SUMMARY
Exome sequencing of 343 families, each with a single child on the autism spectrum and at least one unaffected sibling, reveal de novo small indels and point substitutions, which come mostly from the paternal line in an age-dependent manner. We do not see significantly greater numbers of de novo missense mutations in affected versus unaffected children, but gene-disrupting mutations (nonsense, splice site, and frame shifts) are twice as frequent, 59 to 28. Based on this differential and the number of recurrent and total targets of gene disruption found in our and similar studies, we estimate between 350 and 400 autism susceptibility genes. Many of the disrupted genes in these studies are associated with the fragile X protein, FMRP, reinforcing links between autism and synaptic plasticity. We find FMRP-associated genes are under greater purifying selection than the remainder of genes and suggest they are especially dosage-sensitive targets of cognitive disorders.
INTRODUCTION
Genetics is a major contributor to autism spectrum disorders. The genetic component can be transmitted or acquired through de novo (“new”) mutation. Analysis of the de novo mutations has demonstrated a large number of potential autism target genes (Gilman et al., 2011; Levy et al., 2011; Marshall et al., 2008; Pinto et al., 2010; Sanders et al., 2011; Sebat et al., 2007). Previously cited studies have focused on large-scale de novo copy number events, either deletions or duplications. Because such events typically span many genes, discerning which of the genes in the target region, alone or in combination, contribute to the disorder becomes a matter of educated guessing or network analysis (Gilman et al., 2011). However, with high-throughput DNA sequencing we can readily search for new mutation in single genes by comparing children to both parents. Such mutation is fairly common, on the order of a hundred new mutations per child, with only a few—on the order of one per child—falling in coding regions (Awadalla et al., 2010; Conrad et al., 2011). Thus, candidate gene and exome sequencing, involving the capture of designated coding regions of the genome, are efficient ways to search for new gene mutations in families. Preliminary attempts with these approaches, including studies of autism, have already been published (O’Roak et al., 2011; Schaaf et al., 2011; Vissers et al., 2010; Xu et al., 2011).
Our study is based on 343 families, a subset of the Simons Simplex Collection. In each family, only a single child is on the spectrum, and each has one or more normal siblings. This collection is depleted of multiplex cases where transmission genetics is expected to play a greater role (Fischbach and Lord, 2010). It is enriched for higher-functioning probands. As a result, the gender ratio among probands in this study is roughly 1 female per 6 males. Our focus was to determine first and foremost if the various types of new mutations would have different incidence in affected children than in their sibling controls. Not all types of point mutations are equally likely to be disruptive of gene function, and the contribution of the various types of events to autism incidence could not be evaluated in the absence of knowledge of relative rates, in affected and the sibling controls. Hence, we performed our analysis on family “quads” rather than trios. We rejected the idea that a comparative rate could be obtained by studies of unrelated controls performed at other sequencing centers or even at our own sequencing centers if performed at separate times with ostensibly similar protocols.
We conclude that de novo mutations disrupting gene function, such as indels that cause frame shifts and point mutations that affect splice sites or introduce stop codons, are statistically more likely in children on the autistic spectrum than in their unaffected siblings. In contrast, we see no statistically significant signal from either missense or synonymous mutations. Including de novo copy number variation, the types of mutation we can now detect contribute collectively to about 16% of the cases of simplex autism, undoubtedly an underestimate of the actual contribution. We observe an unusual coincidence between the list of genes with disruptive de novo mutations in children with autism and the list of 842 gene products associated with FMRP (Darnell et al., 2011), itself a target of mutation in ~2% of children with ASD. Within the parental gene pool there are far fewer disruptive variants in FMRP-associated genes than found in typical genes, suggestive of stringent purifying selection acting on FMRP-associated genes.
RESULTS
Scope and Coverage
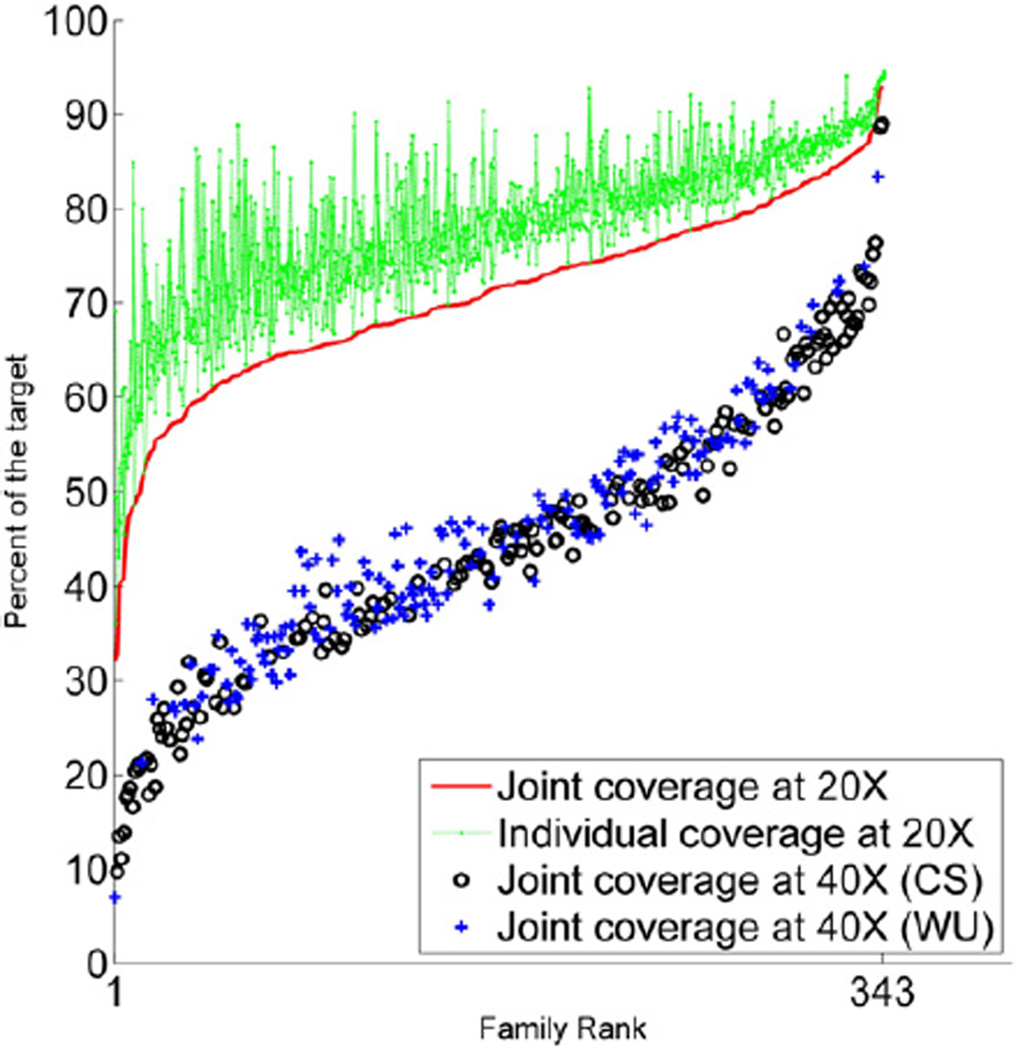
We report on the sequence and analysis of whole exomes from 343 families, each comprising parents and at least two offspring. No families with a member of questionable pedigree were included. To maximize the efficiency and uniformity of sequencing and capture we adopted a barcoding and pooling strategy. We used the NimbleGen SeqCap EZ Exome v2.0 capture reagent (Experimental Procedures). The 36.0 Mb target sequence consists almost entirely of coding exons. On average, individuals had 78% of the target covered at ≥20× and 56% at ≥40×. Since ours is a family study, we define “joint coverage” at a base as the minimum coverage at that base in any individual member of that family. On average, families had 71% joint coverage at ≥20× and 45% at ≥40×. Ninety-six percent of families had fifty percent or greater of target jointly covered at ≥20×. Coverage is presented graphically in Figure 1.
Figure 1. Exome Sequencing Family and Individual Coverage.
For each individual, the proportion of the exome covered in excess of 20× (dotted green line) is plotted horizontally. For each family, the proportion of the exome jointly covered in excess of 20× (solid red line) or 40× (black circle and blue “plus sign” designating sequencing center) in all members of that family is also shown. The family data are ordered by the rank of their joint coverage in excess of 20×.
To improve detection of indels and mutations at potential splice signals, our sequence analysis pipeline included 20 bp flanking each end of the coding exons, bringing our “extended” target to 43.8 Mb. We counted de novo events over the extended region even though coverage was lower than over coding target.
Filters and Validation Testing
We used a new multinomial test to determine likelihood that a mutation was de novo. We also used a chi-square test to exclude loci that did not fit a simple germline model, and we excluded sites that were polymorphic or noisy over the population. We established thresholds for these tests and used additional microassembly criteria, comprising our filters for counting candidate events. We sampled calls for experimental validation testing to determine our false positive rate.
Because the vast majority of false positives originate from the chance undersampling of one parental allele, we made an empirical choice of likelihood thresholds that diminished the frequency with which known polymorphic loci in the population appeared as “de novo” mutations in the children (see Figure S1 available online). These thresholds define part of our “SNV filter.” For each indel call, we also used de Bruijn graph microassembly as a filter (Pevzner et al., 2001) of reads possibly covering candidate regions in each of the family members.
For validation testing, we designed barcoded primers from the reference genome for each mutation examined, individually PCR-amplified DNA from each family member for the locus, pooled by family relation to the proband, made libraries and sequenced pooled products (Experimental Procedures). Validation tests succeeded or failed, and if they succeeded, the results either confirmed or falsified the calls. A summary of results is found in Table 1, for SNVs and indels. The detailed results (including counts) are in Tables S1 and S3. We validated in three batches, each time blind to the gene or affected status. In the first batch, we selected from the SNV data available, picking random calls passing filter. In the other two batches, we focused on indels and nonsense mutations. In all three batches, we tested a few calls close to passing but excluded by our filters.
Table 1.
