Abstract
In eukaryotic cells, proteins and membranes are transported between successive compartments by vesicle trafficking. Since precise protein localization is crucial for a range of cellular functions, it is not surprising that vesicle trafficking plays a role in many processes, including cell division, signaling, development, and even gene expression. We recently found evidence that the yeast secretory pathway directly regulates the dynamics of a key cell survival process, the unfolded protein response (UPR). UPR activation requires the processing of the transcription factor encoding RNA HAC1. We showed that the small yeast GTPase Ypt1, which regulates endoplasmic reticulum-to-Golgi trafficking, associates with and controls the RNA stability of unspliced HAC1 under normal growth conditions. Other small GTPases of the Ypt family also interacted with the unprocessed RNA. Here we speculate about the possible mechanism behind this novel secretory pathway-dependent regulation of endoplasmic reticulum homeostasis.
Keywords: vesicle trafficking, Ypts, UPR, HAC1 RNA, RNA degradation
Introduction
In eukaryotes, ras family GTPases regulate the transport of proteins and membranes in and out of the cell. In the simplest eukaryote, the budding yeast Saccharomyces cerevisiae, these GTPases are called Ypts (yeast protein transport). They are involved in different aspects of intracellular membrane trafficking, including vesicle formation, motility, docking, and membrane fusion and remodeling. There are 11 Ypts, and each of them plays compartment-specific roles in endocytosis (Ypt7, 51/52/53) or exocytosis (Ypt1, Ypt32/32, Sec4). GDP-bound inactive Ypts are found in complex with GDI (GDP-dissociation inhibitor) in the cytosol, and the nucleotide exchange occurs after recruitment to the appropriate membrane. Although we still lack knowledge of a universal mechanism for Ypt targeting to membranes, in vitro assays in mammalian cells showed that a GDI-GTPase complex carries all the information necessary for proper GTPase delivery to the target membrane.1,2 Further, evidence from domain swap experiments suggests that the Ypt C-terminal prenylated domains called hypervariable domains, which associate with GDI,3 can act as localization tags.4,5 These hypervariable domains may also participate in the interactions of Ypts with GDFs (GDI displacement factors),6 the proteins that displace GDI and facilitate the association of Ypts with the target membrane. Since GDFs localize to different compartments, they are likely to provide an additional level of specificity for GTPase targeting (for extensive reviews on GTPase targeting to distinct membranes, see refs. 6−8). Once on a membrane, Ypts undergo a conformational “on” change when stimulated by GEFs (guanine nucleotide exchange factors), and can interact with downstream effector proteins to regulate trafficking. Conversely, when GAPs (GTPase activating proteins) associate with the GTPases, they turn them “off.” As protein transport is crucial for all cellular processes, it is not surprising that vesicle trafficking is coordinated with other processes. Here we discuss novel findings which link protein transport and a key eukaryotic stress survival response, the unfolded protein response (UPR).
