Summary
The mythological story of the Golden Fleece symbolizes the magical regenerative power of skin appendages. Similar to the adventurous pursuit of the Golden Fleece by the multi-talented Argonauts, today we also need an integrated multi-disciplined approach to understand the cellular and molecular processes during development, regeneration and evolution of skin appendages. To this end, we have explored several aspects of skin appendage biology that contribute to the Turing activator / inhibitor model in feather pattern formation, the topo-biological arrangement of stem cells in organ shape determination, the macro-environmental regulation of stem cells in regenerative hair waves, and potential novel molecular pathways in the morphological evolution of feathers. Here we show our current integrative biology efforts to unravel the complex cellular behavior in patterning stem cells and the control of regional specificity in skin appendages. We use feather / scale tissue recombination to demonstrate the timing control of competence and inducibility. Feathers from different body regions are used to study skin regional specificity. Bioinformatic analyses of transcriptome microarrays show the potential involvement of candidate molecular pathways. We further show Hox genes exhibit some region specific expression patterns. To visualize real time events, we applied time-lapse movies, confocal microscopy and multiphoton microscopy to analyze the morphogenesis of cultured embryonic chicken skin explants. These modern imaging technologies reveal unexpectedly complex cellular flow and organization of extracellular matrix molecules in three dimensions. While these approaches are in preliminary stages, this perspective highlights the challenges we face and new integrative tools we will use. Future work will follow these leads to develop a systems biology view and understanding in the morphogenetic principles that govern the development and regeneration of ectodermal organs.
Keywords: feathers, hairs, systems biology, stem cells, regeneration, microarray analyses, imaging, multiphoton microscopy, regional specificity
Introduction
In Greek mythology, Jason launches a journey to pursue the Golden Fleece. The Golden Fleece symbolizes rejuvenation power. In the story, nine headed dragons regenerate their heads instantly when they are severed from the body of the beast. Dragon teeth give rise to warriors when placed in contact with the right soil. In these early days of human civilization, the human mind must have been awed by the robust regenerative power of some ectodermal organ appendages. They could not understand how regeneration occurs and used myths to tell adventurous stories based in part on their observations. In the story, Jason assembled a multi-disciplined, multi-talented team of Argonauts that managed to overcome overwhelming odds to successfully obtain the Golden Fleece. Today, to search for the fundamental principles of morphogenesis and the regenerative power of skin appendages, we will still need to take a multi-disciplined, integrative biological approach.
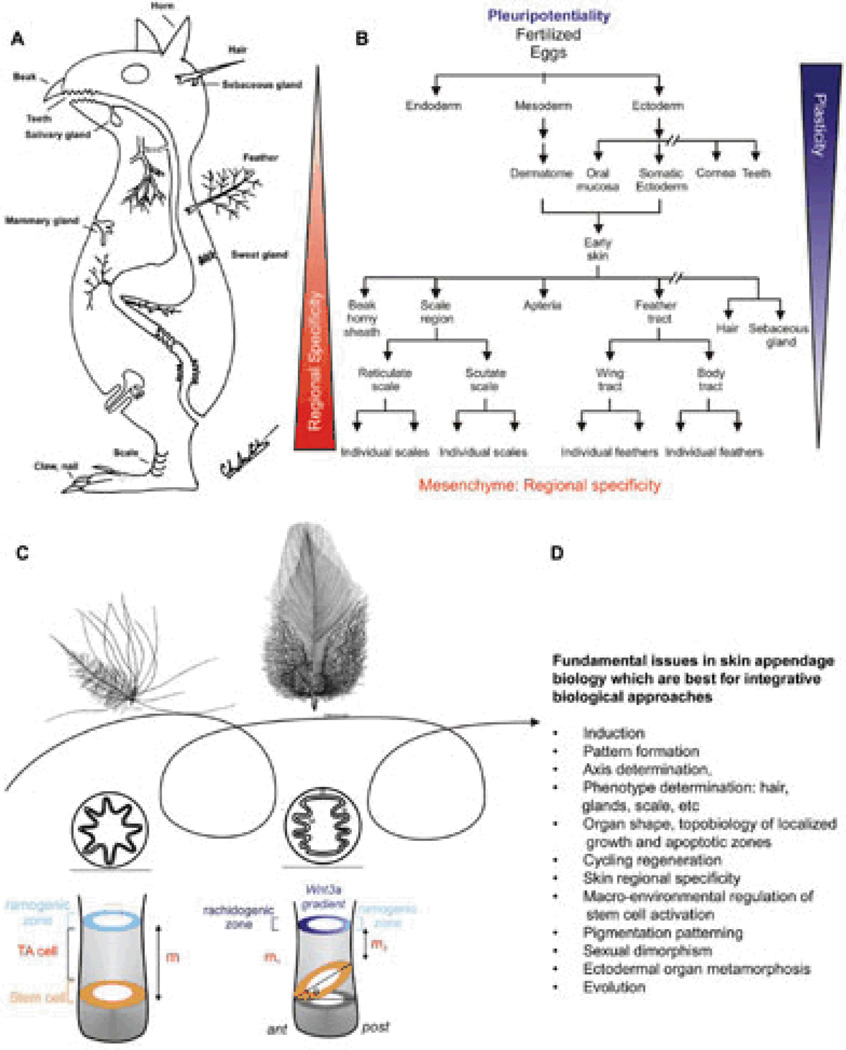
A concept animal with different types of (non-neural) ectodermal organs is shown in Fig. 1A. There are hairs, feathers, teeth, horns, nails, salivary glands, sweat glands, mammary glands, etc. During development, different ectodermal organs share the same developmental origin but become different types of ectodermal organs through interactions between the epithelia and mesenchyme (Fig. 1B).1 Since the integument forms the interface between the body and its external environment, these ectodermal organs have to endure frequent wear and tear and therefore evolved robust healing and regenerative powers. Different modes of regeneration are utilized by different ectodermal organs. Skin epidermis undergoes continuous renewal. Mammary glands undergo involution and growth phases. Hair and feathers are unique in that they undergo cyclic regeneration of the major portion of each organ under physiological conditions. During shedding or molting, the ‘older’ hairs or feathers are shed. Upon initiation of regeneration, the dermal papilla interacts with stem cells and new hairs or feathers (mini-organs) are re-made. Some interesting insights can be gained by comparing apparently different skin appendages. For example, feathers and mammary glands appear to be unrelated organs. Yet a comparison showed that both are ectodermal organs, regulated by sex hormones and each plays a key role in the evolution of the Aves and Mammalia classes, respectively.2 Trans-differentiation of ectodermal organ phenotypes also has been found by tilting the balance of molecular pathways. For example, when BMP activity is reduced in the epithelial-mesenchymal interface in K14 noggin mice, sweat glands and meibomian glands are converted into hairs3 and nipples are also converted into hair-bearing epidermis.4
Fig. 1. Basic concepts in ectodermal organ morphogenesis.
A) Concept animal with different forms of ectodermal skin appendages. Endodermal organs are also shown. Modified from Chuong edit, 1998.1 B) Chart showing the progression of ectodermal development into many different types of ectodermal organs. It also shows that the plasticity in ectoderm derived epithelial cells (i.e., multi-potentiality) gradually decreases, while the complexity of mesenchyme increases. C) Feather follicles undergo cyclic molting and regeneration. The follicles can change phenotypes between cycles. In subsequent generations of feathers, a symmetric downy feather and a contour feather emerge from the same feather follicle. Radial and bilateral feather symmetry can be determined by the topobiologcial arrangement of stem cells. Modified from Yue et al., 200622, Chuong et al., 2000102 D) Fundamental issue in biology that can be addressed by skin appendage model and integrative biology approach.
Skin appendages are ideal models to help answer many fundamental biological issues. These are listed in Fig. 1D. Skin appendages are excellent experimental models because they are at the body surface, accessible to experimentation, and easy to observe. They develop relatively late in embryonic development, and undergo physiological regeneration even in adult life. Because there are many skin appendages on one organism, alterations are less likely to be lethal, allowing more opportunities for perturbation. Grasping this opportunity, we use skin appendages as a Rosetta stone to understand the principles of morphogenesis. We have employed a multi-disciplinary approach using feather and hair models. This integrative biology approach has been fruitful and we have gained new understanding with significance beyond skin appendage biology. The following are some examples of what we have done.
Periodic patterning is a fundamental process in biological development.5 Since the mammalian coat and bird plumages are composed of a population of mini-organs, new properties emerge such as pattern formation (the arrangement, size and number of single skin appendages). We have made progress in applying the Turing reaction-diffusion model to study pattern formation of skin appendage primordia.6 This model has been widely applied to understanding the acquisition of self-organizing, repeated patterns in biological systems. In the Turing model, activators promote while inhibitors block the formation of an organ. Activators and inhibitors are released from the same source. Activators favor the synthesis and release of both activators and inhibitors. Inhibitors suppress the synthesis and release of activators. The model predicts that activators have a locally high effective concentration near their source of release while inhibitors diffuse further and have a higher effective concentration at a distance from the source.7, 8 Cells then migrate toward regions where the effective activator concentration is higher than the effective inhibitor concentration, thus enabling the formation of periodic patterns in biological systems. 9
We studied how stem cells are patterned during feather induction.10 Since the timing of induction in hair and feather placodes occurs relatively late, it allows us to study the pattern determination process from the initial homogenous state. Using tissue reconstitution of feather buds from dissociated cells, we have a system in which the patterning process starts from undetermined epidermal and dermal cells.11 We and others showed that growth factors (FGF / BMP / Wnt) and their inhibitors (ie, Sprouty / Noggin / DKK) fulfill the definition of activators / inhibitors in feathers and hairs, respectively.6, 12, 13 Growth factors are secreted peptides that bind to specific receptors. This in turn signals through independent mechanisms to elicit changes in cell behavior. For example, FGFs are a family of growth factors which bind to FGF receptors. FGF receptors contain a tyrosine kinase domain. Binding of the ligand to the receptor can establish a phosphorylation cascade to affect several aspects of cell behavior (proliferation, differentiation, etc in different cellular contexts.14 BMPs are another family of growth factors which bind to BMP receptors. They were initially identified in bone but have been found since in all types of tissues. BMP receptors have serine/threonine kinase domains. Upon binding of BMP ligands to their receptors, SMAD proteins (1, 5, 8 and the co-SMAD 4) are phosphorylated. SMAD 4 then translocates into the nucleus and activates downstream transcription.15 Wnts are a family of growth factors which bind to Frizzled receptors. This can lead to a stabilization of β-catenin within the cell. β-catenin is then free to move into the nucleus and in conjunction with Lefs/Tcfs promote downstream transcription.16
To fully understand how the homogeneous stem cells are patterned into bud and interbud regions, and how changes in the activator / inhibitor ratios can alter the pattern configuration from spots to stripes, in collaboration with Dr. Philip Maini’s group, we devised a computer simulation model that nicely recapitulates this process.9 These suggest that during periodic patterning, the epidermis initially forms a homogenous feather field in which every cell is equally competent to form feather buds. Mesenchymal cells migrate and sort themselves following principles including the Turing reaction-diffusion mechanism. The chemical patterns thus are consolidated into dermal condensation patterns. In response to the dermal signals, the β-catenin positive, homogenous stem cells in the feather field (basal states) respond to form placodes (state A) with certain sizes, shapes, numbers and inter-bud spacing (state B).17
In the spirit of a multi-disciplined approach, we collaborated with robotics engineers who needed to develop algorithms for the team behavior of swarming robots. We treated each robot in a robot team as a stem cell, and developed a “digital hormone” model. This permitted robot teams to self-organize into certain configurations depending on environmental obstacles that they may encounter.18 These studies developed leads toward the regenerative patterning of swarming robots.19
We next explored possible mechanisms of how individual organs are shaped. The feather model is ideal here because feathers in the adult bird show distinct morphologies to serve different functions in different body regions. These include thermo-regulation (downy in the trunk), communication (contour and tail feathers), and flight (wing feathers) (Fig. 1C, 8B). We found the shape is based on the topological configuration of feather stem cells. Feather stem cells are configured as a ring at the bottom of the follicle.10 Interestingly, this ring is horizontally positioned in radially symmetric feathers, but tilted toward the anterior (rachis) end of bilaterally symmetric feathers. We hypothesize this topological difference leads to the break of symmetry as stem cells progress from transient amplifying cells to differentiated cells (Fig. 1C).20 We then found that there is an anterior-posterior Wnt 3a gradient in the bilaterally symmetric feathers, but not the radially symmetric feathers. By modulating the activity ratio of morphogenesis related molecules (BMP, Wnt 3a, etc) at different times during the growth phase, different feather morphologies can be shaped along the proximal distal axis of the feather shaft.21, 22 Thus feather morphogenesis is determined by micro-environmental (within the follicle) regulation of the stem cell topology. We further explored this interaction through the chimeric recombination of dermal papillae transplanted between wing / body feather follicles. Interestingly, the chimeric feathers show that their new phenotypes are dictated by the origin of the dermal papilla.22
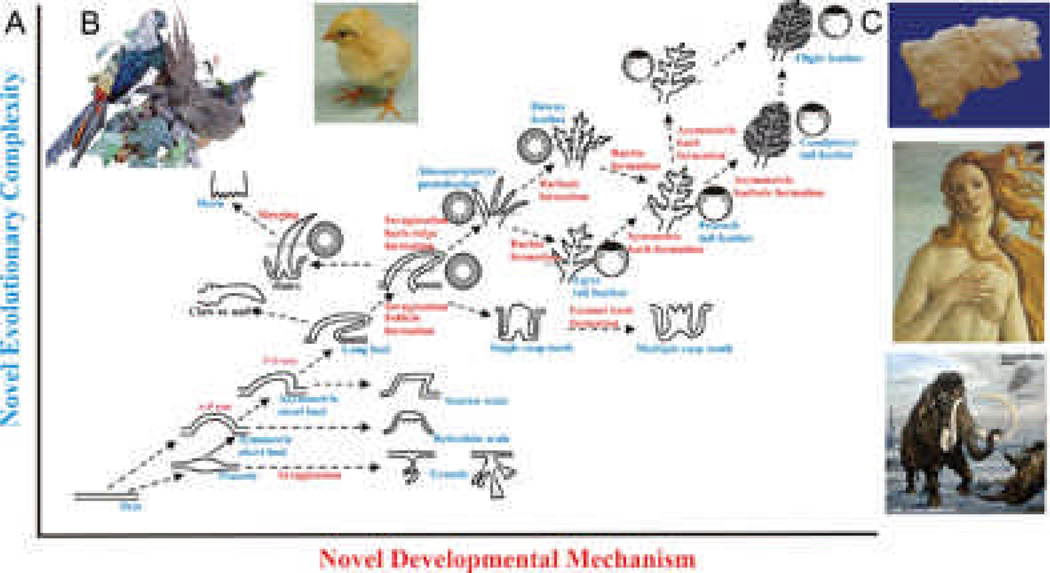
Fig. 8. Evo-Devo of ectodermal appendages.
