miR-612 suppresses local invasion and distant colonization by directly inhibiting AKT2 in HCC.
Abstract
MicroRNAs (miRNAs) play a critical role in tumor metastasis. In this study, we identified a set of 32 miRNAs involved in hepatocellular carcinoma (HCC) metastasis. Among them, miR-612 was shown for the first time to have inhibitory effects on HCC proliferation, migration, invasion, and metastasis. AKT2 was verified to be one of the direct targets of miR-612, through which the epithelial–mesenchymal transition (EMT) and metastasis were inhibited. The level of miR-612 in HCC patients was inversely associated with tumor size, stage, EMT, and metastasis. Of particular importance, miR-612 is involved in both the initial and final steps of the metastatic cascade, by suppressing local invasion and distant colonization. The pleiotropic roles of miR-612 in the HCC metastatic cascade suggest that it could be an effective target for both early and advanced HCC.
As indicated in published statistics, metastases account for 90% of cancer-related deaths (Gupta and Massagué, 2006). Metastasis has always been a bottleneck in tumor prognosis and therapy. The invasive-metastatic cascade is a complex, multistep process that involves the escape of tumor cells from a primary carcinoma, followed by their intravasation into the systemic circulation, survival during transit through the vasculature, extravasation into the parenchyma of distant tissues, establishment of micrometastases, and ultimately outgrowth of macroscopic secondary tumors (Fidler, 2003). Transcriptional expression studies have revealed that sets of genes whose expression profiles in primary tumors are correlated with metastatic relapse and poor survival can be used as prognostic signatures in cancer patients (Ye et al., 2003; Nevins and Potti, 2007). Some of these genes have been validated through functional and clinical studies as candidate mediators of metastasis (Castellano et al., 2008; Sun et al., 2008; Xue et al., 2010). However, few molecules have been implicated in multiple steps of metastatic cascades.
Mature microRNAs (miRNAs) are short, noncoding RNA molecules with 21–24 evolutionarily conserved nucleotides. These small cellular RNAs act as negative regulators of gene expression by binding to the complementary sequences in 3′ untranslated regions (3′UTRs) of specific target mRNAs of protein-coding genes, leading to degradation of the target mRNAs, inhibition of their translation, or both (Ambros, 2004; Bartel, 2004, 2009). Deregulations of miRNAs have been linked to many human diseases, and the central role of miRNAs in the establishment and progression of human tumors has begun to emerge (Voorhoeve et al., 2006; Ma et al., 2007; Tavazoie et al., 2008; Ventura and Jacks, 2009). Recently, high-throughput screening for miRNA expression profiles has revealed that sets of miRNAs (or “signatures”) exist in hepatocellular carcinoma (HCC), which enable clinicians to predict the disease status and prognosis (Ji et al., 2007, 2009; Budhu et al., 2008). Several miRNAs with prometastatic (miR-151, -143, and -30d) or antimetastatic (miR-9, -139, -503, -338, -125, -122, and -101) functions have been identified using HCC cell lines and xenograft animal models (Huang et al., 2009; Li et al., 2009; Tsai et al., 2009; Zhang et al., 2009; Ding et al., 2010; Liang et al., 2010; Tan et al., 2010; Yao et al., 2010; Wong et al., 2011; Zhou and Wang, 2011). However, it remains unclear whether other miRNAs are also involved in the process of HCC invasive-metastatic cascade.
The pleiotropic biological functions of miRNAs, largely attributed to their nonrigorous requirements to bind the sequence motifs of target mRNAs in 3′UTRs, led us to hypothesize that certain miRNAs could be endowed with many different capacities and functions as a crucial modulator in the multistep processes of the invasive-metastatic cascade. To test this, we investigated miRNAs that are differentially expressed between primary tumor and lung-metastasis specimen using HCC xenograft models. We identified for the first time a miRNA, miR-612, with multiple oncobiological functions in the progression of HCC metastasis.
RESULTS
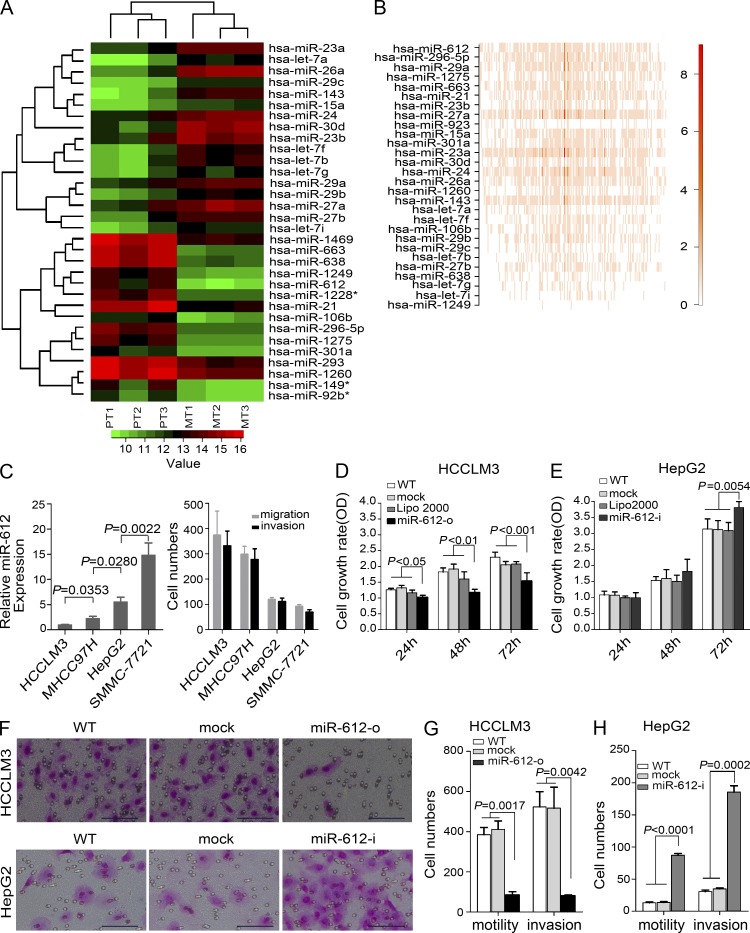
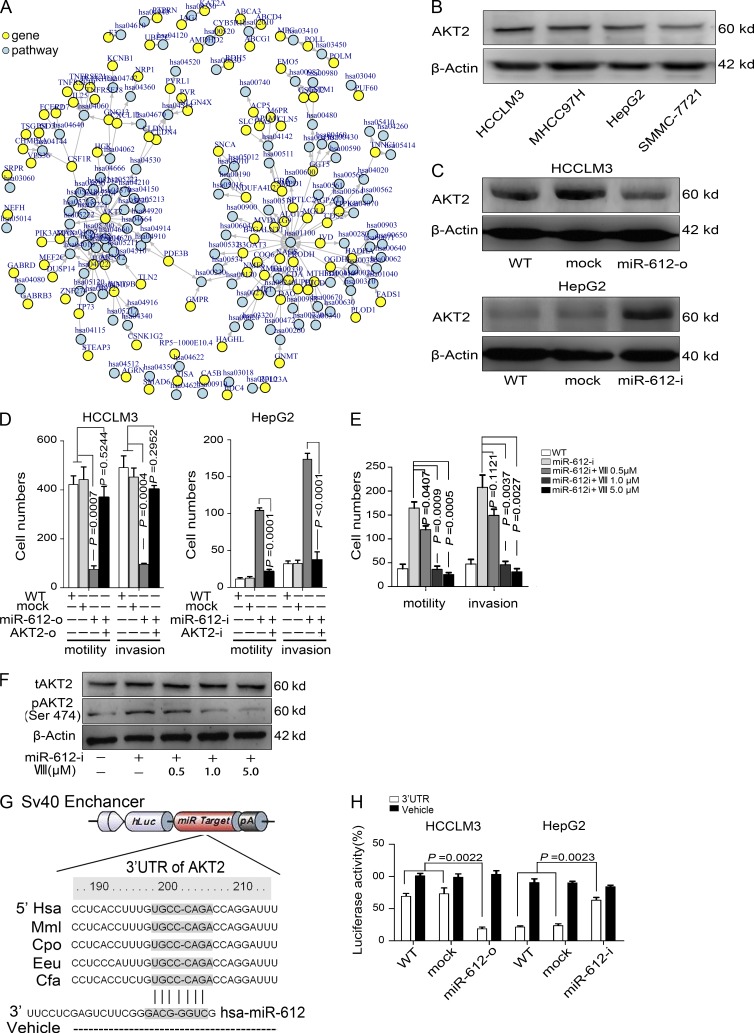
Inverse correlation of miR-612 with HCC metastasis
Array-based human miRNA expression profiles of three primary HCC tissues and their paired lung metastatic tissues from a HCCLM3-RFP xenograft model were determined. Of 1,320 known and predicted human miRNAs, 32 miRNAs were expressed at levels greater than background levels in all specimens and were expressed differentially. Among them, 15 miRNAs were expressed at higher levels in the primary HCC tissues, and 17 miRNAs were expressed at higher levels in the lung metastatic foci. Two distinctive expression clusters of miRNAs were obtained by hierarchical clustering analysis (Fig. 1 A). The five top-ranked miRNAs in the microarray analysis were selected to verify their differential expression by real-time polymerase chain reaction (PCR). miR-612 and miR-296-5p were confirmed to be down-regulated, whereas miR-30d and miR-26a were up-regulated in HCCLM3-RFP lung metastatic foci versus their primary tumor tissues (unpublished data). Bioinformatics analysis of these differentially expressed miRNAs showed that miR-612 was in the top rank of pathway perturbations (Fig. 1 B). Therefore, we focused on whether miR-612 played a pivotal role in HCC invasiveness and metastasis. To answer this question, we investigated the relationship between endogenous miR-612 levels and metastatic potentials. Among four HCC cell lines (SMMC-7721, HepG2, MHCC97H, and HCCLM3) with an ascending order of metastatic potentials, their endogenous level of miR-612 was reversely attenuated (Fig. 1 C, left). These findings indicate that miR-612 may affect HCC metastasis.
