Abstract
Hepatitis C virus (HCV) contributes substantially to human morbidity and mortality world-wide. The development of HCV genomes expressing heterologous proteins has enhanced the ability to study viral infection, but existing systems have drawbacks. Recombinant viruses often require adaptive mutations to compensate for reduced viral titers, or rely on an artificial genomic organization that uncouples viral protein expression from recombinant gene expression. Here, we sought to exploit the viral polyprotein processing machinery to express heterologous proteins within the context of the HCV polyprotein. We show that HCV genotypes 2a and 1b permit insertion of reporter proteins between NS5A and NS5B with minimal impact on viral fitness. Using this strategy we constructed reporter genomes exhibiting a wide dynamic range, simplifying analysis of HCV infection in primary hepatocytes. Expression of heterologous proteins within the HCV genome offers new opportunities to analyze HCV infection in experimental systems without perturbing functions of individual viral proteins.
Keywords: hepatitis C virus, reporter viruses, hepacivirus, polyprotein processing, NS3-4A protease, viral genome organization
INTRODUCTION
Despite the advent of improved antiviral therapy HCV remains a major medical problem with at least 130 million chronic carriers world-wide. Progress in generating more effective therapeutics has been delayed by the difficulty of studying HCV in cell culture. HCV was identified as the etiologic agent causing non-A/non-B hepatitis in 1989 (Choo et al., 1989) but it was not until a decade later that it became possible to study HCV RNA replication in cell culture using HCV replicons, HCV RNA genomes harboring dominant selectable markers (Blight et al., 2000; Lohmann et al., 1999). In 2005 it was demonstrated for the first time that a unique HCV isolate derived from a Japanese patient with fulminant hepatitis, termed JFH1, could complete the entire viral life-cycle in vitro (Lindenbach et al., 2005; Wakita et al., 2005; Zhong et al., 2005). Despite considerable effort, it has proven difficult to identify additional genomes, which, like JFH1, robustly replicate in cell culture. Based on cDNA clones of the HCV Hutchinson strain, which generated RNA transcripts shown to be infectious in vivo (Kolykhalov et al., 1997), a genotype 1a infection system (H77S) has been generated (Yi et al., 2006). The H77S genome contains multiple cell culture-adaptive mutations distributed throughout the nonstructural protein-coding region which permit robust RNA replication in human hepatoma cells and moderate titers of cell culture-infectious virus, although at significantly lower levels than JFH1 (Yi et al., 2006). The most commonly used tissue culture systems supporting HCV infection rely heavily on the human hepatoma cell line Huh7 and its derivatives (Blight et al., 2002a; Friebe et al., 2005; Zhong et al., 2005), originally derived from a 57 year-old Japanese male patient with a well-differentiated liver cancer (Nakabayashi et al., 1982). A few other cell lines support HCV RNA replication to some extent, including LH86 cells (Zhu et al., 2007). Expression of the liver specific microRNA (miR) 122 renders two human hepatoma cell lines, Hep3B and HepG2 supplemented with CD81, and even a non-hepatic 293T cells supplemented with CLDN1, mir122, and apoE permissive to HCV infection (Da Costa et al., 2012; Kambara et al., 2011; Narbus et al., 2011). Hepatoma lines have certainly been instrumental for studies of the HCV lifecycle, but abnormal proliferation and deregulated gene expression of continuously growing cancer cell lines may not reflect the behavior of hepatocytes in vivo. Establishing systems for HCV replication and virus production in primary culture has proven challenging (reviewed in (Sheahan et al., 2010)): primary human hepatocytes quickly dedifferentiate in cell culture, infection is often hampered by a low dynamic range and considerable donor-to-donor variation, and primary liver tissue is scarce and costly.
The study of HCV has been greatly facilitated by the use of reporter systems to monitor various phases of the viral life cycle. This has been accomplished both by the manipulation of HCV-permissive cell culture systems (Jones et al., 2010) and by the generation of recombinant HCV genomes expressing heterologous proteins such as fluorescent markers (Jones et al., 2010), luciferases (Gottwein et al., 2011b), and Cre recombinase (Dorner et al., 2011). Though recombinant genomes are more versatile with respect to the choice of culture system and reporter model, a major caveat is that recombinant HCV genomes are often less fit than wild-type strains, and may require significant adaptation in order to reach sufficiently high titers for experimentation (Gottwein et al., 2011b). It was recently shown that heterologous proteins can be fused within Domain III of NS5A, but only yield sufficient titers when they acquire in-frame adaptive deletions in NS5A Domain II through passaging (Gottwein et al., 2011a). A different approach for overcoming fitness losses conferred by gene insertion employs a bicistronic model in which the viral proteins are translated from an encephalomyocarditis virus internal ribosomal entry site (EMCV-IRES), while the reporter gene is driven from ribosomal binding to the HCV 5’ untranslated region (5’UTR) (Jones et al., 2007). Such bicistronic viruses may not require adaptation, but viral proteins are translated through unnatural non-HCV mechanisms. The artificial nature of HCV genomes requiring either adaptation or the use of a second cistron highlight the need for alternative strategies for generating robust, non-adapted, monocistronic recombinant HCV genomes.
HCV contains a single-stranded plus-strand RNA genome in which the 5’ untranslated region (5’UTR) serves as a ribosomal entry site from which all viral proteins are translated in a single continuous polyprotein in the order NH2-C-E1-E2-p7-NS2-NS3-NS4A-NS4B-NS5A-NS5B-COOH (Bartenschlager and Lohmann, 2000). Polyprotein processing occurs by multiple mechanisms: endogenous signal peptidases mediate cleavage of structural proteins core, E1, E2 and the p7-NS2 junction including signal-peptide peptidase cleavage of core from the E1 signal peptide (Lindenbach and Rice, 2005), while processing of nonstructural proteins occurs by a combination of autoproteolytic and trans-cleavage processes. The present model (reviewed in (Morikawa et al., 2011)) suggests that dimerized NS2, together with the N-terminal domain of NS3, mediates autoproteolytic cleavage of NS2 cleavage from NS3. NS3 in cis cleaves NS4A from itself and NS4B, followed by the association of NS4A with the N-terminus of NS3 to form the NS3-4A protease complex (Kolykhalov et al., 1994). NS3-4A is responsible for cleavage at the NS4B/NS5A and NS5A/NS5B junctions in trans (Grakoui et al., 1993; Kolykhalov et al., 1994). The NS3-4A protease is also known to cleave at least two cellular targets, Toll-like receptor 3 adaptor protein (TRIF) (Li et al., 2005a), mitochondrial antiviral signaling protein (MAVS) (Li et al., 2005b; Meylan et al., 2005), to blunt the innate immune response to viral infection and the action of T-cell protein tyrosine phosphatase (TC-PTP) which results in EGF-induced signal transduction (Brenndorfer et al., 2009). The specific molecular determinants of both cellular and viral cleavage targets of the NS3-4A protease have started to emerge (Bartenschlager et al., 1994; Lin et al., 1994; Romano et al., 2011) (and reviewed in (Morikawa et al., 2011)). We sought to use these known sequences to exploit normal HCV polyprotein processing machinery for expression of heterologous proteins within the context of a monocistronic HCV genome.
Here we report a method for the generation of recombinant HCV genomes on a monocistronic background without the need for viral adaptation through passaging. The NS3-4A protease complex cleaves NS5A from NS5B through specific recognition of the amino acid region spanning the seven C-terminal residues of NS5A and the two N-terminal residues of NS5B (Romano et al., 2011). We show that duplication of this cleavage motif at amino- and carboxy-termini of two reporter proteins, yellow fluorescent protein (YPet) and non-secreted Gaussia luciferase (nsGLuc), inserted between NS5A and NS5B results in normal polyprotein processing and yields high viral titers. We further demonstrate that reporter expression in this model matches or exceeds that of existing reporter genomes on both mono- and bicistronic backgrounds, and show that reporter expression exhibits a wide dynamic range that simplifies longitudinal analysis of HCV infection in primary hepatocytes. Finally, we show that this model is tolerated by multiple genotypes, including the non-JFH1-based HCV Con1 (1b). Expression of heterologous proteins in the context of the HCV polyprotein offers new opportunities to analyze HCV infection in experimental systems without perturbing the functions of individual viral proteins or disrupting normal translational regulation.
