Abstract
Influenza infection is widespread in the United States and the world. Despite low mortality rates due to infection, morbidity is common and little is known about the molecular events involved in recovery. Influenza infection results in persistent distal lung remodeling, and the mechanism(s) involved are poorly understood. Recently IL-22 has been found to mediate epithelial repair. We propose that IL-22 is critical for recovery of normal lung function and architecture after influenza infection. Wild-type and IL-22−/− mice were infected with influenza A PR8/34 H1N1 and were followed up for up to 21 days post infection. IL-22 receptor was localized to the airway epithelium in naive mice but was expressed at the sites of parenchymal lung remodeling induced by influenza infection. IL-22−/− mice displayed exacerbated lung injury compared with wild-type mice, which correlated with decreased lung function 21 days post infection. Epithelial metaplasia was observed in wild-type mice but was not evident in IL-22−/− animals that were characterized with an increased fibrotic phenotype. Gene expression analysis revealed aberrant expression of epithelial genes involved in repair processes, among changes in several other biological processes. These data indicate that IL-22 is required for normal lung repair after influenza infection. IL-22 represents a novel pathway involved in interstitial lung disease.
Influenza viruses are highly contagious RNA viruses in the Orthomyxoviridae family. In humans, influenza A is the most common infectious strain of this family. In the United States, influenza A infects 5% to 20% of the population annually, with approximately 30,000 associated deaths. During the 2009 H1N1 pandemic, as many as 45% of young adults were seropositive for the virus, underscoring the broad scope of infection.1 Recently, outbreaks of more virulent strains have brought to light the serious nature of influenza A and have emphasized the importance of understanding disease pathogenesis.
Influenza A infects epithelial cells of both the upper and lower respiratory tract through binding of viral hemagglutinin to sialic acid moieties on the host cell. Replication occurs rapidly, and viral particles can be released from infected cells as early as 24 hours after infection. Viral replication and subsequent translation can induce endoplasmic reticulum stress in the infected cell,2 and ultimately infected cells die through apoptosis.3 Although the loss of infected cells can be beneficial, because it decreases the ability of the virus to proliferate, it leads to degeneration and desquamation of the epithelium. This airway injury is subsequently followed by spreading of uninfected cells, epithelial proliferation, and mucous cell metaplasia.4 Viral clearance occurs rapidly after infection, and in most cases recovery from viral-associated morbidity occurs within 2 weeks of infection.
Given the high infectivity but low mortality often observed after influenza infection, research has focused on the mechanisms of viral transmission and clearance. However, recent data suggest that influenza infection has long-term immune consequences on the lung. It has been reported that some patients recovering from influenza A outbreaks have bronchiolitis, alveolitis, intimal thickening, and lung fibrosis.5–7 In addition to potentially causing persistent lung remodeling, influenza also affects host defense. By altering T-cell function,8 Toll-like receptor expression,9,10 and pathogen-associated molecular recognition receptors,10 influenza increases the susceptibility of the host to subsequent bacterial pneumonia,11 particularly to Streptococcus pneumoniae12 and Staphylococcus aureus.8 In mouse models, influenza has been found to cause a prolonged period of lung remodeling and alveolitis lasting for up to 180 days post infection (d.p.i.).13–15 With the increasing prevalence of more infective and/or virulent strains of influenza, understanding the impact of the virus on the host epithelium and the processes involved in lung repair are of great importance.
The repair processes in the lung include a complex interaction between the innate immune system and the airway epithelium. Recent studies have identified subsets of innate cells that are in close contact with the mucosal surfaces of the intestine, the skin, and the lung and are required for epithelial maintenance.16 IL-22–producing cells represent a subset of these cells present in the lung. IL-22 is a member of the IL-10 family and signals through the heterodimeric complex of the IL-10R1b and Il-22Ra1 and activates STAT3. Whereas IL-10R1b is ubiquitously expressed, IL-22Ra1 expression is limited to the epithelial cells in digestive, skin, and respiratory tissues.17,18 IL-22 receptor distribution allows IL-22 to have a direct impact on epithelial cells with little or no interaction with immune cells. This one-way communication allows IL-22 to act as a gatekeeper by promoting epithelial integrity and inducing the epithelium to produce a number of antimicrobial peptides, such as β-defensin 2 and 3, S100A7 through 9, RegIIIβ and γ, and lipocalin19–21
An important emerging role of IL-22 is the promotion of epithelial repair. Although IL-22 is thought to be pathogenic and induces psoriasis in the skin,22,23 IL-22 appears beneficial in terms of promoting resolution of injury in both the lung24,25 and intestine.26,27 It has been demonstrated that the production of IL-22 promotes epithelial repair and cellular turnover in a model of inflammatory bowel disease (IBD)27 in a STAT3-dependent manner.28 In the lung, our laboratory has found that IL-22 protects the airways by increasing transepithelial resistance and promoting bronchial epithelial cell proliferation.19 Further, IL-22 has been found to prevent apoptosis through the production of antiapoptotic proteins, such as BclII and Bcl2l1, in a bleomycin model of lung injury.25
Because IL-22 has been suggested to play a role in wound repair in inflammatory disease, we hypothesized that IL-22 plays a critical role in regulating epithelial repair responses during influenza infection and resolution. These data elucidate a novel pathway by which IL-22 is required for appropriate gene transcription and epithelial repair in the lung.
Materials and Methods
Mice
Specific pathogen-free mice were used in all experiments and housed in pathogen-free conditions at Children’s Hospital of Pittsburgh, Pennsylvania. All of the animal studies were conducted with the approval of the University of Pittsburgh Institutional Animal Care and Use Committee. Six- to 8-week-old wild-type (WT) C57BL/6 mice were purchased from Taconic (Germantown, NY). IL-22−/− were bred in house as previously described.8 All of the studies were performed on age-matched male mice.
Influenza A PR/8/34 H1N1 Infection
Influenza A PR/8/34 H1N1 was propagated in chicken eggs as described previously.29 Mice were infected with 100 plaque-forming units of influenza A PR/8/34 H1N1 (in 40 μL of sterile PBS) from a frozen stock or control PBS by oropharyngeal aspiration. Infected mice were incubated for 10 or 21 days, at which time mice lungs were harvested. Viral burden was determined by quantitative real-time RT-PCR on lung RNA for viral matrix protein as described previously.30
Analysis of Lung Inflammation and Lung Injury
At 10 days, bronchoalveolar lavage (BAL) fluid was collected as previously described.31 Tracheas were exposed and cannulated. The BAL fluid was collected in 10 1-mL aliquots of sterile PBS supplemented with 0.6 mmol/L EDTA. The first aliquot was centrifuged for 10 minutes at 500 × g, and the supernatant was collected for protein analysis and lactate dehydrogenase (LDH) activity. The cell pellet was resuspended and combined with the remaining aliquots. Aliquots were combined and centrifuged for 10 minutes at 500 × g. Cells were resuspended in 1 mL of PBS-EDTA and counted. A total of 1 × 105 cells were spun onto a glass slide and stained for differential counting.