Figure 1.
Systematic identification and functional evaluation of HCC metastasis-related miR-612. (A) Hierarchical clustering of 32 differentially expressed miRNAs. The scale bar at the bottom depicts the standard deviation from the mean. PT and MT represent primary tumor and metastatic tumor, respectively. (B) Pathway perturbation assays. The heat map highlights the highest coefficient of miRNA interference on the signal pathway (red bracket). (C) Association between endogenous levels of miR-612 and motility and invasiveness in HCC cells. (C, left) Real-time PCR analysis to quantify the endogenous levels of miR-612 in HCCLM3, MHCC97H, HepG2, and SMMC-7721 cells. U6 was included as a control and data were normalized to the level of HCCLM3 cells. n = 3. (C, right) Transwell analysis to determine motility and invasion abilities of HCC cells. (D and E) Cell proliferation of HCCLM3 and HepG2 after transient transfections with indicated oligonucleotides. n = 6. (F) Representative images of migration and invasion assays. Bars, 100 µm. (G and H) Motility and invasion assays of HCCLM3 and HepG2 cells with indicated treatment. n = 3. WT, Lipo2000, mock, miR-612-o, or miR-612-i were defined as nontransfected, Lipo2000-treated, negative control oligonucleotide transfected, miR-612 mimic transfected, or miR-612 inhibitor transfected, respectively. Data are mean ± SEM and are representative of three independent experiments.
miR-612 negatively regulates HCC proliferation, migration, and invasion
Tumor proliferation, migration, and invasion are the most important steps in the cascade of tumor metastasis. Therefore, we first investigated the effect of miR-612 on HCC proliferation. To optimize our transfection conditions, HCCLM3, HepG2, and SMMC-7721 were treated with different doses of miRIDIAN control oligonucleotide labeled with Dy547. The highest efficiencies were achieved in these cells with a dose at of least 50, 100, and 100 nM, respectively (unpublished data). The level of miR-612 after transient transfection was then evaluated with a luciferase reporter system, which showed that 100 nM of miR-612 mimic (miR-612-o) decreased luciferase activity by more than 2.4 fold in HCCLM3 cells and 100 nM of miR-612 inhibitor (miR-612-i) increased luciferase activity by more than 2.2 and 4.0 fold in HepG2 and SMMC-7721, respectively (unpublished data). The cell proliferation of HCCLM3 and HepG2 was significantly affected by miR-612-o and miR-612-i, respectively. Treatment with miR-612-o resulted in significant inhibition of HCCLM3 proliferation by 32.23%, whereas miR-612-i enabled HepG2 to accelerate its proliferation by 25.77%, compared with the nontransfected (WT) cells. No statistical differences in HCCLM3 and HepG2 proliferation were found in WT, Lipofectamine, and mock groups (Fig. 1, D and E).
Next, we assessed the possible roles of miR-612 on HCC migration and invasion in three independent experiments. First, we investigated the relationship between the endogenous levels of miR-612 and migration and invasion capability in HCCLM3, MHCC97H, HepG2, and SMMC-7721 cells. The results showed that motility and invasiveness were inversely associated with endogenous miR-612 levels (Fig. 1 C, right). Second, we further explored the relationship in nine HCCLM3-RFP–derived monoclonal cells and again found a clear negative correlation between the migration and invasion capabilities and endogenous miR-612 levels (unpublished data). Third, loss- and gain-functions of miR-612 were investigated using a transient transfection strategy with miR-612 mimics or inhibitors. The migration and invasion capabilities of HCCLM3 cells were reduced by more than 4.4 and 6.6 fold, respectively, by miR-612-o compared with the capabilities of control cells (P = 0.0017; P = 0.0042; Fig. 1, F [top] and G). In contrast, in HepG2 cells, migration and invasion increased by more than 6.8 and 6.1 fold with miR-612-i treatment, compared with nontransfected counterparts (P < 0.0001, P = 0.0002; Fig. 1, F [bottom] and H). The promigratory and proinvasive effects of miR-612-i were further confirmed in SMMC-7721 cells (unpublished data). Collectively, our findings suggest that miR-612 negatively regulates tumor migration and invasion of HCC.
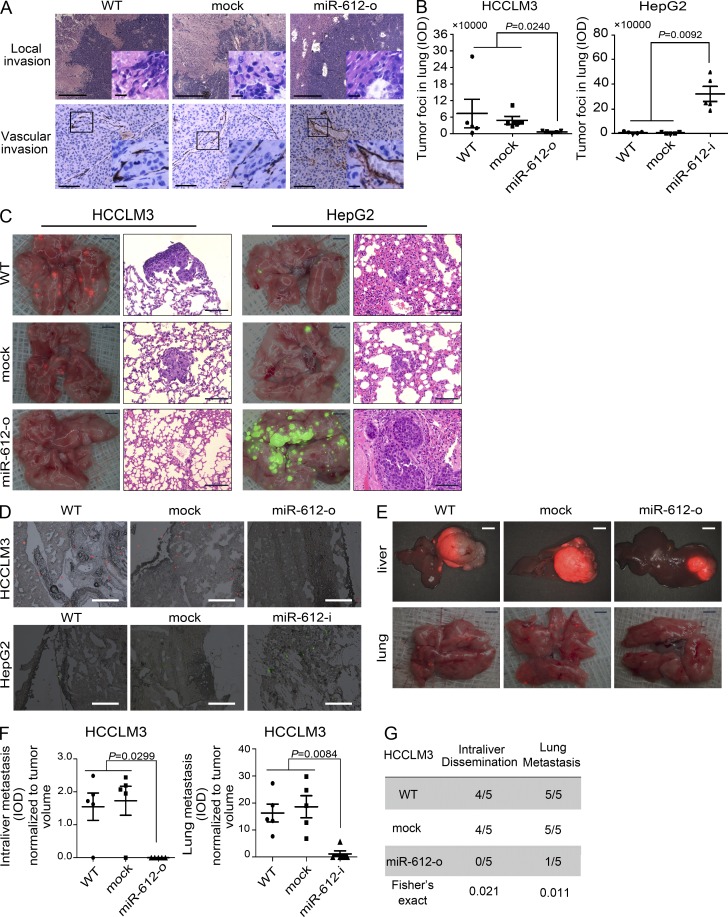
miR-612 suppresses the initial step of tumor metastasis in vivo
Local invasion is the initial step in tumor metastasis. The inhibitory effects of miR-612 on in vitro cell migration and invasion of HCC indicated that miR-612 may also inhibit in vivo metastatic initiation. To address this possibility, we stably overexpressed miR-612 in HCCLM3 (unpublished data), the cell line with the highest metastatic potential, and then established an orthotopic xenograft model in nude mice. After 42 d, WT and mock tumors displayed conspicuous intraliver and microvascular invasion (Fig. 2 A). In stark contrast, miR-612 overexpressed xenograft tumors exhibited almost no local invasion (Fig. 2 A). These findings demonstrate that ectopic forced expression of miR-612 was able to suppress tumor local invasion, the initial step of the metastatic cascade.
Figure 2.
The suppressive effects of miR-612 on HCC invasion, colonization, and distant metastasis. (A) Pathology analysis of tissue sections from recipient mice at week 6 after transplantation. (top) Hematoxylin and eosin (H&E) staining; (bottom) anti–mouse CD34 staining. Boxed area highlights intraliver dissemination and vascular invasion of tumor cells. (B and C) Lung colonization and outgrowth in recipient mice on day 42 after tail vein injection with 1.0 × 105 tumor cells. (B) Quantitation of tumor foci (IOD of fluorescent area) in lung of mice injected HCCLM3 and HepG2 cells. (C) Images showing the colonization and outgrowth foci of HCCLM3-RFP and HepG2-GFP cells in the lung of recipient mice. (D) Images showing the lung colonization in the recipient mice on day 3 after tail vein injection with 1.0 × 105 HCCLM3-RFP and HepG2-GFP cells. (E–G) Intraliver and lung metastasis in the recipient mice on day 42 after HCCLM3-RFP orthotopic transplantation. (E) Images showing metastatic foci in liver (top) and lung (bottom). (F) Quantitation of tumor foci (IOD of fluorescent area) in the liver and lung of recipient mice. (G) Incidence of intraliver dissemination and lung metastasis in WT, mock, and miR-612-o HCCLM3 xenografts. WT, mock, miR-612-o, or miR-612-i was defined as nontransfected, negative control oligonucleotide transfected, miR-612 mimic transfected, or miR-612 inhibitor transfected, respectively. Bars: (A) 100 µm; (C, fluorescent image) 2.0 mm; (C, H&E image) 100 µm; (D) 100 µm; (E, liver) 6.0 mm; (E, lung) 2.0 mm. Data are mean ± SEM (n = 5) and are representative of three independent experiments.
miR-612 suppresses the final step of tumor metastasis in vivo
Colonization and outgrowth at a distant site are the last key step in the metastatic cascade. To test whether miR-612 is involved in this step, a tail vein injection model was used to imitate the pathophysiological process of colonization and outgrowth of disseminated tumor cells. We first injected HCCLM3-RFP cells pretreated with none, mock, or miR-612-o into athymic nude mice via the tail vein. On day 42 after injection, the fluorescent area of tumor foci in the lung of recipient mice was reduced by fourfold in miR-612-o group compared with WT and mock groups (Fig. 2, B and C [left]). We then injected HepG2-GFP cells pretreated with none, mock, or miR-612-i using the same protocol. As expected, inhibition of miR-612 activities resulted in a massive colonization and macrotumor foci of HepG2-GFP in the lung of nude mice (Fig. 2, B and C [right]). Similar results were also found in SMMC-7721, a HCC cell line with a high level of miR-612 and little metastatic potential (unpublished data). To pinpoint the effect of miR-612 on HCC colonization, HCCLM3-RFP and HepG2-GFP cells pretreated as above were injected into the tail vein of nude mice. After 3 d, the number of colonized HepG2-GFP cell blocks in lung was significantly increased in the miR-612-i group compared with controls, whereas the number of HCCLM3-RFP cell blocks was markedly decreased in the miR-612-o treatment group (Fig. 2 D). Thus, miR-612 appears to play a suppressive role in HCC colonization and outgrowth in the last stage of the invasive-metastatic cascade.
