Reduction of the CHOP target Gadd34 restores motor function in P0S63del mice with demyelinating neuropathy.
Abstract
P0 glycoprotein is an abundant product of terminal differentiation in myelinating Schwann cells. The mutant P0S63del causes Charcot-Marie-Tooth 1B neuropathy in humans, and a very similar demyelinating neuropathy in transgenic mice. P0S63del is retained in the endoplasmic reticulum of Schwann cells, where it promotes unfolded protein stress and elicits an unfolded protein response (UPR) associated with translational attenuation. Ablation of Chop, a UPR mediator, from S63del mice completely rescues their motor deficit and reduces active demyelination by half. Here, we show that Gadd34 is a detrimental effector of CHOP that reactivates translation too aggressively in myelinating Schwann cells. Genetic or pharmacological limitation of Gadd34 function moderates translational reactivation, improves myelination in S63del nerves, and reduces accumulation of P0S63del in the ER. Resetting translational homeostasis may provide a therapeutic strategy in tissues impaired by misfolded proteins that are synthesized during terminal differentiation.
ER homeostasis is vital for cellular function and survival. Alterations in the folding capacity of the ER due, for example, to changes in calcium storage, may cause accumulation of unfolded or misfolded proteins in the lumen and ER stress. To safeguard homeostasis, the mammalian ER contains transmembrane proteins that sense stress in the lumen and trigger a complex series of signals collectively termed unfolded protein response (UPR; Harding et al., 2002; Rutkowski et al., 2006; Ron and Walter, 2007). The UPR reduces the load of abnormal protein in the ER and improves the capacity of the ER to fold proteins and degrade terminally misfolded proteins. An adaptive response is achieved through two main mechanisms: translational attenuation, via phosphorylation of the translation initiation factor eIF2α, and activation of transcription factors like ATF6 and XBP-1, which increase the expression of chaperones, such as BiP, and other proteins, such as Derlins, that facilitate ER-associated degradation (ERAD). However, if the accumulation of misfolded proteins persists, the UPR may become maladaptive and activate apoptosis through transcription factors such as CHOP (Zinszner et al., 1998; Tabas and Ron, 2011). What determines the switch from adaptive to maladaptive response is a fundamental unanswered question (Rutkowski et al., 2006); Lin et al., 2007.
Myelin synthesis by oligodendrocytes and Schwann cells requires massive production of lipids and proteins. Not surprisingly, ER stress has been implicated in myelin disorders in both the central nervous system (CNS) and the peripheral nervous system (PNS; D’Antonio et al., 2009; Lin and Popko, 2009; Monk et al., 2013). Myelin protein zero (Mpz; P0) is the major protein synthesized during terminal differentiation of myelinating Schwann cells. It accounts for 20–50% of the total protein in myelin (Kirschner et al., 2004; Patzig et al., 2011) and is essential for its compaction and maintenance (Giese et al., 1992). In humans, diverse mutations in P0, most inherited as dominant traits, cause a range of hereditary neuropathies, suggesting different types of toxic gain of function (Wrabetz et al., 2004). However, the pathogenetic mechanisms remain poorly understood (Wrabetz et al., 2004; Berger et al., 2006; Scherer and Wrabetz, 2008).
Deletion of serine 63 (P0S63del) causes a Charcot-Marie-Tooth type 1B (CMT1B) disease (Kulkens et al., 1993), a demyelinating neuropathy characterized by myelin destruction and failed attempts to remyelinate (onion bulbs; Miller et al., 2012).We have shown in transgenic mice that P0S63del is not detected in the myelin sheath, but is retained in the ER, where its accumulation triggers a canonical UPR, indicating a toxic gain of function (Wrabetz et al., 2006; Pennuto et al., 2008). Notably, in S63del mice the genetic ablation of Chop completely rescues the motor deficit and ameliorates the demyelinating phenotype indicating that the UPR is pathogenetic (Pennuto et al., 2008).
To understand the contribution of the UPR and CHOP to demyelination in S63del nerves, we compared the pool of mRNAs in nerves of WT, S63del, and Chop-null mice at various points in development and found that the UPR is a specific feature of S63del nerves, detectable soon after birth and sustained throughout life. We identify the phosphatase Gadd34, a reactivator of protein translation in the context of the UPR (Novoa et al., 2001), as a target gene of CHOP also in S63del nerve. Genetic and pharmacological assessment of Gadd34 in S63del peripheral neuropathy suggest that regulation of eIF2α and protein translation are candidate therapeutic targets for peripheral neuropathy associated with altered proteostasis and possibly for other misfolded protein diseases.
RESULTS
Although UPR and Chop are pathogenetic in S63del neuropathy, the timing and amount of apoptosis suggest that the death of Schwann cells is a consequence, not cause, of demyelinating neuropathy. Therefore, we speculated that the UPR contributes to demyelination by other maladaptive effects (Pennuto et al., 2008). To further elucidate the molecular mechanisms involved in the neuropathy and in the rescue after the ablation of Chop, we characterized the transcriptome of sciatic nerves from WT, Chop-null, S63del, and S63del/Chop-null mice at three different time-points: (1) postnatal day 5 (P5), when myelination has begun and only the primary effects of the presence of P0S63del should be detected; (2) P28, around the peak of myelination and P0S63del expression, when all targets of CHOP should be activated; and (3) 4 mo, when the motor and morphological rescue caused by the ablation of Chop were clearly detectable (Pennuto et al., 2008).
Expression of P0S63del causes broad transcriptional alterations in nerve
We first characterized the transcriptome of S63del nerves, to understand the pathological events occurring in CMT1B nerves (Fig. 1 A). We observed that P0S63del extensively perturbs gene expression following on the expression of the Mpz-based S63del transgene (Fig. 1 B); in fact, as early as P5, when the transgene is already robustly expressed (Feltri et al., 1999; Wrabetz et al., 2006), 771 genes were found to vary (increased or decreased) by >1.5-fold compared with WT; this number rose to 1,205 genes at P28, when myelination peaks, and decreased to 324 genes at 4 mo, when myelin genes, and thus the MpzS63del transgene, are expressed at lower levels (Wrabetz et al., 2006).
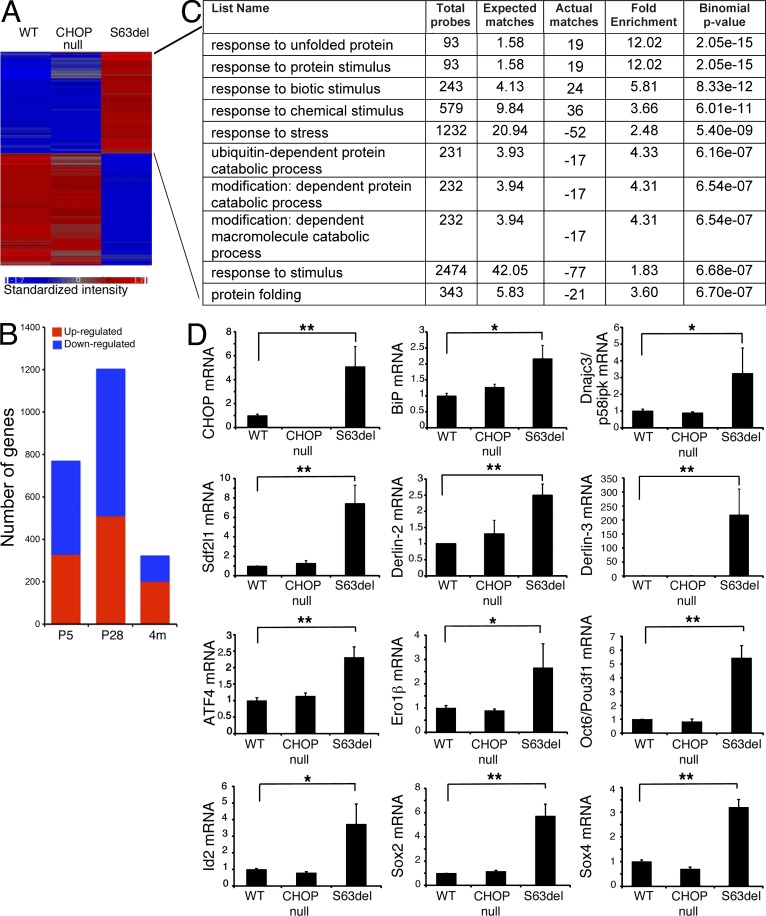
Figure 1.
Transcriptional profiling reveals a robust stress response in S63del nerves. (A) Hierarchical clustering of P5 sciatic nerves from WT, Chop-null, and P0S63del mice. (B) The number of genes that vary in S63del at different stages of myelination. (C) L2L analysis of the S63del group of genes up-regulated in P5 nerves shows an enrichment for the GO categories related to stress response. (D) Quantitative RT-PCR analysis on P28 sciatic nerves to validate a selection of UPR/ERAD-related genes, and early Schwann cell transcription factors. Error bars, SEM; n = 4–6 RT from independent pools of P28 nerves. *, P < 0.05; **, P < 0.01, Student’s t test.
P0S63del activates a robust stress response in transgenic mice
Manual curation of the sets of genes up-regulated in S63del nerves (Table S1) suggested that the expression of P0S63del immediately affects cellular homeostasis. In fact, in S63del nerves at P5, the genes with the highest level of induction were related to the heat shock response, such as Hsp1, 1a, and 1b, along with known members of the UPR (Chop, Atf3, Asns-asparagine synthetase; Barbosa-Tessmann et al., 1999) and of the ER-associated degradation (ERAD) complex (Derl3-Derlin-3; Oda et al., 2006). Other genes were up-regulated that only recently have been associated with the UPR, such as Trb3 (Ohoka et al., 2005) stromal cell–derived factor 2-like 1 (Sdf2l1; Fukuda et al., 2001), and cysteine-rich with EGF-like domains 2 (Creld2; Oh-hashi et al., 2009). Finally, we observed markers of inflammation, such as chemokine (C-X-C motif) ligand 10 and 14 (Cxcl10 and Cxcl14), and the tumor necrosis factor receptor superfamily, member 12a (Tnfrsf12a).
At P28, when MpzS63del is expressed at the highest level, the sets of altered genes were more heterogeneous. Many genes related to UPR/ERAD like Chop, Sdf2l1, and Derl3 were still up-regulated, although the heat shock response was attenuated. More genes related to inflammation, such as serum amyloid A1 (Saa1), Tnfrsf12a, and Cxcl14, and to protein transport, such as members of the sorting nexin family (Snx6 and Snx16), were up-regulated. Interestingly, there was also induction of genes encoding transcription factors that are usually characteristic of pre and pro-myelinating Schwann cells, such as Pou3f1/Oct6, Id2, c-Jun, Sox2, and Sox4 (Fig. 1 and Table S1). Importantly, c-Jun, Sox2, and Id2 are thought to act as negative regulators of myelination (Le et al., 2005; Mager et al., 2008; Parkinson et al., 2008; Arthur-Farraj et al., 2012).
At 4 mo, the expression of MpzS63del is lower, similar to endogenous Mpz, (Feltri et al., 1999), even though demyelinating neuropathy becomes more evident in S63del nerves (Wrabetz et al., 2006), similar to patients with MPZS63del, in whom neuropathy appears gradually over the first two decades of life (Miller et al., 2012). Accordingly, transcriptomic analysis showed that the majority of the up-regulated genes were related to inflammation, a typical consequence of demyelinating neuropathy. Less stress response was detectable (Chop up-regulation was ∼1.6-fold), which is consistent with lower expression of P0S63del. Finally, the early Schwann cell transcription factor genes remained significantly up-regulated.
Although manual curation improves the choice of where to assign genes among Gene Ontology (GO) categories, the different numbers of genes contained among GO categories introduces bias. Therefore, we performed an unbiased analysis using the software L2L (Newman and Weiner, 2005). L2L automates annotation of a list of genes with GO categories, corrects for overrepresentation of the category in the list, and measures statistical significance. This analysis indicated that the GO category named “response to unfolded protein” was highly enriched (12-fold) in P5 nerves from S63del versus WT, confirming that P0S63del quickly activates a robust stress response (Fig. 1 C). Most GO categories enriched at P5 were functionally related, such as stress response, protein folding, or protein catabolism.