Crosstalk between Vesicle Trafficking and Other Cellular Processes
We will start by summarizing examples of crosstalk between the secretory pathway and other processes with special emphasis on the interplay between membrane transport and RNA regulation. Systematic deletion screens performed in Saccharomyces cerevisiae have shown a role for vacuolar protein sorting genes (VPS) in the regulation of telomere length9 and implicated Golgi transport in the correct positioning of the nucleus.10 There is ample evidence that trafficking is coordinated with other cellular processes in higher eukaryotes as well. A comprehensive RNAi screen in Drosophila discovered that COPI coat proteins are important for cell division.11 Also, vesicle trafficking has been linked to development12 and to several signaling cascades.13-16
Accumulating evidence connects vesicular transport and gene expression as components of the trafficking machinery were shown to be necessary for the correct localization of a number of RNAs. Studies in two diverse systems (Drosophila and retrovirus-infected mammalian cells) have implicated a role for Rab11, which regulates traffic from the trans-Golgi to the plasma membrane and through recycling endosomes,17,18 in RNA localization. During Drosophila oocyte development, oskar RNA localizes to the posterior pole and organizes the germ plasm.19 However, in rab11 loss-of-function mutants, oskar RNA does not completely reach the oocyte posterior pole.20,21 Rab11-containing vesicles also transport retroviral RNAs: live imaging of RNAs from murine leukemia virus show co-localization of viral transcripts with the GTPase on recycling endosomes.22 Likewise, components of the yeast secretory pathway are necessary for asymmetric RNA distribution, because mutations in a panel of secretory genes alter the localization of the transcription factor encoding RNA ASH1.23
The phenotypes observed in these studies, however, could be a result of defects in cytoskeleton integrity that arise from perturbations in vesicle trafficking, and therefore may reflect an indirect involvement of the secretory pathway. Indeed, rab11 loss-of-function disrupts the organization of the microtubule plus ends,20,24 and all yeast secretory mutations studied lead to defects in the actin cytoskeleton.23 Recent work provides more convincing evidence for a direct involvement of the secretory pathway in the regulation of gene expression.25 Two independent proteomic screens identified a significant number of trafficking regulators among the proteins associated with yeast total mRNA (Table 1, see legend).25 In reciprocal experiments with two of the transport components (Vtc1 and Ubp3), hundreds of mRNAs were identified by DNA microarrays to be reproducibly associated with each protein.25 Interestingly, a significant number of these RNAs encode proteins with specific sub-cellular localization, which coincides with the localization of the respective trafficking protein. For example, Vtc1, which is found at the vacuole and the endoplasmic reticulum (ER),26 associates with transcripts of ER, membrane, and vacuolar components. Thus, it appears that proteins with distinct roles in membrane trafficking can participate directly in the regulation of gene expression likely by transporting RNAs.
Table 1. Secretory pathway components shown to associate with RNAs.
| Protein Name | Annotated Function* | Annotated Localization* |
|---|---|---|
| Sec1# |
Docking and fusion of exocytic vesicles |
Bud neck; bud tip |
| Sec16# |
COPII vesicle coat component; transport of ER vesicles |
COPII vesicle |
| Sec31# |
COPII vesicle coat component; transport of ER vesicles |
COPII vesicle |
| Sec26# |
COPI vesicle coat component; ER-to-Golgi transport |
COPI vesicle |
| Sec27# |
COPI vesicle coat component; ER-to-Golgi transport |
COPI vesicle |
| Ubp3# |
Ubiquitin-specific protease; ER-Golgi anterograde and retrograde transport |
Cytosol |
| Vtc1# |
Subunit of the vacuolar transporter chaperone (VTC) complex involved in membrane trafficking |
Vacuole; ER |
| Ypt1#,‡ |
Rab family GTPase; ER-to-Golgi step of the secretory pathway |
ER and Golgi |
| Ypt7‡ |
Rab family GTPase; late endosome-to-vacuole sorting |
Vacuole |
| Ypt32‡ |
Rab family GTPase; trans-Golgi-to-plasma membrane sorting |
Endosome and Golgi |
| Rho3‡ | Rho/Rac GTPase; establishment of cell polarity | Cytosol; cell bud; plasma membrane |
Data from the Saccharomyces genome database (www.yeastgenome.org); #Data from Tsvetanova NG, Klass DM, Salzman J and Brown PO (2010); associated with total mRNA; ‡Data from Tsvetanova NG, Riordan DP and Brown PO (2012); associated with HAC1 RNA
In yeast and mammalian cells, the processing of one RNA determines whether or not to activate the UPR, a crucial cell survival signaling cascade. It turns out that the expression of this key transcript in yeast is controlled by the vesicle trafficking machinery.27 Here, we discuss the unexpected GTPase-dependent regulation of the UPR and speculate that it constitutes the biochemical mechanism to explain a functional link known to exist between these two key cellular processes.
