrinogen was purified from human plasma.
Construction of expression vectors
A yeast expression vector RPOT carrying rF13A cDNA was constructed as previously described (25). In vitro mutagenesis was performed by recombinant polymerase chain reaction (PCR) (26) using oligonucleotide primers: for R260C, 5′-ACAGTGGAGCTCCAGGGCGTG-3′ (5′-side, sense), 5′-GACCCC-ACACAGCTGACTTTGATGGGATTC-3′ (middle, antisense), 5′-CAAAGTCAGCTGTGTGGGGTCTGCAATGGT-3′ (middle, sense) and 5′-CATCGCTATTTTCCTGGGGGG-3′ (3′-side, antisense); for IV-del, 5′-TACGTCATTGATGATGCTGTGTATCTGGAC-3′ (sense from exon III) and 5′-ACAGCATCATCAATGACGTATTCCACCCTG-3′ (antisense from exon V). After the mutations were confirmed by dideoxy sequencing, the cDNAs for the wild-type and mutants were released by XhoI from the pBluescriptII vectors and ligated into the yeast expression vector RPOT.
Transformation and cell culture of yeast cells
Each of the expression vectors containing the wild-type rF13A cDNA, or either of R260C or Del-IV was transformed into the yeast strain ZM118 by the lithium acetate method (27), and transformants were selected. Yeast cells were grown for 32 h at 30°C for the proliferation of cells. After washing with sterile water, cells were grown for another 16 h in a non-glucose medium for the induction of rFXIII-A synthesis.
Southern blot analysis
Yeast cells transformed with an rF13A cDNA were treated with a lysis buffer (0.5% sodium dodecyl sulphate [SDS], 10 mM Tris [pH 8.0], 1 mM ethylenediaminetetraacetic acid (EDTA), 0.1 mg/ml proteinase K, 1 mg/ml zymolyase 100 T) and genomic DNA was extracted by a standard phenol/chloroform method. Ten micrograms of genomic DNA was digested with an XhoI restriction endonuclease and were subjected to electrophoresis on a 0.6% agarose gel. The DNA fragments were transferred to a Hybond-N+ nylon membrane (GE healthcare, Amersham, UK). The membrane was hybridized with the part of the FXIII-A cDNA encompassing exons V to IX labelled with digoxigenin (DIG). The rFXIII-A DNA was detected using an anti-DIG antibody conjugated with alkaline phosphatase and a CDP-Star substrate (Roche, Indianapolis, IN).
Reverse transcription PCR assay
Total RNA samples were prepared by the glass beads/phenol/chloroform extraction procedure, and stored at −80°C until use. Reverse transcription of the total RNA (5 µg) was carried out using random primers and Superscript™II RNaseH− (GIBCO-BRL, Gaithersburg, MD). 1/25 (4%) of the synthesized first-strand cDNA or each of its serially diluted samples (1–1/8) was used for PCR in a reaction mixture of 50 µl by employing two pairs of primers separately: for F13-A (exons II-IX), 5′-ACAGTGGAGCTCCAGGGCGTG-3′ (sense) and 5′-CATCGCTATTTTCCTGGGGGG-3′ (antisense); for actin of S. cerevisiae as an internal control, 5′-CTGTTACTAAGTCTCATGTAC-3′ (sense) and 5′-GTAGAAGGTATGATGCCAGATC-3′ (antisense). After 18 cycles (for F13-A) or 26 cycles (for actin), 18 µl of each reaction mixture was applied to a 1.5% agarose gel. The intensity of each band stained with ethidium bromide was quantified using a FLA-2000 Fluoroimage Analyzer (Fuji Photo Film, Tokyo, Japan).
Preparation of yeast cell lysates and centrifugation analysis of rFXIII-As
Yeast cells cultured in 2 ml medium were suspended in a 500 µl lysis buffer (50 mM NaCl, 10 mM Tris and 2 mM EDTA, pH 7.5) containing protease inhibitors (1 mM phenylmethylsulfonyl fluoride [PMSF], 2 µg/µl Pepstatin A, 2 µg/µl Leuptin) with 0.1 g of 0.45 mm glass beads, and disrupted by ultrasound.
The cell lysate was centrifuged at 10,000g for 10 min (low-speed centrifugation) and the supernatant was collected as a ‘non-aggregated’ fraction/form. The pellet (precipitate) was suspended in the lysis buffer as an ‘aggregated’ fraction/form: to avoid confusion in this article, we use these arbitrary terms, when necessary. The protein concentration of a cell lysate, supernatant/non-aggregated fraction or precipitate/aggregated fraction, was determined by BCA Protein Assay Reagent (Pierce, Rockford, IL).
Western blotting for rFXIII-A
To adjust to similar rFXIII-A levels, appropriate amounts of proteins in the yeast cell lysates were electrophoresed on an 8% polyacrylamide gel containing 0.1% SDS, and then transferred to a nitrocellulose membrane (Advantec, Tokyo, Japan). The membrane was incubated with a rabbit anti-human FXIII-A antibody (Calbiochem), and rFXIII-A was visualized using an enhanced chemiluminescence plus western blotting detection kit (GE healthcare, UK) and a Light-Capture apparatus (ATTO, Tokyo, Japan). The signal intensity of each band was quantified using a software CS analyzer 2.0 (ATTO).
Chase experiment in yeast cells
To examine the stability of the intracellular rFXIII-A proteins, the yeast cells were first grown for 32 h in a 0.4% glucose medium and then another 16 h without glucose. The cells were then grown in a 2% glucose medium containing 0.4 mM cycloheximide (CHX) to inhibit the de novo synthesis of rFXIII-A. The cell lysate was analysed by western blotting and ELISA.
Sucrose density-gradient ultra-centrifugation
Five hundred microlitres each of 60, 50, 40, 30, 20 and 10% sucrose solution in 20 mM Tris (pH 7.5), 150 mM NaCl was layered in a tube. 0.1 ml cell lysate (0.45 mg total protein) prepared from yeast cells was overlaid onto the sucrose layers, and then ultra-centrifuged at 100,000g for 1 h at 4°C. 0.5 ml/fraction was collected from top to bottom of tube and the last fraction was added 20 mM Tris (pH 7.5), 150 mM NaCl to 0.5 ml. The rFXIII-As in each fraction were detected by western blotting.
