SUMMARY
The function of the Vibrio 7th pandemic island-1 (VSP-1) in cholera pathogenesis has remained obscure. Utilizing ChIP-seq and RNA-seq to map the regulon of the master virulence regulator ToxT, we identify a TCP island encoded small RNA that reduces the expression of a previously unrecognized VSP-1 encoded transcription factor termed VspR. VspR modulates the expression of several VSP-1 genes including one that encodes a novel class of di-nucleotide cyclase (DncV), which preferentially synthesizes a previously undescribed hybrid cyclic AMP-GMP molecule. We show that DncV is required for efficient intestinal colonization and downregulates V. cholerae chemotaxis, a phenotype previously associated with hyperinfectivity. This pathway couples the actions of previously disparate genomic islands, defines VSP-1 as a pathogenicity island in V. cholerae and implicates its occurrence in 7th pandemic strains as a benefit for host adaptation through the production of a regulatory cyclic di-nucleotide.
INTRODUCTION
The causative agent of cholera, Vibrio cholerae, requires a multifaceted regulatory network involving a hierarchy of transcription factors (TF), small RNAs and signaling molecules to ensure the proper spatial and temporal activation of virulence factor expression (Ritchie and Waldor, 2009). Often referred to as the master virulence regulator, ToxT directly activates expression of several essential virulence genes including those that encode cholera toxin and an essential colonization factor, the toxin co-regulated pilus (TCP) (Lowden et al., 2010). While ToxT is essential for V. cholerae pathogenesis, the complete spectrum of genes it regulates is unknown (DiRita et al., 1991; Weber and Klose, 2011).
ToxT is encoded on the TCP island, a chromosomal segment that encodes many virulence associated genes found in all pandemic strains of V. cholerae (Faruque and Mekalanos, 2003). All pandemic strains also carry the CTX prophage encoding the genes for cholera toxin (ctxAB) as well as the genomes for satellite phages (Hassan et al., 2010). Two V. cholerae biotypes account for the majority of all recorded cholera pandemics. The classical biotype is thought to have caused the first six pandemics (Kaper et al., 1995; Pollitzer, 1959), but was replaced by the 7th pandemic El Tor biotype approximately 50 years ago (Faruque and Mekalanos, 2003). 7th pandemic El Tor strains are genetically distinguishable from classical strains by the presence of two genomic islands, VSP-1 and VSP-2 (Dziejman et al., 2002). These islands are consistently found in clinical El Tor isolates and are thought to be responsible, at least in part, for the success of the 7th pandemic clone (Dziejman et al., 2002; Grim et al., 2010; Rahman et al., 2008; Taviani et al., 2010). However, validation of predicted functions for most ORFs on these islands or evidence for the contribution of VSP-1 or VSP-2 to environmental or host adaptation is lacking (Dziejman et al., 2002). Furthermore, it is unknown how VSP-1 and VSP-2 genes are regulated and whether their products act as isolated modules or integrate into pathways with the ancestral genome or accessory encoded elements such as the TCP island and CTX prophage.
Cyclic di-nucleotides act as intracellular signals, modulating a range of cellular activities (Gomelsky, 2011). In Gram-negative bacteria cyclic-di-GMP (c-di-GMP) is a major regulator of biofilms (Tischler and Camilli, 2004) and flagellum biosynthesis (Lim et al., 2007). Coordination between c-di-GMP and transcription was clearly shown in V. cholerae where c-di-GMP directly binds to and modulates the activity of transcriptional regulator VpsT (Krasteva et al., 2010) and up-regulates another virulence regulator AphA (Srivastava et al., 2011). Gram-positive bacteria also produce cyclic di-AMP (c-di-AMP). Listeria monocytogenes secretes cyclic di-AMP (c-di- AMP), which serves as a recognition signal for the mammalian innate immune system (Woodward et al., 2010) and Staphylococcus aureus uses c-di-AMP to manage cell membrane stress (Corrigan et al., 2011). Surprisingly, cyclic di-nucleotides other than c-di-GMP have not been reported in Gram-negative bacteria including V. cholerae.
Here we use massively parallel DNA sequencing technologies to identify the complete regulon of ToxT. Combining transcriptome profiling (RNA-seq) with binding location analysis (ChIP-seq) we identify all genes directly regulated by ToxT in vivo as well as several indirectly regulated pathways. From this analysis we identified a TCP encoded small RNA (sRNA) that down-regulates expression of a novel VSP-1 encoded transcription factor, which in turn controls the expression of several VSP-1 genes including one that is required for efficient intestinal colonization by V. cholerae. We show that the gene product that stimulates intestinal colonization encodes a di-nucleotide cyclase that preferentially synthesizes a hybrid cyclic AMPGMP molecule. Furthermore, the activity of this enzyme strongly influences the chemotaxtic behavior of V. cholerae thereby associating the action of this enzyme with a process known to influence infectivity and the intestinal colonization process.
RESULTS
ToxT directly activates genes only within the TCP island and CTX prophage and indirectly regulates several other metabolic pathways
ToxT expression increases 500–1000 fold when V. cholerae colonizes a mammalian host (Mandlik et al., 2011). To simulate this expression change we cloned ToxT into a plasmid under the control of an inducible promoter in V. cholerae El Tor strain C6706, an isolate from the 7th pandemic that extended into Peru in 1991 (Mandlik et al., 2011). Using this system we increased ToxT expression ~1000 fold following induction (Table 1). We previously described a method to perform chromatin immunoprecipiation in combination with massively parallel sequencing (ChIP-seq) to identify genome wide binding locations of transcription factors in V. cholerae (Davies et al., 2011). Applying this method, we identified all ToxT binding locations (ChIPpeaks) in the V. cholerae genome under our test conditions (Figure 1A and Table 1).
Table 1.
ChIP-seq and RNA-seq identify ToxT genomic binding locations and transcriptional effects on associated genes in V. cholerae.
| Gene ID | Gene Symbol | Expression Fold Change pToxT/pBADa |
ChIP Peak Coordinatesb | |
|---|---|---|---|---|
| 5′ Range | 3′ Range | |||
| VC0819 | aldA-1* | 25.4 | 876617-876658 | 877119-877227 |
| VC0820 | tagA* | 9.6 | ||
| VC0821 | 4.9 | |||
| VC0822 | 4.4 | |||
| VC0823 | 4.4 | |||
| VC0824 | tagD | 5.5 | ||
| VC0825 | tcpI | 26.7 | 888017-888176 | 889300-889465 |
| VC0826 | tcpP | 11.4 | ||
| VC0827 | tcpH | 14.2 | ||
| VC0828 | tcpA* | 121.0 | 889949-890042 | 890629-890743 |
| VC0829 | tcpB | 83.5 | ||
| VC0830 | tcpQ | 130.0 | ||
| VC0831 | tcpC | 77.4 | ||
| VC0832 | tcpR | 50.2 | ||
| VC0833 | tcpD | 19.4 | ||
| VC0834 | tcpS | 18.3 | ||
| VC0835 | tcpT | 45.2 | ||
| VC0836 | tcpE | 42.7 | ||
| VC0837 | tcpF | 14.5 | ||
| VC0838 | toxT | 1092.8 | ||
| VC0839 | tcpJ | 35.7 | ||
| VC0841 | acfC | 10.7 | ||
| VC0842 | 10.4 | |||
| VC0843 | tagE-1 | 19.3 | ||
| VC0844 | acfA* | 11.8 | 906114-906155 | 906501-906548 |
| VC0845 | acfD* | 12.2 | ||
| VC1456 | ctxB | 14.6 | ||
| VC1457 | ctxA* | 39.7 | 1567651-1567778 | 1568622-1568768 |
| Intergenic VC0845-VC0846 | ------- | 906114-906155 | 910838-910977 | 911831-911899 |
Fold expression change for each gene determined by RNA-seq from three independent samples; FDR corrected p < 0.001.
The location of ToxT peaks were determined from four independent samples (FDR < 0.001). The range of the 5′ and 3′ end coordinate is shown. A ChIP-peak is associated with a gene if it overlaps the start codon.