The UPR and Vesicle Trafficking
The UPR, triggered by accumulation of misfolded proteins in the ER, is an important cellular homeostatic mechanism implicated in a number of human diseases and pathologies such as neurodegeneration, diabetes, autoimmune response, and cancer.28-30 In Saccharomyces cerevisiae UPR, a highly conserved program in eukaryotes, the ER transmembrane kinase-endonuclease Ire1 activates this response via non-canonical splicing of a transcription factor encoding RNA, HAC1.31 The resulting HAC1 junctions are next “ligated” by a tRNA ligase, Rlg1,32 the mature HAC1 mRNA is translated, and activates the expression of UPR target genes.
A number of studies have established a functional relationship between the UPR and vesicle trafficking. Chang et al.33 and Leber et al.34 reported activation of the UPR in the absence of chemical stress in secretory mutants defective in events extending from the ER to distal secretory compartments. Furthermore, when combined with ire1Δ or hac1Δ, the sec mutations were either partially lethal or led to a more extreme growth defect than sec- alone.33 Conversely, overexpression of IRE1 or HAC1 rescued the defects.35,36 Interestingly, mutations in retrograde transport genes had no effect on the UPR.34 One model that can account for all these observations is that mutations affecting exit from the ER and the exocytic pathway will perturb the balance between anterograde and retrograde transport. This will eventually lead to accumulation of Golgi-derived proteins in the ER,37 thus overloading the folding capacity of the organelle (Fig. 1, dotted arrows). The cell would next activate its UPR, which increases the production of genes controlling secretory functions38 in order to cope with the stress (Fig. 1, dotted arrows). Therefore, the functional relationship between trafficking and ER stress has been generally viewed as indirect and somewhat of an uneven dependency, where active UPR allows the cell to compensate for defects in its secretory pathway. However, we recently found evidence for a direct connection between vesicle trafficking and ER homeostasis (Fig. 1, solid arrows). This regulatory interplay is performed by the binding of the small GTPase Ypt1 to unspliced HAC1 RNA and likely helps avert activation of the UPR in the absence of stress.
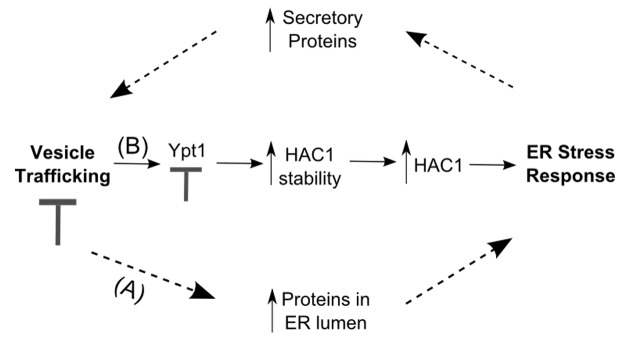
Figure 1. Crosstalk between vesicle trafficking and the unfolded protein response. (A) A block in the secretory pathway can indirectly activate the UPR (dotted arrows) by causing accumulation of proteins in the ER. The UPR, in turn, stimulates the expression of RNAs encoding secretory proteins, and will eventually alleviate the defect in transport. (B) Vesicle trafficking directly regulates the UPR: Ypt1 interacts with unspliced HAC1 and leads to its degradation. A block in Ypt1 function will result in accumulation of HAC1, the RNA will get processed into Hac1 protein and eventually produce enough transcription factor to activate the UPR (solid arrows).