Gel-filtration analyses
A half milligram protein of the cell lysate was ultra-centrifuged at 150,000g for 1 h at 4°C, and the supernatant was applied to a TSK-G3000SW column (7.5 mm I.D. × 60 cm; Tosoh, Tokyo, Japan) equilibrated with a buffer containing 50 mM Tris–HCl, pH 7.5, 0.2 M NaCl and 1 mM EDTA. Elution was performed at a flow rate of 0.5 ml/min, and fractions of 0.5 ml/tube were collected. Ten microlitres of each fraction was subjected to SDS–polyacrylamide gel electrophoresis (SDS–PAGE) and western blotting. For calibration of the column, γ-globulin and BSA were used as molecular weight markers.
Repeated centrifugation analysis of rFXIII-As
On-going aggregation of mutant proteins was investigated before and after a 24-h incubation by separating ‘non-aggregated’ and ‘aggregated’ fractions. First, yeast cell lysate was centrifuged at 10,000g for 10 min. The supernatant was divided into two 30-µl aliquots (1 mg/ml), and further centrifuged at 10,000g for 10 min immediately or 24 h after incubation at 37°C. rFXIII-As in the supernatant and precipitate were detected by western blotting and compared with the original non-incubated samples (set at 100%).
Measurement of FXIII activity by amine-incorporation
The enzymatic activity of rFXIII-A produced by yeast cells was determined by an amine-incorporation assay as described by Lorand et al. (28). Briefly, 80 µg protein in 10 µl of cell lysate were added to 30 µl of a solution containing 7.5 units/ml thrombin in 50 M Tris buffer (pH 7.5). The reaction mixture was incubated at 37°C for 20 min to activate rFXIII-A, then added to a solution containing 6.7 mM CaCl2, 3.3 mM monodansylcadaverin and 0.3% dimethylcasein, 6.7 mM dithiothreitol (a final mixture of 100 μl) and incubated at 37°C for 30 min. The reaction was terminated by the addition of 100 μl of 10% trichloroacetic acid. FXIII activity was measured by fluorescence intensity of the emission at 520 nm with the excitation at 360 nm.
Fibrin cross-linking activity of rFXIII-As
A yeast cell lysate containing either the wild-type (1 µg total protein), mutant rFXIII-As (3 µg total protein) or mock cells (3 µg total protein) was incubated with human fibrinogen (1 mg/ml) in the presence of 5 mM CaCl2, 0.1 M NaCl, 0.1 M Tris–HCl, pH 7.4 and 6 units/ml thrombin for 5, 15 and 60 min at room temperature. The reactions were stopped by the addition of 50 mM EDTA. The fibrin clot was collected by centrifugation at 10,000g for 10 min, washed with 50 mM Tris-buffered saline (pH 7.5), and dissolved in 0.125 M Tris (pH 6.8), 2% SDS, 15% glycerol, 5% β-mercaptoethanol and 0.01% Brome Phenol Blue (a reducing SDS sample buffer) containing 8 M urea, followed by boiling for 5 min. The samples were electrophoresed on an 8% polyacrylamide gel, and stained with Coomassie Brilliant Blue R-250.
Treatment of rFXIII-As by thrombin
A yeast cell lysate (30 µg of protein including rFXIII-A) was incubated with 4 units thrombin in the presence of 5 mM Tris PH 7.4, 10 mM CaCl2 at 37°C for 0, 30, 60 and 120 min. The reaction was stopped by adding the reducing SDS sample buffer. The reaction mixture was electrophoresed on an 8% SDS–PAGE gel and subjected to western blotting.
Molecular modelling
Several crystal structures of human FXIII-A have been reported; all are of dimeric protein in the same overall three-dimensional conformation. The highest resolution structure, that of wild-type FXIII-A bound to calcium ions (accession code 1EVU, 29), was used as a template for structural analysis and modelling. Computational mutagenesis and modelling of the R260C mutant was carried out in the program Coot (30).
Results
Expression and decline of mutant rFXIII-As inside synthesizing yeast cells
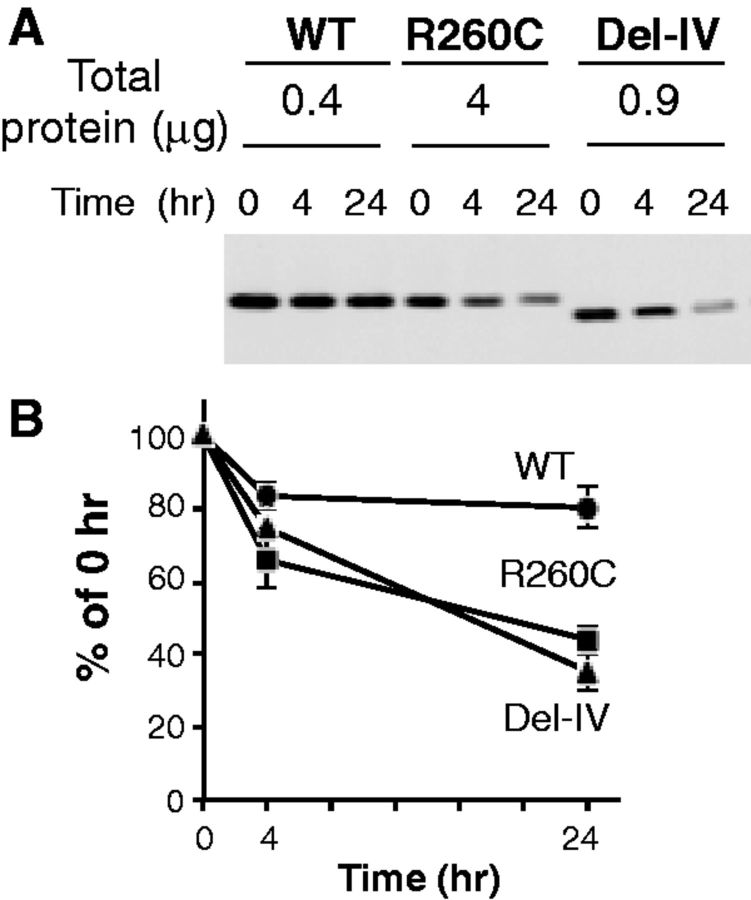
Because two mutant rFXIII-As were produced in significantly lower amounts than the wild-type (Fig. 1), despite their mRNA levels being largely comparable (Supplementary Fig. S1), we examined their intracellular stability by inhibiting the new rFXIII-A synthesis employing a translation inhibitor, CHX. The wild-type was found to be fairly stable inside cells for 24 h, whereas the mutant rFXIII-As decreased much faster than the wild-type (Fig. 2). At 24 h, the amount of the R260C mutant was 44% of 0 h, and the Del-IV mutant decreased to 35%. Thus, the mutant rFXIII-As disappeared much faster than the wild-type, indicating that both mutants were more unstable than the wild-type.