Genes with previously identified/predicted ToxT binding sites in their promoter regions
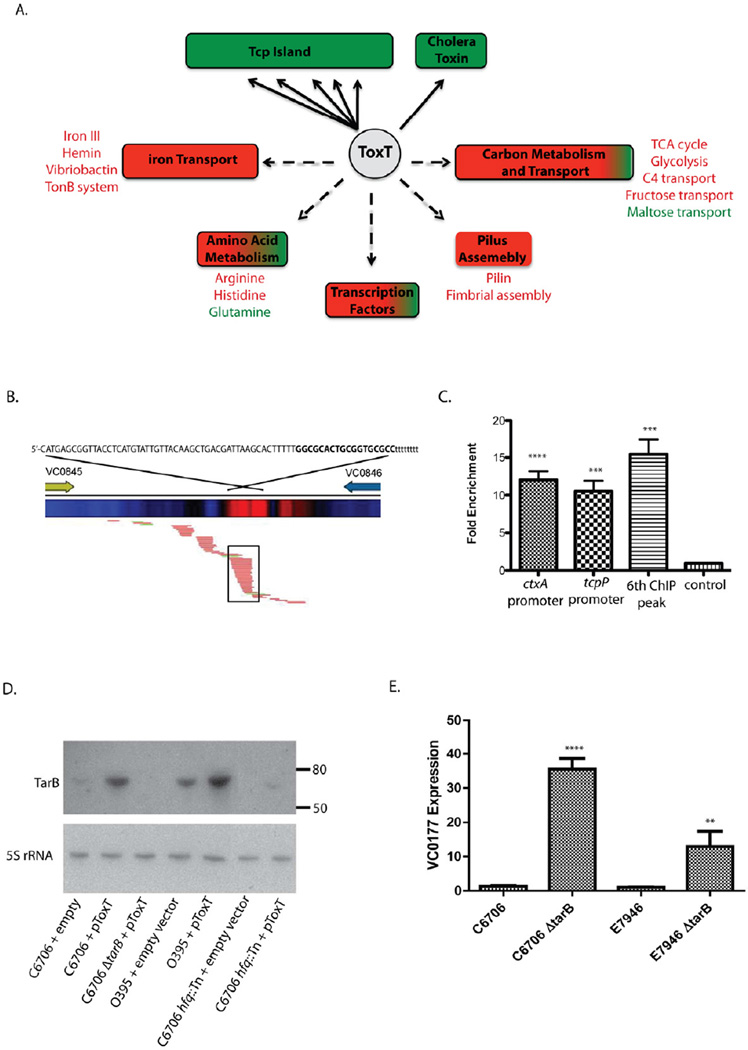
Figure 1.
(A) Schematic outlining ToxT regulon. Major regulatory targets grouped by functional class are shown. Solid arrows denote direct ToxT regulation identified by integrated ChIP-seq and RNA-seq analysis. Dashed arrows denote indirect ToxT regulation identified by RNA-seq alone. Green indicates positive gene expression, red indicates negative gene expression. Specific pathways are indicated beside their functional group and colored to indicate the direction of their regulation. (B) Schematic of the 6th ToxT ChIP-peak site in the Tcp island between VC0845 and VC0846. The heat map shows enrichment (red) of sequenced reads at ToxT ChIP-peak. Below, alignment of RNA-seq reads shows the predicted sRNA outlined by the black box. RNA-seq reads colored red align to the reverse strand of the genome while reads colored green align to the forward strand. The predicted sequence of the sRNA is shown at the top in upper case font from 5′ to 3′. The nucleotides in bold highlight a rho independent hairpin terminator predicted by the ARNold program server. (C) ToxT ChIP enrichment of the promoter regions of ctxA, tcpA and 6th ToxT ChIP peak were determined by qPCR relative to sample DNA input. Enrichment of a non ToxT-dependent promoter of VC1141 is shown as a control. Significance was determined by t-test relative to the VC1141 promoter; *** p < 0.001; **** p < 0.0001 (D) Northern blot of RNA from indicated strains probed for the presence of the sRNA (TarB) as predicted by RNAseq alignment (top panel). The size markers of a ssRNA ladder are indicated. The same samples were probed for 5S rRNA as a loading control (bottom panel). (E) qPCR analysis of the effect of TarB on VC0177 (VspR) expression levels in V. cholerae wild type strains C6706 and E7946 following expression of ToxT from a plasmid. Expression for each sample was determined relative to 16S rRNA and at least 3 independent replicates tested. Fold changes are normalized relative to expression in the wild type strain. Significance was determined by t-test relative to the wild type strain; ** p < 0.01; **** p < 0.0001. See also Table S1.
From each ChIP sequencing run, we aligned an average of 1, 400, 000 short ~36 base reads to the published N16961 V. cholerae genome (Heidelberg et al., 2000). The alignments gave a total average coverage of 13-fold for chromosome 1 and 8-fold coverage of chromosome 2. This depth of coverage allowed us to use a low false discovery rate (FDR) cut-off of 0.001% to call ToxT ChIP peaks. ChIP peaks are called when the sequence coverage of a given genomic region in the experimental sample exceeds the control sample at a rate specified by the FDR (see Experimental Procedures). We compared the peak lists generated from 4 samples and set a limit that a peak must be called in all experiments to be considered a ToxT binding site (Handstad et al., 2011). We then associated a ToxT peak with a gene if the peak overlapped the first codon of the respective gene (Figure 1A; Table 1).
To validate ToxT transcriptional control of associated genes we briefly induced ToxT expression (10 min), isolated and sequenced total mRNA (RNA-seq), and compared the transcriptome profile to a control strain carrying an empty expression vector. We reasoned that direct regulatory targets of ToxT would show the most rapid transcriptional response to brief ToxT induction. Comparison of the transcriptome response with our ChIP-seq analysis uniformly showed strong transcriptional upregulation of the same genes we had associated with ToxT ChIP-peaks supporting our conclusion that these genes are directly activated by ToxT (Figure 1A and Table 1). These results provide the first detailed in vivo map of ToxT binding locations in conjunction with ToxT driven expression alteration.
A ToxT binding motif (Toxbox) has been previously proposed based on the sequences of all known ToxT binding sites (Withey and DiRita, 2006). This Toxbox consensus matches to thousands of potential sites across the V. cholerae genome. Thus we were surprised when our analysis identified only 6 ToxT ChIP-peaks (Table 1). Interestingly all ToxT ChIP-peaks are located in the TCP island and CTX prophage. Importantly, five of the six ChIP-peaks we identified overlap all ToxT binding sites previously identified by direct in vitro ToxT foot printing assays or inferred from genetic analysis (Richard et al., 2010; Withey and DiRita, 2005a, b, 2006) validating our technique to identify authentic ToxT binding locations. Three ToxT ChIP peaks lay between divergently transcribed gene pairs aldA-1/tagA, tcpI/tcpP and acfA/acfD. Each of these genes is known to have its own associated ToxT binding motif(s) (Richard et al., 2010; Withey and DiRita, 2005a, b, 2006), however resolving closely spaced binding sites using ChIPseq is challenging and often results in one overlapping ChIP peak. The confinement of ToxT binding to the TCP island and CTX prophage, despite its predicted statistically common consensus binding motif, suggests factors in addition to nucleotide sequence may influence the stringency of DNA binding selection by ToxT in vivo.
We also identified several additional genes not directly associated with ToxT ChIP-peaks that were differentially regulated following ToxT induction (Figure 1; Table S1). These genes encode components of pathways involved in iron transport, amino acid metabolism, transcriptional regulation, pilus assembly and carbon transport and metabolism (Figure 1). Interestingly, while direct regulatory targets of ToxT were strongly upregulated, the majority of these indirectly regulated genes were downregulated. Among the pathways impacted, iron transport exhibited the largest concerted effect with 19 genes down regulated > 5 fold (Figure 1; Table S1). This includes pathways for the production of vibriobactin, hemin and iron III transporters as well as TonB, a protein that supports transport. Conversely, the largest upregulation was from genes involved in maltose transport (Figure 1A; Table S1). Specifically the maltose ABC transporter and the maltoporin (ompS) all showed increased expression > 12 fold (Table S1). It is unclear how ToxT indirectly impacts the expression of these genes. ToxT does directly increase expression of the transcription factor TcpP encoded by the TCP island, thus it is possible that TcpP directly affects some genes including the 8 additional transcription factors indirectly affected by ToxT (Figure 1A; Table 1). TcpP and these transcriptional regulators may drive changes observed in the ToxT transcriptome profile.
ToxT directly regulates the expression of the TCP Island encoded small RNA TarB
We identified a sixth ToxT ChIP peak located in an intergenic region of the Tcp island between genes VC0845 and VC0846 (Figure 1B). Quantitative PCR (qPCR) validated our sequencing data showing the enrichment of ToxT ChIP DNA at this intergenic site was similar to that of ToxT binding sites upstream of ctxA and tcpP (Figure 1C). Alignment of RNA-seq reads from ToxT expressing V. cholerae with the region between VC0845 and VC0846 revealed a putative small RNA (sRNA) on the reverse strand overlapping the new ToxT binding site (Figure 1B). The sRNA had a clear 5′ start at coordinate 911308 and appeared to terminate at coordinate 911239 following a predicated rho independent hairpin terminator (Figure 1B). Northern blotting using a probe against this region identified a sRNA that was strongly induced by ToxT (Figure 1D). The sRNA migrated slightly below the 80 nucleotides (nt) ssRNA marker agreeing with the 69 nt size predicted from RNA-seq read alignments. An identical sRNA has recently been identified (Bradley et al., 2011) and thus, we called this sRNA ToxT-activated sRNA B (TarB). TarB has also been shown to be strongly upregulated in V. cholerae during infection of an animal model (Mandlik et al., 2011).
