Abstract
RhoG is a member of the Rho family of small GTPases sharing highest sequence similarity with Rac and Cdc42. Mig-2 and Mtl represent the functional equivalents of RhoG in Caenorhabditis elegans and Drosophila, respectively. RhoG has attracted great interest because it plays a central role in the regulation of cytoskeletal reorganization in various physiological and pathophysiological situations. For example, it is fundamental to phagocytotic processes, is able to regulate gene expression, cell survival and proliferation, and is involved in cell migration and in the invasion of pathogenic bacteria. The activation of Rac1 via an ELMO/Dock180 module has been elaborated to be important for RhoG signaling. Although a stimulatory role for neurite outgrowth in the pheochromocytoma PC12 cell line has been assigned to RhoG, the exact function of this GTPase for the development of the processes of primary neurons remains to be clarified. In this view, we discuss the impact of RhoG on axonal and dendritic differentiation, its role as a conductor of Rac1 and Cdc42 activity and the functional regulation of RhoG expression by the microRNA miR-124.
Keywords: RhoG, Rac1, Cdc42, microRNA miR-124, ELMO/Dock180, GEF, axonal branching, dendritic branching, cytoskeletal reorganization
RhoG was originally discovered in a screen for coding sequences specifically accumulated in the late G1 phase of the cell cycle.1 Following research demonstrated that RhoG, in addition to be able to activate gene expression in lymphocytes, is critical for the regulation of the actin cytoskeleton in several organisms and cell types.2 A few examples of the latter function include the phagocytosis of apoptotic cells, the trans-endothelial migration of leucocytes, the T-cell receptor internalization and trogocytosis, the invasion of pathogenic bacteria and the migration of cancer cells.3-8 Previous research on the function of RhoG in neuronal differentiation has been based primarily on experimental investigations using the pheochromocytoma PC12 cell line: here RhoG was found to promote neurite outgrowth.9-11 A major advance in the understanding of how RhoG mediates the regulation of cytoskeletal dynamics was the finding that RhoG can activate Rac1 via an ELMO/Dock180 interaction.3,11 This ELMO/Dock180/Rac1 signaling module has now been established to be crucial not only for RhoG-stimulated neurite outgrowth in PC12 cells, but also for phagocytosis as well as engulfment of apoptotic cells, and the invasion of pathogenic bacteria.
Our interest in RhoG was evoked when performing a screen for genes, whose expression are regulated by the nervous system-specific microRNA miR-124. In this screen, we focused on gene products that potentially could be involved in the establishment of neuronal process complexity. Several algorithms from the public domain predicted RhoG as a target for miR-124-regulated gene expression.12,13 Actually, we could experimentally confirm that endogenously expressed miR-124 regulates the expression of RhoG in primary hippocampal neurons.14 We then set out to elucidate the significance of RhoG for neuronal process differentiation, and came up with a surprising result: RhoG, formerly described to stimulate neurite outgrowth in the PC12 cell line, inhibits axonal and dendritic branching in primary hippocampal neurons. Using the in utero electroporation technique (IUE), we confirmed the in vivo relevance of these results.14,15 At this point, the vitally question regarding the signaling pathway of RhoG-mediated inhibition of axonal and dendritic tree complexity arose. We tackled this question by proving the impact