A) X axis represents addition of new developmental mechanisms. Y axis represents emergence of new phenotypes. Glandular structures, invaginations, protrusion, branching, etc evolved from the flat epidermis. Complex feather forms have come a long way. Modified from Wu et al., 200490 B) Young chicks show downy feathers all over the body, while an adult pheasant shows diverse feather types that are sexually dimorphic. Female pheasant does not show these spectacular feathers (not shown). C) Mammalian ectodermal organs evolve with different emphasis under different physiological conditions as well as in evolutionary time. Top panel shows a fleece. Middle panel: scalp hairs and mammary gland. Bottom panel: long body hairs and tasks.
Another new property that emerges from a population of organs is the coordination of timing in regeneration. It has been known that a single hair follicle goes through regenerative hair cycling continuously during adult life,23 but whether the thousands of hair follicles on one individual cycle randomly, simultaneously, or in coordination is not known. In mice, we observed hair regeneration propagates in waves. Boundaries form because there are refractory regions where the wave cannot pass. We show that intra-follicular Wnt signaling goes up and down, in synchrony with hair cycling. Yet, extrinsic to hair follicles, there is another cyclic molecular change; the oscillation of dermal Bmp signaling, which is asynchronous with hair cycling.24 The interactions of these two rhythms lead to the recognition of refractory and competent phases in telogen, and autonomous and propagating phases in anagen. Boundaries form when propagating anagen waves reach follicles which are in refractory telogen.25 Further, we found hair waves are reset during pregnancy, implying a systemic level of regulation by macro-environmental factors.24 The unexpected link with Bmp2 expression in subcutaneous adipocytes has implications for systems biology and Evo-Devo. Thus, there is a macro-environmental regulation of hair stem cell activities by factors elicited from the surrounding dermis, neighboring follicles, systemic hormones, and external environments (Fig. 8C).25 The macro-environmental factors serve as a bridge between stem cells and the real external environment.
These studies demonstrate that many biological issues in skin appendages are achieved by modulating epithelial stem cell activity with different hierarchical levels of environmental control. This process is modulated from the adjacent dermal papilla, surrounding dermis, and/or systemic physiological conditions. In this regard, Dr. Bissell’s pioneering work has served as an inspiration. Twenty five years ago, using RSV tumor virus as a tool, Dr. Bissell probed the nature of interactions between viral oncogenic activity and the environment. She demonstrated that the embryonic environment restricts the sarcoma forming ability of src.26 Later she further showed that this ability is due to cell types; only endothelial cells could become neoplastic .27
Others have also demonstrated that growth factors, extracellular matrix and proteolytic enzymes present within the stroma are critical for tumorigenesis. Dr. Mintz preimmunized female host mice against the syngeneic male melanotic skin grafts. Subsequent transplants of male skin grafts were delayed in forming tumors. These cells could develop tumors faster when subsequently transplanted to control, non-immunized hosts.28 They interpret the delay as being due to the destruction of the donor male stroma and replacement by the host. The increased growth rate recovered melanotic tumor cells were transplanted was attributed to selection of a faster growing population. Dr. Werb has added major contributions on the role of tumor-stromal interactions in mammary gland development and breast cancer with a particular focus on the extracellular matrix and metalloproteinases. For example, her group found that estradiol can induce amphiregulin 29, which is shed from the epithelium by the action of metalloproteinase and then communicates with the underlying stroma via the FGF Receptor.30. Dr. L. Coussens has examined the role of the immune response and inflammation on tumor formation. The basic concept is that chronic inflammation can promote tumor formation; however, some immune responses may promote while others may fight tumorigenesis. Cytokines, chemokines and mediators of immune response produced within the tumor environment mediate these responses.31, 32
The role of mesenchymal – epithelial interactions in prostate development and disease has been shown by the Cunha group. Androgen signaling through the androgen receptor is essential for prostate development. Recombining epithelium and mesenchyme from wildtype mice and mice with testicular feminization he demonstrated that androgen binds its receptor in the mesenchyme.33 His group later showed that embryonic rat mesenchyme could induce human prostate development from adult bladder epithelium. 34, 35 Furthermore, they showed that androgens plus estrogens could cause an immortalized human prostate cell line to form metastatic tumors when transplanted to a male nude mouse kidney capsule.36
All together, these studies focus more on tumor-stroma interactions and oncogenesis (dys-regulated new growth), these profound observations serve as a reference for us who study the epithelial-mesenchymal interactions in regulated new growth during the development, maintenance and regeneration of organs.37 More recently, Dr. Bissell’s work further demonstrates that normal mammary gland growth and breast cancer are under similar micro- and macro-environmental regulation.38, 39 Conceptually, this is gratifying as we have treated the mammary gland as one of the skin appendages (Fig. 1)1 and now we have come to appreciate similar principles by focusing on different ectodermal organs (mammary glands versus feathers) and different processes (tumorigenesis versus normal development).
Here we present a perspective on how the field has grown to where it is today. To continue the trend of an integrative biology approach, we search for the molecular basis that defines different ectodermal organ phenotypes, feather versus scales, and different types of feathers. We also are eager to visualize the real time processes that occur in the initial phase of periodic patterning of dermal condensations and epithelial placodes. We wish to describe our view of how current methodologies can be applied to skin biology for major advances in future research. We show examples of how these novel methodologies have redefined the classical phenomena at a higher level of resolution. While these data represent work in progress, they reveal unexpected complexity of processes regulating organ morphogenesis and the exciting new potential unraveled by integrative biological approaches.40
RESULTS
Timing of commitment in epithelial – mesenchymal interactions during feather / scale formation
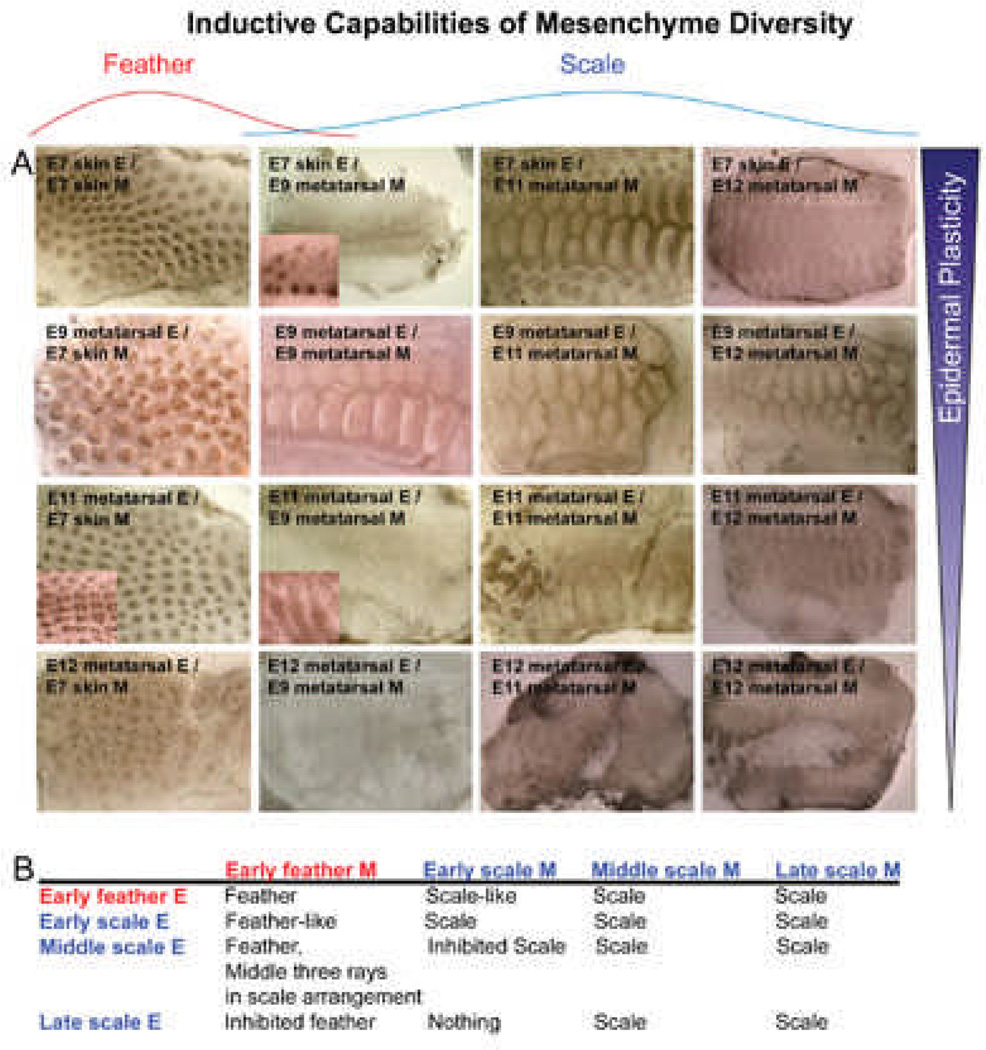
Epithelial appendages are the product of epithelial – mesenchymal interactions. Tissue recombination experiments showed that in general, the dermis determines the phenotype of the epithelial appendage.41, 42 Here we sought to evaluate these abilities by coupling classical tissue recombination experiments with microarray. Chicken dorsal skin epithelium interacts with its underlying mesenchyme to form feathers beginning at E7 (H&H stage 31), while metatarsal scale epithelium interacts with its mesenchyme to form scales beginning at E9 (H&H stage 35) which stabilize around E12 (H&H stage 38). To do this, we designed a set of experiments using E7 dorsal skin (normally feather region), E9, E11 (H&H stage 37) and E12 metatarsal skin (normally scale region). We separated the epithelium and mesenchyme and recombined them to cover interacting components at different ages (Fig. 2). Different epithelia are positioned in different rows, and different dermis in different columns.
Fig. 2. Timing of commitment in feather / scale tissue recombination experiments.
A) Results of chimeric explants. B) Summary of results. Different rows represent different sources of epidermis: E7 dorsal skin epidermis (normally feather forming), E9, E11 and E12 metatarsal skin epidermis (normally scale forming). Different columns represent different sources of mesenchyme: E7 dorsal skin mesenchyme (normally feather inducing), E9, E11 and E12 metatarsal skin mesenchyme (normally scale inducing). We can observe the gradual restriction of epidermal plasticity and the beginning of dermal complexity, echoing what we see in Fig. 1B. Red, feather derived tissue; blue, scale derived tissue.
When E7 feather epithelium is combined with E7 feather mesenchyme, feathers form. However, when this epithelium is recombined with different stage scale mesenchyme (E9-E12), scales form to varying degrees. E11 scale mesenchyme had the highest capacity to induce scale formation from E7 epithelium, while both E9 and E12 scale mesenchyme are weaker in their inducing ability. These findings suggest that the inducing ability of the mesenchyme is transient (Fig. 2, 1st row). Also, as animal develops, different inductive mesenchymal signals arise, forming diverse types of ectodermal organs.