TarB represses the expression of a V. cholerae VSP-1 gene
The small regulatory protein Hfq binds sRNAs, stabilizing and assisting them for targeting and promoting degradation of specific mRNAs (Brennan and Link, 2007). TarB levels were greatly decreased in a V. cholerae C6706 hfq::Tn mutant (Figure 1D) suggesting that TarB likely interacts with Hfq and thus may help to negatively regulate target mRNAs. We reasoned that if TarB negatively regulated a gene, the expression of the target gene should be higher in a ΔtarB strain compared to the wild type when ToxT is expressed. We took a genetic approach to identify potential mRNA targets of TarB by using RNA-seq to compare the expression profiles of wild type and ΔtarB strains expressing ToxT. We identified only two genes, VC0177 and VC2706, as having significantly increased expression levels in the ΔtarB strain suggesting that expression of TarB may decrease their stability (VC0177 expression ΔtarB/WT = 21.5 fold, p < 1e−5; VC2706 expression ΔtarB/WT = 4.5 fold, p < 1e−5). Interestingly, the most strongly up-regulated gene in the ΔtarB mutant, VC0177, is part of V. cholerae 7th pandemic island -1, VSP-1. qPCR confirmed the RNA-seq analysis showing that VC0177 expression was significantly higher in the ΔtarB strain than the wild type (Figure 1E). We also showed that this pathway was active in a second clinical El Tor strain, E7946 (Figure 1E). This indicates that the ToxT-TarB-VC0177 pathway is not strain specific. However, TarB does appear to have a more profound effect on VC0177 in strain C6706 compared to E7946 suggesting some variability in the magnitude of the effect of this pathway in different strains.
The interaction between TarB and VSP-1 encoded VC0177 is reminiscent of the coordinated control of the TCP island and prophage CTXΦ. Both VSP-1 and prophage CTXΦ are acquired horizontally and separately from the TCP island, yet genes from both VSP-1 and phage CTXΦ are ultimately regulated by TCP island encoded ToxT. VC0177 expression formally joins the ToxT regulon through its dependence on ToxT regulated TarB induction. This association suggests that the co-occurrence of at least VSP-1 in 7th pandemic strains of V. cholerae may have been driven by its regulatory integration with other accessory elements (TCP and CTX) encoding pathogenic adaptation.
VC0177 is a transcription factor that represses several VSP-1 genes
Annotated as a hypothetical protein, VC0177 does not share sequence homology with known proteins by basic alignment searches (BLAST). However, HHPRED analysis (Biegert et al., 2006), which uses protein structure prediction, indicates that VC0177 shares structural homology with metallo-regulator repressor proteins. Because ToxT directly regulates genes located predominantly in the island in which it is encoded, we reasoned that VC0177 might have similar restrictions and repress genes within VSP-1, which spans loci VC0175 –VC0185. Agreeing with the predicted transcription repressor role for VC0177, we found that expression of four VSP-1 genes was significantly increased in a VC0177::Tn mutant (Figure 2A). Furthermore, we found the expression of same four genes was similarly affected in strains carrying an in-frame deletion of VC0177 indicating the regulation control was not due to the transposon insertion (Figure S1A).
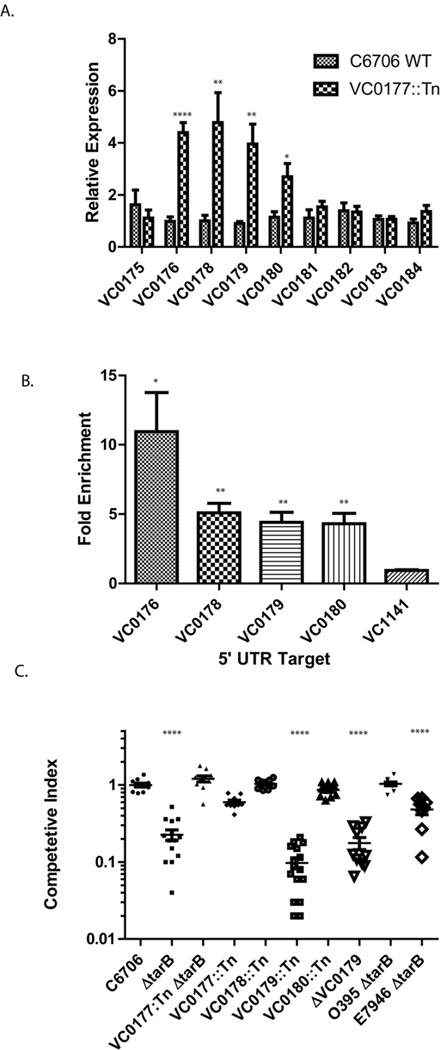
Figure 2.
(A) Expression analysis of VSP-1 genes in WT and VC0177::Tn C6706 strains. The expression of each gene was determined relative to total 16S rRNA. 8 replicates were sampled for each gene. Significance was determined by t-test; * p < 0.05; ** p < 0.01; **** p < 0.0001. (B) VC0177 (VspR) ChIP enrichment of the 5′ UTR regions of VC0176, VC0178, VC0179 and VC0180 was determined by qPCR relative to sample DNA input. Enrichment of the promoter region of non VSP-1 gene VC1141 is shown as a control. Significance was determined by t-test relative to control enrichment; * p < 0.05; ** p < 0.01 (C) In vivo competition experiments measuring the ability of mutant strains to colonize the infant mouse intestine compared to the parental strain. Significance was determined by t-test relative to colonization ratio of parental strains wild type C6706 vs. C6706 ΔlacZ; **** p < 0.0001. See also Figure S1.
To test whether VC0177 was directly involved in the regulation of these genes, we performed ChIP with VC0177 and assayed for enrichment in regions directly upstream of VC0177 repressed genes by qPCR. We identified enrichment of 5′ UTR regions directly upstream of VC0176, VC0178, VC0179 and VC0180 supporting a direct role for VC0177 as a transcriptional repressor of these genes (Figure 2B). Therefore we called the product of VC0177 the V. cholerae 7th pandemic regulator (VspR).
The TarB regulatory circuit and VSP-1 ORF VC0179 influence V. cholerae intestinal colonization
To determine if the TarB-VspR regulatory circuit is involved in pathogenesis, we used the infant mouse model to measure V. cholerae intestinal colonization. To our knowledge the effect of genes in VSP-1 on intestinal colonization has not been investigated. We found that the V. cholerae C6706 ΔtarB mutant showed a significant decrease in ability to colonize the small intestine compared to the wild type strain in competition assays (Figure 2C).
Combining the ΔtarB deletion with a VC0177::Tn mutant rescued the ΔtarB-dependent colonization defect (Figure 2C). This suggests that TarB likely influences colonization by down-regulation of the VC0177 encoded regulator VspR. Because expression of VC0178, VC0179 and VC0180 increased in the V. cholerae VC0177::Tn VspR mutant strain, we tested V. cholerae transposon mutants disrupted in these genes for intestinal colonization defects. Remarkably, only disruption of VC0179 caused a significant defect in intestinal colonization (Figure 2C). An in-frame deletion mutant of VC0179 (ΔVC0179) also showed a similar in vivo colonization defect (Figure 2C). These results identify VC0179 as a direct target of VspR repression in VSP-1, and thus an indirect target of TarB mediated induction.
TarB is well conserved among all sequenced strains of V. cholerae including the classical strain O395 even though classical strains lack VSP-1 and therefore lack VspR. Northern blots show that ToxT also induces TarB in an O395 strain (Figure 1D); however, we did not anticipate a critical in vivo role for TarB O395 due to the lack of VSP-1. Agreeing with this we found that deletion of TarB does not affect O395 colonization of the infant mouse intestine in competition assays (Figure 2C). TarB may regulate additional genes in O395 strains that do not influence intestinal colonization.
During the preparation of this manuscript another group published the presence of TarB in a different El Tor strain, E7946, as part of a genome-wide study identifying ToxT induced sRNAs (Bradley et al., 2011). While agreeing with our observation, Bradley et al. also reported a variable colonization phenotype for a ΔtarB mutant in strain E7946 that depended on initial growth conditions of the bacterium. We found a consistent intestinal colonization defect of an E7946 ΔtarB mutant with our growth conditions (Figure 2C). This defect was less severe than in strain C6706, which may be due to the difference in down-regulation of VspR by TarB between these strains (Figure 1E). Using a computational approach to identify TarB mRNA targets, Bradley et al. reported the most significant effect of TarB is a ~2 fold repression of colonization factor TcpF mRNA. The authors noted that repression of a colonization factor by a ToxT activated sRNA was counterintuitive. In contrast, we did not detect an effect on TcpF expression in our genetic approach to identify TarB targets, but did show that TarB down regulates VspR expression ~ 12 fold in strain E7946 indicating the ToxT-TarB-VspR pathway is active in E7946 and not specific to strain C6706 (Figure 1E). Computational prediction indicates that TarB has a more favorable and a simpler base paring configuration with the 5′ untranslated region of VspR than TcpF providing an explanation as to why TarB regulates VspR more strongly (Figure S1B and C).