of the ELMO/Dock180/Rac1 signaling pathway using three different dominant-negative constructs and two different specific knockdown constructs, all of which are well established from previous research (Fig. 1).11,16-18 The results clearly showed that the ELMO/Dock180/Rac1 signaling pathway is functionally important to reduce axonal branching. To our best knowledge, this is the first demonstration that Rac1 is inhibitory to the establishment of axonal branching in hippocampal neurons. By epistasis analysis we further found that the inhibition of axonal branching elicited by the constitutively active RhoG-G12V can be revoked by the knockdown of endogenous Rac1 expression. In this way, our results point to the view that RhoG reduces axonal branching via ELMO/Dock180/Rac1 signaling (Fig. 2). However, in contrast to stimulate axonal branching, Rac1 knockdown reduced dendritic branching. Additionally, the RhoG-F37A mutant, shown to strongly increase axonal branching, also reduced dendritic branching. This mutant is notably because of two reasons: (1) RhoG-F37A cannot interact with the downstream signaling molecule ELMO, but very probably is able to bind guanine nucleotide exchange factors (GEFs) and that way compete with endogenous RhoG for activating GEFs. Here, RhoG-F37A operates in a dominant-negative manner with respect to the RhoG/ELMO/Dock180/Rac1 signaling pathway. (2) Deduced from studies of a F37A mutant of Rac1, which is structurally similar to RhoG, it is very likely that RhoG-F37A may be able to activate downstream signaling molecules different from the ELMO/Dock180/Rac1 module. In this case, overexpression of RhoG-F37A functionally mimics an overexpression of RhoG. Therefore, in addition to the results obtained by Rac1 knockdown, the diverging effects of the RhoG-F37A mutant on axonal vs. dendritic branching strongly imply that RhoG does not inhibit dendritic tree complexity through ELMO/Dock180/Rac1 signaling but, most likely, activates other downstream signaling molecules.14 Motivated by early data of Gauthier-Rouvière et al. indicating that RhoG can independently activate Rac1 and Cdc42, we then analyzed the relevance of Cdc42 for dendritic branching.14,19 The results of these experiments clearly demonstrated that knockdown of Cdc42 increases dendritic tree complexity, and furthermore that knockdown of Cdc42 precludes the inhibition of dendritic branching by RhoG. Based on these results we propose that RhoG inhibits dendritic branching via Cdc42 signaling (Fig. 2).
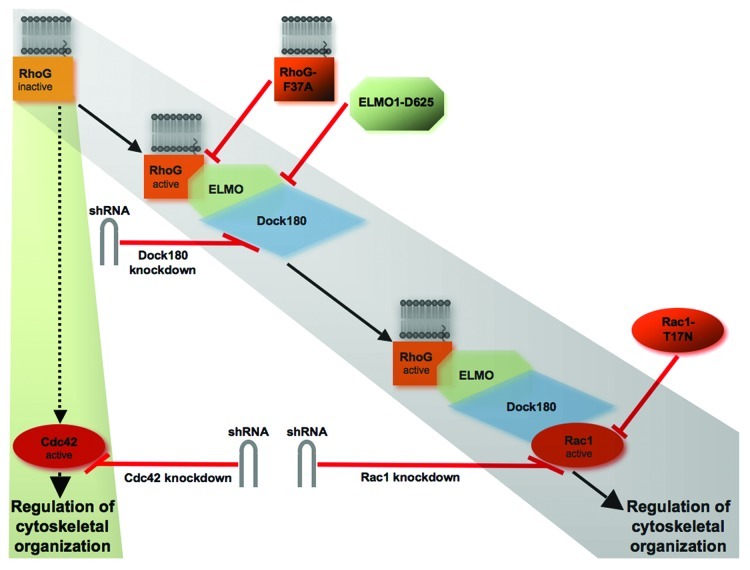
Figure 1. Two signaling pathways involving either the ELMO/Dock180/Rac1 module or Cdc42 may be activated by RhoG to regulate cytoskeletal reorganization in neuronal process differentiation. RhoG can activate Rac1 by ELMO/Dock180 signaling to regulate cytoskeletal organization (gray track). We used three dominant-negative constructs, RhoG-F37A, ELMO1-D625 and Rac1-T17N, to explore the relevance of ELMO/Dock180/Rac1 signaling for the regulation of axonal branching. RhoG-F37A harbors a mutation in the effector region which prevents binding to ELMO. ELMO1-D625 cannot bind to Dock180, and Rac1-T17N is an established dominant-negative construct for inhibiting Rac1 activity. In addition, the endogenous expression of Dock180 as well as Rac1 was reduced by specific shRNA-mediated knockdown of Dock180 and Rac1, respectively. RhoG may also signal via Cdc42 to regulate cytoskeletal organization (green track). To analyze the impact of Cdc42 for the regulation of dendritic branching, we reduced the endogenous expression of Cdc42 by a knockdown approach with a shRNA construct specific to Cdc42.