miR-612 suppresses metastasis of HCC in vivo
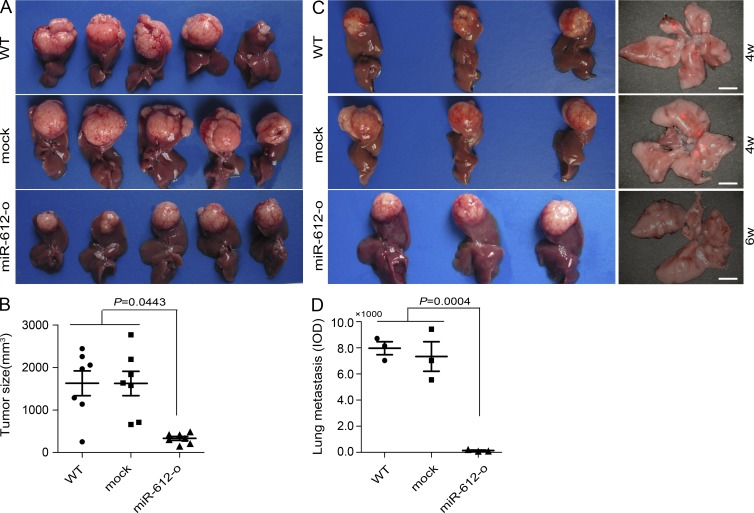
The preceding results indicated that miR-612 could suppress both the initial invasion and final colonization of HCC tumor cells. Thus, we speculated that miR-612 would suppress HCC metastasis in an orthotopic xenograft model. Again, a miR-612 stably overexpressed HCCLM3 xenograft model was used. On day 42 after transplantation, miR-612-overexpressed xenograft tumors displayed significantly less intraliver dissemination and lung metastasis in nude mice compared with WT and mock xenograft tumors (Fig. 2, E–G). The number and size (integrated optical density [IOD] of fluorescence) of metastatic foci normalized to primary tumor volume both in liver and lung of recipient mice were significantly decreased by ectopic overexpression of miR-612 (Fig. 2, E and F). In addition, the ratio of intraliver and lung metastasis in the miR-612 overexpressed group was dramatically lower than that of controls (Fig. 2 G). The average size of miR-612 overexpressed tumors was smaller than that of control tumors (P = 0.0443; Fig. 3, A and B). To account for this difference, we reevaluated the number of lung metastases in HCCLM3-RFP xenograft model with a tumor volume–matched endpoint. The results showed that miR-612 overexpression resulted in a dramatic decrease in lung metastasis (Fig. 3, C and D).
Figure 3.
The suppressive roles of miR-612 on tumor growth and lung metastasis. (A) Images of primary HCCLM3 tumor in liver of recipient mice. (B) Quantitation of tumor sizes in recipient mice at week 6 after indicated treatments. (C and D) Lung metastasis in the recipient mice with the similar tumor volume in HCCLM3-RFP orthotopic xenografts. (C) Images showing the primary tumor volume (left) and metastatic foci in the lung of the recipient mice (right). Bars, (right) 3.0 mm. (D) Quantitation of lung metastatic foci (IOD of fluorescent area) of recipient mice at the similar tumor size endpoint. WT, mock, or miR-612-o were defined as nontransfected, negative control oligonucleotide transfected, or miR-612 overexpressed, respectively. Data are mean ± SEM (n = 5) and are representative of three independent experiments.
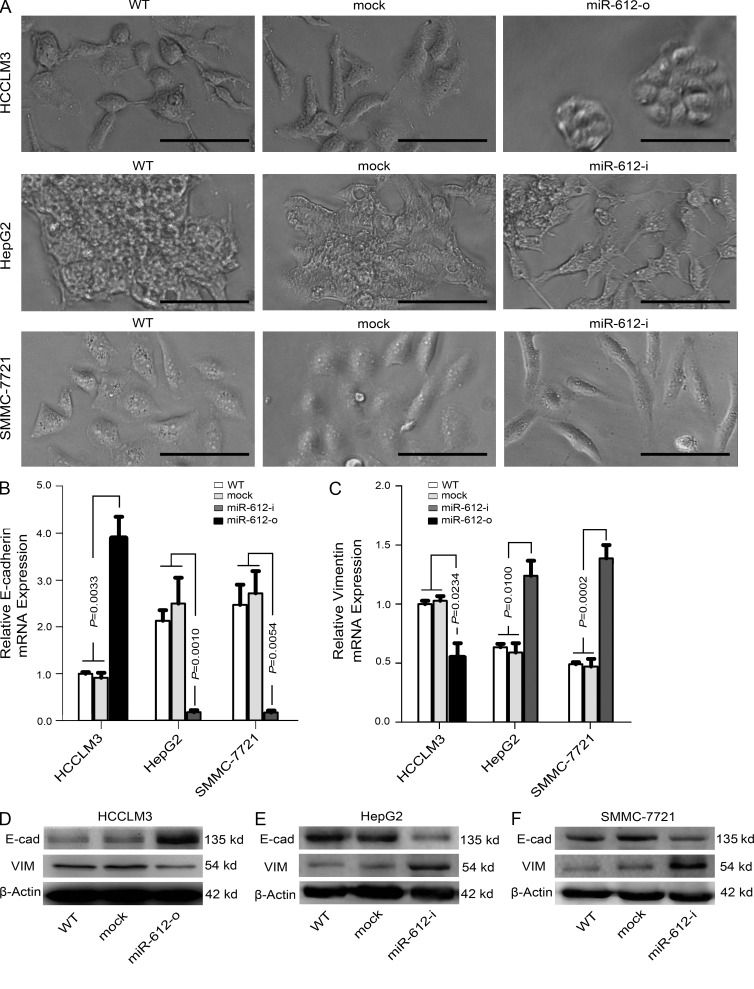
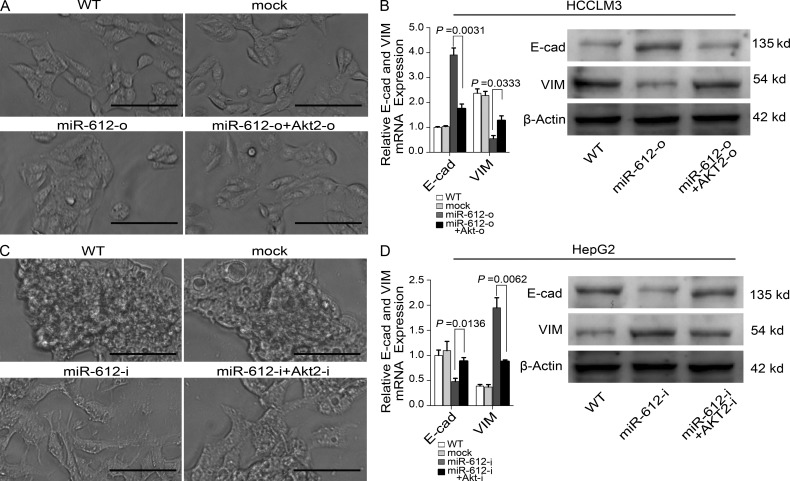
miR-612 suppresses the epithelial–mesenchymal transition (EMT)
A typical morphological change of HCCLM3, HepG2, and SMMC-7721 was noted after treatment with miR-612 mimic or inhibitor. Compared with WT cells, HepG2 and SMMC-7721 cells treated with miR-612-i had more changes resembling the EMT, in which the cells scattered and assumed a spindly or star-like morphology, whereas HCCLM3 cells treated with miR-612-o condensed and assumed a cobblestone-like morphology (Fig. 4 A). These phenomena suggested that miR-612 is involved in the EMT of HCC. Therefore, several putative EMT-related markers, such as E/N-cadherin, β-catenin, and vimentin were evaluated by real-time PCR. The mRNA levels of E-cadherin and vimentin were significantly increased and decreased, respectively, in miR-612-o-treated HCCLM3 cells (Fig. 4, B and C). In contrast, the mRNA levels of E-cadherin and vimentin were significantly decreased and increased in both miR-612-i–treated HepG2 and SMMC-7721 cells (Fig. 4, B and C). No significant differences in the expression of other genes were found (unpublished data). The protein changes of these genes were further confirmed by Western blots (Fig. 4, D–F). These observations indicate that miR-612 can suppress the EMT of HCC.
Figure 4.
The inhibitory roles of miR-612 on EMT of HCC cells. (A) Image showing the morphological changes of HCCLM3, HepG2, and SMMC-7721 after treatment with indicated reagent at hour 72. Bars: (HCCLM3 and SMMC-7721 cell) 100 µm; (HepG2 cell) 200 µm. (B and C) Real-time PCR analysis to quantify the level of E-cadherin and vimentin in HCCLM3, HepG2, and SMMC-7721 cells. β-Actin was included as a loading control and all data were normalized by WT cells. (D–F) Western blot analysis to evaluate the protein levels of E-cadherin (E-cad) and vimentin (VIM) in HCCLM3, HepG2, and SMMC-7721. β-Actin was used as a loading control. WT, mock, miR-612-o, or miR-612-i were defined as nontransfected, negative control oligonucleotide transfected, miR-612 mimic transfected, or miR-612 inhibitor transfected, respectively. Data are mean ± SEM (n = 3) and are representative of three independent experiments.