RESULTS
Recombinant HCV Genome Development
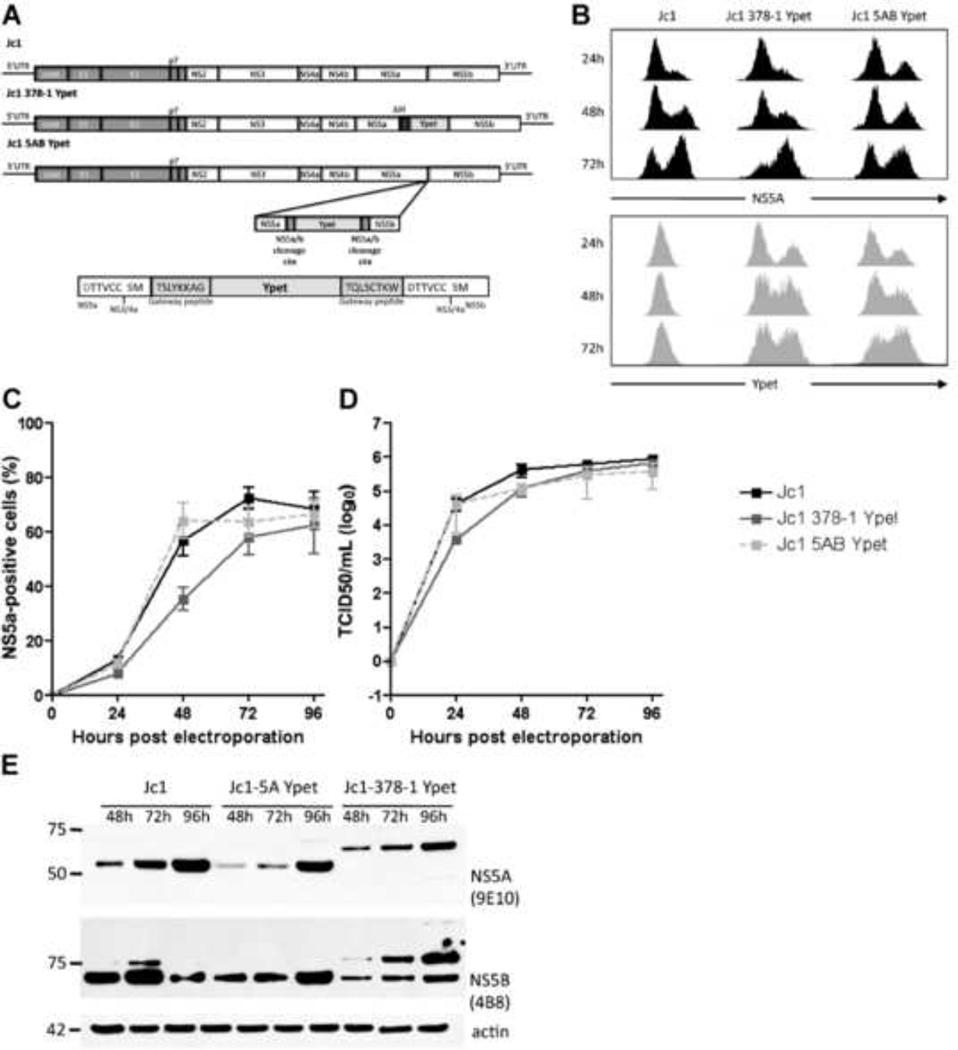
We hypothesized that duplication of the NS3-4A-dependent proteolytic cleavage site between NS5A and NS5B would permit insertion of genes of interest into the HCV genome without diminishing viral fitness. Due to restriction site limitations in JFH1-based vectors, we constructed a Gateway®-based destination vector, Jc1-5AB-DEST, from the parental Jc1 (Pietschmann et al., 2006) to facilitate gene insertion at the NS5A/5B site. The nine-amino acid sequence spanning P7-P2’ of the NS3-4A-dependent proteolytic cleavage site between NS5A and NS5B is encoded at both ends of the destination cassette (Fig.1a). All excess amino acid residues associated with Gateway® sites and gene insertion are internal to the cleavage sequences, such that NS5A and NS5B proteins should be identical to wild-type following NS3-4A-mediated polyprotein processing. From Jc1-5AB-DEST, we constructed recombinant HCV(2a) genomes of J6/JFH, Jc1, and J6/JFH Clone2 containing the enhanced yellow fluorescent protein variant YPet (Nguyen and Daugherty, 2005). To assess whether insertion of the 5AB YPet expression cassette affected viral fitness, we compared viral growth kinetics of Jc1 5AB YPet to the parental Jc1 genome. As a control we also compared another recombinant monocistronic Jc1 genome described elsewhere (Oh and Jones et al.) in which YPet is fused to NS5A Domain III and harbors an adaptive deletion of 34 amino acids in NS5A Domain II (Jc1(Δ34-378-YPet)). While the number of HCV NS5A-positive cells was similar for all three genomes one day after electroporation, the Jc1 5AB YPet genome exhibited fewer NS5A-positive cells on day two but reached levels similar to Jc1 and Jc1(Δ34-378-YPet) by day four (Fig. 1b). This suggested that insertion of YPet between NS5A and NS5B minimally impairs replicative fitness. Encouragingly, however, viral titers were very similar for Jc1, Jc1(Δ34-378-YPet) and Jc1 5AB YPet, peaking between 1×105–106 TCID50/ml (Fig. 1c). YPet expression was readily detectable by flow cytometry within 24 hours post electroporation in Huh-7.5 cells harboring the Jc1(Δ34-378-YPet) or Jc1 5AB YPet genomes (Fig. 1e). Expression of endogenous NS5A (Fig. 1d) and YPet (Fig. 1e) were similar for both reporter genomes. To determine whether NS5A and NS5B are properly processed in the Jc1 5AB reporter genome we visualized both proteins by western blot in cell lysates at 24, 48, and 72 hours following electroporation. NS5A and NS5B migrated with the expected molecular mass of approximately 56 kDa and 65 kDa, respectively (Fig. 1f). Bands indicative of incompletely processed NS5A and/or NS5B bound to YPet were not detectable for either Jc1 or Jc1 5AB Ypet. Expectedly, Jc1(Δ34-378-YPet) NS5A ran at higher molecular weight due to its fusion to YPet. These data show that YPet is efficiently cleaved from both flanking HCV proteins NS5A and NS5B in the Jc1 5AB YPet genome, indicating that YPet insertion does not impair polyprotein processing or affect the steady state levels of the flanking nonstructural proteins.
Figure 1. Gene insertion between NS5A and NS5B does not impair viral fitness.
(a) Genome structure of the Jc1(Δ34-378-YPet) and Jc1 5AB YPet illustrating the insertion of YPet either as a fusion protein with NS5A with a subsequent adaptive deletion in domain II (II) or by duplicating the NS3-4A protease cleavage site between NS5A and NS5B (5A/B CS) flanking the YPet insertion. (b) HCV replication following electroporation of in vitro transcribed RNA of Jc1, Jc1(Δ34-378-YPet) or Jc1 5AB YPet into Huh-7.5 cells as measured by flow cytometry. (c) Longitudinal virus production as measured by end-point limiting dilution following electroporation of Jc1, Jc1(Δ34-378-YPet) or Jc1 5AB YPet RNA into Huh-7.5 cells. Results shown are means ± SD from three independent experiments (b, c). (d, e) Histograms of (d) NS5A- and (e) YPet expression following electroporation of Huh-7.5.1 cells with Jc1, Jc1(Δ34-378-YPet)), or Jc1 5AB YPet. (f) NS5A/5B cleavage at different times post electroporation of Jc1, Jc1(Δ34-378-YPet) or Jc1 5AB YPet into Huh-7.5 cells as measured by Western blot against either NS5A or NS5B.