Ypt1-Dependent Active Control of the UPR
We were interested in identifying HAC1-interacting proteins to gain new insights into UPR regulation, so we performed an protein microarray in vitro screen of the yeast proteome for HAC1-binding factors. We were surprised to find the RNA associating almost exclusively with small GTPases (Table 1, see legend).27 Among the strongest interactors were three members of the Ypt family- Ypt1, Ypt7, and Ypt32. Since Ypt1 regulates ER-to-Golgi transport, perturbations of which trigger the UPR, we focused on the association between Ypt1 and HAC1. We confirmed the in vitro result by showing that the GTPase also interacts with unspliced HAC1 in vivo, and observed that the interaction happens only in the absence of ER stress. The Ypt1-HAC1 association could no longer be detected, once cells were treated with the UPR-inducing chemical DTT. These initial results suggested that there must be a more intimate link between vesicle trafficking and the UPR than previously anticipated. Further confirming a direct crosstalk were our findings that Ypt1 and HAC1 do not associate in mutant strains lacking two genes required for proper UPR initiation, IRE1 and ADA5,31,39 and that Ypt1 interacts with Ada5.27 Ada5 and Ire1 form a complex,39 and Ire1 is an ER transmembrane protein; thus, we think it is very likely that an Ire1/Ada5/Ypt1-HAC1 complex forms near the ER membrane. Consistent with this model, deleting the HAC1 3′UTR, which is required for efficient ER localization,40 abolishes the Ypt1-HAC1 interaction.27 The precise roles of Ire1 and Ada5 in the GTPase-RNA association are currently unknown, but we speculate that these proteins may participate in the UPR-vesicle trafficking crosstalk by recruiting HAC1 in proximity to Ypt1.
To determine the functional role of the Ypt1-HAC1 interaction, we examined HAC1 RNA levels in a YPT1 knockdown strain. Unspliced HAC1 copy number was ~2.5-fold higher in this mutant compared with wild type, and we found that this increase in expression is due to stabilization of the RNA.27 The GTPase is also involved in recovery from ER stress, because cells with compromised YPT1 expression recovered slower compared with wild type cells once the source of UPR was removed.27 Therefore, there are physiological consequences of disturbing the Ypt1-HAC1 interaction.
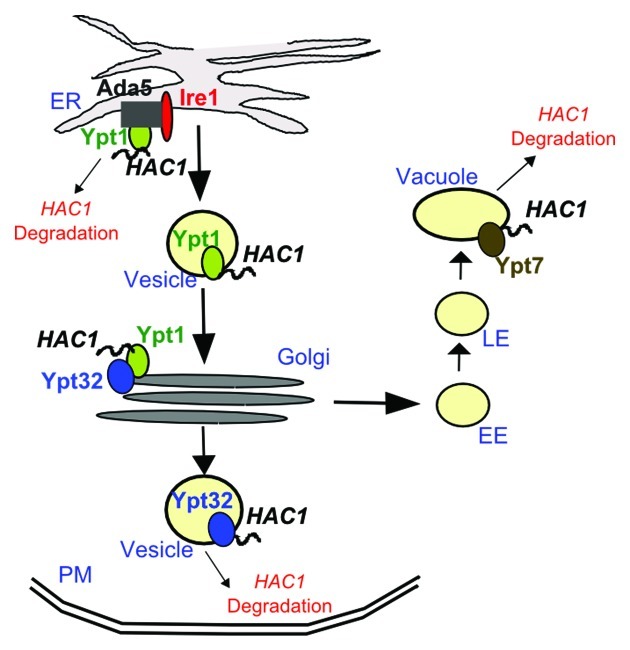
What could be the mechanism behind the destabilization of HAC1 RNA by Ypt1? One possibility is that the RNA is degraded directly at the site of Ypt1-HAC1 interaction, which we believe is the ER membrane (discussed above). An equally likely and not mutually exclusive model is that Ypt1-HAC1 binding at the ER is followed by transport of the RNA away from the ER-localized processing machinery (Ire1 and Ada5) and close to RNA decay factors elsewhere in the cell (Fig. 2). We favor the second model, because it is consistent with our findings that two other Ypts (Ypt7 and 32), involved in trafficking to distal cellular compartments, also interact with unsliced HAC1 RNA. Ypt7 orchestrates late endosome-to-vacuole trafficking and Ypt32 controls the intra-Golgi and post-Golgi steps of exocytosis. While we have not examined the interactions between HAC1 and Ypts 7 and 32 beyond the initial screen, it is possible that Ypts 1/7/32 work together to coordinate the regulation of HAC1 (Fig. 2). In support of this model, biochemical and genetic data suggest that small GTPases couple discrete vesicle trafficking steps. This GTPase crosstalk is established via interactions with common GEFs or downstream effectors (for extensive review, see refs. 41 and 42).