Fig. 1.
Expression of mutant rFXIII-A proteins in yeast cells. The indicated amounts of total protein for yeast cell lysates were subjected to SDS-PAGE and western blotting. The band for rFXIII-A was detected by employing an anti-human FXIII-A polyclonal antibody. W, wild-type; R, R260C mutant; D, del-IV mutant.
Fig. 2.
Decline of mutants inside synthesizing yeast cells. (A) rFXIII-As in yeast cells were measured by western blotting at the indicated time periods after CHX treatment to block new protein synthesis. (B) rFXIII-As of WT (circle), R260C (square), and Del-IV (triangle) were quantified by densitometry, and means and SD of three independent experiments are shown.
Abnormally aggregated mutant rFXIII-A proteins
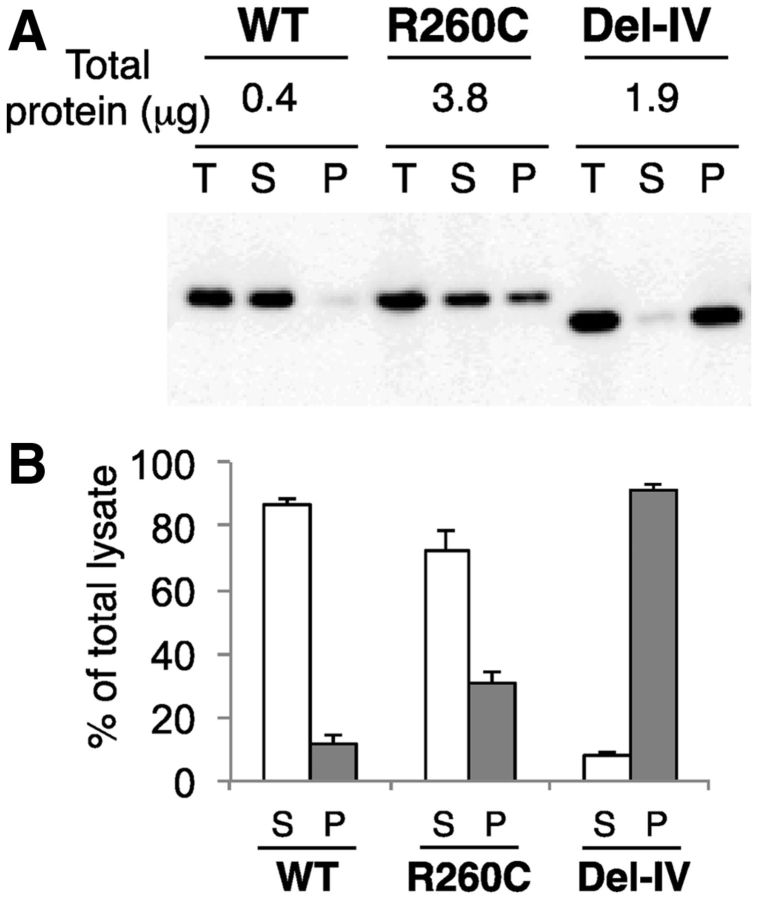
When these mutants were centrifuged at low speed, ∼90% of the wild-type remained in a supernatant fraction, whereas 30 and 90% of R260C and Del-IV precipitated, respectively (Fig. 3), suggesting that at least a part of mutant proteins were aggregated.
Fig. 3.
Aggregation of mutant rFXIII-As inside synthesizing yeast cells detected by low-speed centrifugation. (A) The aggregation of rFXIII-As was analysed by western blotting before and after centrifugation at 10,000 g for 10 min (T, total yeast cell lysate; S, supernatant; P, precipitate). (B) Amounts of non-aggregated (S; open bars) and aggregated forms (P; filled bars) were quantified by densitometry and results for each rFXIII-A are shown as percentage of total yeast cell lysate.