VC0179 is the first member of a new family of di-nucleotide cyclases (DncV)
Disruption of VSP-1 encoded VC0179 caused a significant defect in intestinal colonization (Figure 2C). VC0179 is a hypothetical protein and does not share overall sequence homology with known proteins. HHPRED analysis shows that VC0179 shares spatial alignment of key active site residues found in 2′–5′-oligoadenylate synthetases (OAS1) and polyA polymerase both of which are part of the nucleotidyl transferase superfamily (Holm and Sander, 1995) (Figure S2). The placement of these active site residues in VC0179 match the consensus sequence (G[G/S]x9–13Dx[D/E]) of the nucleotidyl transferase superfamily (NTS) (Holm and Sander, 1995). OAS1 and polyA polymerase catalyze the polymerization of oligoadenylates from ATP (Hartmann et al., 2003). The structural similarity with OAS1 and polyA polymerase suggested that VC0179 might act catalytically on ATP.
To test this hypothesis, we purified VC0179, incubated it with α-P32 ATP and separated the products on a denaturing polyacrylamide gel. VC0179 produced a single product that migrated slightly above the ATP precursor (Figure 3A). As a control, we mutated the aspartate residues of the predicted VC0179 Dx[D/E] catalytic center to alanine residues (D131A and D133A - Figure S2) and purified the mutant protein. When incubated with ATP, the D131A/D133A VC0179 mutant protein did not produce the VC0179 product from ATP (Figure 3A) indicating activity from the WT protein is dependent upon the predicted NTS catalytic residues and is not due to a contaminate.
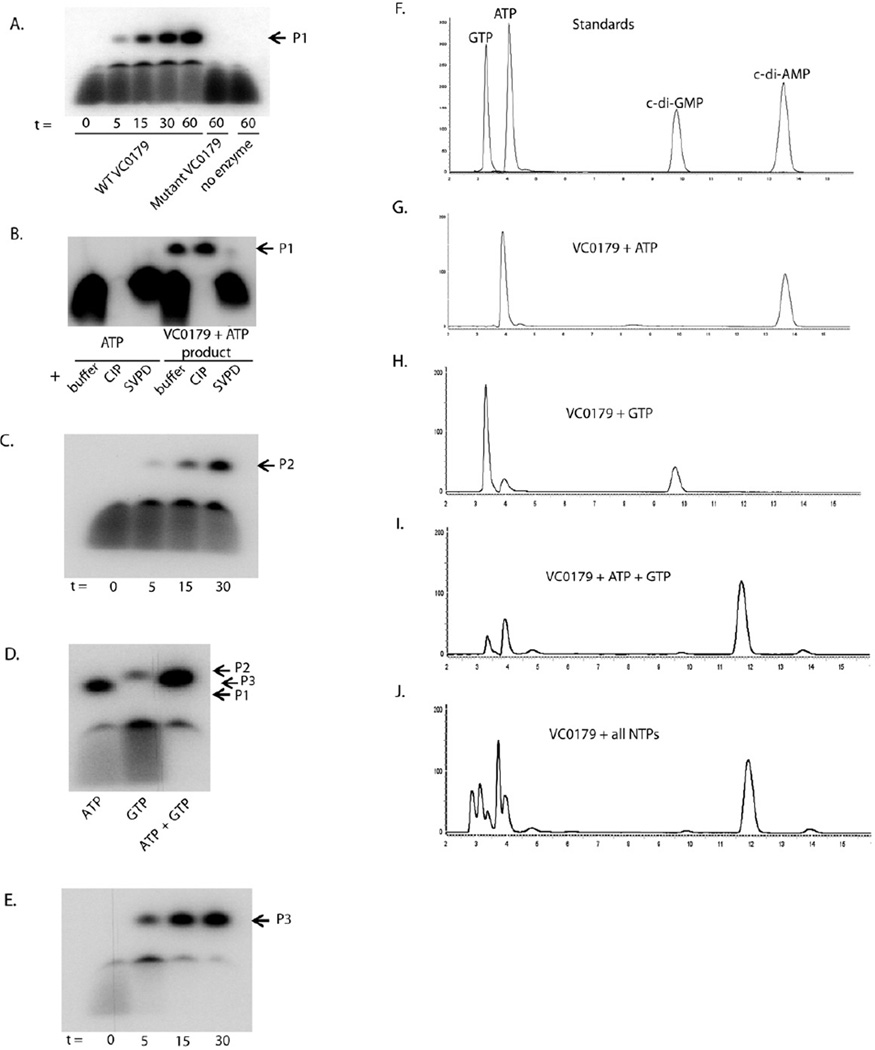
Figure 3.
Di-nucleotide cyclase activity of VC0179 (DncV). (A) Purified wild type or mutated VC0179 was incubated with 1 mM ATP and 10 µCi α-P32 ATP for the indicated time (t in min) and the products (P1) separated by denaturing PAGE. (B) The product from the reaction between VC0179 and ATP (P1) was incubated with calf intestinal phosphatase (CIP) or snake venom phosphodieasterase (SVPD) for 30 min and separated by denaturing PAGE. (C) Purified wild type VC0179 was incubated with 1 mM GTP and 10 µCi α-P32 GTP for the indicated time (t in min) and the products (P2) separated by denaturing PAGE. (D) VC0179 was incubated with ATP or GTP as described above or with 1mM GTP and 10 µCi α-P32 ATP and the products (P1–3) separated by denaturing PAGE. (E) Purified wild type VC0179 was incubated with 1 mM GTP and 10 µCi α-P32 ATP for the indicated time (t in min) and the products (P3) separated by denaturing PAGE. P1, c-di-AMP; P2, c-di-GMP; P3, c-AMP-GMP. (F) Commercial standards of ATP, GTP, c-di-AMP and c-di-GMP were fractionated by RP-HPLC. The peaks are labeled by their respective compound. VC0179 incubated with 2mM ATP (G), 2mM GTP (H), 1mM ATP + 1mM GTP (I), or 1mM of each of the 5 NTPs (J) for 30 min at 37 °C was fractionated by RPHPLC. 50 nM of VC0179 (DncV) wild type or mutant enzyme was used per reaction. See also Figure S2, S3, S4, S5; Table S2, S3.
The product produced from ATP by VC0179 is insensitive to calf intestinal phosphatase (CIP) indicating that it does not contain 5´ or 3´ terminal phosphate groups (Figure 3B). The product is sensitive to snake venom phosphodiesterase and nuclease P1 indicating that it contains 3′–5 ′ phosphate linkages (Figure 3B and data not shown). These results suggested that the VC0179 product was a cyclic nucleotide. HPLC analysis showed that the VC0179 product eluted with the same retention time as a commercial cyclic di-AMP (c-di-AMP) standard (Figure 3F and G). Furthermore, LCMS and MS/MS analysis showed that the VC0179 product and c-di- AMP have identical molecular masses and fragmentation fingerprint (Table S2 and S3; Figure S3 and S4) confirming that VC0179 synthesizes c-di-AMP. To our knowledge this is the first report of c-di-AMP synthesis by a Gram-negative bacterium enzyme and the first adenylate cyclase based on a non-DU147 protein domain structure (Romling, 2008).
We tested the catalytic ability of VC0179 with additional nucleotides. We found that VC0179 is not active against dATP (data not shown). When incubated with GTP, VC0179 produces cyclic di-GMP (c-di-GMP) (Figure 3C) that was confirmed by HPLC, LCMS and MS/MS comparison with a commercial c-di-GMP standard (Figure 3F and H; Table S2 and S3; Figure S3 and S4). Interestingly, both PAGE and HPLC analysis indicated more c-di-AMP production than c-di-GMP per unit time suggesting VC0179 synthesis c-di-AMP more efficiently than c-di-GMP. Prior to these results, only proteins containing GGDEF domains were thought to be able to produce c-di-GMP in bacteria (Ryjenkov et al., 2005). We also tested whether VC0179 could enzymatically modify TTP, UTP and CTP but did not detect significant activity (data not shown) suggesting VC0179 preferentially uses purine nucleotide triphosphates as substrates.