Figure 2. A putative mechanism for Ypt-dependent regulation of HAC1 stability. Under normal growth conditions, Ypts mediate vesicle-assisted trafficking of unspliced HAC1 away from the ER and the ER-localized Ire1/Ada5 complex to prevent unnecessary activation of the UPR. HAC1 degradation may also be happening directly adjacent to the ER. Ypt GTPases ‘communicate’ with each other via interactions with common protein factors (e.g., GEFs, effectors, etc. not shown) and may utilize this crosstalk to direct HAC1 localization in proximity to RNA decay factors for degradation. Additional adaptor proteins linking the HAC1 RNA to the lipid membrane of the vesicle or mediating the Ypt-RNA interactions may be present but are not shown for simplicity. The HAC1 RNA is depicted as a squiggly line. EE, early endosomes; LE, late endosomes; ER, endoplasmic reticulum; PM, plasma membrane.
Secretory vesicles associated with a motor protein would provide a fast and efficient way to transport HAC1 to distal cellular compartments. Since Ypt GTPases are key regulators of vesicle formation, trafficking, and docking, we anticipate that they could play a role in multiple aspects of HAC1 trafficking. As no RNAs have been shown to interact directly with lipid membranes, the vesicle loading and trafficking of HAC1 likely requires one or more adaptor proteins. Ypt GTPases, which are anchored to lipids via their di-prenylated C-termini, or Ypt protein interactors that have the capacity to interact with both nucleic acids and membranes are natural candidates for such mediators (it should be noted that none of the currently documented Ypt binding partners have been shown to associate with both membranes and RNA). In addition to acting as adaptors stabilizing the HAC1-vesicle interaction, Ypts may also be directly involved in the recruitment of motor proteins that transport the RNA-containing vesicle. Indeed, one of the three Ypts associating with HAC1, Ypt32, interacts with the class V myosin motor Myo2 during secretion.43 Directed vesicle-mediated transport of HAC1 away from the ER aided by HAC1-interacting GTPases would provide an efficient means for the cell to keep its UPR “off” in the absence of stress stimuli.
Our model (Fig. 2) would predict that defects in vesicle trafficking would prevent Ypt-dependent HAC1 RNA localization to decay factors and lead to stabilization and accumulation of unspliced HAC1 near the ER and the ER-localized HAC1 splicing machinery (Fig. 1, solid arrows). Amassed HAC1 RNA would then be processed by Ire1 to produce functional Hac1 protein that will eventually reach a critical threshold and activate the UPR. In agreement with this scenario, we observed increased amounts of spliced HAC1 RNA, when we knocked down YPT1.27 Such direct regulatory relationship between vesicle trafficking and ER stress would provide an efficient way for the cell to communicate defects in its secretory pathway to ER surveillance and enable a robust response to perturbations of cellular homeostasis.