To measure lung injury, total protein in the first milliliter of BAL fluid was measured using the BCA Protein Assay Kit (Pierce Chemical, Rockford, IL). LDH activity was measured using the LDH assay (Sigma-Aldrich, St. Louis, MO). Both assays were performed in a 96-well plate for 30 minutes according to the manufacturer’s instructions and analyzed using the Benchmark Plus plate reader (Bio-Rad, Hercules, CA).
Histologic Analysis
Animals were sacrificed by intraperitoneal injection of 0.9 mL/kg of ketamine and xylazine followed by exsanguination through the renal artery. After exposing the chest cavity, the right main bronchus was sutured at the base of the main stem, and the right lung was excised and placed in Trizol for RNA extraction. The left lung was inflated with 10% neutral-buffered formalin (Sigma Chemical Co, St Louis, MO) at a pressure of 25 cm H2O for 10 minutes, removed from the animal, and placed in fresh 10% neutral-buffered formalin for 24 hours before processing and paraffin embedding. For histologic analysis, 5 μm sections were cut and stained with H&E. Histologic scoring was performed by a pathologist reviewer (D.A.P.) on blinded lung sections. Lung injury induced by influenza was assessed by a numerical scoring scale from 0 to 3 (increasing in severity). Regions of lung injury in sections were scored for the extent of intimal thickening, alveolitis, and the presence of proteinaceous material in the alveolar space. Epithelial denudation was quantified by counting the number of denuded airways per section.
Immunohistochemistry for IL-22Ra1
Lung sections were deparaffinized in xylene (3 × 10 minutes) and rehydrated through sequential washings of 100%, 95%, and 75% ethanol (2 × 10 minutes). Antigen retrieval was performed by boiling for 10 minutes in 10 mmol/L citrate buffer followed by 30 minutes at room temperature. After peroxidase blocking (3% hydrogen peroxide for 10 minutes), slides were blocked following the Vectastat ABC blocking protocol for Rat IgG (Vector Laboratories, Burlingame, CA). IL-22Ra1 was visualized using Rat anti-mouse IL-22Ra1 (clone 496514; R&D Systems, Minneapolis, MN) at a dilution of 1:100.
Picrosirius Red Staining and Collagen Quantitation
After deparaffinization and rehydration, nuclei were stained with Wiegert’s hematoxylin for 10 minutes followed by a 10-minute wash in running tap water. Collagen was stained in picrosirius red for 1 hour followed by two washes in acidified water. Slides were dehydrated, cleared in xylene, and mounted.
Collagen was quantified microscopically. To be considered for analysis, each field had to be >90% parenchyma. Five fields of influenza affected (altered epithelial morphologic features and inflammation) and five fields of unaffected lung (normal alveolar pathologic features) were identified at ×200 magnification. After identification under bright field, images were taken under polarized light. NIS Elements software version 3.2 (Nikon Instruments, Melville, NY) was used to calculate tissue volume density (TVD) defined as the percentage of each microscopic field that is lung tissue and the percentage of each tissue-containing field that is collagen. Values for all fields were averaged to yield a single TVD per animal. TVD values per animal were then averaged to yield a group average. Unaffected areas were used as a negative control for each group and showed no differences between groups.
RNA Isolation and Microarray Analysis of Gene Expression
RNA was isolated from the right lung using Agilent Absolutely RNA kit (Agilent, Foster City, CA). RNA quality was determined using a Nanodrop-1000 spectrophotometer (Nanodrop Technologies, Wilmington, DE) and Agilent 2100 Bioanalyzer (Agilent, Foster City, CA). All microarray analysis was performed at the University of Pittsburgh Genomics and Proteomics Core Laboratory using the Illumina mouse WG6 array (Illumina, San Diego, CA) according to the manufacturer protocol.
Microarray data obtained from whole lung RNA were analyzed by the J5 score threshold as previously published.32 The threshold value is the absolute value of the J5 score that a gene must be above to be considered differentially expressed. The J5 score is calculated by dividing the mean difference between group 1 and 2 by the averaged absolute mean difference for all genes in the data set. Its sign indicates the directionality. For example, positive J5 means overexpressed and negative means underexpressed. Microarray data are deposited at the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo; accession number GSE41549).
To develop functional profiles of the microarray data, gene ontology analysis was performed using Onto-Express, and biological pathways were identified using Pathway-Express,33,34 both of which are parts of Onto-Tools version 3 (http://vortex.cs.wayne.edu/projects.htm; Detroit, MI; registration required).35 Pathway-level impact analysis was implemented as described by Draghici et al,33 and the results are summarized as impact score and P values.
Lung Function Testing
Pulmonary function was measured by the forced oscillation technique using the FlexiVent apparatus (Scireq, Montreal, QC, Canada) as previously described.36 Mice were tracheotomized with an 18-gauge cannula while under anesthesia with 90 mg/kg of pentobarbital-sodium. Ventilation was conducted at 200 breaths per minute at a tidal volume of 0.2 mL with a positive end-expiratory pressure of 3 cm H2O. Multiple linear regression was used to fit pressure and volume measurements to the model of linear motion of the lung. Model fits <0.80 were excluded. Baseline measurements of airway resistance, tissue resistance, and tissue elastance were made. In addition, pressure-volume analysis of the lung was conducted using stepwise volume inflation and deflation of the lung to a maximum pressure of 30 cm H2O. The pressure-volume data were used to calculate static lung compliance.
Statistical Analysis
All data are represented as the mean ± SEM. Significance was determined using either an unpaired t-test when comparing two groups or a one-way analysis of variance with Bonferroni adjustment when comparing multiple groups. All statistics were calculated using computer software (GraphPad Prism version 4, San Diego, CA).
Results
Previously, we have reported that IL-22 signals via STAT3 in mouse tracheal epithelial cultures and human bronchial epithelial cells and induces genes that encode antimicrobial proteins and cell cycle genes.19 Although these data suggest IL-22 has an impact on the airway epithelium, little is known about the distribution of the IL-22 receptor in vivo. To examine this, immunohistochemical analysis was performed in the lungs of naive C57BL/6 mice for high-affinity IL-22Ra1. IL-22Ra1 was distributed on epithelial cells of the large and small airways with no identifiable staining in the lung parenchyma or endothelium in naive mice (Figure 1). This receptor distribution suggests IL-22 interacts mainly with the bronchial epithelial cells in the absence of lung injury.
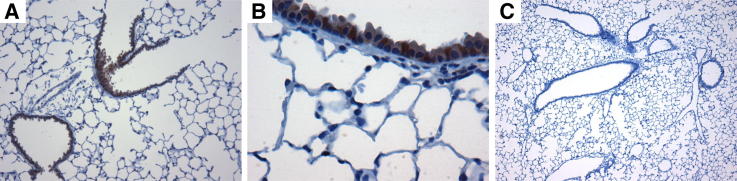
Figure 1.
A and B: IL-22Ra1 is distributed on epithelial cells of the large and small airways of naive C57BL/6 mice. IL-22Ra1 was immune-localized in formalin-fixed, paraffin-embedded lung tissue. These representative photomicrographs demonstrate the IL-22Ra1 is localized to the airway epithelial cells with no staining in the lung parenchyma. C: Control without primary antibody. Original magnification: ×100 (A); ×400 (B).