Validation of De Novo Calls
| Passes (Filter) | Confirmed | Falsified |
|---|---|---|
| SNVs (99/105 Successfully Tested) | ||
| No (SNV) | 7 | 3 |
| Yes (SNV) | 89 | 0 |
| Nonsense (22/24 Successfully Tested) | ||
| No (SNV) | 1 | 0 |
| Yes (SNV) | 21 | 0 |
| Indels (49/49 Successfully Tested) | ||
| No (SNV or indel) | 0 | 1 |
| Yes (SNV only) | 1 | 0 |
| Yes (Indel only) | 9 | 0 |
| Yes (SNV and indel) | 38 | 0 |
Two filters were used for counting calls, an SNV and indel filter. We list all calls tested and the number of successful tests, separated into de novo SNVs (top) and indels (bottom). The middle subtable is the subset of SNVs that created stop codons (nonsense mutations), which are included in the overall number in the top subtable. Calls fell either within (“Yes”) or outside (“No”) the thresholds of the filter (first column), and were either confirmed or falsified as a de novo variants (second and third columns).
We sought to produce a list of autism candidates with as few false positives as possible and to be able to make the strongest statistical evaluation of the differential rates of de novo mutation between affecteds and siblings. We confirmed all 137 calls passing filters that we successfully tested. We subsequently loosened some and tightened other criteria for indels (the “indel filter”) to capture more events, as described below.
De Novo SNV Rates
We first investigated de novo SNVs. We counted 754 candidate de novo events passing our SNV filter (summarized in Table 2; complete list with details in Table S1). The distribution of events in families closely fit a Poisson model. Events were classified by affected status, gender, location (within exon, splice site, intron, 5′UTR, and 3′UTR) and type of coding mutation (synonymous, missense, or nonsense). The specific position of the mutation and the resulting coding change, if any, are also listed. In all cases examined, microassembly qualitatively validated the de novo SNV calls. Every de novo SNV candidate that passed filter and was successfully tested was confirmed present in the child and absent in the parents (89/89; Table 1 and Table S1).
Table 2.
Summary of De Novo Single Nucleotide Variants (SNVs) in 343 SSC Families
| SNV Effect | 40× (High) Coverage |
All Loci | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Proband | Sibling | Proband | Sibling | Proband F (29) | Proband M (314) | Sibling F (182) | Sibling M (161) | Both | Total | |
| Splice site | 4 | 1 | 6 | 3 | 1 | 5 | 1 | 2 | 0 | 9 |
| Nonsense | 15 | 7 | 19 | 9 | 3 | 16 | 6 | 3 | 2 | 30 |
| Missense | 125 | 121 | 207 | 207 | 19 | 188 | 116 | 91 | 3 | 417 |
| Synonymous | 53 | 42 | 79 | 69 | 8 | 71 | 43 | 26 | 4 | 152 |
| Promoter | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 2 |
| UTR | 5 | 7 | 8 | 9 | 0 | 8 | 3 | 6 | 0 | 17 |
| Intron | 34 | 35 | 59 | 64 | 5 | 54 | 38 | 26 | 1 | 124 |
| Intergenic | 0 | 2 | 1 | 2 | 0 | 1 | 2 | 0 | 0 | 3 |
| Total | 236 | 216 | 380 | 364 | 36 | 344 | 209 | 155 | 10 | 754 |
De novo SNVs were tabulated according to affected status, gender, and type of mutation. Data under “40×coverage” indicate variants in the subset of the exome target region in which all members of a given family were covered by at least 40 sequence reads. The power to detect de novo variants in children from this well-covered portion of the target is very high, and we found no bias in coverage between affected and unaffected children. No significant difference was seen for missense mutations (125 in probands to 121 in unaffected siblings), but larger ratios of nonsense (15:7) and splice site (4:1) mutations were observed in probands relative to unaffected siblings. When we expanded our set of calls to include every variant that passed our thresholds (under “all loci”; see Experimental Procedures), similar ratios were observed. Probands and unaffected siblings are further subdivided based on gender: “proband F” indicates an affected female; “proband M” an affected male; “sibling F” an unaffected female; and “sibling M” an unaffected male. In parentheses, we indicate the number of children with the corresponding affected status and gender. The “both” column shows de novo SNVs that were shared by both siblings from the same family.
Because variation in the number of mutations detected could be a function of variable sequence coverage in probands versus siblings, we also determined counts of mutation equalized by high coverage, assessing only regions where the joint coverage was at least 40×. At such high coverage, less than 5% of true de novo SNVs would be missed (as judged by simulations). We then determined the de novo SNV mutation rate by summing the total number of de novo SNVs in these 40× joint regions from all individual children, then dividing by the sum of base pairs within these regions in these children. The rate was 2.0 * 10−8 (±10−9) per base pair, or about 120 mutations per diploid genome per generation (6 * 109 * 2 * 10−8), consistent with a range of estimates obtained by others (Awadalla et al., 2010; Conrad et al., 2011).
Table 2 contains a summary of our findings. The number of de novo SNVs only in probands versus the number only in their siblings is not significantly different than expected from the null hypothesis of equal rates between probands and siblings, whether counting all SNVs (380 versus 364), synonymous (79 versus 69), or missense (207 versus 207). Ten de novo variants occurred in both proband and sibling. The balance does not change if we examine only regions of joint coverage ≥40×. Applying additional filters for amino acid substitutions (conservative versus nonconservative) or genes expressed in brain also did not substantively change this conclusion (Table S1). However, this study lacks the statistical power to reject the hypothesis that missense or synonymous mutations make a major contribution (see Discussion).
We did see a differential signal when comparing the numbers of nonsense mutations (19 versus 9) and point mutations that alter splice sites (6 versus 3). Such mutations could reasonably be expected to disrupt protein function, and in the following we refer to such mutations as ‘likely gene disruptions’ (LGD). The LGD targets and the specifics of the mutations in the affected population are listed in Table 3, and more details for all children are provided in Table S2. The sum of LGDs caused by point mutation was 25 in affecteds to 12 in siblings, with a p value of 0.047 by the two-sided binomial test. Every nonsense candidate that passed filter and was successfully tested was confirmed present in the child and absent in the parents (21/21; Tables 1 and S1).
Table 3.
Likely Gene-Disrupting (LGD) Mutations in Affected Children
| Family ID | Gender | Variant | Effect | Gene | Amino Acid Position | FMRP Target |
|---|---|---|---|---|---|---|
| 12221 | M | sub(C →T) | N | FAM91A1 | 72/839 | Yes |
| 12501 | M | sub(A →T) | N | NRXN1 | 587/1,177 | Yes |
| 12645 | M | sub(C →T) | N | ANK2 | 104/1,049 | Yes |
| 12764 | F | sub(C →A) | N | NCKAP1 | 1,082/1,129 | Yes |
| 12840 | M | sub(C→T) | N | ATP1B1 | 143/299 | Yes |
| 12867 | F | sub(G →A) | N | TRIP12 | 35/1,723 | Yes |
| 13094 | M | sub(T→A) | N | WDFY3 | 978/3,527 | Yes |
| 12683 | M | del(1) | F | KDM6B | 192/1,683 | Yes |
| 12705 | M | ins(C) | F | DIP2C | 657/1,557 | Yes |
| 12952 | M | del(1) | F | MLL5 | 1,066/1,859 | Yes |
| 13012 | M | ins(CTGGTCT) | F | DIP2A | 608/1,568 | Yes |
| 13092 | M | ins(AGGTCAG) | F | LMTK3 | 307/1,490 | Yes |
| 13612 | M | del(4) | F | DST | 3,268/5,172 | Yes |
| 12969 | M | ins(C) | S | MED13L | 1,789/2,211 | Yes |
| 12409 | M | sub(G →A) | N | LRP2 | 3,184/4,656 | No |
| 12463 | M | sub(C →T) | N | UNC80 | 518/1,765 | No |
| 12653 | M | sub(C →T) | N | KIAA0232 | 118/1,396 | No |
| 12669 | M | sub(C →T) | S | RABGGTA | UTR | No |
| 12792 | M | sub(C →T) | N | ANO5 | 420/913 | No |
| 12840 | M | sub(C →T) | S | TM4SF19 | 68/210 | No |
| 12864 | F | sub(C →T) | S | SUV420H1 | 86/646 | No |
| 13010 | M | sub(G →C) | S | TBC1D23 | 293/700 | No |
| 13042 | M | sub(G →A) | N | THSD7A | 879/1,658 | No |
| 13125 | M | sub(C →T) | N | RAD21L1 | 54/557 | No |
| 13197 | M | sub(G →T) | N | NR3C2 | 701/868 | No |
| 13234 | M | sub(G →A) | N | CBX4 | 131/561 | No |
| 13349 | M | sub(G →A) | N | NUAK1 | 433/662 | No |
| 13364 | M | sub(G →A) | N | ECM2 | 182/678 | No |
| 13506 | M | sub(C →A) | N | SCUBE2 | 80/1,000 | No |
| 13513 | M | sub(G →A) | S | ZMYND11 | 239/455 | No |
| 13526 | M | sub(A /G) | S | CACNA2D3 | 592/939 | No |
| 13670 | F | sub(C →T) | N | NFIA | 30/502 | No |
| 12323 | M | del(1) | F | PHF2 | 1,088/1,097 | No |
| 12652 | M | del(1) | F | SPATA13 | 127/1,278 | No |
| 12653 | M | ins(A) | F | DLL1 | 431/724 | No |
| 12773 | M | del(4) | F | PCDHA13 | 678/951 | No |
| 12826 | F | del(4) | F | TROVE2 | 253/539 | No |
| 12858 | F | del(1) | F | PAX5 | 111/392 | No |
| 12939 | M | del(2) | F | SLC25A39 | 104/352 | No |
| 12950 | M | del(4) | F | UBN2 | 1,063/1,348 | No |
| 13018 | M | del(1) | F | PCOLCE | 101/450 | No |
| 13070 | M | del(2) | F | CTTNBP2 | 760/1,664 | No |
| 13096 | M | del(1) | F | GIMAP8 | 149/666 | No |
| 13162 | M | ins(A) | F | RIMS1 | 196/1,693 | No |
| 13168 | F | del(1) | F | MFRP | 342/580 | No |
| 13176 | F | del(1) | F | ZFYVE26 | 397/2,540 | No |
| 13183 | M | ins(G) | F | BCL11A | 265/836 | No |
| 13398 | M | ins(CGTCATCA) | F | POGZ | 1,108/1,316 | No |
| 13439 | M | ins(A) | F | CSTF2T | 354/617 | No |
| 13471 | M | del(2) | F | FLG | 147/4,062 | No |
| 13537 | M | del(4) | S | TUBGCP4 | 578/667 | No |
| 13548 | F | del(1) | F | GALNTL4 | 521/608 | No |
| 13552 | M | del(1) | F | DYRK1A | 487/755 | No |
| 13585 | M | ins(G) | F | ACACB | 114/2,459 | No |
| 13586 | M | ins(G) | F | TRIM17 | 274/478 | No |
| 13590 | M | ins(A) | F | MTHFS | 147/147 | No |
| 13590 | M | del(1) | F | EFCAB5 | 848/857 | No |
| 13616 | M | ins(G) | F | ATP10D | 1,001/1,427 | No |
| 13646 | M | del(5) | F | VCP | 515/807 | No |
SNV and indel variants from affected children that are likely to disrupt the function of the corresponding proteins are listed. The “family ID” column indicates the SSC ID of the relevant family. Under “gender,”Mstands for males and F for females. The “variant” column shows detail for reconstructing the haplotype around the de novo variant relative to the reference genome as follows: “sub(R/A)” represents a substitution of the reference allele to an alternative allele; “ins(seq)” indicates an insertion of the provided sequence “seq” and “del(N)” denotes a deletion of N nucleotides. Chromosomal locations for events can be found in Table S2. Under “effect,” “N” stands for nonsense, “S” for splice site, and “F” for frame shift events. The “amino acid position” column shows the position of the first incorrectly encoded amino acid (or nonsense codon) within the encoded protein/the length of the protein. When a mutation affects multiple isoforms of a transcript, the earliest proportionate coordinate is given. “FMRP target” indicates whether the corresponding gene’s RNA was found to physically associate with FMRP (Darnell et al., 2011). See “Recurrence and Overlaps with FMRP-Associated Genes” in the Results as well as Tables 5 and 6 for additional details.