In a reciprocal experimental design, when E7 feather dermis is recombined with different stage scale epithelium (Fig. 2, 1st column), feathers are produced with E9 scale epidermis, but this ability declines with advancing age of the scale epidermis. With E12, the chimeric feather / scale appendages are arranged in a scale pattern. With E11 scale epidermis, feather buds form, but are in scale pattern in the 3 rows around the midline which matures faster than the flanking regions. We also examined the inducing ability of scale dermis (E9-E12) to induce scale epithelium (E9-E12). It shows similar trends. Thus the competence of epidermis, i.e., the multi-potential ability of the epidermal progenitors, to respond to the inducing signal and become specific types of skin appendages is also transient, higher in the earlier stages of the epidermis. This ability is gradually restricted with developmental timing. We wondered what molecules might regulate competence vs non-competence in developing tissues. We also wondered what molecules might underlie regional specificity, in other words, why some regions develop in to feathers and others into scales.
In search of molecular pathways involved in tissue competence and regional specificity
To search for molecules involved in competence we chose to perform expression studies on competent E7 and non-competent at E9 feather forming skin. We previously had shown that differences in wing or body feathers were based on differences within the dermal papilla, while epidermal cells in the proximal follicle represent stem cells that can be modulated into different types of feathers.22 To examine differences in regional specificity we compared the above results with results from competent E9 and non-competent E11 meta-tarsal scale forming regions as well as the proximal follicle epidermis and dermal papilla in adult wing, body and tail feather follicles. These time points were chosen based on the results from experiments described in Fig. 2.
Microarray analysis
Embryonic and adult chicken tissues were micro-dissected, RNA was extracted and then probed onto GeneChip Chicken Genome Arrays (Affymetrix). Array Assist, Partek Genomic Suite, and Ingenuity Pathway Analysis software were utilized to detect differential gene expression patterns and significant pathway enrichments. After pre-processing the data, the 55 tissue samples formed five groups according to principal component analysis (PCA) (Fig. S1A-D). PCA predominantly grouped the samples according to age and tissue type; 1) adult epithelium from proximal feather follicle (small blue spheres), 2) adult mesenchyme, i.e., dermal papilla from feathers in different body regions, (small green spheres), 3) embryonic epithelium (large blue spheres), 4) embryonic mesenchyme (large green spheres), and 5) embryonic tissue containing both epithelium and mesenchyme (large red spheres). Analysis of Variance (ANOVA) supported the five PCA groups by demonstrating that most of the variance in the data set can be attributed to the factors of age and tissue type (Fig. S1E).
Molecular differences of embryonic versus adult tissues
In order to gauge important molecular expression differences between embryonic and adult tissues, we looked at a time course experiment comparing epithelium or mesenchyme at different time points (Fig. S2). Differential gene expression analysis detected numerous changes in gene expression. In the feather forming epithelium, EphA3 and Bmp3 are down-regulated while Msx2 (muscle specific homeobox gene) and Wnt2B are up-regulated (Fig. S2A). In the feather forming mesenchyme, Hoxa11 and PitX1 are down-regulated while WFIKKN2 and Sox9 are up-regulated (Fig. S2B). Two dimensional hierarchical clustering based on the original 55 gene chips identified genes that were up-regulated in the adult tissues and down-regulated in embryonic tissues (Fig. S2C). Entering these gene lists into IPA yielded a biological function chart. Tissue development, organ development and connective tissue development and function were statistically enriched in our data sets comparing embryonic to adult tissues (Fig. S2D).
Molecular differences of tissues with different competence and inductive abilities to form feathers and scales
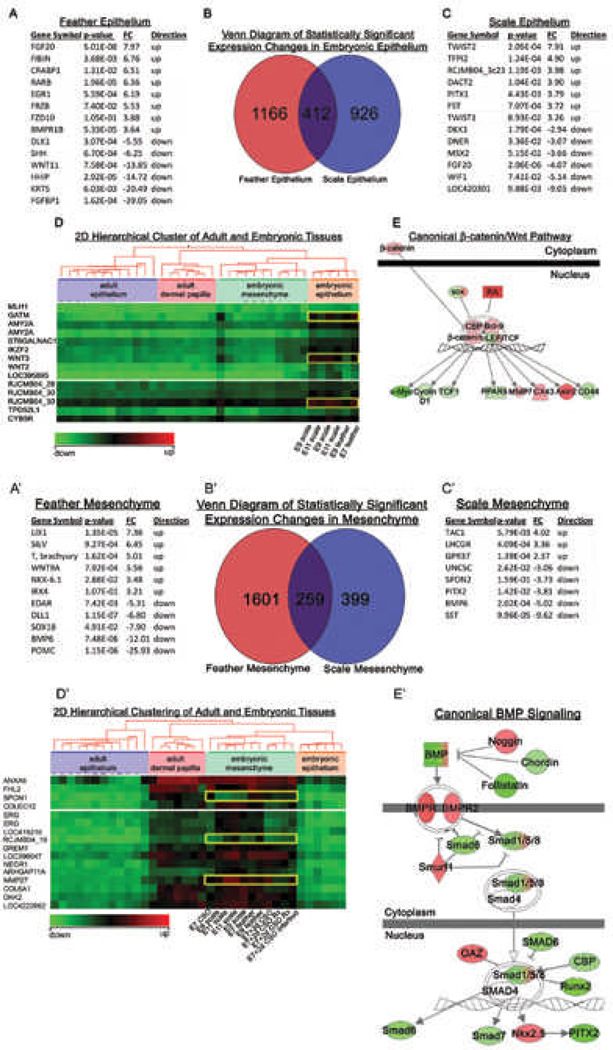
We wanted to know what changes in gene expression are associated with epithelial competency and mesenchymal induction capability (Fig. 3). Based on the previous tissue recombination experiment, we compared E7 feather forming epithelium to E9 feather forming epithelium, and E9 scale forming epithelium to E11 scale forming epithelium (Fig. 3A-E). ANOVA identified numerous changes in gene expression. Up-regulating Fgf20, Frz10 and BmpR1b, while simultaneously down-regulating of SHH, Wnt11 and Krt 5 are associated with E7 feather epithelium formation (Fig. 3A). Up-regulating TWIST1, TWIST2 and DACT, while simultaneously down-regulating of Dkk1, Msx2 and Fgf20 are associated with E9 scale epithelium formation (Fig. 3C). Two dimensional hierarchical clustering aided our study in identifying interesting changes in gene expression. The cluster analysis suggested that GATM and WNT3 may play a role in modulating epithelial competency (Fig. 3D). The systems biology approach identified putative biologically relevant molecular pathways. Genes that are shared between scale forming and feather forming epithelium in the Venn diagram (Fig. 3B) are likely to be involved in promoting tissue competency in both the scale and feather epithelia. Entering the common gene list into IPA generated canonical pathways and networks that demonstrated how epithelial competency might be regulated and that β-catenin plays a key role (Fig. 3E). The IPA pathway generation algorithm calculates the statistical significance of association between the genes and the canonical pathway by the Fisher’s exact test resulting in a score P-value.43
Figure 3. Differential gene expression analysis of embryonic chicken feather / scale regions.
A-E) Differential gene expression analysis of embryonic epithelium. The gene expression of E7 feather was compared to E9 feather forming epithelium, and E9 scale was compared to E11 scale forming epithelium. B) Venn diagram separates the genes that contribute to scale forming, feather forming, or non-specific forming epithelium. A representative list of genes that contribute to the formation of scale (A) or feather epithelium (C) are listed. D) Two dimensional hierarchical cluster exhibiting differential gene expression for embryonic epithelium. E) Example showing genes involved in epithelium formation were enriched in the canonical β-catenin/Wnt pathway. Red is up-regulated and green is down-regulated gene expression.
A'-E') Differential gene expression analysis of embryonic mesenchyme. The gene expression of E7 feather was compared to E9 feather forming mesenchyme, and E9 scale was compared to E11 scale forming mesenchyme. B’) Venn diagram separates the genes that contribute to scale forming, feather forming, or non-specific forming mesenhcyme. Representative lists of genes contributing to the formation of feather (A') or scale mesenchyme (C'). D’) Two dimensional hierarchical cluster exhibiting differential gene expression for embryonic mesenchyme. E') Genes involved in mesenchyme formation were enriched in the canonical BMP pathway. Red is up-regulated and green is down-regulated gene expression.
The mesenchymal tissues of E7 and E9 feather forming, as well as E9 and E11 scale forming were also compared (Fig. 3A’-E’). According to the ANOVA performed, WNT9A, NKX-6.1 and IRX4 were up-regulated while EDAR, SOX18 and BMP6 were down-regulated in the feather forming mesenchyme (Fig. 3A’). The scale forming mesenchyme exhibited an up-regulation of TAC1 and GPR37 while UNC5C, PITX2 and BMP6 were down-regulated (Fig. 3C’). A Venn diagram demarcates gene lists that may be playing a role in feather or scale mesenchyme inducing ability. Two dimensional hierarchical clustering analysis suggested that SPON1 and MMP27 play a role in modulating mesenchyme inducing ability (Fig. 3D’). Entering the common gene list into IPA generated canonical pathways and networks that demonstrated how mesenchymal inducing capability might be regulated (Fig. 3E’). In this case BMP pathway activity appears to play an important role.
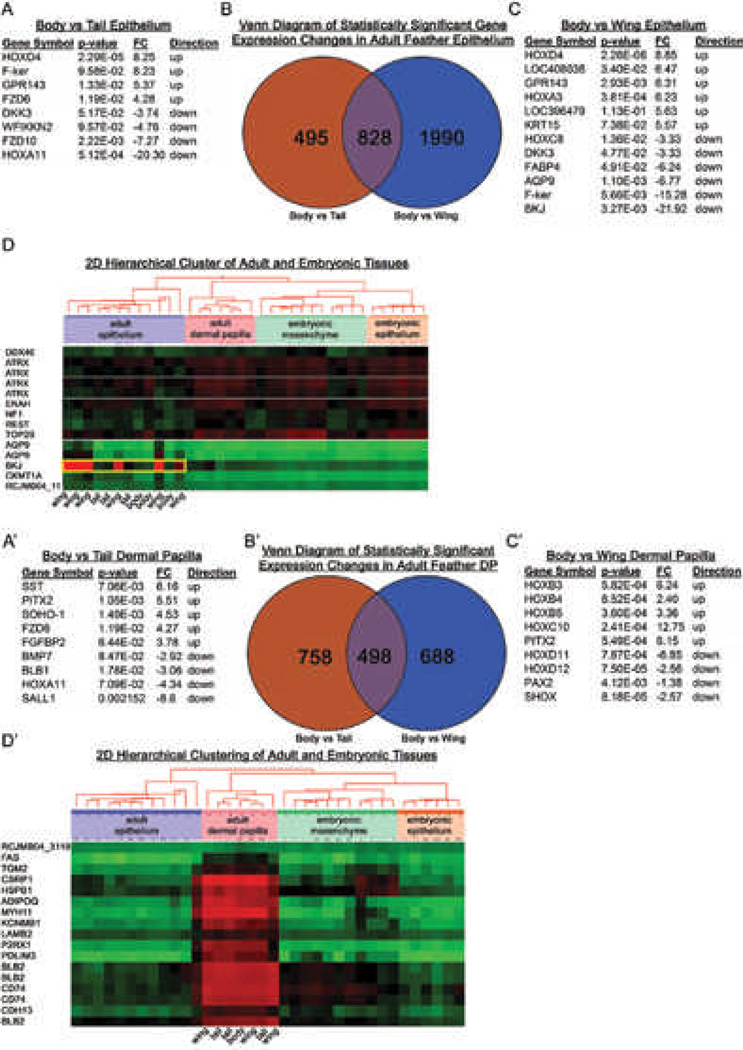
Molecular differences of dermal papilla and proximal follicle epidermis in feathers from different body regions
In the adult chicken feather, we wanted to know what changes in gene expression associate with body feather, wing feather and tail feather regional specificity (Fig. 4). We focused our attention on the adult feather epithelium. ANOVA identified HOXd4, F-Ker and FRZD6 as up-regulated and DKK3, FRZD10 and HOXa11 as down-regulated in the body feather when compared to the tail feather (Fig. 4A). The ANOVA identified HOXd4, HOXa3 and KRT15 as up-regulated and HOXc8, FABP4 and BKJ as down-regulated in the body feather compared to the wing feather (Fig. 4C). Most interestingly, two dimensional hierarchical clustering suggested that BKJ is specifically up-regulated only in the wing feather epithelium when compared to the tail and body feathers (Fig. 4D).
Figure 4. Differential gene expression analysis of feather follicles from different body regions of adult chickens.
A-D) Differential gene expression analysis of adult chicken epithelium. The gene expression of body feather epithelium was compared to wing feather or tail feather epithelium. Representative gene lists that contribute to the formation of tail epithelim (A) and wing epithelium (C) are listed. B) Venn diagram separates the genes that contribute to form wing feather or tail feather epithelium. D) Two dimensional hierarchical cluster exhibits up-regulation of genes in the wing epithelium and their respective down-regulation in other tissues and ages. Red is up-regulated and green is down-regulated gene expression.