In P28 sciatic nerves, L2L analysis again showed enrichment for the GO categories ER–nuclear signaling pathways (11-fold) and the response to unfolded protein (5.7 fold; Fig. S1). Activation of genes involved in UPR and ERAD was confirmed by real-time PCR (Fig. 1 D). In agreement with manual curation, inflammation appeared in the neuropathy at P28, as indicated by the strong enrichment of GO categories related to cytokine secretion (15-fold).
By 4 mo, the GO categories related to inflammation were most represented, such as antigen processing and presentation (12.5-fold) and immune response (3.2-fold; Fig. S1). The UPR was less prominent compared with P5 and P28, with a relative enrichment of threefold (unpublished data).
P0S63del is associated with down-regulation of the lipid synthesis program
The presence of P0S63del was also associated with down-regulation of a large number of genes. At P5, the down-regulation affected mainly genes encoding for enzymes involved in lipid metabolism, such as lanesterol synthase (Lss), Acyl-coA synthetase (Acss2), and farnesyl diphosphate farnesyl transferase 1 (Fdft1), and in transport, like the solute carriers Slc15a2 and Slc6a15 (Table S2).
At P28 most of these genes were still down-regulated, but there was also a striking down-regulation of cytoskeletal genes, such as actin and myosin. At 4 mo, fewer genes were down-regulated, although primarily related to lipid metabolism and cytoskeletal reorganization (Table S2).
Application of L2L analysis to down-regulated genes identified the highest enrichment in GO categories related to lipid/sterol/cholesterol biosynthesis at all time-points (Fig. 2 A and Fig. S2). The down-regulation of the genes encoding for the key enzymes in cholesterol biosynthesis was confirmed via real-time qPCR (Fig. 2 B). Given that ∼70% of myelin is constituted by lipids, and the importance of cytoskeletal remodeling during myelination, these observations may be relevant to the hypomyelinating phenotype of S63del mice.
Figure 2.
The lipid/cholesterol biosynthetic program is strongly down-regulated in S63del nerves. (A) L2L analysis at P5 of genes down-regulated in S63del nerves identified an enrichment for GO categories related to lipid metabolism. (B) Quantitative RT-PCR for a selection of enzymes involved in the cholesterol biosynthetic pathway. Error bars, SEM; n = 4 RT from independent pools of nerves. *, P < 0.05, **, P < 0.01 by Student’s t test; n.s. = not significant.
Surprisingly, the same down-regulation was not observed for myelin structural protein genes. The mRNAs encoding myelin basic protein (MBP) and peripheral myelin protein 22 (PMP22) were unchanged relative to WT, whereas the mRNAs for noncompact myelin components, such as Connexin32, Periaxin, and myelin-associated glycoprotein (MAG) were only mildly down-regulated (Table S2 and not depicted). Moreover, the transcription factor Egr2/Krox20, which is a key regulator of myelination (Topilko et al., 1994; Svaren and Meijer, 2008) that drives synthesis of both myelin proteins and lipids (Leblanc et al., 2005; Nagarajan et al., 2001) was expressed at levels comparable to WT (Fig. 2 B). Myelin protein and lipid synthesis is uncoordinated in S63del nerves, despite normal levels of Krox20.
UPR is a unique feature of S63del nerves
To address which transcriptional changes are mutation- and neuropathy-specific, we analyzed the transcriptome of P28 sciatic nerves from mice overexpressing P0wt at levels comparable to P0S63del (P0oe line 80.4; Wrabetz et al., 2000). P0oe mice manifest a hypomyelinating neuropathy, similar to S63del mice, but without signs of demyelination or onion bulbs. L2L analysis for up-regulated genes (listed in Table S3) showed the highest enrichment for categories related to mRNA metabolism and processing (Fig. S3). Some of these categories were enriched also in S63del nerves (unpublished data), indicating that P0 mRNA or protein overexpression perturbs general transcription. On the contrary, and confirming our previous observations (Pennuto et al., 2008), P0oe nerves did not show an enrichment for genes related to UPR, suggesting that the ER stress is not caused by the overexpression of P0 by itself, but to the MpzS63del mutation. Surprisingly, despite the fact that the P0oe nerves are hypomyelinated (Wrabetz et al., 2000), we did not see a significant down-regulation of genes related to lipid or cholesterol biosynthesis (Table S4); the main down-regulation was for genes involved in cell growth and morphogenesis (Fig. S3), confirming that developmental hypomyelination in P0oe and S63del mice result from different pathomechanisms.
Collectively, these data confirm that the UPR is a specific feature of S63del nerves, which is already active as myelination begins, whereas inflammation and cell death are secondary effects of the neuropathy. In parallel, there is a significant increase in genes related to Schwann cell dedifferentiation, accompanied by a dramatic decrease in the levels of lipid/cholesterol synthetic genes (Table 1).
Table 1.
Summary of the main pathways dysregulated in S63del nerves
| P5 | P-value | P28 | P-value | 4 mo | P-value | |
| Up-regulated pathways | UPR (stress) | 2.1e-15 | UPR (stress) | 1.8e-7 | Inflammation | 2.7e-6 |
| Cell death | 2.9e-5 | Cell death | 4.4e-4 | |||
| UPR (stress) | 2.1e-3 | |||||
| Early TFa (Oct6, Id2, Sox2) | Early TFa (Oct6, Id2, Sox2, c-Jun) | Early TF* (Oct6, Id2, Sox2, c-Jun) | ||||
| Down-regulated pathways | Lipid biosynthesis | 1.4e-35 | Lipid biosynthesis | 1.8e-7 | Lipid biosynthesis | 8.9e-4 |
| Sterol biosynthesis | 5.6e-28 | Sterol biosynthesis | 2.7e-7 | Sterol biosynthesis | 4.7e-3 | |
| Cholesterol biosynthesis | 3.0e-26 | Cholesterol biosynthesis | 1.6e-4 | Cholesterol biosynthesis | 6.6e-3 |
Results are based on manual categorization and L2L analysis.
Arbitrary category, not present in L2L or GO. For this reason, Early TF have no p-values.
Genes rescued in S63del/Chop-null mice
The UPR is a generally adaptive response that, in some circumstances, can become maladaptive. Ablation of the transcription factor Chop from S63del mice produced a complete rescue of the motor capacity, and the number of demyelinating fibers was halved (Pennuto et al., 2008), indicating that CHOP plays a maladaptive role in CMT1B neuropathy. We have previously shown that Chop-null mice are morphologically and behaviorally identical to WT (Pennuto et al., 2008). Accordingly, comparison of WT and Chop-null transcriptomes revealed very few differences (Fig. 1 A and Table S5) that could obscure a comparison of S63del to S63del/Chop-null nerves.
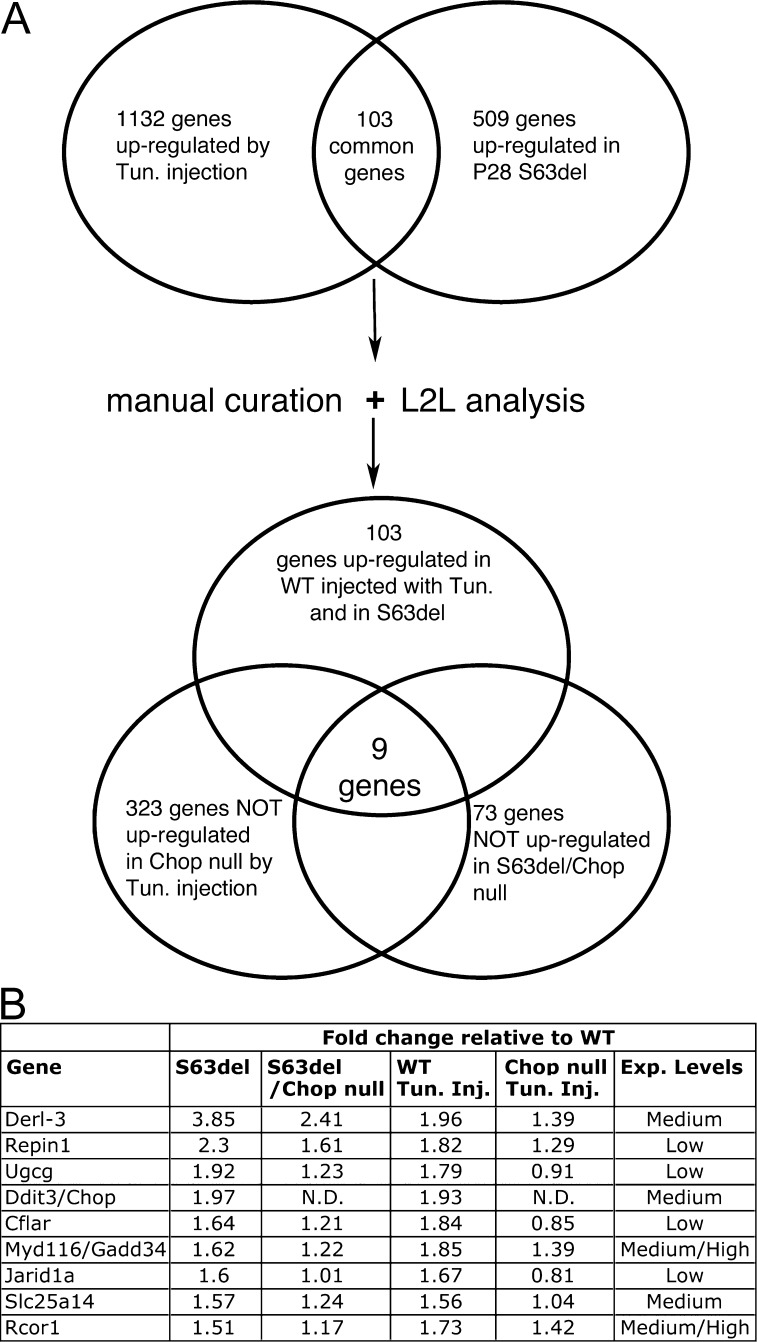
Therefore, to identify a molecular basis for the rescue, we compared the transcriptomes of S63del and S63del/Chop-null nerves. A search for genes that were up-regulated in S63del versus WT (see above), and returned toward normal levels after the ablation of Chop revealed a total of 157 genes (24 at P5, 73 at P28, and 60 at 4 mo; Table S6). However, there were no common genes among the three different time points, apart from CHOP itself. The P5 group was the smallest, suggesting the possibility that many targets of CHOP had not yet been activated by the neuropathic process. Moreover, most of the genes in this group were expressed at very low levels and were not expressed in the later time points (unpublished data), making it unlikely for them to have a role in the rescue. We also reasoned that at 4 mo, most of the rescued genes could be a consequence of secondary effects of the attenuated neuropathy. Hence, we focused on P28, where we identified 73 genes that were significantly less up-regulated in the absence of CHOP (Table S6). To choose those that were more likely to be targets of CHOP, we injected P28 WT and Chop-null mice with tunicamycin, a potent activator of the UPR, and performed transcriptomic analysis on sciatic nerves harvested after 48 h, when CHOP mRNA was up-regulated to a level similar to that seen in P28 S63del nerves (by real-time qPCR; unpublished data). Surprisingly, the transcriptional alteration caused by tunicamycin injection in WT mice was much broader than the one caused by the chronic expression of S63del. More than 1,000 genes were up-regulated at least 1.5-fold by tunicamycin compared with the 509 up-regulated in P28 S63del nerves (Fig. 3 A). Of these, 103 were up-regulated in common, and L2L analysis confirmed that they were mainly stress-related genes (Fig. S4 and not depicted). Finally, we examined how many of these 103 genes returned toward WT levels in both the nerves from S63del/Chop-null and tunicamycin-injected Chop-null mice. The result was eight genes (excluding CHOP itself), which are all candidate CHOP target genes (Fig. 3 B).
Figure 3.