De Novo Indel Rates
Further support for the hypothesis that LGDs contribute to ASD comes from counting small insertions or deletions (indels) within coding regions. Small indels were ascertained using a simple protocol. This protocol works best for indels less than six base pairs: we surveyed all reads that required a gap to align to the reference genome, and marked where the gap was placed. After eliminating all gap positions that are common in the population, we again used our SNV filter: multinomial sampling to estimate the likelihood that a gap in the child was not inherited from either parent, and used a chi-square test for a germline model (Experimental Procedures). We set the same thresholds as used for SNVs. Microassembly excluded ten presumptive indel loci as inconsistent, failing either because of low count for confirmatory reads, absence of an indel, or finding the nonreference allele in a parent. For two loci, the sizes of deletions were corrected by microassembly. We tested 49 candidate de novo indels, and all 39 that passed the SNV filter were confirmed. Incidence of indels in families again followed a Poisson model.
It was quite clear from validation testing that many candidate indels excluded by the SNV filter were true positives. There was clearly allele imbalance favoring the reference allele over the indel in the exome sequencing, but this bias was absent in the validation testing (Table S2). Because of the importance of indels, we wished to establish an “indel filter” that diminished false negatives, so we lowered our chi-square stringency (from 10−4 to 10−9) and multinomial threshold (from 60 to 30). To guard against false positives resulting from undersampling the parents, we excluded any locus at which the variant allele was seen fewer than six times in the child, or appeared even once in the parents, and insisted on certain lower limits of coverage, all of which was done without respect to affected status (Experimental Procedures). Of the 49 tested loci, 47 passed this new filter and confirmed (Table 1).
With the indel filter, we detected 53 indels in probands and 32 in siblings (p value=0.03). Of these, 32 in probands and 15 in siblings caused frame shifts (p value = 0.02; see Table 4 for summary and Table S3 for complete list). Frame shift mutations, like nonsense and splice mutations, can cause severe disruption of coding capacity and hence we classify them as LGDs. Three more indels (2 in probands and 1 in siblings) are likely to be LGDs, as they either introduce stop codons or disrupt a splice site. It is likely that even in-frame insertions or deletions are more disruptive to a peptide than a mere substitution, and we refer later to an interesting example, but we do not count them as LGDs.
Table 4.
Summary of De Novo Indels in 343 SSC Families
| Indel Effect | Proband | Sibling | Proband F | Proband M | Sibling F | Sibling M | Both | Total |
|---|---|---|---|---|---|---|---|---|
| Splice site | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 2 |
| Frame shift | 32 | 15 | 5 | 27 | 4 | 11 | 0 | 47 |
| Nonsense | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 1 |
| No frame shift | 7 | 7 | 0 | 7 | 3 | 4 | 0 | 14 |
| UTR | 2 | 1 | 0 | 2 | 1 | 0 | 0 | 3 |
| Intron | 10 | 8 | 0 | 10 | 4 | 4 | 1 | 19 |
| Total | 53 | 32 | 5 | 48 | 13 | 19 | 1 | 86 |
The detected de novo indels were tabulated based on affected status and gender and stratified by the effects the events are likely to have on the corresponding genes. De novo indels that are likely to severely disrupt the encoded proteins—by causing frame shifts, destroying splice sites, or introducing nonsense codons—are significantly more abundant in affected children than in unaffected siblings. See Table 2 for details of the column headings.
All LGD targets are listed in Table 3, with further details in Table S2. In summary, using our filters for SNVs and indels, we observe 59 LGDs in probands versus 28 in siblings (p value of 0.001).
Gender and Phenotype
The de novo LGD incidence by gender and status can be summarized from Tables 2 and 4. We observe 9 de novo LGD events in 29 females on the spectrum, and 50 in 314 males. Although only marginally statistically significant (p value = 0.07), the higher incidence in females matches the higher incidence of de novo CNVs seen in females on the spectrum (Levy et al., 2011), and does not reflect a higher rate of de novo mutations in females overall: we detected 12 in 182 female siblings and 16 in 161 male siblings. We observed no significant difference with respect to verbal or nonverbal IQ, or overall severity in children with or without detectable de novo LGDs.
Origin of Mutations
Our data are consistent with a paternal origin for variation of the type we detect. From the original sequencing and validation of our data, we were able to ascertain the parental haplotype for some de novo mutations, i.e., those that were linked to a polymorphism found in only one of the two parents. We found that the father is more frequently the parent of origin than the mother: 50/17 for SNVs and 6/1 for indels (Table S1), with a combined p value of 10−5. Although this was previously known for SNVs, or at least suspected (Conrad et al., 2011), our results suggest it is true for small indels as well. Because it is implausible that the origin of a parental haplotype should influence its global mutation rate in the child, we conclude that most of the de novo variants passing our filters originated in the parent.
Parental age also appears to play a role in mutation rate, further evidence of the parental origin of the mutations we observe. We divided all the data of de novo SNV mutations from the 40× joint family coverage into three bins nearly equal in base pairs covered, separated by the age of the father at child’s birth, and then counted de novo SNVs in all three bins. The bins spanned fathers from 16.1 to 30.9 (mean of 27.3), 30.9 to 35.9 (mean of 33.4), and 35.9 to 58.0 (mean of 39.6) years old. There was no significant difference in overall SNV rate between probands and siblings; hence, we utilized both children. We measured the counts of de novo mutation in the three bins as 136, 139, and 181, respectively. The hypothesis that the counts for de novo SNVs in children with the youngest fathers and in those with the oldest arose from equal mutation rates has a p value of 0.013. Performing the same computation for mothers, we compute a p value of 0.002.
Our de novo filters are biased against somatic mutation, as our likelihood models are based on germline mutation. However, there are a handful of loci where the evidence is consistent with a somatic origin: diminished ratios of variant to reference allele in the child both in the exome coverage and in the counts from the PCR validation tests. Moreover, in one case where it could be discerned, the maternal haplotype was seen in association with both the variant and reference allele. All these examples, however, are also consistent with multiple copies of the loci in question. Our de novo filters are also biased against mosaicism in the blood of the parent. Nevertheless, we see two examples where deep sequencing of the PCR test revealed the presence of the variant in the parent: one SNV in mother (1,308 counts of reference to 28 counts of the variant) and one indel from the father (15,399 to 79). Not surprisingly, neither variant was observed in the parent in the sparser exome data.
Altogether, with the filters we use, the de novo events we report are largely and perhaps almost entirely germline in origin and this affects our assessment of the contribution of new mutation to autism.
Recurrence and Overlaps with FMRP-Associated Genes
We searched for recurrences and overlaps between the 59 LGD target genes and other gene lists (Tables 3 and 5), including genes struck by de novo missense or present in de novo CNVs from previous studies. There are no recurrences among our LGD targets (but see Discussion). Given the large number of potential autism target genes, failure to observe overlap in this small list is not surprising. There are two overlaps with the 72 most likely candidate genes from our previous CNV study: NRXN1 and PHF2. The former is considered to be casual for ASD (Ching et al., 2010). A few overlaps of the LGD targets and targets of missense mutations were observed, two in siblings and one in probands, but this is well within random expectation.
Table 5.
Overlaps with FMRP-Associated Genes
| Gene List | Number | FMRP Targets | p Value |
|---|---|---|---|
| LGD in probands | 59 | 14 | 0.006 |
| CNV candidates | 72 | 13 | 0.0004 |
| LGD + CNV candidates | 129 | 26 | <10−13 |
| LGD in siblings | 28 | 2 | 0.8 |
| Missense in probands | 207 | 22 | 0.9 |
| Missense in siblings | 207 | 30 | 0.1 |
The overlap of six different gene lists with the set of FMRP-associated genes (Darnell et al., 2011) is shown. “LGD in probands” is the list of genes affected by de novo LGDs in affected children; “CNV candidates”are the best gene candidates for ASD derived from de novo CNVs analyzed in a previous study (Gilman et al., 2011); “LGD + CNV candidates”is the union of the previous two categories; “LGD in siblings” is the list of genes affected by de novo LGD in unaffected siblings; and “missense in probands” and “missense in siblings” are the lists of genes affected by de novo missense variants in probands and unaffected siblings, respectively. For each list, we show the total number of genes, the number of genes that are associated with FMRP, and the p value under the hypothesis that the overlap is random. See Experimental Procedures for details of the p value computation.
