A novel pre-TCRα (pTα) reporter mouse reveals that expression of pTα is confined to the T lineage and does not occur on prethymic progenitors.
Abstract
Expression of the pre–T cell receptor α (pTα) gene has been exploited in previous studies as a molecular marker to identify tiny cell populations in bone marrow (BM) and blood that were suggested to contain physiologically relevant thymus settling progenitors (TSPs). But to what extent these cells genuinely contribute to thymopoiesis has remained obscure. We have generated a novel pTαiCre knockin mouse line and performed lineage-tracing experiments to precisely quantitate the contribution of pTα-expressing progenitors to distinct differentiation pathways and to the genealogy of mature hematopoietic cells under physiological in vivo conditions. Using these mice in combination with fluorescent reporter strains, we observe highly consistent labeling patterns that identify pTα expression as a faithful molecular marker of T lineage commitment. Specifically, the fate of pTα-expressing progenitors was found to include all αβ and most γδ T cells but, in contrast to previous assumptions, to exclude B, NK, and thymic dendritic cells. Although we could detect small numbers of T cell progenitors with a history of pTα expression in BM and blood, our data clearly exclude these populations as physiologically important precursors of thymopoiesis and indicate that they instead belong to a pathway of T cell maturation previously defined as extrathymic.
The pre-TCRα (pTα) chain is an essential and invariant subunit of the pre-TCR (von Boehmer, 2005). The only known physiological function of pTα protein is to associate with nascent TCRβ chains in committed T lineage progenitors to form a functional pre-TCR, which provides essential signals to promote development of αβ thymocytes and to attune αβ/γδ lineage choice. In line with this highly restricted function, pTα expression is largely confined to immature thymocytes. However, pTα message has also been detected in lineage-negative (Lin−) BM cells of wild-type and athymic nude mice (Bruno et al., 1995), which has given rise to the idea that pTα expression in BM may mark the enigmatic progenitors destined for settling the thymus. So far, neither identity nor full characteristics of thymus settling progenitors (TSPs) have been determined with certainty, leaving an embarrassing gap in our current schemes of T lymphopoiesis (Petrie and Kincade, 2005; Bhandoola and Sambandam, 2006; Bhandoola et al., 2007; Zlotoff and Bhandoola, 2011). Characterization of pTα-expressing BM cells, which seem to proffer tantalizing TSP candidates, thus appears imperative.
Cell surface expression of pTα depends on the presence of a functional TCRβ chain and members of the CD3 complex, which may not be available for complex formation at early developmental stages. Moreover, physiological pre-TCR surface expression is too weak to allow purification and further characterization of pre-TCR–positive cells. In an early attempt to overcome this obstacle, a transgenic mouse line was generated, which expressed a human CD25 surface marker (hCD25) under the control of a short regulatory element from the pTα-encoding Ptcra locus (Gounari et al., 2002). The analysis of such pTα/hCD25 reporter mice resulted in several high-profile publications (Gounari et al., 2002; Martin et al., 2003; Krueger and von Boehmer, 2007) reporting the identification and characterization of the common lymphoid progenitor 2 (CLP-2) and the circulating T cell progenitor (abbreviated CTP by Krueger and von Boehmer [2007]), which were commended to comprise physiologically relevant TSPs in BM and blood, respectively. However, these conclusions were based on experiments that did not provide information on to what extent pTα-expressing cells in BM and blood would genuinely contribute to thymopoiesis under in vivo steady-state conditions. Moreover, although a live marker like hCD25 can be useful to identify individual cells with active pTα expression, it does not allow the elucidation of in vivo differentiation pathways and precursor-product relationships. To directly quantify the contribution of pTα-expressing progenitor cells to thymopoiesis and to determine their in vivo commitment status, we have generated a novel knockin mouse line expressing an improved version of Cre recombinase (iCre) under the control of the endogenous Ptcra locus. In combination with fluorescent reporter mice, Ptcra-controlled iCre expression results in irreversible activation of fluorescent protein expression, providing a heritable lineage marker that indicates current and past Ptcra activity. Analysis of our pTαiCre reporter mice revealed highly consistent labeling patterns with recombination of floxed reporter alleles in αβ T lineage cells at near 100% efficiency. Using this in vivo fate mapping system, we reveal a previously unappreciated restriction in the developmental fate of pTα-expressing progenitor populations, arguing against a physiologically relevant CLP-2 stage in T lymphopoiesis. In fact, our data contest any appreciable contribution of cells with a history of pTα expression from BM or blood to canonical pathways of thymopoiesis and thus refute key conclusions from previous studies using conventional pTα/hCD25 reporter mice (Gounari et al., 2002; Martin et al., 2003; Krueger and von Boehmer, 2007).
RESULTS
Generation of pTαiCre knockin mice for lineage-tracing experiments
In vivo lineage tracing based on Cre/loxP technology provides a powerful genetic marking method, which can be used to permanently label specific cell subsets and to directly visualize cell fate decisions in a noninvasive manner within the intact organism (Hadjantonakis et al., 2003; Branda and Dymecki, 2004). To exploit this technique for fate mapping of pTα-expressing progenitor populations, we introduced a mini-gene carrying a codon-optimized version of Cre recombinase (iCre) into the first exon of the pTα-encoding Ptcra gene locus (Fig. 1 A). Particular attention was attached to the design of the construct to assure maximal Cre recombinase expression and activity, while strictly preserving pTα-specific regulation, as described in detail in Materials and methods.
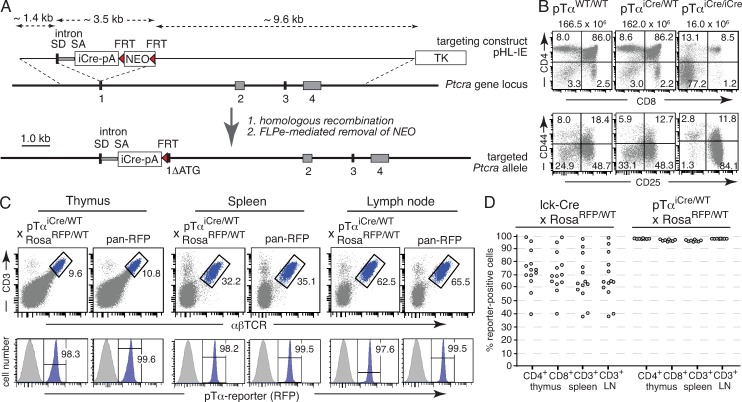
Figure 1.
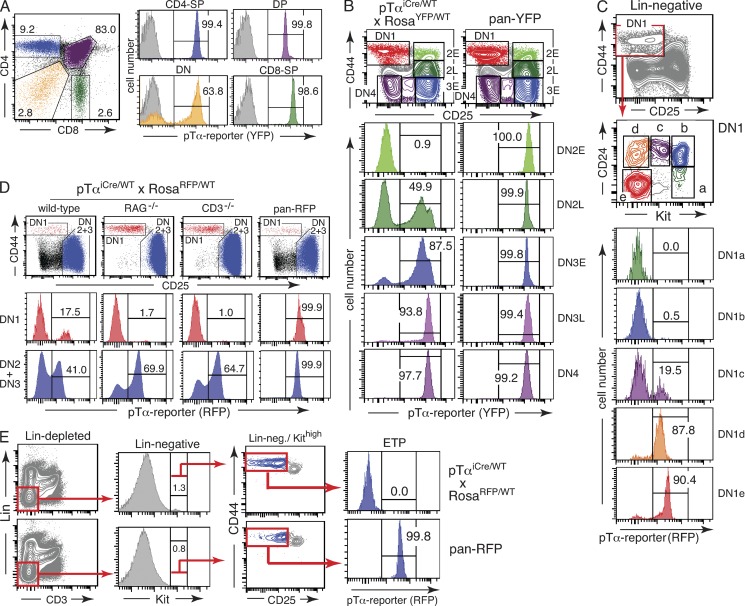
Generation of pTαiCre knockin mice. (A) Targeting strategy. Gray rectangles represent exons; red triangles represent FRT sites. TK indicates the thymidine kinase gene and NEO an expression cassette encoding neomycin resistance. SD and SA mark splice donor and acceptor sites flanking an intron, all derived from the rabbit β-globin locus; ΔATG indicates deletion of the Ptcra start codon upon targeted insertion of the iCre-pA cassette. (B) Thymocytes were isolated from mice of the indicated genotypes and analyzed by flow cytometry. Top panels show representative CD4/CD8 profiles of total thymocytes, and bottom panels show corresponding CD44/CD25 profiles of thymocytes pregated on Lin− cells. The total number of thymocytes is shown on top of each panel. Numbers in dot plots indicate percentages of cells in each quadrant. (C) Thymocytes, splenocytes, and lymph node cells were isolated from a pTαiCre/WT × RosaRFP/WT mouse and a pan-RFP control animal, and expression of the pTα reporter was determined by flow cytometry. Gates in the top panels identify αβ T cells, and blue histograms in the panels below depict pTα reporter expression in gated cells. The gray histograms correspond to αβ T cells from a RosaRFP/RFP mouse lacking iCre expression as negative control. Pan-RFP refers to a positive control mouse carrying one constitutively activated RosaRFP allele. Numbers indicate percentages of cells in each gate. (D) Comparison of labeling efficiencies in T cell compartments of conventional lck-Cre transgenic (n = 13) and newly generated pTαiCre knockin mice (n = 8) both intercrossed with RosatdRFP reporter mice. Circles correspond to data from individual animals.
Insertion of the iCre-encoding DNA sequences into the first exon of Ptcra was predicted to prevent expression of a functional pTα chain. In line with this prediction, homozygous pTαiCre mice exhibited the same characteristic defects in thymopoiesis (Fig. 1 B) as described previously for mice lacking functional pTα chains (Fehling et al., 1995). Importantly, heterozygous pTαiCre mice were phenotypically indistinguishable from wild-type littermates (Fig. 1 B and not depicted), confirming the well-established notion that loss of one Ptcra allele does not affect T cell development (Fehling et al., 1995).
To visualize iCre expression, pTαiCre knockin mice were intercrossed with RosaEYFP (Srinivas et al., 2001) or RosatdRFP (Luche et al., 2007) reporter lines, the choice dependent on compatibility with fluorescent antibodies in multicolor FACS experiments. Both reporter lines gave equivalent results. Cre-mediated excision of a transcriptional stopper resulted in irreversible activation of the respective reporter gene. Importantly, because of the irreversibility of the recombination event, Cre-expressing cells are heritably marked, i.e., all descending cells remain labeled, irrespective of their extant Cre expression status. The majority of αβ T cells depend on pre-TCR–mediated β-selection during their development in the thymus. However, some αβ T cells can mature in the absence of a functional pTα chain (Fehling et al., 1995). Thus, it remained to be determined how many αβ T progenitors normally pass through a pTα-expressing stage. Analysis of pTαiCre reporter mice revealed essentially complete labeling of CD3+TCRαβ+ cells in thymus, spleen, and lymph node (Fig. 1 C). Pan-RFP or pan-YFP mice carrying a constitutively active reporter allele (see Materials and methods) served as positive control, highlighting near saturation of labeling frequencies in αβ T cells from pTαiCre reporter mice. Importantly, there was no significant variation in labeling efficiencies between individual reporter mice. This is most obvious in comparison with conventional Lck-Cre transgenics (Hennet et al., 1995), which may exhibit strong interindividual variation in recombination efficiencies (Fig. 1 D) and even in lineage fidelity (not depicted), most likely as the result of variegated transgene expression, a phenomenon typical for multicopy transgenes inserted outside their natural genomic context (Martin and Whitelaw, 1996). Interestingly, much less variation has been observed in a recent systematic study using the same lck-Cre transgenic and Rosa-tdRFP reporter strains (Shi and Petrie, 2012), suggesting a particular vulnerability of the transgenic system to as yet undefined experimental settings. Our pTαiCre knockin mice proved devoid of such problems and thus well suited for lineage-tracing experiments.