Gadd34 is a target of CHOP in S63del nerves. (A) A Venn diagram of genes up-regulated both in sciatic nerves from P28 WT mice injected with tunicamycin and from P28 S63del mice identified 103 genes in common. Of these, nine returned to WT levels in Chop-null mice injected with tunicamycin and in S63del/Chop-null nerves. (B) Expression of CHOP and the eight CHOP target genes. Exp. Levels = raw level of expression in S63del nerves.
The gene Myd116/Gadd34 was the most interesting in the group. It was expressed most highly, and in support of our data, it had been shown to be a target of CHOP in cultured cells treated with tunicamycin (Marciniak et al., 2004). Real-time qPCR analysis confirmed that Gadd34 mRNA was up-regulated ∼2.5-fold in S63del and returned to near normal levels in S63del/Chop-null nerves (Fig. 4 A). Gadd34 is the activating subunit of the PP1–phosphatase complex that dephosphorylates eIF2α and reverses translational attenuation in the context of UPR (Novoa et al., 2001). Western blot analysis showed that the lack of up-regulation of Gadd34 protein in S63del/Chop-null (Fig. 4, B and C) resulted in a sustained up-regulation of eIF2α phosphorylation compared with S63del (Fig. 4, B and D).
Figure 4.
Reduction of Gadd34 levels results in increased eIF2α phosphorylation. (A) Quantitative RT-PCR for Gadd34 mRNA from P28 sciatic nerves. Error bars, SEM; *, P < 0.05; n = 4 RT from independent pools of nerves. (B) Western blot analysis on P28 sciatic nerve lysates was performed for Gadd34 and phosphorylated eIF2α; β-tubulin provides a control for loading. One representative experiment of four is shown. Numbers indicate relative molecular weights. (C) Gadd34 and (D) phosphorylated eIF2α protein levels as determined by densitometry. Error bars, SEM; **, P < 0.01, ***, P < 0.001; n = 4.
Could this difference in Gadd34 activation contribute to the beneficial effects of Chop deletion in the S63del mice? Phosphorylation of eIF2α blocks general protein translation but activates translation of specific mRNAs, such as ATF4 and Gadd34 itself (Harding et al., 2000; Lee et al., 2009). We hypothesized that the relative increase in P-eIF2α in S63del/Chop-null nerves would limit reactivation of translation that, in turn, could reduce the load of misfolded P0S63del in the ER, a benefit for the Schwann cell.
Inactivation of Gadd34 rescues the S63del phenotype
To further investigate the role of Gadd34, we crossed S63del mice with mice in which the Gadd34 protein lacks the domain that interacts with the catalytic unit of PP1c phosphatase (Novoa et al., 2003), which acts on P-eIF2α. Gadd34Δc/Δc mice are healthy and fertile, manifested normal motor capacity (Fig. 5 A), and no morphological abnormality when compared with WT sciatic nerve (not depicted).
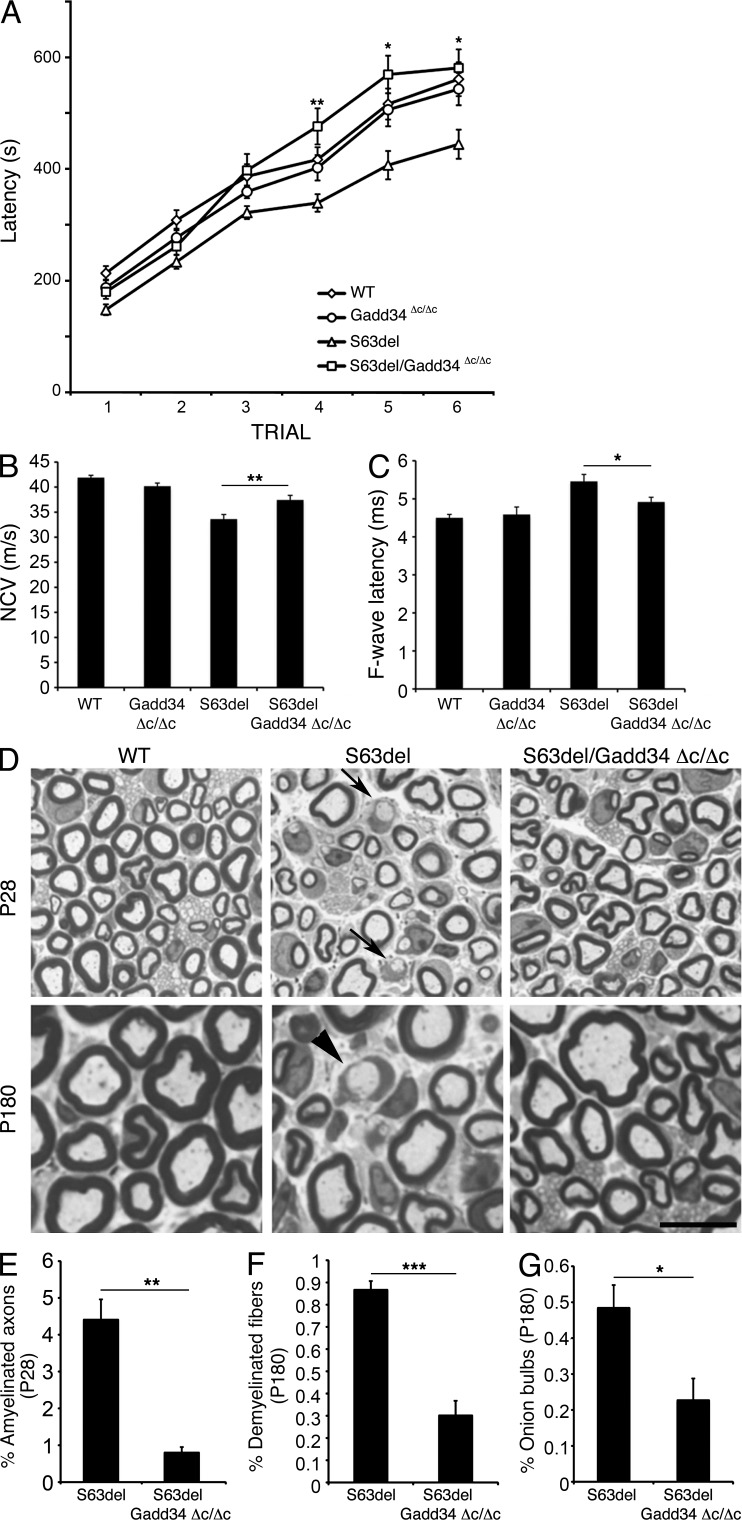
Figure 5.
Inactivation of Gadd34 restores motor function and ameliorates the neurophysiological and morphological deficits in S63del mice. (A) Rotarod analysis of motor function in WT, Gadd34Δc/Δc, S63del and S63del/Gadd34Δc/Δc. Error bars, SEM; *, P < 0.05; **, P < 0.01 for S63del/Gadd34Δc/Δc relative to S63del; n = 20–25 animals. (B and C) Electrophysiological analysis showing NCV and F-wave latency; n = 10–12 animals; *, P < 0.05; **, P < 0.01, relative to S63del by Student’s t test. NCV, nerve conduction velocity. (D) Semithin sections stained with toluidine blue from P28 and P180 WT, S63del, and S63del/Gadd34Δc/Δc sciatic nerves. Arrows, amyelinated axons in P28 S63del nerve. Arrowhead, demyelinated fiber/onion bulb in 6-mo-old S63del nerve. Bar, 10 µm. (E) Number of amyelinated axons at P28, and of demyelinated fibers (F) and onion bulbs (G) at 6 mo. Error bars, SEM; *, P < 0.05; **, P < 0.01; ***, P < 0.001, by Student’s t test; n = 20–35 microscopic fields from 4–6 animals per genotype.
S63del mice show reduced motor capacity as measured by the rotarod test. The genetic ablation of Chop was able to rescue the motor deficit (Wrabetz et al., 2006; Pennuto et al., 2008). Therefore, we asked whether the inactivation of Gadd34 could have a similar effect. Rotarod analysis on 4-mo-old mice showed that S63del/Gadd34Δc/Δc mice regained normal motor capacity (Fig. 5 A). In S63del/Chop-null mice, the motor rescue was accompanied by a reduction by half in demyelinating fibers, a small reduction in onion bulbs, and a rescue in F-wave latency, despite the fact that NCV remained impaired (Pennuto et al., 2008). The rescue after Gadd34 inactivation from S63del mice was strikingly more extensive: nerve conduction velocities (NCVs) improved from 33.6 ± 0.9 m/s in S63del to 37.4 ± 0.9 m/s in S63del/Gadd34Δc/Δc, and F-wave latency returned to almost WT levels (Fig. 5, B and C). Moreover, morphological rescue was more complete. P28 S63del nerves show a significant number of amyelinated axons, which are larger than 1 mm in diameter and in 1:1 relationship with a Schwann cell but do not have a myelin sheath. Inactivation of Gadd34 resulted in a fivefold reduction of these fibers (Fig. 5 E). Finally, in 6-mo-old mice, the lack of Gadd34 activity produced a significant reduction in both demyelinated fibers and in onion bulbs (Fig. 5, F and G).
The reduction in amyelinated axons in P28 nerves prompted us to perform a morphometric analysis to measure myelin thickness. In fact, it appeared that whereas ablation of Chop had little effect on the hypomyelination phenotype of S63del mice (Pennuto et al., 2008), the inactivation of Gadd34 might have a positive effect. Indeed, hypomyelination was improved, as measured by g-ratio, at both P28 and 6 mo (Fig. 6, A and B), although it did not return fully to WT levels. Interestingly, the g-ratio rescue was more evident in small-caliber axons, whereas the axonal numbers and distribution appeared to be normal among the genotypes (g-ratio scatterplots and histograms of axonal diameter are available upon request). An improvement in myelin thickness should result in an increased amount of myelin proteins in bulk myelin. Accordingly, Western analysis showed a significant increase in MBP in S63del/Gadd34Δc/Δc as compared with S63del nerves, and a small increase also in the amount of PMP22, whereas MAG, a component of noncompact myelin, was not increased (Fig. 6, C–E).
Figure 6.
Inactivation of Gadd34 ameliorates the hypomyelination in S63del mice. (A) Semithin sections stained with toluidine blue from P28 and P180 WT, S63del, and S63del/Gadd34Δc/Δc sciatic nerves. Bar, 20 µm. (B) G-ratios in 6-mo old nerves. **, P < 0.01, relative to S63del, by Student’s t test; 8–10 microscopic fields per mouse were analyzed, from 5 mice per genotype. (C) Western blot analysis was performed on sciatic nerve lysates for the myelin proteins MAG, PMP22, and MBP. Numbers indicate relative molecular weights. One representative experiment of three is shown. (D) MBP and (E) PMP22 protein levels as measured by densitometric analysis and E. Error bars, SEM; **, P < 0.01; n = 3 repeat blots.
Inactivation of Gadd34 limits reactivation of translation in S63del nerves
Gadd34 acts to reverse translational attenuation caused by eIF2α phosphorylation during ER stress. Western blot analysis showed that inactivation of Gadd34 in S63del nerves significantly increased eIF2α phosphorylation and, as a consequence, the level of ATF4 (Fig. 7, A and B). We therefore hypothesized that blocking Gadd34 activity would keep translation attenuated, reducing the toxicity associated with expression of high levels of P0S63del. To measure this directly, we explanted sciatic nerves in culture and performed metabolic labeling of total protein synthesis. Accumulation of labeled proteins proceeded linearly for at least 2 h after nerve dissection (unpublished data), whereas adding an agent (DTT) that elicited a strong UPR nearly completely blocked translation (Fig. 7 C). Metabolic labeling of total protein showed a ∼20% reduction in S63del, and a more pronounced 35–40% reduction in S63del/Gadd34Δc/Δc as compared with WT nerves (Fig. 7, C and D). S63del/Chop-null nerves also translated fewer proteins than the S63del, but more than the S63del/Gadd34Δc/Δc nerves (Fig. 7, E and F), consistent with the existence of CHOP-independent activation of Gadd34 expression (Marciniak et al., 2004).
Figure 7.