To investigate the functional importance of IL-22 in the airways after viral injury, IL-22−/− mice were challenged with a sublethal dose of 100 plaque-forming units of influenza A PR/8/34 H1N1.30 There were no differences in viral clearance between C57BL/6 and IL-22−/− mice because the viral matrix protein was undetectable by quantitative real-time PCR in both strains of mice at 10 days (data not shown). Because viral burden is relatively low 10 d.p.i., we measured virus levels on day 7. Relative expression of influenza matrix protein was 1.00 ± 0.76 in C57BL/6 mice compared with 1.34 ± 0.23 in IL-22−/− mice (n = 4), further confirming no differences in viral load. Lung injury was assed 10 d.p.i.by measuring total protein and LDH, a marker of cell lysis, in BAL fluid. IL-22−/− mice demonstrated more severe injury and greater lung edema because they both had significantly higher LDH levels and more total protein in the BAL fluid (Figure 2, A and B) than WT controls. These differences in lung injury did not correspond with weight loss because there were no significant differences in weight loss when comparing WT mice and IL-22−/− mice at this time point (data not shown).
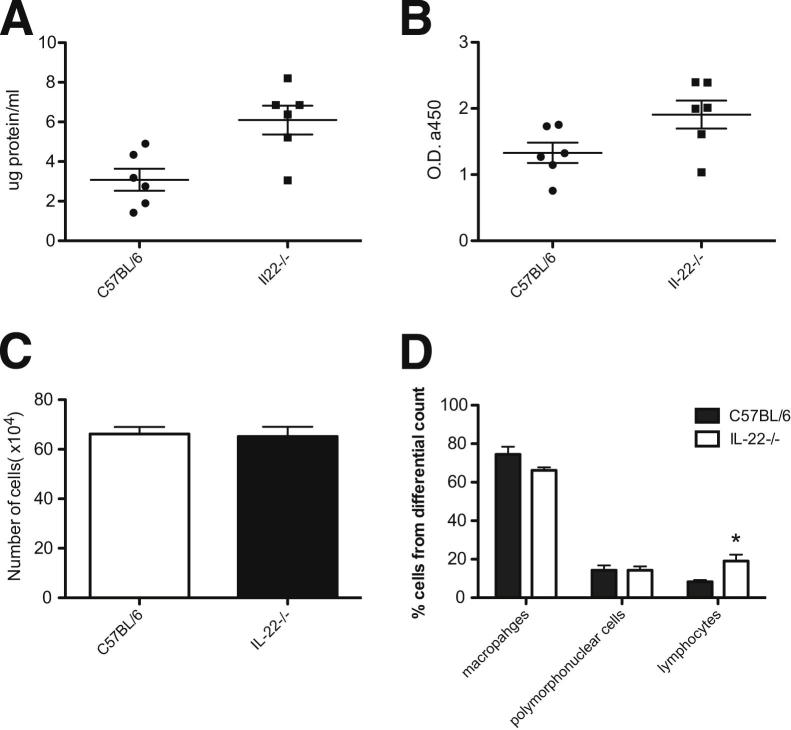
Figure 2.
IL-22−/− mice have greater injury and altered inflammatory response to influenza infection. BAL fluid was collected and analyzed from influenza infected mice 10 d.p.i. A: Total protein was measured using the BCA protein assay kit. P < 0.01. B: Cell cytotoxicity was detected by measuring lactate dehydrogenase. P < 0.05. Each circle and square in A and B represents an individual mouse. C: Total cell counts in BAL fluid. D: Differential cell counts preformed on BAL cells. Results are representative of two separate experiments, and values are the means ± SD (n = 6). *P < 0.05.
Although IL-22Ra1 may not be found on inflammatory cells, IL-22 has been shown to have immunosuppressive effects.37 To characterize the inflammatory role of IL-22 during influenza infection, total and differential cell counts were performed on BAL fluid. There were no differences in total cell counts between WT and IL-22−/− mice (Figure 2C). Further, whereas both IL-22−/− and WT mice had similar percentages of macrophages and polymorphonuclear cells, IL-22−/− had significantly greater numbers of lymphocytes (Figure 2D). These data indicate that the loss of IL-22 results in worsened lung injury, despite small changes in lung inflammation and no differences in viral clearance.
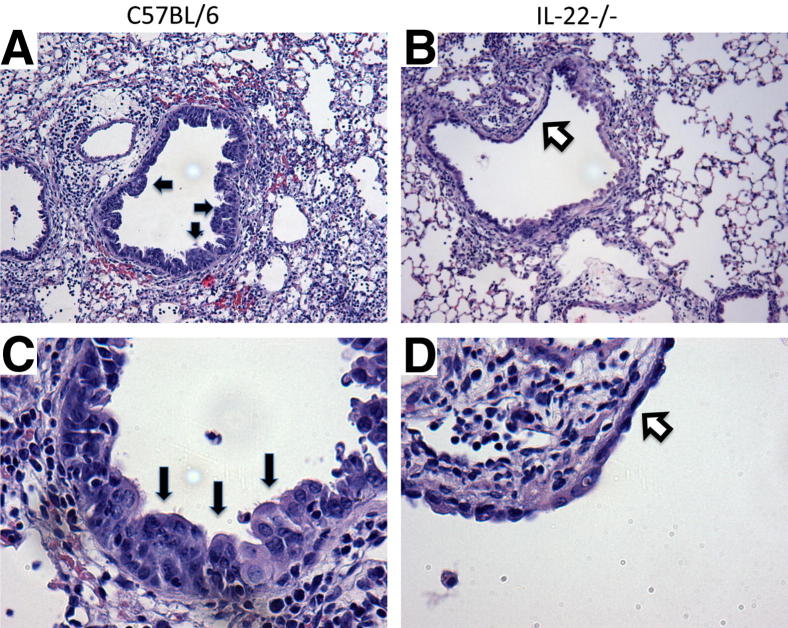
Influenza infection is known to induce the development of epithelial hyperplasia and mucous metaplasia during recovery from injury.4 Histopathologic analysis at 10 d.p.i. revealed stark differences between the two strains. IL-22−/− mice demonstrated epithelial degeneration with flattened airway cells and little to no airway mucous cell metaplasia and hyperplasia in contrast to the typical regeneration seen in the WT controls (Figure 3, A–D). Denudation of the airways was not observed in C57BL/6 mice but was present in IL-22−/− animals. Quantification of epithelial denudation revealed its presence in 2.29 ± 0.28 airways per section in IL-22−/− mice (n = 7). These data suggest that IL-22 is required for airway epithelial homeostasis and repair after influenza infection. Because IL-22Ra1 is expressed on the epithelium of naive mice, we examined the epithelium of naive C57BL/6 and IL-22−/− mice to determine whether IL-22 regulates lung homeostasis. We did not find structural abnormalities in the large or small airways of IL-22−/− mice (Supplemental Figure S1). These data indicate that IL-22 is likely to play a critical role in epithelial repair.