Instability of HCV reporter genomes over serial passage
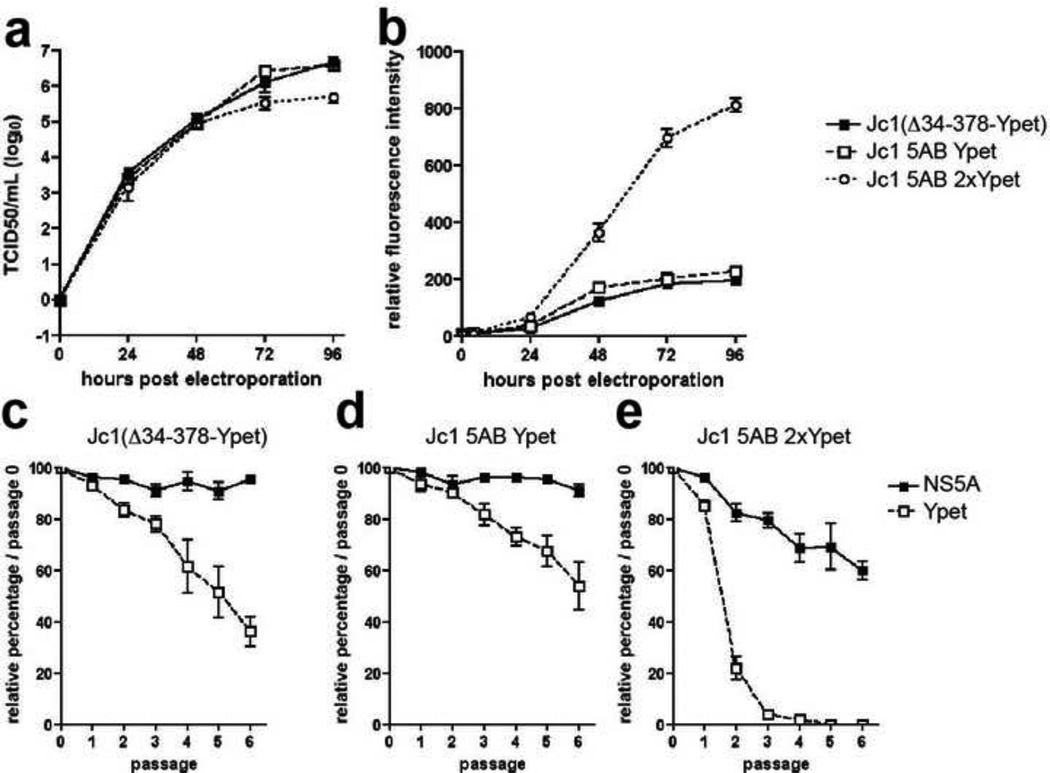
Next, we aimed to determine the stability of heterologous protein expression in recombinant monocistronic genomes through serial passage. Jc1 5AB genomes were generated expressing either one or two YPet proteins, which increased the size of the insert from 700 bp to 1500 bp (230 aa to 500 aa). Both Jc1 5AB YPet and Jc1 5AB 2xYPet genomes replicated equally efficiently (data not shown) and produced similar viral titers reaching approximately 1×106 TCID/ml (Fig. 2a). Expression of two copies of YPet increased the sensitivity of the reporter by ca. 5-fold over Jc1 5AB YPet or the Jc1(Δ34-378-YPet) – each harboring a single copy of YPet per genome - in electroporated Huh-7.5 cells (Fig. 2b). To test the stability of recombinant HCV genomes containing heterologous proteins we serially passaged Huh-7.5 cells electroporated with each of the three YPet-containing genomes. While the frequency of NS5A positive cells remained stable over time for Jc1 (Δ34-378-YPet) (Fig. 2c) and Jc1 5AB YPet (Fig. 2d), and only slightly decreased in cells infected with the Jc1 5AB 2xYPet genome (Fig. 2e), the frequency of YPet positive cells decreased rapidly after the second passage of Huh-7.5 cells harboring the Jc1 5AB 2xYPet genome. YPet fluorescence was almost completely lost by the third passage of Huh-7.5 cells replicating Jc1 5AB 2xYPet (Fig. 2e), compared with only a ca. 2-fold reduction in reporter expression for both Jc1 (Δ34-378-YPet) (Fig. 2c) and Jc1 5AB YPet after six passages (Fig. 2d). These data show that YPet insertion into the HCV genome on a monocistronic background is unstable through serial passage, and that duplication of the heterologous insert drastically increases reporter expression at the cost of substantially greater genomic instability. This finding highlights the potential utility of reporter gene duplication for expanding dynamic range and signal-to-noise ratios in single-round viral infection settings. Taken together these data suggest that despite limited genomic stability following serial passage, gene insertion at the NS5A/5B junction as we describe permits robust heterologous protein expression during short-term infection.
Figure 2. Ypet insertion into the HCV genome is unstable through serial passage.
(a) Longitudinal virus production in Huh-7.5 cells following electroporation of Jc1(Δ34-378-YPet), Jc1 5AB YPet or Jc1 5AB 2xYPet in which two copies of YPet separated by a GS-linker are inserted between NS5A and NS5B. (b) Mean fluorescence intensity of YPet following electroporation of Jc1(Δ34-378-YPet), Jc1 5AB YPet or Jc1 5AB 2xYPet into Huh-7.5 cells. (c–e) Genomic stability of (c) Jc1(Δ34-378-YPet), (d) Jc1 5AB YPet and (e) Jc1 5AB 2xYPet over multiple passages. Electroporated cells were passaged and cells were analyzed for the expression of NS5A and YPet by flow cytometry at the indicated time points. Results shown are means ± SD of three independent experiments.
Insertion of reporter genes between HCV NS5A and NS5B maintains the viral phenotype of parental genomes
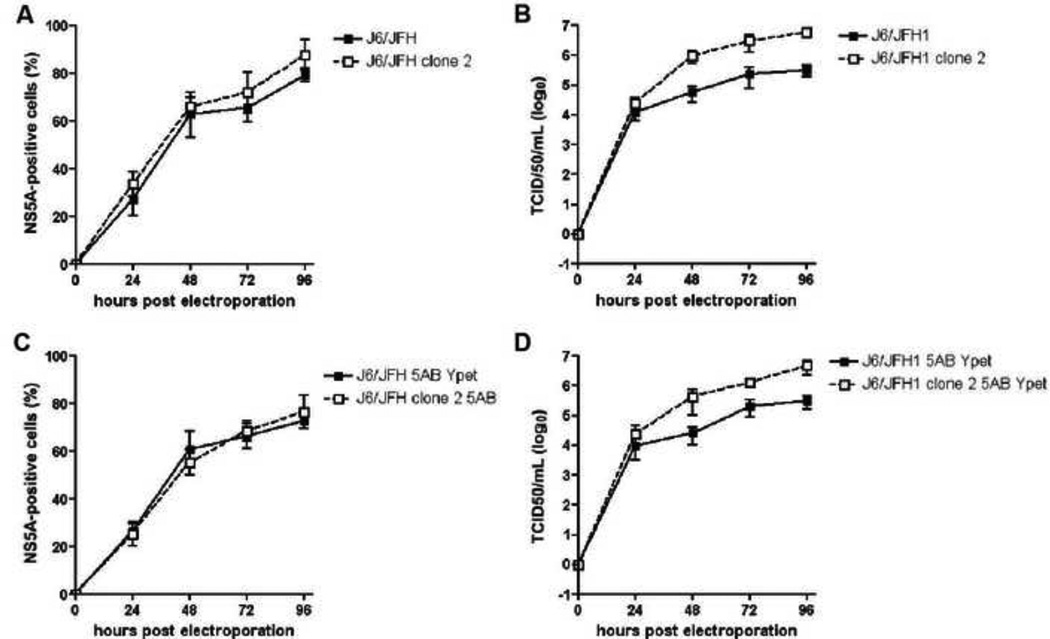
Insertion of heterologous proteins into monocistronic HCV genomes often decreases viral fitness by affecting replication efficiency, production of infectious particles and the ability to spread. Our data demonstrate that the viral fitness of the Jc1 genome is only marginally affected by insertion of YPet between NS5A and NS5B. To test whether this strategy would allow us to preserve distinct features of HCV genomes we inserted the 5AB YPet cassette into the parental J6/JFH1(Lindenbach et al., 2005) and its cell culture-adapted derivative, termed J6/JFH1 clone 2, which was shown to produce higher viral titers and is believed to spread more efficiently than J6/JFH1 or Jc1 in cell culture (Andrus et al., 2011; Walters et al., 2009). The J6/JFH1 and J6/JFH1 clone 2 genomes replicate equally efficiently following electroporation into Huh-7.5 cells irrespective of heterologous reporter gene insertion between NS5A and NS5B (Fig. 3a and c). As previously reported (Andrus et al., 2011; Walters et al., 2009) J6/JFH1 clone 2 produces approximately 10-fold higher titers than the parental J6/JFH1 genome (Fig. 3b). Importantly, the insertion of YPet between NS5A and NS5B in either J6/JFH1 or J6/JFH1 clone 2 resulted in infectivity titers similar to parental genomes, peaking around 1×105 and 1×106, respectively (Fig. 3d). J6/JFH1 clone 2 5AB YPet preserved the increased fitness of the clone 2 variant over J6/JFH1 5AB YPet, suggesting that the insertion of reporter genes between NS5A and NS5B does not affect the viral phenotype of parental genomes.
Figure 3. Gene insertion between NS5A/5B does not alter the viral phenotypes of distinct HCV genomes.
(a, c) Replication, as measured by flow cytometry of NS5A and (b, d) longitudinal virus production of J6/JFH1, J6/JFH1 clone 2, J6/JFH1 5AB YPet and J6/JFH1 clone 2 5AB YPet. Results shown are means ± SD of three independent experiments.