A'-D') Differential gene expression analysis of adult chicken feather mesenchyme. The gene expression of body feather dermal papilla was compared to wing feather or tail feather dermal papilla. Representative gene lists that contribute to the formation of tail dermal papillae (A') and wing dermal papilla (C') are tabulated. B’) Venn diagram separates the genes that contribute to form wing feather or tail feather dermal papillae. D’) Two dimensional hierarchical cluster exhibits up-regulation of genes in the dermal papillae and their respective down-regulation in other tissues and ages. Red is up-regulated and green is down-regulated gene expression.
We next looked at the feather dermal papillae (Fig. 4A’-D’) because data from recombination experiments and the literature demonstrate that the mesenchyme determines what skin organ develops (Fig. 2).41 Using the body feather as the control or non-specialized feather, we compared body to wing feather dermal papillae gene expression patterns (Fig. 4C). ANOVA identified numerous changes in gene expression. Up-regulating Hoxb3, Hoxb4 and PITX2, while simultaneously down-regulating of Hoxd11, Hoxd12 and PAX2 are associated with dermal papillae in the body region (Fig. 4A). Two dimensional hierarchical clustering identified genes that were commonly up-regulated in the adult dermal papilla (Fig. 4D).
We then used qPCR to validate a subset of 5 genes that showed significant differences in expression levels as determined by microarray (Table I). Our qPCR study confirmed that these genes were expressed to significantly different levels as a function of competence vs determination or of regional specificity.
Table I.
Verifying microarray analysis by qPCR
| Comparison | Gene | Regulation |
|---|---|---|
| E7 Epithelium vs E9 Epithelium | RARb | Upregulated |
| E9 Scale Epithelium vs E11 Scale Epithelium | Tac1 | Upregulated |
| Body Feather Epithelium vs Tail Feather Epithelium | HoxD4 | Upregulated |
| Body Feather Epithelium vs Wing Feather Epithelium | BKJ | Down regulated |
| Body Feather Epithelium vs Tail Feather Epithelium | Pitx2 | Upregulated |
Several genes shown to be up or downregulated by microarrays were chosen and qPCR was performed with the samples indicated.
Next we compared gene lists derived from our microarray study on dermal papilla with those from mouse hair dermal papilla (Table 2).44 We next compared a gene list derived from wing growth feather collar epithelium (the site of feather precursor cells)10 with the gene list from hair bulge enriched genes (Table 3).45 We found many genes in common, highlighting their fundamental importance in these two skin appendages that evolved convergently. These molecules will be the target for future investigations.
Table 2.
Gene Homology for Feather vs Hair Dermal Papilla
| Growing Wing Feather Dermal Papilla | Hair Dermal Papilla Genes |
|---|---|
| Frizzled 2 | Frzd Related Protein, Secreted Frzd Related Protein 2 |
| Hox D4 | Hox C8 |
| FGFR Activation Protein 1 | FRGR1 |
| Melanocortin Receptor 5 | FGFR Activation Protein 1 |
| Solute Carrier 16 | Solute Carrier 16 |
| Insulin-like Growth Factor Binding Protein 1 | Insulin-like Growth Factor Binding Protein 3 |
| Potassium Voltage-gated Channel Shaker | Potassium Voltage-gated Channel Shaker |
| Related Family, Member 2 | Related Family, Member 2 |
Information of hair dermal papilla are from Rendl et al., 2005.
Table 3.
Gene Homology for Feather vs Hair Putative Stem Cell Populations
| Growing Wing Feather Collar Epithelium | K15 Positive Hair Bulge Cell Enriched Genes |
|---|---|
| G protein coupled receptor family C, Group 6, member A | G protein-coupled receptor 49 |
| Potassium channel subfamily K, member 2 | Potassium channel subfamily K, member 2 |
| FGF2, FGF7, FGF10 | FGF1 |
| Frizzled Homolog 4 | Frizzled Homolog 2 |
| Frizzled Related Protein | Secreted Frizzled Related Protein 1 |
| TNF, member 13b | TNF Receptor 11b |
| Col1A2, Col11A1, ColA52 | Col5 alpha2 |
| Annexin A6 | Annexin A6 |
| Tenascin C | Tenascin C |
| CD34 | CD34 |
| Solute Carrier, Family 4, 7, 18 | Solute Carrier, Family 29 |
Information for K15 positive hair bulge cells are from Morris et al., 2004.
Homeobox gene expression in developing chicken skin
Our microarray data suggest that homeobox genes show differential expression pattern from different skin regions (body vs wing; body vs tail). Hox proteins have been implicated in embryonic axial development and morphogenesis.46 Their expression patterns help to establish spatial identity, such that the term Hox code was invoked.47 Hoxc8, d9, d11 and d13 has been shown to be expressed in specific patterns in developing limb buds.48 We rationalized that Hox expression in adults should be set during embryonic development before different tract regions were defined.
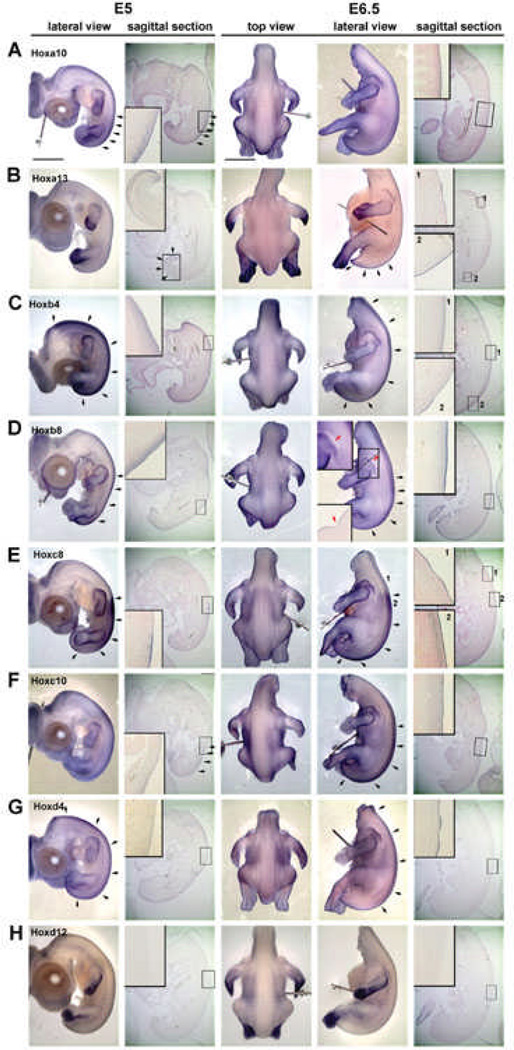
We performed whole mount in situ hybridization using two Hox genes in every cluster to examine the expression in H&H stage 26 (E5) and 29 (E6.5) chicken embryos (Fig. 5). Sagittal sections and some cross sections were collected from the whole mount in situ hybridization samples. The feather placodes started to form in dorsal tract at E6.5. Our purpose is to examine the role of Hox genes in regional specificity leading to pteric versus apteric regions, short feather versus long feather regions, why feathers grow on the wing versus scale growth on the leg, etc. The results show some interesting regional differences. Overall, the body skin did not strictly follow the co-linear expression patterns with lower numbers of Hox genes in the anterior trunk or proximal limbs. This result is similar to that of Reid and Gaunt49 which only studied sagittal sections. We observed some hox genes at E6.5, such as Hoxa13 (Fig. 5B) and Hoxc8 (Fig. 5E) expressed in the feather placode at the region which did not have expression at E5, suggested that these genes may take part in future feather morphogenesis. Some hox, such as Hoxb8, is absent at the future apteric region (Fig. 5D, red arrow) suggesting it may play some role in regional specification. Currently, the data is limited at this stage and more work on hox gene expression patterns and functional studies are required to investigate their role in regional specification.
Fig. 5. Expression of Hox genes in chicken embryonic skin at E5 (stage 26) and E6.5 (stage 29) as determined by wholemount in situ hybridization.
A. Hoxa10 is expressed in the epithelium and mesenchyme at E5 but only in the epithelium at E6.5. B. Hoxa13, is expressed in the distal limb bud and tail region epithelium and mesenchyme at E5. However, feather placodes in the upper dorsal tract (insert 1) start to express Hoxa13 at E6.5 only in the epithelium. C. Hoxb4 and D, Hox b8 are expressed in the epithelium and dermis at E5 and E6.5. However, Hoxb4 expression extends more proximally than Hoxb8. At E6.5, there is a Hoxb8 negative region between the scapular and dorsal feather tract that extends posteriorly until the boundary between the femoral and dorsal tract (red arrow). The dashed line shows the plane of section shown in the inset. This non-Hoxb8 area may be related to the apteric regionof the chicken skin. E. Hoxc8 expression is the same at E5 and E6.5. However, in region 1, Hoxc8 is expressed in the epidermis of feather placodes. This region did not have strong staining under the skin (compare to region 2). F. Hoxc10 did not show a clear staining pattern in skin as compared to Hoxc8, but it can be found in the skin from the sagittal section at both E5 and E6.5. G. Hoxd4 is weakly expressed in all dorsal epidermis at E5. Staining is stronger in the epithelium of bud and interbud regions at E6.5. H. Hoxd12 is expressed in the distal and ventral limb bud but not in the skin at both E5 and E6.5. Black arrows indicate the intense expression region in the sample. Scale bar: 2mm
Visualizing cellular events during feather morphogenesis
The exciting aspect of using an integrative biology approach to study the morphogenesis of skin appendages is that it offers an opportunity to understand the clearly visible skin appendage phenotype from the level of molecules, cells, tissue interactions, all the way to organ shape. Heterotypic recombination defines the tissue interaction question, illustrating that organ phenotype is established by the epidermis and dermis together. Microarray analyses help us identify critical molecular signaling pathways involved in this process. Our current understanding of these processes is depicted in Fig. 6A. In order to understand the cellular events that lead to feather morphogenesis at a deeper level, we need to visualize cell movements in real time and observe specific interactions among cells during the process of skin morphogenesis. Here we apply several types of modern imaging technologies for this purpose.
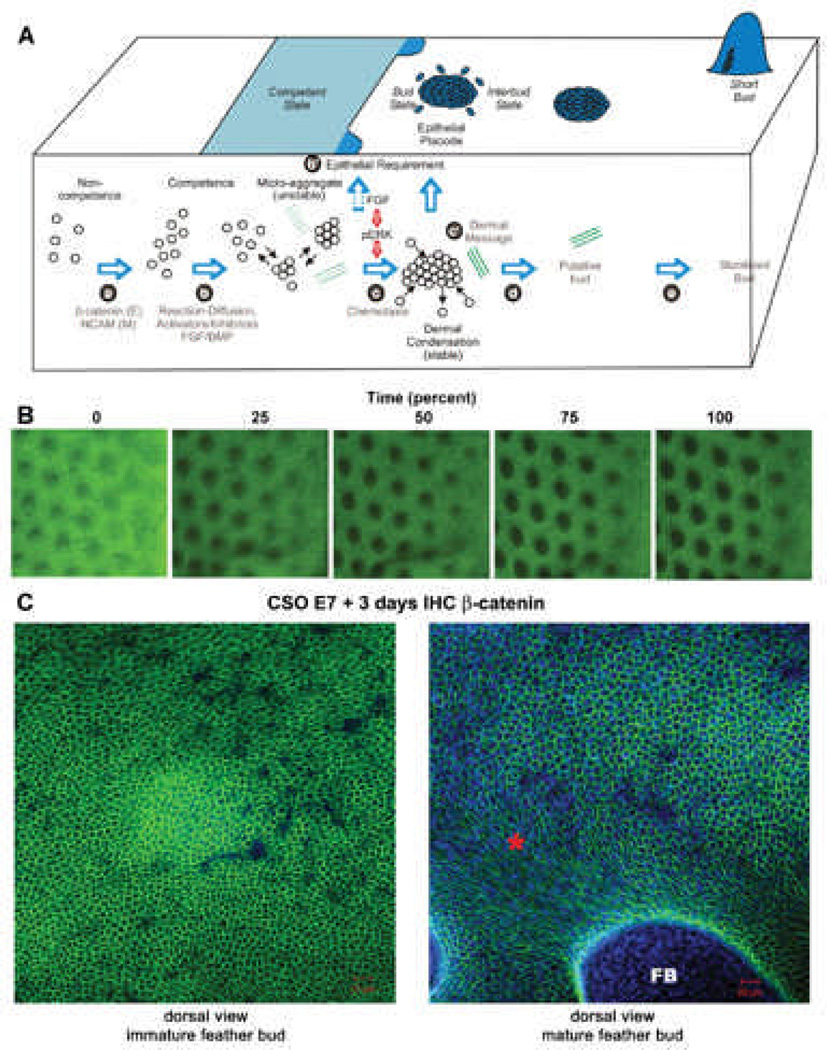
Fig. 6. Video microscopy imaging of developing chicken skin explant cultures.