Increased translational attenuation in S63del/Gadd34Δc/Δc nerves. (A) Western blot analysis on P28 sciatic nerves for phosphorylated eIF2α and ATF4. Numbers indicate relative molecular weights. One representative experiment of five is shown. (B) Phosphorylated eIF2α levels, as measured by densitometric analysis. Error bars, SEM; n = 5 repeat blots. (C) Measurement of newly synthesized proteins via S35 incorporation (metabolic labeling) in explanted P28 WT, S63del, and S63del/Gadd34Δc/Δc sciatic nerves. (E) Metabolic labeling measuring newly synthesized proteins in explanted sciatic nerves from P28 WT, Chop-null, and S63del/Chop-null nerves. 1 representative blot of 10 is shown for each experiment. (D and F). Quantification of newly synthesized proteins (via S35 incorporation) by densitometric analysis. (F) Error bars, SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001, n.s., not significant, by Student’s t test; n = 10 independent experiments.
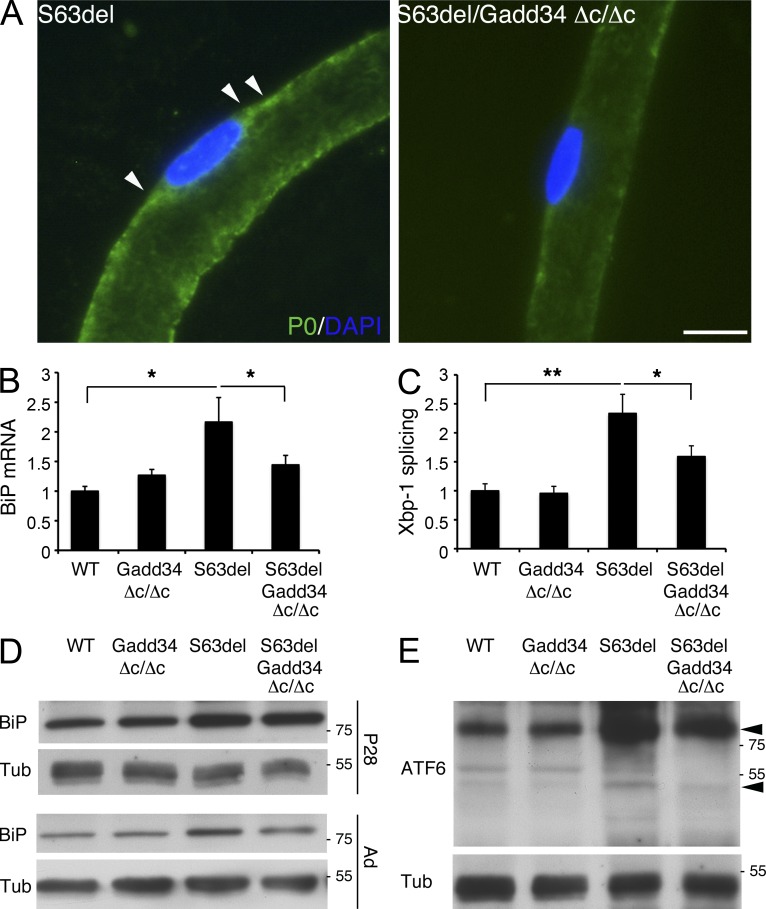
Less protein synthesis would be predicted to lower the load of unfolded proteins in the ER of the S63del/Gadd34Δc/Δc compared with the S63del nerves and lead to a commensurate reduction in UPR activity. Accordingly, immunostaining of teased fibers from S63del and S63del/Gadd34Δc/Δc sciatic nerves showed that in the absence of Gadd34 activity, P0S63del accumulates much less in the perinuclear area of Schwann cells (Fig. 8 A). This is further supported by real-time PCR analysis for markers of the UPR that showed, as early as P28, a significant reduction in BiP mRNA and in Xbp-1 splicing in S63del/Gadd34Δc/Δc compared with S63del (Fig. 8, B and C). Moreover, Western analysis showed that BiP protein is lower in 6-mo-old S63del/Gadd34Δc/Δc compared with S63del nerves, and this was paralleled by a significant decrease in the cleaved form of ATF6. Therefore, all three arms of the UPR (Ron and Walter, 2007) are less active, consistent with a general reduction in ER stress in S63del nerves lacking Gadd34 activity.
Figure 8.
Attenuation of ER stress in S63del/Gadd34Δc/Δc nerves. (A) Immunostaining of teased fibers reveals P0 (green) staining in the perinuclear area (arrowheads) of S63del Schwann cells; nuclei were stained with DAPI (blue). n = 3 mice per genotype were analyzed. Bar, 10 µm. (B and C) mRNA levels for BiP and spliced Xbp-1 were measured by quantitative RT-PCR in P28 sciatic nerves. Error bars, SEM. *, P < 0.05; **, P < 0.01, Student’s t test, n = 6 RT from independent pools of nerves (D and E) Western blot analysis on sciatic nerve lysates for BiP and ATF6. Arrowheads in E indicate the 90-kD precursor and the 50-kD active form of ATF6. Numbers indicate relative molecular weights. One representative experiment of four is shown.
eIF2α phosphorylation as a therapeutic target
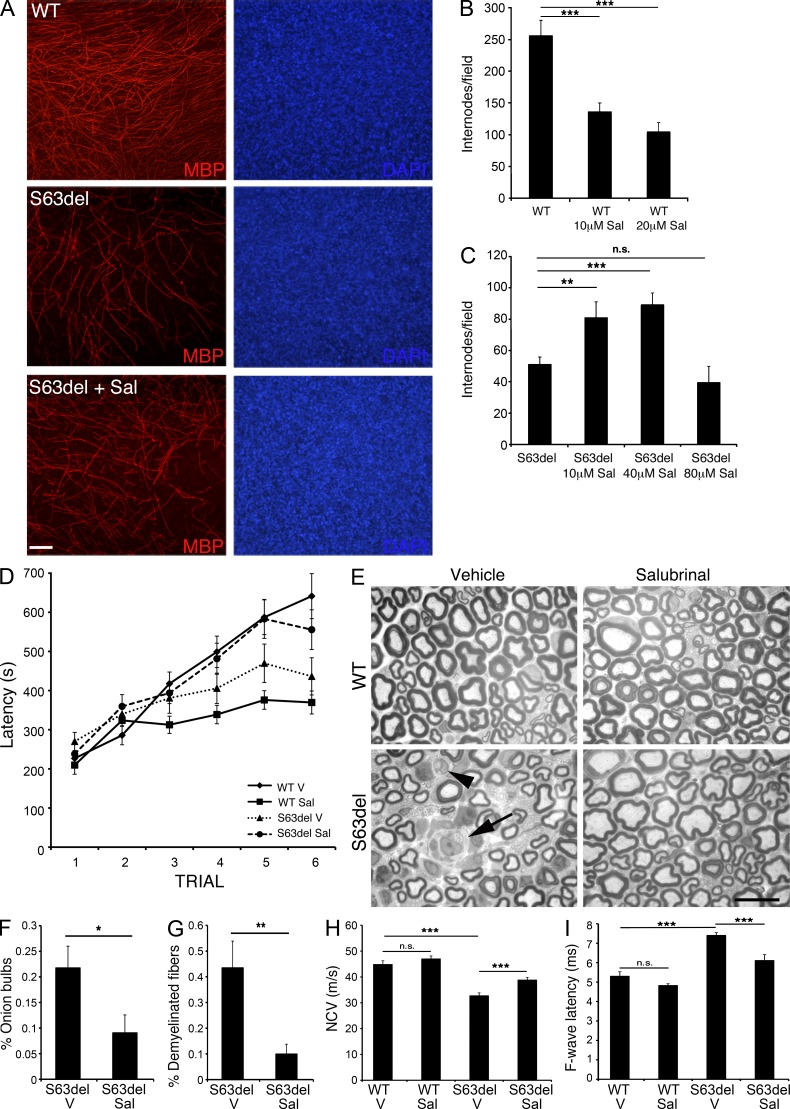
The effective rescue of the CMT1B phenotype by Gadd34 inactivation, suggested it could be a therapeutic target. Salubrinal is a recently identified small molecule that inhibits the Gadd34-mediated dephosphorylation of eIF2α (Boyce et al., 2005). To provide proof of principle for salubrinal as a therapeutic agent, we took advantage of myelinating dorsal root ganglia (DRG)–explant cultures. S63del DRG explant cultures myelinate 80% less than WT DRG explant cultures (Fig. 9 A). Salubrinal is known to be toxic in some contexts, and accordingly, treatment of WT DRG cultures with salubrinal reduced myelination, but by <80% (Fig. 9 B and not depicted). However, at concentrations between 20 and 40 µM, salubrinal significantly improved myelination in S63del DRG cultures, as measured by the number of MBP-positive internodes (Fig. 9, A and C). Strikingly, at these concentrations of salubrinal, the number of internodes in S63del was very similar to WT, suggesting an almost complete rescue, whereas at higher concentrations, salubrinal appeared to be toxic also in S63del DRG cultures (Fig. 9 C). CMT1B associated with P0S63del manifests both developmental hypomyelination (amyelinated fibers from early in development), and even more predominant adult demyelinating neuropathy (myelin destruction and onion bulbs; Miller et al., 2012), but demyelination is poorly represented in DRG-explant cultures. Therefore, to evaluate the effect of salubrinal on demyelinating neuropathy, we treated S63del mice, starting at P30, when myelination is nearly complete, and then measured motor capacity at 4 mo (90 d of treatment) and morphology and electrophysiology at 6 mo (150 d of treatment). Strikingly, S63del mice treated with salubrinal regained almost normal motor capacity in rotarod analysis (Fig. 9 D), and this was accompanied by a rescue of morphological and electrophysiological abnormalities. In particular, demyelinated fibers and onion bulbs were significantly reduced as compared with S63del treated with vehicle (Fig. 9, E–G). Moreover, both NCV and F-wave latency were significantly ameliorated in salubrinal-treated S63del nerves (Fig. 9, H and I). In contrast, little amelioration was observed in the hypomyelination of S63del nerves (Fig. 9 E and not depicted), consistent with the fact that the treatment started when myelination was nearly complete. These data suggest that targeting eIF2α phosphorylation could be a therapeutic strategy for neuropathies related to misfolded proteins.
Figure 9.
Salubrinal improves myelination in S63del DRG explant cultures and reduces demyelination in S63del mice. (A) Dorsal root ganglia (DRG) were dissected from E14 WT and S63del embryos and myelination was induced with 50 µg/ml ascorbic acid. Myelin internodes were detected with antibodies against MBP (red). Nuclei were visualized with DAPI (blue). Bar, 100 µm. (B and C) Number of myelinating internodes in WT and S63del DRG treated for 3 wk with salubrinal at the indicated concentrations; n = 25–50 fields from 6–8 DRG per condition. Error bars, SEM; **, P < 0.01; ***, P < 0.001. (D) Rotarod analysis of WT and S63del mice treated for 90 d with either salubrinal (Sal) or vehicle (V). Error bars, SEM; n = 12 mice per condition. (E) Semithin sections stained with toluidine blue from P180 sciatic nerves from WT and S63del mice treated with either vehicle (V) or salubrinal (Sal) for 150 d. S63del nerves show several demyelinated fibers (arrowhead) and onion bulbs (arrow). Nerves from 10 mice per genotype were analyzed; bar, 10 µm. (F–G) Percentage of onion bulbs and demyelinated fibers in sciatic nerves from 6-mo-old S63del mice treated with Sal for 150 d. Error bars, SEM; *, P < 0.05; **, P < 0.01 by Student’s t test. 8 microscopic fields from each mouse were analyzed, n = 10 mice per condition. (H and I). Electrophysiological analysis of NCV and F-wave latency in 6-mo-old mice (150 d of treatment with Sal or vehicle). Error bars, SEM; ***, P < 0.001; n.s. = not significant by Student’s t test. n = 12–18 nerves per condition.