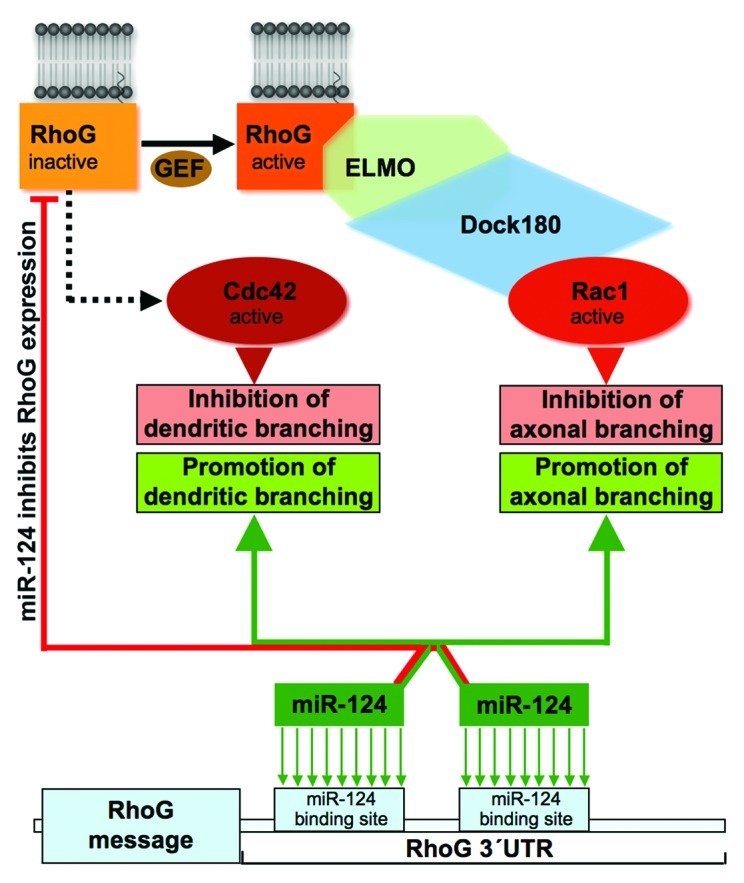
Figure 2. miR-124 reduces the expression of RhoG, which inhibits axonal and dendritic branching via ELMO/Dock180/Rac1 and Cdc42 signaling, respectively. RhoG stimulates Rac1 via the ELMO/Dock180 module. This leads to an inhibition of axonal branching. The guanine nucleotide exchange factor (GEF), which activates RhoG in this case, is currently not known. This GEF, however, has a major role in the ELMO/Dock180/Rac1 signaling cascade for reducing axonal branching as it seems to be rate-limiting for cascade activation in neurons during times of axogenesis. In addition to inhibiting axonal branching, RhoG reduces dendritic tree complexity. This inhibition of dendritic branching is dependent on Cdc42 but not on ELMO/Dock180/Rac1 signaling. Currently, the signaling intermediates connecting RhoG and Cdc42 are unidentified. The 3′UTR of the RhoG gene comprises two binding sites for miR-124. This microRNA inhibits RhoG expression and, this way, promotes axonal and dendritic branching.