Many questions remain to be addressed. As Ypt1-dependent recruitment of HAC1 to decay factors is a plausible mechanism of how the GTPase regulates RNA stability, it will be important to test if Ypt1 interacts physically and/or genetically with known members of the yeast decay machinery, in order to start dissecting the precise features of the process. Also, the recruitment of HAC1 could be either active, i.e., involving a physical association of the RNA with the GTPase and subsequent directional localization of the RNA, or indirect, in which case the RNA ‘hitches’ a ride on a secretory vesicle along with other cargo trafficked between the ER and Golgi. Our data do not distinguish between the two possibilities, since we purified the proteins used for generating protein microarrays for the initial screen from yeast25 and cannot exclude co-purification of proteins. Further, we suspect that the Ypt1-HAC1 interaction takes place close to the ER membrane and is assisted by the Ire1/Ada5 complex, but this model remains to be tested. Also, the regulatory roles of the other secretory proteins identified in the screen (e.g., Ypt7 and Ypt32) should be established. If these GTPases are important in regulating the RNA stability of HAC1 similar to Ypt1, it will be interesting to determine if Ypts communicate with each other to control the fate of HAC1. Lastly, Ypts 1, 7 and 32 share 60–70% homology with and function in the same transport compartments as mammalian Rabs 1, 7 and 11, respectively.41 Future studies should investigate whether mammalian Rabs play a direct role in UPR regulation analogous to their yeast counterparts by testing for interactions between Rabs and the mammalian ortholog of HAC1, XBP1. Despite the open questions, it is clear that there is a direct link between the secretory pathway and the UPR and that vesicle trafficking plays an active role in the regulation of ER homeostasis.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/smallgtpases/article/22599
References
- 1.Soldati T, Shapiro AD, Svejstrup AB, Pfeffer SR. Membrane targeting of the small GTPase Rab9 is accompanied by nucleotide exchange. Nature. 1994;369:76–8. doi: 10.1038/369076a0. [DOI] [PubMed] [Google Scholar]
- 2.Ullrich O, Horiuchi H, Bucci C, Zerial M. Membrane association of Rab5 mediated by GDP-dissociation inhibitor and accompanied by GDP/GTP exchange. Nature. 1994;368:157–60. doi: 10.1038/368157a0. [DOI] [PubMed] [Google Scholar]
- 3.Rak A, Pylypenko O, Durek T, Watzke A, Kushnir S, Brunsveld L, et al. Structure of Rab GDP-dissociation inhibitor in complex with prenylated YPT1 GTPase. Science. 2003;302:646–50. doi: 10.1126/science.1087761. [DOI] [PubMed] [Google Scholar]
- 4.Chavrier P, Gorvel JP, Stelzer E, Simons K, Gruenberg J, Zerial M. Hypervariable C-terminal domain of rab proteins acts as a targeting signal. Nature. 1991;353:769–72. doi: 10.1038/353769a0. [DOI] [PubMed] [Google Scholar]
- 5.Stenmark H, Valencia A, Martinez O, Ullrich O, Goud B, Zerial M. Distinct structural elements of rab5 define its functional specificity. EMBO J. 1994;13:575–83. doi: 10.1002/j.1460-2075.1994.tb06295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.ten Klooster JP, Hordijk PL. Targeting and localized signalling by small GTPases. Biol Cell. 2007;99:1–12. doi: 10.1042/BC20060071. [DOI] [PubMed] [Google Scholar]
- 7.Pfeffer S. A model for Rab GTPase localization. Biochem Soc Trans. 2005;33:627–30. doi: 10.1042/BST0330627. [DOI] [PubMed] [Google Scholar]
- 8.Pfeffer S, Aivazian D. Targeting Rab GTPases to distinct membrane compartments. Nat Rev Mol Cell Biol. 2004;5:886–96. doi: 10.1038/nrm1500. [DOI] [PubMed] [Google Scholar]
- 9.Rog O, Smolikov S, Krauskopf A, Kupiec M. The yeast VPS genes affect telomere length regulation. Curr Genet. 2005;47:18–28. doi: 10.1007/s00294-004-0548-y. [DOI] [PubMed] [Google Scholar]