Since VC0179 can use ATP and GTP substrates, but appears to be more efficient in its use of ATP, we asked if it preferred one substrate to the other when given the option. We incubated VC0179 with equal molar amounts of GTP and ATP and analyzed the products by PAGE, HPLC, LCMS and MS/MS. Remarkably we found the dominant reaction product migrated between c-di-AMP and c-di-GMP by PAGE (Figure 3D and E). This new product was observed in much greater amounts than either c-di-AMP or c-di-GMP as confirmed by HPLC (Figure 3F and I). This suggested that VC0179 preferentially synthesizes a hybrid c-AMP-GMP molecule from ATP and GTP. High resolution LCMS analysis confirmed that the mass of the VC0179 reaction product incubated with ATP and GTP was in agreement with a c-AMP-GMP molecule (Table S2 and Figure S3). MS/MS was used to fragment this product into masses accurate for AMP and GMP further confirming the hybrid structure (Table S3 and Figure S4). As with ATP, the D131A/D133A VC0179 mutant protein did not produce any product when incubated with GTP or an ATP/GTP mixture (Figure S5A).
Given that in metabolically active cells there is a mixture of all NTPs, we tested the preference of VC0179 in a mixture that included equal amounts of all 5 NTPs. We again found that VC0179 produced substantially more of the hybrid c-AMP-GMP molecule than any other cyclic-dinucleotide (Figure 3J). To our knowledge this is the first report of an enzyme that can produce a hybrid cyclic di-nucleotide and the first enzyme capable of producing three different cyclic di-nucleotides. Therefore we have designated the product of VC0179 di-nucleotide cyclase Vibrio or DncV. Using BLAST and ClustalW we identified and align homologs that were at least 20% similar to DncV and had the conserved G[G/S]x9–13Dx[D/E] motif (Figure S5B). DncV homologs can be identified in a wide range of bacteria, several of which are pathogenic.
c-AMP-GMP is the dominate DncV product in vivo
Since DncV is capable of synthesizing three different cyclic di-nucleotides in vitro, we assayed its activity in vivo. We addressed this question using two systems. We first used a heterologous system to induce high level expression of WT DncV or the D131A/D133A DncV mutant from a plasmid in E. coli. Analysis of cytosolic samples by LCMS showed a strong signal for c-AMPGMP in E. coli expressing WT DncV but not in E. coli expressing D131A/D133A DncV mutant (Table 2; Figure S6A and B). We also observed a c-di-AMP signal in E. coli expressing WT DncV but again not in E. coli expressing D131A/D133A DncV mutant (Table 2; Figure S6C and D). We observed c-di-GMP in both WT and mutant DncV expressing E. coli that most likely represents endogenous production (data not shown). Thus we cannot conclude that the c-di-GMP detected in this sample was a product of DncV under this condition.
Table 2.
Expected and determined masses of cyclic di-nucleotides identified in E. coli and V. cholerae strains expressing WT DncV from LCMS analysis.
| Sample | Expected Mass | Determined Mass | ppm Error |
|---|---|---|---|
| E coli c-AMP-GMP | 673.0927 | 673.0922 | 0.7428397 |
| E. coli c-di-AMP | 657.0978 | 657.0972 | 0.9131061 |
| V. cholera c-AMP-GMP | 673.0927 | 673.0936 | 1.3371115 |
We next examined cytosolic samples from a V. cholerae ΔdncV strain expressing WT DncV or D131A/D133A DncV mutant from an inducible plasmid. LCMS analysis identified c- AMP-GMP in the WT DncV expressing strain but not in the D131A/D133A DncV mutant expressing strain (Table 2; Figure 4A). Interestingly we could not detect c-di-AMP of c-di-GMP in any of the V. cholerae samples. However we cannot rule out that DncV does produce c-di- AMP and/or c-di-GMP in V. cholerae at levels below our detection limit. These results indicate that DncV can synthesize c-AMP-GMP and c-di-AMP in vivo but that c-AMP-GMP is the major product. Equal amounts of culture where processed for each sample, and total ion count analyzed for each sample is shown as an analysis control (Figure S7A, B).
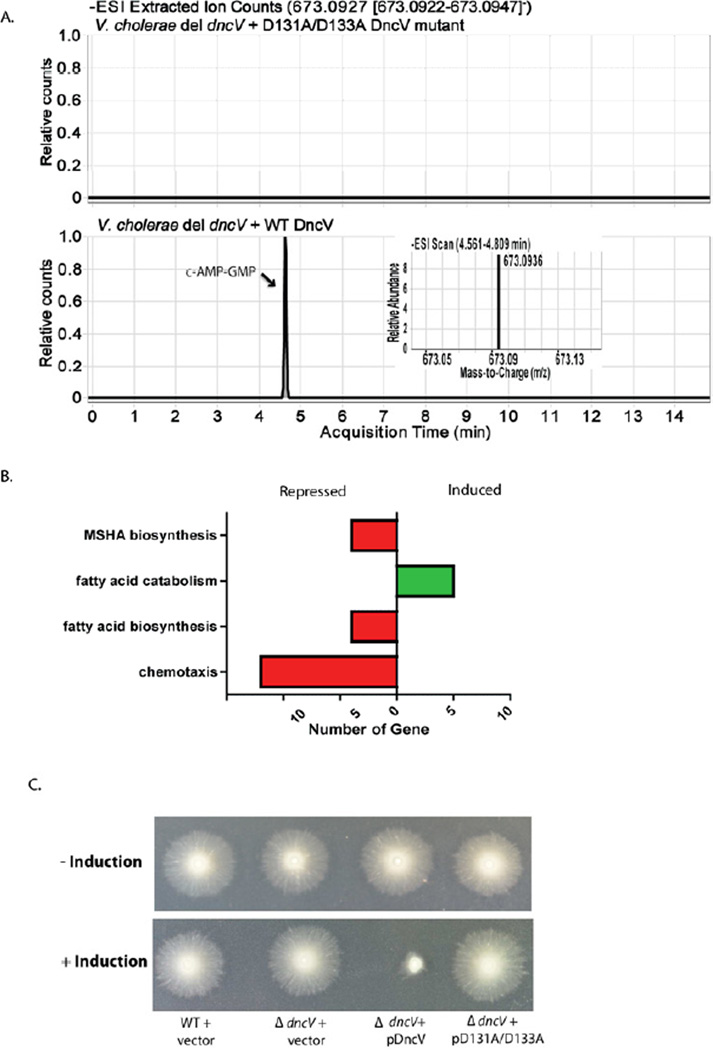
Figure 4.
(A) LCMS chromatogram trace corresponding to the ion extraction of c-AMP-GMP (theoretical mass [M-H] = 673.0927 Da; ion extraction: 673.0922 – 673.0947 Da) in cell lysate from V. cholerae ΔdncV strain expressing plasmid borne WT or D131A/D133A mutant DncV. High resolution mass found under the peak eluting at approximately 4.75 min in chromatogram A corresponding to c-AMP-GMP from V. cholerae ΔdncV strain expressing WT DncV is shown on the inset. This peak eluted with the same retention time as in vitro generated c-AMP-GMP. (B) Functional categories of differentially expressed genes in response to expression of DncV. The number of genes induced or repressed in response to DncV expression is presented. Green indicates genes in the respective category are induced > 2 fold, red indicated genes in respective category are repressed > 2 fold (FDR corrected p-value < 0.01). (C) Examination of chemotactic behavior of V. cholerae wild type, ΔdncV mutant, and ΔdncV mutant expressing wild type or D131A/D131A mutant DncV from an arabinose inducible plasmid, with and without induction by arabinose. See also Figure S6, S7; Table S4; Movie S1, S2.
DncV activity modulates V. cholerae chemotaxis and does not affect c-di-GMP-dependent regulation
We did not notice a significant growth difference between V. cholerae wild type and ΔdncV mutant strains in rich or minimal medium (data not shown). To determine biological effects of DncV expression, we used RNA-seq to compare the transcriptome profiles of a ΔdncV mutant and a ΔdncV mutant in which DncV expression was induced from a plasmid for 15 min. From this analysis, we observed coordinated differential expression of groups of genes that clustered into four biological processes (Figure 4B; Table S4). The largest concerted regulatory effect showed many chemotactic genes downregulated following DncV induction. Signal-transducing chemotactic proteins are known to bind purines (Stock et al., 1987) suggesting the products of DncV could directly interact and modulate the activity of these proteins. We tested of the chemotactic abilities of the wild type, ΔdncV mutant and ΔdncV mutant expressing DncV from a plasmid using semisolid agar chemotaxis plates and found that increased expression of DncV severely inhibited V. cholerae chemotaxis (Figure 4C). We also found that expressing the DncV D131A/D133A mutant did not affect chemotaxis indicating the catalytic activity of DncV is required for this phenotype (Figure 4C). WT and D131A/D133A mutant proteins showed similar expression and degradation patterns following induction, indicating that the observed effects were not due to a difference in protein level (Figure S7C).