History of pTαiCre expression in γδ T cells
Approximately 10–20% of γδ T cells express functional TCRβ chains in the cytoplasm (Dudley et al., 1995; Burtrum et al., 1996; Wilson and MacDonald, 1998), which have the capacity to associate with pTα chains to form a signaling-competent pre-TCR (Bosco et al., 2008). This observation is not easily compatible with models of αβ/γδ lineage determination, which postulate that successful formation of a pre-TCR in uncommitted precursor cells provides an important signal for adoption of an αβ lineage fate (Lee et al., 2010). One possibility to reconcile this inconsistency would be the ad hoc assumption that at least some γδ T cells develop from a pTα-negative precursor population, which cannot form a functional pre-TCR irrespective of TCRβ expression. In fact, there is no need for pTα chains in the γδ lineage, as γδ T cell development proceeds unperturbed in pTα knockout mice (Fehling et al., 1995). Although indirect evidence for pTα expression in the γδ lineage exists (Dudley et al., 1995; Aifantis et al., 1998; Wilson and MacDonald, 1998), the precise number of γδ T cells, which pass through a pTα-expressing developmental stage under physiological in vivo conditions, has not been determined yet.
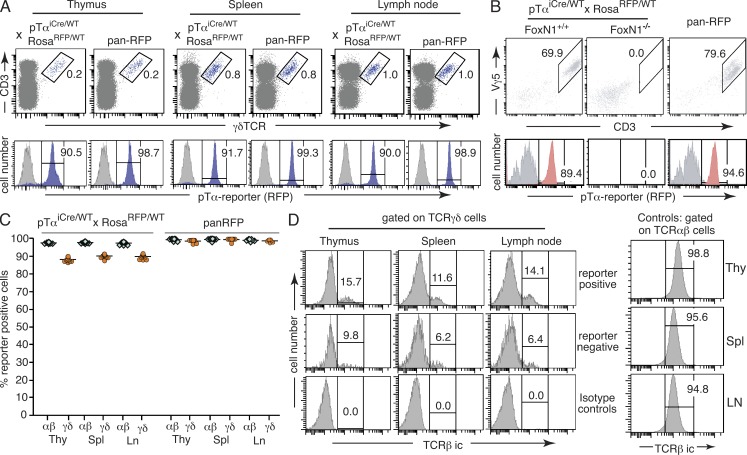
As shown in Fig. 2 A, ∼90% of CD3+TCRγδ+ cells in thymus, spleen, and lymph node are consistently labeled in pTαiCre reporter mice and thus have a history of pTα expression. Approximately the same percentage of reporter-positive cells is found among dendritic epidermal T cells (DETCs; Fig. 2 B), a specialized γδ T cell population in murine skin (Hayday, 2009; Havran and Jameson, 2010), which can be readily identified because of the expression of a largely invariant Vγ5Vδ1 TCR (nomenclature according to Hayday et al. [1985]). The fact that the percentage of DETCs with a history of pTαiCre expression is not significantly reduced in comparison with conventional γδ T cells is noteworthy, as DETCs develop in a single wave from a fetal precursor population beginning on day 14 of gestation (Havran and Allison, 1990), and thus well before pTα-controlled differentiation of αβ thymocytes.
Figure 2.
Quantification of past pTα expression in TCRγδ cells. (A) Dot plots in top row show the gating scheme to identify TCRγδ–expressing cells; the histograms below show the percentage of gated TCRγδ+ cells with a history of pTαiCre expression. The underlying gray histograms represent TCRγδ cells from a RosaRFP/RFP mouse lacking iCre expression as negative control. A pan-RFP mouse (carrying one constitutively active RosaRFP allele) was used as positive control. Numbers indicate percentages of cells in each gate. (B) DETCs were isolated from skin as CD3+TCRVγ5+ cells, and expression of the pTα reporter was assessed by flow cytometry. A nude mouse (FoxN1−/−) lacking DETCs was included as specificity control for CD3/Vγ5 stainings; other controls as in A. Populations were pregated on CD45+ cells. Red histograms refer to pTα reporter expression in gated CD45+CD3+TCRVγ5+ cells. Numbers indicate percentages of cells in each gate. Equivalent results were obtained in two independent experiments. (C) The frequency of reporter-positive cells was compared between TCRαβ+ and TCRγδ+ cells in thymus (Thy), spleen (Spl), and lymph nodes (Ln). Relevant cell populations were identified by flow cytometry as shown in Fig. 1 C and panel A. Each data point represents an individual mouse, with green diamonds referring to TCRαβ+ and orange circles to TCRγδ+ cells. Data were obtained from 12 pTαiCre/WT × RosaRFP/WT reporter and 8 pan-RFP control mice. (D) The frequency of TCRγδ+ cells with intracellular (ic) TCRβ expression in reporter-positive and -negative populations was determined by flow cytometry. Gating for TCRγδ+ cells as in A. Numbers indicate percentages of cells in each gate. Data are representative of three independent experiments.
Although the overwhelming majority of γδ T cells is derived from a pTα-expressing precursor population, we consistently observed reduced labeling frequencies in γδ compared with αβ T cells. In contrast to αβ T cells, which are in essence fully labeled, we found within γδ T cell populations from all organs analyzed a discrete reporter-negative subset of ∼10%. This difference in reporter expression was not caused by lack of Rosa26 transcription in a specific cell subset, as virtually all γδ T cells were labeled in pan-RFP control mice carrying a constitutively active RosatdRFP allele (Fig. 2, A and C). Intracellular staining for TCRβ chains revealed that TCRβ-positive γδ T cells were enriched in the reporter-positive rather than reporter-negative γδ T cell subset (Fig. 2 D). Lack of pTα expression in γδ precursor cells can thus be formally excluded as an explanation for the presence of functional TCRβ chains in a significant percentage of γδ T cells (Bosco et al., 2008).
pTαiCre expression is confined to T progenitors lacking B cell, NK cell, and myeloid lineage potential
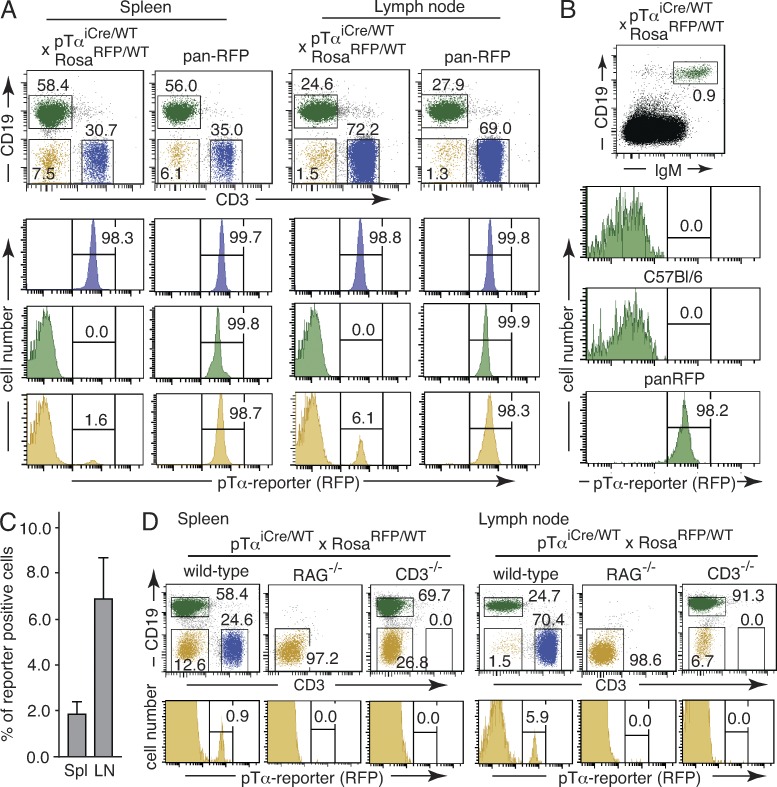
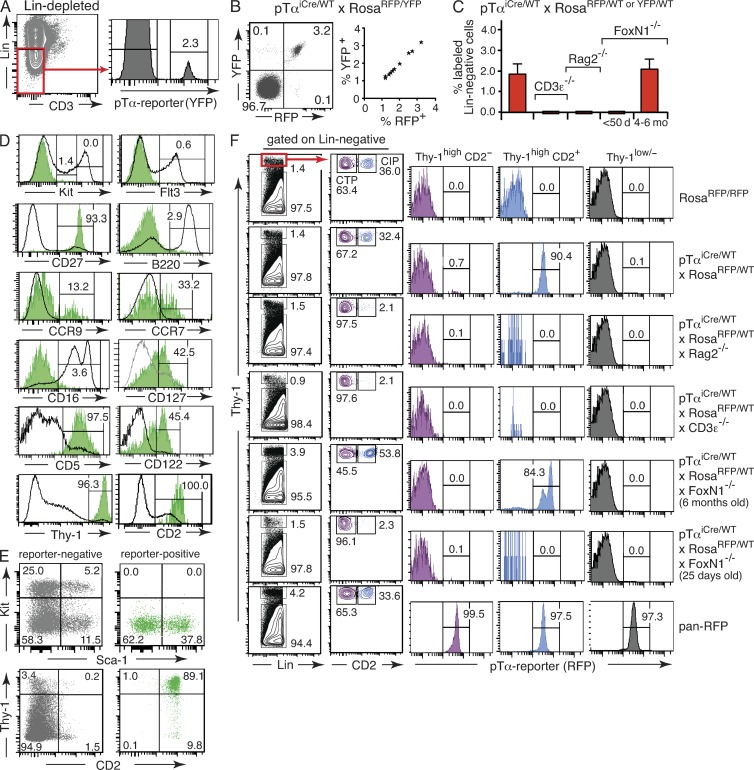
If pTα was expressed in a T lineage precursor population with physiologically relevant CLP activity, as suggested previously (Gounari et al., 2002; Martin et al., 2003), one should find a significant fraction of B cells, NK cells, and possibly DCs permanently labeled in pTαiCre reporter mice. Although mature CD3+ T cells in lymph node and spleen were reporter positive, no cells in the CD19+ B cell compartment were labeled (Fig. 3 A). Even in the thymus, where some B lymphopoiesis occurs in close proximity to T cell development (Akashi et al., 2000), the tiny population of mature B cells was consistently reporter negative (Fig. 3 B). These in vivo findings are incompatible with the view that B cells pass through a pTαiCre-expressing precursor stage during normal, steady-state hematopoiesis.