DISCUSSION
We showed previously that the CMT1B mutant P0S63del is retained in the ER of myelinating Schwann cells where it elicits a dose-dependent UPR. Ablation of Chop partially rescues the demyelination (Pennuto et al., 2008; Wrabetz et al., 2006), indicating that the PERK/CHOP arm of the UPR is pathogenetic in this context. In the last decade, the transcriptomes of PNS myelination, axonal degeneration/regeneration, and neuropathy have revealed complex molecular mechanisms and several interesting signaling pathways, but not a direct link with nerve pathology (Araki et al., 2001; Kubo et al., 2002; Nagarajan et al., 2002; Verheijen et al., 2003; Giambonini-Brugnoli et al., 2005; D’Antonio et al., 2006; Le et al., 2005; Bosse et al., 2006; Ten Asbroek et al., 2006; Jesuraj et al., 2012). Here, through transcriptomic and bioinformatic analysis, we show that the activation of a UPR is the earliest and predominant molecular event in the P0S63del neuropathy, and identify the gene Gadd34 as an important pathogenetic target of CHOP. Accordingly, inactivation of Gadd34 in S63del mice ameliorates the CMT phenotype more effectively than ablation of Chop, resets translational homeostasis toward attenuation, and reduces accumulation of P0S63del in the ER of Schwann cells. Finally, we show that the eIF2α–Gadd34 interaction can be pharmacologically targeted to improve myelination in S63del DRG-explants and mice. Although Gadd34 is a target of CHOP in cultured cells treated with tunicamycin (Marciniak et al., 2004), this is the first time to our knowledge that it has been shown to be regulated by CHOP in an authentic disease model.
The PERK-CHOP arm of the UPR is pathogenetic in S63del mice
Transcriptomic analysis of CMT1B nerves confirmed and expanded our previous observation that the UPR induced by ER retention of P0S63del is pathogenetic (Pennuto et al., 2008). First, the UPR was dramatically activated very early in development, in association with hypomyelination. The transcriptional profile of Schwann cells in S63del nerves revealed that by P5, and persisting through to P28, the largest changes were in pathways related to the response to unfolded protein, as well as to protein folding and degradation. Instead genes and pathways related to inflammation and cell death become prominent only later, after the peak of myelination, and likely reflect secondary events in the neuropathy. Second, the UPR is persistent throughout the life of S63del animals in association with later demyelination. Third, the activation of the UPR requires the P0 mutation. The transcriptome of nerves from transgenic mice that overexpress P0wt at levels similar to P0S63del (Wrabetz et al., 2000) did not reveal any increase in genes related to ER stress. Fourth, the UPR has been identified in other demyelinating neuropathies, including a mouse model of the P0R98C mutation which causes congenital hypomyelination in humans (Saporta et al., 2012) and in the PMP22 Trembler (Tr or TrJ) mouse where an increase of some stress response genes, such as Hsp1a and Hsp1b (Tr; Giambonini-Brugnoli et al., 2005), as well as UPR (TrJ; Khajavi, M., and Lupski, J.R., personal communication) have been detected, probably related to the impaired intracellular trafficking and misfolding of the mutant PMP22 proteins (Fortun et al., 2003; Naef et al., 1997). The UPR is not only closely associated with neuropathy, but in the context of a misfolding-prone P0 mutation, ablation of UPR target genes Chop, and even more so Gadd34, had a strong salutary effect on the associated neuropathy. Thus, comprehensive expression and functional analysis indicate that the UPR, the PERK pathway in particular, is pathogenetic in neuropathy associated with protein misfolding in the ER of Schwann cells.
The microarray analysis of CMT1B nerves suggests that the sustained activation of the PERK/CHOP arm of the UPR does not primarily result in Schwann cell death in S63del nerves (Rutkowski et al., 2006; Pennuto et al., 2008). In fact, we did not detect a significant increase in apoptotic genes until 4 mo after birth. Moreover, the known effectors of ER stress–related cell death such as Bax, Bak, and caspase-12 (Nakagawa et al., 2000; Wei et al., 2001; Ron and Walter, 2007) are not significantly induced in S63del nerves. These observations further support the observation that the detrimental effects of persistent PERK activation through CHOP are context dependent (Gow and Wrabetz, 2009).
CHOP targets translational homeostasis in CMT1B nerves through Gadd34
The immediate consequence of PERK activation in the UPR is to attenuate translation through phosphorylation of eIF2α, whereas protein folding and degradation capacity increase (Scheuner et al., 2001; Ron and Walter, 2007; Back et al., 2009). This is followed by induction of a feedback mechanism via eIF2α/ATF4/CHOP/Gadd34 by which translation is reactivated. Gadd34 is the regulatory subunit of the PP1 phosphatase complex that dephosphorylates eIF2α to reactivate translation in UPR (Novoa et al., 2001; Kojima et al., 2003), and this occurs also in S63del nerves.
However, by favoring lower levels of phosphorylated eIF2α, Gadd34 enhances the burden of unfolded proteins, which can be detrimental in persistent ER stress conditions. For example, Gadd34-null cells in culture are more resistant to ER stress (Marciniak et al., 2004). Here, we show in S63del neuropathy, an authentic model of CMT1B, that Gadd34 is detrimental, as myelination improves in S63del/Gadd34Δc/Δc nerves. Moreover, we identify the beneficial effect in S63del/Gadd34Δc/Δc Schwann cells, as the accumulation of mutant P0S63del in their ER was clearly reduced (Fig. 8 A).
This also provides further evidence that myelinating cells are particularly sensitive to alterations in translation, especially relating to eIF2 regulation (D’Antonio et al., 2009; Lin and Popko, 2009). By dephosphorylating eIF2α, Gadd34 promotes the GTP exchange by eIF2B on eIF2. Not only does Gadd34 moderate CMT1B neuropathy in the PNS, and interferon-γ–induced demyelination in the CNS (Lin et al., 2008), but also mutations in various subunits of eIF2B cause vanishing white matter disease in the CNS, which is often associated with diffuse oligodendrocyte death (van der Knaap et al., 2006).
We hypothesize that the excessive load of misfolded protein in the ER of S63del Schwann cells is a direct consequence of Gadd34 reactivation of translation too aggressively toward normal rates. Directly inactivating Gadd34, or diminishing its up-regulation as in S63del/Chop-null mice, maintains an intermediate level of eIF2α phosphorylation that limits the reactivation of translation (Marciniak et al., 2004) and achieves a better compromise between the load of misfolded protein and the need for translation to produce and maintain myelin. Accordingly, we found that in both S63del/Chop-null and S63del/Gadd34Δc/Δc nerves, eIF2α phosphorylation was significantly higher than in S63del nerves, and global protein translation was reduced as compared with S63del (Fig. 7). In particular, in S63del/Gadd34Δc/Δc mice, the accumulation of mutant P0S63del in the Schwann cell ER was clearly reduced (Fig. 8 A). Finally, we detected a reduction in UPR markers such as BiP chaperone, spliced Xbp-1 and active ATF6, indicating less ER stress (Fig. 8).
Gadd34 versus Chop inactivation in S63del myelination
In S63del mice, the inactivation of Gadd34 more effectively rescues myelination than ablation of Chop, indicating that Gadd34 is more detrimental in S63del Schwann cells. As with Chop ablation (Pennuto et al., 2008), we found restoration of the motor capacity to WT levels, and a remarkable reduction in the number of demyelinated fibers. However, in S63del/Gadd34Δc/Δc mice we also found a significant improvement in nerve conduction velocity, a reduction in onion bulb formation, and a more substantial amelioration of the hypomyelinating phenotype, as measured by the number of amyelinated axons, g-ratio, and myelin protein levels, both at P28 and in the adult.
Possible explanations for this difference include that the block in global protein synthesis remains more pronounced with inactivation of Gadd34 (Fig. 7; Marciniak et al., 2004), whereas ablation of Chop impedes Gadd34 up-regulation, but the remaining basal level of Gadd34 activity could still partially dephosphorylate eIF2α and reactivate translation. Alternatively, inactivation of Gadd34 may more directly correct the imbalance within the program of myelin proteins and lipids that led to dysmyelination and demyelination Dosage of genes encoding myelin proteins and their stoichiometry in compact myelin are very precisely regulated in normal myelination (Scherer and Wrabetz, 2008). Finally, although Gadd34 is a maladaptive target of Chop in S63del Schwann cells, CHOP mediates a balance of both maladaptive and adaptive targets. For example, derlin-3 is significantly less up-regulated in S63del/Chop-null compared with S63del nerves (Fig. 3), and derlin-3 is a component of a complex that functions in ER-associated degradation (ERAD) of misfolded proteins—an adaptive role of the UPR (Oda et al., 2006). Thus, impairing Chop may remove not only maladaptive, but also adaptive effectors and produce a less complete rescue of myelination. This idea is particularly appealing because it could explain the context-dependent behavior of CHOP (Gow and Wrabetz, 2009), that is detrimental in Schwann cells (Pennuto et al., 2008), but beneficial in oligodendrocytes in a model of Pelizeus-Merzbacher disease (Southwood et al., 2002). The balance of adaptive and maladaptive CHOP targets may simply differ between the two cell types.
UPR-limited Schwann cell differentiation as an adaptive strategy in S63del neuropathy
Limited differentiation is likely to be adaptive in S63del Schwann cells, as P0 is the most abundant terminal differentiation product in myelinating Schwann cells (8% of mRNA and >20% of protein). Moreover increasing the dose of P0S63del also increases activation of the UPR (Pennuto et al., 2008). Therefore, we speculate that altering translational homeostasis through Gadd34 manipulation reduces the level of the misfolded P0S63del, whereas permitting a slow rate of accumulation of P0wt in myelin and some myelin associated function (note that CMT1B is a dominant disease where both a misfolded and a normal version of P0 are synthesized). This idea is nicely supported by the observation that less total P0 accumulates in the perinuclear region of S63del/Gadd34Δc/Δc-teased fibers, and this probably represents P0S63del because P0wt is not detected by immunofluorescence in the same area of WT-teased fibers. In support of our speculation, limited differentiation may play an adaptive role in other cells where misfolded proteins include terminal differentiation products (e.g., collagen in chondrocytes; Tsang et al., 2007).
Interestingly, we identified other examples of limited terminal differentiation in S63del Schwann cells. However, their effects may be detrimental. For example, in all time points analyzed, we found a remarkable down-regulation of genes and pathways related to lipid and cholesterol biosynthesis, previously reported also in other hypomyelination neuropathies (Giambonini-Brugnoli et al., 2005). The UPR may directly interfere with the lipid synthesis program as both ATF6 and sterol regulatory element binding protein 2 (SREBP2) transcription factors are activated via proteolytic cleavage by S1P and S2P proteases in the Golgi, and it has been recently proposed that conditions of stress that activate ATF6 also suppressed SREBP2 activity (Zeng et al., 2004). We also found up-regulation of early Schwann cell transcription factors, such as Oct6, Id2, Sox4, Sox2, and c-Jun in S63del nerves. Some of these factors, in particular c-Jun, Sox2, and Id2, have been suggested to act as negative regulators of myelination (Le et al., 2005; Mager et al., 2008; Parkinson et al., 2008).
Pharmacological targeting of eIF2α
Protein quality control is a potential therapeutic target in myelin diseases. Autophagy, stimulated via fasting or rapamycin, or an increase in the heat shock response, can improve the phenotype of some PMP22-related neuropathies (Fortun et al., 2007; Rangaraju et al., 2008, 2010; Madorsky et al., 2009), and salubrinal enhancement of the integrated stress response (ISR) ameliorated hypomyelination and oligodendrocyte loss in cultured hippocampal slices exposed to IFN-γ (Lin et al., 2008).
We have shown here that Gadd34 is detrimental in S63del nerves, and that its inhibition could be an effective therapeutic strategy. Salubrinal blockade of eIF2α dephosphorylation significantly improves hypomyelination in S63del DRG explants. More importantly, it restores motor function and ameliorates morphological and electrophysiological parameters of demyelination in S63del mice. Unfortunately, treatment with salubrinal reduced motor capacity in WT mice. However, the normal morphology and electrophysiology in these mice suggest that poor rotarod performance may be caused by an effect outside of nerve, on muscle for example.