AKT2 is a direct target of miR-612
To elucidate the underlying mechanisms of the suppressive effects of miR-612 on EMT and metastasis of HCC, we used several bioinformatics methods to help identify the target human genes of miR-612. Among the 167 targets predicted by the search programs of miRanda, TargetScan, and miRTarget2, AKT2 was determined to be the gene that localized at one of two main centers of the net comprised of 104 predicted genes and pathways (Fig. 5 A). This finding suggested that AKT2 is a key target of miR-612. Interestingly, the basal levels of AKT2 in several HCC cell lines were negatively correlated with the endogenous levels of miR-612 (Fig. 1 C [left] and Fig. 5 B). Furthermore, the level of AKT2 in HCCLM3 cells was dramatically reduced by miR-612-o, whereas that in HepG2 (Fig. 5 C) and SMMC-7721 (unpublished data) cells was significantly promoted by miR-612-i. The promoting effects of miR-612-i on HepG2 and SMMC-7721 migration and invasion was mostly compromised by AKT2 knockdown mediated by shRNA (AKT2-i) (Fig. 5 D), whereas restoration of AKT2 (AKT2-o) with the expression vector lacking the miR-612 target site in the 3′UTR increased the motility and invasiveness of miR-612-o–treated HCCLM3 cells (Fig. 5 D). In addition, we found one AKT2 inhibitor (AKT inhibitor VIII, VIII) that could abolish the pro-metastatic role of miR-612-i on HepG2 cells in vivo in a dose-dependent manner, and that was notably associated with the attenuated level of phosphorylated AKT2 (Ser 474) in a dose-dependent manner without affecting the total level of AKT2 (Fig. 5, E and F). In other words, AKT2 is a functional target gene of miR-612.
Figure 5.
Direct regulation of miR-612 on AKT2 expression. (A) Bioinformatics analysis to predict target gene of miR-612 and assay pathway perturbation. (B) Western blot analysis to quantify endogenous levels of AKT2 in HCCLM3, MHCC97H, HepG2, and SMMC-7721 cells. (C) Protein levels of AKT2 in HCCLM3 and HepG2 cells after indicated treatments were detected by Western blots. (D) Motility and invasion assays for HCCLM3 (left) and HepG2 (right) cells. (E) Motility and invasion abilities of HepG2 treated with 0.5– 5.0 µM of AKT2 inhibitor VIII (VIII). (F) The levels of total and phosphorylated AKT2 (t-and p-AKT2) in HepG2 cells after treatment with AKT2 inhibitor were detected by Western blots. β-actin was a loading control. (G) Schematic diagram of the dual luciferase miRNA target reporter vector. The conserved seed sequence (gray) in the 3′UTR of AKT2 was cloned into pmirGLO dual luciferase reporter system. Vehicle vector was included as a negative control. (H) Luciferase activity was assayed in miR-612-o–transfected HCCLM3 cells and miR-612-i–transfected HepG2 cells. The amount was normalized to WT cells. WT, mock, miR-612-o, miR-612-i, AKT2-o, or AKT2-i were defined as nontransfected, negative control oligonucleotide transfected, miR-612 mimic transfected, miR-612 inhibitor transfected, AKT2 overexpressed, or AKT2 knocked-down, respectively. Data are mean ± SEM (n = 3) and are representative of three independent experiments.
To test whether AKT2 is a direct target gene of miR-612, we cloned a 3′UTR element of AKT2, which has a conserved sequence of mRNA orthologous in Hsa, Mml, Cpo, Eeu, and Cfa and is complementary to the seed sequence of miR-612 (Fig. 5 G), into a dual luciferase reporter, and then transfected it into HCCLM3 and HepG2 cells. The basal levels of luciferase activity in HepG2 cells were lower than those in HCCLM3 cells, and the levels of luciferase activity in HCCLM3 and HepG2 cells could be reversed by miR-612-o and miR-612-i, respectively (Fig. 5 H). Thus, we conclude that AKT2 is a direct target gene of miR-612.
miR-612 negatively regulates EMT through the AKT2 pathway
To test whether miR-612 suppresses EMT through the AKT2 pathway, we evaluated the relationship between AKT2 and miR-612–associated EMT. Restoration of miR-612 expression in HCCLM3 cells caused a morphological change from spindly to cobblestone-like (Fig. 6 A), whereas inhibition of miR-612 expression caused EMT changes in HepG2 (Fig. 6 C). Through the knock-down of AKT2 with shRNAs (AKT2-i) in miR-612-i-treated HepG2 cells, mesenchyma-like cells returned to the epithelial phenotype, whereas co-transfection of miR-612-o with an AKT2 expression vector lacking the miR-612 target site in the 3′UTR abolished the cobblestone-like change in the phenotype of HCCLM3 cells (Fig. 6, A and C). The morphological changes were followed by changes in the mRNA and protein levels of E-cadherin and vimentin (Fig. 6, B and D). These findings suggest that miR-612 regulates the EMT of HCC cells through suppression of AKT2.
Figure 6.
Modulations of miR-612 on EMT through AKT2 pathway. (A) Image showing the morphological changes of HCCLM3 after indicated treatment at hour 72. (B) Real-time PCR and Western blot analysis to quantify the mRNA and protein levels of E-cadherin (E-cad) and Vimentin (VIM) in HCCLM3 cells. β-Actin was a loading control, and the amount was normalized by WT cells in PCR experiments (WT). (C) Image showing the morphological changes of HepG2 after treatment with indicated reagent at hour 72. (D) Real-time PCR and Western blot analysis to quantify the mRNA and protein levels of E-cadherin (E-cad) and Vimentin (VIM) in HepG2 cells. β-Actin was used as a loading control, and the amount was normalized by WT cells in PCR experiments (WT). WT, mock, miR-612-o, miR-612-i, AKT2-o, or AKT2-i were defined as nontransfected, negative control oligonucleotide transfected, miR-612 mimic transfected, miR-612 inhibitor transfected, AKT2 overexpressed, or AKT2 knocked-down, respectively. Bars: (A) 100 µm; (C) 200 µm. Data are mean ± SEM (n = 3) and are representative of three independent experiments.
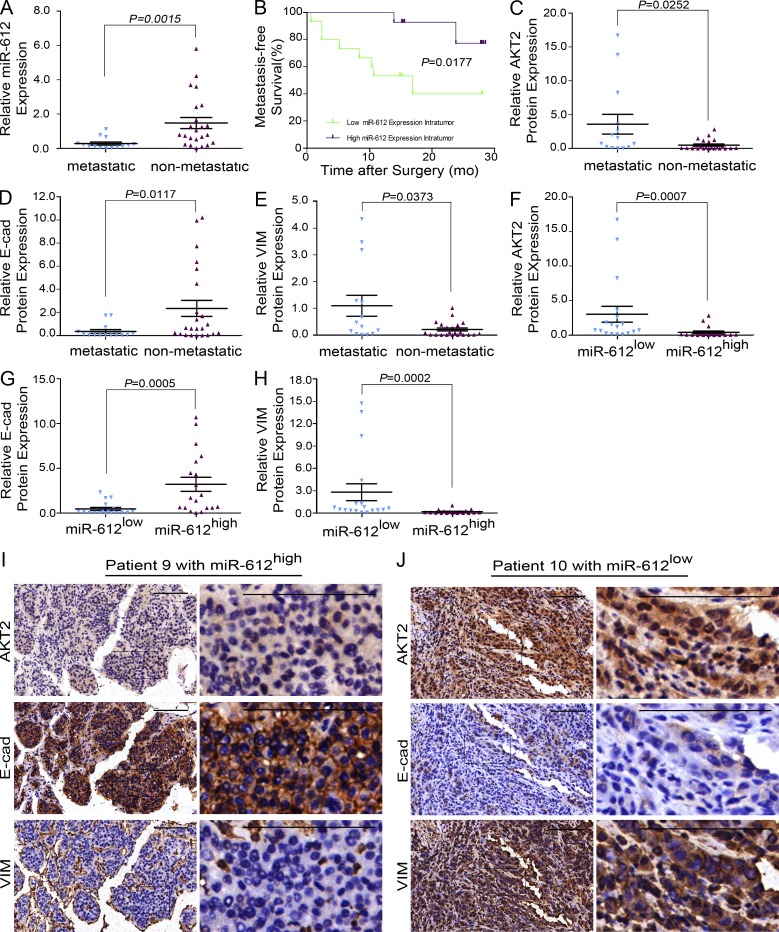
miR-612, AKT2, E-cadherin, and vimentin expression in HCC patients
Based on the similar results of experiments using HCC cell lines and xenograft models, it appears that the decreased expression of miR-612 would induce the EMT, and thus promote HCC invasion and metastasis through the AKT2 pathway. To investigate whether this is true in HCC patients, the relationship between miR-612 level and metastatic status was first tested in 37 archived primary liver tumors using real-time PCR. The clinicopathological features of these patients are listed in Table S1. Tumors from 14 patients who had portal vein thrombosis, microvascular invasion, and intrahepatic dissemination tended to express lower levels of miR-612 compared with those from nonmetastatic patients (0.2771 ± 0.0886 vs. 1.475 ± 0.3217; P = 0.0015; Fig. 7 A). The low expression levels of miR-612 were correlated with poor metastasis-free survival (P = 0.0177; Fig. 7 B). Four of the 18 patients with low levels of miR-612 developed lung (3 patients) and bone (1 patient) metastasis within 1 yr, whereas no distant metastases were found in the 19 patients with high levels of miR-612 (4/18 vs. 0/19; P = 0.046). In addition, the level of miR-612 had a significant inverse correlation with tumor size and stage, intrahepatic metastasis, and microvascular invasion (Table S2). These findings revealed a significant association between the low expression of miR-612 and HCC local dissemination, as well as distant metastasis.