Figure 3.
pTαiCre expression is confined to the T lineage. (A) Splenocytes and lymph node cells were isolated from a pTαiCre/WT × RosaRFP/WT mouse and a pan-RFP control animal, and expression of the pTα reporter was assessed by flow cytometry. Dot plots on top provide the gating scheme for the histograms depicted below. CD3+ T cells are shown in blue, CD19+ B-cells in green, and cells lacking surface expression of both CD3 and CD19 in yellow. Numbers indicate percentages of cells in each gate. The figure is representative of 13 individual pTαiCre/WT × RosaRFP/WT and 5 pan-RFP mice. (B) Mature B lymphocytes in lineage-depleted thymic cell populations (see Materials and methods) were identified as CD19+IgM+ cells (dot plot on top). The histograms below show reporter expression in gated CD19+IgM+ cells from thymi of a pTαiCre/WT × RosaRFP/WT mouse (top), a C57BL/6 negative control (middle), and a pan-RFP positive control mouse (bottom). Numbers indicate percentages of cells in each gate. Data are representative of three separate experiments with five individual pTαiCre/WT × RosaRFP/WT reporter mice. (C) Bar graph indicates the mean percentage of reporter-positive splenocytes (Spl) and lymph node cells within the CD3−CD19− population of pTαiCre/WT × RosaRFP/WT reporter mice (n = 13). Error bars indicate SD. (D) Splenocytes and lymph node cells from pTαiCre/WT × RosaRFP/WT reporter mice on the indicated genetic backgrounds were analyzed for RFP expression. Dot plots on top show the gating scheme, and enlarged histograms below show reporter expression in the corresponding CD3−CD19− populations. Numbers indicate percentages of cells in each gate. Data are representative of five separate experiments with a total of five mice of each mutant genotype.
In the CD3−CD19− compartment of spleen and lymph node, which contains NK cells and cells of myeloid descent, we consistently found a small, but discrete population of reporter-positive cells (Fig. 3, A and C). Importantly, the labeled population completely disappeared both in spleen and lymph nodes when pTαiCre reporter mice were bred on RAG−/− or CD3−/− backgrounds (Fig. 3 D), formally demonstrating that the development of labeled CD3−CD19− cells is entirely dependent on V(D)J recombination and the presence of a functional CD3–TCR complex. A comprehensive phenotypic characterization of the labeled CD3−CD19− population for expression of informative cell surface markers and cytoplasmic TCR–CD3 complexes is shown in Fig. S1. Staining patterns overlap significantly with those on mature, peripheral αβ T cells, suggesting that reporter-positive CD3−CD19− cells in spleen and lymph node may contain a considerable fraction of bona fide T cells, which have internalized their TCR–CD3 complex. A similar explanation has been proposed in a previous study to account for the presence of a reporter-positive, but surface CD3–negative cell population in peripheral lymphoid organs of GFP/TCRδ knockin reporter mice (Prinz et al., 2006). However, the observed marker expression is also compatible with the view that a significant fraction of reporter-positive CD3−CD19− cells may comprise circulating counterparts of BM T lineage committed intermediate progenitors (CIPs) to be described below. Collectively, the absence of labeled cells in spleen and lymph nodes of RAG- and CD3-deficient pTαiCre reporter mice demonstrates that neither NK cells nor myeloid cells normally pass through a pTαiCre-expressing precursor stage.
No evidence for a “lymphoid past” of thymic DCs
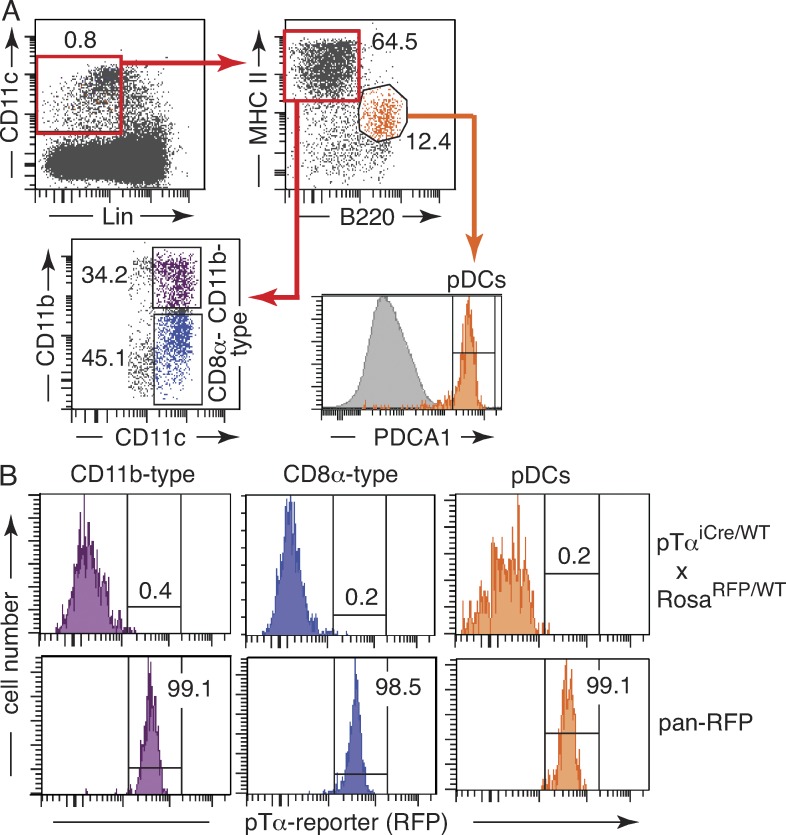
The developmental origin of DC subsets, and in particular their relationship to the lymphoid lineage, has remained a complex and controversial issue (Shortman and Naik, 2007; Liu and Nussenzweig, 2010). For instance, detection of pTα transcripts by RT-PCR in purified mouse plasmacytoid cells and in a subset of thymic DCs has been proposed to indicate a lymphoid past, i.e., a lymphoid developmental origin of these cells (Corcoran et al., 2003). The analysis of pTαiCre reporter mice does not support such speculations. In the thymus, neither CD11c+CD11b+ CD11b-type, nor CD11c+CD11b− CD8α-type, nor CD11c+B220+PDCA1+ plasmacytoid DC subpopulations contained a significant fraction of reporter-positive cells (Fig. 4, A and B). Equivalent results were obtained upon analysis of splenic DC subsets (not depicted). Our data demonstrate that pTα expression is no appropriate molecular marker to distinguish DC subpopulations and that DCs are not derived from a pTαiCre-expressing precursor population. These observations are fully congruent with recent fate mapping experiments using either LangerinDTR-EGFP or IL-7RiCre knockin mice, which both provided compelling in vivo evidence for a nonlymphoid origin of thymic DCs (Schlenner et al., 2010; Luche et al., 2011).
Figure 4.
No evidence for current or past pTαiCre expression in thymic DCs. (A) Gating scheme to distinguish thymic DC subsets. After digestion of thymi (see Materials and methods), conventional thymic DCs were identified in the resultant single cell suspensions as Lin−CD11c+MHC class IIhighB220− cells and further fractionated in CD11b-type DCs (purple) and CD8a-type DCs (blue) based on CD11b expression. Plasmacytoid DCs (pDCs; orange) were identified as Lin−CD11c+MHC class IIlowB220+ cells expressing plasmacytoid DC antigen-1 (PDCA1). The underlying gray histogram refers to PDCA1 staining of total thymocytes. Numbers indicate percentages of cells in each gate. (B) Reporter expression in CD11b-type, CD8a-type, and plasmacytoid DCs isolated from thymi of a pTαiCre/WT × RosaRFP/WT mouse and a pan-RFP control animal. Numbers indicate percentages of cells in each gate. Equivalent results were obtained in three independent experiments with a total of seven pTαiCre/WT × RosaRFP/WT and three pan-RFP mice.
Onset of pTαiCre expression in early thymopoiesis
The observed T lineage–restricted labeling pattern in pTαiCre reporter mice unveils a previously unappreciated link between pTα expression and T lineage commitment, i.e., the complete extinction of all alternative differentiation fates in pTα-expressing progenitor cells. By carefully monitoring reporter expression during early thymopoiesis, we can thus directly visualize the developmental stage at which individual cells have completed T lineage commitment. As shown in Fig. 5 A, essentially all immature CD4+CD8+ double-positive (DP) and more mature CD4+ or CD8+ single-positive thymocytes are reporter positive, demonstrating that all of these αβ T lineage committed cells have gone through a pre-TCR–dependent β-selection step (Hayday and Pennington, 2007). In contrast, the CD4−CD8− double-negative (DN) compartment contains a significant fraction of cells lacking signs of present or past pTα expression (Fig. 5 A). Based on CD25 and CD44 expression, the DN population can be subdivided into four developmentally successive stages (DN1–4), with DN2 and DN3 often being further subdivided into developmentally early (E) and late (L) fractions (Ceredig and Rolink, 2002; Rothenberg et al., 2008). Analysis of these DN subsets in pTαiCre reporter mice reveals a continuous increase in the percentage of labeled cells with progressing maturity (Fig. 5 B). Although few cells in the DN2E compartment exhibit the pTα expression mark, the number of labeled cells gradually increases to >97% in the post–β-selection DN4 subset. This labeling pattern demonstrates that T lineage commitment can be completed in some cells as early as DN2. In contrast, most cells in this compartment are reporter negative, consistent with experimental evidence that DN2 contains cells, which, when tested in vitro or in cell transfer assays, can still differentiate along the NK or DC lineages (Shen et al., 2003; Rothenberg et al., 2010; Yui et al., 2010). The observed labeling pattern in pTαiCre reporter mice is fully in line with microarray data provided by the Immunological Genome Project, which identify DN2L cells (Immgen nomenclature, preT.DN2-3) as the first DN subset with detectable pTα message and DN3E (Immgen nomenclature, preT.DN3A) as the subset with highest pTα expression levels.
Figure 5.
Onset of pTαiCre expression in thymopoiesis. (A) Total thymocytes of a pTαiCre/WT × RosaYFP/WT mouse were analyzed by flow cytometry for reporter expression in CD4/CD8 thymocyte subsets. Single-positive (SP), DP, and DN thymocytes were delineated as shown in the dot plot on the left. The underlying gray histograms correspond to thymocytes from a RosaYFP/YFP mouse lacking iCre expression analyzed in the same experiment with identical gates. (B) Lin− thymocytes (lineage depleted and electronically gated; see Materials and methods) were separated into developmentally successive subpopulations based on CD25 and CD44 expression (top). Color-coded histograms (bottom) show pTα reporter expression in the respective CD25/CD44 subsets. (C) DN1 thymocytes (Lin−CD44+CD25−) were separated into five distinct subsets based on CD24 and Kit expression. Color-coded histograms refer to the respective Kit/CD24 subset in the dot plot above. Combined DN1a + DN1b subsets correspond to ETPs. (D) DN1 and combined DN2 + DN3 thymocyte compartments (top) of pTαiCre/WT × RosaRFP/WT mice on indicated genetic backgrounds were analyzed by flow cytometry to determine the percentage of reporter-positive cells. Color-coded histograms (bottom) show pTα reporter expression in the respective subsets. Numbers indicate percentages of cells in each gate. (E) Flow cytometric analysis of ETPs for pTα reporter expression using an alternative gating scheme. Arrows indicate the gating hierarchy to identify ETPs, defined as Lin−KithighCD44highCD25−/low cells. Numbers indicate percentages of cells in each gate. All data are representative of at least three independent experiments.
The DN1 subpopulation constitutes a heterogeneous mixture of cells, which can be divided into five distinct subsets (DN1a–e) based on surface expression of CD117 (Kit) and CD24 (HSA; Porritt et al., 2004). Canonical T cell progenitors are confined to the CD117high subsets DN1a and DN1b (Porritt et al., 2004), which correspond to early T lineage progenitors (ETPs), generally considered the earliest intrathymic T lineage precursors (Allman et al., 2003; Benz et al., 2008; Luc et al., 2012). Consistent with lack of reporter expression in DN2E, the DN1a+b/ETP ancestors are reporter negative (Fig. 5, C and E). In contrast, the vast majority of more mature DN1d and DN1e cells are labeled, and DN1c cells contain reporter-positive and -negative subsets (Fig. 5 C), indicative of their heterogeneous composition (Porritt et al., 2004; Luche et al., 2011). Importantly, when pTαiCre reporter mice were bred on RAG−/− or CD3ε−/− backgrounds, the fraction of labeled cells in DN1 dropped dramatically, whereas the labeling index in combined stages DN2 + DN3 remained high (Fig. 5 D), indicating that the vast majority of labeled cells within the DN1 compartment are developmentally advanced populations, which are dependent on V(D)J recombination and a functional TCR–CD3 complex. The finding that DN1a+b/ETP populations are homogenously reporter negative (Fig. 5, C and E) is of key importance because this result effectively excludes all pTαiCre-expressing prethymic progenitors as physiologically relevant ETP precursors. The absence of labeling among ETPs is again supported by microarray data from the Immunological Genome Project, which mark the ETP population as negative for pTα expression.