Salubrinal has also been successfully used in a model of ALS to reset homeostasis in motor neurons with SOD1 mutations (Saxena et al., 2009). However, a note of caution is raised by prion-induced neurodegeneration, where Gadd34 overexpression is protective and salubrinal treatment in mouse models actually impairs neuronal survival (Moreno et al., 2012). Finally, compounds like guanabenz may provide less toxic alternatives for modulation of eIF2α phosphorylation in animal models of disease (Tsaytler et al., 2011).
Relevance for neuropathy and other misfolded protein diseases
Mutations in >30 genes account for about one-half of CMT hereditary neuropathies (Wrabetz et al., 2004; Scherer and Wrabetz, 2008), but the pathomechanisms are still poorly understood and at present there is no cure. ER retention of CMT-related mutant proteins is not unique to P0S63del; we have identified at least five other P0 mutants that are retained in the ER and elicit a UPR (Pennuto et al., 2008; Saporta et al., 2012), and protein misfolding and altered trafficking have been implicated in the pathogenesis of other CMT neuropathies as a result of mutations in PMP22 and Cx32 (Colby et al., 2000; Kleopa et al., 2002; Yum et al., 2002). Moreover, protein misfolding and ER stress have been associated with various diseases, ranging from neurodegenerative diseases (Alzheimer disease, Parkinson disease, and ischemic insults) to cancer and diabetes, suggesting that this study may have broad therapeutic implications.
MATERIALS AND METHODS
Animals.
All experiments involving animals were performed in accord with experimental protocols approved by the San Raffaele Scientific Institute Animal Care and Use Committee. P0S63del transgenic mice (Wrabetz et al., 2006), P0-overexpressing (oe) mice (line 80.4; Wrabetz et al., 2000), Chop-null mice (Zinszner et al., 1998), and Gadd34Δc/Δc mice (Novoa et al., 2003) have been described previously. P0S63del, P0oe, and Chop-null mice were maintained on the FVB/N background, whereas Gadd34Δc/Δc mice, initially on a C57BL/6 background, were backcrossed for at least six generations onto the FVB/N background before initiation of experiments. In all the experiments, littermates were used as controls.
Tunicamycin and salubrinal injections.
Where indicated, WT and Chop-null mice were injected i.p. with 1 mg tunicamycin/g body weight, in 150 mM dextrose. 48 h later, mice were killed by CO2 inhalation, and sciatic nerves were harvested for RNA preparation. Salubrinal (Tocris) was reconstituted at 2 mM in PBS, 0.1% BSA with 10% DMSO, and i.p injected every alternate day for 150 d (P30–P180) at 1 mg/kg of body weight.
RNA extraction and hybridization of arrays.
Total RNA from at least 15 (P5) or 10 (P28 or 4 mo) desheathed sciatic nerves for each of WT, Chop-null, S63del, S63del/Chop-null mice; from 5 desheathed sciatic nerves from WT and Chop-null, tunicamycin-injected mice; and 10 sciatic nerves from P0oe mice were extracted using TRIzol (Roche) and purified with RNeasy kit (QIAGEN), as previously described (D’Antonio et al., 2006). Biotin-labeled cRNA targets were synthesized starting from 5 µg of total RNA. RNA quality was tested with an Agilent Bioanalyzer. Double-stranded cDNA synthesis was performed with Gibco SuperScript Custom cDNA Synthesis kit, and biotin-labeled antisense RNA was transcribed in vitro using Ambion’s in vitro Transcription System, including Bio-11-UTP and Bio-11-CTP (PerkinElmer) in the reaction. All steps of the labeling protocol were performed as suggested by Affymetrix. The size and the accuracy of quantitation of targets were confirmed by agarose gel electrophoresis of 2 mg aliquots, before and after fragmentation. After fragmentation, targets were applied to Affymetrix Mouse GeneChipMOE430A 2.0. Two replicates for each genotype at each time point were prepared from completely independent pools of nerves (biological replicas).
Data Analysis.
Images were scanned with an Affymetrix GeneChip Scanner3000 7G, using default parameters. The resulting images were analyzed using GeneChip Operating Software v1.4 (GCOS1.4). Report files were extracted for each chip, and performance of labeled targets was evaluated on the basis of several values (scaling factor, background and noise levels, percentage of present calls, average signal value, 5′-3′ values). Data were then imported onto GeneSpring (Agilent) and Partek for further analysis.
Average signal intensity of transcripts from WT nerves was used to normalize the corresponding signal intensity in all other conditions. Replicate experiments were highly reproducible, with pairwise correlation coefficients mostly ranging from 0.95 to 0.99. Genes that had a correlation coefficient <0.95 were eliminated. Taking into account that CHOP mRNA had signal intensity of ∼80–100 in the Chop-null extracts, genes with a signal intensity value <100 in all the genotypes were discarded.
Starting from this gene list, a selection of regulated transcripts in comparison to the WT baseline was performed with the fold-change cutoff set to 1.5. Only a few genes were different between WT and Chop-null, and therefore we considered them to be identical. We focused our attention on genes that were up- or down-regulated in S63del as compared with WT and then examined whether or not they returned toward WT levels in S63del/Chop-null nerves. All of the gene-lists created (Tables S1–S6) were then loaded onto L2L software to search for enriched GO categories (binomial p-value test; P < 0.05). Data combined with experimental information in MIAME compliant format have been submitted to Gene Expression Omnibus (GEO, accession no. GSE40610).
TaqMan quantitative PCR analysis.
Total RNA was prepared as described above, and reverse transcription was performed as described previously (Wrabetz et al., 2006). Quantitative PCR was performed on an ABI PRISM 7700 sequence detection system (Applied Biosystems Instruments) according to the manufacturer’s instructions (TaqMan; Applied Biosystems). The relative standard curve method was applied using WT mice as reference. Normalization was performed using either 18S rRNA or phosphoglycerate 1 (Pgk1) as reference genes. Target and reference gene PCR amplification were performed in separate tubes with Assays on Demand (Applied Biosystems Instruments): 18s, Hs99999901_s1; Pgk1, Mm00435617_m1; Ddit3/Chop, Mm00492097_m1; Myd116/Gadd34, Mm00435119_m1; ATF4, Mm00515324_m1; ATF3, Mm00476032_m1; Xbp-1u, Mm00457357_m1; Xbp-1s, Mm03464496_m1; Derl2, Mm00504288_m1; Derl3, Mm00508292_m1; Ero1b, Mm00470754_m1; Dnajc3/p58ipk, Mm01226332_m1; Hspa5/BiP, Mm00517691_m1; Ppp1r15b/CReP, Mm00551747_m1; Sdf2l1, Mm00452079_m1; Snx6, Mm00459049_m1; Snx16, Mm00511049_m1; Fdft1, Mm00815354_s1; Hmgcr, Mm01282499_m1; Hmgcs2, Mm00550050_m1; Mvd, Mm00507014_m1; Sqle, Mm00436772_m1; Egr2/Krox20, Mm00456650_m1; Pou3f1/Oct6, Mm00843534_s1; Sox2, Mm00488369_s1; Sox4, Mm00486320_m1; Id2, Mm00711781_m1; c-Jun, Mm00495062_s1.
Western blot.
Sciatic nerves from WT and transgenic mice were dissected and immediately frozen in liquid nitrogen. Proteins were extracted and analyzed as previously described (Wrabetz et al., 2000). Mouse monoclonal antibodies recognized β-tubulin (1:500; Sigma-Aldrich), ATF6 (1:1,000; Imgenex,), MBP (1:5,000; Covance), and MAG (1:1,000; EMD Millipore). Rabbit polyclonal antibodies recognized Grp78/BiP (1:1,000; Stressgen), ATF4 (1:500; Santa Cruz Biotechnology), phosphorylated eIF2α (Ser51; 1:500; Cell Signaling Technology), PMP22 (Abcam), eIF2α (1:1,000; the anti-serum was raised in a rabbit injected with purified mouse eIF2α [peptide 1–185] produced as a HisX6 fusion protein in E. coli), and Gadd34 (Novoa et al., 2003). Peroxidase conjugated anti–mouse, anti–rabbit (1:2,000; Sigma-Aldrich), or anti–protein A (1:5,000; Invitrogen) secondary antibodies were visualized using ECL Plus method with autoradiography films (GE Healthcare).
Metabolic labeling.
Sciatic nerves from P28 WT and transgenic mice were dissected, desheathed, and placed in DMEM media lacking methionine and cysteine (Gibco) supplemented with 1% FCS, glutamine, and pyruvic acid, for 30 min at 37°C (starvation period), and then pulsed with 35S protein labeling mix (PerkinElmer) for 2 h. Proteins were extracted, quantified, and resolved by SDS-PAGE. The gel was then dried (Bio-Rad) and exposed with Biomax autoradiography films (GE Healthcare). Densitometric quantification was performed with ImageJ.
Myelinating DRG explant cultures.
DRG were dissected from E13.5 WT and S63del embryos, and maintained in culture as previously described (Taveggia et al., 2005). Myelination was induced with 50 µg/ml ascorbic acid (Sigma-Aldrich). Salubrinal (Tocris) was applied at the indicated concentration for 2–3 wk in parallel to the induction of myelination with ascorbic acid. The number of MBP-positive (see below) myelinated internodes was counted from 8–10 fields per DRG, from 6–8 DRG cultures per genotype.
Behavioral, morphological, and electrophysiological analyses.
Motor capacity was assessed in 4-mo-old mice by rotarod analysis, as previously described (Wrabetz et al., 2006), on 20–25 mice per genotype. Electrophysiology was performed on 6-mo-old mice, as previously described (Wrabetz et al., 2006), on 8–12 animals per genotype. The number of amyelinated axons at P28, and demyelinated axons and onion bulbs at 6 mo were counted blind to genotype in semithin sections stained with toluidine blue in images acquired with a 100× objective as previously described (Quattrini et al., 1996). We analyzed 2,000–3,000 fibers in 20–35 fields for each genotype in six WT, four Gadd34Δc/Δc, six S63del, and six S63del/Gadd34Δc/Δc nerves. Semiautomated computer-based morphometry was performed on semithin sections of P28 and 6-mo-old sciatic nerve to determine the G-ratio and the distribution of fiber diameters for myelinated axons as described (Pennuto et al., 2008). 6–15 microscopic fields from nerves of 4–6 animals per genotype were analyzed.
Immunofluorescent staining on sciatic nerve sections, teased fibers, and myelinating DRG explant cultures was performed as previously described (Wrabetz et al., 2006; Nodari et al., 2007) The following antibodies were used: chicken anti-P0 (1:2,000; Aves laboratories), mouse anti-KDEL (1:500; Stressgen), rat anti-MBP (1:5), rabbit anti-NF-H (1:1000; EMD Millipore); and TRITC- and FITC-conjugated secondary antibodies (1:200; Southern Biotech or Cappel). Specimens were incubated with DAPI (1:10,000), rinsed, mounted with VectaShield (Vector Laboratories), and viewed with a Leica DM5000B microscope. Images were collected with a Leica DFC480 digital color camera and processed with Adobe Photoshop CS4.
Online supplemental material.
Fig. S1 shows the bioinformatic (GO) analyses performed with L2L for the genes up-regulated in S63del nerves at P28 and 4-months. Fig. S2 shows further GO analysis for the genes down-regulated in S63del nerves at P28 and 4-months. Fig. S3 presents the results of the GO analysis of the genes dysregulated in WT MPZ overexpressing (P0oe) sciatic nerves. Fig. S4 shows the results of the GO analysis of the 103 genes in the Venn diagram in Fig. 3. Table S1 lists the genes up-regulated in the mutant mice compared with WT at all the time-points analyzed. Table S2 lists the genes down-regulated in the mutant mice compared with WT at all the time-points analyzed. Table S3 lists the group of genes up-regulated in P0oe mice compared with WT at P28. Table S4 lists the group of genes down-regulated in P0oe mice compared with WT at P28. Table S5 lists the short group of genes that differ in expression between WT and Chop-null nerves. Table S6 lists the genes that are up-regulated in the mutant nerves and return toward WT levels after the ablation of Chop (“rescued” genes). Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20122005/DC1.