Figure 7.
Clinical validation of miR-612, AKT2, E-cadherin, and vimentin. (A) Real-time PCR analysis to quantify the endogenous levels of miR-612 in HCC patients with or without local dissemination/metastasis. U6 was used as a loading control. The amount of miR-612 was normalized to the mean value of all patients. (B) Kaplan-Meier curves of 37 HCC patients after stratification by the level of miR-612 were used for depicting metastasis-free survival. (C–E) Relative levels of AKT2, E-cadherin, and vimentin in HCC patients with or without local dissemination/metastasis. (F–H) Relative levels of AKT2, E-cadherin, and vimentin in patients with low and high miR-612 expression levels. (I and J) Images to visualize positive staining of AKT2, E-cadherin, and vimentin in patients with low and high miR-612 expression levels. The slides are constructed by serial section method. Bars: (left) magnification ×100, (right) magnification ×400. Data are mean ± SEM and are representative of three independent experiments.
Next, we investigated whether miR-612 level was associated with the protein levels of AKT2 and EMT biomarkers in these patients. Immunohistochemical assays revealed distinct expression patterns of AKT2, E-cadherin, and vimentin in the 14 metastatic and 23 nonmetastatic patients. Higher protein levels of AKT2 and vimentin were found in the metastatic patients, whereas a higher protein level of E-cadherin was found in the nonmetastatic patients (Fig. 7, C–E). Furthermore, tumors with a high level of miR-612 tended to express low levels of AKT2 and vimentin and a high level of E-cadherin (as illustrated in patient 9 with no metastasis), whereas tumors with a low level of miR-612 (as seen in patient 10 with local metastasis) tended to express high levels of AKT2 and vimentin and a low level of E-cadherin (Fig. 7, F–J). Hence, it appears that miR-612 negatively regulates the EMT and tumor metastasis in HCC patients by targeting AKT2.
DISCUSSION
Genome-wide miRNA analysis with the miRAGE method revealed that miR-612 was down-regulated in tumors versus nontumorous colorectal tissues (Cummins et al., 2006). However, the biological function of miR-612, especially in the progression of cancer, was unknown. In this study, we revealed for the first time that miR-612 has pleiotropic inhibitory effects on cell proliferation, migration, invasion, and metastasis of HCC. We also verified that the levels of miR-612 were inversely associated with tumor size and stage, local invasion, and distant metastasis in HCC patients, but were positively correlated with metastasis-free survival. These data indicate that miR-612 suppresses metastasis in HCC, but it is unclear whether miR-612 plays an antimetastatic role in other types of tumors. Therefore, we modulated miR-612 level in the colon cancer cell line HCT116 and found that overexpression of miR-612 reduced liver metastasis of colon cancer in a HCT116 spleen xenograft mice model and vice versa (Unpublished data). In the present study, we further verified that AKT2 is one of direct targets of miR-612, and its suppressive role on HCC metastasis was facilitated by EMT inhibition through the AKT2 pathway.
The complex invasive-metastatic cascade can be conceptually simplified into two major phases: one is the physical translocation of cancer cells from the primary tumor to a distant tissue, and the other is the colonization and outgrowth of translocated cells within a secondary organ (Chaffer and Weinberg, 2011). We scrutinized the roles of miR-612 in the HCC metastatic cascade step by step. For successful translocation, HCC cells must acquire the ability of migration and invasion in the initial stage. Indeed, up-regulation of miR-612 was able to suppress HCC cell invasion into the surrounding normal liver and microvascular tissue in nude mice model. Clinical data also confirmed that miR-612 levels were inversely associated with intrahepatic metastasis and microvascular invasion of patients. These findings suggest that miR-612 plays a suppressive role in the initial step of the HCC metastatic cascade.
Tumor colonization is an utterly inefficient and rate-limiting step in the metastatic cascade, in which millions of cancerous cells may be released by a primary tumor into systemic circulation every day, but only a small minority of these cells will colonize a distant organ (Gupta and Massagué, 2006). This study also underscored the influence of miR-612 during this stage using a tail vein injection model, which is theoretically analogous to the colonization and outgrowth phase of the tumor metastatic cascade. Our results showed that overexpression of miR-612 strikingly diminished the number of lung tumor foci in HCCLM3-RFP xenograft model, and the loss of function of miR-612 markedly enhanced lung colonization in HepG2. These results suggest that miR-612 plays a pivotal role in multiple rate-limiting steps in the metastatic cascade.
Obtaining an understanding of the process of metastatic dissemination of cancer cells is in sight. The EMT appears to be essential for cancerous cells to acquire the capability of migration and invasion and is a key driver of tumor cell translocation (Polyak and Weinberg, 2009). This idea is supported by our finding that miR-612 negatively regulated EMT, local invasion, and dissemination of HCC. As tumor colonization and outgrowth in a distant organ seems to represent a far more complex set of phenomena with a relatively small number of unifying, generalizable principles (Chaffer and Weinberg, 2011), we cannot yet define the underlying mechanism of miR-612 in this process. However, a cancer stem cell (CSC)–like state of HCC cells induced by EMT, accompanied by gaining the traits of self-renewal and apoptosis resistance (Charafe-Jauffret et al., 2009; Thiery et al., 2009), may provide a reasonable explanation of the role of miR-612 in this stage. This hypothesis is based on the following observations: (a) overexpression of miR-612 greatly inhibited in vitro cell proliferation and suppressed in vivo tumor growth of HCC in nude mice; (b) restoration of miR-612 first reduced HCCLM3 cells blocks in lung and then inhibited the outgrowth to a second tumor, whereas the inhibition of miR-612 in HepG2 cells resulted in the opposite results; and (c) tumors of patients with high levels of miR-612 were two times smaller than those of patients with low levels of miR-612. Therefore, we speculate that miR-612 may interfere with the initiation of a CSC-like state of HCC through the inhibition of the EMT, which thus suppresses tumor growth and metastasis. Although the detail mechanisms need to be further investigated, miR-612 did suppress both the initial and final steps of the invasive-metastatic cascade of HCC.
The full spectrum of signaling agents that contribute to EMTs of carcinoma cells remains unclear. Evidence indicates that activation of the EMT process during tumorigenesis and progression most often requires signals from reactive stroma (Yang and Weinberg, 2008). Recently, the involvement of miRNAs in the EMT process was demonstrated (Bracken et al., 2008; Gregory et al., 2008; Park et al., 2008). For example, miR-205 and miRNAs from the miR-200 family (miR-200a, -200b, -200c, -141, and -429) are closely related with regulation of the EMT. ZEB1 and ZEB2/SIP1 are subsequently recruited by these miRNAs during the process. Here, we reported a novel miRNA, miR-612, whose expression level was inversely correlated with EMT phenotype. Forced down-regulation or a low endogenous level of miR-612 expression was sufficient to induce the EMT of HCC cells, which were observed in our in vitro system (without stroma), xenograft model of nude mice (with immunocompromised stroma), and clinical HCC samples (with immunocompetent stroma), suggesting that the genetic alteration during tumor progression made HCC cells more aggressive in a stroma-independent manner.
Collectively, the findings of the present study have significant implications regarding our understanding of the pathogenesis of this high-grade malignancy. The pleiotropic roles of miR-612 on the early and final steps of the HCC invasive/metastatic cascade suggest that it could be a useful target for interfering with HCC metastasis in clinical practice.
MATERIALS AND METHODS
Cell lines.
Four human hepatoma cell lines and nine monoclonal cell strains from HCCLM3-RFP cells were used in this study. HepG2 and SMMC-7721 cells were purchased from the Shanghai cell bank, Chinese Academy of Sciences. MHCC97H, HCCLM3, and its red fluorescence protein (RFP)–expressing derivatives were described previously (Sun et al., 1999; Li et al., 2004; Yang et al., 2008). HepG2-GFP cells were established previously by our group (Yang et al., 2008). Human colon cancer cell HCT116 was provided by C.H. Tang (Minghong Biotechnology Inc., Shanghai, China). All these cells were cultured under standard conditions.
Constructs, oligonucleotides, and transfection.
Three GV248 lentiviral particles containing short hairpin sequences targeting the human AKT2 gene and the GV142 vector with human AKT2 complementary DNA without miR-612 targeting sites in 3′UTR, as well as the negative plasmids, were purchased from GeneChem. The three pooled shRNA sequences that yielded the greatest knockdown were: 5′-GCACAGGTTCTTCCTCAGCATCTCGAGATGCTGAGGAAGAACCTGTGC-3′, 5′-ACAAGGTACTTCGATGATGAACTCGAGTTCATCATCGAAGTACCTTGT-3′, and 5′-CCCTTAAACAACTTCTCCGTACTCGAGTACGGAGAAGTTGTTTAAGGG-3′. Primers for amplification of human AKT2 were as follows: 5′-CCAACTTTGTGCCAAAAAGTTGGC-3′ (sense) and 5′-AATGCCAACTCTGTCCAAGAAAG-3′ (antisense). The oligonucleotides including miRIDIAN miR-612 hairpin inhibitor, mimic, and negative control (Thermo Fisher Scientific) for inhibition and restoration of miR-612 were used in this study. All these constructs and oligonucleotides were transfected into HCC cells using Lipofectamine 2000 according to the product manual (Invitrogen). A luciferase reporter system that contains putative recognition elements of miR-612 was used to evaluate miR-612 levels, and Western blot was used to determine the level of AKT2 after oligonucleotide transfection.
Luciferase reporter assays.