Figure 3.
IL-22−/− mice display altered airway epithelial activation after influenza infection. Lungs from mice infected with influenza for 10 days were inflated to 25 cm H2O by intratracheal installation of 10% neutral buffered formalin (n = 7). Lung sections were stained with H&E. A and C: WT, C57BL/6 mice demonstrated inflammation and regenerative epithelium with mucous cell metaplasia (black arrows) within the airways. B and D: IL-22−/− mice had reduced epithelial regeneration with flattened epithelial cells (white arrows) and in some cases denuded airway epithelium. Original magnification: ×100 (A and B); ×400 (C and D).
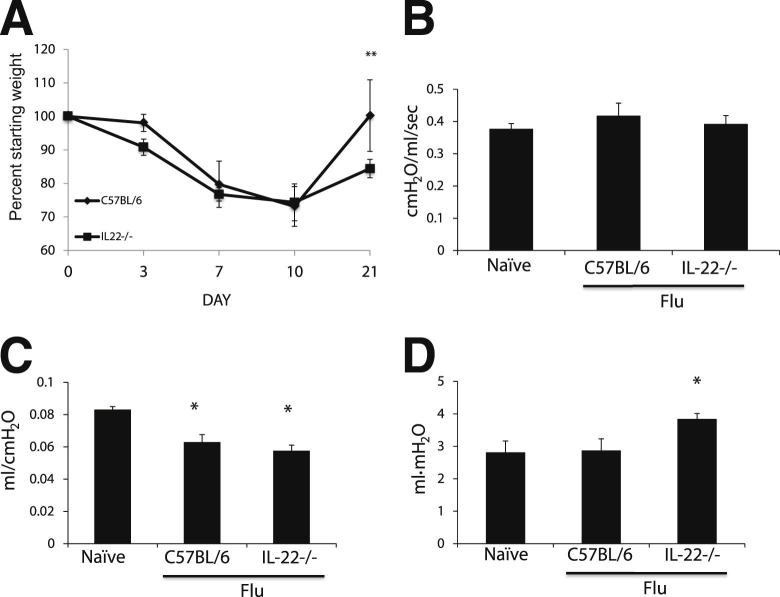
Although influenza virus is typically cleared 7 d.p.i., it has been found that influenza A PR/8/34 H1N1 causes long-term changes (up to 180 d.p.i.) in the lung parenchyma, with persistent inflammation and patches of metaplastic epithelium in the lung parenchyma.13–15 Given the phenotypic differences seen at 10 d.p.i., we next wished to determine whether mice lacking IL-22 recovered from influenza infection similar to controls. Mice were challenged with influenza A PR/8/34 H1N1, and their weight was followed for 21 days as a measure of morbidity. Although there were no differences in weight at the early time points, IL-22−/− had a significantly poorer weight recovery (P < 0.0001) compared with C57BL/6 mice (84.42% ± 2.73% versus 100.52% ± 10.68%) (Figure 4A). Neither WT nor IL-22−/− mice had detectable influenza virus gene expression 21 d.p.i., indicating no defect in viral clearance (data not shown). To determine whether IL-22−/− mice had functional differences after influenza infection, lung mechanics were measured in mechanically vented mice 21 d.p.i. Mice infected with influenza had no significant differences in baseline airway resistance compared with naive controls (Figure 4B). Consistent with the injury phenotypes seen, both flu (0.063 ± 0.0128 cm H2O/mL/second) and IL-22−/− mice (0.057 ± 0.0104 cm H2O/mL/second) had significantly decreased compliance (P < 0.001) compared with naive controls (0.083 ± 0.0059 cm H2O/mL/second) (Figure 4C). Further, IL-22−/− mice had significantly increased hysteresis (3.83 ± 0.515 mL/cm H2O) compared with both the naive (2.86 ± 1.025 mL/cm H2O) and WT plus flu (3.033 ± 0.345 mL/cm H2O) groups (Figure 4D). Decreased compliance and increased hysteresis indicate a stiffening of the lung.
Figure 4.
IL-22−/− mice have greater morbidity and altered lung function during recovery from influenza infection. WT (n = 7) and IL-22−/− (n = 8) mice were infected with influenza for 21 days. A: Weight loss shown as a percentage of the original starting weight. B: Airway resistance. C: Lung compliance measured by pressure-volume analysis. D: Hysteresis (area) of the pressure volume curve. *P < 0.05 versus naive control; **P < 0.05 versus WT mice.
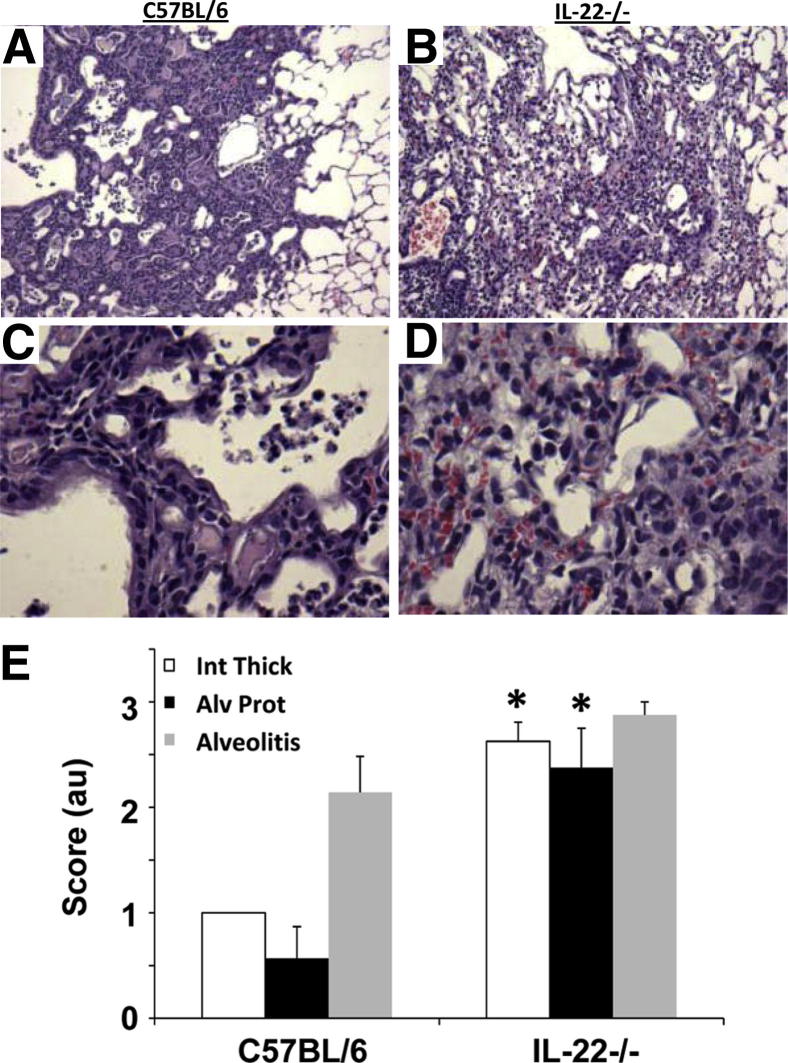
Lung stiffening may be due to decreased epithelial repair and subsequent collagen deposition. Consistent with these data, histopathologic analysis revealed at 21 d.p.i. WT mice had patchy inflammation and developed alveolar metaplasia with activated epithelium (Figure 5, A and C). In contrast, IL-22−/− mice displayed regions lacking in this metaplasia (Figure 5, B and D). Instead, they developed a diffuse inflammation and diffuse alveolar injury with little metaplasia evident. Lung injury was measured by examining the extent of intimal thickening, alveolitis, and the presence of proteinaceous material in the alveolar space. IL-22−/− mice had significantly higher intimal thickening and proteinaceous accumulation compared with WT mice (Figure 5E). To determine whether there were differences in WT and IL-22−/− mice regarding collagen deposition, picrosirius red staining was performed on tissues 21 d.p.i. Collagen was visualized using polarized light, and images were quantified for percentage of collagen per ×200 field. Histologic analysis revealed IL-22−/− mice had increased collagen deposition compared with WT controls (Figure 6, A and B). Digital quantification confirmed that IL-22−/− had a significantly greater percentage of collagen staining in the lung parenchyma (Figure 6C).