By contrast, we saw unexpected overlap between the LGD targets, CNV-derived autism candidate genes and the set of 842 FMRP-associated genes. This last set of genes corresponds to mRNAs whose translation may be controlled by the fragile X mental retardation gene product FMRP (Darnell et al., 2011). Microsatellite expansion in the X-linked FMR1 gene is an established cause of autism spectrum disorders. Significant overlap of the 842 FMRP-associated genes with autism candidate genes has been previously suggested (Darnell et al., 2011). 14 of our 59 LGD targets and 13 of 72 CNV target genes, with one in common, overlap with the 842 FMRP-associated genes. We calculate the p values to be 0.006 and 0.0004, respectively. The first p value is calculated relative to the cumulative gene length of FMRP-associated genes, whereas the second is more related to gene number and is determined by simulation (Experimental Procedures). Altogether, the observation of 26 genes (14 plus 13 minus one in common) out of 129 (59 plus 72 with two in common) overlapping with the 842 FMRP-associated genes has a p value of <10−13 (calculated on a per-gene basis). In contrast, we see no significant overlap between FMRP-associated genes with LGD targets from siblings (2 of 28) or with de novo missense mutations in probands (22 of 207) or siblings (30 of 207), all squarely within expectation given the size of the FMRP-associated genes. All data on overlaps are summarized in Table 5.
Absent Statistical Signal from Inheritance
This study lacks the power to discover small effects due to inheritance (see Discussion). Nevertheless, we sought evidence for large effects. From 686 parents, we enumerated all rare synonymous, missense, nonsense, and splice site variants in the parents, over a set of well-annotated genes (the set of ~18,000 CCDS genes; Pruitt et al., 2009), and the intersection of that set with candidate genes from previous CNV studies (Gilman et al., 2011; Levy et al., 2011), candidate genes from the present study of de novo LGDs, and all FMRP-associated genes. We considered only rare variants (defined as occurring only once in the population), eliminating the polymorphic variants so that all variants were on an equal footing. We then examined transmission to children, by affected status. We observed no statistically significant transmission bias of either missense or LGDs (nonsense plus splice variants) in any gene set to either probands or siblings. There was, in fact, slightly lower transmission to the affected population than to the siblings (Tables 6A and 6B). None of these statements change if we look specifically at variants carried by the mother.
Table 6.
Lack of Segregation Distortion
| Gene Group | Proband | Sibling | Proband & Sibling | Neither | Total |
|---|---|---|---|---|---|
| (A) Segregation of Rare Nonsense and Splice Site Variants | |||||
| FMRP-associated | 9 | 15 | 16 | 20 | 60 |
| CNV candidates | 3 | 1 | 3 | 0 | 7 |
| De novo LGD | 0 | 6 | 4 | 3 | 13 |
| All of the above | 12 | 22 | 20 | 23 | 77 |
| All well-annotated | 666 | 726 | 750 | 692 | 2,834 |
| (B) Segregation of Rare Missense Variants | |||||
| FMRP-associated | 1,784 | 1,808 | 1,924 | 1,591 | 7,107 |
| CNV candidates | 99 | 99 | 116 | 108 | 422 |
| De novo LGD | 165 | 189 | 200 | 214 | 768 |
| All of the above | 1,973 | 2,017 | 2,168 | 1,833 | 7,991 |
| All well-annotated | 22,267 | 22,379 | 24,255 | 20,875 | 89,776 |
| (C) Genes with Compound Heterozygosity (Rare Nonsense, Splice Site, and Missense) | |||||
| FMRP-associated | 28 | 31 | 59 | ||
| CNV candidates | 1 | 0 | 1 | ||
| De novo LGD | 4 | 3 | 7 | ||
| All of the above | 32 | 33 | 65 | ||
| All well-annotated | 242 | 224 | 466 | ||
Rare SNVs (those observed only once in the 686 parents in this study) are classified based on the children to whom they were transmitted: only to the proband (“proband”); only to the unaffected sibling (“sibling”); to both children (“proband & sibling”) or to “neither” child.
(A) We classified rare variants by the gene they target. The segregation of several target groups of rare nonsense and splice site variants is indicated. The “FMRP-associated” group is all variants that occur within the coding region of FMRP-associated genes; “CNV candidates” are defined as in Table 5; “de novo LGD” are the variants that occur in the 59 genes affected by de novo LGD in probands from this study; “all of the above” represents the variants in at least one of the previous three target sets; and “all well-annotated” are variants that target well-annotated CCDS genes (Pruitt et al., 2009).
(B) Segregation of rare missense variants categorized as in (A).
(C) Compound heterozygosity in probands and unaffected siblings. A compound heterozygote was defined as an instance in which a gene was affected by two different rare variants—one from the mother and one from the father—in the same child. Genes are classified as in (A) and (B).
We examined as well the prevalence of compound heterozygotes of rare LGD variants, where an offspring receives one rare variant from each parent, and again we see no statistically significant difference between probands and unaffected siblings (Table 6C). In this case, however, there is a slight increase in the number of compound heterozygotes of well-annotated genes in probands compared to siblings (242 versus 224).
We specifically examined the possibility of compound heterozygosity in offspring at loci hit by de novo LGDs, caused by transmission of rare missense or LGD mutations. We observed nine such events in probands and twelve in siblings, all but one in each group a combination of the de novo LGD event and a rare missense variant. Thus, there is no differential signal for compound heterozygosity and no evidence that the de novo event in the affected created a homozygous null.
Extreme Scarcity of LGDs in FMRP-Associated Genes
In the course of the above work, we did make an unexpected and striking observation. The number of rare nonsense or splice site variants over the FMRP-associated genes was much lower than expected given the abundance of these variants found in the CCDS genes (Table 7). We observed 2,192 rare nonsense variants in all genes, of which 55 fell within FMRP-associated genes—a proportion of 0.025. We observed 63,080 synonymous rare variants with 7,051 falling within FMRP-associated genes, a proportion of 11.18. The proportion of all synonymous variants falling within in FMRP-associated genes is roughly equal to the sum of the lengths of all FMRP-associated genes divided by the sum of lengths of all well-annotated genes. But the proportion of nonsense variants is one-fourth of this cumulative length proportion. Using the proportion of synonymous variants in CCDs as an unbiased measure of opportunity for mutation between sets, we calculate a p value of <10−50 that the variants in FMRP-associated genes are under the same degree of purifying selection as are the well-annotated genes. Because FMRP-associated genes are on average longer than the “typical” gene, we also computed the proportion of genes in a given set that are ever observed with a variant of a specified type. Qualitatively, we see the same pattern. We see an even stronger decrease in variants that disrupt splice sites within the FMRP-associated genes.
Table 7.
Incidence of Rare SNVs in the Parental Gene Pool
| Effect | CCDS (18,168) | FMRP-Associated Genes (790 in CCDS) |
Disease Genes (244 in CCDS) | Essential Genes (1,672 in CCDS) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Number | Number | Percent | Ratio to Syn. | Number | Percent | Ratio to Syn. | Number | Percent | Ratio to Syn. | |
| (A) Per Position | ||||||||||
| Missense | 104,637 | 8,573 | 8.19 | 0.73 | 2,023 | 1.93 | 0.97 | 10,874 | 10.39 | 0.84 |
| Nonsense | 2,192 | 55 | 2.51 | 0.22 | 38 | 1.73 | 0.87 | 109 | 4.97 | 0.40 |
| Splice site | 1042 | 19 | 1.82 | 0.16 | 15 | 1.44 | 0.72 | 55 | 5.28 | 0.42 |
| Synonymous | 63,080 | 7,051 | 11.18 | 1.00 | 1,260 | 2.00 | 1.00 | 7,849 | 12.44 | 1.00 |
| (B) Per Gene | ||||||||||
| Missense | 16,232 | 755 | 4.65 | 0.91 | 221 | 1.36 | 0.96 | 1,509 | 9.30 | 0.96 |
| Nonsense | 1,887 | 44 | 2.33 | 0.46 | 34 | 1.80 | 1.26 | 95 | 5.03 | 0.52 |
| Splice site | 980 | 17 | 1.73 | 0.34 | 13 | 1.33 | 0.93 | 49 | 5.00 | 0.52 |
| Synonymous | 15,160 | 776 | 5.12 | 1.00 | 216 | 1.42 | 1.00 | 1,464 | 9.66 | 1.00 |
(A) Rare SNVs (defined as in Table 6) were classified according to their predicted mutational effect and whether they fell within four different sets of genes. The mutational consequences of these variants have been determined using well-annotated CCDS gene models (Pruitt et al., 2009), first column. “FMRP-associated” is the list of 842 FMRP-associated genes; “disease” is the list of 256 positionally cloned human disease genes (Feldman et al., 2008), and “essential” is the list of 1,750 human orthologs of mouse genes that have been associated with lethality in the Mouse Genome Database (Blake et al., 2011). The numbers in parentheses next to each gene set label show the numbers of genes from each category that are found among the well-annotated CCDS set. For each mutational effect and for each gene set, the “number” column indicates how many variants occur in that set, the “percent” column shows the percentage of all variants with that effect that fall within the gene set, and the “ratio to syn” column shows the percentage normalized by the percent for the synonymous variants. Nonsense (2.51%) and splice site (1.82%) variants were substantially underrepresented among FMRP-associated genes when compared to the percent of synonymous variants (11.18%).
(B) The “number” column refers to the number of genes in the given set affected by rare SNVs of the given mutational effect. The percent of these, and the normalized percent, as defined in (A) above, are also shown.
On the other hand, missense variants show a much less extreme depletion in the FMRP-associated genes. This is consistent with the view that while missense mutations can create hypo- or hypermorphic alleles, they generally do not have the impact of a disruption.