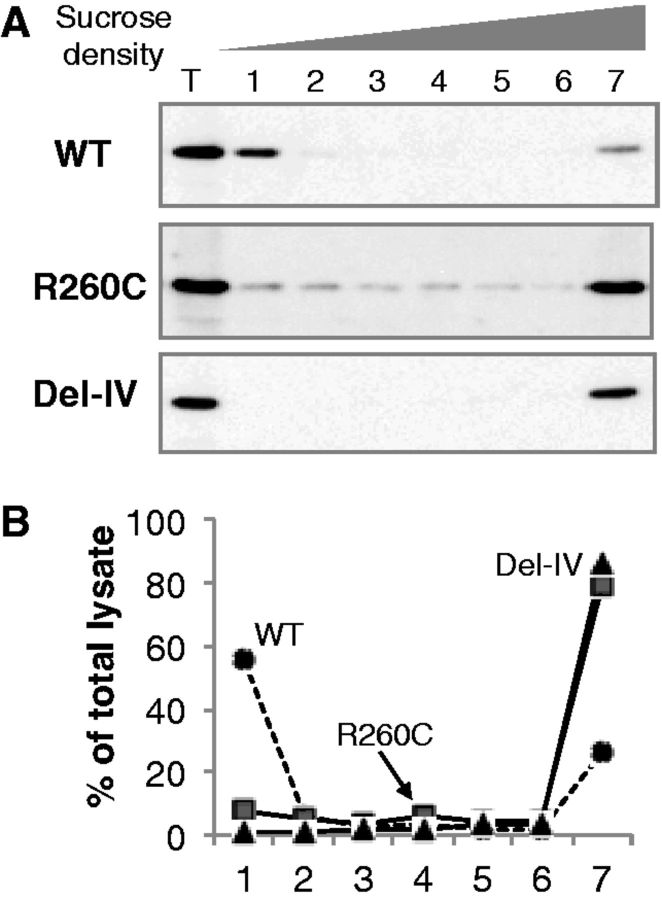
Fractionation of the total yeast cell lysate by a sucrose density-gradient ultra-centrifugation demonstrated that ∼60% of the wild-type was found in the lowest-density fraction (Fig. 4). On the other hands, 80% of R260C was detected in the highest density fraction, and its small amounts were present in intermediate fractions 1–6. Essentially, all Del-IV was present in the highest density fraction. These results indicated that Del-IV was of an extremely high-molecular weight and was in highly aggregated forms, whereas R260C existed broadly as aggregates of various degrees as well as extra-large aggregates.
Fig. 4.
Highly aggregated forms of intracellular mutants detected by density-gradient ultra-centrifugation. (A) Total yeast cell lysate containing rFXIII-As (T) was subjected to sucrose density-gradient ultra-centrifugation, and each fraction was analyzed by western blotting. (B) The intensities of rFXIII-A’s bands were quantified by densitometry, and results for each rFXIII-A are shown as percentage of total cell lysate. WT, circle; R260C, square; Del-IV, triangle.
Impaired subunit assembly of mutant rFXIII-A proteins
Because a part of R260C was found in the lowest density fraction, gel-filtration analysis was performed after ultra-centrifugation to remove aggregated forms of two mutants, to examine the subunit structure of the mutant rFXIII-As, especially with regard to normal dimer assembly. A peak of the wild-type rFXIII-A was observed in an elution volume of 16 ml, the molecular mass of which was estimated to be ∼150 kDa, corresponding to a dimer (Fig. 5). In contrast, a peak of the R260C mutant was detected in the fraction of 17 ml where a small molecular weight standard, BSA, was eluted, whereas no peak was seen for the Del-IV mutant. These results indicated that at least a part of the R260C mutant was in monomer form, and the rest was in the forms of high-molecular weight, and that no homodimer of rFXIII-A formed for both mutants.
Fig. 5.
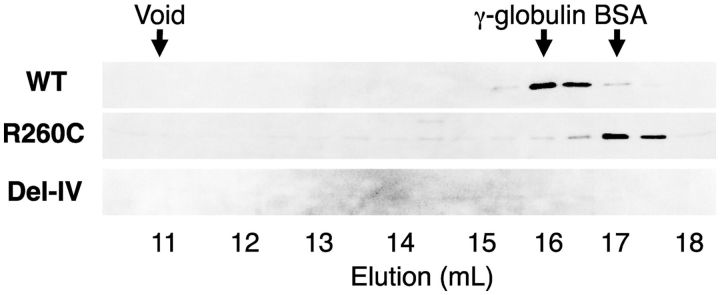
Gel-filtration analysis of ultra-centrifuged rFXIII-As. The supernatant fraction of ultra-centrifugation was subjected to gel-filtration on a TSK-G3000SW column. Fractions of 0.5 ml/tube were collected. BSA (68 kDa) and γ-globulin (160 kDa) were used as molecular weight markers.
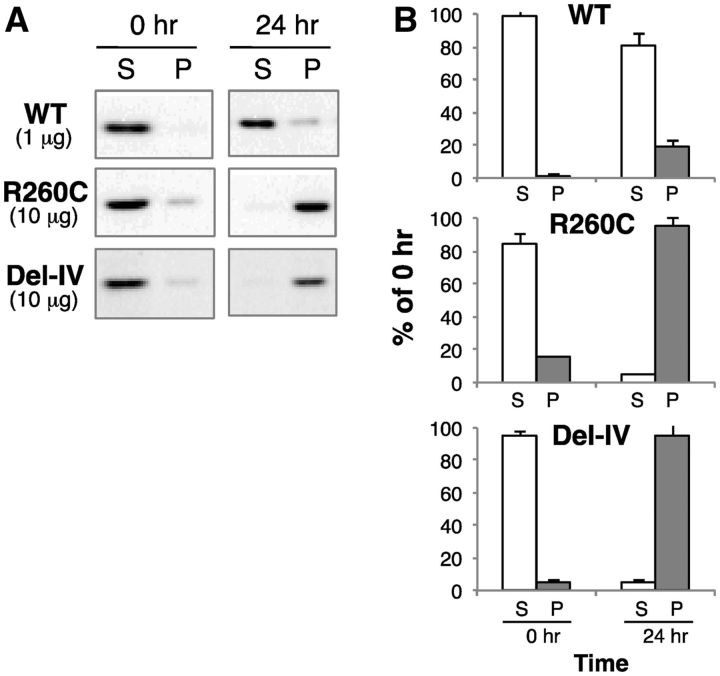
To explore whether non-aggregated forms of mutant rFXIII become aggregated with time, a repeated centrifugation analysis was carried out. This study on the on-going/progressive aggregation of rFXIII-As in vitro showed that the supernatants of both mutant proteins became precipitated after incubation at 37°C for 24 h (Fig. 6). These results strongly suggest that the aggregation of both mutants is very likely due to their having an abnormal structure compared with the wild-type.
Fig. 6.
On-going/progressive aggregation of non-aggregated mutants in vitro. (A) The non-aggregated/supernatant fraction of each rFXIII-A (set at 100%) obtained by low-speed centrifugation of total yeast cell lysate was incubated at 37°C for 24 h. Before and after incubation, the samples were further separated into supernatant (S, open bars) and precipitate (P, filled bars) fractions by low-speed centrifugation, and analysed by western blotting. (B) Protein bands were quantified by densitometry.