Taken together, our data demonstrating that RhoG not only inhibits axonal branching via ELMO/Dock180/Rac1 signaling, but also inhibits dendritic branching dependent on Cdc42 are somewhat surprising and probably stimulating for the following reasons: (1) Up to now, only promoting effects on neuronal process differentiation have been attributed to RhoG as it was shown that RhoG stimulates neurite outgrowth in PC12 cells and in superior cervical ganglion (SCG) neurons.9-11,20 It may be that RhoG functions in a lineage-specific manner as SCG neurons and PC12 cells both are derived from the sympathoadrenal lineage.21,22 Here, it will be interesting to analyze neurite outgrowth of SCG neurons expressing reduced amounts of RhoG due to RhoG knockdown. (2) According to current textbook knowledge, the main function of Rac1 and Cdc42 is to promote cellular process development by stimulating lamellipodia and filopodia formation, respectively.23 Here, our results clearly demonstrate that Rac1 acts inhibitory on axonal branching and that Cdc42 is inhibitory for dendritic branching. Other recent publications present data demonstrating that Rac1 as well as Cdc42 can be inhibitory for process formation. For example, both dominant-negative Cdc42 and Cdc42 knockout led to increased branching of the endfeet of radial glial cells.24 Additionally, using biosensors for Rac1 and Cdc42 activity, it was recently shown that in mouse embryonic fibroblasts Rac1 and Cdc42 activity is temporally and spatially less coupled to cell protrusion but, surprisingly, maintain a significant level of activation during the retraction phase.25 The authors’ interpretation of these data are that the primary role of Rac1 and Cdc42 in this case may not be to initiate protrusion but rather to regulate adhesion dynamics. Thus, the spatiotemporal activation state and the strength of activation of RhoG, Rac1 and Cdc42 may determine whether these GTPases stimulate or inhibit neuronal process formation. Consequently, a major future goal will be to elucidate the spatiotemporal activation profile of RhoG and its downstream targets Rac1 and Cdc42 during neuronal process formation. (3) Our epistasis analysis support the view that RhoG, via different signaling pathways, conducts Rac1 and Cdc42 activity to inhibit axonal and dendritic branching. Interestingly, the results of our study suggest that RhoG-activating guanine nucleotide exchange factors (GEFs) are limiting for RhoG-driven activation of the ELMO/Dock180/Rac1 signaling module, at least during axogenesis. Therefore, a second major aim will be the identification of the GEFs which activate RhoG to either drive ELMO/Dock180/Rac1 or Cdc42 signaling during axogenesis and dendritogenesis, respectively. (4) We performed Sholl analysis for more precisely analyzing the impact of RhoG signaling on dendritic branching. The Sholl analysis was done by constructing a series of concentric circles with gradually increasing radius around the neuronal cell body and then counting how many times the dendrites intersect the circumference of these concentric circles. Finally, the intersection number was graphically represented as a function of the circle radius. The results of our Sholl analysis for characterizing dendritic tree complexity indicate that RhoG inhibits dendritic branching predominantly in the proximal but not in the distal part of the dendritic trees of hippocampal neurons. According to our data obtained from Sholl analysis it could be presumed that this kind of specificity is caused by a combination of RhoG-driven Cdc42 signaling to reduce dendritic branching in the proximal and distal parts of the dendritic tree and RhoG-driven Rac1 signaling to stimulate dendritic branching mainly in the distal part of the dendritic tree. Thus, the amounts of combined Rac1 and Cdc42 activity may determine the specificity of the functional outcome. This working hypothesis is strengthened by a study of loss-of-function phenotypes of the Drosophila GTPases Rac1, Rac2 and Mtl.26 The results of this study showed that axon growth, guidance and branching require increasing amounts of combined Rac1, Rac2 and Mtl activity. The relevance of C. elegans Mig-2 for the regulation of axonal branching in the context of axon pathfinding was also indicated by Lundquist et al.27
A different manner to modulate RhoG-driven cytoskeletal reorganization is the regulation of the amount of expressed RhoG. When RhoG was originally described, it was found to be a gene the expression of which can be regulated by growth factors. We now discovered a novel way to affect the amount of RhoG: the expression of this gene is suppressed by the nervous system-specific microRNA miR-124. Furthermore, this regulation of RhoG expression by miR-124 could be connected to the stimulation of axonal and dendritic tree complexity (Fig. 2). MicroRNAs are versatile means highly qualified to contribute to the specificity of functional outcome of signaling cascades, because they can operate in specific subcellular compartments (e.g., axons, dendrites or spines) as well as regulate the expression of different gene products at the same time. miR-124, for example, in addition to be localized to the neuronal cell body, was also found in the synaptodendritic compartment.28,29 Interestingly, miR-124, shown by us to downregulate RhoG expression, was also described in a microarray analysis to upregulate the expression of its downstream effector Cdc42.30 In the same microarray analysis, also an increased amount of message coding for Trio, a potential RhoG GEF, was detected. Therefore, it is tempting to speculate that miR-124 (and this may apply for other microRNAs as well), by reducing or increasing gene expression of different members of the same signaling cascade concurrently, may participate in the precise adjustment of the functional outcome of this signaling pathway. In summary, we suggest at least four levels of regulation of RhoG signaling in neurons: (1) General regulation of expression of members of RhoG signaling cascades (e.g., by transcription factors); (2) spatiotemporally coordinated expression regulation of some of these members by miR-124; (3) regulation of the amount of active RhoG by different GEFs; (4) spatiotemporally coordinated activity of different RhoG effectors, which may include the combinations of activities of different RhoG effectors at the same time.