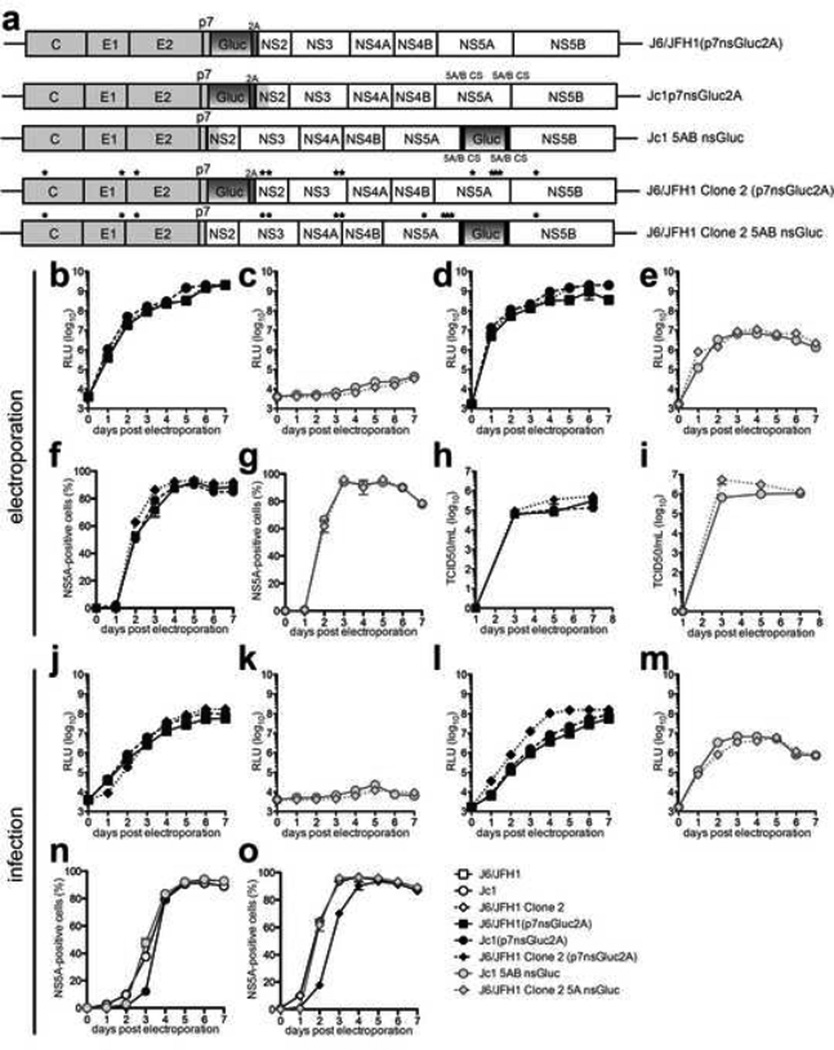
To further explore the utility of this genome configuration we compared genomes expressing Gaussia luciferase (Gluc) between either p7 and NS2 (Jc1(p7nsGluc2A) (Marukian et al., 2008), J6/JFH1 (p7nsGluc2A), J6/JFH1 clone 2(p7nsGluc2A)) or between NS5A and NS5B (Jc1 5AB nsGluc, J6/JFH1 clone 2 5AB nsGluc) (Fig. 4a). Following electroporation of RNA into Huh-7.5 cells the frequency of NS5A positive cells was indistinguishable between the three p7nsGluc2A genomes (Fig. 4f) and all had similar infectivity titers peaking around 1×105 – 1×106 TCID50/ml (Fig. 4h). The percentage of NS5A positive cells was also similar for Huh-7.5 cells electroporated with Jc1 5AB nsGluc or J6/JFH1 clone 2 5AB nsGluc RNA (Fig. 4g) but reached higher peak titers than the p7nsGluc2A genomes, ranging from 1×106 – 1×107 TCID50/ml (Fig. 4i). Very robust Gluc activity was detectable in both the supernatants (Fig. 4b) and lysates (Fig. 4d) of cells electroporated with all of the p7nsGluc2A genomes and increased over time proportionally to the number of NS5A+ cells (Fig. 4f). In cells replicating the 5AB nsGluc genomes reporter activity was only detectable in the cell lysates (Fig. 4e) but not in the supernatants (Fig. 4c). These results were largely upheld following infection of Huh-7.5 cells with a multiplicity of infection (MOI) of 0.1 of each of the respective genomes (Fig. 4j–o). Interestingly, however, the frequency of NS5A positive cells was lower at days 1 to 3 for J6/JFH1 clone 2(p7nsGluc2A) relative to both J6/JFH1 clone 2 and J6/JFH1 clone 2 5AB nsGluc (Fig. 4o) indicating that the p7nsGluc2A insertion leads to a loss of the clone 2-associated viral phenotype. This suggests that insertion of heterologous proteins between NS5A and NS5B may be more suitable than insertions at other positions of the genome for maintaining the specific viral characteristics of a given HCV genome.
Figure 4. Insertion of Gaussia luciferase between NS5A/5B retains characteristics of parental genomes.
(a) Schematic representations of reporter genomes harboring Gaussia luciferase (Gluc) either followed by a FMDV 2A site inserted between p7 and NS2 (p7nsGluc2A) or Gluc between NS5A/5B (5AB nsGluc). (b–e, j–m) Gaussia luciferase expression in (b, c, j, k) supernatants or (d, e, l, m) cell lysates, (f, g, n, o) NS5A-expression and (h, i) titers of the indicated genomes following (b–i) electroporation of Huh-7.5.1 cells with in vitro transcribed RNA or (j–o) infection of Huh-7.5 cells with MOI=0.1. Data shown are mean ± SD of three to four independent experiments.
Characterization of luminescent 5AB reporter genomes in human primary hepatocyte cultures
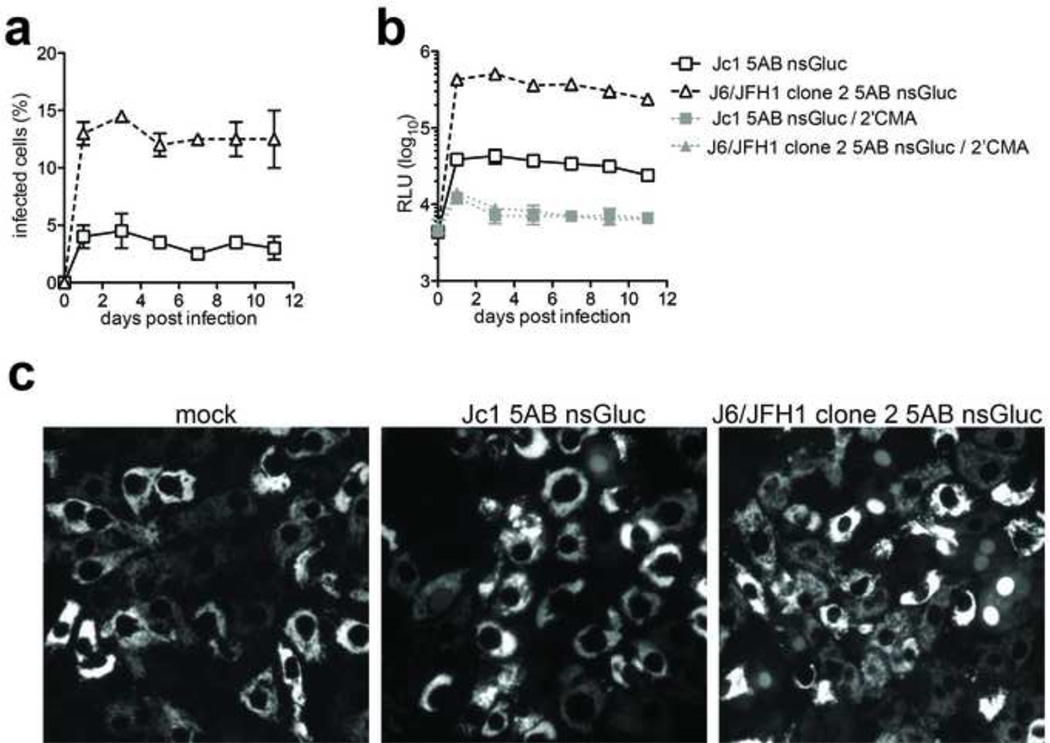
To further analyze the utility of the Gaussia luciferase 5AB reporter insertion we monitored infection in primary hepatocyte cultures. We have previously demonstrated that a monocistronic secreted Gluc reporter inserted between p7 and NS2 (Jc1p7nsGluc2A) (Marukian et al., 2011) enables sensitive detection of HCV RNA replication in both micropatterned primary hepatocyte cocultures (Jones et al., 2010; Ploss et al., 2010) and human fetal liver cultures (Andrus et al., 2011; Marukian et al., 2011) despite the low infection frequency of 1–3% with the Jc1 genome (Jones et al., 2010; Ploss et al., 2010). J6/JFH1 clone 2 is capable of spreading more robustly than Jc1 in primary hepatocytes as visualized in real time (Andrus et al., 2011) by monitoring of infection using a cell-based fluorescent reporter system that allows sensitive detection of individual HCV-infected cells (Jones et al., 2010).
Based on our previous data, we hypothesized that the difference in the ability to spread in primary hepatocyte cultures between Jc1 and J6/JFH1 clone 2 should be reflected by a greater dynamic range of a luminescent reporter when inserted between NS5A and NS5B (Fig. 5a). HCV infections in primary hepatocytes even with highly sensitive reporter genomes are usually characterized by a low signal to noise ratio in part due exacerbated antiviral signaling, which limits the number of infected cells in a culture and presumably keeps the level of replication per cell at a low level (Andrus et al., 2011; Marukian et al., 2011). In contrast to the minimal differences observed between the Jc1 and J6/JFH1 clone 2 genomes harboring the 5AB nsGluc insertions in highly permissive Huh-7.5 cells (Fig. 4n and o), J6/JFH1 clone 2 5AB nsGluc infected 3–4 fold more cells than Jc1 5AB nsGluc in human fetal liver cells transduced with the TagRFPnlsMAVS reporter (Jones et al., 2010) as measured by nuclear translocation of the TagRFP signal (Fig. 5a and c). This higher infection frequency resulted in a ca. 10-fold difference in luciferase signal between the two genomes (Fig. 5b). A signal-to-noise ratio of ca. 50:1 was observed for J6/JFH1 clone 2 5AB nsGluc, compared with 5:1 for Jc1 5AB nsGluc, both measured against infected cultures treated with the NS5B polymerase inhibitor 2’ C-methyl adenosine (2’CMA) (Fig. 5b).
Figure 5. Viral characteristics of HCV with Gaussia luciferase inserted between NS5A and NS5B are maintained during infection of primary human hepatocytes.