To understand better the significance of the results just described, we examined the same statistics for two other genes sets (Table 7). The first is a set of “disease genes,” ~250 human genes linked to known genetic disorders, the majority of which are severely disabling (Feldman et al., 2008). In this set, variants of all types behaved much the same as the synonymous variants. The second set, “essential genes,” were the human orthologs of ~1,700 murine genes. The murine genes were extracted by us (combining automated and manual methods) from a set of genes annotated by the Jackson Laboratory, with annotations based on breeding and transgenic experiments. The distribution of variants in the “essential genes” closely resembles the distribution in the FMRP-associated genes.
DISCUSSION
From previous genetic studies, we expected that de novo mutation plays a large role in autism incidence and introduces variation that is short-lived in the human gene pool because such variation is deleterious and highly penetrant. Sequencing reveals the type and rates of small-scale mutation and pinpoints the responsible gene targets more definitively than does copy number or karyotypic analysis. Our study is a partial confirmation of our expectations, provides sources and rates of some classes of mutation, and strengthens the notion that a convergent set of events might explain a good portion of autism: a class of neuronal genes, defined empirically as FMRP-associated genes, overlap significantly with autism target genes.
Our data set is the largest set of family exome data to be reported so far, and it is derived from whole-blood DNA to avoid the perils of immortalized cell lines. While we focused on the role of de novo mutation of different types in autistic spectrum disorders, we have looked at additional questions related to new mutation. We project overall rates of de novo mutation to be 120 per diploid genome per birth. Most small-scale de novo mutation comes from fathers, and is related to parental age. Per event (and probably en masse), missense mutations have far less impact on the individual than do gene-disrupting mutations such as nonsense, splice variants, and frame shifts. This is evident both in overall differential in de novo mutations, but also from the effects of purifying selection on sets of genes (Table 7).
Differential Signal from De Novo Missense and Gene-Disrupting Mutations
Missense mutation should contribute to autism to some degree, as gene function can be severely altered by single-amino-acid substitutions. However, we see no statistical evidence in our work for the hypothesis that de novo missense mutations contribute to autism. The number of de novo missense events we observe is not greater in probands than in siblings. Moreover, the ratio of numbers of missense mutations in probands to siblings is not significantly different than the observed ratio of numbers of synonymous mutations. Even when we filter for genes expressed in brain, count missense mutations that cause nonconservative amino acid changes, or count missense mutations at positions conserved among vertebrates (Table S1, columns BA–BJ), we see no statistical evidence for contribution from this type of mutation. This is also true when we look for overlap of de novo missense mutations with FMRP-associated genes (Table 5). The lack of signal is not attributable to the type of population we study, as we observe de novo copy number imbalance of the expected magnitude in this very same population (Levy et al., 2011; Sanders et al., 2011). But given the size of the population and background mutation rate, we are unable to find signal in the present study. A simple power calculation indicates that we cannot rule out confidently even a 20% contribution to autism from de novo missense mutation. Despite these caveats, it is worth considering that de novo mutation causing merely amino acid substitution may only rarely create a dominant allele of strong effect.
We make a strikingly different observation for mutations that are likely to disrupt gene function. In contrast to de novo missense mutation, we do get signal from de novo mutations likely to severely disrupt coding: mutations at splice sites, nonsense mutations, and small indels, particularly indels that cause frame shifts. We observe 59 likely gene disruptions (LGD) in affected and 28 in siblings, a ratio of two to one. We note that girls on the autistic spectrum have a higher rate (9/29) than boys (50/314), a bias we have previously noted for de novo CNV events. The total contribution from LGD mutations can be estimated as 31 events in 343 families (59 events in probands minus 28 events in siblings), or roughly 10% of affected children.
Germline Origin: Rates, Parental Age, and Paternal Variation
We observed de novo point mutations in children at the rate expected from other studies (Awadalla et al., 2010; Conrad et al., 2011), about 120 point mutations per genome per generation. We observe that the frequency of de novo mutation is dependent on parental age, and know this with a high degree of statistical certainty. This observation is in keeping with, and potentially explains, other studies that have shown increased incidence of certain genetic disorders in the progeny of older parents, including ASD (Saha et al., 2009).
From sequencing adjacent linked polymorphisms in children and parents, we infer that on the order of 3/4 of new point mutations (50 of 67) derive from the father’s germline. Although we have less data, this conclusion holds as well for de novo small indels (6 of 7). These data confirm the paternal line is the main source for these types of new human variation. The data also indicate that the majority of the de novo calls in this study are not somatic in origin, but occur prior to conception. We infer this by assuming that after zygote formation, the mother’s and father’s genomes are equally vulnerable to subsequent somatic mutation. By contrast, a previous study indicated that for de novo copy number variation both parents contribute almost equally (Sanders et al., 2011).
We observe very few cases where two siblings share the same de novo mutation, about one for every fifty occurrences, suggesting that the parent is rarely a broad mosaic. However, this conclusion could be an ascertainment bias, because our operational identification of “de novo” precludes observing the mutation in the parent at levels higher than expected from sequencing error. As presented, we do observe some evidence of parental mosaicism, and this is a subject of ongoing scrutiny using enhanced statistical modeling and validation.
Total Contribution from De Novo Mutation
Finding the correct contribution from each genetic mechanism is critical for understanding the nature of the factors causing autistic spectrum disorders. Adding the 6% differential for large-scale de novo copy number mutation previously observed (Levy et al., 2011; Sanders et al., 2011) to the 10% differential for LGDs, we reach a total differential of 16% between affected children and siblings. This is far less than our predictions, based on modeling the AGRE population (Zhao et al., 2007), that causal de novo mutations would occur in about 50% of the SSC. This gap could be attributable to having modeled a more severely affected population. The SSC is skewed to higher functioning cases with a male to female ratio of 6:1 (Fischbach and Lord, 2010), so there may be more borderline cases in that collection than in the AGRE collection (male to female ratio of 3:1), from which we built our model (Zhao et al., 2007).
But our differential must underestimate the contribution from de novo events. First, we use extremely stringent criteria meant to eliminate false positives, and we fail to detect many true positives as a consequence. Second, even among the de novo events we do observe, we may be missing gene-disruptive events, for example, mutations outside the consensus that disrupt splicing and in-frame indels that disrupt the spacing of the peptide backbone. It would not be unlikely to miss even a 5% differential from de novo missense mutation in a study of this size, given the high background rate of neutral missense mutation. Third, our coverage of the genome is incomplete. Some of this arises by chance, and some is systematic due to the exome capture reagents or errors in the reference genome. Fourth, large classes of mutations are eliminated by our filters, such as those that originate in a parent who is a mosaic, and in children who suffer somatic mutation early after zygote formation. Fifth, there are biases in correctly mapping reads covering regions of the genome that are highly rearranged in the child. Sixth, we have not implemented tools that can reliably detect large indels and rearrangements. Our present tool is efficient only for small indels, less than seven base pairs. Seventh, an entire class of events involving repetitive elements is presently unexplored by us because we currently demand that reads have unique mappings. Eighth, we make calls from only coding regions and thus are not able to assess noncoding events that might affect RNA expression or processing. From all these presently hidden sources, the contribution of de novo mutation could easily double or more.
While there is still a gap between the incidence of de novo gene disrupting events and our expectations from population analysis—especially in males—this gap may yet be filled by deeper coverage, more refined genomic tools, and whole-genome sequencing. Interpretation of a richer data set will undoubtedly require a greater understanding of biology, such as the role for noncoding RNAs and how transcript expression and processing are controlled. By contrast, the differential incidence of de novo mutation in females is very strong, and from CNV and exome sequencing data, runs at nearly twice the differential as in males.
Transmission Genetics and Gene Dosage
We find almost no evidence of a role for transmission genetics. We do not think the present study of only 343 families would display statistical evidence for any of the plausible models of contribution from transmission. Such studies will require greater power, and previous larger copy number studies of the SSC have found such evidence (Levy et al., 2011). There is, however, a weak signal from the increased ratio of compound heterozygotes of rare coding variants in probands to siblings (242 versus 224). This would be consistent with a 5% contribution from this genetic mechanism, but is also consistent with virtually no contribution (p value = 0.4). We can virtually rule out that such events are contributory in more than 20% of children on the spectrum. Fortunately, even a modestly larger study will resolve the strength of contribution from this source.
We do not find evidence of compound heterozygosity at the vast majority of loci where one allele was hit by a disruptive mutation. These events are thus likely to have high impact by altering gene dosage, although we cannot rule out at present that the mutant allele acts by dominant interference.
Individual Vulnerability to New Mutation
Conceptually, any individual of a given genetic lineage has a “vulnerability” to a disorder caused by new mutation in that lineage. We can speak of the “naive genetic lineage” of the zygote as that which is inherited from the grandparents before the action of any mutation acquired during passage through the parental germline. We then define the number of individual vulnerability genes as the number of genes which if disrupted (either in the parental germline or by early somatic mutation after the zygote is formed) will result in the development of the disorder. The size of individual vulnerability is not the same as the target size of autism genes because the former depends on genetic background and future history. Children do not necessarily have the same set of vulnerability genes. The average individual vulnerability over a population can be measured from the ratio of number of de novo LGD events in probands and siblings, as follows.
We will solve for the general case. Assume the rate for a given mutation class in unaffecteds is R, and the rate in probands is AR. In a population of size P, roughly RP mutations of that class will occur, neglecting the small surplus coming from the small number of affected individuals. The number of affected individuals will be P / N, where 1 / N is the incidence in the population. Thus, ARP / N mutations of the class will be found in affecteds. RP / N of these will be present by chance and not contributory, whereas (A – 1)RP / N events are contributory. Thus the proportion of all de novo mutations in a population of size P that contribute to the condition is
S is the probability that a de novo mutation of the particular class will contribute to the condition, and S is a function only of AandN.
If each of G total genes had a uniform probability of being a target for a de novo mutation, and T was the mean number of vulnerability genes per affected, and mutations of the class were completely penetrant, we also have S = T / G, so
Now, for LGD in autism, taking N = 150, A = 2 and G = 25,000, we can compute the average individual vulnerability per child as 167 genes.
This of course is only a crude argument because genes do not have a uniform mutation rate, and not every LGD in a target gene will have complete penetrance. Nevertheless we make note that the size of individual vulnerability appears to be roughly half the target size of all autism genes (see last section of the Discussion).