Acknowledgments
We thank the DFG for funding of research in the Schumacher lab (grants SFB 665-A2 to S.S., and SCHU 1406/3-1 to S.S.).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/smallgtpases/article/22922
References
- 1.Vincent S, Jeanteur P, Fort P. Growth-regulated expression of rhoG, a new member of the ras homolog gene family. Mol Cell Biol. 1992;12:3138–48. doi: 10.1128/mcb.12.7.3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vigorito E, Billadeu DD, Savoy D, McAdam S, Doody G, Fort P, et al. RhoG regulates gene expression and the actin cytoskeleton in lymphocytes. Oncogene. 2003;22:330–42. doi: 10.1038/sj.onc.1206116. [DOI] [PubMed] [Google Scholar]
- 3.deBakker CD, Haney LB, Kinchen JM, Grimsley C, Lu M, Klingele D, et al. Phagocytosis of apoptotic cells is regulated by a UNC-73/TRIO-MIG-2/RhoG signaling module and armadillo repeats of CED-12/ELMO. Curr Biol. 2004;14:2208–16. doi: 10.1016/j.cub.2004.12.029. [DOI] [PubMed] [Google Scholar]
- 4.van Buul JD, Allingham MJ, Samson T, Meller J, Boulter E, García-Mata R, et al. RhoG regulates endothelial apical cup assembly downstream from ICAM1 engagement and is involved in leukocyte trans-endothelial migration. J Cell Biol. 2007;178:1279–93. doi: 10.1083/jcb.200612053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martínez-Martín N, Fernández-Arenas E, Cemerski S, Delgado P, Turner M, Heuser J, et al. T cell receptor internalization from the immunological synapse is mediated by TC21 and RhoG GTPase-dependent phagocytosis. Immunity. 2011;35:208–22. doi: 10.1016/j.immuni.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patel JC, Galán JE. Differential activation and function of Rho GTPases during Salmonella-host cell interactions. J Cell Biol. 2006;175:453–63. doi: 10.1083/jcb.200605144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roppenser B, Röder A, Hentschke M, Ruckdeschel K, Aepfelbacher M. Yersinia enterocolitica differentially modulates RhoG activity in host cells. J Cell Sci. 2009;122:696–705. doi: 10.1242/jcs.040345. [DOI] [PubMed] [Google Scholar]
- 8.Hiramoto-Yamaki N, Takeuchi S, Ueda S, Harada K, Fujimoto S, Negishi M, et al. Ephexin4 and EphA2 mediate cell migration through a RhoG-dependent mechanism. J Cell Biol. 2010;190:461–77. doi: 10.1083/jcb.201005141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Katoh H, Yasui H, Yamaguchi Y, Aoki J, Fujita H, Mori K, et al. Small GTPase RhoG is a key regulator for neurite outgrowth in PC12 cells. Mol Cell Biol. 2000;20:7378–87. doi: 10.1128/MCB.20.19.7378-7387.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Estrach S, Schmidt S, Diriong S, Penna A, Blangy A, Fort P, et al. The Human Rho-GEF trio and its target GTPase RhoG are involved in the NGF pathway, leading to neurite outgrowth. Curr Biol. 2002;12:307–12. doi: 10.1016/S0960-9822(02)00658-9. [DOI] [PubMed] [Google Scholar]
- 11.Katoh H, Negishi M. RhoG activates Rac1 by direct interaction with the Dock180-binding protein Elmo. Nature. 2003;424:461–4. doi: 10.1038/nature01817. [DOI] [PubMed] [Google Scholar]
- 12.Krek A, Grün D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 13.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 14.Franke K, Otto W, Johannes S, Baumgart J, Nitsch R, Schumacher S. miR-124-regulated RhoG reduces neuronal process complexity via ELMO/Dock180/Rac1 and Cdc42 signalling. EMBO J. 2012;31:2908–21. doi: 10.1038/emboj.2012.