A) Schematic drawing on our current concept of periodic patterning process (from Lin et al.9). This concept is based on current biochemical and functional perturbation data. However, the detailed cellular processes and tissue interactions remain to be elucidated. The microarray data earlier showed differences in molecular expression. The following imaging data give us a glimpse on the complexity of the cellular behavior and redefine this classical phenomenon. Green dotted and solid lines represent the development of extracellular matrices.
B) Time lapse videomicroscopy. Movie is in supplement. E9 skin explants were photographed every 15 minutes for 17 hours to track the process of early feather morphogenesis. Representative images are taken from the movie. Each black spot represent a bud, which is about 10 cells long in diameter. The dorsal skin midline lies to the left of the images, and buds to the right side of the panel are in earlier stage than those in the left. Size bar = 500um.
C) E7 chicken skin organs were cultured for 3 days. Immunohistochemistry with antibodies against β-catenin was used. A) Cells within the immature feather bud exhibit a rather homogenous hexagonal cell shape. B) In the more mature bud region (*), the cell shape changes as the feather bud (FB) forms and elongates. Size bars: 20 microns. Beta-catenin stains Green, DAPI stains blue.
Time-lapse videomicroscopy of cultured developing skin explants
Our first method uses visible light and low power videomicroscopy to observe changes in cell density and changes in feather formation which take place during early stages of feather formation. In this time lapse movie, E9 feather forming dorsal skin was grown for 17 hours in a humidified culture chamber (Wafergen Smart Slide) at 37°C. HEPES buffer (10 uM) was used to control the pH. Time-lapse video pictures of the explants were taken at 15 minute intervals to assess cell movements and changes in cell density during feather bud development. Feathers start to develop along the midline of dorsal (back) skin about this time. The first visible signs of feather development both in vitro and in vivo are the appearance of dermal condensations, seen as dark circles on the skin. Subsequent rows of feathers develop bilaterally from the midline. The midline is to the left of the panel and the less developed feather buds are present at the lateral edges (to the right of the panel, Fig. 6B). As the movie progresses, feather buds toward the lateral edge are being consolidated as those toward the midline elongate (supplemental data, movie). The interbud cell density maintains a similar density across the whole field. Frames from this movie representing time 0h (0%), 4.25h (25%) , 8.5h (50%), 12.75h (75%) and 17 hours (100%) are shown (Fig. 6B and movie in supplement). While the still shots do not give one a sense of cell movement, they do show how feather structures change over this time interval. Using this method we can get a view into the dynamic events required to form feather buds. This approach gives us a nice overview of feather formation. It demonstrates early changes which take place in the skin as feathers begin to become periodically patterned and is highly suggestive of the complex cellular flow involved in feather formation although specific cell movements are not visible at this magnification.
Confocal microscopy
In order to study how cell shape plays a role in skin organ morphogenesis, we looked at β-catenin protein expression in immature and mature feather buds (Fig. 6C). A) In the immature feather bud the β-catenin protein expression is predominantly located at the cell surface rendering a pentagonal cell shape. The epithelial cells forming interbud and the feather bud have very similar shapes at this stage. B) As the feather bud matures and elongates, the epithelial cells forming the feather bud alter their shape to become more oblong. However, the interbud cells maintain the same pentagonal shape similar to their immature state. These data show that the regions between feather buds are not in fact uniform. Rather, cells in specific regions become elongated while in others retain their pentagonal shape. In scales, the dynamic cellular shape changes and flow will follow a different pattern (not shown). These results suggest that complex and yet specific interactions, either within the epithelium and / or between the epithelulm and mesenchyme, remain to be solved.
Multiphoton microscopy to visualize 3D movement and extracellular matrix remodeling
Since organs develop in 3-dimensional space, we need a tool that provides a means to analyze these events. Conventionally, characterization of the temporal and spatial developmental events in feather morphogenesis relies on tissue sections from tissues of different developmental stages. Since the tissue of interest is prepared for histological examination, dynamic analysis of cell reorganization and extracellular remodeling within the same field of interest is hindered. In recent years, the minimally invasive imaging technology of multiphoton microscopy has gained popularity in skin research to probe the dynamic three-dimensional distribution and organization of cells and extracellular matrix50–56,50–56 Multiphoton microscopy utilizes infrared laser as light source and the excitation of fluorophores requires the simultaneous absorption of two or more photons of lower energy. 56, 57 For efficient multiphoton excitation, ultrashort femtosecond pulses with high peak power are required. In comparison with confocal microscopy and conventional fluorescence microscopy utilizing light in the visible light spectrum for excitation, the longer wavelength of the light source is advantageous in its lower phototoxicity and higher tissue penetration. Its high efficiency in exciting autofluorescence in biological specimens also renders it a unique ability to visualize cells and certain extracellular matrix without fluorescent labeling. 51, 52, 56, 58, 59
For example, cells can be visualized without staining by the autofluorescent NAD(P)H in the cytoplasm. In skin, keratin and elastic fibers also have unique fluorescent signatures that can help to identify specific cells and extracellular matrix networks.56 Using the ultrashort femtosecond laser, a non-linear polarization effect of second harmonic generation (SHG) can also be effectively achieved for imaging.51, 58, 60,51, 58, 60 The interaction of incident light with biological structures of non-centrosymmetry, including collagen and myosin, can produce photons of exactly half the wavelength of the incident photons. Since SHG is a direct polarization effect without absorption of incident photons, there is no heat generated in the process and the photodamage to the targets of interest is minimized. Under the same incident laser, the autofluorescence wavelength is longer than the SHG. Hence, signals from autofluorescence and SHG can be easily spectrally separated for imaging. For example, autofluorescent elastic fibers and SHG-generating collagen fibers can be imaged at the same time by use of multiphoton microscopy.55 Furthermore, since SHG is structurally sensitive, it can be used to analyze the structural transition and denaturation of collagen fibers under various physiological and pathological conditions. 56, 58, 59, 61, 62
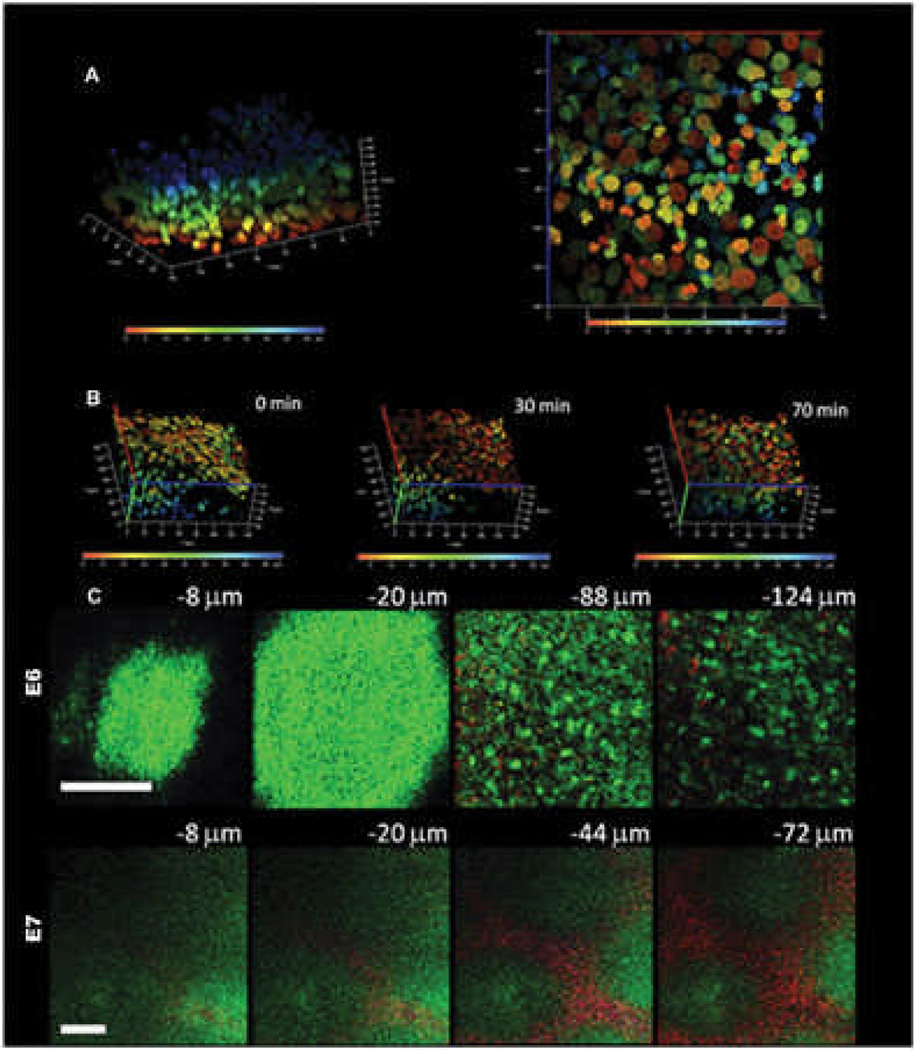
We have employed multiphoton microscopy to analyze the dynamic cell rearrangements In Figure 7A, B, the embryonic skin specimen is labeled with the Hoechst nuclear dye and cultured as an explant. Serial en face mulitphoton images are taken from the surface down to the bottom and a three-dimensional image can be reconstructed from the serial images. To differentiate cells at different depths and to facilitate single cell tracing, cells at depth from the epidermal surface (blue) to the bottom of the dermis (red) are graded by pseudocolor. Reconstituted three-dimensional images that were taken at various time points during explant culture allowed us to analyze the dynamics of cell rearrangements during feather morphogenesis. In Figure 7B, we can see the trend of dermal cell movement toward the right and also toward the bottom of the dermis from time zero to 70 min. For easier analysis of three-dimensional cell movement, we can project the image to either X-Y, X-Z or Y-Z planes and calculate the cell movement vector on each plane. For example, the X-Y projection image of the depth-graded pseudocolor image allows us to trace single cell movement on the X-Y plane (Fig. 7A, right panel).
Fig. 7. Multiphoton microscope imaging of developing chicken skin explant cultures.
A) The embryonic E6 skin is labeled with Hoechst 33342 dye and cultured as an explant. Serial images from the surface to the bottom are reconstituted into a three-dimensional image. The nuclei at different depths from the surface are graded with pseudocolor from blue (epithelial surface) to red (50 µm from surface) to facilitate single cell tracing. The left picture shows the three-dimensional distributions of cells and the right panel shows a bottom view of the nuclei in an en face projection to X-Y plane. The X or Y axis is 140 mm and the Z axis is 50 mm.
B) Time-lapse multiphoton tracing of cell rearrangement. To highlight the mesenchymal cell movement, the dermal side is on the top and epidermal side at the bottom. The depth-graded pseudocolor helps to delineate individual cells and facilitate single cell tracing. There is a trend of movement toward the right hand side of the figure and to the dermal side. The X or Y axis is 140 mm and the Z axis is 50 mm.
C) Multiphoton auto-fluorescence and second harmonic generation (SHG) images of unstained developing feather bud. The upper panel shows the images of E6 skin at different depths before dermal condensation formation. In E6, the dermal cells have an even cell distribution and there is scanty SHG signals from collagen (−88 and −124 mm). The epithelial cells can also be visualized with an autofluorescent cytoplasm and a nuclear halo (−8 and −20 mm).The lower panel shows the images of E7 skin at different depths when dermal condensates appear. In the lower power view of E7 skin, single cells cannot be delineated. The dermal condensates have higher autofluorescence due to the higher cell density and interbud area is rich in SHG signals from the collagen. The low power image clearly demonstrates the preferential cell and collagen distribution in the developing skin. Autofluorescence is green and second harmonic generation is red; bars: 100 mm.
Another strength of multiphoton microscopy is the ability to acquire autofluorescence and SHG signals simultaneously.51, 52, 59 Since the cells are rich in autofluorescent cytoplasmic NAD(P)H and collagen fibers are an effective SHG generator, we can visualize dynamic three-dimensional changes of cell and collagen organization within the developing tissue. The nature and organization of the extracellular matrix is another macro-environmental factor that can influence cell behavior. In Figure 7C, we use unlabeled embryonic chicken skin for multiphoton imaging. We found that the autofluorescence signals (green color) from the cytoplasm allow us to visualize both the epithelial and mesenchymal cells. From the epithelial surface down to 20 µm in depth, the epithelial cells have autofluorescent cytoplasm and the nuclei appear as halos since they lack NAD(P)H. Further down into the dermis, the dermal cells as well as the collagen networks can be seen. On E6, before dermal condensations appear there is no preferential distribution of collagen (Fig. 7C, upper panel). Dot-like short collagen fibers are scattered in the dermis. On E7 when dermal condensates start to build up, there is preferential distribution of collagen in the interbud region and cell density is higher in the dermal condensate (Fig. 7C, lower panel). The collagen fibers are longer and interwoven into connecting networks at this stage.