Candidate Genes
Other than NRXN1, we did not see any genes among the detected de novo LGD targets that had been conclusively linked to ASD (independent of FMR1 association), although CTTNBP2 (encoding a cortactin-binding protein) was suggested as a potential candidate for the autism susceptibility locus (AUTS1) at 7q31 (Cheung et al., 2001). We now provide evidence, based on a de novo 2 bp frame shift deletion, that mutations in CTTNBP2 may cause ASD. In addition, a number of other candidates stood out as being potentially causal due to a combination of provocative expression patterns, known roles in human disease and suggestive mouse mutant phenotypes. Among these were RIMS1, a Ras superfamily member necessary for presynaptic long-term potentiation (Castillo et al., 2002). A targeted Rims1 mutation in the mouse leads to increased postsynaptic density and impaired associative learning as well as memory and cognition deficits (Powell et al., 2004; Schoch et al., 2002), and the frame shift allele we found may lead to a similarly severe condition. Another intriguing candidate was the serine/threonine-specific protein kinase DYRK1A, which is located within the Down syndrome critical region of chromosome 21 and believed to underlie at least some of the pathogenesis of Down syndrome as a consequence of increased dosage. Several reports of likely inactivating mutations in DYRK1A result in symptoms including developmental delay, behavioral problems, impaired speech and mental retardation (Møller et al., 2008; van Bon et al., 2011), and a heterozygous knockout in the mouse also led to developmental delay and increased neuronal densities (Fotaki et al., 2002). Truncating mutations in ZFYVE26 (encoding a zinc finger protein) are known to cause autosomal recessive spastic paraplegia-15, consisting of lower limb spasticity, cognitive deterioration, axonal neuropathy and white matter abnormalities (Hanein et al., 2008). It is possible that a heterozygous truncating mutation such as the de novo frame shift allele found in our study might cause a less severe version of this condition resulting in an ASD diagnosis. Other de novo mutations of interest were a 4 bp deletion in DST (encoding the basement membrane glycoprotein dystonin), which is associated with FMRP (Darnell et al., 2011) and produces a neurodegeneration phenotype when inactivated in the mouse, and a nonsense mutation in ANK2 (an ankyrin protein involved in synaptic stability [Koch et al., 2008]). A nonsense mutation in UNC80 has been linked to control of “slow” neuronal excitability (Lu et al., 2010).
We also note that thirteen of the 59 LGD candidates appear to be involved in either transcription regulation or chromatin remodeling. Among the latter are three proteins involved in epigenetic modification of histones: ASH1L, a histone H3/H4 methyltransferase that activates transcription (Gregory et al., 2007); KDM6B, a histone H3 demethylase implicated in multiple developmental processes (Swigut and Wysocka, 2007), and MLL5, a histone H3 methyltranserase involved in cell lineage determination (Fujiki et al., 2009). These three are also FMRP-associated genes.
Relation of Candidate Genes to FMRP-Associated Genes
Fragile X syndrome (FXS) is one of the most common genetic causes of intellectual disability, with up to 90% of affected children exhibiting autistic symptoms. This has suggested overlaying recent understanding of FXS biology onto candidate ASD genes (Darnell et al., 2011). The FMR1 gene is expressed in neurons and controls the translation of many products. A set of 842 FMRP-associated genes has been enumerated by cross-linking, immunoprecipitation, and high-throughput sequencing (HITS-CLIP), and this set was previously noted to overlap candidate genes from de novo CNVs (Darnell et al., 2011). Hence, we checked the list of FMRP-associated genes with our lists of 59 LGD targets and 72 most likely autism candidate genes from de novo CNVs, and found a remarkable overlap: 14 and 13 with one in common, thus 26/129, with a p value of 10−13 determined on a per gene basis (842 FMRP-associated genes out of 25,000 genes). This overlap is remarkable because half of the LGD targets should not be ASD related, and probably a similar number of the most likely CNV genes. We found no unusual overlap between the FMRP-associated genes and de novo LGD targets in unaffected siblings, or between FMRP-associated genes and de novo missense targets in either affected or unaffected children.
As a follow-up to this striking observation, we searched for de novo mutations in targets upstream of FMR1 and found an intriguing one: GRM5. It is hit by a deletion that is not a frame shift but removes a single amino acid and causes an additional substitution at the deletion site. GRM5 encodes mGluR5, a glutamate receptor coupled to a G protein (Bear et al., 2004). Defects in mGluR5 compensate for some of the fragile X symptoms in mice (Dölen et al., 2007), and mGluR5 antagonists are currently in clinical trial (Jacquemont et al., 2011).
Lack of LGD Variants in FMRP-Associated Genes in the Population
FMRP has been proposed to inhibit protein translation of certain critical transcripts involved in neuroplasticity, the coordinated sensitization or desensitization of neurons in response to activity. Hence, it is reasonable to suppose that the physiological mechanisms modulated by FMRP depend on protein concentration, which in turn might be sensitive to gene dosage.
Direct support for this idea comes from surveying the entire parental population for carriers of potentially disruptive gene variants. Using a well-annotated set of human genes as controls, FMRP-associated genes are strongly depleted for mutations that affect splicing or introduce stop codons. The statistical significance of the numbers is striking, whether computed as a rate relative to synonymous mutations or on a per gene basis. We see a similar depletion of LGDs in a set of human orthologs of mouse genes that are enriched for essential genes but we do not see this extreme depletion in a set of 250 genes linked to known disabling genetic disorders. This difference may reflect the strong purifying selection in humans against disruptions of even a single allele of genes in this set. The hypothesis that the majority of the FMRP-associated genes are dosage-sensitive requires a more thorough analysis.
Mediators of Neuroplasticity in Cognitive and Behavioral Disorders
FMRP may act as one component of a central regulator of synaptic plasticity, among others such as TSC2 (Darnell et al., 2011; Auerbach et al., 2011). Impairment of its function, or the components it regulates, or other regulators like it, might produce a deficit in human adaptive responses. This study shows these components may be dosage-sensitive targets in autism. By extension, neuroplasticity, the hallmark function of our nervous system that enables learning and adaption in responses to stimulation, might have a general vulnerability to mutation affecting gene dosage. Mediators of neuroplasticity could be searched profitably for involvement in other cognitive disorders.
Three Recent Studies
While our manuscript has been under review, three similar but smaller studies were published: Neale et al., 2012 (N), O’Roak et al., 2012 (O), and Sanders et al., 2012 (S). Each reported exome sequence of about 200 family trios (N) or a mixture of trios and quads (O and S). (O) and (S) report of families from the SSC collection. None of the SSC samples overlapped with ours, but unlike our random selection from the SSC, (O) was enriched for females and severely affected children, and (S) was enriched for families with > 1 normal sibling.
We summarize the findings in these papers that overlap ours: more de novo point mutation in children with older parents (all three), higher incidence in female than male probands (N), paternal origin of most de novo mutations (O), an elevated ratio (≥2:1) of de novo gene disruptions in probands versus siblings (S), no segregation distortion of rare polymorphisms from parents (S), and a de novo point mutation rate of about 2.0 × 10−8 per base pair per generation (O and N). The single point of slight disagreement concerns differential signal from de novo missense mutation, which is marginal in (S) and not evident in our data.
All groups report de novo gene disruptions (nonsense, splice, and frame shifts) in probands, 18 in (N), 33 in (O), and 17 in (S), for a total of 68. With the 59 from this study, a total of 127 hits in probands have been found. Judging from our two-fold differential rate in probands and siblings, we expect that at least half of the 127 hits, about 65, are causal. Five genes were hit twice. DYRK1A and POGZ are the new recurrences found by combining our data with theirs. With our projected differential between probands and sibling controls, these five genes that are recurrent targets of de novo disruptions in probands are almost certainly autism targets.
From our estimate of 65 causal gene disruptions and 5 recurrent gene targets, we project that the total number of dosagesensitive targets for autism is about 370 genes. We made a similar estimate from de novo CNVs (Levy et al., 2011; see Recurrence Analysis in Supplemental Experimental Procedures). With this target size, and an expected 50% increase in rate of discovery of de novo gene disruptions, similar studies of all 2800 SSC families should yield about 116 autism genes, thereby identifying unequivocally about a third of the dosage-sensitive gene targets.
The other groups did not report on the number of gene disruptions occurring within the FMRP-associated genes. However, 15 of their 68 do hit these genes, a rate similar to what we observed (14 of 59). Combining data, we now compute a p value of 2 × 10−4 that this is mere coincidence. We project that nearly half of autism target genes will be among the list of FMRP-associated genes.
EXPERIMENTAL PROCEDURES
Sample Collection
The Simons Simplex Collection (SSC) was assembled at 13 clinical centers, accompanied by detailed and standardized phenotypic analysis. The institutional review board of Cold Spring Harbor Laboratory approved this study, and written informed consent from all subjects was obtained by SFARI. Families with single probands, usually with unaffected siblings, were preferentially recruited, and families with two probands were specifically excluded (Fischbach and Lord, 2010). Bloods, drawn from parents and children (affected and unaffected) were sent to the Rutgers University Cell and DNA Repository (RUCDR) for DNA preparation. DNAs from 357 families (of 2,800 total in the collection) were used in this study for exome capture, sequencing, and analysis. We used family sets of four individuals (father, mother, proband, one unaffected sibling), referred to as “quads,” for all analyses in this study. Of a starting total of 357 families, 173 were sent to the Genome Center at Washington University (St. Louis, MO, USA) for exome capture and sequencing; the remaining 184 were processed and sequenced at CSHL. Three hundred forty-three families met coverage targets and passed gender, pedigree, and sample integrity checks. Only those 343 families were considered in this report.
Exome Capture and Sequencing
Sequence capture was performed with NimbleGen SeqCap EZ Exome v2.0, representing 36.0 Mb (approximately 300,000 exons) of the human genome (hg19 build). We used standard protocols from NimbleGen (http://www.nimblegen.com/products/lit/06403921001.pdf) with minor changes as per published procedures (Hodges et al., 2009). We made additional modifications as follows: 1 µg genomic DNA was sonicated from each individual on a Covaris E210 instrument (300 bp setting). Barcoded sequencing adapters were ligated prior to capture to allow multiplexing of samples. A total of 96 different barcodes were used; eight pools of twelve 8 nt barcodes each were created, and one pool applied to each individual. This allowed us to sequence two families (or 8 individuals) per sequencing lane. Following adaptor ligation, DNAs were purified using 0.4 volumes of AMPure XP beads (Agencourt). DNAs were then amplified for 8 cycles, and family sets (250 ng of DNA from each of 4 individuals) were pooled and captured in the same reaction. Post-capture DNAs were amplified for 15 cycles. Samples were quantitated after pre- and postcapture PCR on the Agilent 2100 Bioanalyzer and diluted to 10 nM concentration prior to loading on sequencing flow cells. All sequencing was performed on the Illumina HiSeq 2000 platform using paired-end 100 bp reads.