The binding sites for miR-612 in the 3′UTR sequence of AKT2 were cloned into the pmirGLO Dual-Luciferase miRNA Target Expression Vector (Promega). The miR-612 recognition elements were as follows: 5′-AAACTAGCGGCCGCCCTCACCTTTGTGCCCAGACCAGGATTTT-3′ (sense) and 5′-CTAGAAAATCCTGGTCTGGGCACAAAGGTGAGGGCGGCCGCTAGTTT-3′ (antisense). AKT2 complementary DNA was purchased from Fulengen Co. Ltd. These constructs (1.0 µg) were co-transfected into target cells in 24-well plates together with 100 nM of miR-612 hairpin inhibitors or mimic using Lipofectamine 2000. Luciferase activity was measured 24 h after transfection using the Dual-Luciferase Reporter Assay System (Promega). The levels of firefly luciferase activities were obtained by normalizing to Renilla luciferase activities and relative to a control, as previously reported (Tavazoie et al., 2008).
Stable expression of miR-612 with lentiviral vector.
Human miR-612 gene was PCR amplified from normal genomic DNA and cloned into pEZX-AM03-mCFP (mCherry Fluorescent Protein) for ectopic expression of miR-612. Primers used for amplification were 5′-CAACAGAAGGCTCGAGTCAGGATGAGTAGAAGGCAC-3′ (sense) and 5′-AATATACCTTCTCGAGACACCACTGCTGTTGGCTC-3′ (antisense; XhoI site marked as bold). The following primers were used for PCR to confirm the insertion: 5′-ATGAGGCTTCAGTACTTTACAG-3′ (MIR30-F) and 5′-CATAGCGTAAAAGGAGCAACA-3′ (WPRE-R). A scrambled shRNA clone (mock) (CmiR-AN0001-AM03, GeneCopoeia, Inc.) was used as a negative control. The lentiviral particles were prepared as described previously (Lois et al., 2002). HCCLM3 cells were infected with mock or miR-612–overexpression virus and selected by Ampicillin (Stewart et al., 2003). The mCFP+ cells accounted for >90% of infected HCCLM3 cells.
Analysis of miRNA expression.
For analysis of miRNA expression, we used the CapitalBio Mammalian miRNA microarray and the service to process the samples (CapitalBio Inc.). The primary tumors and their paired lung metastatic specimens were isolated from orthotopic xenograft models of HCCLM3-RFP cells (Sun et al., 1996; Yang et al., 2008). Total RNAs were prepared with TRIzol Reagent (Invitrogen). miRNAs were purified by precipitation methods with PEG solutions (Thomson et al., 2004; Watanabe et al., 2005). A total of 4 µg RNA from each sample was labeled with Cy3 (Thermo Fisher Scientific), as described previously (Thomson et al., 2004), and hybridized to mammalian miRNA chips containing capture probes against all known human miRNAs (Sanger miRBASE version 12.0). After hybridization, signals were scanned, quantified, and normalized with a global mean algorithm. Differentially expressed miRNAs were selected according to the following criteria: the change fold was no less than two times and Q value was set at 5% (SAM, version 2.1; He et al., 2005). Hierarchical clustering of variance-normalized expression values was performed with Cluster 3.0 software for Windows using the metric of Euclidean distance and average linkage. The raw and normalized microarray data have been deposited in the National Institutes of Health NCBI Gene Expression Omnibus under accession no. GSE26323.
RNA extraction and real-time PCR assays for miRNA and mRNA detection.
Total RNA was extracted from cultured cells or freshly iced HCC tissues with TRIzol Reagent (Invitrogen). The quality of RNA was examined by A260 absorption, and then 2 µg of total RNA were used for first-strand DNA synthesis. Real-time PCR was performed in triplicate by the SYBR Green PCR method using an All-in-One miRNA qPCR Detection kit (GeneCopoeia Inc.). The following forward primers were synthesized by GeneCopoeia: 5′-TTCAAGTAATCCAGGATAGGCTAAA-3′ for hsa-miR-26a, 5′-GCCCCCCCTCAATCCTGTAA-3′ for hsa-miR-296-5p, 5′-AACATCCCCGACTGGAAGAA-3′ for hsa-miR-30d, 5′-GGGCGGGTGGCGGCCTAAA-3′ for hsa-miR-638, 5′-GCAGGGCTTCTGAGCTCCTTAA-3′ for hsa-miR-612, and 5′-CAAATTCGTGAAGCGTTCCATAT-3′ for U6 small nuclear RNA (U6). The common reverse primer was purchased from the same company. For mRNA detection, 500 ng of total RNA were used for complementary DNA synthesis with a PrimeScript RT reagent kit (Takara Bio, Inc.). Real-time PCR was performed in triplicate using SYBR Premix Ex Taq (Takara Bio, Inc.). The primers for the genes of interest (E/N-cadherin, β-catenin, vimentin) were synthesized by Sangon Biotech Co., Ltd., as follows: 5′-CTGCAGGTCTCATCATGGA-3′ (forward) and 5′-ACCTGTAGACCTCGGCACTG-3′ (reverse) for E-cadherin; 5′-CCGTGAATGGGCAGATCACT-3′ (forward) and 5′-TAGGCGGGATTCCATTGTCA-3′ (reverse) for N-cadherin; 5′-GAGTCCACTGAGTACCGGAGAC-3′ (forward) and 5′-TGTAGGTGGCAATCTCAATGTC-3′ (reverse) for vimentin; 5′-ATAGCACAGCCTGGATAGCAACGTAC-3′ (forward) and 5′-CACCTTCTACAATGAGCTGCGTGTG-3′ (reverse) for β-catenin; and 5′-CACCATGAAGATCAAGATCATTGC-3′ (forward) and 5′-GGCCGGACTCATCGTACTCCTGC-3′ (reverse) for β-actin. The U6 small nuclear RNA and β-actin were used, respectively, as internal control for miRNAs and mRNAs assays. The threshold cycle (Ct) values were analyzed using the comparative Ct (–ΔCt) method adapted as previously reported (Landgraf et al., 2007). The level of targets was obtained by normalizing to the endogenous reference and relative to a control.
Inhibition of AKT2 activation.
AKT inhibitor VIII (Isozyme-selective, AKTi-1/2) (EMD Millipore) was used to inhibit AKT2 activation and its corresponding downstream signal pathway. Final doses of AKT inhibitor VIII ranging from 0.5 to 5 µM were used in the experiments.
In vitro cell proliferation assays.
Cell proliferation was determined by the Cell Counting kit-8 (Dojin Laboratories) assay. In brief, HCCLM3 and HepG2 cells were seeded into 96-well plates at an initial number of 2,000 cells per well. At 24, 48, or 72 h after oligonucleotide transfection, 10 µl of the kit reagent was added to each well, and 2 h later all plates were scanned by a microplate reader (Thermo Fisher Scientific) at 450 nm. Cell proliferation was calculated on the basis of absorbency.
In vitro migration and invasion assays.
Cell migration and invasion were analyzed by a Transwell Permeable Supports system with 8-µm pores (Corning). For motility assays, 5 × 104 cells were seeded into upper uncoated inserts; for invasion assays, 1.0 × 105 cells were seeded into upper inserts with a Matrigel-coated membrane (BD). Cells were seeded in 1% serum medium and translocated to 10% serum media for 24 or 48 h. After removal of the nonmigrating or noninvading cells, the remaining cells were fixed, stained, and analyzed by inverted microscopy. A total of 100 nM miR-612 inhibitors, mimics, or mocks were transfected 60 h before plating using Lipofectamine 2000, as well as AKT2 shRNA (60 nM) or expression vector (80 nM). AKT inhibitor VIII at a concentration ranging from 0.5 to 5.0 µM was used to treat cells for 60 h before seeding.
miRNA-target prediction, pathway perturbation, and miRNA ranking
In silico target genes of all human miRNAs were predicted by miRanda, TargetScan, and miRTarget2 algorithms. The predicted gene accordance in at least two algorithms were retained and used for KEGG pathway annotation, which was obtained from the “has_pathway.list” file downloaded from KEGG ftp (ftp://ftp.genome.jp/pub/kegg/). Next, the miRNA pathway relationships were achieved according to the relationships between miRNAs and genes and between genes and the pathway (namely the earlier annotated KEGG pathway of the genes). A miRNA-pathway-weighted matrix MP (m, n) was composed to denote the relationships between all m miRNAs and n pathways. Furthermore, pathway perturbation by differentially expressed miRNAs (*) was evaluated, and the pseudo pathway expression profile was constructed. The GlobalAncova test (Hummel et al., 2008) was performed on the pseudo pathway expression profile to estimate the perturbation degree in samples from primary tumor to lung metastasis. The perturbation degree of each pathway was judged by the resultant F value. Finally, miRNAs were ranked according to their contributions to pathway perturbation. All statistical analyses were performed using R/Bioconductor software (http://www.bioconductor.org).
Protein levels detected by Western blot analysis.
Lysates were obtained from cultured cells with a mixture of ProteoJET Mammalian Cell Lysis Reagent (Fermentas) and phenylmethanesulfonyl fluoride (Roche) and PhosSTOP (Roche). Proteins were separated by sodium dodecyl sulfate-PAGE and transferred onto polyvinylidene fluoride membranes. After being blocked in 5% nonfat milk or bovine serum albumin; probed with antibodies against human AKT2 (1:1,000; Cell Signaling Technology), phospho-AKT2 (S474) (1:1,000; Abcam), E-cadherin (1:1,000; Cell Signaling Technology), vimentin (1:500; Bioworld Technology Inc.), and β-actin (1:10,000; Abcam); incubated with goat anti–rabbit or anti–mouse IgG (1:10,000 for both; Jackson ImmunoResearch Laboratories); and detected with enhanced chemiluminescence reagents (Thermo Fisher Scientific). Bands were acquired by Molecular Imager ChemiDox XRS+ Imaging System with Quantity One Image software (Bio-Rad Laboratories) and quantified with Gel-Pro Analyzer (United Bio).