Figure 5.
Influenza induces alveolar metaplasia in WT mice but not IL-22−/− mice. WT (n = 7) and IL-22−/− (n = 8) mice were infected with influenza for 21 days. A: Representative histologic stain shows WT mice have organized epithelial regeneration occurring in the alveolar spaces. C: Epithelial cells appear metaplastic. Histologic stains in IL-22−/− mice where epithelial cells are less organized in appearance (B) and epithelium have a more injured fibrotic appearance with no evidence of metaplasia (D). E: Lung injury scoring for intimal thickening, alveolitis, and the presence of proteinaceous material in the alveolar space (n = 7 and 8, respectively). *P < 0.05 versus WT mice. Original magnification, ×100 (A and B); ×400 (C and D)
Figure 6.
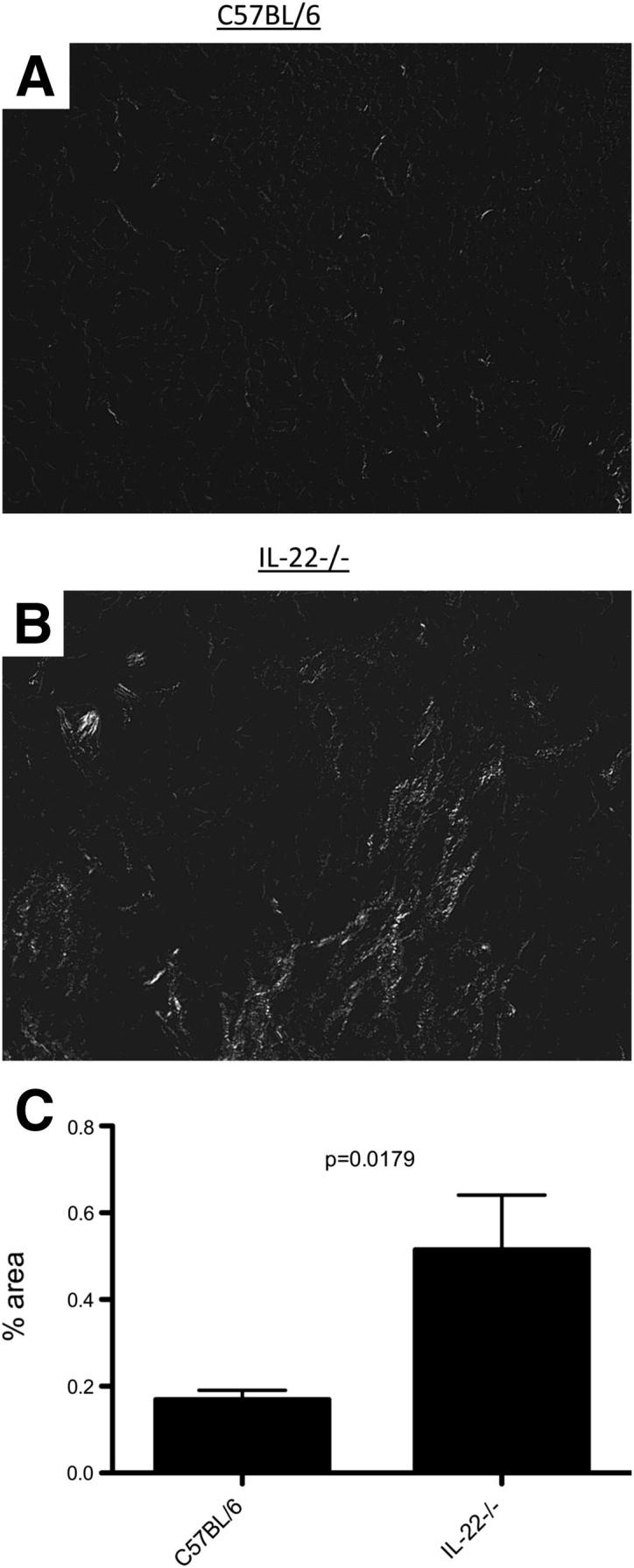
IL-22−/− mice have greater collagen deposition after influenza infection. Sections were stained for picrosirius red and analyzed by bright field microscopy. Areas undergoing injury and repair were chosen and imaged under polarized light. IL-22−/− mice (A) had more collagen in affected areas than WT mice (B). C: Digital quantification of staining (n = 6). White (collagen) versus black (area) was quantified as percent area.
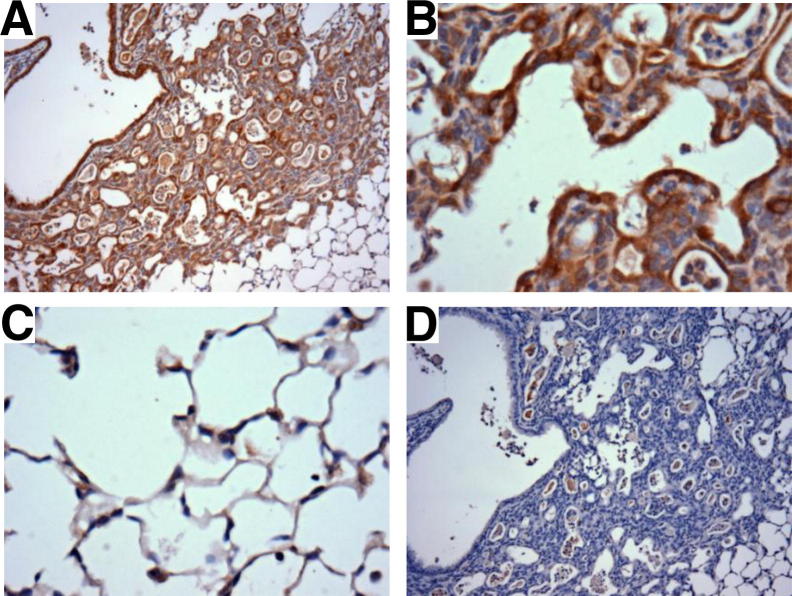
These findings in the lung parenchyma were interesting because we found IL-22 receptor only in the airways of naive mice. If IL-22 is directly involved in alveolar repair, the receptor must be up-regulated in these cells after injury. To test our hypothesis, we stained for the IL-22Ra1 in the lung from C57BL/6 mice 21 after influenza infection. Results indicate that although the receptor is not normally found in the parenchyma (Figure 1), it is expressed in the repairing alveoli after injury (Figure 7, A and B), including type II cells adjacent to areas of remodeling (Figure 7C). Antibody control revealed specific epithelial cell staining because only airspace debris stained positive (Figure 7D). IL-22Ra1 expression was also increased in the areas of lung injury in IL-22−/− mice (data not shown). These novel findings suggest IL-22Ra1 can be up-regulated at sites of injury and further emphasize the importance of this potent cytokine in pulmonary epithelial repair.