Validation
Candidate de novo variants were confirmed via a PCR and pooled highthroughput sequencing procedure. For each event, primers were designed using BatchPrimer3 (http://probes.pw.usda.gov/batchprimer/) according to the following conditions: primers were 18–27 nt in length; amplicons were 300 bp; and the optimal Tm of the primers was 62°C. The specificity of each primer pair was assessed by BLAST against the human genome reference sequence (hg19 build). Primers were synthesized by Integrated DNA Technologies (Coralville, IA, USA). For each of the target mutations (and for only those families of children with the candidate de novo event), we conducted four discrete PCR reactions (corresponding to the four family members). After PCR, we created four product pools, one each for fathers, mothers, probands and unaffected siblings, by combining the respective products from different families. A single adenosine nucleotide was added to the 3′ end of PCR amplicons, followed by ligation of barcoded adapters to each pool of 3′-adenylated products. All pools were then combined and enriched by 8 cycles of PCR. The resulting libraries of pooled validation products were quantitated (Agilent 2100) Bioanalyzer), loaded onto a MiSeq instrument (Illumina), and analyzed by 150 bp paired-end reads. After deconvolution of barcodes and mapping, reads were binned according to genomic location and family member.
Computational Pipeline
Additional detail for each of the following paragraphs is found in the Supplemental Information.
Sequence Analysis Pipeline
We used the standard Illumina analysis pipeline (CASAVA) with custom additions to deconvolute and trim our barcodes. We used BWA for alignment (Li and Durbin, 2009) and GATK (McKenna et al., 2010) for refinements. SNV and indel variant callers were based on the same core statistical model, the Multinomial Model. We established databases of parental genotypes and all allele read counts. Filter thresholds were the parameter settings for the multinomial model. Local microassembly was added for further computational validation.
We used two databases of gene models to assign mutational effects to the identified de novo and inherited variants: the UCSC genes and the CCDS sets of gene models, both downloaded from the UCSC Genome Bioinformatics website (http://genome.ucsc.edu). The UCSC database was more comprehensive but with potentially more noise, as opposed to the CCDS that contained fewer but high-confidence gene models. Variants that altered the 2 bp at the beginning or end of each intron were classified as “splice sites.”
Multinomial Model
Our approach has been to genotype the members of a family all at the same time, not individually, thus using all the family information and the exploiting the uniformity of data that results from barcoding and pooled capture and sequencing. In our computations, we use a simple error model in which the allele in a read may be incorrectly called once per hundred times, and is assumed independent for family member and position. The genotypes are assigned based on the likelihood of the 81 possible states given a two allele standard autosomal model, and fewer states when assessing the X chromosome. The de novo score (denovoScr) is based on an aggregation of the posterior probabilities of the states which obey Mendelian segregation rules, and is scaled as the negative log10. This score is used for one filter threshold. A p value is also determined at every locus for the best state from a “goodness of fit” χ2 test, based on an analytical multinomial distribution with 8 outcomes. The p value is used as another filter threshold.
Counting SNVs/Indels
For the de novo and Mendelian SNV genotype calls, we only considered reads that had been mapped with quality of at least 30 (Li and Durbin, 2009) and that had not been flagged as PCR duplicates, and we counted only bases whose recalibrated base quality was at least 20. The reference allele was set to the nucleotide found in the reference genome, and the alternative allele was set to the non-reference allele with the largest count.
For every read that BWA aligned with a gap, we generated one or more candidate indel variants. A candidate indel is characterized with a position in the reference genome coordinates, whether an insertion or a deletion, as well as a length. We then counted the number of reads that supported a particular candidate indel variant and the number of reads that overlapped the candidate position but did not support the same variant.
Filters: De Novo SNV Genotype Calls
We only considered de novo candidates with denvoScr > = 60 and . The choice of 60 was dictated by a desire to keep false positives to a minimum, and we determined it by computing the proportion of polymorphic loci that appear as de novo candidates as a function of the score (Figure S1). We introduced additional filter criteria to suppress false positives: we only accepted candidates for which the parents were homozygous for the reference allele and that were not at polymorphic or noisy positions. This comprises our “SNV filter.” Further details are found in the Supplemental Information.
Filters: De Novo Indel Genotype Calls
We applied two filters for the indel caller. For the “SNV filter” applied to indels, we used the same settings for denovoScr and , but to control for polymorphism and noise, we used a simple approach of filtering candidates for which the same indel was seen in more than 200 reads from the entire data set. For the “Indel filter,” we substantially relaxed the denovoScr and requirements to 30 and 10−9 but insisted on having clean counts: parents were not allowed to have any reads with the candidate indel and were required to have at least 15 reads supporting the reference allele. At least one of the children had to have 6 or more reads with the candidate allele comprising at least 5% of her reads. We also strengthened the population requirement by filtering positions with more than 100 reads containing the candidate indel in the entire data set outside of the family.
Microassembly Validation Pipeline
Most de novo SNV mutations and all indels passing the filters were tested using the microassembly pipeline. The basic steps of the microassembly method are as described by Pevzner et al., 2001 (using de Bruijn graphs). Reads were decomposed into overlapping k-mers, and directed edges were added between k-mers that were consecutive within any read. All reads mapping within 100 bp of the candidate mutation, as well as reads that did not map to the assembly but were indicated to have been within 100 bp of the candidate mutation by their mates, were included. Paths were partitioned by family member.
Significance Tests for Overlap with FMRP-Associated Genes
Likely Gene Disruptors (LGD)
Under the assumption that SNV and indel variants occur randomly across coding regions, larger genes will be more likely to accumulate higher numbers of such variants. In addition, (1) the proportion of a gene that is included in the design of the capture reagents and (2) nonuniform capture coverage across the target will influence the expected numbers of variants of a gene or group of genes. To address these issues, we based our expectation of number of variants per gene (or group of genes) on the distribution of observed rare synonymous mutations in the 686 parents. Specifically, 7,051 of the 63,080 (or 11.18%) of all rare synonymous SNVs fell within the FMRP-associated genes (Table 6). We set 11.18% as the expected proportion for LGD variants in FMRP-associated genes and used a binomial test to assign p values to the observed overlap between FMRP-associated genes and LGD variants in probands and their unaffected siblings and assigned p values for the overlap with missense variants similarly (Table 5).
CNV Candidates
CNV candidate genes (Gilman et al., 2011) were obtained through a greedy optimization procedure that selected the most interconnected (in the context of a whole genome molecular network) subset of genes from the set of genes affected by a de novo deletion or duplication in autistic probands. The molecular network utilized cumulative expert and experimental knowledge that was heavily biased toward what had been studied such that it was difficult to accurately quantify. To measure the significance of the observed overlap between the 72 CNV candidates and the FMRP-associated genes, we performed a permutation test: random CNV regions were selected, preserving the number of genes as in the real CNVs, the 72 most interconnected genes were identified using the greedy optimization (allowing at most 2 genes per CNV region), and the overlap with FMRP-associated genes was recorded (Gilman et al., 2011). We repeated this procedure 10,000 times and built an empirical distribution for the number of FMRP-associated genes if the CNVs were taken as random. Only 4 of 10,000 permutations produced an overlap equal to or larger than the observed 13 FMRP-associated genes.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the Simons Foundation (SF51 and SF235988) to M.W. and by a grant from the NIH (5RC2MH090028-02) to M.W. and W.R.M. We are grateful to all of the families at the participating SFARI Simplex Collection (SSC) sites, as well as the principal investigators (A. Beaudet, R. Bernier, J. Constantino, E. Cook, E. Fombonne, D. Geschwind, D. Grice, A. Klin, R. Kochel, D. Ledbetter, C. Lord, C. Martin, D. Martin, R. Maxim, J. Miles, O. Ousley, B. Pelphrey, B. Peterson, J. Piggot, C. Saulnier, M. State, W. Stone, J. Sutcliffe, C. Walsh, and E. Wijsman). The DNA samples used in this work are included within SSC release 13. Approved researchers can obtain the SSC population dataset described in this study by applying at https://base.sfari.org. We also thank Gerald Fischbach, Marian Carlson, Cori Bargmann, Richard Axel, Mark Bear, Catherine Lord, Matthew State, Stephan Sanders, Seungtai Yoon, David Donoho, and Jim Simons for helpful discussions.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes one figure, three tables, and Supplemental Experimental Procedures and can be found with this article online at doi:10.1016/j.neuron.2012.04.009.