(a) Infection frequency of primary human fetal hepatocytes with Jc1 5AB nsGluc or J6/JFH1 Clone 2 5AB nsGluc as determined by nuclear translocation in cells expressing tagRFPnlsMAVS. (b) Gaussia luciferase expression in cell lysates following infection of hepatocytes with Jc1 5AB nsGluc or J6/JFH1 Clone 2 5AB nsGluc with or without the NS5B inhibitor 2’CMA. (c) Representative images of infected hepatocytes indicated by nuclear translocation of the tagRFPnlsMAVS reporter. Data shown are mean ± SD of four independent experiments.
Characterization of an HCV genotype 1b NS5A/5B Gaussia luciferase reporter genome
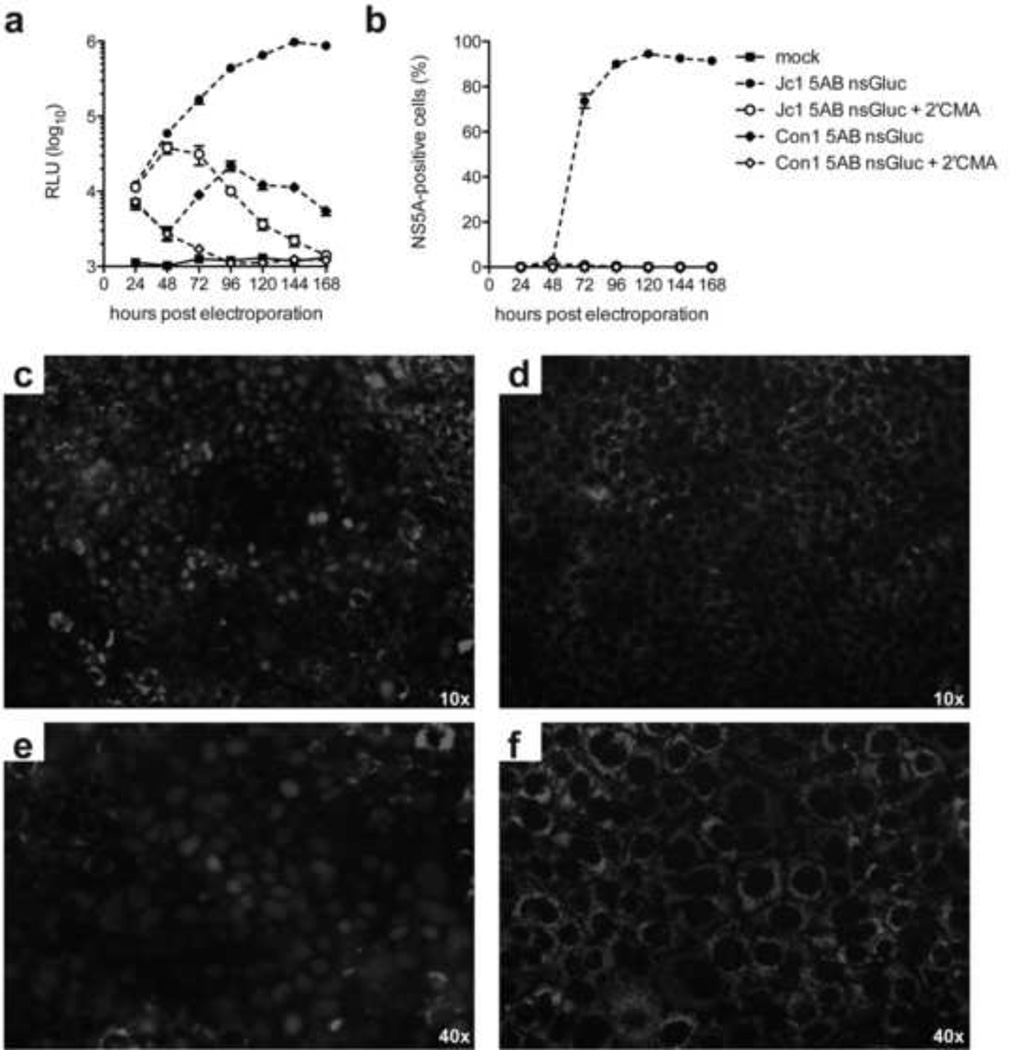
To explore whether gene insertion between NS5A/5B is tolerated by multiple HCV genotypes, we generated a recombinant full-length HCV Con1(1b) genome expressing nsGluc between duplicate Con1(1b) NS5A/5B cleavage sites (termed Con1-5AB-nsGluc) as was done for Jc1-5AB-nsGluc. Jc1 and Con1 nsGluc genomes were electroporated into Huh7.5 cells and cultured with or without 2’CMA. Luciferase expression for Con1-5AB-nsGluc was significantly elevated over the 2’CMA treated cultures (ca. 20-fold) at 2–3 days post-electroporation, indicating that the recombinant genome was able to replicate successfully. However, Con1 replicated only transiently as Gluc activity returned to background levels at day 5 post electroporation, in contrast to the Jc1-5AB-nsGluc genome which robustly replicated throughout the duration of the experiment (Fig. 6b). Given the extremely low levels of Con1 HCV RNA replication we were not able to detect HCV NS5A antigen by flow-cytometry (Fig 6a) or nuclear translocation of the TagRFPnlsMAVS reporter (Fig. 6c) both of which readily allow to quantify HCV replication in Jc1-5AB-nsGluc electroporated cells. These data demonstrate that the insertion of nsGluc in a monocistronic configuration provides a uniquely sensitive readout to monitor HCV RNA replication even in non-JFH1 derived genomes.
Figure 6. Sensitive detection of HCV RNA replication of a monocistronic HCV genotype 1b genome containing a Gaussia luciferase gene between NS5A and NS5B.
HCV replication following electroporation of in vitro transcribed RNA of Jc1 5AB nsGluc or Con1 5AB ns Gluc into Huh-7.5 cells in the presence or absence of the NS5B polymerase inhibitor 2’CMA as measured by luciferase activity in cell lysates (a) or NS5A staining and flow cytometry (b). Representative images of Huh7.5 clone 8 cells electroporated with Jc1 5AB nsGluc (c, e) or Con1 5AB ns Gluc (d, f) indicated by nuclear translocation of the tagRFPnlsMAVS reporter at 100 (c, d) or 400 (d, f) fold magnification. Data shown are mean ± SD of three independent experiments.
In summary, these data demonstrate that viral fitness is not dramatically impaired by the insertion of YPet or nsGluc between NS5A and NS5B. Importantly, expression of these heterologous proteins within the context of the viral polyprotein did not alter the distinct virological features of J6/JFH1 and its derivates Jc1 and J6/JFH1 clone 2, which could facilitate analysis of viral determinants that contribute to HCV’s ability to spread and establish persistent infection in physiological primary cell culture systems. Additionally, gene insertion between NS5A/5B was tolerated for both JFH-1 based HCV(2a) and full-length HCV Con1(1b) genomes, suggesting that this strategy is not genotype-restricted.
DISCUSSION
The present study demonstrates that native viral polyprotein processing machinery can be successfully harnessed for the purpose of generating robust recombinant HCV genomes. We have shown that monocistronic HCV(2a) genomes on the JFH1 background are permissive for gene insertion between NS5A and NS5B by duplication of the NS3-4A-dependent proteolytic cleavage site. Incorporation of heterologous proteins into the HCV polyprotein in this way gives rise to expression levels that are stoichiometrically linked to polyprotein expression. Viral protein processing was shown to occur normally in this setting, with all fusion products resulting from heterologous protein insertion being efficiently cleaved from both NS5A and NS5B by action of the NS3-4A protease.
This strategy for introducing heterologous proteins into the HCV genome may be applicable for a variety of insert types beyond the reporter molecules we have used in this study. If, as with the reporter proteins we have used here, the fusion products resulting from gene insertion are readily cleaved during polyprotein processing, insertion at the NS5A/5B locus would have significant advantages over other gene insertion methods (e.g. fusion to NS5A): inserts would be free to interact with other cellular or viral elements outside of a fusion protein context, and would be more likely to retain normal folding and function because of the lack of steric interference from fusion counterparts. It may also be possible to introduce dominant selectable markers, such as neo or blasticidin S-deaminase, into the HCV genome at the NS5A/5B locus for in vitro selection of HCV-replicating cell populations as with the replicon system. As selectable marker expression in this context would be coupled to viral polyprotein translation, such an approach could have added utility over bicistronic full-length or subgenomic selectable genomes in priming cell populations to replicate monocistronic HCVs.
Admittedly, some gene insertions may be poorly tolerated at the NS5A/5B locus. Protein inserts which occlude the NS5A/5B cleavage domain immediately upstream of NS5B may prevent proper cleavage and thereby impair polymerase activity, as could occur in the case of large proteins with sequestered C-termini or for inserts which multimerize in such a way that cleavage sites are sterically blocked. Additionally, any insert that leads to significant decompartmentalization of NS5B during initial translation, such as one that includes cellular targeting signals, may prevent sufficient interaction with viral RNAs to achieve stable replication.