In vivo tumor invasion, lung colonization, and metastasis assays.
Male athymic BALB/c nude mice (4 wk old) were used for animal studies. An orthotopic human HCC xenograft model was established for in vivo tumor invasion and metastasis analysis (Thomson et al., 2004). HCCLM3-mCFP cells with stably overexpressed miR-612 were established and transplanted into livers of nude mice. The experiments were designed to have two independent parts with the corresponding endpoints of 42 d and similar tumor volume. At the endpoint, all mice were sacrificed. Tumor sizes were evaluated by the formula: Volume (mm3) = [width2 (mm2) × length (mm)]/2. Intraliver dissemination and lung metastasis were determined by fluorescent areas using Image-Pro Plus software 6.0 (Media Cybernetics Inc.), as described previously (Yang et al., 2008), and by pathological examinations. The vascular in tumor microenvironments was labeled with antibody against mice CD34, and tumor cell invasion was evaluated by pathological examinations. A tail vein injection model was used for lung colonization assays (Tavazoie et al., 2008). The experiment also contained two independent parts with endpoints of 42 d and 3 d later. In brief, mice were injected with 105 viable HCCLM3-RFP, HepG2-GFP, or SMMC-7721-GFP cells via a lateral tail vein. miRIDINA miR-612 hairpin inhibitor, mimic, and mock with final concentrations of 100 nM was transfected two consecutive times before injections using an adapted method (Gregory et al., 2008). Lung colonization and outgrowth was quantified by fluorescent area and pathological examination. The animals used in a single experiment were littermates. All procedures were approved by the Animal Care and Use Committee of Shanghai, China.
Patient selection, evaluation, and clinical validation of miR-612.
A total of 37 archived consecutive patients who underwent curative liver resection with pathologically confirmed HCC were randomly enrolled. The procedures were approved by the Zhongshan Hospital Research Ethics Committee. Informed consent was obtained from each patient according to the committee’s regulations. Tumor stage was determined according to the 2002 International Union against Cancer TNM classification system. Patients were followed until 5 July 2011, with a median follow up of 15.6 mo. Metastasis was detected by computed tomography scanning or magnetic resonance imaging and confirmed by pathological examinations. Metastasis-free survival was defined as the interval between surgery and metastasis. Low/high expression of miR-612 was classified by the median. Specimens were collected, snap-frozen immediately, and stored at −80°C. Total RNA was isolated from frozen tissues with TRIzol reagent (Invitrogen), and real-time PCR was performed as described above.
Immunohistochemistry assays.
Immunohistochemistry for the target molecules was performed on single serial sections made from surgical samples. The slides were probed with a primary antibody against AKT2 (1:200; Cell Signaling Technology), E-cadherin (1:100; Cell Signaling Technology), vimentin (1:100; Bioworld Technology Inc.), and CD34 (1:400; Santa Cruz Biotechnology, Inc.), and then incubated with horseradish peroxidase–conjugated IgG (1:500; Invitrogen), and the proteins in situ were visualized with 3,3′-diaminobenzidine. Density of target proteins, except CD34 used for vascular labeling, was determined as previously reported (Zhu et al., 2008).
Statistical analysis.
Data were analyzed using GraphPad Prism 5 software (Tavazoie et al., 2008). Quantitative variables were expressed as means ± SEM and analyzed by one-way ANOVA, Student’s t test, Kruskal-Wallis test, or Mann–Whitney test. Qualitative variables were compared using Pearson χ2 test or Fisher exact test. The log-rank test was used to determine the statistical significance of the differences between metastasis-free survival curves. R/Bioconductor software was used for all bioinformatics analysis. Results were considered statistically significant at P < 0.05.
Online supplemental material.
Table S1 shows clinicopathological features of HCC patients. Table S2 shows the relationships between intratumoral levels of miR-612 and clinicopathological features of HCC patients. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20120153/DC1.
Supplementary Material
Acknowledgments
The authors would like to thank all members of the Animal Department of Liver Cancer Institute of Fudan University for nude mouse maintenance and xenograft establishment. We thank Dr. Chun-Hua Tang from Shanghai Minghong Biotechnology Inc. for providing human colon cancer cells, luciferase activity evaluations, and vector constructions. We also thank Dr. Wei-De Zhang for providing clinical data of HCC patients. A special thanks to Dr. Xiao-Dong Zhu for his informative discussions and technical assistance in tail vein injection.
This work was jointly funded by grants from the National Natural Science Foundation of China (81071904 and 81272437), the Science and Technology Commission of Shanghai Municipality (1140902501), China National Key Projects for Infectious Disease (2012ZX10002-012), and 973 Basic Research Program of China (2010CB834305).
The authors have no conflicts of interest to disclose.
Author contributions: Z.-H. Tao, J.-L. Wan, and W.-Z. Wu conceived and designed the study. Z.-H. Tao, L.-Y. Zeng, and W.-Z. Wu prepared the manuscript. Z.-H. Tao and J.-L. Wan performed in vitro and in vivo experiments. H.-C. Sun, L. Wang, J. Zhou, Z.-G. Ren, L.-X. Qin, Y.-X. Li, and J. Fan participated in study design. L.-Y. Zeng and L. Xie performed the miRNA expression array analyses and bioinformatics analyses. H.-C. Sun provided valuable discussions with regard to clinical correlates.
Footnotes
Abbreviations used:
- 3′UTR
- 3′ untranslated region
- EMT
- epithelial–mesenchymal transition
- HCC
- hepatocellular carcinoma
- IOD
- integrated optical density
- miRNA
- microRNA
References
- Ambros V. 2004. The functions of animal microRNAs. Nature. 431:350–355 10.1038/nature02871 [DOI] [PubMed] [Google Scholar]
- Bartel D.P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 116:281–297 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- Bartel D.P. 2009. MicroRNAs: target recognition and regulatory functions. Cell. 136:215–233 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken C.P., Gregory P.A., Kolesnikoff N., Bert A.G., Wang J., Shannon M.F., Goodall G.J. 2008. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 68:7846–7854 10.1158/0008-5472.CAN-08-1942 [DOI] [PubMed] [Google Scholar]
- Budhu A., Jia H.L., Forgues M., Liu C.G., Goldstein D., Lam A., Zanetti K.A., Ye Q.H., Qin L.X., Croce C.M., et al. 2008. Identification of metastasis-related microRNAs in hepatocellular carcinoma. Hepatology. 47:897–907 10.1002/hep.22160 [DOI] [PubMed] [Google Scholar]
- Castellano G., Malaponte G., Mazzarino M.C., Figini M., Marchese F., Gangemi P., Travali S., Stivala F., Canevari S., Libra M. 2008. Activation of the osteopontin/matrix metalloproteinase-9 pathway correlates with prostate cancer progression. Clin. Cancer Res. 14:7470–7480 10.1158/1078-0432.CCR-08-0870 [DOI] [PubMed] [Google Scholar]
- Chaffer C.L., Weinberg R.A. 2011. A perspective on cancer cell metastasis. Science. 331:1559–1564 10.1126/science.1203543 [DOI] [PubMed] [Google Scholar]
- Charafe-Jauffret E., Ginestier C., Birnbaum D. 2009. Breast cancer stem cells: tools and models to rely on. BMC Cancer. 9:202 10.1186/1471-2407-9-202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins J.M., He Y., Leary R.J., Pagliarini R., Diaz L.A., Jr, Sjoblom T., Barad O., Bentwich Z., Szafranska A.E., Labourier E., et al. 2006. The colorectal microRNAome. Proc. Natl. Acad. Sci. USA. 103:3687–3692 10.1073/pnas.0511155103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J., Huang S., Wu S., Zhao Y., Liang L., Yan M., Ge C., Yao J., Chen T., Wan D., et al. 2010. Gain of miR-151 on chromosome 8q24.3 facilitates tumour cell migration and spreading through downregulating RhoGDIA. Nat. Cell Biol. 12:390–399 10.1038/ncb2039 [DOI] [PubMed] [Google Scholar]
- Fidler I.J. 2003. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat. Rev. Cancer. 3:453–458 10.1038/nrc1098 [DOI] [PubMed] [Google Scholar]
- Gregory P.A., Bert A.G., Paterson E.L., Barry S.C., Tsykin A., Farshid G., Vadas M.A., Khew-Goodall Y., Goodall G.J. 2008. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 10:593–601 10.1038/ncb1722 [DOI] [PubMed] [Google Scholar]
- Gupta G.P., Massagué J. 2006. Cancer metastasis: building a framework. Cell. 127:679–695 10.1016/j.cell.2006.11.001 [DOI] [PubMed] [Google Scholar]