In BM and blood, pTαiCre-labeled cells comprise T lineage precursors of an extrathymic maturation pathway
pTα-expressing cells in BM and blood have been promoted as physiologically relevant TSP candidates (Gounari et al., 2002; Martin et al., 2003; Krueger and von Boehmer, 2007). In accordance with published data on pTα mRNA expression (Bruno et al., 1995), we found a small population of reporter-positive cells (1.9 ± 0.5%) within the Lin− BM population of pTαiCre reporter mice (Fig. 6, A–C). For analysis of BM and blood, we frequently had to switch between RosaRFP and RosaYFP reporter mice to conform to available fluorescent antibodies. Equivalence of both reporter systems was demonstrated with pTαiCre mice carrying both a RosaRFP and a RosaYFP allele (Fig. 6 B and not depicted). As pointed out above, reporter-positive cells cannot be precursors of the thymic ETP population because the latter is homogeneously reporter negative (Fig. 5, C and E). Although most experimental data favor ETPs as the earliest canonical intrathymic T progenitor population (Allman et al., 2003; Porritt et al., 2004; Benz et al., 2008), the formal possibility remains that prethymic cells with a history of pTα expression contribute substantially to thymopoiesis by bypassing the ETP stage. Does the analysis of reporter-positive cells in BM and blood support such a view?
Figure 6.
Characterization of reporter-positive cells in BM. (A) Gating scheme to identify reporter-positive cells in Lin− BM. The number in the histogram refers to the percentage of reporter-positive cells. (B) Flow cytometric analysis of dual reporter expression in Lin− BM cells from pTαiCre/WT × RosaRFP/YFP mice, harboring both an RFP and an YFP reporter allele. Dot plot to the left shows labeling pattern in a representative animal, and the graph to the right summarizes data from 11 animals; each star corresponds to an individual mouse; pregating as in A. (C) Mean percentage of reporter-positive cells in Lin− BM of mice with different genetic backgrounds. Designations below graphs refer to the age of FoxN1−/− mice in the respective cohort. Error bars denote SD (pTαiCre/WTxRosaRFP/WT or YFP/WT on WT background, n = 15; on CD3ε−/− background, n = 16; on Rag2−/− background, n = 11; on FoxN1−/− background, younger than 50 d, n = 7; on FoxN1−/− background, 4–6 mo old, n = 3). (D) Cell surface phenotype of reporter-positive Lin− cells (green histograms). Black line histograms refer to the expression pattern of the respective surface marker on total Lin− cells (red gate in A). Numbers indicate percentages of cells in each gate. Each histogram is representative of three independent experiments. (E) Kit/Sca-1 and Thy-1/CD2 cell surface phenotype of reporter-positive and -negative Lin− cells. Numbers indicate percentages of cells in each quadrant. Dot plots are representative of two independent experiments. (F) Reporter expression in Lin−Thy-1low/− and previously described CTP and CIP populations (García-Ojeda et al., 2005). Color codes refer to gated Thy-1/CD2 subsets, as indicated on top of each histogram. Numbers indicate percentages of cells in each gate. Data are representative of at least five animals of each genotype.
Chemokine receptors CCR7 and CCR9 are critical for efficient homing of TSPs to the thymus (Schwarz et al., 2007; Krueger et al., 2010; Zlotoff et al., 2010). Moreover, all TSP activity in BM and blood has been shown to reside in a Lin− cell subset coexpressing CD27 and CD135 (Flk2/Flt3; Serwold et al., 2009; Saran et al., 2010) but lacking Thy-1high and B220+ cells (Serwold et al., 2009). A significant fraction of fluorescently labeled cells in pTαiCre reporter mice is positive for CCR7 and/or CCR9, and the majority of reporter-positive cells express CD27, as predicted for cells with presumed TSP activity (Fig. 6 D). However, >99% of reporter-positive cells lack Flt3 expression, leaving at best spurious numbers of reporter-positive cells as TSP candidates.
pTαiCre-labeled Lin− BM cells are ∼99% positive for CD2, and most reporter-positive cells express Thy-1 at very high levels (Fig. 6, D and E). Lin−, Thy-1high BM cells have been described extensively before (Dejbakhsh-Jones and Strober, 1999; Dejbakhsh-Jones et al., 2001; Chatterjea-Matthes et al., 2003; García-Ojeda et al., 2005). These cells contain firmly T lineage committed progenitors, which have been shown in adoptive transfer experiments to adhere to an extrathymic pathway of T cell maturation (Dejbakhsh-Jones et al., 2001; García-Ojeda et al., 2005). Based on CD2 surface expression, Thy-1high cells are divided into two developmentally successive subsets, a CD2−CD5−CD16+CD122− population, termed committed T cell progenitor (CTP), which gives rise in vivo and in vitro within hours to the CD2+CD5+CD16−CD122+ subset, termed CIP (García-Ojeda et al., 2005). The vast majority of pTαiCre-labeled cells reproduce the complete Thy-1highCD2+CD5+CD16−CD122+ phenotype of CIPs, indicating identity with CIPs.
In pTαiCre reporter mice, ∼0.5–1.0% of CTPs and ∼90% of CIPs are fluorescently labeled (Fig. 6 F, histograms in second row), a labeling pattern consistent with the reported precursor-progeny relationship between both cell subsets. Moreover, the number of CIPs has been reported to be dramatically reduced in Rag2−/− and young athymic nude (FoxN1−/−) mice (Chatterjea-Matthes et al., 2003). Remarkably alike, pTαiCre-labeled cells are virtually absent in Rag2−/−, CD3ε−/− and young athymic nude reporter mice (Fig. 6 F) but gradually reappear on nude background with increasing age, coincident with the reappearance of an increasingly prominent CIP population (Fig. 6 F, second row from bottom). The absence of labeled cells in pTαiCre reporter mice on CD3ε−/− and Rag2−/− backgrounds indicates that survival of pTα-expressing BM progenitors is contingent on prompt pre-TCR–mediated β-selection within the BM, consistent with their proposed extrathymic maturation program.
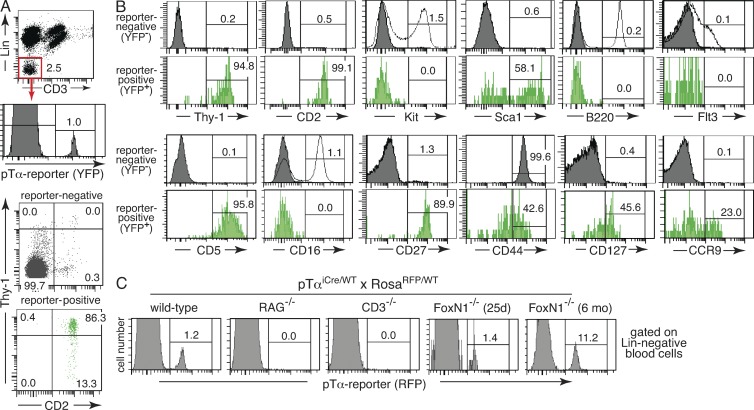
Analogous observations were made when analyzing pTα reporter expression in peripheral blood of pTαiCre mice. The Lin− blood fraction consistently contained ∼1.0% of reporter-positive cells, which were largely confined to a discrete Thy-1highCD2+ population with a cell surface phenotype closely resembling the CIP population in BM (Fig. 7, A and B). Of note, this reporter-positive population was absent in Rag2- and CD3ε-deficient pTαiCre mice (Fig. 7 C). In blood of athymic nude mice, the number of both Thy-1highCD2+ and pTαiCre-labeled cells was again, like in BM, age dependent (Fig. 7 C).
Figure 7.
Analysis of peripheral blood from adult pTα reporter mice. (A) Gating scheme to identify reporter-positive cells within the Lin− population (top two panels). Numbers refer to the percentages of cells in the respective gates. Dot plots (bottom) reveal the Thy-1/CD2 surface phenotype of reporter-positive and -negative Lin− blood cells. The numbers refer to the percentage of cells in each quadrant. Data were obtained with pooled blood from eight pTαiCre/WT × RosaYFP/WT reporter mice and are representative of two independent experiments. (B) Cell surface phenotype of reporter-positive and -negative cell subsets, pregated as shown in A. The open lines in some histograms refer to the expression pattern of the respective surface marker (Kit, B220, Flt3, CD16) on Lin− BM cells, which were stained in the same experiment to provide positive controls. Numbers refer to the percentage of blood cells positive for the respective surface marker. Each histogram is derived from pooled blood of two to three animals and representative of three independent staining experiments. (C) Flow cytometric analysis of pTα reporter expression in Lin− peripheral blood of pTαiCre/WT × RosaRFP/WT mice bred on the indicated genetic backgrounds. The information in brackets refers to the age of the particular mouse. All cells were pregated on the Lin− population as shown in A. Numbers indicate percentages of cells in each gate. Data are representative of three independent experiments, including all mouse genotypes shown.
BM progenitors with a history of pTαiCre expression are devoid of TSP activity
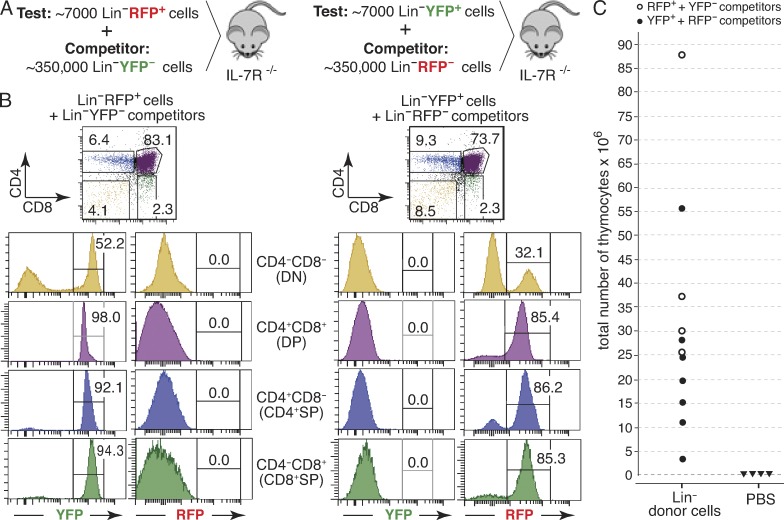
The CD135-negative phenotype of reporter-positive cells and their virtual absence in pTαiCre mice on RAG- and CD3ε-deficient backgrounds argue against cells with a history of pTα expression as physiologically important TSPs. To directly assay for TSP activity, we performed competitive complementation transfer assays. Our experimental approach, outlined in Fig. 8 A, took advantage of the availability of pTαiCre mice with distinct reporter alleles (RosaRFP and RosaYFP). In brief, Lin−RFP+ and Lin−YFP+ cells were sorted from BM of pTαiCre/WT × RosaRFP/WT and pTαiCre/WT × RosaYFP/WT reporter mice, respectively. For subsequent use as competitor population, we also sorted the complementary Lin− BM fractions completely devoid of RFP+ or YFP+ cells, respectively (for sorting gates, see Fig. S2). Approximately 7,000 sorted RFP+ cells were mixed with 350,000 Lin−YFP− competitor cells and injected i.v. into nonirradiated Il7ra-deficient mice, whose thymi have been shown to be highly receptive for T progenitor settling independent of irradiation (Fig. 8 A, left; Prockop and Petrie, 2004). In an analogous fashion, we tested sorted Lin−YFP+ BM cells in competition with Lin−RFP− BM cells (Fig. 8 A, right). Importantly, the injected ratio between test and competitor population (7,000:350,000 = 2%) was chosen to appropriately reflect the actual abundance of each subset in BM of pTαiCre reporter mice (Fig. 6, B and C). The relative contribution of test versus competitor population to thymopoiesis was evaluated by analyzing the origin of DP thymocytes 2 wk after transfer, a time point sufficiently early to allow direct capture of the most proximal progenitor activity, as established in previous studies with Il7ra-deficient recipients (Serwold et al., 2009; Saran et al., 2010). As DP thymocytes exhibit a history of pTα expression (Fig. 5 A), donor-derived DP thymocytes can be easily identified as fluorescent cells. Moreover, the observed ratio between RFP+ and YFP+ DP cells directly reveals the relative contribution of test versus competitor population. Two independent transfer experiments gave exactly the same results (Fig. 8, B and C). In all recipient mice successfully reconstituted with RFP+ test/YFP− competitor cells, 100% of donor-derived DP thymocytes were YFP+, whereas no RFP+ thymocytes were detectable, indicating exclusive contribution of reporter-negative BM cells to thymopoiesis. An analogous, complementary set of data were obtained in mice injected with a mixture of YFP+ test and RFP− competitor cells, again demonstrating exclusive contribution of reporter-negative BM cells to thymopoiesis (Fig. 8, B and C). These results effectively refute the possibility that cells with a history of pTαiCre expression contain physiologically relevant TSP activity.