Discussion
Just as Jason and his Argonauts possessed different indispensible skills in their pursuit of the Golden Fleece, today’s research science also requires a multi-disciplinary approach that is a hallmark of integrative biology. We have applied this approach to understand the intricate cellular and molecular interactions that lead to specific skin appendage formation. In this study we integrate data from many sources to paint a picture of feather morphogenesis based upon tissue interactions, and molecular profiling that may be behind the tissue interactions.
Microarray transcriptome analyses help identify candidate molecules, but we are still in search of the molecular basis of competency and regional specificity in feather development / regeneration
Microarray analyses allow investigators to examine the complete transcriptome from a small number of cells at certain cellular states. By analyzing transcriptomes from different cellular states, one may gain unbiased clues as to which genes and pathways may be involved either as the cause or the consequence cellular state changes. These cellular fates can change during normal development; tissues can obtain different fates in different body regions (Fig. 1B), and normal fates can be trans-differentiated when different mesenchyme are imposed or molecular pathway activities are tilted. For example, rabbit cornea can be forced to form hairs63 and chicken oral mucosa can be induced to form tooth like appendages.64 It also has been possible to achieve this by modulating molecular pathway activities. Classical experiments demonstrated that retinoic acid can convert hair follicles into glands65 in the mouse, and feather buds to form on chicken scales.66 This scale – feather transforming ability is also observed in response to over-expression of delta,67 a BMP dominant negative receptor68,68 and β-catenin. 69 In mice, by enhancing noggin expression and reducing BMP 2, 4, 7 activities at the epithelial-mesenchymal junction, we were able to convert sweat glands and sebaceous glands into hairs.3 In these mice, nipples are also converted into hair forming epithelia. 4 Here we use two examples for microarray analyses: the first one is the determination of feather and scale fate, the second one is the determination of wing and body feathers.
Tissue recombination experiments demonstrated embryonic chicken skin epithelial competency is restricted to a specific time during feather bud morphogenesis (Fig. 3). E9 feather forming epithelium and E11 scale forming epithelium responded poorly to inducing signals arising from the underlying mesenchyme. In order to discover which genes and their relevant pathways were modulating this competency, we integrated microarray analysis with tissue recombination experiments. The result was a comparison of transcriptomes between competent and non-competent tissues, E7 vs E9 feather forming and E9 vs E11 scale forming epithelia (Fig. 3). We observed the classical genes involved in tissue development such at Fgf, Wnt pathway members (Dkk, Frz) and SHH. These were all differentially expressed during modulation of epithelial tissue competency. We also found some interesting putative players in the role of tissue competency. Msx2 was found to be significantly suppressed during scale epithelial tissue competency (Fig. 3A). Further, Msx2 was found to be enriched in a network with Msx1, Bmp3 and Dkk (data not shown). Msx2 has been shown to be a downstream effector of the Bmp pathway70 and Msx2 has been shown to work alongside Msx1 during tissue development.71 The surprising interaction suggested by the network is Msx2 and Dkk (data not shown). Msx2 is known to activate the Wnt pathway during bone anabolsim,72 and Msx2 is activated by the Wnt pathway during stem cell neural crest induction.73 Our data suggests that Msx2 is also involved in epithelial tissue competency and may be working in concert with the Wnt pathway. There are many other candidate pathways. However, the analyses of these data also suffer from a lack of annotation of chicken genes even though efforts to re-annotate continue.74 When more annotation data become available, we will revisit our database and deduce additional relevant pathways.
The mechanism of region specific gene expression has tantalized scientists for decades. The adult chicken provides a fantastic scientific model to explore this question. In the young chick, all feathers appear to be of the same downy type. Yet different morphologies of feathers can emerge from the same feather follicle in the adult (Fig. 8B). To study the molecular mechanism of this process, we first want to know the transcriptome difference among feathers from different body regions. The flight feathers develop on the wings, the tail feathers develop on the tail, and contour feathers develop on the body. We applied microarray analysis to micro-dissected adult chicken feather tissues (Fig. 4A-C; A’-C’). Two dimensional hierarchical clustering yielded some genes specific to the dermal papilla (Fig. 4D, D’). They also yield some difference in Hox genes.
In the vertebrae and in the limb Hox genes were found to be expressed in a collinear pattern.75, 76 This led to the concept that Hox codes specify skeletal identities. 77, 78 Based on this finding and our own observations in the skin,78 we have proposed the “Skin Hox code hypothesis”,79 proposing that combinatorial Hox expression might be involved in determining skin specification (i.e., apteric or pteric, anterior or posterior, medial or lateral, scale or feather). In the developing skin and in the dermal papilla from wing and body feathers, we found some distinct Hox expression patterns and some that are region specific (Fig. 5). Hoxb8 was not expressed at the apetric region at E6.5 suggesting that Hoxb8 may take part in the determining the pteric and apteric regions of the skin. As more research goes on, we can test our prediction that more hox genes are involved in this developmental process.
Dhouailly’s group examined the expression of Hoxc8, d9, d11 and d13 in the dermis of developing limb and found some region specific expression patterns.48 These are in general consistent with our finding. In cultured fibroblasts derived from different parts of the human body, microarray analysis shows specific HOX expression related to their topographic origin.80, 81 Combined, these studies show there is region specific Hox expression in the skin. Complexity is added when we consider skin as being composed of epithelium and mesenchyme, and that it is a two dimensional plane, in contrast to the one dimensional spine or limb axis. Some Hox genes may also be involved in different functions such as growth control82, 83, 84 or other molecules may have to work together to establish skin regional specificity. This complex issue will require further investigation.
Imaging technology revealed complex cellular flow and matrix organization in the developing skin explants
Our molecular expression studies suggest that there are temporal and spatial differences which may guide cells toward specific locations and to specific cell fates. To trace cell movements we have turned to modern imaging technologies. The developing chicken skin explants are unique because the patterning events take place in flat skin composed of a single layer of epidermis and dermis which is about 10 cell layers thick. This unique system offers a wonderful opportunity to explore cell migration during organ (feather) morphogenesis.
We previously had addressed this issue using replication defective spleen necrosis virus to deliver beta-galactosidase to the developing skin and feather buds.85 Based on still images we found that cells seemed to migrate from the midline across the dorsal feather tract. As feather buds formed they contained a mixture of labeled and unlabeled cells suggesting that feathers are derived from a multiple cell lineage. We also used DiI labeling to track the motility of cells in different regions within the feather bud and interbud. With DiI labeling, we examined the role of the p-ERK signaling pathway in cellular chemotaxis.9 Suppression of p-ERK signaling with U0126 led to a more rapid dispersion of dermal cells and a loss of feather bud boundaries, leading to feather bud fusion.
Here, we continued along this line of study using several imaging modalities to learn more about the roles of cell motility and changes of cell shapes during feather morphogenesis. If one were to try to imagine the rules of a football game from watching still images taken at different time points during a game, the task would be very difficult. However, by watching the players in action, it becomes more manageable. Similarly, understanding the dynamic process of tissue morphogenesis from fixed still images is difficult. By capturing the skin appendage morphogenetic events as a movie, it makes the whole process easier to comprehend. We used time-lapse video visible light microscopy to demonstrate the cell movements that are necessary to form the early feather buds. This enables us to begin to see patterns that may be crucial to proper organ formation. Do cells enter the forming buds from all directions or from specific directions? Once in the buds do they stay or can they leave? Do they stay at the base of the bud or do they migrate toward the distal tip? These questions will take some time to answer. Since feathers are made of epithelium and mesenchyme, the formation of epithelial placodes and dermal condensations are in different, albeit related, tissue layers. Along these lines we developed techniques to trace individual cell movements within the epidermis or dermis alone. This technical advance will help us view the relative mobility of cells located at different locations in the morphogenetic explants.
Feather buds, after all, are not as flat as a single layer. To capture these events we turned to confocal and multi-photon microscopy. Confocal microscopy allows a three dimensional reconstruction of tissues and their corresponding molecular expression patterns. Feather bud morphogenesis begins with a uniform field of epithelium overlying mesenchyme at E7. At this stage the epidermal cells are multi-potential, with an equal chance to become bud or interbud epidermis).9, 11 Across this stem cell field, β-catenin expression in the epithelium is uniform and is located at each cell’s membrane. The result is a pentagonal or hexagonal cell shape pattern (Fig. 6C). As feather buds develop, the suprabasal layer expresses β-catenin on the lateral sides but not at the apical side of the cell. The basal layers have a more complex expression pattern with β-catenin expression at the cell membrane, in the cytoplasm, and in the nucleus (data not shown). In the patterning stage, β-catenin is localized to the membrane and thus can help us to visualize changes in cell shape. After H&H stage 31, explants are cultured for 3 days. During this growth period feather buds form and are starting to elongate. Clear distinctions can be seen between the shapes of cells in the interbud zone from those within the feather buds. The β-catenin expression pattern of the suprabasal layer near the base of the feather bud has changed from pentagonal to oblong and diamond shaped, with predominantly four sides (Fig. 6C). This change in cell shape is integral to feather morphogenesis and may be caused by tensile forces. This tension probably derives from the upward growth of the feather bud pulling on the epithelium. These forces result in tension on each of these cells, causing the cells to become elongated and narrow. Conversely, the cells in the interbud region remain pentagonal in shape due to minimal forces acting on them.
Multiphoton microscopy allows for deep imaging without damaging the tissues to be characterized. Auto-fluorescence captured by mulitphoton microscopy enables us to discern differences in cell matrix, keratins and elastic fibers without a need for molecular staining or external illumination. We have shown that a reaction-diffusion mechanism is involved in determining the initial spacing and size of a dermal condensation.9, 12 It has been shown that dermal cell proliferation stops for about 24 hours in the early stage of dermal condensation formation.86 Hence, active cell reorganization should happen during this process and the cell movement should be non-random. However, the events of dermal condensation have not been captured in high resolution, in terms of spatial rearrangements and time intervals. Time-lapse multiphoton images will greatly enhance our appreciation of this patterning process. Here we show two examples on how it starts to change our understanding of the system. First, dermal condensation used to be considered as centripetal migration of dermal cells toward the center of the condensation. Preliminary data here revealed that there is also a non-random Z axis movement which should be taken into consideration in constructing a model. Second, in the developing skin, birefringence has been thought to be derived from dermal collagen. Analysis of birefringence led to the suggestion that a lattice-like system of collagen tracts could have played a guidance role for the alignment and migration of mesenchymal cells during the process of dermal condensation formation.87 Our observation here showed that these collagens are unorganized at the time of patterning and get organized later when feather buds form. It appears that the collagen becomes excluded from the dense dermal condensates. Therefore, the collagen lattice is the consequence, not the cause of the dermal condensation process. It is the dermal condensations that appear first and instruct collagen matrix formation that feeds back to regulate subsequent organization of the dermis. It is interesting that at the same time fibronectin accumulates within the dermal condensations.88 What’s the possible role of the dynamic rearrangement of extracellular matrix in this process? Our unpublished data (SJL) suggests that fibronectin is able to down regulate Bmp2 and Bmp4 expression in mesenchymal cells. Since Bmp proteins are inhibitors for feather bud formation, the selective down regulation of BMP by fibronectin may help to sharpen the ratio of activator/inhibitors in feather buds against the interbud area. Hence, the dynamic arrangement of extracellular matrix during feather bud formation may have a role in stabilizing the initial feather buds.
Cycling skin appendages as a model for systems biology research
In general, over the past several decades developmental biology took a reductionist approach, by taking the system apart and identifying individual components. While these analytical approaches are insightful in providing clues on what pathways are involved, we now aspire to achieve a holistic understanding of how the whole system works. Systems biology represents the trend to understand the function of a whole system by studying the interactions of its different components. This complexity can be appreciated at molecular, structural, temporal, emergence and algorithmic levels. Through these studies, cycling appendages are argued to be an ideal model for systems biological research.89
With the ambitious goal of using system biology to understand complexity, we have employed an integrative biology approach that integrates different, complementary disciplines. Based on our molecular data, we have collaborated with mathematical biologists to develop computer simulation models that can describe the system behavior and also identify some molecular bases of model parameters. One model is the periodic patterning behavior in forming spots and stripes based on Turing activator / inhibitor and chemotaxis.6, 9 The other model is the behavior of the regenerative hair wave.24 These approaches have been fruitful and indeed bring our understanding of the whole system to a higher level.