- He H., Jazdzewski K., Li W., Liyanarachchi S., Nagy R., Volinia S., Calin G.A., Liu C.G., Franssila K., Suster S., et al. 2005. The role of microRNA genes in papillary thyroid carcinoma. Proc. Natl. Acad. Sci. USA. 102:19075–19080 10.1073/pnas.0509603102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X.H., Wang Q., Chen J.S., Fu X.H., Chen X.L., Chen L.Z., Li W., Bi J., Zhang L.J., Fu Q., et al. 2009. Bead-based microarray analysis of microRNA expression in hepatocellular carcinoma: miR-338 is downregulated. Hepatol. Res. 39:786–794 10.1111/j.1872-034X.2009.00502.x [DOI] [PubMed] [Google Scholar]
- Hummel M., Meister R., Mansmann U. 2008. GlobalANCOVA: exploration and assessment of gene group effects. Bioinformatics. 24:78–85 10.1093/bioinformatics/btm531 [DOI] [PubMed] [Google Scholar]
- Ji J., Shi J., Budhu A., Yu Z., Forgues M., Roessler S., Ambs S., Chen Y., Meltzer P.S., Croce C.M., et al. 2009. MicroRNA expression, survival, and response to interferon in liver cancer. N. Engl. J. Med. 361:1437–1447 10.1056/NEJMoa0901282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H.L., Ye Q.H., Qin L.X., Budhu A., Forgues M., Chen Y., Liu Y.K., Sun H.C., Wang L., Lu H.Z., et al. 2007. Gene expression profiling reveals potential biomarkers of human hepatocellular carcinoma. Clin. Cancer Res. 13:1133–1139 10.1158/1078-0432.CCR-06-1025 [DOI] [PubMed] [Google Scholar]
- Landgraf P., Rusu M., Sheridan R., Sewer A., Iovino N., Aravin A., Pfeffer S., Rice A., Kamphorst A.O., Landthaler M., et al. 2007. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 129:1401–1414 10.1016/j.cell.2007.04.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Tian B., Yang J., Zhao L., Wu X., Ye S.L., Liu Y.K., Tang Z.Y. 2004. Stepwise metastatic human hepatocellular carcinoma cell model system with multiple metastatic potentials established through consecutive in vivo selection and studies on metastatic characteristics. J. Cancer Res. Clin. Oncol. 130:460–468 10.1007/s00432-004-0564-9 [DOI] [PubMed] [Google Scholar]
- Li S., Fu H., Wang Y., Tie Y., Xing R., Zhu J., Sun Z., Wei L., Zheng X. 2009. MicroRNA-101 regulates expression of the v-fos FBJ murine osteosarcoma viral oncogene homolog (FOS) oncogene in human hepatocellular carcinoma. Hepatology. 49:1194–1202 10.1002/hep.22757 [DOI] [PubMed] [Google Scholar]
- Liang L., Wong C.M., Ying Q., Fan D.N., Huang S., Ding J., Yao J., Yan M., Li J., Yao M., et al. 2010. MicroRNA-125b suppressesed human liver cancer cell proliferation and metastasis by directly targeting oncogene LIN28B2. Hepatology. 52:1731–1740 10.1002/hep.23904 [DOI] [PubMed] [Google Scholar]
- Lois C., Hong E.J., Pease S., Brown E.J., Baltimore D. 2002. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 295:868–872 10.1126/science.1067081 [DOI] [PubMed] [Google Scholar]
- Ma L., Teruya-Feldstein J., Weinberg R.A. 2007. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 449:682–688 10.1038/nature06174 [DOI] [PubMed] [Google Scholar]
- Nevins J.R., Potti A. 2007. Mining gene expression profiles: expression signatures as cancer phenotypes. Nat. Rev. Genet. 8:601–609 10.1038/nrg2137 [DOI] [PubMed] [Google Scholar]
- Park S.M., Gaur A.B., Lengyel E., Peter M.E. 2008. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 22:894–907 10.1101/gad.1640608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polyak K., Weinberg R.A. 2009. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat. Rev. Cancer. 9:265–273 10.1038/nrc2620 [DOI] [PubMed] [Google Scholar]
- Stewart S.A., Ben-Porath I., Carey V.J., O’Connor B.F., Hahn W.C., Weinberg R.A. 2003. Erosion of the telomeric single-strand overhang at replicative senescence. Nat. Genet. 33:492–496 10.1038/ng1127 [DOI] [PubMed] [Google Scholar]
- Sun F.X., Tang Z.Y., Lui K.D., Ye S.L., Xue Q., Gao D.M., Ma Z.C. 1996. Establishment of a metastatic model of human hepatocellular carcinoma in nude mice via orthotopic implantation of histologically intact tissues. Int. J. Cancer. 66:239–243 [DOI] [PubMed] [Google Scholar]
- Sun F.X., Sasson A.R., Jiang P., An Z., Gamagami R., Li L., Moossa A.R., Hoffman R.M. 1999. An ultra-metastatic model of human colon cancer in nude mice. Clin. Exp. Metastasis. 17:41–48 10.1023/A:1026442321295 [DOI] [PubMed] [Google Scholar]
- Sun B.S., Dong Q.Z., Ye Q.H., Sun H.J., Jia H.L., Zhu X.Q., Liu D.Y., Chen J., Xue Q., Zhou H.J., et al. 2008. Lentiviral-mediated miRNA against osteopontin suppresses tumor growth and metastasis of human hepatocellular carcinoma. Hepatology. 48:1834–1842 10.1002/hep.22531 [DOI] [PubMed] [Google Scholar]
- Tan H.X., Wang Q., Chen L.Z., Huang X.H., Chen J.S., Fu X.H., Cao L.Q., Chen X.L., Li W., Zhang L.J. 2010. MicroRNA-9 reduces cell invasion and E-cadherin secretion in SK-Hep-1 cell. Med. Oncol. 27:654–660 10.1007/s12032-009-9264-2 [DOI] [PubMed] [Google Scholar]
- Tavazoie S.F., Alarcón C., Oskarsson T., Padua D., Wang Q., Bos P.D., Gerald W.L., Massagué J. 2008. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 451:147–152 10.1038/nature06487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery J.P., Acloque H., Huang R.Y., Nieto M.A. 2009. Epithelial-mesenchymal transitions in development and disease. Cell. 139:871–890 10.1016/j.cell.2009.11.007 [DOI] [PubMed] [Google Scholar]
- Thomson J.M., Parker J., Perou C.M., Hammond S.M. 2004. A custom microarray platform for analysis of microRNA gene expression. Nat. Methods. 1:47–53 10.1038/nmeth704 [DOI] [PubMed] [Google Scholar]
- Tsai W.C., Hsu P.W., Lai T.C., Chau G.Y., Lin C.W., Chen C.M., Lin C.D., Liao Y.L., Wang J.L., Chau Y.P., et al. 2009. MicroRNA-122, a tumor suppressor microRNA that regulates intrahepatic metastasis of hepatocellular carcinoma. Hepatology. 49:1571–1582 10.1002/hep.22806 [DOI] [PubMed] [Google Scholar]
- Ventura A., Jacks T. 2009. MicroRNAs and cancer: short RNAs go a long way. Cell. 136:586–591 10.1016/j.cell.2009.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorhoeve P.M., le Sage C., Schrier M., Gillis A.J., Stoop H., Nagel R., Liu Y.P., van Duijse J., Drost J., Griekspoor A., et al. 2006. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell. 124:1169–1181 10.1016/j.cell.2006.02.037 [DOI] [PubMed] [Google Scholar]
- Watanabe T., Takeda A., Mise K., Okuno T., Suzuki T., Minami N., Imai H. 2005. Stage-specific expression of microRNAs during Xenopus development. FEBS Lett. 579:318–324 10.1016/j.febslet.2004.11.067 [DOI] [PubMed] [Google Scholar]
- Wong C.C., Wong C.M., Tung E.K., Au S.L., Lee J.M., Poon R.T., Man K., Ng I.O. 2011. The microRNA miR-139 suppresses metastasis and progression of hepatocellular carcinoma by down-regulating Rho-kinase 2. Gastroenterology. 140:322–331 10.1053/j.gastro.2010.10.006 [DOI] [PubMed] [Google Scholar]
- Xue Y.H., Zhang X.F., Dong Q.Z., Sun J., Dai C., Zhou H.J., Ren N., Jia H.L., Ye Q.H., Qin L.X. 2010. Thrombin is a therapeutic target for metastatic osteopontin-positive hepatocellular carcinoma. Hepatology. 52:2012–2022 10.1002/hep.23942 [DOI] [PubMed] [Google Scholar]
- Yang J., Weinberg R.A. 2008. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev. Cell. 14:818–829 10.1016/j.devcel.2008.05.009 [DOI] [PubMed] [Google Scholar]
- Yang B.W., Liang Y., Xia J.L., Sun H.C., Wang L., Zhang J.B., Tang Z.Y., Liu K.D., Chen J., Xue Q., et al. 2008. Biological characteristics of fluorescent protein-expressing human hepatocellular carcinoma xenograft model in nude mice. Eur. J. Gastroenterol. Hepatol. 20:1077–1084 10.1097/MEG.0b013e3283050a67 [DOI] [PubMed] [Google Scholar]
- Yao J., Liang L., Huang S., Ding J., Tan N., Zhao Y., Yan M., Ge C., Zhang Z., Chen T., et al. 2010. MicroRNA-30d promotes tumor invasion and metastasis by targeting Galphai2 in hepatocellular carcinoma. Hepatology. 51:846–856 [DOI] [PubMed] [Google Scholar]
- Ye Q.H., Qin L.X., Forgues M., He P., Kim J.W., Peng A.C., Simon R., Li Y., Robles A.I., Chen Y., et al. 2003. Predicting hepatitis B virus-positive metastatic hepatocellular carcinomas using gene expression profiling and supervised machine learning. Nat. Med. 9:416–423 10.1038/nm843 [DOI] [PubMed] [Google Scholar]
- Zhang X., Liu S., Hu T., Liu S., He Y., Sun S. 2009. Up-regulated microRNA-143 transcribed by nuclear factor kappa B enhances hepatocarcinoma metastasis by repressing fibronectin expression. Hepatology. 50:490–499 10.1002/hep.23008 [DOI] [PubMed] [Google Scholar]
- Zhou J., Wang W. 2011. Analysis of microRNA expression profiling identifies microRNA-503 regulates metastatic function in hepatocellular cancer cell. J. Surg. Oncol. 104:278–283 10.1002/jso.21941 [DOI] [PubMed] [Google Scholar]
- Zhu X.D., Zhang J.B., Zhuang P.Y., Zhu H.G., Zhang W., Xiong Y.Q., Wu W.Z., Wang L., Tang Z.Y., Sun H.C. 2008. High expression of macrophage colony-stimulating factor in peritumoral liver tissue is associated with poor survival after curative resection of hepatocellular carcinoma. J. Clin. Oncol. 26:2707–2716 10.1200/JCO.2007.15.6521 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.