Lost enzymatic capability of mutant rFXIII-As
The rapid on-going aggregation of mutant rXIII-As also suggested that these mutant proteins were already degenerated, and thus they were further characterized by their biochemical properties. An amine-incorporation assay, using a low-molecular weight substrate, clearly demonstrated that the wild-type rFXIII-A had a high TGase activity after activation by thrombin (Fig. 7A). However, the same amount of cell lysates of both mutants showed an extremely low activity (1.7% of the wild-type) for the R260C mutant, and no TGase activity for the Del-IV mutant rFXIII-A.
Fig. 7.
Lost enzymatic activity of mutants. (A) TGase activity of rFXIII-As measured by an amine incorporation assay. TGase activity of each yeast cell lysate was determined by employing monodansylcadaverin and dimethylcasein as substrates. The results represent mean values and SD of three parallel measurements. (B) Fibrin cross-linking activity of rFXIII-As. The indicated amounts of yeast cell lysates for rFXIII-As were incubated with 0.5 mg/ml of fibrinogen, 10 mM CaCl2, and 20 units/ml of thrombin for 5, 15 and 60 min at room temperature.
We also tested the capability of rFXIII-As to cross-link fibrin, a physiological macromolecular substrate in plasma (Fig. 7B). In the case of the wild-type rFXIII-A, the γ−γ dimer formed within 5 min and the α-chain disappeared as the α-polymer formed. However, an extremely faint band of the γ−γ dimer was detected for the R260C mutant at 60 min, but none for the Del-IV mutant. In addition, no α-polymer was ever seen for either mutant. These results indicated that the mutants also lost the enzymatic activity.
Altered susceptivity of mutant rFXIII-As to thrombin
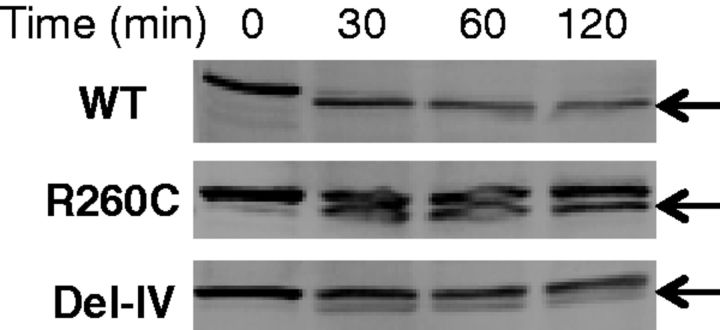
Since neither mutant rFXIII-A showed substantial enzymatic activity, each was examined next for the activation cleavage. When each rFXIII-A was incubated with thrombin, the wild-type was fully converted into the activated form of a smaller molecular weight within 30 min, whereas the mutants were converted only partially even after 120 min (Fig. 8), confirming that both mutants have an abnormal conformation(s), which prevents thrombin from accessing the activation cleavage site of rFXIII-A.
Fig. 8.
Altered susceptivity of mutants to proteolytic cleavage by thrombin. Each yeast cell lysate was incubated with four units thrombin for 30, 60 and 120 min at 37°C. The samples were subjected to western blotting. Arrows indicate cleaved forms of rFXIII-As.
Tertiary structure around R260C predicted by molecular modelling
Analysis of the rFXIII-A crystal structure provides a basis for understanding the possible effects of the R260C mutation on protein structure and consequently on enzyme function. Each FXIII-A subunit folds into four sequential domains: the N-terminal sandwich, a central core which contains the catalytic triad residues and calcium binding site, and two barrels at the C-terminus (Supplementary Fig. S2A) (29, 31). The FXIII-A dimer interface is formed primarily by the central core domains, with the sandwich domains contributing as well. In the wild-type FXIII-A dimer, the central core residue R260 is surrounded by amino acids from both monomers, and thus inaccessible to bulk solvent and unavailable for binding to other proteins (Supplementary Fig. S2A). The large positively charged R260 side chain participates in both intramolecular and intermolecular hydrogen-bonding interactions (Supplementary Fig. S2B). Within the same subunit, the R260 side chain interacts with the side chain of D427 and the main chain carbonyl oxygen of G410. Between subunits, the R260 side chain interacts with the side chain of D404 across the dimer interface. Substitution of the large positively charged arginine side chain with a much shorter neutral cysteine in the R260C mutant results in loss of both intramolecular and intermolecular hydrogen bonds (Supplementary Fig. S2C).
Discussion
A number of mutations in the gene for FXIII-A have been identified in patients with its deficiency (12, 13, 18, 32). These natural mutations are highly heterogeneous. Nevertheless, their effects on FXIII-A biosynthesis have been confirmed in only limited cases, by expressing mutant proteins in yeast (19, 33, 34), COS (14, 17, 18) and BHK cells (15), or Meg-01 cells (16). These reports concluded that the mutants have very low enzymatic activity, are remarkably unstable, and rapidly disappear in the synthesizing cells. The last mentioned conclusion had been obtained through pulse-chase experiments using 35S because of its high sensitivity as a tracer (14–17). To our best knowledge, however, none of the natural mutant rFXIII-As in these reports has been analysed in detail with regard to biochemical properties, mainly because of the low efficiency of their mammalian expression systems.
In this study, we expressed the R260C and Del-IV mutants in yeast cells by taking advantage of its ability for mass protein expression. In fact, the mutant rFXIII-A was expressed about 50 times more than mammalian BHK cells (unpublished data).
It is important to note that large portions of both mutant proteins were found to be in very high-molecular weight forms as determined by sucrose-density-gradient ultra-centrifugation. In addition, large portions of both mutant proteins were precipitated by low-speed centrifugation. These results clearly indicate that none of the mutant proteins can assemble to form a stable conformation, suggesting rather that they were apt to aggregate. Unfortunately, the on-going/progressive aggregation process itself could not be demonstrated by regular biochemical techniques, e.g. light scattering or turbidity measurements, because mutant rFXIII-As could not be purified because of their instability.