REFERENCES
- Auerbach BD, Osterweil EK, Bear MF. Mutations causing syndromic autism define an axis of synaptic pathophysiology. Nature. 2011;480:63–68. doi: 10.1038/nature10658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awadalla P, Gauthier J, Myers RA, Casals F, Hamdan FF, Griffing AR, Côté M, Henrion E, Spiegelman D, Tarabeux J, et al. Direct measure of the de novo mutation rate in autism and schizophrenia cohorts. Am. J. Hum. Genet. 2010;87:316–324. doi: 10.1016/j.ajhg.2010.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear MF, Huber KM, Warren ST. The mGluR theory of fragile X mental retardation. Trends Neurosci. 2004;27:370–377. doi: 10.1016/j.tins.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Blake JA, Bult CJ, Kadin JA, Richardson JE, Eppig JT Mouse Genome Database Group. The Mouse Genome Database (MGD): premier model organism resource for mammalian genomics and genetics. Nucleic Acids Res. 2011;39(Database issue):D842–D848. doi: 10.1093/nar/gkq1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo PE, Schoch S, Schmitz F, Südhof TC, Malenka RC. RIM1alpha is required for presynaptic long-term potentiation. Nature. 2002;415:327–330. doi: 10.1038/415327a. [DOI] [PubMed] [Google Scholar]
- Cheung J, Petek E, Nakabayashi K, Tsui LC, Vincent JB, Scherer SW. Identification of the human cortactin-binding protein-2 gene from the autism candidate region at 7q31. Genomics. 2001;78:7–11. doi: 10.1006/geno.2001.6651. [DOI] [PubMed] [Google Scholar]
- Ching MS, Shen Y, Tan WH, Jeste SS, Morrow EM, Chen X, Mukaddes NM, Yoo SY, Hanson E, Hundley R, et al. Children’s Hospital Boston Genotype Phenotype Study Group. Deletions of NRXN1 (neurexin-1) predispose to a wide spectrum of developmental disorders. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2010;153B:937–947. doi: 10.1002/ajmg.b.31063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad DF, Keebler JE, DePristo MA, Lindsay SJ, Zhang Y, Casals F, Idaghdour Y, Hartl CL, Torroja C, Garimella KV, et al. 1000 Genomes Project. Variation in genome-wide mutation rates within and between human families. Nat. Genet. 2011;43:712–714. doi: 10.1038/ng.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dölen G, Osterweil E, Rao BS, Smith GB, Auerbach BD, Chattarji S, Bear MF. Correction of fragile X syndrome in mice. Neuron. 2007;56:955–962. doi: 10.1016/j.neuron.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman I, Rzhetsky A, Vitkup D. Network properties of genes harboring inherited disease mutations. Proc. Natl. Acad. Sci. USA. 2008;105:4323–4328. doi: 10.1073/pnas.0701722105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach GD, Lord C. The Simons Simplex Collection: a resource for identification of autism genetic risk factors. Neuron. 2010;68:192–195. doi: 10.1016/j.neuron.2010.10.006. [DOI] [PubMed] [Google Scholar]
- Fotaki V, Dierssen M, Alcántara S, Martínez S, Martí E, Casas C, Visa J, Soriano E, Estivill X, Arbonés ML. Dyrk1A haploinsufficiency affects viability and causes developmental delay and abnormal brain morphology in mice. Mol. Cell. Biol. 2002;22:6636–6647. doi: 10.1128/MCB.22.18.6636-6647.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki R, Chikanishi T, Hashiba W, Ito H, Takada I, Roeder RG, Kitagawa H, Kato S. GlcNAcylation of a histone methyltransferase in retinoic-acid-induced granulopoiesis. Nature. 2009;459:455–459. doi: 10.1038/nature07954. [DOI] [PubMed] [Google Scholar]
- Gilman SR, Iossifov I, Levy D, Ronemus M, Wigler M, Vitkup D. Rare de novo variants associated with autism implicate a large functional network of genes involved in formation and function of synapses. Neuron. 2011;70:898–907. doi: 10.1016/j.neuron.2011.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory GD, Vakoc CR, Rozovskaia T, Zheng X, Patel S, Nakamura T, Canaani E, Blobel GA. Mammalian ASH1L is a histone methyltransferase that occupies the transcribed region of active genes. Mol. Cell. Biol. 2007;27:8466–8479. doi: 10.1128/MCB.00993-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanein S, Martin E, Boukhris A, Byrne P, Goizet C, Hamri A, Benomar A, Lossos A, Denora P, Fernandez J, et al. Identification of the SPG15 gene, encoding spastizin, as a frequent cause of complicated autosomal-recessive spastic paraplegia, including Kjellin syndrome. Am. J. Hum. Genet. 2008;82:992–1002. doi: 10.1016/j.ajhg.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges E, Rooks M, Xuan Z, Bhattacharjee A, Benjamin Gordon D, Brizuela L, Richard McCombie W, Hannon GJ. Hybrid selection of discrete genomic intervals on custom-designed microarrays for massively parallel sequencing. Nat. Protoc. 2009;4:960–974. doi: 10.1038/nprot.2009.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquemont S, Curie A, des Portes V, Torrioli MG, Berry-Kravis E, Hagerman RJ, Ramos FJ, Cornish K, He Y, Paulding C, et al. Epigenetic modification of the FMR1 gene in fragile X syndrome is associated with differential response to the mGluR5 antagonist AFQ056. Sci. Transl. Med. 2011;3:64ra61. doi: 10.1126/scitranslmed.3001708. [DOI] [PubMed] [Google Scholar]
- Koch I, Schwarz H, Beuchle D, Goellner B, Langegger M, Aberle H. Drosophila ankyrin 2 is required for synaptic stability. Neuron. 2008;58:210–222. doi: 10.1016/j.neuron.2008.03.019. [DOI] [PubMed] [Google Scholar]
- Levy D, Ronemus M, Yamrom B, Lee YH, Leotta A, Kendall J, Marks S, Lakshmi B, Pai D, Ye K, et al. Rare de novo and transmitted copy-number variation in autistic spectrum disorders. Neuron. 2011;70:886–897. doi: 10.1016/j.neuron.2011.05.015. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B, Zhang Q, Wang H, Wang Y, Nakayama M, Ren D. Extracellular calcium controls background current and neuronal excitability via an UNC79-UNC80-NALCN cation channel complex. Neuron. 2010;68:488–499. doi: 10.1016/j.neuron.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, Skaug J, Shago M, Moessner R, Pinto D, Ren Y, et al. Structural variation of chromosomes in autism spectrum disorder. Am. J. Hum. Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller RS, Kübart S, Hoeltzenbein M, Heye B, Vogel I, Hansen CP, Menzel C, Ullmann R, Tommerup N, Ropers HH, et al. Truncation of the Down syndrome candidate gene DYRK1A in two unrelated patients with microcephaly. Am. J. Hum. Genet. 2008;82:1165–1170. doi: 10.1016/j.ajhg.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale BM, Kou Y, Liu L, Ma’ayan A, Samocha KE, Sabo A, Lin C-F, Stevens C, Wang L-S, Makarov V, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012 doi: 10.1038/nature11011. in press. 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Roak BJ, Deriziotis P, Lee C, Vives L, Schwartz JJ, Girirajan S, Karakoc E, Mackenzie AP, Ng SB, Baker C, et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat. Genet. 2011;43:585–589. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, Levy R, Ko A, Lee C, Smith JD, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012 doi: 10.1038/nature10989. in press. 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pevzner PA, Tang H, Waterman MS. An Eulerian path approach to DNA fragment assembly. Proc. Natl. Acad. Sci. USA. 2001;98:9748–9753. doi: 10.1073/pnas.171285098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, Conroy J, Magalhaes TR, Correia C, Abrahams BS, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell CM, Schoch S, Monteggia L, Barrot M, Matos MF, Feldmann N, Südhof TC, Nestler EJ. The presynaptic active zone protein RIM1alpha is critical for normal learning and memory. Neuron. 2004;42:143–153. doi: 10.1016/s0896-6273(04)00146-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, Harrow J, Harte RA, Wallin C, Diekhans M, Maglott DR, Searle S, Farrell CM, Loveland JE, Ruef BJ, et al. The consensus coding sequence (CCDS) project: Identifying a common protein-coding gene set for the human and mouse genomes. Genome Res. 2009;19:1316–1323. doi: 10.1101/gr.080531.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S, Barnett AG, Foldi C, Burne TH, Eyles DW, Buka SL, McGrath JJ. Advanced paternal age is associated with impaired neurocognitive outcomes during infancy and childhood. PLoS Med. 2009;6:e40. doi: 10.1371/journal.pmed.1000040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, Ercan-Sencicek AG, Hus V, Luo R, Murtha MT, Moreno-De-Luca D, Chu SH, Moreau MP, Gupta AR, Thomson SA, et al. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron. 2011;70:863–885. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, Willsey AJ, Ercan-Sencicek AG, DiLullo NM, Parikshak NN, Stein JL, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012 doi: 10.1038/nature10945. in press. 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaf CP, Sabo A, Sakai Y, Crosby J, Muzny D, Hawes A, Lewis L, Akbar H, Varghese R, Boerwinkle E, et al. Oligogenic heterozygosity in individuals with high-functioning autism spectrum disorders. Hum. Mol. Genet. 2011;20:3366–3375. doi: 10.1093/hmg/ddr243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch S, Castillo PE, Jo T, Mukherjee K, Geppert M, Wang Y, Schmitz F, Malenka RC, Südhof TC. RIM1alpha forms a protein scaffold for regulating neurotransmitter release at the active zone. Nature. 2002;415:321–326. doi: 10.1038/415321a. [DOI] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, Yamrom B, Yoon S, Krasnitz A, Kendall J, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swigut T, Wysocka J. H3K27 demethylases, at long last. Cell. 2007;131:29–32. doi: 10.1016/j.cell.2007.09.026. [DOI] [PubMed] [Google Scholar]
- van Bon BW, Hoischen A, Hehir-Kwa J, de Brouwer AP, Ruivenkamp C, Gijsbers AC, Marcelis CL, de Leeuw N, Veltman JA, Brunner HG, de Vries BB. Intragenic deletion in DYRK1A leads to mental retardation and primary microcephaly. Clin. Genet. 2011;79:296–299. doi: 10.1111/j.1399-0004.2010.01544.x. [DOI] [PubMed] [Google Scholar]
- Vissers LE, de Ligt J, Gilissen C, Janssen I, Steehouwer M, de Vries P, van Lier B, Arts P, Wieskamp N, del Rosario M, et al. A de novo paradigm for mental retardation. Nat. Genet. 2010;42:1109–1112. doi: 10.1038/ng.712. [DOI] [PubMed] [Google Scholar]
- Xu B, Roos JL, Dexheimer P, Boone B, Plummer B, Levy S, Gogos JA, Karayiorgou M. Exome sequencing supports a de novo mutational paradigm for schizophrenia. Nat. Genet. 2011;43:864–868. doi: 10.1038/ng.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Leotta A, Kustanovich V, Lajonchere C, Geschwind DH, Law K, Law P, Qiu S, Lord C, Sebat J, et al. A unified genetic theory for sporadic and inherited autism. Proc. Natl. Acad. Sci. USA. 2007;104:12831–12836. doi: 10.1073/pnas.0705803104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.