Figure 8.
pTαiCre-labeled BM cells lack TSP activity. (A) Outline of the competitive complementation transfer experiment. Approximately 7,000 Lin−RFP+ cells sorted from pooled BM of female pTαiCre/WT × RosaRFP/WT mice were mixed with ∼350,000 Lin−YFP− cells sorted from pooled BM of female pTαiCre/WT × RosaYFP/WT mice and injected i.v. into an unconditioned IL-7Rα−/− female mouse (left). In the reciprocal experiment, ∼7,000 Lin−YFP+ cells sorted from pooled BM of the pTαiCre/WT × RosaYFP/WT mice were mixed with ∼350,000 Lin−RFP− cells sorted from pooled BM of the pTαiCre/WT × RosaYFP/WT mice and also injected i.v. into an unconditioned IL-7Rα−/− female mouse (right). On day 14 after injection, thymocytes of recipient mice were isolated and analyzed for the ratio of red versus yellow donor cells in CD4/CD8 thymic subsets. (B) Cytofluorometric analysis of recipient thymi from two representative IL-7R−/− mice, reconstituted i.v. with a mixture of RFP+/YFP− (left) or YFP+/RFP− (right) Lin− donor BM cells 14 d earlier. Total numbers of recovered thymocytes were 37 × 106 and 28.4 × 106, respectively. Two independent experiments gave identical results: fluorescently labeled thymocytes were derived exclusively (100%) from the reporter-negative donor cell fraction in 4/4 recipients of RFP+/YFP− and in 7/7 recipients of YFP+/RFP− Lin− cells. (C) Total number of thymocytes in IL-7R−/− recipients 14 d after receiving RFP+/YFP− (open circles) or YFP+/RFP− Lin− cells (closed circles). Control recipients were injected with PBS only (closed triangles).
DISCUSSION
Detection of pTα message in Lin− BM cells of wild-type and athymic nude mice has given rise to speculation that such cells may belong to the long-sought, but still elusive TSP population (Bruno et al., 1995). Several publications, all based on a single line of transgenic reporter mice, seemed to provide experimental support to this view (Gounari et al., 2002; Martin et al., 2003; Krueger and von Boehmer, 2007). The reporter, encoding an hCD25 surface marker under the control of a short (9 kb) pTα promoter fragment randomly inserted into the mouse genome as multicopy transgene, was found to mark small cell subsets in BM and blood, which were proposed to represent physiologically meaningful maturation stages between the CLP and early intrathymic precursor populations. However, the conclusions were based on cell transfer and in vitro differentiation assays and did not provide information to what extent pTα-expressing cells in BM and blood genuinely contribute to thymopoiesis under in vivo steady-state conditions. The present lineage-tracing study was designed to fill this knowledge gap. To our surprise, our data turned out to refute major findings and key conclusions of the aforementioned studies.
In mature cells, we found pTαiCre-mediated labeling strictly confined to the T lineage. No labeled cells were observed in lymph node, spleen, BM, and blood when reporter mice were bred on RAG- or CD3ε-deficient genetic backgrounds, demonstrating complete lack of pTαiCre expression in cells with in vivo NK or myeloid differentiation fates. Also plasmacytoid and thymic DCs were reporter negative, and no labeled B lymphocytes were observed. Our fate mapping data thus highlight pTα as an exquisitely T lineage–specific marker, whose expression indicates firm commitment, perfectly consistent with its restricted physiological function in T committed precursor cells.
The failure to detect labeled cells outside the T lineage necessarily implies the absence of pTαiCre expression in cells with physiologically relevant CLP activity. This result is in striking contrast to the concept of a pTα-expressing CLP-2 stage in early T lymphopoiesis. CLP-2 cells have been identified in BM of pTα/hCD25 transgenic mice as an hCD25+B220+Kit− population with efficient thymic immigration ability and robust CLP activity (Gounari et al., 2002; Martin et al., 2003). Based on these data, we expected to find at least some labeled B, NK, or DCs in pTαiCre reporter mice, which was clearly not the case. We conclude that either B220+Kit− CLP-2 cells do not exhibit CLP activity in vivo or cells with CLP activity within the CLP-2 population do not express pTα at significant levels, which would seem paradoxical because pTα/hCD25 reporter expression is the defining feature of these cells and led to their identification in the first place.
What could be the basis for this striking discrepancy between our results and data obtained with conventional pTα/hCD25 reporter mice? Evidence suggests that aberrant expression of the transgenic hCD25 reporter may account for much of the confusion. Although we have targeted our sensitive enzymatic reporter (iCre) into the endogenous Ptcra gene locus to preserve all known and unknown genetic elements possibly controlling physiological pTα expression, the hCD25 reporter was inserted randomly into the mouse genome as multicopy transgene. Moreover, reporter expression was controlled by just 9 kb of pTα 5′-flanking sequence, unlikely to contain all cis-regulatory elements required for faithful genetic and epigenetic regulation of pTα expression at all developmental stages from HSCs to mature T cells. Transgene copy number, genomic insertion site and a phenomenon termed transgene variegation are infamous for insidiously perturbing expression of conventional transgenes (Martin and Whitelaw, 1996; Montoliu et al., 2000). With regard to the CLP-2, one can thus imagine a scenario in which a few marker-positive cells, devoid of CLP activity, coexpress pTα, accounting for positive RT-PCR results at the population level, whereas the majority of hCD25+ CLP-2 cells are aberrantly labeled, lacking endogenous pTα expression, but exhibiting CLP activity. The reported aberrant expression of the huCD25 reporter in 30% of thymic TCRγδ cells and in B lineage committed CD19+ cells, clearly negative for endogenous pTα message (Gounari et al., 2002; Martin et al., 2003), supports this explanation.
Importantly, and in agreement with hCD25 reporter studies (Gounari et al., 2002; Martin et al., 2003), we do find Lin− cells with a history of pTα expression in BM of pTαiCre reporter mice, which are mostly Kit− and a few of which even express B220 (Fig. 6 D), reminiscent of the CLP-2 surface phenotype. However, labeled cells are absent in mice on Rag2−/− or CD3ε−/− genetic backgrounds, indicating that BM cells with a history of pTα expression pass through an obligatory extrathymic β-selection stage and thus cannot serve as precursors of early thymopoiesis. We further validated this conclusion in competitive complementation transfer assays, which demonstrated that pTαiCre-labeled BM cells do not contribute to thymopoiesis to any measurable extent (Fig. 8). In fact, our data strongly suggest that BM cells with a history of pTαiCre expression are largely identical with CIPs, which arise from CTPs and represent a T lineage committed population that belongs to an extrathymic pathway of maturation (García-Ojeda et al., 2005).
In agreement with data obtained from the analysis of huCD25 reporter mice (Krueger and von Boehmer, 2007), we do find Lin− cells with a history of pTα expression in peripheral blood of adult pTαiCre reporter mice on wild-type as well as nude background. These cells closely resemble in surface phenotype pTαiCre-labeled cells in BM, suggesting that they represent circulating counterparts of CIPs. The vast majority of labeled cells in adult blood belong to a discrete Thy-1highCD2+ population (Fig. 7 A). Although labeled cells completely lack B220 and Flt3 surface expression, a substantial fraction stains positive for IL-7Rα and Sca-1 (Fig. 7 B). A population of Thy-1highSca-1+IL-7Rα+B220−Flt3− cells has been identified in pTα/hCD25 transgenic mice based on hCD25 reporter expression. The cells, termed circulating T cell progenitors (also abbreviated CTPs) were shown to be T committed and claimed to represent “T cell precursors linking extrathymic with intrathymic lymphopoiesis in adult mice” (Krueger and von Boehmer, 2007). Our data do not support this conclusion because all cells with a history of pTαiCre expression disappear in knockin reporter mice when bred on Rag2−/− or CD3ε−/− backgrounds. Circulating T cell progenitors (also abbreviated CTPs) described by Krueger and von Boehmer (2007) and CTPs described by Dejbakhsh-Jones and Strober (1999), Dejbakhsh-Jones et al. (2001), and García-Ojeda et al. (2005) may thus not just share acronyms, but represent cells of the same extrathymic maturation pathway. Collectively, our data refute the concept of pTα expression as a marker for cells with physiologically relevant TSP activity tout ensemble. Instead, we propose that pTα-expressing cells in BM and adult blood belong to an extrathymic pathway of T cell maturation, similar to pTα-expressing cells in gut and liver (Bruno et al., 1995), which would also not be considered TSP candidates. To what extent T lineage committed progenitors in fetal blood (Rodewald et al., 1994), which contain pTα-expressing cells (Bruno et al., 1995), are en route to the thymus remains to be investigated.
Two more issues may merit brief discussion. First, our lineage-tracing study provides conclusive evidence that ∼90% of CD3+TCRγδ+ cells in thymus, spleen, and lymph nodes pass through a pTα-expressing developmental stage. The demonstration of pTα expression in γδ progenitors as such may not come as a surprise because earlier experiments have provided correspondent hints. For instance, an augmented frequency of in-frame TCRβ rearrangements in γδ T cells has been suggested to indicate pre-TCR–mediated β-selection in the γδ lineage (Dudley et al., 1995). In line, intracellular TCRβ expression in a fraction of TCRγδ+ thymocytes was reported to correlate with increased proliferation (Wilson and MacDonald, 1998). Although the latter finding has been disputed in another study using pTα−/− mice (Aifantis et al., 1998), pTα deficiency was observed to correlate with an increased percentage of intracellular TCRβ+ γδ cells, again suggesting pre-TCR–mediated effects on the γδ lineage. Although all these studies point to pTα expression in at least some γδ progenitors, our data provide not only direct proof in mice with unperturbed T lymphopoiesis, but also for the first time a precise in vivo quantification. Whether the consistently observed absence of pTαiCre labeling in ∼10% of TCRγδ cells delineates a functionally distinct γδ subset or simply indicates stochastic deviation of a fixed percentage of γδ progenitors from the common αβ/γδ developmental path before initiation of pTα expression is currently under investigation.