While we did not observe any changes in the expression of flagellum biogenesis genes, it is possible that the effect we observed on the semisolid agar were due to defects in flagellum biogenesis or function. To test this possibility we extracted wild type + vector and ΔdncV mutant + pDncV from the plate containing arabinose shown in Figure 4C and checked for motility. Phase contrast microscopy showed that increased expression of DncV did not change overall motility of V. cholerae agreeing with our transcriptome analysis that expression of flagellum components were unaffected (Movie S1 and S2). Interestingly, repressing chemotaxis has been shown to significantly enhance V. cholerae intestinal colonization (Butler et al., 2006). Thus, expression of VC0179 may enhance colonization by suppressing chemotactic behavior.
We compared our transcriptome results to recently published microarray analysis of the regulatory effects of increased c-di-GMP production in V. cholerae (Beyhan et al., 2006). The most prominent regulatory effects of increased c-di-GMP levels included increased expression of MSHA, the extracellular protein secretion system (EPS) and Vibrio polysaccharide (VPS) biogenesis genes, as well as decreased expression of flagellum biogenesis genes. In contrast our transcriptome analysis of DncV induction showed decreased expression of MSHA biosynthesis genes and no effect on EPS, VPS or flagellum biogenesis genes (Table S4). Furthermore, increased c-di-GMP levels were shown to drastically reduce the number of motile bacterium and increase biofilm production (Beyhan et al., 2006), whereas expression of DncV does not affect V. cholerae motility (Movie S1 and S2) or biofilm production (unpublished results). c-AMP-GMP is the dominant molecule produced by DncV in vivo (Figure 4A; Figure S6). The lack of gene regulatory and phenotypic overlap between increased DncV expression and increased c-di-GMP levels suggest that c-AMP-GMP and c-di-GMP have different regulatory roles in the cell.
DISCUSSION
The VSP islands are the most pronounced genetic difference between classical and El tor strains of V. cholerae (Dziejman et al., 2002), however any fitness benefit they offered El tor strains was unknown. Our results describe a multifaceted regulatory cascade driven by ToxT and mediated through the TCP island encoded sRNA, TarB, that down-regulates expression of VSP-1 transcription factor VspR (Figure 5). Loss of VspR derepresses VSP-1 genes, including VC0179, which encodes a novel di-nucleotide cyclase, DncV, which is required for efficient V. cholerae intestinal colonization and regulates the colonization influencing process of chemotaxis. DncV is the first reported enzyme capable of synthesizing c-di-AMP in Gram-negative bacteria and the first enzyme capable of synthesizing the previously undescribed hybrid c-AMP-GMP.
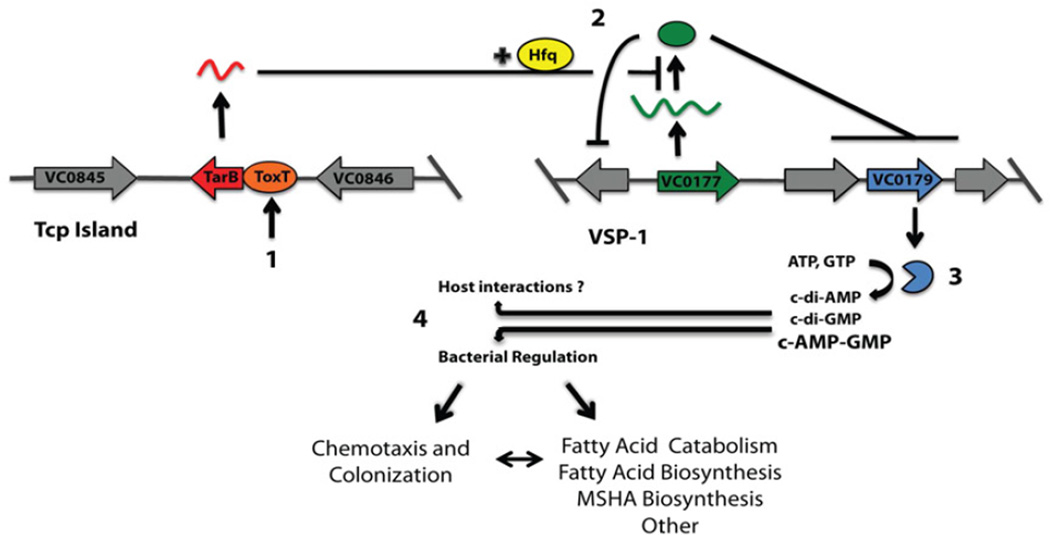
Figure 5.
Model for ToxT-dependent TarB-mediated control of VSP-1. (1) Host signals induce ToxT activity resulting in transcription of TarB from the Tcp Island. (2) Stabilized by Hfq, TarB downregulates expression of transcriptional repressor VC0177 (VspR) resulting in de-repression of VSP-1 genes including VC0179 (DncV). (3) DncV activity increases cellular concentration of c-AMP-GMP (and possibly c-di-AMP and c-di-GMP) that affects chemotactic behavior and other metabolic aspects of 7th pandemic V. cholerae.
It was previously shown that VC0179 expression was up-regulated in the rabbit intestine supporting a role for VC0179 in intestinal colonization (Xu et al., 2003). The connection between induction of the master virulence regulator ToxT and VSP-1 regulation strongly suggested that genes encoded by VSP-1 were expressed in vivo and likely contribute to pathogenesis or other virulence associated properties ( e.g. transmission or infectivity). The association of VSP-1 with V. cholerae colonization offers the first evidence that the evolutionary success of V. cholerae El Tor strains carrying VSP-1 islands may have been driven by increased fitness within the host and thus implies that VSP-1 is indeed a pathogenicity island. It is possible that VSP-1 contributes to properties that might improve the fitness of 7th pandemic El Tor strains of V. cholerae in the aquatic environment since some VSP-1 genes have been recently found in environmental non-O1, non-O139 V. cholerae strains (Grim et al., 2010). However, VSP-1 was universally present in TCP+ O1 and O139 strains (Grim et al., 2010; Rahman et al., 2008), suggesting that the regulatory linkage of the TCP and VSP-1 islands documented here may be selecting for their co-inheritance in pathogenic 7th pandemic strains.
Comparative genome analysis shows that DncV homologs are found in pathogenic islands of other disease causing bacteria including the High Pathogenicity Island (HPI) of Enterobacteriaceae strains (Paauw et al., 2010; Schubert et al., 2004). Interestingly, DncV homologs in HPI are often found associated with the closely neighboring VSP-1 gene VC0181. VC0181 is annotated as an integrative and conjugative element suggesting it may help DncV recombine into new genomes and genomic islands. There are additional VSP-1 genes that did not appear in our network that may also influence V. cholerae colonization. These genes might still be regulated by VspR but our conditions may not have favored their expression. VspR might also play a regulator role outside of VSP-1 that future ChIP-seq analysis will illuminate. The neighboring gene, VC0176, is annotated as a putative CI/Cro like regulator. Since VspR appears to affect VC0176 expression there may be regulatory interplay between these two factors that modulates VSP-1 activity or other genes outside of the island. Further characterization of the hypothetical proteins encoded in VSP-1 and VSP-2, may offer significant insights into differences into host adaptation of the 7th pandemic El Tor lineage of V. cholerae.
In bacteria, c-di-GMP is synthesized by proteins containing a GGDEF motif, which are found in the majority of bacterial genomes (Ryjenkov et al., 2005). c-di-AMP is synthesized in gram-positive bacteria by proteins containing the unrelated DUF147 domain (Witte et al., 2008). Remarkably, the active site of DncV is able to produce both c-di-AMP and c-di-GMP cyclic compounds as well as a previously undescribed hybrid c-AMP-GMP. Our initial biochemical analysis suggests that DncV has a strong product preference for synthesis of c-AMP-GMP followed by c-di-AMP and c-di-GMP. These results agree with our finding that c-AMP-GMP is the dominant DncV product in vivo. Future structural analysis of DncV will be particularly interesting to determine its mode of substrate selection and catalytic action. While DncV appears to have a product preference, the balance of product it produces in vivo would be heavily influenced by its local intracellular NTP concentration. Energetic shifts that alter the cytosolic ATP concentration could also impact DncV product production.
Cyclic nucleotides act as intracellular signals, modulating a range of cellular activities (Gomelsky, 2011). In Gram-negative bacteria c-di-GMP is a major regulator of biofilms (Tischler and Camilli, 2004) and flagellum biosynthesis (Lim et al., 2007), and in Gram-positive bacteria, c-di-AMP reports on DNA integrity and cell membrane stress (Corrigan et al., 2011; Woodward et al., 2010). Since DncV can produce both molecules, its activity may influence all of these processes. The hybrid c-AMP-GMP molecule may function in both c-di-GMP and c-di- AMP pathways and/or completely new processes specific to a hybrid interaction. Expression comparison showed that regulatory events mediated by DncV induction are not likely due to increased c-di-GMP levels. Increased expression of DncV strongly inhibits V. cholerae chemotaxis but the bacteria are still highly motile. Interestingly, V. cholerae shed from humans also show decreased chemotaxis and normal motility (Merrell et al., 2002). Repression of chemotaxis is thought to contribute to a hyperinfectious state of V. cholerae and indeed repressing chemotaxis has been shown to significantly enhance V. cholerae intestinal colonization (Butler et al., 2006). These observations suggest a mechanism for the necessity of DncV for efficient V. cholerae intestinal colonization. We propose that the in vivo environment induces ToxT activity which in turn results in derepression of DncV via the TarB-VspR pathway and an inhibition of chemotaxis via the accumulation of cyclic di-nucleotides. Reduced chemotaxis might not only stimulate colonization but also influence infectivity long after V. cholerae exits the ToxT inducing environment of the small intestine (Butler et al., 2006; Merrell et al., 2002).