It is unlikely that the extensive aggregation of multiple mutant rFXIII-As is mainly mediated by disulphide-bond formation, because there were one or two minor bands of high-molecular weight rFXIII-As for both non-aggregated wild-type and extensively aggregated mutants under the non-reduced conditions of SDS–PAGE analyses (unpublished data). These high-molecular weight forms of rFXIII-As disappeared under the reduced conditions, suggesting that the high-molecular weight bands were disulphide-bonded rFXIII-As between their multiple monomers or with another unknown protein(s).
Both of these mutant proteins were also shown to be more resistant to proteolytic cleavage by thrombin of the activation peptide than the wild-type. It is highly likely that the abnormal conformation of the mutants made their R37-G38 cleavage site less accessible for thrombin. It is very unlikely that the R260C mutant in a monomer form has a substantial enzymatic activity in vivo, because a Japanese case in a hemizygous state (compound heterozygote with a null allele), an Iranian homozygote and a Polish homozygote of this mutation showed <1%, 4% and <1.5% of the normal FXIII activity, respectively (20, 22, 23). This is consistent with the fact that there was essentially no FXIII activity (1.7% of the wild-type) after thrombin treatment, as demonstrated in our present in vitro study (Figs 7 and 8).
It was predicted that the R260C mutation would lead to a reduction of the electrostatic interaction between two monomers, resulting in destabilization of the molecule’s dimer, in our previous study (20). Here, we tested this assumption predicted by molecular modelling regarding the effect(s) of the mutation on the structure/function of FXIII-A, especially their theoretical stability and its capability to form a homodimer, in this study. The mutant rFXIII-A was expressed less efficiently and more short-lived than the wild-type in yeast cells, and it demonstrated essentially no catalytic activity. In addition, gel-filtration analysis revealed that a portion of the R260C mutant was a monomer, while the wild-type rFXIII-A formed a homodimer, like native FXIII-A. This is exactly what was predicted again by molecular modelling in this study: It is interesting to note that R260 is conserved among all eight known human TGase enzymes. Computational modelling indicates that the R260C mutant cannot form the intermolecular hydrogen bonds with D404, across the dimer interface. D404 is not conserved in any of the other human TGases; four different hydrophilic residues are found in this position in the other enzymes. This is in keeping with the unique dimeric nature of FXIII-A, whereas the other TGases are primarily monomeric. In contrast, the residues with which R260 forms intramolecular interactions, D427 and G410, are conserved in all eight human TGases. This strict conservation of not only R260 but also its intramolecular interaction partners suggests that loss of the intramolecular hydrogen bonds in the R260C variant may compromise the folding/stability of the FXIII-A monomer. Thus the computational structural modelling indicates that the R260C variant may have two consequences: it may impair not only FXIII-A dimer formation but also the folding and/or stability of the monomer. This is in agreement with the observations that unlike wild-type FXIII-A, the R260C mutant is expressed as a monomeric protein, and that the mutant is much more prone to aggregate, likely due to protein misfolding/instability.
The inability of FXIII-A mutants to form a dimer would make the mutant monomer susceptible to aggregation and degeneration. This may be a common mechanism for FXIII-A deficiency since the presence of monomeric FXIII-A has been found in at least one other natural Y283C mutant (16). This mechanism may be applicable to other mutants, because there are 33 natural missense mutations identified in the FXIII-A gene, including R77C, Y167C and W375C (http://www.f13-database.de). The possibility that the R260C mutant binds to FXIII-B cannot be excluded because similar Y283C mutant formed a hetero-tetramer with FXIII-B at least in part in vitro (16). However, it would not happen in vivo, because this mutant FXIII-As degrades inside the synthesizing bone-marrow-derived cells, and are never secreted outside to meet FXIII-B, which is synthesized by hepatocytes. This is consistent with the fact that a patient possessing this R260C mutation lacked both enzymatic activity of FXIII and the antigen of FXIII-A in plasma (20).
In contrast, the G501R mutation has been reported to form a normal dimer by a gel-filtration analysis and to be as active as the wild-type by an amine incorporation assay (19). This variant protein may cause a relatively mild conformational change in the FXIII-A molecule and result in a mild reduction of the plasma level (35) due to its thermal instability at a relatively high temperature of 55°C (19). However, all other missense mutants clearly verified marked protein instability by (pulse-)chase experiments (14–17).
In this study, we also examined a deletion mutation, Del-IV, which was identified in a Japanese patient with complete FXIII-A deficiency (21). The Del-IV mutant was in fact detected at a low steady-state level and degraded at a rate faster than the wild-type in yeast cells, as discussed earlier. Gel-filtration analysis revealed that no dimer was present for this mutant, either.
As expected, the stable wild-type rFXIII-A expressed by yeast cells proved to have a long-half life as native FXIII in vivo as well, i.e. in both healthy volunteers (36) and patients with congenital FXIII deficiency (8), and thus has more recently been used in a phase III prophylaxis trial for congenital FXIII deficiency (37).
In conclusion, the results reported here clearly indicate that these mutations truly impair the structure of the FXIII-A molecule. Thus, we propose the impaired dimer assembly and the aggregation of mutant FXIII-As as a novel molecular basis for FXIII deficiency.
Supplementary Data
Supplementary Data are available at JB Online.
Authorship Contributions
S.M. and W.Z. performed the research, analyzed data, and drafted the manuscript. M.S. and V.C.Y. performed the research, and analysed data. A.I. designed the research, analysed data and wrote the manuscript.
Funding
Ministry of Education, Science and Culture, Japan; the Uehara Memorial Foundation (Japan); the Ryoichi Naito Foundation for Medical Research (Japan). U.S. National Institutes of Health grants DK075897 and GM61388 (to V.C.Y.).
Conflicts of Interest
None declared.
Acknowledgements
The authors thank Drs R. Seal and D. Foster for providing yeast expression vectors, RPOT and pRS215, and the yeast strain S. cerevisiae, ZM118; Dr H. Kaetsu for providing purified plasma FXIII; Drs N. Takahashi, T. Yamazaki, T. Izumi and H. Iwata for helpful discussion and Ms L. Boba for her help in preparation of this article.