Figure 7.
IL-22Ra1 is found in areas of epithelial metaplasia after influenza infection. Representative immunohistochemical staining for IL-22Ra1 reveals expression of the receptor in the alveoli 21di. A and B: Images demonstrate strong staining of the alveolar epithelial cells. C: Type II cells adjacent to the injury also demonstrate receptor staining. D: IgG control. Original magnification: ×100 (A); ×400 (B and C).
These data suggest that IL-22 is required for proper epithelial maintenance and repair in both the airways and parenchyma. To better define the cellular and molecular processes involved in epithelial remodeling 21 d.p.i., genome-wide mRNA expression profiling was performed. RNA was screened by oligonucleotide arrays. Bioinformatic analysis using a threshold of >5 identified 1205 differentially expressed genes between influenza infected IL-22−/− mice and WT mice. Of these, 643 were transcriptionally up-regulated, and 562 genes were negatively regulated. Table 1 lists the top 20 differentially regulated genes.
Table 1.
Top 20 Differentially Regulated and Identified Non-mitochondrial Genes between IL-22−/−- and C57BL/6 (WT) Mice 21 d.p.i.
| Gene | Accession no. | J5 score |
|---|---|---|
| Lactotransferrin (Ltf) | NM_008522.3 | 187.302 |
| Heat shock protein 8 (Hspa8) | NM_031165.4 | −103.969 |
| Thymosin, beta 4, X chromosome (Tmsb4x) | NM_021278.2 | −97.382 |
| Glutathione peroxidase 1 (Gpx1) | NM_008160.5 | 70.805 |
| PRED: similar to MHC class II antigen beta chain (LOC641240) | XM_918601.3 | 69.453 |
| Procollagen, type IV, alpha 1 (Col4a1) | NM_009931.1 | −68.738 |
| Ribosomal protein L27 (Rpl27) | NM_011289.1 | 68.074 |
| Glutathione S-transferase omega 1 (Gsto1) | NM_010362.2 | 66.784 |
| Surfactant associated protein D (Sftpd) | NM_009160.1 | −65.692 |
| PRED: similar to Ig kappa chain V-V region L7 precursor (LOC636875) | XM_992953.1 | 63.438 |
| CD74 antigen (Cd74), transcript variant 1 | NM_001042605.1 | 63.417 |
| PRED: similar to ribosomal protein L3 (LOC433745) | XM_001475733.1 | 62.876 |
| Eukaryotic translation initiation factor 3, subunit F (Eif3f) | NM_025344.1 | 61.929 |
| Cytochrome P450, family 2, subfamily f, polypeptide 2 (Cyp2f2) | NM_007817.2 | 60.559 |
| Coagulation factor II (thrombin) receptor (F2r) | NM_010169.3 | −59.797 |
| Ribosomal protein L35 (Rpl35) | NM_025592.3 | 57.276 |
| Histocompatibility 2, class II antigen A, beta 1 (H2-Ab1) | NM_207105.2 | 56.000 |
| Palate, lung, and nasal epithelium associated (Plunc) | NM_011126.2 | 55.965 |
| Caveolin 1, caveolae protein (Cav1) | NM_007616.3 | −55.597 |
| Ribosomal protein S5 (Rps5) | NM_009095.1 | 55.142 |
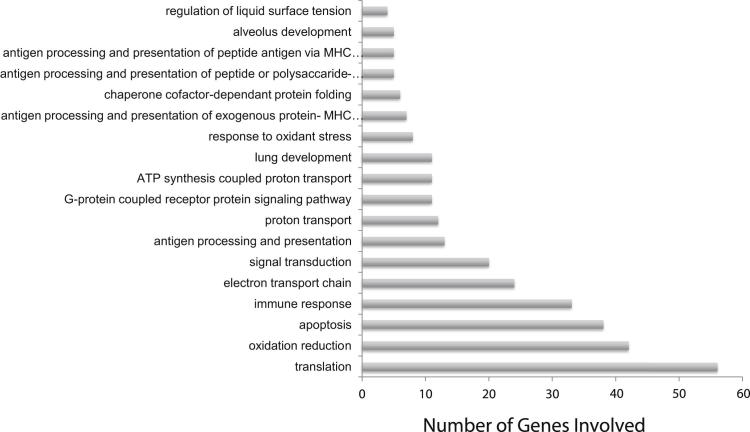
Gene ontology analysis of these differentially regulated genes revealed enrichment of genes associated with translation, oxidation reduction, apoptosis, the immune response, and antigen processing and presentation (Figure 8). Further, genes involved in lung development, alveolus development, and regulation of surface tension were also differentially regulated. Of the epithelial genes, surfactant protein D and surfactant protein A1 were down-regulated (−65.69 and −47.02), and surfactant proteins B and C were up-regulated (19.11 and 32.35) in IL-22−/− mice versus controls. Tables 2 and 3 list the relevant epithelial and basement membrane markers. These data suggest differences in epithelial regeneration between IL-22−/− and WT mice.
Figure 8.
Functional analysis of gene array data comparing WT and IL-22−/− mice. Functional annotation analysis (Onto-Tools) was performed on genes found to be significantly different (increased or decreased) at a J5 threshold >5 (n = 4). The graph represents the 18 processes found to be significantly different (P < 0.01).
Table 2.
Differentially Regulated Epithelial Genes
| Lung epithelial genes | Accession no. | J5 score |
|---|---|---|
| Surfactant associated protein B (Sftpb) | NM_147779.1 | 32.346 |
| Surfactant associated protein C (Sftpc) | NM_011359.1 | 19.118 |
| Surfactant associated protein D (Sftpd) | NM_009160.1 | −65.692 |
| Surfactant associated protein A1 (Sftpa1) | NM_023134.4 | −47.018 |
| Secretoglobin, family 1A, member 1 (uteroglobin) (Scgb1a1) | NM_011681.2 | −51.336 |
Table 3.