The finding that heterologous protein expression is steadily lost through passage despite normal viral replication and virus particle production highlights the fragility of artificially manipulated HCV genomes. The extremely high error rate of HCV replication is likely to give rise to genomic transcripts lacking functional reporter genes, which could rapidly come to predominate. This might occur if the reporter gene confers some minor (perhaps undetectable) fitness cost, and reporter-dysfunctional mutants are selected, or because the reporter is non-essential to the viral life cycle and becomes progressively diluted by transcripts harboring loss-of-function mutations or deletions. The latter hypothesis may well explain the gradual loss of reporter strength (2-fold reduction after 6 passages) without visible fitness defects seen for both Jc1-5AB-Ypet and Jc1 (Δ34-378-YPet). In the case of Jc1-5AB-2xYpet, reporter expression is lost so quickly that the dilution hypothesis is unlikely, while multiple factors may contribute to a fitness cost. First, the sheer size of the 2xYpet insertion may reduce the efficiency of viral genome packaging and/or transcription relative to mutant transcripts that have kicked out the insert; and second, the insertion of a dimerizing fusion protein such as 2xYpet may disrupt local polyprotein folding at the NS5A/NS5B locus and thereby reduce the efficiency of proteolytic cleavage. The fitness cost hypothesis is further supported by the fact that the 2xYpet genome achieves a 10 fold lower TCID50 at 96 days relative to parental and single-Ypet genomes, and results in progressive loss of NS5A-positive cells through passage.
Although we have explored only a limited number of inserts within genotypes 2a and 1b, we have shown that recombinant genome fitness matches parental genome fitness, and that this holds across genomes with varying fitness phenotypes. Other JFH1-based genomes, such as the intergenotypic chimaeras (Gottwein et al., 2009; Scheel et al., 2008), may behave similarly to the genotype 2a intragenotypic variants tested here, and it is likely that non-JFH1-based genomes from other genotypes would be permissive for NS5A/5B gene insertion as we showed for Con1-5AB-nsGluc, The successful generation of this genotype 1 reporter genome may assist in the study of Con1(1b) and other non-2a genotypes, which have been notoriously difficult to manipulate in tissue culture with few exceptions (Yi et al., 2006). The strategy presented here may also be applicable to viruses other than HCV, such as other Flaviviridae with genomic organizations similar to that of HCV. The exploitation of viral processing mechanisms as we have shown here for HCV may prove more inocuous than existing methods for generating recombinant genomes on some virus backgrounds. In sum, the present work outlines a versatile new tool for expression of heterologous proteins in the context of monocistronic HCV genomes, one that is likely to be applicable beyond the limited examples provided in this study.
MATERIALS & METHODS
Culture of cell lines
Huh-7.5 (Blight et al., 2002b) Huh7.5 Clone 8 (Jones et al., 2010) and Huh-7.5.1 (Zhong et al., 2005) were maintained in Dulbecco’s Modified Eagle Medium (DMEM) (Invitrogen, Life Technologies, Carlsbad, CA) supplemented with 5% heat inactivated fetal bovine serum (FBS, Hyclone, Thermo Scientific, Waltham, MA), 0.1 mM nonessential amino acids (Invitrogen, Life Technologies, Carlsbad, CA).
Human Subjects
All protocols involving human tissue were reviewed and exempted by the Rockefeller University Institutional Review Board.
Isolation and Culture of human fetal liver cells (HFLCs)
Human fetal liver cells were prepared as described previously (Andrus et al., 2011). Briefly, de-identified fetal livers (16–24 weeks gestation) were procured through Advanced Bioscience Resources (ABR; Alameda, CA) or the Human Fetal Tissue Repository of the Albert Einstein College of Medicine (AECOM; Bronx, NY). Livers received on ice were washed with hepatocyte wash buffer (HWB) consisting of Williams' E Medium (WEM) plus 10 mM HEPES, 50 µg/mL gentamicin, 100 U/mL penicillin, and 100 µg/mL streptomycin (Invitrogen, Life Technologies, Carlsbad, CA). Tissue was minced then resuspended in 20–40 mL warm digestion buffer consisting of Hanks Balanced Salt Solution plus 40 mM HEPES, 3.26 mM CaCl2, 2 U/mL DNase I Grade II (Roche), and 0.2% Collagenase type IV (Sigma). Tissue was digested for 30 minutes at 37°C, then diluted 1:1 with HWB and gently pushed through 70 µm cell-strainers (BD Biosciences, San Diego, CA). The suspension was centrifuged at 100 × g for 3 minutes and the cell pellet containing large hepatocytes was washed twice by resuspension in 50 mL HWB and centrifugation at 100 × g for 4 minutes. Hepatocytes were enriched by 1 × g sedimentation in 25 mL HWB for 1 hour at room temperature, followed by additional washing. In some experiments hepatocytes were further enriched by centrifugation through lymphocyte separation medium (Cellgro, Manassas, VA) as described (Schmelzer et al., 2007). Hepatocyte yields ranged from 0.5 to 4 × 107 cells per tissue and cells were generally >80% viable as assessed by Trypan blue exclusion and collagen attachment. Hepatocytes were plated at ≈ 1 × 105/cm2 on 96-well collagen I-coated plates (BD Biosciences, San Diego, CA) in WEM containing 10% fetal bovine serum (FBS) (Omega Scientific, Tarzana, CA), 2 mM L-glutamine (Invitrogen, Life Technologies, Carlsbad, CA), 1 × ITS Plus (BD Biosciences, San Diego, CA) and antibiotics. After overnight incubation, adherent cells were washed with WEM, then maintained in Hepatocyte Defined Medium (HDM; BD Biosciences, San Diego, CA) plus L-glutamine and antibiotics. The culture medium was aspirated and replaced every 2 days.
Transduction of HFLC with TagRFPnlsMAVS HCV infection reporter
Construction of the TRIP lentiviral vector expressing the TagRFPnlsMAVS HCV reporter was described previously (Jones et al., 2010). Pseudoparticles (PPs) were generated by co-transfection of 293T cells with TRIP-TagRFPnlsMAVS, HIV gag-pol, and vesicular stomatitis virus envelope protein G (VSV-G) plasmids using a ratio of 1:0.8:0.2, as described previously (Ploss et al., 2009). HFLCs were transduced 1–3 days post-plating by incubation for 3–6 hours with PP stocks diluted 1:3 in HDM supplemented with 20 mM HEPES and 4 ug/ml polybrene, then washed and fed with HDM.
Generation of Recombinant HCV Plasmids
J6/JFH1, Jc1 and Jc1FLAG2(p7-nsGluc2A) are fully-infectious HCVcc viruses that have been previously described (Lindenbach et al., 2005; Marukian et al., 2008; Pietschmann et al., 2006). J6/JFH Clone2 is a passaged derivative of J6/JFH that contains a number of adaptive mutations that increase infectious titers (Walters et al., 2009). Con1/FL is a full-length HCV genotype 1b virus described previously (Lohmann et al., 1999).
J6/JFH1 clone2 p7nsGluc2A
To construct the reporter virus genome J6/JFH1 clone2 (p7nsGluc2A), we used the secreted Gaussia luciferase (Gluc) cassette from the Jc1FLAG(p7-nsGluc2A) genome (Marukian et al., 2008), in which the Gluc reporter, in tandem with the foot and mouth disease autoproteolytic peptide sequence (2A) is inserted between p7 and NS2. Briefly, a unique silent restriction site was introduced between the p7 and NS2 coding sequence at position 2784 (MluI) in J6/JFH1 clone 2 using overlapping PCR mutagenesis. This site was used to insert the 573 bp DNA cassette encoding the p7nsGluc2A cassette excised from Jc1FLAG(p7-nsGluc2A) into J6/JFH1 clone 2 by digestion of both vectors with MluI enzyme. Bacterial plasmid clones were screened for the correct orientation of the Gaussia luciferase cassette.
Jc1(Δ34-378-YPet)
A detailed description of the is prepared elsewhere (Oh and Jones et al. manuscript in preparation).
HCVcc containing heterologous proteins between NS5A and NS5B
We generated Gateway®-compatible destination vectors (Invitrogen, Life Technologies, Carlsbad, CA) based upon the Jc1 and Con1/FL HCV genomes, Jc1-5AB-DEST and Con1-5AB-DEST, for insertion of reporter genes between NS5A and NS5B. The 9-amino acid region spanning P7-P2’ of the NS3-4A proteolytic cleavage site between NS5A and NS5B was positioned on both ends of the destination cassette. Jc1-5AB-DEST was generated by PCR amplification of the Gateway® destination cassette, DEST (Invitrogen, Life Technologies, Carlsbad, CA), and insertion into the DraIII restriction site at the 3’ end of Jc1(2a) NS5A using standard molecular cloning techniques. Con1-5AB-DEST was similarly generated by amplification of DEST, PCR ligation to the flanking NS5A/NS5B regions of Con1/FL, and inserted into Con1/FL using available restriction sites and standard molecular cloning techniques. Jc1-5AB-YPet, Jc1-5AB-nsGLuc, Jc1-5AB-2xYPet and Con1-5AB-nsGluc were generated by PCR amplification of YPet, nsGLuc or two YPet molecules separated by a glycine-serine linker, with primers containing AttB sites for Gateway®-mediated insertion into pDONR™221, and subsequent BP and LR gateway reactions were performed using the appropriate destination vector. J6/JFH-5AB-YPet, J6/JFH Clone2-5AB-YPet, and J6/JFH Clone2-5AB-nsGLuc were generated from Jc1-5AB-YPet and Jc1-5AB-nsGLuc using compatible restriction sites.