Supplementary Material
Acknowledgments
We thank Stefania Saccucci, Desiree Zambroni and Simone Minardi for excellent technical assistance.
This work was supported by grants from the National Institutes of Health (R01-NS55256 to L. Wrabetz; R01-NS45630 to M.L. Feltri), Telethon, Italy (GGP071100 to L. Wrabetz, GGP08021 to M.L. Feltri, and GPP10007 to L. Wrabetz and M.L. Feltri) and the European Community (FP7/2007-1013 under Grant Agreement HEALTH-F2-2008-201535). M. D’Antonio is the recipient of a Giovane Ricercatore Award from the Italian Ministry of Health (GR-2009-1548255).
None of the authors have competing financial interests to disclose.
Footnotes
Abbreviations used:
- ATF6
- activating transcription factor
- Bak
- BCL2-antagonist/killer
- Bax
- BCL2-associated X protein
- BiP
- binding immunoglobulin protein
- Chop
- CCAAT/enhancer-binding protein homologous protein
- CMT1B
- Charcot-Marie-Tooth disease type 1B
- DRG
- dorsal root ganglia
- eIF2a
- eukaryotic initiation factor 2α
- ERAD
- ER-associated degradation
- GO
- gene ontology
- Hsp1
- heat shock protein 1
- ISR
- integrated stress response
- MAG
- myelin-associated glycoprotein
- MBP
- myelin basic protein
- NCV
- nerve conduction velocity
- P-eIF2a
- phosphorylated eIF2a
- PERK
- protein kinase RNA-like endoplasmic reticulum kinase
- PMP22
- peripheral myelin protein 22 kD
- SREBP2
- sterol regulatory element binding protein 2
- UPR
- unfolded protein response
- XBP-1
- X-box binding protein 1
References
- Araki T., Nagarajan R., Milbrandt J. 2001. Identification of genes induced in peripheral nerve after injury. Expression profiling and novel gene discovery. J. Biol. Chem. 276:34131–34141 10.1074/jbc.M104271200 [DOI] [PubMed] [Google Scholar]
- Arthur-Farraj P.J., Latouche M., Wilton D.K., Quintes S., Chabrol E., Banerjee A., Woodhoo A., Jenkins B., Rahman M., Turmaine M., et al. 2012. c-Jun reprograms Schwann cells of injured nerves to generate a repair cell essential for regeneration. Neuron. 75:633–647 10.1016/j.neuron.2012.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Back S.H., Scheuner D., Han J., Song B., Ribick M., Wang J., Gildersleeve R.D., Pennathur S., Kaufman R.J. 2009. Translation attenuation through eIF2alpha phosphorylation prevents oxidative stress and maintains the differentiated state in beta cells. Cell Metab. 10:13–26 10.1016/j.cmet.2009.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa-Tessmann I.P., Chen C., Zhong C., Schuster S.M., Nick H.S., Kilberg M.S. 1999. Activation of the unfolded protein response pathway induces human asparagine synthetase gene expression. J. Biol. Chem. 274:31139–31144 10.1074/jbc.274.44.31139 [DOI] [PubMed] [Google Scholar]
- Berger P., Niemann A., Suter U. 2006. Schwann cells and the pathogenesis of inherited motor and sensory neuropathies (Charcot-Marie-Tooth disease). Glia. 54:243–257 10.1002/glia.20386 [DOI] [PubMed] [Google Scholar]
- Bosse F., Hasenpusch-Theil K., Küry P., Müller H.W. 2006. Gene expression profiling reveals that peripheral nerve regeneration is a consequence of both novel injury-dependent and reactivated developmental processes. J. Neurochem. 96:1441–1457 10.1111/j.1471-4159.2005.03635.x [DOI] [PubMed] [Google Scholar]
- Boyce M., Bryant K.F., Jousse C., Long K., Harding H.P., Scheuner D., Kaufman R.J., Ma D., Coen D.M., Ron D., Yuan J. 2005. A selective inhibitor of eIF2alpha dephosphorylation protects cells from ER stress. Science. 307:935–939 10.1126/science.1101902 [DOI] [PubMed] [Google Scholar]
- Colby J., Nicholson R., Dickson K.M., Orfali W., Naef R., Suter U., Snipes G.J. 2000. PMP22 carrying the trembler or trembler-J mutation is intracellularly retained in myelinating Schwann cells. Neurobiol. Dis. 7:561–573 10.1006/nbdi.2000.0323 [DOI] [PubMed] [Google Scholar]
- D’Antonio M., Michalovich D., Paterson M., Droggiti A., Woodhoo A., Mirsky R., Jessen K.R. 2006. Gene profiling and bioinformatic analysis of Schwann cell embryonic development and myelination. Glia. 53:501–515 10.1002/glia.20309 [DOI] [PubMed] [Google Scholar]
- D’Antonio M., Feltri M.L., Wrabetz L. 2009. Myelin under stress. J. Neurosci. Res. 87:3241–3249 10.1002/jnr.22066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feltri M.L., D’antonio M., Quattrini A., Numerato R., Arona M., Previtali S., Chiu S.Y., Messing A., Wrabetz L. 1999. A novel P0 glycoprotein transgene activates expression of lacZ in myelin-forming Schwann cells. Eur. J. Neurosci. 11:1577–1586 10.1046/j.1460-9568.1999.00568.x [DOI] [PubMed] [Google Scholar]
- Fortun J., Dunn W.A., Jr, Joy S., Li J., Notterpek L. 2003. Emerging role for autophagy in the removal of aggresomes in Schwann cells. J. Neurosci. 23:10672–10680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortun J., Verrier J.D., Go J.C., Madorsky I., Dunn W.A., Notterpek L. 2007. The formation of peripheral myelin protein 22 aggregates is hindered by the enhancement of autophagy and expression of cytoplasmic chaperones. Neurobiol. Dis. 25:252–265 10.1016/j.nbd.2006.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda S., Sumii M., Masuda Y., Takahashi M., Koike N., Teishima J., Yasumoto H., Itamoto T., Asahara T., Dohi K., Kamiya K. 2001. Murine and human SDF2L1 is an endoplasmic reticulum stress-inducible gene and encodes a new member of the Pmt/rt protein family. Biochem. Biophys. Res. Commun. 280:407–414 10.1006/bbrc.2000.4111 [DOI] [PubMed] [Google Scholar]
- Giambonini-Brugnoli G., Buchstaller J., Sommer L., Suter U., Mantei N. 2005. Distinct disease mechanisms in peripheral neuropathies due to altered peripheral myelin protein 22 gene dosage or a Pmp22 point mutation. Neurobiol. Dis. 18:656–668 10.1016/j.nbd.2004.10.023 [DOI] [PubMed] [Google Scholar]
- Giese K.P., Martini R., Lemke G., Soriano P., Schachner M. 1992. Mouse P0 gene disruption leads to hypomyelination, abnormal expression of recognition molecules, and degeneration of myelin and axons. Cell. 71:565–576 10.1016/0092-8674(92)90591-Y [DOI] [PubMed] [Google Scholar]
- Gow A., Wrabetz L. 2009. CHOP and the endoplasmic reticulum stress response in myelinating glia. Curr. Opin. Neurobiol. 19:505–510 10.1016/j.conb.2009.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding H.P., Novoa I., Zhang Y., Zeng H., Wek R., Schapira M., Ron D. 2000. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell. 6:1099–1108 10.1016/S1097-2765(00)00108-8 [DOI] [PubMed] [Google Scholar]
- Harding H.P., Calfon M., Urano F., Novoa I., Ron D. 2002. Transcriptional and translational control in the Mammalian unfolded protein response. Annu. Rev. Cell Dev. Biol. 18:575–599 10.1146/annurev.cellbio.18.011402.160624 [DOI] [PubMed] [Google Scholar]
- Jesuraj N.J., Nguyen P.K., Wood M.D., Moore A.M., Borschel G.H., Mackinnon S.E., Sakiyama-Elbert S.E. 2012. Differential gene expression in motor and sensory Schwann cells in the rat femoral nerve. J. Neurosci. Res. 90:96–104 10.1002/jnr.22752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner D.A., Wrabetz L., Feltri M.L. 2004. The P0 Gene. Myelin Biology and Disorders. Lazzarini R.A., editor Elsevier Academic Press, S. Diego: 523–546 [Google Scholar]
- Kleopa K.A., Yum S.W., Scherer S.S. 2002. Cellular mechanisms of connexin32 mutations associated with CNS manifestations. J. Neurosci. Res. 68:522–534 10.1002/jnr.10255 [DOI] [PubMed] [Google Scholar]
- Kojima E., Takeuchi A., Haneda M., Yagi A., Hasegawa T., Yamaki K., Takeda K., Akira S., Shimokata K., Isobe K. 2003. The function of GADD34 is a recovery from a shutoff of protein synthesis induced by ER stress: elucidation by GADD34-deficient mice. FASEB J. 17:1573–1575 10.1096/fj.02-0462com [DOI] [PubMed] [Google Scholar]
- Kubo T., Yamashita T., Yamaguchi A., Hosokawa K., Tohyama M. 2002. Analysis of genes induced in peripheral nerve after axotomy using cDNA microarrays. J. Neurochem. 82:1129–1136 10.1046/j.1471-4159.2002.01060.x [DOI] [PubMed] [Google Scholar]
- Kulkens T., Bolhuis P.A., Wolterman R.A., Kemp S., te Nijenhuis S., Valentijn L.J., Hensels G.W., Jennekens F.G., de Visser M., Hoogendijk J.E., et al. 1993. Deletion of the serine 34 codon from the major peripheral myelin protein P0 gene in Charcot-Marie-Tooth disease type 1B. Nat. Genet. 5:35–39 10.1038/ng0993-35 [DOI] [PubMed] [Google Scholar]
- Le N., Nagarajan R., Wang J.Y., Araki T., Schmidt R.E., Milbrandt J. 2005. Analysis of congenital hypomyelinating Egr2Lo/Lo nerves identifies Sox2 as an inhibitor of Schwann cell differentiation and myelination. Proc. Natl. Acad. Sci. USA. 102:2596–2601 10.1073/pnas.0407836102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leblanc S.E., Srinivasan R., Ferri C., Mager G.M., Gillian-Daniel A.L., Wrabetz L., Svaren J. 2005. Regulation of cholesterol/lipid biosynthetic genes by Egr2/Krox20 during peripheral nerve myelination. J. Neurochem. 93:737–748 10.1111/j.1471-4159.2005.03056.x [DOI] [PubMed] [Google Scholar]
- Lee Y.Y., Cevallos R.C., Jan E. 2009. An upstream open reading frame regulates translation of GADD34 during cellular stresses that induce eIF2alpha phosphorylation. J. Biol. Chem. 284:6661–6673 10.1074/jbc.M806735200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W., Popko B. 2009. Endoplasmic reticulum stress in disorders of myelinating cells. Nat. Neurosci. 12:379–385 10.1038/nn.2273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J.H., Li H., Yasumura D., Cohen H.R., Zhang C., Panning B., Shokat K.M., Lavail M.M., Walter P. 2007. IRE1 signaling affects cell fate during the unfolded protein response. Science. 318:944–949 10.1126/science.1146361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W., Kunkler P.E., Harding H.P., Ron D., Kraig R.P., Popko B. 2008. Enhanced integrated stress response promotes myelinating oligodendrocyte survival in response to interferon-gamma. Am. J. Pathol. 173:1508–1517 10.2353/ajpath.2008.080449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madorsky I., Opalach K., Waber A., Verrier J.D., Solmo C., Foster T., Dunn W.A., Jr, Notterpek L. 2009. Intermittent fasting alleviates the neuropathic phenotype in a mouse model of Charcot-Marie-Tooth disease. Neurobiol. Dis. 34:146–154 10.1016/j.nbd.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mager G.M., Ward R.M., Srinivasan R., Jang S.W., Wrabetz L., Svaren J. 2008. Active gene repression by the Egr2.NAB complex during peripheral nerve myelination. J. Biol. Chem. 283:18187–18197 10.1074/jbc.M803330200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marciniak S.J., Yun C.Y., Oyadomari S., Novoa I., Zhang Y., Jungreis R., Nagata K., Harding H.P., Ron D. 2004. CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev. 18:3066–3077 10.1101/gad.1250704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller L.J., Patzko A., Lewis R.A., Shy M.E. 2012. Phenotypic presentation of the Ser63Del MPZ mutation. J. Peripher. Nerv. Syst. 17:197–200 10.1111/j.1529-8027.2012.00398.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk K.R., Voas M.G., Franzini-Armstrong C., Hakkinen I.S., Talbot W.S. 2013. Mutation of sec63 in zebrafish causes defects in myelinated axons and liver pathology. Dis. Model Mech. 6:135–145 10.1242/dmm.009217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno J.A., Radford H., Peretti D., Steinert J.R., Verity N., Martin M.G., Halliday M., Morgan J., Dinsdale D., Ortori C.A., et al. 2012. Sustained translational repression by eIF2α-P mediates prion neurodegeneration. Nature. 485:507–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naef R., Adlkofer K., Lescher B., Suter U. 1997. Aberrant protein trafficking in Trembler suggests a disease mechanism for hereditary human peripheral neuropathies. Mol. Cell. Neurosci. 9:13–25 10.1006/mcne.1997.0604 [DOI] [PubMed] [Google Scholar]