Differentially Regulated Basement Membrane Genes
| Basement membrane genes | Accession no. | J5 score |
|---|---|---|
| Procollagen, type IV, alpha 1 (Col4a1) | NM_009931.1 | −68.739 |
| Collagen, type IV, alpha 2 (Col4a2) | NM_009932.2 | −29.891 |
| Procollagen, type VI, alpha 3 (Col6a3) | NM_009935.1 | −8.381 |
| Procollagen, type VI, alpha 1 (Col6a1) | NM_009933.2 | −7.574 |
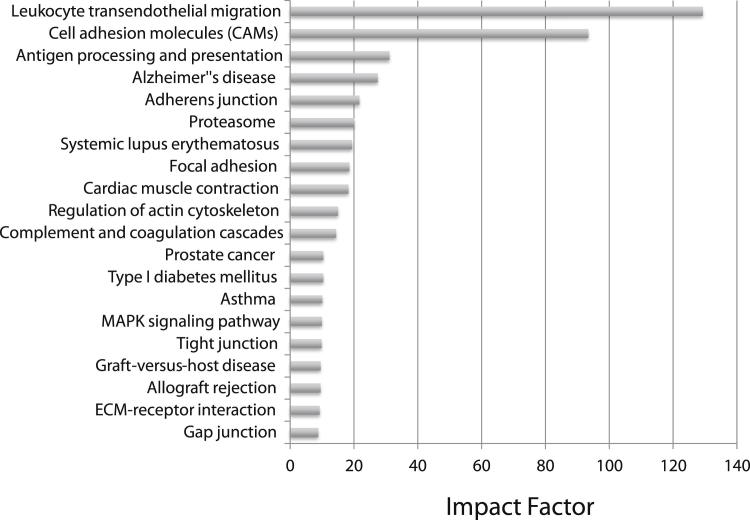
To identify deregulated KEGG pathways, Onto-Tools Pathway Express analysis was performed on all genes with a threshold difference of >5. Figure 9 shows the significantly affected pathways. The top three pathways ranked by impact factor were leukocyte adhesion molecules (impact factor = 129.19, P = 1.94 × 10−5), cellular adhesion molecules (impact factor = 93.57, P = 9.48 × 10−6), and antigen processing (impact factor = 31.06, P = 4.44 × 10−7). In these pathways, the genes that are up-regulated are all involved in cell-cell interaction and cellular adhesion. These data support the pathologic data and suggest that mice lacking IL-22 have increased inflammation and slower wound healing after H1N1 infection.
Figure 9.
Pathway analysis of genes found to be significantly different in IL-22−/− mice 21 d.p.i. Pathway analysis was performed using Pathway Express (Onto-Tools) on genes found to be significantly different (increased or decreased) at a J5 threshold >5 (n = 4). The graph represents the impact factor of the top 20 significantly affected pathways.
Discussion
Influenza viruses are among the most common cause of respiratory disease. Although influenza generally has a low mortality, it can induce acute respiratory distress syndrome38 associated with diffuse alveolar damage39,40 and even pulmonary fibrosis.13 With the recent H1N1 pandemic and the outbreaks of more virulent strains, it is increasingly important to understand the injury caused by influenza infection and the mechanisms involved in pulmonary repair and recovery after infection. In this study, we demonstrate the importance of IL-22 in the resolution and repair of lung injury after influenza-induced pneumonia. We found that IL-22Ra1 is expressed in the airways but not the parenchyma of naive mice. Through the use of IL-22−/− mice, we demonstrated that IL-22 promotes epithelial repair in the airways after infection because IL-22−/− mice had significantly more damage and less repair at 10 days. Interestingly, by 21 d.p.i., we found that IL-22Ra1 was expressed in the parenchyma in areas of epithelial metaplasia in WT mice, suggesting a role for IL-22 in parenchymal repair. This observation was supported by histopathologic evidence in which IL-22−/− had less epithelial repair in the parenchyma, which coincided with greater collagen deposition and significantly increased lung stiffness as measured by FlexiVent. Further, microarray analysis demonstrated differences in the regulation of epithelial genes. These data demonstrated that IL-22 is critical in the initial response to airway injury because IL-22−/− mice had increased airway epithelial injury.
IL-22 was initially thought to be a signature TH17 cytokine. However, recent investigations have found that IL-22 is produced by multiple T-cell subsets during both innate and adaptive immune responses. In the lung, IL-22 is produced by TH17 cells,19 γδ T cells,41 innate lymphoid cells,42 and natural killer T cells.43,44 In response to influenza, IL-22 is produced by natural killer cells,44 invariant natural killer cells, T cells,45 and CD8T (Tc)17 effector cells.46
Although a number of inflammatory cells produce IL-22, its mode of action is limited because of its receptor distribution. IL-22 signals through the combination of the IL-10R2 and IL-22Ra1. Although IL-10R2 is widely expressed, IL-22Ra1 expression is limited predominantly to epithelial cells of the skin,22 lung,25,44 and intestine,47 as well as being basally expressed in the pancreas48 and liver.49 In this study, we found that IL-22Ra1 is produced by both Clara and ciliated cells of the airway epithelium of naive mice but is not constitutively produced by epithelial cells in the parenchyma. This expression pattern suggests the primary role of IL-22 is in the protection and maintenance of the airway epithelium, which is the primary target of the influenza virus. Through the use of IL-22−/− mice, we were able to demonstrate the importance of IL-22 in airway repair because IL-22−/− mice had greater injury markers and limited epithelial repair with limited airway cell metaplasia as observed by histologic analysis.
Because influenza targets airway epithelial cells where it induces endoplasmic reticulum stress2 and apoptosis,3 the mode of protection for IL-22 can be either antiviral or directly protecting the epithelium. IL-22 is induced in response to both bacterial19 and viral44,45 infections. IL-22 is integral in the clearance of various bacterial infections; however, in the case of viral infections, the role of IL-22 is less obvious. IL-22 is up-regulated in hepatitis B50,51 and hepatitis C52 infection but has not been found to have any antiviral properties. Similarly, in the lung, although IL-22–producing cells are found in response to influenza infection, we and others have found that IL-22 does not affect viral titer.44,45 These observations suggest that the benefits of IL-22 in influenza infection are not due to lessened viral burden but due to direct interaction with epithelial cell repair.
Although IL-22 appears important in promoting airway repair, airway epithelial cells appeared similar in WT and IL-22−/− mice by 21 d.p.i. This is not surprising because a number of factors, such as hepatocyte growth factor53 and IL-33,18,54 are known to play a role in airway repair. More surprising were findings that IL-22 may have a role in parenchymal repair after infection. Previously, others have reported that influenza infection leads to an acute respiratory distress syndrome–like disease in mice,55 characterized alveolar destruction similar to diffuse alveolar damage,15,55 and even fibrotic changes.13–15 In our model, we found WT C57BL/6 mice had alveolar injury and areas of repairing alveoli characterized by areas of epithelial metaplasia or “activated” epithelial cells. Intriguingly the repairing parenchyma also stained for the IL-22Ra1. These data were in contrast to naive parenchyma in which the IL-22Ra1 was not detected. Previously increased induction of IL-22Ra1 has been associated with psoriasis,22,56 and increased receptor levels have been found in patients with Crohn’s disease57 and in mouse models of ConA hepatitis.49 IL-22Ra1 expression was also elevated in regions of lung injury in IL-22−/− mice. This is not surprising because IL-22Ra1 is used by additional ligands IL-20 and IL-24, the role of which is not currently known in the lung.