Finally, the strict T lineage specificity of pTαiCre expression commends our knockin mice as a novel tool for the conditional genetic modification of αβ T lineage cells. So far, T lineage–specific gene inactivation has been dependent on the availability of mice carrying randomly inserted Cre transgenes, like lck-Cre transgenics (Hennet et al., 1995), which suffer from problems inherent in classical transgenesis. In contrast, pTαiCre knockin mice afford consistent recombination efficiencies of essentially 100%, do not violate lineage fidelity, and do not exhibit significant interindividual variation neither in efficacy nor pattern of recombination. This advantageous combination of favorable features should make pTαiCre knockin mice a preferred tool for studies requiring faithful and proficient T lineage–specific gene modification.
MATERIALS AND METHODS
Generation of pTαiCre knockin mice.
To achieve optimal concordance between pTα expression and Cre activity, we opted for a knockin approach and constructed a targeting vector, which upon homologous recombination would insert a Cre recombinase–encoding expression cassette into the first exon of the pTα-encoding Ptcra locus (Fig. 1 A, top). To optimize pTα-controlled expression of Cre recombinase, the expression cassette was designed as a mini-gene, containing a splice donor site (SD), a short intron, a splice acceptor site (SA), a cDNA encoding Cre recombinase with its translational stop codon, and an SV40-derived polyadenylation signal (pA) for efficient termination of transcription. The short intronic sequence derived from the rabbit β-globin locus along with appendant splice sites (Kouskoff et al., 1993) was included to allow splicing of the primary transcript, which is known to boost transcription, translation (Moore and Proudfoot, 2009), and nuclear export (Luo and Reed, 1999) of eukaryotic mRNA. A codon-optimized, improved version of Cre recombinase, termed iCre, was chosen to further enhance pTα-controlled marker activity (Shimshek et al., 2002). The SV40-derived polyadenylation signal was inserted immediately downstream of the translational termination codon to impede transcription of subsequent pTα exons and formation of an pTαiCre chimeric message, prone to become susceptible to nonsense-mediated decay caused by the position of the iCre stop codon upstream of exon/exon junctions (Chang et al., 2007). A restriction map and the complete nucleotide sequence of the final targeting vector (pHL-IE), which was constructed using classical recombinant DNA technology, can be obtained from H.J. Fehling upon request. Gene targeting experiments were performed in embryonic day (E) 14.1 embryonic stem cells using G418/Ganciclovir double selection as described previously (Madan et al., 2009). Five independent embryonic stem cell clones with correctly targeted Ptcra alleles were identified after screening a total of 400 doubly resistant colonies by PCR. Correct homologous recombination was confirmed by Southern blotting with a probe located outside of the targeting construct. To exclude unpredictable effects of the Neo gene and its associated strong enhancer/promoter elements on the expression pattern of the targeted Ptcra locus, the FRT-flanked Neomycin selection cassette was excised in vitro by Flp-mediated recombination in one of the correctly targeted clones (E14-IE44). Embryonic stem cells of a Neo-deficient subclone (E14-IE44ΔNeo2) were used to generate pTαiCre knockin mice according to conventional methodology. Unless stated otherwise, all data shown are from animals 6–14 wk old and backcrossed for at least 10 generations onto C57BL/6 background.
Other mice.
Lck-Cre mice (B6.Cg-TgN(Lck-Cre)548Jxm (Hennet et al., 1995) were purchased from the Jackson Laboratory, IL-7Rα−/− mice (B6.129S7-Il7rtm1Imx/J) from Charles River, CD3ε−/− mice (C57BL/6-Cd3εtm1Mal/Orl; EMMA ID: EM:00047) from the European Mouse Mutant Archive, and Rag2−/− mice (B6.SJL(129S6)-PtprcaRag2tm1Fwa/Boy/Cr/Tac) from Taconic. RosatdRFP and RosaEYFP reporter mice have been described previously (Srinivas et al., 2001; Luche et al., 2007). Pan-RFP and pan-YFP mice carry constitutively active reporter alleles and were generated by intercrossing RosatdRFP or RosaEYFP reporter mice with animals from a germline Cre-deleter strain (Schwenk et al., 1995). Offspring in which the Rosa26-driven fluorescent reporter had been activated irreversibly as the result of Cre/loxP-mediated recombination in the germline were backcrossed for several generations onto C57BL/6, thereby eliminating the Cre recombinase transgene. Animals carrying one constitutively active reporter allele were used as controls in all experiments to indicate the maximal percentage of labeled cells detectable within the cell population of interest and to reveal any potential toxicity of the fluorescent markers for the particular cell subset of interest. All mice were bred and housed in individually ventilated cages in the mouse facility of the University of Ulm. All experiments involving the manipulation of mice were approved by the Regierungspräsidium Tübingen.
Flow cytometry.
Single cell suspensions from thymus, spleen, lymph nodes, and BM were prepared by mechanical dissociation of respective tissues and passage of cells through a 70-µm filter. For spleen and BM, red blood cells were lysed with ACK solution (150 mM ammonium chloride, 1 mM potassium bicarbonate, and 0.1 mM EDTA). Fc receptors were blocked by incubating cells in staining buffer (PBS supplemented with 5% heat-inactivated FCS) with anti-CD16/CD32 antibodies (Fc-Block; BD) and purified mouse IgG (Jackson ImmunoResearch Laboratories, Inc.). All stainings were performed in staining buffer on ice for 30 min with optimal dilutions of commercially prepared antibodies. Intracellular staining for TCRβ, TCRγδ, and CD3ε was performed using the Cytofix/Cytoperm kit (BD) according to the manufacturer’s instruction. A complete list of all antibodies used in the study is shown in Table S1. In all experiments, Sytox Blue (Invitrogen) was added before analysis to mark dead cells. Cells were acquired on a FACSCanto II (BD) or FACSAria II (BD) with electronic gates set on live cells by a combination of forward/side scatter and Sytox Blue exclusion. Doublets were systematically excluded based on forward and side scatter area versus width parameters. Data were analyzed using mostly Diva (BD) and sometimes (for overlays) FlowJo software (Tree Star). Specific procedures for the isolation and cytofluorometric analysis of particular cell subsets are given below.
Analysis of thymic DCs.
DCs were isolated as published (Feyerabend et al., 2009). In brief, thymi were minced with scissors into small pieces and digested in 1 ml PBS containing 0.2 mg/ml Collagenase D (Roche), 0.1 mg/ml Dispase I (Roche), and 25 mg/ml DNase I (Sigma-Aldrich) for 10 min at 37°C while shaking in a thermomixer at 800 rpm. The supernatant containing released cells was collected, and enzymatic activities were inhibited by adding an equal volume of PBS containing 5% FCS and 5 mM EDTA. Additional cells were collected by four to five rounds of repeated tissue digestion and harvest. Pooled cells were counted, and three million cells were stained with a mixture of biotinylated antibodies directed against CD3 (500A2), CD4 (GK1.5), CD19 (1D3), TER119, and CD49b (DX5) and the following DC-specific antibodies: anti–CD11c-APC-Cy7 (N418), anti–CD11b-PE-Cy7, anti–MHC class II–APC (M5/114.15.2), anti–B220-PE-Cy5.5 (RA3-6B2), and anti–PDCA1-FITC (eBio-927). Cells stained with biotinylated antibodies were revealed by secondary staining with Streptavidin–eFluor 450 and electronically excluded. DC subsets were identified within the eFluor 450–negative cell fraction as CD11c+MHC class IIhighB220−CD11b− lymphoid DCs (lyDCs), CD11c+MHC class IIhighB220−CD11b+ myeloid DCs (myDCs) and CD11c+MHC class IIlowB220+PDCA1+ plasmacytoid DCs according to reported surface phenotypes (Colonna et al., 2004; Wu and Shortman, 2005). The designation lyDC or myDC originally referred to the suggested origin of these DC populations from lymphoid or myeloid pathways, respectively. LyDCs are also named CD8+ and myDCs CD8− conventional DCs (Shortman and Heath, 2010).
Analysis of DETCs.
DETCs were isolated from mouse ears, which were minced with scissors into small pieces and digested in 1 ml protease solution (200 mg/ml Collagenase D1, 2 mg/ml Dispase I, and 5 mg/ml DNase I in PBS) for 15 min at 37°C while shaking in a thermomixer at 800 rpm. Supernatant containing released cells was harvested and kept on ice. Remaining tissue was subject to a second round of digestion with fresh protease solution, again for 15 min at 37°C while shaking at 800 rpm. Supernatants of successive digests were pooled and adjusted to 5 mM EDTA to antagonize cell aggregation. After a 5-min incubation period on ice, cells were passed through a 70-µm filter, washed once, and resuspended in staining buffer. Cells were stained with antibodies specific for CD45 (30-F11), CD3 (145-2C11), and Vγ5 (F536).
Analysis of thymic B cells.
To enrich for thymic B cells, total thymocytes were stained with biotinylated rat antibodies against CD4 (GK1.5) and CD8 (53-6.7) and depleted using anti–rat IgG-conjugated magnetic Dynabeads (Invitrogen) according to the manufacturer’s instruction. Remaining cells were stained with Streptavidin–APC-Cy7 (BD), anti–CD19-FITC (1D3), and anti-IgM (1B4B1) antibodies. Mature thymic B cells were identified in the pregated APC-Cy7–negative population as CD19+IgM+ cells.
Analysis of DN thymocytes.
To enrich for most immature thymocyte populations, total thymocytes were stained with a mixture of biotinylated antibodies directed against CD3ε (500A2), CD8α (53-6.7), CD11b (M1/70), CD19 (1D3), NK1.1 (PK136), Gr1 (RB6-8C5), TCRβ (H57-597), TCRγδ (GL3), TER119, CD11c (HL3), and B220 (RA-6B2), followed by depletion with Dynabeads according to the manufacturer’s instruction. Remaining cells were stained with directly conjugated antibodies against relevant surface molecules and always with a second anti-CD3ε–specific antibody (145-2C11) as well as with Streptavidin-QDot605 to facilitate gating on the Lin− population and to permit electronic exclusion of remaining Lin+ cells.
Analysis of Lin− BM cells.
To enrich for Lin− progenitor populations, BM cells were stained with a mixture of biotinylated antibodies directed against CD3ε (500A2), CD8α (53-6.7), CD11b (M1/70), CD19 (1D3), NK1.1 (PK136), GR1 (RB6-8C5), TCRβ (H57-597), TCRγδ (GL3), TER119, and CD11c (HL3), followed by depletion with Dynabeads according to the manufacturer’s instruction. Antibodies against CD4 and B220 were deliberately omitted from the lineage mix as relevant precursor populations, like the CLP-2, have been reported to express these surface markers (Martin et al., 2003). Remaining cells were stained with a mixture of directly labeled antibodies against relevant surface molecules, Strepavidin-QDot605, and a second anti-CD3ε antibody (145-2C11) conjugated to APC or APC-Cy7. Streptavidin-QDot605 and the additional anti-CD3ε antibody were used to rigorously exclude Lin+ cells, in particular mature T cells, and to facilitate electronic gating on the Lin− population.
Analysis of progenitors in peripheral blood.