- Nagarajan R., Svaren J., Le N., Araki T., Watson M., Milbrandt J. 2001. EGR2 mutations in inherited neuropathies dominant-negatively inhibit myelin gene expression. Neuron. 30:355–368 10.1016/S0896-6273(01)00282-3 [DOI] [PubMed] [Google Scholar]
- Nagarajan R., Le N., Mahoney H., Araki T., Milbrandt J. 2002. Deciphering peripheral nerve myelination by using Schwann cell expression profiling. Proc. Natl. Acad. Sci. USA. 99:8998–9003 10.1073/pnas.132080999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T., Zhu H., Morishima N., Li E., Xu J., Yankner B.A., Yuan J. 2000. Caspase-12 mediates endoplasmic-reticulum-specific apoptosis and cytotoxicity by amyloid-beta. Nature. 403:98–103 10.1038/47513 [DOI] [PubMed] [Google Scholar]
- Newman J.C., Weiner A.M. 2005. L2L: a simple tool for discovering the hidden significance in microarray expression data. Genome Biol. 6:R81 10.1186/gb-2005-6-9-r81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nodari A., Zambroni D., Quattrini A., Court F.A., D’Urso A., Recchia A., Tybulewicz V.L., Wrabetz L., Feltri M.L. 2007. Beta1 integrin activates Rac1 in Schwann cells to generate radial lamellae during axonal sorting and myelination. J. Cell Biol. 177:1063–1075 10.1083/jcb.200610014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novoa I., Zeng H., Harding H.P., Ron D. 2001. Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2alpha. J. Cell Biol. 153:1011–1022 10.1083/jcb.153.5.1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novoa I., Zhang Y., Zeng H., Jungreis R., Harding H.P., Ron D. 2003. Stress-induced gene expression requires programmed recovery from translational repression. EMBO J. 22:1180–1187 10.1093/emboj/cdg112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda Y., Okada T., Yoshida H., Kaufman R.J., Nagata K., Mori K. 2006. Derlin-2 and Derlin-3 are regulated by the mammalian unfolded protein response and are required for ER-associated degradation. J. Cell Biol. 172:383–393 10.1083/jcb.200507057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh-hashi K., Koga H., Ikeda S., Shimada K., Hirata Y., Kiuchi K. 2009. CRELD2 is a novel endoplasmic reticulum stress-inducible gene. Biochem. Biophys. Res. Commun. 387:504–510 10.1016/j.bbrc.2009.07.047 [DOI] [PubMed] [Google Scholar]
- Ohoka N., Yoshii S., Hattori T., Onozaki K., Hayashi H. 2005. TRB3, a novel ER stress-inducible gene, is induced via ATF4-CHOP pathway and is involved in cell death. EMBO J. 24:1243–1255 10.1038/sj.emboj.7600596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson D.B., Bhaskaran A., Arthur-Farraj P., Noon L.A., Woodhoo A., Lloyd A.C., Feltri M.L., Wrabetz L., Behrens A., Mirsky R., Jessen K.R. 2008. c-Jun is a negative regulator of myelination. J. Cell Biol. 181:625–637 10.1083/jcb.200803013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patzig J., Jahn O., Tenzer S., Wichert S.P., de Monasterio-Schrader P., Rosfa S., Kuharev J., Yan K., Bormuth I., Bremer J., et al. 2011. Quantitative and integrative proteome analysis of peripheral nerve myelin identifies novel myelin proteins and candidate neuropathy loci. J. Neurosci. 31:16369–16386 10.1523/JNEUROSCI.4016-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennuto M., Tinelli E., Malaguti M., Del Carro U., D’Antonio M., Ron D., Quattrini A., Feltri M.L., Wrabetz L. 2008. Ablation of the UPR-mediator CHOP restores motor function and reduces demyelination in Charcot-Marie-Tooth 1B mice. Neuron. 57:393–405 10.1016/j.neuron.2007.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrini A., Previtali S., Feltri M.L., Canal N., Nemni R., Wrabetz L. 1996. Beta 4 integrin and other Schwann cell markers in axonal neuropathy. Glia. 17:294–306 [DOI] [PubMed] [Google Scholar]
- Rangaraju S., Madorsky I., Pileggi J.G., Kamal A., Notterpek L. 2008. Pharmacological induction of the heat shock response improves myelination in a neuropathic model. Neurobiol. Dis. 32:105–115 10.1016/j.nbd.2008.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangaraju S., Verrier J.D., Madorsky I., Nicks J., Dunn W.A., Jr, Notterpek L. 2010. Rapamycin activates autophagy and improves myelination in explant cultures from neuropathic mice. J. Neurosci. 30:11388–11397 10.1523/JNEUROSCI.1356-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D., Walter P. 2007. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 8:519–529 10.1038/nrm2199 [DOI] [PubMed] [Google Scholar]
- Rutkowski D.T., Arnold S.M., Miller C.N., Wu J., Li J., Gunnison K.M., Mori K., Sadighi Akha A.A., Raden D., Kaufman R.J. 2006. Adaptation to ER stress is mediated by differential stabilities of pro-survival and pro-apoptotic mRNAs and proteins. PLoS Biol. 4:e374 10.1371/journal.pbio.0040374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saporta M.A., Shy B.R., Patzko A., Bai Y., Pennuto M., Ferri C., Tinelli E., Saveri P., Kirschner D., Crowther M., et al. 2012. MpzR98C arrests Schwann cell development in a mouse model of ear.onset Charcot-Marie-Tooth disease type 1B. Brain. 135:2032–2047 10.1093/brain/aws140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena S., Cabuy E., Caroni P. 2009. A role for motoneuron subtype-selective ER stress in disease manifestations of FALS mice. Nat. Neurosci. 12:627–636 10.1038/nn.2297 [DOI] [PubMed] [Google Scholar]
- Scherer S.S., Wrabetz L. 2008. Molecular mechanisms of inherited demyelinating neuropathies. Glia. 56:1578–1589 10.1002/glia.20751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuner D., Song B., McEwen E., Liu C., Laybutt R., Gillespie P., Saunders T., Bonner-Weir S., Kaufman R.J. 2001. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol. Cell. 7:1165–1176 10.1016/S1097-2765(01)00265-9 [DOI] [PubMed] [Google Scholar]
- Southwood C.M., Garbern J., Jiang W., Gow A. 2002. The unfolded protein response modulates disease severity in Pelizaeus-Merzbacher disease. Neuron. 36:585–596 10.1016/S0896-6273(02)01045-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svaren J., Meijer D. 2008. The molecular machinery of myelin gene transcription in Schwann cells. Glia. 56:1541–1551 10.1002/glia.20767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabas I., Ron D. 2011. Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nat. Cell Biol. 13:184–190 10.1038/ncb0311-184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taveggia C., Zanazzi G., Petrylak A., Yano H., Rosenbluth J., Einheber S., Xu X., Esper R.M., Loeb J.A., Shrager P., et al. 2005. Neuregulin-1 type III determines the ensheathment fate of axons. Neuron. 47:681–694 10.1016/j.neuron.2005.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ten Asbroek A.L., Van Ruissen F., Ruijter J.M., Baas F. 2006. Comparison of Schwann cell and sciatic nerve transcriptomes indicates that mouse is a valid model for the human peripheral nervous system. J. Neurosci. Res. 84:542–552 10.1002/jnr.20966 [DOI] [PubMed] [Google Scholar]
- Topilko P., Schneider-Maunoury S., Levi G., Baron-Van Evercooren A., Chennoufi A.B., Seitanidou T., Babinet C., Charnay P. 1994. Krox-20 controls myelination in the peripheral nervous system. Nature. 371:796–799 10.1038/371796a0 [DOI] [PubMed] [Google Scholar]
- Tsang K.Y., Chan D., Cheslett D., Chan W.C., So C.L., Melhado I.G., Chan T.W., Kwan K.M., Hunziker E.B., Yamada Y., et al. 2007. Surviving endoplasmic reticulum stress is coupled to altered chondrocyte differentiation and function. PLoS Biol. 5:e44 10.1371/journal.pbio.0050044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsaytler P., Harding H.P., Ron D., Bertolotti A. 2011. Selective inhibition of a regulatory subunit of protein phosphatase 1 restores proteostasis. Science. 332:91–94 10.1126/science.1201396 [DOI] [PubMed] [Google Scholar]
- van der Knaap M.S., Pronk J.C., Scheper G.C. 2006. Vanishing white matter disease. Lancet Neurol. 5:413–423 10.1016/S1474-4422(06)70440-9 [DOI] [PubMed] [Google Scholar]
- Verheijen M.H., Chrast R., Burrola P., Lemke G. 2003. Local regulation of fat metabolism in peripheral nerves. Genes Dev. 17:2450–2464 10.1101/gad.1116203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei M.C., Zong W.X., Cheng E.H., Lindsten T., Panoutsakopoulou V., Ross A.J., Roth K.A., MacGregor G.R., Thompson C.B., Korsmeyer S.J. 2001. Proapoptotic BAX and BAK: a requisite gateway to mitochondrial dysfunction and death. Science. 292:727–730 10.1126/science.1059108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrabetz L., Feltri M.L., Quattrini A., Imperiale D., Previtali S., D’Antonio M., Martini R., Yin X., Trapp B.D., Zhou L., et al. 2000. P(0) glycoprotein overexpression causes congenital hypomyelination of peripheral nerves. J. Cell Biol. 148:1021–1034 10.1083/jcb.148.5.1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrabetz L., Feltri M.L., Kleopa K.A., Scherer S.S. 2004. Inherited neuropathies: clinical, genetic and biological features. Myelin Biology and Disorders. Lazzarini R.A., editor Elsevier Academic Press, S. Diego: 905–051 [Google Scholar]
- Wrabetz L., D’Antonio M., Pennuto M., Dati G., Tinelli E., Fratta P., Previtali S., Imperiale D., Zielasek J., Toyka K., et al. 2006. Different intracellular pathomechanisms produce diverse Myelin Protein Zero neuropathies in transgenic mice. J. Neurosci. 26:2358–2368 10.1523/JNEUROSCI.3819-05.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yum S.W., Kleopa K.A., Shumas S., Scherer S.S. 2002. Diverse trafficking abnormalities of connexin32 mutants causing CMTX. Neurobiol. Dis. 11:43–52 10.1006/nbdi.2002.0545 [DOI] [PubMed] [Google Scholar]
- Zeng L., Lu M., Mori K., Luo S., Lee A.S., Zhu Y., Shyy J.Y. 2004. ATF6 modulates SREBP2-mediated lipogenesis. EMBO J. 23:950–958 10.1038/sj.emboj.7600106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinszner H., Kuroda M., Wang X., Batchvarova N., Lightfoot R.T., Remotti H., Stevens J.L., Ron D. 1998. CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev. 12:982–995 10.1101/gad.12.7.982 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.