RNA transcription
In vitro transcripts were generated as previously described (Lindenbach et al., 2005). Briefly, plasmid DNA was linearized by XbaI and purified by using a Minelute column (Qiagen, Valencia, CA). RNA was transcribed from 1 µg of purified template by using the T7 Megascript kit (Ambion, Austin, TX) or the T7 RNA polymerase kit (Promega, Madison, WI). Reaction mixtures were incubated at 37°C for 3 h, followed by a 15-min digestion with 3 U of DNase I (Ambion, Austin, TX). RNA was purified by using an RNeasy kit (Qiagen) with an additional on-column DNase treatment. RNA was quantified by absorbance at 260 nm and diluted to 0.5 µg/µl. Prior to storage at −80°C, RNA integrity was determined by agarose gel electrophoresis and visualization by ethidium bromide staining.
RNA electroporation
Huh-7.5 cells were electroporated with RNA as previously described (Lindenbach et al., 2005). Briefly, Huh-7.5.1 cells were treated with trypsin, washed twice with ice-cold RNase-free AccuGene phosphate-buffered saline (PBS) (Bio-Whittaker, Rockland, ME), and resuspended at 1.75 × 107 cells/ml in PBS. Then, 5 µg of each RNA was combined with 0.4 ml of cell suspension and immediately pulsed using a BTX ElectroSquare Porator ECM 830 (820 V, 99 µs, five pulses). Electroporated cells were incubated at room temperature for 10 min prior to resuspension in 15 ml or 30 ml complete medium for nonreporter and reporter constructs, respectively. Resuspended cells were plated into 24-well, 6-well, and P100 tissue culture dishes.
HCV infection assays
Infectious units (as median tissue culture infective dose [TCID50]) were quantified by limiting dilution titration on naïve Huh-7.5 cells (Lindenbach et al., 2005). For infection experiments in primary human fetal hepatocytes, cells were seeded in 96 well plates. On the third or fourth day after plating, human fetal hepatocytes (1 × 105/cm2) were infected with 3 × 106 TCID50 units/well of HCVcc. Inocula were left in place for 6 hours and then removed with three washes (200 µL) of Williams E media without serum and then maintained in hormonally defined medium containing indicated amounts of DMSO or 2’CMA. Replication of Gaussia luciferase expressing HCV genomes was monitored by measurement of secreted Gluc in the supernatants or Gluc in cell lysates.
Flowcytometric analysis
For flowcytometric analysis, cells were harvested using AccuMax (eBioscience) and fixed using Fixation/Permeabilization buffer (BD Biosciences, San Diego, CA) for 10 min at 4°C. Fixed cells were washed with BD Perm/Wash buffer (BD Biosciences, San Diego, CA), incubated 30 min at RT with AlexaFluor-647-conjugated 9E10 antibody (1:4000 in BD Perm/Wash buffer), washed twice with BD Perm/Wash buffer and once with FACS buffer (PBS/3%FBS) prior to analysis using a BD LSR II flow cytometer and BD FACSDiva software. Analysis of was performed using FlowJo software.
Western blotting
Cells were lysed at the indicated times using modified RIPA buffer containing 50mM Tris-HCl pH 7.4, 1% v/v NP-40, 0.25% v/w Na-deoxycholate, 150mM NaCl, 1mM EDTA, 1mM PMSF, 1mM Sodium orthovanadate and 1mM NaF for 20 minutes on ice. 15 µg of protein lysate was separated on 4–12% Bis/Tris NuPage polyacrylamide gels (Invitrogen). Proteins were transferred to nitrocellulose membranes and entry factors were detected using antibodies against NS5A (9E10; 1:5000) (Lindenbach et al., 2005), NS5B (4B8; 1:100) (Biofront Technologies) or eta-actin (AC15, 1:10000) (Sigma). Following secondary antibody staining with Peroxidase-AffiniPure Donkey Anti-Mouse IgG (H+L) (JIR, 1:10000) (Jackson Immuno Research). Western blots were visualized using SuperSignal West Pico (Thermo Scientific).
Fluorescence microscopy
Images were captured on an Axioplan 2 imaging fluorescence microscope (Zeiss, Thornwood, NY) using Metavue Software (Molecular Devices, Sunnyvale, CA). Images were processed using ImageJ software (NIH, Bethesda, MD).
Highlights.
Novel recombinant HCV genomes exploit viral polyprotein processing
Reporter expression between NS5A-B does not impair viral fitness
Recombinant NS5A-B HCV genomes do not require adaptive mutations
Insertions within the HCV genome maintain virological phenotypes of the parent
New luminescent reporters replicate robustly in primary hepatocytes
Acknowledgements
The authors thank Dr. Julia Sable, Ellen Castillo, and Brenna Flatley for excellent technical support. This study was supported in part by grants from the National Institutes of Health National Cancer Institute (R01 CA057973), the NIH Directors Office (R01 DK085713-01), The Greenberg Medical Research Institute, and the Starr Foundation. J.A.H. was supported by the David Rockefeller Graduate Program at The Rockefeller University. M.D. was supported by a postdoctoral fellowship from the German Research Foundation. A.P. is a recipient of the Astella Young Investigator Award from the Infectious Disease Society of America and a Liver Scholar Award from the American Liver Foundation. The funding sources were not involved in the study design, collection, analysis and interpretation of data and in the writing of the report.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions:
Author contributions: J.A.H., M.D., C.M.R., and A.P. designed research; J.A.H., M.D., T.F., B.M.D., A.V., J.L., T.O. and A.P. performed research; J.A.H., M.D., T.F., B.M.D., A.V., C.M.R and A.P. analyzed data; and J.A.H., M.D. and A.P. wrote the paper.
The authors declare the following conflicts of interests, which are managed under University policy: C.M.R. has equity in Apath, LLC, which holds commercial licenses for the Huh-7.5 cell line and the HCVcc cell culture system and A.P. is a consultant for Apath, LLC.
References
- Andrus L, Marukian S, Jones CT, Catanese MT, Sheahan TP, Schoggins JW, Barry WT, Dustin LB, Trehan K, Ploss A, Bhatia SN, Rice CM. Expression of paramyxovirus V proteins promotes replication and spread of hepatitis C virus in cultures of primary human fetal liver cells. Hepatology. 2011;54:1901–1912. doi: 10.1002/hep.24557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartenschlager R, Ahlborn-Laake L, Mous J, Jacobsen H. Kinetic and structural analyses of hepatitis C virus polyprotein processing. Journal of virology. 1994;68:5045–5055. doi: 10.1128/jvi.68.8.5045-5055.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartenschlager R, Lohmann V. Replication of hepatitis C virus. J Gen Virol. 2000;81:1631–1648. doi: 10.1099/0022-1317-81-7-1631. [DOI] [PubMed] [Google Scholar]
- Blight KJ, Kolykhalov AA, Rice CM. Efficient initiation of HCV RNA replication in cell culture. Science. 2000;290:1972–1974. doi: 10.1126/science.290.5498.1972. [DOI] [PubMed] [Google Scholar]
- Blight KJ, McKeating JA, Rice CM. Highly permissive cell lines for hepatitis C virus genomic and subgenomic RNA replication. J. Virol. 2002a;76:13001–13014. doi: 10.1128/JVI.76.24.13001-13014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blight KJ, McKeating JA, Rice CM. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J Virol. 2002b;76:13001–13014. doi: 10.1128/JVI.76.24.13001-13014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenndorfer ED, Karthe J, Frelin L, Cebula P, Erhardt A, Schulte am Esch J, Hengel H, Bartenschlager R, Sallberg M, Haussinger D, Bode JG. Nonstructural 3/4A protease of hepatitis C virus activates epithelial growth factor-induced signal transduction by cleavage of the T-cell protein tyrosine phosphatase. Hepatology. 2009;49:1810–1820. doi: 10.1002/hep.22857. [DOI] [PubMed] [Google Scholar]
- Choo Q-L, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- Da Costa D, Turek M, Felmlee DJ, Girardi E, Pfeffer S, Long G, Bartenschlager R, Zeisel MB, Baumert TF. Reconstitution of the entire hepatitis C virus life cycle in non-hepatic cells. Journal of virology. 2012 doi: 10.1128/JVI.01066-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorner M, Horwitz JA, Robbins JB, Barry WT, Feng Q, Mu K, Jones CT, Schoggins JW, Catanese MT, Burton DR, Law M, Rice CM, Ploss A. A genetically humanized mouse model for hepatitis C virus infection. Nature. 2011;474:208–211. doi: 10.1038/nature10168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friebe P, Boudet J, Simorre JP, Bartenschlager R. Kissing-loop interaction in the 3' end of the hepatitis C virus genome essential for RNA replication. J Virol. 2005;79:380–392. doi: 10.1128/JVI.79.1.380-392.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottwein JM, Jensen TB, Mathiesen CK, Meuleman P, Serre SB, Lademann JB, Ghanem L, Scheel TK, Leroux-Roels G, Bukh J. Development and application of hepatitis C reporter viruses with genotype 1 to 7 core-nonstructural protein 2 (NS2) expressing fluorescent proteins or luciferase in modified JFH1 NS5A. Journal of virology. 2011a;85:8913–8928. doi: 10.1128/JVI.00049-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottwein JM, Jensen TB, Mathiesen CK, Meuleman P, Serre SBN, Lademann JB, Ghanem L, Scheel TKH, Leroux-Roels G, Bukh J. Development and Application of Hepatitis C Reporter Viruses with Genotype 1 to 7 Core-Nonstructural Protein 2 (NS2) Expressing Fluorescent Proteins or Luciferase in Modified JFH1 NS5A. J Virol. 2011b;85:8913–8928. doi: 10.1128/JVI.00049-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottwein JM, Scheel TK, Jensen TB, Lademann JB, Prentoe JC, Knudsen ML, Hoegh AM, Bukh J. Development and characterization of hepatitis C virus genotype 1–7 cell culture systems: role of CD81 and scavenger receptor class B type I and effect of antiviral drugs. Hepatology. 2009;49:364–377. doi: 10.1002/hep.22673. [DOI] [PubMed] [Google Scholar]