Glossary
Abbreviations
- CHX
cycloheximide
- Del-IV
deletion of exon IV
- DIG
digoxigenin
- EDTA
ethylenediaminetetraacetic acid
- FXIII
coagulation factor XIII
- FXIII-A
FXIII A subunit
- FXIII-B
FXIII B subunit
- PCR
polymerase chain reaction
- R260C
substitution of Arg260 by Cys
- rFXIII-A
recombinant FXIII-A
- SDS–PAGE
sodium dodecyl sulphate–polyacrylamide gel electrophoresis
- TGase
transglutaminase
References
- 1.Lorand L, Losowsky MS, Miloszewski KJ. Human FXIII: fibrin-stabilizing factor. Prog. Hemost. Thromb. 1980;5:245–290. [PubMed] [Google Scholar]
- 2.Muszbek L, Yee VC, Hevessy Z. Blood coagulation factor XIII: structure and function. Thromb. Res. 1999;94:271–305. doi: 10.1016/s0049-3848(99)00023-7. [DOI] [PubMed] [Google Scholar]
- 3.Ichinose A. Physiopathology and regulation of factor XIII. Thromb. Haemost. 2001;86:57–65. [PubMed] [Google Scholar]
- 4.Seitz R, Duckert F, Lopaciuk S, Muszbek L, Rodeghiero F, Seligsohn U. ETRO Working Party on factor XIII questionnaire on congenital factor XIII deficiency in Europe: status and perspectives. Study Group. Semin. Thromb. Hemost. 1996;22:415–418. doi: 10.1055/s-2007-999040. [DOI] [PubMed] [Google Scholar]
- 5.Mary A, Achyuthan KE, Greenberg CS. b-Chains prevent the proteolytic inactivation of the a-chains of plasma factor XIII. Biochim. Biophys. Acta. 1988;966:328–335. doi: 10.1016/0304-4165(88)90082-7. [DOI] [PubMed] [Google Scholar]
- 6.Souri M, Kaetsu H, Ichinose A. Sushi domains in the B subunit of factor XIII responsible for oligomer assembly. Biochemistry. 2008;47:8656–8664. doi: 10.1021/bi8006143. [DOI] [PubMed] [Google Scholar]
- 7.Souri M, Koseki-Kuno S, Takeda N, Degen JL, Ichinose A. Administration of factor XIII B subunit increased plasma factor XIII A subunit levels in factor XIII B subunit knock-out mice. Int. J. Hematol. 2008;87:60–68. doi: 10.1007/s12185-007-0005-z. [DOI] [PubMed] [Google Scholar]
- 8.Lovejoy AE, Reynolds TC, Visich JE, Butine MD, Young G, Belvedere MA, Blain RC, Pederson SM, Ishak LM, Nugent DJ. Safety and pharmacokinetics of recombinant factor XIII-A2 administration in patients with congenital factor XIII deficiency. Blood. 2006;108:57–62. doi: 10.1182/blood-2005-02-0788. [DOI] [PubMed] [Google Scholar]
- 9.Ichinose A, Izumi T, Hashiguchi T. The normal and abnormal genes of the a and b subunits in coagulation FXIII. Semin. Thromb. Hemost. 1996;22:385–391. doi: 10.1055/s-2007-999036. [DOI] [PubMed] [Google Scholar]
- 10.Ichinose A, Davie EW. Characterization of the gene for the a subunit of human FXIII (plasma transglutaminase), a blood coagulation factor. Proc. Natl. Acad. Sci. USA. 1988;85:5829–5833. doi: 10.1073/pnas.85.16.5829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ichinose A, Hendrickson LE, Fujikawa K, Davie EW. Amino acid sequence of the a subunit of human FXIII. Biochemistry. 1986;25:6900–6906. doi: 10.1021/bi00370a025. [DOI] [PubMed] [Google Scholar]
- 12.Ivaskevicius V, Seitz R, Kohler HP, Schroeder V, Muszbek L, Ariens RA, Seifried E, Oldenburg J Study Group. International registry on factor XIII deficiency: a basis formed mostly on European data. Thromb. Haemost. 2007;97:914–921. [PubMed] [Google Scholar]
- 13.Ivaskevicius V, Biswas A, Bevans C, Schroeder V, Kohler HP, Rott H, Halimeh S, Petrides PE, Lenk H, Krause M, Miterski B, Harbrecht U, Oldenburg J. Identification of eight novel coagulation factor XIII subunit A mutations: implied consequences for structure and function. Haematologica. 2010;95:956–962. doi: 10.3324/haematol.2009.017210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mikkola H, Muszbek L, Haramura G, Hamalainen E, Jalanko A, Palotie A. Molecular mechanisms of mutations in FXIII A-subunit deficiency: in vitro expression in COS-cells demonstrates intracellular degradation of the mutant proteins. Thromb. Haemost. 1997;77:1068–1072. [PubMed] [Google Scholar]
- 15.Takahashi N, Tsukamoto H, Umeyama H, Castaman G, Rodeghiero F, Ichinose A. Molecular mechanisms of type II FXIII deficiency: novel Gly562-Arg mutation and C-terminal truncation of the A subunit cause FXIII deficiency as characterized in a mammalian expression system. Blood. 1998;91:2830–2838. [PubMed] [Google Scholar]
- 16.Souri M, Ichinose A. Impaired protein folding, dimer formation, and heterotetramer assembly cause intra- and extracellular instability of a Y283C mutant of the A subunit for coagulation factor XIII. Biochemistry. 2001;40:13413–13420. doi: 10.1021/bi0111449. [DOI] [PubMed] [Google Scholar]
- 17.Vysokovsky A, Rosenberg N, Dardik R, Seligsohn U, Inbal A. Effect of four missense mutations in the factor XIII A-subunit gene on protein stability: studies with recombinant proteins. Blood Coagul. Fibrinolysis. 2006;17:125–130. doi: 10.1097/01.mbc.0000214707.65750.f4. [DOI] [PubMed] [Google Scholar]