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brandt N, Franke K, Rasin MR, Baumgart J, Vogt J, Khrulev S, et al. The neural EGF family member CALEB/NGC mediates dendritic tree and spine complexity. EMBO J. 2007;26:2371–86. doi: 10.1038/sj.emboj.7601680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meller J, Vidali L, Schwartz MA. Endogenous RhoG is dispensable for integrin-mediated cell spreading but contributes to Rac-independent migration. J Cell Sci. 2008;121:1981–9. doi: 10.1242/jcs.025130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leemhuis J, Bouché E, Frotscher M, Henle F, Hein L, Herz J, et al. Reelin signals through apolipoprotein E receptor 2 and Cdc42 to increase growth cone motility and filopodia formation. J Neurosci. 2010;30:14759–72. doi: 10.1523/JNEUROSCI.4036-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim JY, Oh MH, Bernard LP, Macara IG, Zhang H. The RhoG/ELMO1/Dock180 signaling module is required for spine morphogenesis in hippocampal neurons. J Biol Chem. 2011;286:37615–24. doi: 10.1074/jbc.M111.268029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gauthier-Rouvière C, Vignal E, Mériane M, Roux P, Montcourier P, Fort P. RhoG GTPase controls a pathway that independently activates Rac1 and Cdc42Hs. Mol Biol Cell. 1998;9:1379–94. doi: 10.1091/mbc.9.6.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.May V, Schiller MR, Eipper BA, Mains RE. Kalirin Dbl-homology guanine nucleotide exchange factor 1 domain initiates new axon outgrowths via RhoG-mediated mechanisms. J Neurosci. 2002;22:6980–90. doi: 10.1523/JNEUROSCI.22-16-06980.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anderson DJ. Molecular control of cell fate in the neural crest: the sympathoadrenal lineage. Annu Rev Neurosci. 1993;16:129–58. doi: 10.1146/annurev.ne.16.030193.001021. [DOI] [PubMed] [Google Scholar]
- 22.Unsicker K. The chromaffin cell: paradigm in cell, developmental and growth factor biology. J Anat. 1993;183:207–21. [PMC free article] [PubMed] [Google Scholar]
- 23.Nobes CD, Hall A. Rho, rac and cdc42 GTPases: regulators of actin structures, cell adhesion and motility. Biochem Soc Trans. 1995;23:456–9. doi: 10.1042/bst0230456. [DOI] [PubMed] [Google Scholar]
- 24.Yokota Y, Eom TY, Stanco A, Kim WY, Rao S, Snider WD, et al. Cdc42 and Gsk3 modulate the dynamics of radial glial growth, inter-radial glial interactions and polarity in the developing cerebral cortex. Development. 2010;137:4101–10. doi: 10.1242/dev.048637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Machacek M, Hodgson L, Welch C, Elliott H, Pertz O, Nalbant P, et al. Coordination of Rho GTPase activities during cell protrusion. Nature. 2009;461:99–103. doi: 10.1038/nature08242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ng J, Nardine T, Harms M, Tzu J, Goldstein A, Sun Y, et al. Rac GTPases control axon growth, guidance and branching. Nature. 2002;416:442–7. doi: 10.1038/416442a. [DOI] [PubMed] [Google Scholar]
- 27.Lundquist EA, Reddien PW, Hartwieg E, Horvitz HR, Bargmann CI. Three C. elegans Rac proteins and several alternative Rac regulators control axon guidance, cell migration and apoptotic cell phagocytosis. Development. 2001;128:4475–88. doi: 10.1242/dev.128.22.4475. [DOI] [PubMed] [Google Scholar]
- 28.Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, et al. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–9. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- 29.Siegel G, Obernosterer G, Fiore R, Oehmen M, Bicker S, Christensen M, et al. A functional screen implicates microRNA-138-dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nat Cell Biol. 2009;11:705–16. doi: 10.1038/ncb1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27:435–48. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]