Peripheral blood was drawn from tail veins into EDTA-containing microtubes (Sarstedt). Blood from several mice was pooled, diluted 1:1 with PBS, and carefully underlaid with Ficoll-Paque Plus (GE Healthcare) at a ratio of 3:1 (vol/vol). Samples were centrifuged in a swing-out bucket for 30 min at 20°C. The buffy coat layer was collected and transferred into 10 ml of staining buffer. Cells were pelleted by centrifugation, resuspended in staining buffer, and counted, excluding dead cells with Trypan Blue. Cells were stained with a mixture of biotinylated antibodies directed against the lineage markers CD3 (500A2), CD8α (53-6.7), CD11b (M1/70), CD19 (1D3), NK1.1 (PK136), GR1 (RB6-8C5), TCRβ (H57-597), TCRγδ (GL3), TER119, and CD11c (HL3) and directly labeled antibodies against specific cell surface molecules. Lin+ cells were revealed by secondary staining with Streptavidin-QDot605 and electronically excluded. To rigorously exclude reporter-positive T cells and to facilitate electronic gating on Lin− cells, a second anti-CD3 antibody (145-2C11) conjugated to APC was included in each staining. Antibodies against CD4 and B220 were deliberately omitted from the lineage mix.
Adoptive cell transfer into Il-7Rα−/− recipients (competitive complementation transfer assay).
BM cells from eight female pTαiCre/WT × RosaRFP/WT and eight female pTαiCre/WT × RosaYFP/WT mice were stained with the following mixture of biotinylated, lineage-specific antibodies: anti-CD3 (500A2), CD19 (1D3), CD11c (HL3), TCRγδ (GL3), NK1.1 (PK136), TER119, Gr1 (RB6-8C5), and CD11b (M1/70). Stained cells were depleted using anti–rat IgG-conjugated magnetic Dynabeads according to the manufacturer’s instruction. Antibodies against CD4 and B220 were deliberately omitted from the lineage mix. Cells from all pTαiCre/WT × RosaRFP/WT mice remaining after depletion were pooled, as were remaining cells from all pTαiCre/WT × RosaYFP/WT mice. Both samples were then stained with Streptavidin-QDot605 and CD3-PECy7 (145-2C11) to visualize remaining Lin+ cells and to facilitate gating on the Lin− population, respectively. Stained cells from both samples were sorted on a FACSAria IIu, the first sample for Lin−RFP+ and Lin−RFP− cells, the second sample for Lin−YFP+ and Lin−YFP− cells. A small fraction of sorted cells was reanalyzed to ascertain purity. Representative sorting gates and postsort data are shown in Fig. S2. The following number of cells was obtained in two independent experiments: first experiment, 18 × 103 Lin−RFP+ cells and 0.8 × 106 Lin−RFP− cells; 14 × 103 Lin−YFP+ cells and 1.2 × 106 Lin−YFP− cells; second experiment, 32 × 103 Lin−RFP+ cells and 1.2 × 106 Lin−RFP− cells; 25 × 103 Lin−YFP+ cells and 1.6 × 106 Lin−YFP− cells. Lin−RFP+ cells from pTαiCre/WT × RosaRFP/WT mice were mixed with Lin−YFP− cells from pTαiCre/WT × RosaYFP/WT mice, and Lin−YFP+ cells from pTαiCre/WT × RosaYFP/WT were mixed with Lin−RFP− cells from pTαiCre/WT × RosaRFP/WT mice, each time at a ratio of 1/50 (2%) to correctly reflect the actual abundance of each subset in BM of reporter mice. Mixed tester and competitor cells were injected i.v. into young (6–12 wk old), untreated IL-7Rα−/− recipient females. In the first experiment, we injected each of three recipients with 6 × 103 Lin−YFP+ + 300 × 103 Lin−RFP− cells, and each of two recipients with 7 × 103 Lin−RFP+ + 350 × 103 Lin−YFP− cells. In the second experiment, we injected each of four recipients with 8 × 103 Lin−YFP+ + 400 × 103 Lin−RFP− cells and each of three recipients with 8 × 103 Lin−RFP+ + 400 × 103 Lin−YFP− cells. In each experiment, two recipients were injected with PBS only as negative control. Reconstituted mice were analyzed on day 14 after injection, counting the day of injection as day 0.
Online supplemental material.
Fig. S1 shows a comprehensive cytofluorometric characterization of pTαiCre reporter–positive cells residing within the splenic and lymph node CD3−CD19− population. Fig. S2 depicts the sorting strategy and postsort data for tester and competitor cell subsets, which were used as donor populations for thymic reconstitution of IL-7Rα−/− recipients (competitive complementation assay). Table S1 describes all antibodies used for flow cytometry. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20122609/DC1.
Supplementary Material
Acknowledgments
We thank Ramona Syhachak and the Ulm Tierforschungszentrum for expert care of our mouse colony.
This work was supported by grants from the Interdisziplinären Zentrum für Klinische Forschung (Ulm), LFS “Defekte bei Aufbau und Erhalt von Immunfunktionen”, and Deutsche Forschungsgemeinschaft (FE 578/3-1).
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- CIP
- committed intermediate progenitor
- CLP
- common lymphoid progenitor
- CTP
- committed T cell progenitor
- DETC
- dendritic epidermal T cell
- DN
- double negative
- DP
- double positive
- ETP
- early T lineage progenitor
- lyDC
- lymphoid DC
- myDC
- myeloid DC
- TSP
- thymus settling progenitor
References
- Aifantis I., Azogui O., Feinberg J., Saint-Ruf C., Buer J., von Boehmer H. 1998. On the role of the pre-T cell receptor in alphabeta versus gammadelta T lineage commitment. Immunity. 9:649–655 10.1016/S1074-7613(00)80662-7 [DOI] [PubMed] [Google Scholar]
- Akashi K., Richie L.I., Miyamoto T., Carr W.H., Weissman I.L. 2000. B lymphopoiesis in the thymus. J. Immunol. 164:5221–5226 [DOI] [PubMed] [Google Scholar]
- Allman D., Sambandam A., Kim S., Miller J.P., Pagan A., Well D., Meraz A., Bhandoola A. 2003. Thymopoiesis independent of common lymphoid progenitors. Nat. Immunol. 4:168–174 10.1038/ni878 [DOI] [PubMed] [Google Scholar]
- Benz C., Martins V.C., Radtke F., Bleul C.C. 2008. The stream of precursors that colonizes the thymus proceeds selectively through the early T lineage precursor stage of T cell development. J. Exp. Med. 205:1187–1199 10.1084/jem.20072168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhandoola A., Sambandam A. 2006. From stem cell to T cell: one route or many? Nat. Rev. Immunol. 6:117–126 10.1038/nri1778 [DOI] [PubMed] [Google Scholar]
- Bhandoola A., von Boehmer H., Petrie H.T., Zúñiga-Pflücker J.C. 2007. Commitment and developmental potential of extrathymic and intrathymic T cell precursors: plenty to choose from. Immunity. 26:678–689 10.1016/j.immuni.2007.05.009 [DOI] [PubMed] [Google Scholar]
- Bosco N., Engdahl C., Bénard A., Rolink J., Ceredig R., Rolink A.G. 2008. TCR-beta chains derived from peripheral gammadelta T cells can take part in alphabeta T-cell development. Eur. J. Immunol. 38:3520–3529 10.1002/eji.200838668 [DOI] [PubMed] [Google Scholar]
- Branda C.S., Dymecki S.M. 2004. Talking about a revolution: The impact of site-specific recombinases on genetic analyses in mice. Dev. Cell. 6:7–28 10.1016/S1534-5807(03)00399-X [DOI] [PubMed] [Google Scholar]
- Bruno L., Rocha B., Rolink A., von Boehmer H., Rodewald H.R. 1995. Intra- and extra-thymic expression of the pre-T cell receptor alpha gene. Eur. J. Immunol. 25:1877–1882 10.1002/eji.1830250713 [DOI] [PubMed] [Google Scholar]
- Burtrum D.B., Kim S., Dudley E.C., Hayday A.C., Petrie H.T. 1996. TCR gene recombination and alpha beta-gamma delta lineage divergence: productive TCR-beta rearrangement is neither exclusive nor preclusive of gamma delta cell development. J. Immunol. 157:4293–4296 [PubMed] [Google Scholar]
- Ceredig R., Rolink T. 2002. A positive look at double-negative thymocytes. Nat. Rev. Immunol. 2:888–897 10.1038/nri937 [DOI] [PubMed] [Google Scholar]
- Chang Y.F., Imam J.S., Wilkinson M.F. 2007. The nonsense-mediated decay RNA surveillance pathway. Annu. Rev. Biochem. 76:51–74 10.1146/annurev.biochem.76.050106.093909 [DOI] [PubMed] [Google Scholar]
- Chatterjea-Matthes D., García-Ojeda M.E., Dejbakhsh-Jones S., Jerabek L., Manz M.G., Weissman I.L., Strober S. 2003. Early defect prethymic in bone marrow T cell progenitors in athymic nu/nu mice. J. Immunol. 171:1207–1215 [DOI] [PubMed] [Google Scholar]
- Colonna M., Trinchieri G., Liu Y.J. 2004. Plasmacytoid dendritic cells in immunity. Nat. Immunol. 5:1219–1226 10.1038/ni1141 [DOI] [PubMed] [Google Scholar]
- Corcoran L., Ferrero I., Vremec D., Lucas K., Waithman J., O’Keeffe M., Wu L., Wilson A., Shortman K. 2003. The lymphoid past of mouse plasmacytoid cells and thymic dendritic cells. J. Immunol. 170:4926–4932 [DOI] [PubMed] [Google Scholar]
- Dejbakhsh-Jones S., Strober S. 1999. Identification of an early T cell progenitor for a pathway of T cell maturation in the bone marrow. Proc. Natl. Acad. Sci. USA. 96:14493–14498 10.1073/pnas.96.25.14493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejbakhsh-Jones S., Garcia-Ojeda M.E., Chatterjea-Matthes D., Zeng D., Strober S. 2001. Clonable progenitors committed to the T lymphocyte lineage in the mouse bone marrow; use of an extrathymic pathway. Proc. Natl. Acad. Sci. USA. 98:7455–7460 10.1073/pnas.131559798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley E.C., Girardi M., Owen M.J., Hayday A.C. 1995. Alpha beta and gamma delta T cells can share a late common precursor. Curr. Biol. 5:659–669 10.1016/S0960-9822(95)00131-X [DOI] [PubMed] [Google Scholar]
- Fehling H.J., Krotkova A., Saint-Ruf C., von Boehmer H. 1995. Crucial role of the pre-T-cell receptor alpha gene in development of alpha beta but not gamma delta T cells. Nature. 375:795–798 10.1038/375795a0 [DOI] [PubMed] [Google Scholar]
- Feyerabend T.B., Terszowski G., Tietz A., Blum C., Luche H., Gossler A., Gale N.W., Radtke F., Fehling H.J., Rodewald H.R. 2009. Deletion of Notch1 converts pro-T cells to dendritic cells and promotes thymic B cells by cell-extrinsic and cell-intrinsic mechanisms. Immunity. 30:67–79 10.1016/j.immuni.2008.10.016 [DOI] [PubMed] [Google Scholar]
- García-Ojeda M.E., Dejbakhsh-Jones S., Chatterjea-Matthes D., Mukhopadhyay A., BitMansour A., Weissman I.L., Brown J.M., Strober S. 2005. Stepwise development of committed progenitors in the bone marrow that generate functional T cells in the absence of the thymus. J. Immunol. 175:4363–4373 [DOI] [PubMed] [Google Scholar]
- Gounari F., Aifantis I., Martin C., Fehling H.J., Hoeflinger S., Leder P., von Boehmer H., Reizis B. 2002. Tracing lymphopoiesis with the aid of a pTalpha-controlled reporter gene. Nat. Immunol. 3:489–496 10.1038/ni778 [DOI] [PubMed] [Google Scholar]