To completely understand a system, we need to know its origin and how it is built. Here, we want to know the Evo-Devo of ectodermal organs (Fig 8A, C). Since an organism tends to have thousands of hairs or feathers which are individually dispensable, they may be lost or altered without lethal effects. This may provide one path for evolutionary change in hair / feather structures resulting in the acquisition of different functions. Because of the plasticity of the ectodermal organs, we can have variations in lengths and shapes. This variation may provide a small percentage of organisms with an advantage for their particular niche. Natural selection working on those organisms over time may have produced regional specificity. In the last two decades, different fossils of feathered dinosaurs and Mesozoic birds unearthed in the Jehol Biota of China90–92 have provided valuable information that inspires our thoughts on how feathers evolved.90, 93, 94 Indeed, feathers have come a long way in the evolution of ectodermal appendages (Fig. 8). Through novel molecular pathways and cellular processes (invagination, branching, etc.), localized growth and apoptotic zones95, 96 work together to sculpt out different forms of skin appendages.
What the Golden Fleece represents is the distilled essence of the principles of morphogenesis that allow ectodermal organs to generate and regenerate a myriad of forms. The pursuit of the Golden Fleece is fruitful as we have already been enlightened by many unexpected new things on the journey. Even if we do not find the ultimate answer, as Denis Duboule says our scientific journey represents “an intellectual adventure, a trip into ourselves”.97 While this paper represents the status of a work in progress, using an integrative biology approach, we expect many more fascinating facets of the Golden Fleece will be unraveled from different perspectives. We anticipate that this approach will enable us to gain a holistic understanding of ectodermal organ morphogenesis and regeneration in the years to come.
Materials and Methods
Chicken embryos
Chicken embryos for epidermis/dermis recombination and whole mount in situ hybridization were from SPAFAS/Charles River Laboratories and were staged according to Hamburger and Hamilton (1951).98
Embryonic skin explant culture
Embryonic skin from the indicated days were dissected from the body, placed on a culture insert (Falcon, Becton Dickinson) and grown for the indicated time in DMEM containing 10% fetal bovine serum.
Epidermis and dermis recombination
Dorsal skin from E7-E9 and metatarsal skin from E9-E12 were used for epidermis/dermis recombination. Skins were dissected in HBSS. 2XCMF in 0.25% EDTA, pH 7.5 was used to treat the skin for 10 min on ice to separate the epidermis and dermis. The recombined epidermis and dermis were cultured on inserts (Falcon, Becton Dickinson) for 4–6 days in DMEM containing 10% fetal bovine serum.
Whole mount in situ hybridization
Hox RNA probes are from Dr. Tabin.99 Our whole mount in situ hybridization method was performed according to the method of Jiang et al (1998).100 Paraffin sections (15 µm) were prepared from the whole mount in situ hybridization samples and faintly counterstained with eosin.
Time lapse video microscopy
An E9 chicken skin explant was grown on a culture insert (Falcon, Becton Dickinson) within a culture chamber (SmartSlide, Wafergen) for 17 hours. Brightfield images were collected every 15 minutes with an inverted microscope (Olympus) and assembled into a time lapse movie. Explants were grown in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10 % fetal bovine serum plus gentamycin (diluted 1:1000). The bottom plate of the culture chamber was set to 37°C. The cover plate was heated to 37.5°C to avoid condensation which would obscure our view.
Multiphoton microscopy
Multiphoton microscopy was performed as described.59 Imaging was performed on E6 and E7 skin explants.
RNA collection
Dorsal feather forming and ventral scale forming skins from embryonic day 7, day 9, and day 11 (E7, E9 and E11) white leghorn embryos were dissected in HBSS. The epithelium and mesenchyme were separated as described above. Total RNA was extracted from the specific tissue types: E7 feather forming epithelium and mesenchyme, E9 feather forming epithelium and mesenchyme, E9 scale forming epithelium and mesenchyme, and E11 scale forming epithelium and mesenchyme. Each RNA sample contains five (5) pooled tissues.
Microarray Analysis
The chicken RNA samples were probed onto GeneChip Chicken Genome Arrays (Affymetrix). Replicates were performed for each tissue type for a sample size of three (n=3). The raw data was uploaded into Partek Genomic Suite (PGS) using the .cel files. Robust Multi-chip Analysis for background adjustment, Quantile normalization and Robust Linear Model summarization of raw data were performed using the default mode of PGS. Each of the cluster figures (fig. 3D&D’, 4d&D’) are actually subsets of the same, single, and much larger cluster that is based on the initial normalized (RMA) data and not on differential expression gene lists. The cluster was generated with the Pearson Centered distance metric and Centroid linkage. A 4 way Analysis of Variance (ANOVA) was performed to detect statistically different gene expression patterns and generate gene lists. The gene lists were uploaded into Ingenuity Pathway Analysis (IPA) to examine gene enrichment in pathways and gene ontology. Candidate genes were verified by whole mount in situ hybridization and by qPCR.
Quantitative PCR
Quantitative PCR (qPCR) was performed using the Applied Biosciences Power SYBR® Green RNA-to-CT™ 1-Step Kit following the manufacturers protocol. Briefly, replicate RNA samples from the microarray assays were aliquoted onto a 96 well plate. The RT-PCR (reverse transcriptase) master mix, QPCR (quantitative) master mix, and dH20 were added to each sample replicate. Primers designed (QuantPrime) for specific genes were added to the samples.101 The qPCR machine (Stratagene Mx3000p) was programmed with an annealing temperature of 60 degrees for 40 amplification cycles. The threshold cycle (Ct) was used to determine gene expression differences.
Supplementary Material
Acknowledgement
This work is supported by US NIH grant AR42177, AR47364, (to CMC), AR 060306 (CMC, TXJ, RW), and Taiwan National Science Council (to SJ L). SJ Lin is also supported by physician scientist award from National Health Research Institutes (NHRI), Taiwan. AL is supported by pre-doctoral fellowship of California Institute of Regenerative Medicine training grant. Microscopy was performed by the Cell and Tissue Imaging Core of the USC Research Center for Liver Diseases, NIH grant No. P30 DK048522.
References
- 1.Chuong CM. In: Molecular Basis of Epithelial Appendage Morphogenesis. Chuong CM, editor. Austin: Landes Bioscience; 1998. pp. 3–14. [Google Scholar]
- 2.Widelitz RB, Veltmaat JM, Mayer JA, Foley J, Chuong CM. Semin. Cell Dev. Biol. 2007;18:255–266. doi: 10.1016/j.semcdb.2007.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Plikus M, Wang WP, Liu J, Wang X, Jiang TX, Chuong CM. Am. J. Pathol. 2004;164:1099–1114. doi: 10.1016/S0002-9440(10)63197-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mayer JA, Foley J, De La Cruz D, Chuong CM, Widelitz R. Am. J. Pathol. 2008;173:1339–1348. doi: 10.2353/ajpath.2008.070920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chuong CM, Richardson MK. Int. J. Dev. Biol. 2009;53:653–658. doi: 10.1387/ijdb.082594cc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maini PK, Baker RE, Chuong CM. Science. 2006;314:1397–1398. doi: 10.1126/science.1136396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turing AM. Bull. Math. Biol. 1990;52:153–197. doi: 10.1007/BF02459572. [DOI] [PubMed] [Google Scholar]
- 8.Kondo S, Miura T. Science. 2010;329:1616–1620. doi: 10.1126/science.1179047. [DOI] [PubMed] [Google Scholar]
- 9.Lin CM, Jiang TX, Baker RE, Maini PK, Widelitz RB, Chuong CM. Dev. Biol. 2009;334:369–382. doi: 10.1016/j.ydbio.2009.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yue Z, Jiang TX, Widelitz RB, Chuong CM. Nature. 2005;438:1026–1029. doi: 10.1038/nature04222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang TX, Jung HS, Widelitz RB, Chuong CM. Development. 1999;126:4997–5009. doi: 10.1242/dev.126.22.4997. [DOI] [PubMed] [Google Scholar]
- 12.Jung HS, Francis-West PH, Widelitz RB, Jiang TX, Ting-Berreth S, Tickle C, Wolpert L, Chuong CM. Dev. Biol. 1998;196:11–23. doi: 10.1006/dbio.1998.8850. [DOI] [PubMed] [Google Scholar]
- 13.Schlake T, Sick S. Cell. Adh Migr. 2007;1:149–151. doi: 10.4161/cam.1.3.5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turner N, Grose R. Nat. Rev. Cancer. 2010;10:116–129. doi: 10.1038/nrc2780. [DOI] [PubMed] [Google Scholar]
- 15.Blitz IL, Cho KW. Dev. Dyn. 2009;238:1321–1331. doi: 10.1002/dvdy.21984. [DOI] [PubMed] [Google Scholar]
- 16.van Amerongen R, Nusse R. Development. 2009;136:3205–3214. doi: 10.1242/dev.033910. [DOI] [PubMed] [Google Scholar]
- 17.Jiang TX, Widelitz RB, Shen WM, Will P, Wu DY, Lin CM, Jung HS, Chuong CM. Int. J. Dev. Biol. 2004;48:117–135. doi: 10.1387/ijdb.041788tj. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen W, Will P, Galstyan A, Chuong C-M. Autonomous Robots. 2004;17:93. [Google Scholar]
- 19.Rubenstein M, Sai Y, Chuong CM, Shen WM. Int. J. Dev. Biol. 2009;53:869–881. doi: 10.1387/ijdb.092937mr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chuong CM, Wu P, Plikus M, Jiang TX, Bruce Widelitz R. Curr. Top. Dev. Biol. 2006;72:237–274. doi: 10.1016/S0070-2153(05)72005-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu M, Wu P, Widelitz RB, Chuong CM. Nature. 2002;420:308–312. doi: 10.1038/nature01196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yue Z, Jiang TX, Widelitz RB, Chuong CM. Proc. Natl. Acad. Sci. U. S. A. 2006;103:951–955. doi: 10.1073/pnas.0506894103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stenn KS, Paus R. Physiol. Rev. 2001;81:449–494. doi: 10.1152/physrev.2001.81.1.449. [DOI] [PubMed] [Google Scholar]
- 24.Plikus MV, Mayer JA, de la Cruz D, Baker RE, Maini PK, Maxson R, Chuong CM. Nature. 2008;451:340–344. doi: 10.1038/nature06457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Plikus MV, Widelitz RB, Maxson R, Chuong CM. Int. J. Dev. Biol. 2009;53:857–868. doi: 10.1387/ijdb.072564mp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dolberg DS, Bissell MJ. Nature. 1984;309:552–556. doi: 10.1038/309552a0. [DOI] [PubMed] [Google Scholar]
- 27.Stoker AW, Hatier C, Bissell MJ. J. Cell Biol. 1990;111:217–228. doi: 10.1083/jcb.111.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mintz B, Silvers WK. Cancer Res. 1996;56:463–466. [PubMed] [Google Scholar]