- 18.Schroeder V, Meili E, Cung T, Schmutz P, Kohler HP. Characterisation of six novel A-subunit mutations leading to congenital factor XIII deficiency and molecular analysis of the first diagnosed patient with this rare bleeding disorder. Thromb. Haemost. 2006;95:77–84. [PubMed] [Google Scholar]
- 19.Coggan M, Baker R, Miloszewski K, Woodfield G, Board P. Mutations causing coagulation FXIII subunit A deficiency: characterization of the mutant proteins after expression in yeast. Blood. 1995;85:2455–2460. [PubMed] [Google Scholar]
- 20.Ichinose A, Tsukamoto H, Izumi T, Yamazaki T, Togashi M, Takamatsu J, Saito H, Umeyama H. Arg260-Cys mutation in severe FXIII deficiency: conformational change of the A subunit is predicted by molecular modelling and mechanics. Br. J. Haematol. 1998;101:264–272. doi: 10.1046/j.1365-2141.1998.00698.x. [DOI] [PubMed] [Google Scholar]
- 21.Izumi T, Nagaoka U, Saito T, Takamatsu J, Saito H, Ichinose A. Novel deletion and insertion mutations cause splicing defects, leading to severe reduction in mRNA levels of the A subunit in severe FXIII deficiency. Thromb. Haemost. 1998;79:479–485. [PubMed] [Google Scholar]
- 22.Peyvandi F, Tagliabue L, Menegatti M, Karimi M, Komáromi I, Katona E, Muszbek L, Mannucci PM. Phenotype-genotype characterization of 10 families with severe a subunit factor XIII deficiency. Hum. Mutat. 2004;23:98. doi: 10.1002/humu.9206. [DOI] [PubMed] [Google Scholar]
- 23.Ivaskevicius V, Windyga J, Baran B, Schroeder V, Junen J, Bykowska K, Seifried E, Kohler HP, Oldenburg J. Phenotype-genotype correlation in eight Polish patients with inherited factor XIII deficiency: identification of three novel mutations. Haemophilia. 2007;13:649–657. doi: 10.1111/j.1365-2516.2007.01517.x. [DOI] [PubMed] [Google Scholar]
- 24.Castaman G, Giacomelli SH, Schroeder V, Sanna S, Valdrè L, Morfini M, Banov L, Kohler HP, Rodeghiero F. Further evidence of heterogeneity of gene defects in Italian families with factor XIII deficiency. Haemophilia. 2011;18:e6–e8. doi: 10.1111/j.1365-2516.2011.02622.x. [DOI] [PubMed] [Google Scholar]
- 25.Bishop PD, Teller DC, Smith RA, Lasser GW, Gilbert T, Seale RL. Expression, purification, and characterization of human FXIII in Saccharomyces cerevisiae. Biochemistry. 1990;29:1861–1869. doi: 10.1021/bi00459a028. [DOI] [PubMed] [Google Scholar]
- 26.Du Z, Regier DA, Desrosiers RC. Improved recombinant PCR mutagenesis procedure that uses alkaline- denatured plasmid template. Biotechniques. 1995;18:376–378. [PubMed] [Google Scholar]
- 27.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lorand L, Lockridge OM, Campbell LK, Myhrman R, Bruner-Lorand J. Transamidating enzymes. II. A continuous fluorescent method suited for automating measurements of FXIII in plasma. Anal. Biochem. 1971;44:221–231. doi: 10.1016/0003-2697(71)90363-0. [DOI] [PubMed] [Google Scholar]
- 29.Fox BA, Yee VC, Pedersen LC, Le Trong I, Bishop PD, Stenkamp RE, Teller DC. Identification of the calcium binding site and a novel ytterbium site in blood coagulation factor XIII by X-ray crystallography. J. Biol. Chem. 1999;274:4917–4923. doi: 10.1074/jbc.274.8.4917. [DOI] [PubMed] [Google Scholar]
- 30.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 31.Yee VC, Pedersen LC, Le Trong I, Bishop PD, Stenkamp RE, Teller DC. Three-dimensional structure of a transglutaminase: human blood coagulation factor XIII. Proc. Natl. Acad. Sci. USA. 1994;19:7296–7300. doi: 10.1073/pnas.91.15.7296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ichinose A, Souri M, Izumi T, Takahashi N. Molecular and genetic mechanisms of FXIII A subunit deficiency. Semin. Thromb. Hemost. 2000;26:5–10. doi: 10.1055/s-2000-9795. [DOI] [PubMed] [Google Scholar]
- 33.Kangsadalampai S, Chelvanayagam G, Baker RT, Yenchitsomanus P, Pung-amritt P, Mahasandana C, Board PG. A novel Asn344 deletion in the core domain of coagulation FXIII A subunit: its effects on protein structure and function. Blood. 1998;92:481–487. [PubMed] [Google Scholar]
- 34.Kangsadalampai S, Chelvanayagam G, Baker R, Tiedemann K, Kuperan P, Board PG. Identification and characterization of two missense mutations causing FXIIIA deficiency. Br. J. Haematol. 1999;104:37–43. doi: 10.1046/j.1365-2141.1999.01145.x. [DOI] [PubMed] [Google Scholar]
- 35.Castle S, Board PG, Anderson RA. Genetic heterogeneity of factor XIII deficiency: first description of unstable A subunits. Br. J. Haematol. 1981;48:337–342. [PubMed] [Google Scholar]
- 36.Reynolds TC, Butine MD, Visich JE, Gunewardena KA, MacMahon M, Pederson S, Bishop PD, Morton KM. Safety, pharmacokinetics, and immunogenicity of single-dose rFXIII administration to healthy volunteers. J. Thromb. Haemost. 2005;3:922–928. doi: 10.1111/j.1538-7836.2005.01224.x. [DOI] [PubMed] [Google Scholar]
- 37.Inbal A, Oldenburg J, Carcao M, Rosholm A, Tehranchi R, Nugent D. Recombinant factor XIII: a safe, and novel treatment for congenital factor XIII deficiency. Blood. 2012;119:5111–5117. doi: 10.1182/blood-2011-10-386045. [DOI] [PubMed] [Google Scholar]