- Hadjantonakis A.K., Dickinson M.E., Fraser S.E., Papaioannou V.E. 2003. Technicolour transgenics: imaging tools for functional genomics in the mouse. Nat. Rev. Genet. 4:613–625 10.1038/nrg1126 [DOI] [PubMed] [Google Scholar]
- Havran W.L., Allison J.P. 1990. Origin of Thy-1+ dendritic epidermal cells of adult mice from fetal thymic precursors. Nature. 344:68–70 10.1038/344068a0 [DOI] [PubMed] [Google Scholar]
- Havran W.L., Jameson J.M. 2010. Epidermal T cells and wound healing. J. Immunol. 184:5423–5428 10.4049/jimmunol.0902733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayday A.C. 2009. Gammadelta T cells and the lymphoid stress-surveillance response. Immunity. 31:184–196 10.1016/j.immuni.2009.08.006 [DOI] [PubMed] [Google Scholar]
- Hayday A.C., Pennington D.J. 2007. Key factors in the organized chaos of early T cell development. Nat. Immunol. 8:137–144 10.1038/ni1436 [DOI] [PubMed] [Google Scholar]
- Hayday A.C., Saito H., Gillies S.D., Kranz D.M., Tanigawa G., Eisen H.N., Tonegawa S. 1985. Structure, organization, and somatic rearrangement of T cell gamma genes. Cell. 40:259–269 10.1016/0092-8674(85)90140-0 [DOI] [PubMed] [Google Scholar]
- Hennet T., Hagen F.K., Tabak L.A., Marth J.D. 1995. T-cell-specific deletion of a polypeptide N-acetylgalactosaminyl-transferase gene by site-directed recombination. Proc. Natl. Acad. Sci. USA. 92:12070–12074 10.1073/pnas.92.26.12070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouskoff V., Fehling H.J., Lemeur M., Benoist C., Mathis D. 1993. A vector driving the expression of foreign cDNAs in the MHC class II-positive cells of transgenic mice. J. Immunol. Methods. 166:287–291 10.1016/0022-1759(93)90370-M [DOI] [PubMed] [Google Scholar]
- Krueger A., von Boehmer H. 2007. Identification of a T lineage-committed progenitor in adult blood. Immunity. 26:105–116 10.1016/j.immuni.2006.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger A., Willenzon S., Lyszkiewicz M., Kremmer E., Förster R. 2010. CC chemokine receptor 7 and 9 double-deficient hematopoietic progenitors are severely impaired in seeding the adult thymus. Blood. 115:1906–1912 10.1182/blood-2009-07-235721 [DOI] [PubMed] [Google Scholar]
- Lee S.Y., Stadanlick J., Kappes D.J., Wiest D.L. 2010. Towards a molecular understanding of the differential signals regulating alphabeta/gammadelta T lineage choice. Semin. Immunol. 22:237–246 10.1016/j.smim.2010.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K., Nussenzweig M.C. 2010. Origin and development of dendritic cells. Immunol. Rev. 234:45–54 10.1111/j.0105-2896.2009.00879.x [DOI] [PubMed] [Google Scholar]
- Luc S., Luis T.C., Boukarabila H., Macaulay I.C., Buza-Vidas N., Bouriez-Jones T., Lutteropp M., Woll P.S., Loughran S.J., Mead A.J., et al. 2012. The earliest thymic T cell progenitors sustain B cell and myeloid lineage potential. Nat. Immunol. 13:412–419 10.1038/ni.2255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luche H., Weber O., Nageswara Rao T., Blum C., Fehling H.J. 2007. Faithful activation of an extra-bright red fluorescent protein in “knock-in” Cre-reporter mice ideally suited for lineage tracing studies. Eur. J. Immunol. 37:43–53 10.1002/eji.200636745 [DOI] [PubMed] [Google Scholar]
- Luche H., Ardouin L., Teo P., See P., Henri S., Merad M., Ginhoux F., Malissen B. 2011. The earliest intrathymic precursors of CD8α(+) thymic dendritic cells correspond to myeloid-type double-negative 1c cells. Eur. J. Immunol. 41:2165–2175 10.1002/eji.201141728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M.J., Reed R. 1999. Splicing is required for rapid and efficient mRNA export in metazoans. Proc. Natl. Acad. Sci. USA. 96:14937–14942 10.1073/pnas.96.26.14937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madan V., Madan B., Brykczynska U., Zilbermann F., Hogeveen K., Döhner K., Döhner H., Weber O., Blum C., Rodewald H.R., et al. 2009. Impaired function of primitive hematopoietic cells in mice lacking the Mixed-Lineage-Leukemia homolog MLL5. Blood. 113:1444–1454 10.1182/blood-2008-02-142638 [DOI] [PubMed] [Google Scholar]
- Martin C.H., Aifantis I., Scimone M.L., von Andrian U.H., Reizis B., von Boehmer H., Gounari F. 2003. Efficient thymic immigration of B220+ lymphoid-restricted bone marrow cells with T precursor potential. Nat. Immunol. 4:866–873 10.1038/ni965 [DOI] [PubMed] [Google Scholar]
- Martin D.I., Whitelaw E. 1996. The vagaries of variegating transgenes. Bioessays. 18:919–923 10.1002/bies.950181111 [DOI] [PubMed] [Google Scholar]
- Montoliu L., Chávez S., Vidal M. 2000. Variegation associated with lacZ in transgenic animals: a warning note. Transgenic Res. 9:237–239 10.1023/A:1008995730285 [DOI] [PubMed] [Google Scholar]
- Moore M.J., Proudfoot N.J. 2009. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 136:688–700 10.1016/j.cell.2009.02.001 [DOI] [PubMed] [Google Scholar]
- Petrie H.T., Kincade P.W. 2005. Many roads, one destination for T cell progenitors. J. Exp. Med. 202:11–13 10.1084/jem.20050990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porritt H.E., Rumfelt L.L., Tabrizifard S., Schmitt T.M., Zúñiga-Pflücker J.C., Petrie H.T. 2004. Heterogeneity among DN1 prothymocytes reveals multiple progenitors with different capacities to generate T cell and non-T cell lineages. Immunity. 20:735–745 10.1016/j.immuni.2004.05.004 [DOI] [PubMed] [Google Scholar]
- Prinz I., Sansoni A., Kissenpfennig A., Ardouin L., Malissen M., Malissen B. 2006. Visualization of the earliest steps of gammadelta T cell development in the adult thymus. Nat. Immunol. 7:995–1003 10.1038/ni1371 [DOI] [PubMed] [Google Scholar]
- Prockop S.E., Petrie H.T. 2004. Regulation of thymus size by competition for stromal niches among early T cell progenitors. J. Immunol. 173:1604–1611 [DOI] [PubMed] [Google Scholar]
- Rodewald H.R., Kretzschmar K., Takeda S., Hohl C., Dessing M. 1994. Identification of pro-thymocytes in murine fetal blood: T lineage commitment can precede thymus colonization. EMBO J. 13:4229–4240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg E.V., Moore J.E., Yui M.A. 2008. Launching the T-cell-lineage developmental programme. Nat. Rev. Immunol. 8:9–21 10.1038/nri2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg E.V., Zhang J., Li L. 2010. Multilayered specification of the T-cell lineage fate. Immunol. Rev. 238:150–168 10.1111/j.1600-065X.2010.00964.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saran N., Łyszkiewicz M., Pommerencke J., Witzlau K., Vakilzadeh R., Ballmaier M., von Boehmer H., Krueger A. 2010. Multiple extrathymic precursors contribute to T-cell development with different kinetics. Blood. 115:1137–1144 10.1182/blood-2009-07-230821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlenner S.M., Madan V., Busch K., Tietz A., Läufle C., Costa C., Blum C., Fehling H.J., Rodewald H.R. 2010. Fate mapping reveals separate origins of T cells and myeloid lineages in the thymus. Immunity. 32:426–436 10.1016/j.immuni.2010.03.005 [DOI] [PubMed] [Google Scholar]
- Schwarz B.A., Sambandam A., Maillard I., Harman B.C., Love P.E., Bhandoola A. 2007. Selective thymus settling regulated by cytokine and chemokine receptors. J. Immunol. 178:2008–2017 [DOI] [PubMed] [Google Scholar]
- Schwenk F., Baron U., Rajewsky K. 1995. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res. 23:5080–5081 10.1093/nar/23.24.5080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serwold T., Ehrlich L.I., Weissman I.L. 2009. Reductive isolation from bone marrow and blood implicates common lymphoid progenitors as the major source of thymopoiesis. Blood. 113:807–815 10.1182/blood-2008-08-173682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H.Q., Lu M., Ikawa T., Masuda K., Ohmura K., Minato N., Katsura Y., Kawamoto H. 2003. T/NK bipotent progenitors in the thymus retain the potential to generate dendritic cells. J. Immunol. 171:3401–3406 [DOI] [PubMed] [Google Scholar]
- Shi J., Petrie H.T. 2012. Activation kinetics and off-target effects of thymus-initiated cre transgenes. PLoS ONE. 7:e46590 10.1371/journal.pone.0046590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimshek D.R., Kim J., Hübner M.R., Spergel D.J., Buchholz F., Casanova E., Stewart A.F., Seeburg P.H., Sprengel R. 2002. Codon-improved Cre recombinase (iCre) expression in the mouse. Genesis. 32:19–26 10.1002/gene.10023 [DOI] [PubMed] [Google Scholar]
- Shortman K., Heath W.R. 2010. The CD8+ dendritic cell subset. Immunol. Rev. 234:18–31 10.1111/j.0105-2896.2009.00870.x [DOI] [PubMed] [Google Scholar]
- Shortman K., Naik S.H. 2007. Steady-state and inflammatory dendritic-cell development. Nat. Rev. Immunol. 7:19–30 10.1038/nri1996 [DOI] [PubMed] [Google Scholar]
- Srinivas S., Watanabe T., Lin C.S., William C.M., Tanabe Y., Jessell T.M., Costantini F. 2001. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 1:4 10.1186/1471-213X-1-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Boehmer H. 2005. Unique features of the pre-T-cell receptor alpha-chain: not just a surrogate. Nat. Rev. Immunol. 5:571–577 10.1038/nri1636 [DOI] [PubMed] [Google Scholar]
- Wilson A., MacDonald H.R. 1998. A limited role for beta-selection during gamma delta T cell development. J. Immunol. 161:5851–5854 [PubMed] [Google Scholar]
- Wu L., Shortman K. 2005. Heterogeneity of thymic dendritic cells. Semin. Immunol. 17:304–312 10.1016/j.smim.2005.05.001 [DOI] [PubMed] [Google Scholar]
- Yui M.A., Feng N., Rothenberg E.V. 2010. Fine-scale staging of T cell lineage commitment in adult mouse thymus. J. Immunol. 185:284–293 10.4049/jimmunol.1000679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlotoff D.A., Bhandoola A. 2011. Hematopoietic progenitor migration to the adult thymus. Ann. N. Y. Acad. Sci. 1217:122–138 10.1111/j.1749-6632.2010.05881.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlotoff D.A., Sambandam A., Logan T.D., Bell J.J., Schwarz B.A., Bhandoola A. 2010. CCR7 and CCR9 together recruit hematopoietic progenitors to the adult thymus. Blood. 115:1897–1905 10.1182/blood-2009-08-237784 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.