- Grakoui A, McCourt DW, Wychowski C, Feinstone SM, Rice CM. Characterization of the hepatitis C virus-encoded serine proteinase: determination of proteinase-dependent polyprotein cleavage sites. J Virol. 1993;67:2832–2843. doi: 10.1128/jvi.67.5.2832-2843.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CT, Catanese MT, Law LM, Khetani SR, Syder AJ, Ploss A, Oh TS, Schoggins JW, MacDonald MR, Bhatia SN, Rice CM. Real-time imaging of hepatitis C virus infection using a fluorescent cell-based reporter system. Nat Biotechnol. 2010;28:167–171. doi: 10.1038/nbt.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CT, Murray CL, Eastman DK, Tassello J, Rice CM. Hepatitis C virus p7 and NS2 proteins are essential for infectious virus production. J Virol. 2007 doi: 10.1128/JVI.00690-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambara H, Fukuhara T, Shiokawa M, Ono C, Ohara Y, Kamitani W, Matsuura Y. Establishment of a novel permissive cell line for propagation of hepatitis C virus by the expression of microRNA122. J Virol. 2011 doi: 10.1128/JVI.06242-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolykhalov AA, Agapov EV, Blight KJ, Mihalik K, Feinstone SM, Rice CM. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science. 1997;277:570–574. doi: 10.1126/science.277.5325.570. [DOI] [PubMed] [Google Scholar]
- Kolykhalov AA, Agapov EV, Rice CM. Specificity of the hepatitis C virus NS3 serine protease: effects of substitutions at the 3/4A, 4A/4B, 4B/5A, and 5A/5B cleavage sites on polyprotein processing. J Virol. 1994;68:7525–7533. doi: 10.1128/jvi.68.11.7525-7533.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Foy E, Ferreon JC, Nakamura M, Ferreon ACM, Ikeda M, Ray SC, Gale M, Jr, Lemon SM. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc Natl Acad Sci U S A. 2005a;102:2992–2997. doi: 10.1073/pnas.0408824102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X-D, Sun L, Seth RB, Pineda G, Chen ZJ. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc Natl Acad Sci U S A. 2005b;102:17717–17722. doi: 10.1073/pnas.0508531102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Pragai BM, Grakoui A, Xu J, Rice CM. Hepatitis C virus NS3 serine proteinase: trans-cleavage requirements and processing kinetics. Journal of virology. 1994;68:8147–8157. doi: 10.1128/jvi.68.12.8147-8157.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach BD, Evans MJ, Syder AJ, Wolk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA, Rice CM. Complete Replication of Hepatitis C Virus in Cell Culture. Science. 2005;309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- Lindenbach BD, Rice CM. Unravelling hepatitis C virus replication from genome to function. Nature. 2005;436:933–938. doi: 10.1038/nature04077. [DOI] [PubMed] [Google Scholar]
- Lohmann V, Korner F, Koch JO, Herian U, Theilmann L, Bartenschlager R. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- Marukian S, Andrus L, Sheahan TP, Jones CT, Charles ED, Ploss A, Rice CM, Dustin LB. Hepatitis C virus induces interferon-lambda and interferon-stimulated genes in primary liver cultures. Hepatology. 2011 doi: 10.1002/hep.24580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marukian S, Jones CT, Andrus L, Evans MJ, Ritola KD, Charles ED, Rice CM, Dustin LB. Cell culture-produced hepatitis C virus does not infect peripheral blood mononuclear cells. Hepatology. 2008;48:1843–1850. doi: 10.1002/hep.22550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- Morikawa K, Lange CM, Gouttenoire J, Meylan E, Brass V, Penin F, Moradpour D. Nonstructural protein 3-4A: the Swiss army knife of hepatitis C virus. Journal of viral hepatitis. 2011;18:305–315. doi: 10.1111/j.1365-2893.2011.01451.x. [DOI] [PubMed] [Google Scholar]
- Nakabayashi H, Taketa K, Miyano K, Yamane T, Sato J. Growth of human hepatoma cells lines with differentiated functions in chemically defined medium. Cancer Res. 1982;42:3858–3863. [PubMed] [Google Scholar]
- Narbus CM, Israelow B, Sourisseau M, Michta ML, Hopcraft SE, Zeiner GM, Evans MJ. HepG2 cells expressing microRNA miR-122 support the entire hepatitis C virus life cycle. J Virol. 2011;85:12087–12092. doi: 10.1128/JVI.05843-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen AW, Daugherty PS. Evolutionary optimization of fluorescent proteins for intracellular FRET. Nature biotechnology. 2005;23:355–360. doi: 10.1038/nbt1066. [DOI] [PubMed] [Google Scholar]
- Pietschmann T, Kaul A, Koutsoudakis G, Shavinskaya A, Kallis S, Steinmann E, Abid K, Negro F, Dreux M, Cosset FL, Bartenschlager R. Construction and characterization of infectious intragenotypic and intergenotypic hepatitis C virus chimeras. Proc Natl Acad Sci U S A. 2006;103:7408–7413. doi: 10.1073/pnas.0504877103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploss A, Evans MJ, Gaysinskaya VA, Panis M, You H, de Jong YP, Rice CM. Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature. 2009;457:882–886. doi: 10.1038/nature07684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploss A, Khetani SR, Jones CT, Syder AJ, Trehan K, Gaysinkaya VA, Mu K, Ritola K, Rice CM, Bhatia SN. Persistent hepatitis C virus infection in microscale primary human hepatocyte cultures. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.0915130107. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano KP, Laine JM, Deveau LM, Cao H, Massi F, Schiffer CA. Molecular mechanisms of viral and host cell substrate recognition by hepatitis C virus NS3/4A protease. J Virol. 2011;85:6106–6116. doi: 10.1128/JVI.00377-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheel TK, Gottwein JM, Jensen TB, Prentoe JC, Hoegh AM, Alter HJ, Eugen-Olsen J, Bukh J. Development of JFH1-based cell culture systems for hepatitis C virus genotype 4a and evidence for cross-genotype neutralization. Proc Natl Acad Sci U S A. 2008;105:997–1002. doi: 10.1073/pnas.0711044105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelzer E, Zhang L, Bruce A, Wauthier E, Ludlow J, Yao HL, Moss N, Melhem A, McClelland R, Turner W, Kulik M, Sherwood S, Tallheden T, Cheng N, Furth ME, Reid LM. Human hepatic stem cells from fetal and postnatal donors. J Exp Med. 2007;204:1973–1987. doi: 10.1084/jem.20061603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan T, Jones CT, Ploss A. Advances and challenges in studying hepatitis C virus in its native environment. Expert Rev Gastroenterol Hepatol. 2010;4:541–550. doi: 10.1586/egh.10.53. [DOI] [PubMed] [Google Scholar]
- Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Krausslich HG, Mizokami M, Bartenschlager R, Liang TJ. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters KA, Syder AJ, Lederer SL, Diamond DL, Paeper B, Rice CM, Katze MG. Genomic analysis reveals a potential role for cell cycle perturbation in HCV-mediated apoptosis of cultured hepatocytes. PLoS Pathog. 2009;5:e1000269. doi: 10.1371/journal.ppat.1000269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi M, Villanueva RA, Thomas DL, Wakita T, Lemon SM. Production of infectious genotype 1a hepatitis C virus (Hutchinson strain) in cultured human hepatoma cells. Proc Natl Acad Sci U S A. 2006;103:2310–2315. doi: 10.1073/pnas.0510727103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, Wieland SF, Uprichard SL, Wakita T, Chisari FV. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci U S A. 2005;102:9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Dong H, Eksioglu E, Hemming A, Cao M, Crawford JM, Nelson DR, Liu C. Hepatitis C virus triggers apoptosis of a newly developed hepatoma cell line through antiviral defense system. Gastroenterology. 2007;133:1649–1659. doi: 10.1053/j.gastro.2007.09.017. [DOI] [PubMed] [Google Scholar]