- 10.Ohya Y, Sese J, Yukawa M, Sano F, Nakatani Y, Saito TL, et al. High-dimensional and large-scale phenotyping of yeast mutants. Proc Natl Acad Sci USA. 2005;102:19015–20. doi: 10.1073/pnas.0509436102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eggert US, Kiger AA, Richter C, Perlman ZE, Perrimon N, Mitchison TJ, et al. Parallel chemical genetic and genome-wide RNAi screens identify cytokinesis inhibitors and targets. PLoS Biol. 2004;2:e379. doi: 10.1371/journal.pbio.0020379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Entchev EV, Schwabedissen A, González-Gaitán M. Gradient formation of the TGF-beta homolog Dpp. Cell. 2000;103:981–91. doi: 10.1016/S0092-8674(00)00200-2. [DOI] [PubMed] [Google Scholar]
- 13.Alvarez-Dominguez C, Stahl PD. Interferon-gamma selectively induces Rab5a synthesis and processing in mononuclear cells. J Biol Chem. 1998;273:33901–4. doi: 10.1074/jbc.273.51.33901. [DOI] [PubMed] [Google Scholar]
- 14.Barbieri MA, Kohn AD, Roth RA, Stahl PD. Protein kinase B/akt and rab5 mediate Ras activation of endocytosis. J Biol Chem. 1998;273:19367–70. doi: 10.1074/jbc.273.31.19367. [DOI] [PubMed] [Google Scholar]
- 15.Barbieri MA, Roberts RL, Gumusboga A, Highfield H, Alvarez-Dominguez C, Wells A, et al. Epidermal growth factor and membrane trafficking. EGF receptor activation of endocytosis requires Rab5a. J Cell Biol. 2000;151:539–50. doi: 10.1083/jcb.151.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lanzetti L, Rybin V, Malabarba MG, Christoforidis S, Scita G, Zerial M, et al. The Eps8 protein coordinates EGF receptor signalling through Rac and trafficking through Rab5. Nature. 2000;408:374–7. doi: 10.1038/35042605. [DOI] [PubMed] [Google Scholar]
- 17.Chen W, Feng Y, Chen DY, Wandinger-Ness A. Rab11 is required for trans-golgi network-to-plasma membrane transport and a preferential target for GDP dissociation inhibitor. Mol Biol Cell. 1998;9:3241–57. doi: 10.1091/mbc.9.11.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin SXH, Teter K, Young JJ, Moore HP. Role of rab11 in TF recycling and transport between TGN and recycling endosomes. Mol Biol Cell. 1996;7:3438. [Google Scholar]
- 19.Ephrussi A, Dickinson LK, Lehmann R. Oskar organizes the germ plasm and directs localization of the posterior determinant nanos. Cell. 1991;66:37–50. doi: 10.1016/0092-8674(91)90137-N. [DOI] [PubMed] [Google Scholar]
- 20.Dollar G, Struckhoff E, Michaud J, Cohen RS. Rab11 polarization of the Drosophila oocyte: a novel link between membrane trafficking, microtubule organization, and oskar mRNA localization and translation. Development. 2002;129:517–26. doi: 10.1242/dev.129.2.517. [DOI] [PubMed] [Google Scholar]
- 21.Jankovics F, Sinka R, Erdélyi M. An interaction type of genetic screen reveals a role of the Rab11 gene in oskar mRNA localization in the developing Drosophila melanogaster oocyte. Genetics. 2001;158:1177–88. doi: 10.1093/genetics/158.3.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Basyuk E, Galli T, Mougel M, Blanchard JM, Sitbon M, Bertrand E. Retroviral genomic RNAs are transported to the plasma membrane by endosomal vesicles. Dev Cell. 2003;5:161–74. doi: 10.1016/S1534-5807(03)00188-6. [DOI] [PubMed] [Google Scholar]
- 23.Aronov S, Gerst JE. Involvement of the late secretory pathway in actin regulation and mRNA transport in yeast. J Biol Chem. 2004;279:36962–71. doi: 10.1074/jbc.M402068200. [DOI] [PubMed] [Google Scholar]