Interestingly DncV expression induced fatty acid catabolism and repressed fatty acid biosynthesis. Metabolomic analysis of cecal fluid harvested from V. cholerae infected rabbits has recently revealed that the intestinal milieu contains high levels of long chain fatty acids compared to laboratory medium (Mandlik et al., 2011). Fatty acids were recently shown modulate ToxT activity and indeed a 16-carbon fatty acid cis-palmitoleate, crystallizes with ToxT when expressed and purified from E. coli (Lowden et al., 2010). It will be interesting to investigate if DncV activity creates regulatory signals that modulate ToxT activity in vivo by specifically stimulating the breakdown of host lipids and transport of fatty acids. However, a role for cyclic di-nucleotides in regulating fatty acid metabolism has not yet been demonstrated in V. cholerae.
The association of DncV expression with ToxT activity suggests that the cellular concentration of c-di-AMP, c-di-GMP and/or c-AMP-GMP may increase significantly when V. cholerae enters the host intestinal environment. Interestingly, c-di-AMP has recently been shown to be secreted by multidrug efflux pumps from L. monocytogenes (Woodward et al., 2010). In the context of L. monocytogenes infection, c-di-AMP alters the mammalian host immune response (Woodward et al., 2010), and recently cyclic di-nucleotides have been identified as key agonists of the STING innate immune signaling pathway (Burdette et al., 2011). Purinergic receptors known to modulate pH and electrolyte secretion are found extensively on the apical membrane of intestinal epithelial cells where V. cholerae colonizes (Dho et al., 1992; Kaunitz and Akiba, 2011). We are currently investigating whether V. cholerae may also secrete c-AMP-GMP and/or c-di-AMP, as these cyclic di-nucleotides could interact with the host cell receptors to modulate intestinal cell electrophysiology or innate immunity. Such an effect could be beneficial to a range of intestinal pathogens that carry DncV homologs. Thus, secreted c-AMPGMP and/or c-di-AMP could act as bacterial toxins that affect disease progression. The discovery of DncV produced cyclic di-nucleotides adds another layer of complexity to V. cholerae pathogenesis and implicates these novel small molecules in the disease process.
EXPERIMENTAL PROCEDURES
Chromatin Immunoprecipitation Sequencing (ChIP-seq) and Transcriptome profiling (RNA-seq)
ChIP, sample preparation, sequencing and data processing was performed as previously described (Davies et al., 2011). Experiments were repeated in quadruplicate. Samples were sequenced using the HeliScope™ Single Molecule Sequencer at the Molecular Biology Core Facility in the Dana-Farber Cancer Institute.
For ToxT expression studies RNA was converted to cDNA and subsequent sample preparation and sequencing was performed as described for ChIP-seq. For VC0179 expression studies RNA samples were processed for Illumina multiplex sequencing as described by the manufacturer. Sequencing data was aligned to the V. cholerae N16961 genome using CLC genomic workbench software using RPKM for expression values. The statistical test performed was a Baggerley’s test on proportion of counts in each group of samples to generate a p-value associated with the weighted proportion fold change between experiment and control groups for each gene. The result is a weighted t-type test statistic. We then used a cut-off of 4-fold weighted proportions absolute change with a false-discovery rate corrected p-value of ≤ 0.01.
RP-HPLC, LCMS and LC-MS/MS
Reverse-phase high performance liquid chromatography (RP-HPLC) was performed as previously described (Oppenheimer-Shaanan et al., 2011). High resolution LCMS analysis was performed on an Agilent 6520 Accurate-Mass Q-TOF mass spectrometer using an electrospray (ESI) ionization source in negative mode. Tandem MS/MS analysis was performed using collision induced dissociation (CID). Ions were selected using a medium iso width (approx 4 m/z) and CID energy set to 20. Parent masses and fragment ions are listed in Tables S2 and S3.
In vivo detection of cyclic di-nucleotides
Wild type and D131A/D133A mutant VC0179 were expressed in BL21 E. coli or V. cholerae. Clarified cell supernatant was passed over a Waters Sep-Pak C18 cartridge by gravity flow. Bound material was eluted, resuspended in 50 % acetonitrile and analyzed by LCMS as described.
Di-nucleotide cyclase assay
50 nM of VC0179-6XHis or D131A/D133A mutant VC0179-6XHis enzyme was incubated with the indicated NTPs at 37 °C for the indicated time. Samples were processed for RP-HPLC, LCMS, MS/MS or separated by denaturing PAGE and exposed to film.
Supplementary Material
Highlights.
Integrated ChIP-seq and RNA-seq analysis characterize the ToxT regulon in V. cholerae
TCP island small RNA regulates gene expression the Vibrio 7th pandemic island-1 genes
VSP-1 island encoded di-nucleotide cyclase affects chemotaxis and host colonization
V. cholerae di-nucleotide cyclase synthesizes c-AMP-GMP, c-di-AMP and c-di-GMP
ACKNOWLEDGMENTS
We thank Dr. D. Ewen Cameron for support in use of the V. cholerae transposon library. B. W. D. was supported by the Jane Coffin Childs Memorial Fund for Medical Research and by the National Research Council of Canada H.L. Holmes Award. This work was supported by the National Institutes of Health Grants AI-018045 to J. J. M.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Beyhan S, Tischler AD, Camilli A, Yildiz FH. Transcriptome and phenotypic responses of Vibrio cholerae to increased cyclic di-GMP level. J Bacteriol. 2006;188:3600–3613. doi: 10.1128/JB.188.10.3600-3613.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biegert A, Mayer C, Remmert M, Soding J, Lupas AN. The MPI Bioinformatics Toolkit for protein sequence analysis. Nucleic Acids Res. 2006;34:W335–W339. doi: 10.1093/nar/gkl217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley ES, Bodi K, Ismail AM, Camilli A. A Genome-Wide Approach to Discovery of Small RNAs Involved in Regulation of Virulence in Vibrio cholerae. PLoS Pathog. 2011;7:e1002126. doi: 10.1371/journal.ppat.1002126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan RG, Link TM. Hfq structure, function and ligand binding. Curr Opin Microbiol. 2007;10:125–133. doi: 10.1016/j.mib.2007.03.015. [DOI] [PubMed] [Google Scholar]
- Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, Vance RE. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011 doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler SM, Nelson EJ, Chowdhury N, Faruque SM, Calderwood SB, Camilli A. Cholera stool bacteria repress chemotaxis to increase infectivity. Mol Microbiol. 2006;60:417–426. doi: 10.1111/j.1365-2958.2006.05096.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrigan RM, Abbott JC, Burhenne H, Kaever V, Grundling A. c-di-AMP Is a New Second Messenger in Staphylococcus aureus with a Role in Controlling Cell Size and Envelope Stress. PLoS Pathog. 2011;7:e1002217. doi: 10.1371/journal.ppat.1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies BW, Bogard RW, Mekalanos JJ. Mapping the regulon of Vibrio cholerae ferric uptake regulator expands its known network of gene regulation. Proc Natl Acad Sci U S A. 2011;108:12467–12472. doi: 10.1073/pnas.1107894108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dho S, Stewart K, Foskett JK. Purinergic receptor activation of Cl- secretion in T84 cells. Am J Physiol. 1992;262:C67–C74. doi: 10.1152/ajpcell.1992.262.1.C67. [DOI] [PubMed] [Google Scholar]
- DiRita VJ, Parsot C, Jander G, Mekalanos JJ. Regulatory cascade controls virulence in Vibrio cholerae. Proc Natl Acad Sci U S A. 1991;88:5403–5407. doi: 10.1073/pnas.88.12.5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziejman M, Balon E, Boyd D, Fraser CM, Heidelberg JF, Mekalanos JJ. Comparative genomic analysis of Vibrio cholerae: genes that correlate with cholera endemic and pandemic disease. Proc Natl Acad Sci U S A. 2002;99:1556–1561. doi: 10.1073/pnas.042667999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faruque SM, Mekalanos JJ. Pathogenicity islands and phages in Vibrio cholerae evolution. Trends Microbiol. 2003;11:505–510. doi: 10.1016/j.tim.2003.09.003. [DOI] [PubMed] [Google Scholar]