- 29.Sternlicht MD, Kouros-Mehr H, Lu P, Werb Z. Differentiation. 2006;74:365–381. doi: 10.1111/j.1432-0436.2006.00105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sternlicht MD, Sunnarborg SW, Kouros-Mehr H, Yu Y, Lee DC, Werb Z. Development. 2005;132:3923–3933. doi: 10.1242/dev.01966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mantovani A, Allavena P, Sica A, Balkwill F. Nature. 2008;454:436–444. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 32.Coussens LM, Werb Z. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cunha GR, Donjacour AA, Cooke PS, Mee S, Bigsby RM, Higgins SJ, Sugimura Y. Endocr. Rev. 1987;8:338–362. doi: 10.1210/edrv-8-3-338. [DOI] [PubMed] [Google Scholar]
- 34.Hayward SW, Haughney PC, Rosen MA, Greulich KM, Weier HU, Dahiya R, Cunha GR. Differentiation. 1998;63:131–140. doi: 10.1046/j.1432-0436.1998.6330131.x. [DOI] [PubMed] [Google Scholar]
- 35.Aboseif S, El-Sakka A, Young P, Cunha G. Differentiation. 1999;65:113–118. doi: 10.1046/j.1432-0436.1999.6520113.x. [DOI] [PubMed] [Google Scholar]
- 36.Ricke WA, Ishii K, Ricke EA, Simko J, Wang Y, Hayward SW, Cunha GR. Int. J. Cancer. 2006;118:2123–2131. doi: 10.1002/ijc.21614. [DOI] [PubMed] [Google Scholar]
- 37.Mayer JA, Chuong CM, Widelitz R. Differentiation. 2004;72:474–488. doi: 10.1111/j.1432-0436.2004.07209003.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weigelt B, Bissell MJ. Semin. Cancer Biol. 2008;18:311–321. doi: 10.1016/j.semcancer.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.LaBarge MA, Nelson CM, Villadsen R, Fridriksdottir A, Ruth JR, Stampfer MR, Petersen OW, Bissell MJ. Integr. Biol. (Camb) 2009;1:70–79. doi: 10.1039/b816472j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bissell M. Integr. Biol. (Camb) 2010;2:9. doi: 10.1039/b923760g. [DOI] [PubMed] [Google Scholar]
- 41.Prin F, Dhouailly D. Int. J. Dev. Biol. 2004;48:137–148. doi: 10.1387/ijdb.15272378. [DOI] [PubMed] [Google Scholar]
- 42.Dhouailly D. Int. J. Dev. Biol. 2009;53:775–782. doi: 10.1387/ijdb.072552cc. [DOI] [PubMed] [Google Scholar]
- 43.Kupershmidt I, Su QJ, Grewal A, Sundaresh S, Halperin I, Flynn J, Shekar M, Wang H, Park J, Cui W, Wall GD, Wisotzkey R, Alag S, Akhtari S, Ronaghi M. PLoS One. 2010;5:e13066. doi: 10.1371/journal.pone.0013066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rendl M, Lewis L, Fuchs E. PLoS Biol. 2005;3:e331. doi: 10.1371/journal.pbio.0030331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morris RJ, Liu Y, Marles L, Yang Z, Trempus C, Li S, Lin JS, Sawicki JA, Cotsarelis G. Nat. Biotechnol. 2004;22:411–417. doi: 10.1038/nbt950. [DOI] [PubMed] [Google Scholar]
- 46.Lewis EB. Nature. 1978;276:565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- 47.Kessel M, Gruss P. Science. 1990;249:374–379. doi: 10.1126/science.1974085. [DOI] [PubMed] [Google Scholar]
- 48.Kanzler B, Viallet JP, Le Mouellic H, Boncinelli E, Duboule D, Dhouailly D. Int. J. Dev. Biol. 1994;38:633–640. [PubMed] [Google Scholar]
- 49.Reid AI, Gaunt SJ. Int. J. Dev. Biol. 2002;46:209–215. doi: 10.1387/ijdb.011495. [DOI] [PubMed] [Google Scholar]
- 50.Brown EB, Campbell RB, Tsuzuki Y, Xu L, Carmeliet P, Fukumura D, Jain RK. Nat. Med. 2001;7:864–868. doi: 10.1038/89997. [DOI] [PubMed] [Google Scholar]
- 51.Zoumi A, Yeh A, Tromberg BJ. Proc. Natl. Acad. Sci. U. S. A. 2002;99:11014–11019. doi: 10.1073/pnas.172368799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zipfel WR, Williams RM, Christie R, Nikitin AY, Hyman BT, Webb WW. Proc. Natl. Acad. Sci. U. S. A. 2003;100:7075–7080. doi: 10.1073/pnas.0832308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee JN, Jee SH, Chan CC, Lo W, Dong CY, Lin SJ. J. Invest. Dermatol. 2008;128:2240–2247. doi: 10.1038/jid.2008.82. [DOI] [PubMed] [Google Scholar]
- 54.Li FC, Liu Y, Huang GT, Chiou LL, Liang JH, Sun TL, Dong CY, Lee HS. Am. J. Physiol. Gastrointest. Liver Physiol. 2009;296:G1091–G1097. doi: 10.1152/ajpgi.90681.2008. [DOI] [PubMed] [Google Scholar]
- 55.Tsai TH, Jee SH, Chan JY, Lee JN, Lee WR, Dong CY, Lin SJ. J. Biomed. Opt. 2009;14:024034. doi: 10.1117/1.3116711. [DOI] [PubMed] [Google Scholar]
- 56.Tsai TH, Jee SH, Dong CY, Lin SJ. J. Dermatol. Sci. 2009;56:1–8. doi: 10.1016/j.jdermsci.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 57.Denk W, Strickler JH, Webb WW. Science. 1990;248:73–76. doi: 10.1126/science.2321027. [DOI] [PubMed] [Google Scholar]
- 58.Lin SJ, Hsiao CY, Sun Y, Lo W, Lin WC, Jan GJ, Jee SH, Dong CY. Opt. Lett. 2005;30:622–624. doi: 10.1364/ol.30.000622. [DOI] [PubMed] [Google Scholar]
- 59.Lin SJ, Wu R, Jr, Tan HY, Lo W, Lin WC, Young TH, Hsu CJ, Chen JS, Jee SH, Dong CY. Opt. Lett. 2005;30:2275–2277. doi: 10.1364/ol.30.002275. [DOI] [PubMed] [Google Scholar]
- 60.Campagnola PJ, Loew LM. Nat. Biotechnol. 2003;21:1356–1360. doi: 10.1038/nbt894. [DOI] [PubMed] [Google Scholar]
- 61.Lin SJ, Jee SH, Kuo CJ, Wu RJ, Lin WC, Chen JS, Liao YH, Hsu CJ, Tsai TF, Chen YF, Dong CY. Opt. Lett. 2006;31:2756–2758. doi: 10.1364/ol.31.002756. [DOI] [PubMed] [Google Scholar]
- 62.Lin SJ, Lo W, Tan HY, Chan JY, Chen WL, Wang SH, Sun Y, Lin WC, Chen JS, Hsu CJ, Tjiu JW, Yu HS, Jee SH, Dong CY. J. Biomed. Opt. 2006;11:34020. doi: 10.1117/1.2209959. [DOI] [PubMed] [Google Scholar]
- 63.Pearton DJ, Yang Y, Dhouailly D. Proc. Natl. Acad. Sci. U. S. A. 2005;102:3714–3719. doi: 10.1073/pnas.0500344102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen Y, Zhang Y, Jiang TX, Barlow AJ, St Amand TR, Hu Y, Heaney S, Francis-West P, Chuong CM, Maas R. Proc. Natl. Acad. Sci. U. S. A. 2000;97:10044–10049. doi: 10.1073/pnas.160245097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hardy MH, Dhouailly D, Torma H, Vahlquist A. J. Exp. Zool. 1990;256:279–289. doi: 10.1002/jez.1402560307. [DOI] [PubMed] [Google Scholar]
- 66.Dhouailly D, Hardy MH, Sengel P. J. Embryol. Exp. Morphol. 1980;58:63–78. [PubMed] [Google Scholar]
- 67.Crowe R, Niswander L. Dev. Biol. 1998;195:70–74. doi: 10.1006/dbio.1997.8844. [DOI] [PubMed] [Google Scholar]
- 68.Zou H, Niswander L. Science. 1996;272:738–741. doi: 10.1126/science.272.5262.738. [DOI] [PubMed] [Google Scholar]
- 69.Widelitz RB, Jiang TX, Lu J, Chuong CM. Dev. Biol. 2000;219:98–114. doi: 10.1006/dbio.1999.9580. [DOI] [PubMed] [Google Scholar]
- 70.Brugger SM, Merrill AE, Torres-Vazquez J, Wu N, Ting MC, Cho JY, Dobias SL, Yi SE, Lyons K, Bell JR, Arora K, Warrior R, Maxson R. Development. 2004;131:5153–5165. doi: 10.1242/dev.01390. [DOI] [PubMed] [Google Scholar]
- 71.Reginelli AD, Wang YQ, Sassoon D, Muneoka K. Development. 1995;121:1065–1076. doi: 10.1242/dev.121.4.1065. [DOI] [PubMed] [Google Scholar]
- 72.Cheng SL, Shao JS, Cai J, Sierra OL, Towler DA. J. Biol. Chem. 2008;283:20505–20522. doi: 10.1074/jbc.M800851200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hussein SM, Duff EK, Sirard C. J. Biol. Chem. 2003;278:48805–48814. doi: 10.1074/jbc.M305472200. [DOI] [PubMed] [Google Scholar]
- 74.van den Berg BH, McCarthy FM, Lamont SJ, Burgess SC. PLoS One. 2010;5:e10642. doi: 10.1371/journal.pone.0010642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Iimura T, Pourquie O. Nature. 2006;442:568–571. doi: 10.1038/nature04838. [DOI] [PubMed] [Google Scholar]
- 76.Morgan BA, Tabin CJ. Curr. Opin. Genet. Dev. 1993;3:668–674. doi: 10.1016/0959-437x(93)90105-x. [DOI] [PubMed] [Google Scholar]
- 77.Zakany J, Duboule D. Curr. Opin. Genet. Dev. 2007;17:359–366. doi: 10.1016/j.gde.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 78.Chuong CM, Oliver G, Ting SA, Jegalian BG, Chen HM, De Robertis EM. Development. 1990;110:1021–1030. doi: 10.1242/dev.110.4.1021. [DOI] [PubMed] [Google Scholar]
- 79.Chuong CM. Bioessays. 1993;15:513–521. doi: 10.1002/bies.950150804. [DOI] [PubMed] [Google Scholar]
- 80.Chang HY, Chi JT, Dudoit S, Bondre C, van de Rijn M, Botstein D, Brown PO. Proc. Natl. Acad. Sci. U. S. A. 2002;99:12877–12882. doi: 10.1073/pnas.162488599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chang HY. Science. 2009;326:1206–1207. doi: 10.1126/science.1175686. [DOI] [PubMed] [Google Scholar]
- 82.Godwin AR, Capecchi MR. J. Investig. Dermatol. Symp. Proc. 1999;4:244–247. doi: 10.1038/sj.jidsp.5640221. [DOI] [PubMed] [Google Scholar]
- 83.Rieger E, Bijl JJ, van Oostveen JW, Soyer HP, Oudejans CB, Jiwa NM, Walboomers JM, Meijer CJ. J. Invest. Dermatol. 1994;103:341–346. doi: 10.1111/1523-1747.ep12394888. [DOI] [PubMed] [Google Scholar]
- 84.Godwin AR, Capecchi MR. Genes Dev. 1998;12:11–20. doi: 10.1101/gad.12.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chuong CM, Jung HS, Noden D, Widelitz RB. Biochem. Cell Biol. 1998;76:1069–1077. doi: 10.1139/o99-015. [DOI] [PubMed] [Google Scholar]
- 86.Wessells NK. Dev. Biol. 1965;12:131–153. doi: 10.1016/0012-1606(65)90025-4. [DOI] [PubMed] [Google Scholar]
- 87.Stuart ES, Moscona AA. Science. 1967;157:947–948. doi: 10.1126/science.157.3791.947-a. [DOI] [PubMed] [Google Scholar]
- 88.Mauger A, Demarchez M, Herbage D, Grimaud JA, Druguet M, Hartmann D, Sengel P. Dev. Biol. 1982;94:93–105. doi: 10.1016/0012-1606(82)90072-0. [DOI] [PubMed] [Google Scholar]
- 89.Al-Nuaimi Y, Baier G, Watson RE, Chuong CM, Paus R. Exp. Dermatol. 2010;19:707–713. doi: 10.1111/j.1600-0625.2010.01114.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wu P, Hou L, Plikus M, Hughes M, Scehnet J, Suksaweang S, Widelitz R, Jiang TX, Chuong CM. Int. J. Dev. Biol. 2004;48:249–270. doi: 10.1387/ijdb.041825pw. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Prum RO, Brush AH. Q. Rev. Biol. 2002;77:261–295. doi: 10.1086/341993. [DOI] [PubMed] [Google Scholar]
- 92.Hou L, Chuong C, Yang A, Zheng XL, Hou JF. Fossil Birds of China. China: Yunnan Science and Technology; 2003. [Google Scholar]
- 93.Chuong CM, Wu P, Zhang FC, Xu X, Yu M, Widelitz RB, Jiang TX, Hou L. J. Exp. Zoolog B. Mol. Dev. Evol. 2003;298:42–56. doi: 10.1002/jez.b.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Prum RO. J. Exp. Zool. B. Mol. Dev. Evol. 2005;304:570–579. doi: 10.1002/jez.b.21073. [DOI] [PubMed] [Google Scholar]
- 95.Chodankar R, Chang CH, Yue Z, Jiang TX, Suksaweang S, Burrus L, Chuong CM, Widelitz R. J. Invest. Dermatol. 2003;120:20–26. doi: 10.1046/j.1523-1747.2003.12008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chang CH, Yu M, Wu P, Jiang TX, Yu HS, Widelitz RB, Chuong CM. J. Invest. Dermatol. 2004;122:1348–1355. doi: 10.1111/j.0022-202X.2004.22611.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Duboule D. Int. J. Dev. Biol. 2009;53:717–723. doi: 10.1387/ijdb.072558mr. [DOI] [PubMed] [Google Scholar]
- 98.Hamburger V, Hamilton HL. J. Morphol. 1951;88:49–92. [PubMed] [Google Scholar]
- 99.Nelson CE, Morgan BA, Burke AC, Laufer E, DiMambro E, Murtaugh LC, Gonzales E, Tessarollo L, Parada LF, Tabin C. Development. 1996;122:1449–1466. doi: 10.1242/dev.122.5.1449. [DOI] [PubMed] [Google Scholar]
- 100.Jiang TX, Stott S, Widelitz RB, Chuong CM. Chuong CM. Molecular Basis of Epithelial Appendage Morphogenesis. Austin, TX: Landes Bioscience; 1998. p. 359. [Google Scholar]
- 101.Arvidsson S, Kwasniewski M, Riano-Pachon DM, Mueller-Roeber B. BMC Bioinformatics. 2008;9:465. doi: 10.1186/1471-2105-9-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chuong CM, Chodankar R, Widelitz RB, Jiang TX. Curr. Opin. Genet. Dev. 2000;10:449–456. doi: 10.1016/s0959-437x(00)00111-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.