- 24.Zhang H, Squirrell JM, White JG. RAB-11 permissively regulates spindle alignment by modulating metaphase microtubule dynamics in Caenorhabditis elegans early embryos. Mol Biol Cell. 2008;19:2553–65. doi: 10.1091/mbc.E07-09-0862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsvetanova NG, Klass DM, Salzman J, Brown PO. Proteome-wide search reveals unexpected RNA-binding proteins in Saccharomyces cerevisiae. PLoS ONE. 2010;5:5. doi: 10.1371/journal.pone.0012671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cherry JM, Ball C, Weng S, Juvik G, Schmidt R, Adler C, et al. Genetic and physical maps of Saccharomyces cerevisiae. Nature. 1997;387(Suppl):67–73. doi: 10.1038/43025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsvetanova NG, Riordan DP, Brown PO. The yeast Rab GTPase Ypt1 modulates unfolded protein response dynamics by regulating the stability of HAC1 RNA. PLoS Genet. 2012;8:e1002862. doi: 10.1371/journal.pgen.1002862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Austin RC. The unfolded protein response in health and disease. Antioxid Redox Signal. 2009;11:2279–87. doi: 10.1089/ars.2009.2686. [DOI] [PubMed] [Google Scholar]
- 29.Zhang K, Kaufman RJ. The unfolded protein response: a stress signaling pathway critical for health and disease. Neurology. 2006;66(Suppl 1):S102–9. doi: 10.1212/01.wnl.0000192306.98198.ec. [DOI] [PubMed] [Google Scholar]
- 30.Kaufman RJ. Orchestrating the unfolded protein response in health and disease. J Clin Invest. 2002;110:1389–98. doi: 10.1172/JCI16886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sidrauski C, Walter P. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell. 1997;90:1031–9. doi: 10.1016/S0092-8674(00)80369-4. [DOI] [PubMed] [Google Scholar]
- 32.Sidrauski C, Cox JS, Walter P. tRNA ligase is required for regulated mRNA splicing in the unfolded protein response. Cell. 1996;87:405–13. doi: 10.1016/S0092-8674(00)81361-6. [DOI] [PubMed] [Google Scholar]
- 33.Chang HJ, Jesch SA, Gaspar ML, Henry SA. Role of the unfolded protein response pathway in secretory stress and regulation of INO1 expression in Saccharomyces cerevisiae. Genetics. 2004;168:1899–913. doi: 10.1534/genetics.104.032961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leber JH, Bernales S, Walter P. IRE1-independent gain control of the unfolded protein response. PLoS Biol. 2004;2:E235. doi: 10.1371/journal.pbio.0020235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Higashio H, Kohno K. A genetic link between the unfolded protein response and vesicle formation from the endoplasmic reticulum. Biochem Biophys Res Commun. 2002;296:568–74. doi: 10.1016/S0006-291X(02)00923-3. [DOI] [PubMed] [Google Scholar]
- 36.Sato M, Sato K, Nakano A. Evidence for the intimate relationship between vesicle budding from the ER and the unfolded protein response. Biochem Biophys Res Commun. 2002;296:560–7. doi: 10.1016/S0006-291X(02)00922-1. [DOI] [PubMed] [Google Scholar]
- 37.Cole NB, Ellenberg J, Song J, DiEuliis D, Lippincott-Schwartz J. Retrograde transport of Golgi-localized proteins to the ER. J Cell Biol. 1998;140:1–15. doi: 10.1083/jcb.140.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–58. doi: 10.1016/S0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- 39.Welihinda AA, Tirasophon W, Kaufman RJ. The transcriptional co-activator ADA5 is required for HAC1 mRNA processing in vivo. J Biol Chem. 2000;275:3377–81. doi: 10.1074/jbc.275.5.3377. [DOI] [PubMed] [Google Scholar]
- 40.Aragón T, van Anken E, Pincus D, Serafimova IM, Korennykh AV, Rubio CA, et al. Messenger RNA targeting to endoplasmic reticulum stress signalling sites. Nature. 2009;457:736–40. doi: 10.1038/nature07641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Segev N. Ypt/rab gtpases: regulators of protein trafficking. Sci STKE. 2001;2001:re11. doi: 10.1126/stke.2001.100.re11. [DOI] [PubMed] [Google Scholar]
- 42.Segev N. Ypt and Rab GTPases: insight into functions through novel interactions. Curr Opin Cell Biol. 2001;13:500–11. doi: 10.1016/S0955-0674(00)00242-8. [DOI] [PubMed] [Google Scholar]
- 43.Lipatova Z, Tokarev AA, Jin Y, Mulholland J, Weisman LS, Segev N. Direct interaction between a myosin V motor and the Rab GTPases Ypt31/32 is required for polarized secretion. Mol Biol Cell. 2008;19:4177–87. doi: 10.1091/mbc.E08-02-0220. [DOI] [PMC free article] [PubMed] [Google Scholar]