- Gomelsky M. cAMP, c-di-GMP, c-di-AMP and now cGMP: bacteria use them all! Mol Microbiol. 2011;79:562–565. doi: 10.1111/j.1365-2958.2010.07514.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grim CJ, Choi J, Chun J, Jeon YS, Taviani E, Hasan NA, Haley B, Huq A, Colwell RR. Occurrence of the Vibrio cholerae seventh pandemic VSP-I island and a new variant. OMICS. 2010;14:1–7. doi: 10.1089/omi.2009.0087. [DOI] [PubMed] [Google Scholar]
- Handstad T, Rye MB, Drablos F, Saetrom P. A ChIP-Seq Benchmark Shows That Sequence Conservation Mainly Improves Detection of Strong Transcription Factor Binding Sites. PLoS One. 2011;6:e18430. doi: 10.1371/journal.pone.0018430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann R, Justesen J, Sarkar SN, Sen GC, Yee VC. Crystal structure of the 2'-specific and double-stranded RNA-activated interferon-induced antiviral protein 2'-5'-oligoadenylate synthetase. Mol Cell. 2003;12:1173–1185. doi: 10.1016/s1097-2765(03)00433-7. [DOI] [PubMed] [Google Scholar]
- Hassan F, Kamruzzaman M, Mekalanos JJ, Faruque SM. Satellite phage TLCphi enables toxigenic conversion by CTX phage through dif site alteration. Nature. 2010;467:982–985. doi: 10.1038/nature09469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidelberg JF, Eisen JA, Nelson WC, Clayton RA, Gwinn ML, Dodson RJ, Haft DH, Hickey EK, Peterson JD, Umayam L, et al. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406:477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Sander C. DNA polymerase beta belongs to an ancient nucleotidyltransferase superfamily. Trends Biochem Sci. 1995;20:345–347. doi: 10.1016/s0968-0004(00)89071-4. [DOI] [PubMed] [Google Scholar]
- Kaper JB, Morris JG, Jr, Levine MM. Cholera. Clin Microbiol Rev. 1995;8:48–86. doi: 10.1128/cmr.8.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaunitz JD, Akiba Y. Purinergic regulation of duodenal surface pH and ATP concentration: implications for mucosal defence, lipid uptake and cystic fibrosis. Acta Physiol (Oxf) 2011;201:109–116. doi: 10.1111/j.1748-1716.2010.02156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasteva PV, Fong JC, Shikuma NJ, Beyhan S, Navarro MV, Yildiz FH, Sondermann H. Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science. 2010;327:866–868. doi: 10.1126/science.1181185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim B, Beyhan S, Yildiz FH. Regulation of Vibrio polysaccharide synthesis and virulence factor production by CdgC, a GGDEF-EAL domain protein, in Vibrio cholerae. J Bacteriol. 2007;189:717–729. doi: 10.1128/JB.00834-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowden MJ, Skorupski K, Pellegrini M, Chiorazzo MG, Taylor RK, Kull FJ. Structure of Vibrio cholerae ToxT reveals a mechanism for fatty acid regulation of virulence genes. Proc Natl Acad Sci U S A. 2010;107:2860–2865. doi: 10.1073/pnas.0915021107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandlik A, Livny J, Robins WP, Ritchie JM, Mekalanos JJ, Waldor MK. RNA-Seq-Based Monitoring of Infection-Linked Changes in Vibrio cholerae Gene Expression. Cell Host Microbe. 2011;10:165–174. doi: 10.1016/j.chom.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrell DS, Butler SM, Qadri F, Dolganov NA, Alam A, Cohen MB, Calderwood SB, Schoolnik GK, Camilli A. Host-induced epidemic spread of the cholera bacterium. Nature. 2002;417:642–645. doi: 10.1038/nature00778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheimer-Shaanan Y, Wexselblatt E, Katzhendler J, Yavin E, Ben-Yehuda S. c-di-AMP reports DNA integrity during sporulation in Bacillus subtilis. EMBO Rep. 2011;12:594–601. doi: 10.1038/embor.2011.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paauw A, Leverstein-van Hall MA, Verhoef J, Fluit AC. Evolution in quantum leaps: multiple combinatorial transfers of HPI and other genetic modules in Enterobacteriaceae. PLoS One. 2010;5:e8662. doi: 10.1371/journal.pone.0008662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollitzer R. Cholera. Geneva: World health Organization; 1959. [Google Scholar]
- Rahman MH, Biswas K, Hossain MA, Sack RB, Mekalanos JJ, Faruque SM. Distribution of genes for virulence and ecological fitness among diverse Vibrio cholerae population in a cholera endemic area: tracking the evolution of pathogenic strains. DNA Cell Biol. 2008;27:347–355. doi: 10.1089/dna.2008.0737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard AL, Withey JH, Beyhan S, Yildiz F, DiRita VJ. The Vibrio cholerae virulence regulatory cascade controls glucose uptake through activation of TarA, a small regulatory RNA. Mol Microbiol. 2010;78:1171–1181. doi: 10.1111/j.1365-2958.2010.07397.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie JM, Waldor MK. Vibrio cholerae interactions with the gastrointestinal tract: lessons from animal studies. Curr Top Microbiol Immunol. 2009;337:37–59. doi: 10.1007/978-3-642-01846-6_2. [DOI] [PubMed] [Google Scholar]
- Romling U. Great times for small molecules: c-di-AMP, a second messenger candidate in Bacteria and Archaea. Sci Signal. 2008;1:pe39. doi: 10.1126/scisignal.133pe39. [DOI] [PubMed] [Google Scholar]
- Ryjenkov DA, Tarutina M, Moskvin OV, Gomelsky M. Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: insights into biochemistry of the GGDEF protein domain. J Bacteriol. 2005;187:1792–1798. doi: 10.1128/JB.187.5.1792-1798.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert S, Dufke S, Sorsa J, Heesemann J. A novel integrative and conjugative element (ICE) of Escherichia coli: the putative progenitor of the Yersinia highpathogenicity island. Mol Microbiol. 2004;51:837–848. doi: 10.1046/j.1365-2958.2003.03870.x. [DOI] [PubMed] [Google Scholar]
- Srivastava D, Harris RC, Waters CM. Integration of Cyclic di-GMP and Quorum Sensing in the Control of vpsT and aphA in Vibrio cholerae. J Bacteriol. 2011 doi: 10.1128/JB.05167-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock A, Mottonen J, Chen T, Stock J. Identification of a possible nucleotide binding site in CheW, a protein required for sensory transduction in bacterial chemotaxis. J Biol Chem. 1987;262:535–537. [PubMed] [Google Scholar]
- Taviani E, Grim CJ, Choi J, Chun J, Haley B, Hasan NA, Huq A, Colwell RR. Discovery of novel Vibrio cholerae VSP-II genomic islands using comparative genomic analysis. FEMS Microbiol Lett. 2010;308:130–137. doi: 10.1111/j.1574-6968.2010.02008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischler AD, Camilli A. Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Mol Microbiol. 2004;53:857–869. doi: 10.1111/j.1365-2958.2004.04155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber GG, Klose KE. The complexity of ToxT-dependent transcription in Vibrio cholerae. Indian J Med Res. 2011;133:201–206. [PMC free article] [PubMed] [Google Scholar]
- Withey JH, DiRita VJ. Activation of both acfA and acfD transcription by Vibrio cholerae ToxT requires binding to two centrally located DNA sites in an inverted repeat conformation. Mol Microbiol. 2005a;56:1062–1077. doi: 10.1111/j.1365-2958.2005.04589.x. [DOI] [PubMed] [Google Scholar]
- Withey JH, Dirita VJ. Vibrio cholerae ToxT independently activates the divergently transcribed aldA and tagA genes. J Bacteriol. 2005b;187:7890–7900. doi: 10.1128/JB.187.23.7890-7900.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Withey JH, DiRita VJ. The toxbox: specific DNA sequence requirements for activation of Vibrio cholerae virulence genes by ToxT. Mol Microbiol. 2006;59:1779–1789. doi: 10.1111/j.1365-2958.2006.05053.x. [DOI] [PubMed] [Google Scholar]
- Witte G, Hartung S, Buttner K, Hopfner KP. Structural biochemistry of a bacterial checkpoint protein reveals diadenylate cyclase activity regulated by DNA recombination intermediates. Mol Cell. 2008;30:167–178. doi: 10.1016/j.molcel.2008.02.020. [DOI] [PubMed] [Google Scholar]
- Woodward JJ, Iavarone AT, Portnoy DA. c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science. 2010;328:1703–1705. doi: 10.1126/science.1189801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Dziejman M, Mekalanos JJ. Determination of the transcriptome of Vibrio cholerae during intraintestinal growth and midexponential phase in vitro. Proc Natl Acad Sci U S A. 2003;100:1286–1291. doi: 10.1073/pnas.0337479100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.