The finding that IL-22Ra1 was induced after parenchymal injury suggests that IL-22 is an important cytokine in alveolar epithelial repair. A number of studies have demonstrated the importance of IL-22 after parenchymal injury. Simonian et al40 demonstrated that mice lacking IL-22–producing γδ T cells developed a more severe fibrosis after chronic bacterial infection. Further, although IL-22 was not found to reduce fibrosis, it protected epithelial cells from bleomycin-induced apoptosis.25 In our study, we found that IL-22−/− mice had less activated epithelium and poorly organized repair. The reduced epithelial repair leads to increased collagen deposition and greater lung stiffness. Consistent with this, functional analysis of gene array data demonstrated that genes in pathways regulating surface tension and genes involved in alveolar development were significantly different in IL-22−/− mice. Secretoglobin family 1A, member 1 (Scgb1a1), which is specific for Clara cells,58 and caveolin 1 (Cav1), which is an alveolar type I and endothelial cell marker,59 were decreased, whereas surfactant protein C (Sftpc), a type II cell marker,60 was increased. Decreased alveolar type I cell markers and increased type II markers suggest less cell differentiation in IL-22−/− mice because of decreased repair and a persistent injured state.
Most influenza A infections are not fatal, and the symptoms of influenza infection last only a few days. Consequently, the more chronic effects of influenza infection tend to be overlooked. Our laboratory and others have found that influenza infection enhances the susceptibility to secondary bacterial8,12 and fibrotic13,14 challenges. These studies demonstrate the importance of understanding the more long-term consequences of influenza challenge. In the work presented here, we report that IL-22 is an important cytokine in pulmonary epithelial repair after influenza infection. The therapeutic potential of manipulating IL-22 and its receptor offers a target to promote epithelial recovery after influenza infection and ultimately may improve patient outcome in regard to secondary challenges.
Acknowledgments
Microarray data analysis was conducted by Drs. Tamanna Sultana and James Lyons-Weiler (Genomics and Proteomics Core Bioinformatics Analysis Core, University of Pittsburgh, Pittsburgh, PA).
Footnotes
Supported in part by NIH grants HL079142 (J.K.K.) and HL107380 (J.F.A.) and Parker B. Francis Foundation Fellowship (J.F.A.).
Supplemental Data
Epithelial morphologic findings in naive C57BL/6 and IL-22−/− mice (n = 4). Large and small airways. Original magnification, ×600.
References
- 1.Ross T.M., Hairong L., Chia B.S., Hill E., Weirback H., Zimmer S. Prevalence of antibodies against seasonal influenza A and B viruses during the 2009-2010 and 2010-2011 influenza seasons in residents of Pittsburgh PA, USA. PLoS Curr. 2011;3:RRN1265. doi: 10.1371/currents.RRN1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roberson E.C., Tully J.E., Guala A.S., Reiss J.N., Godburn K.E., Pociask D.A., Alcorn J.F., Riches D.W., Dienz O., Janssen-Heininger Y.M., Anathy V. Influenza induces endoplasmic reticulum stress, caspase-12-dependent apoptosis, and c-Jun N-terminal kinase-mediated transforming growth factor-beta release in lung epithelial cells. Am J Respir Cell Mol Biol. 2012;46:573–581. doi: 10.1165/rcmb.2010-0460OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ludwig S., Pleschka S., Planz O., Wolff T. Ringing the alarm bells: signalling and apoptosis in influenza virus infected cells. Cell Microbiol. 2006;8:375–386. doi: 10.1111/j.1462-5822.2005.00678.x. [DOI] [PubMed] [Google Scholar]
- 4.Buchweitz J.P., Harkema J.R., Kaminski N.E. Time-dependent airway epithelial and inflammatory cell responses induced by influenza virus A/PR/8/34 in C57BL/6 mice. Toxicol Pathol. 2007;35:424–435. doi: 10.1080/01926230701302558. [DOI] [PubMed] [Google Scholar]
- 5.Li P., Su D.J., Zhang J.F., Xia X.D., Sui H., Zhao D.H. Pneumonia in novel swine-origin influenza A (H1N1) virus infection: high-resolution CT findings. Eur J Radiol. 2011;80:e146–e152. doi: 10.1016/j.ejrad.2010.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mineo G., Ciccarese F., Modolon C., Landini M.P., Valentino M., Zompatori M. Post-ARDS pulmonary fibrosis in patients with H1N1 pneumonia: role of follow-up CT. Radiol Med. 2012;117:185–200. doi: 10.1007/s11547-011-0740-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nin N., Sanchez-Rodriguez C., Ver L.S., Cardinal P., Ferruelo A., Soto L., Deicas A., Campos N., Rocha O., Ceraso D.H., El-Assar M., Ortin J., Fernandez-Segoviano P., Esteban A., Lorente J.A. Lung histopathological findings in fatal pandemic influenza A (H1N1) Med Intensiva. 2012;36:24–31. doi: 10.1016/j.medin.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 8.Kudva A., Scheller E.V., Robinson K.M., Crowe C.R., Choi S.M., Slight S.R., Khader S.A., Dubin P.J., Enelow R.I., Kolls J.K., Alcorn J.F. Influenza A inhibits Th17-mediated host defense against bacterial pneumonia in mice. J Immunol. 2011;186:1666–1674. doi: 10.4049/jimmunol.1002194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miettinen M., Sareneva T., Julkunen I., Matikainen S. IFNs activate toll-like receptor gene expression in viral infections. Genes Immun. 2001;2:349–355. doi: 10.1038/sj.gene.6363791. [DOI] [PubMed] [Google Scholar]
- 10.Didierlaurent A., Goulding J., Patel S., Snelgrove R., Low L., Bebien M., Lawrence T., van Rijt L.S., Lambrecht B.N., Sirard J.C., Hussell T. Sustained desensitization to bacterial Toll-like receptor ligands after resolution of respiratory influenza infection. J Exp Med. 2008;205:323–329. doi: 10.1084/jem.20070891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morens D.M., Taubenberger J.K., Fauci A.S. Predominant role of bacterial pneumonia as a cause of death in pandemic influenza: implications for pandemic influenza preparedness. J Infect Dis. 2008;198:962–970. doi: 10.1086/591708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shahangian A., Chow E.K., Tian X., Kang J.R., Ghaffari A., Liu S.Y., Belperio J.A., Cheng G., Deng J.C. Type I IFNs mediate development of postinfluenza bacterial pneumonia in mice. J Clin Invest. 2009;119:1910–1920. doi: 10.1172/JCI35412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jakab G.J. Sequential virus infections, bacterial superinfections, and fibrogenesis. Am Rev Respir Dis. 1990;142:374–379. doi: 10.1164/ajrccm/142.2.374. [DOI] [PubMed] [Google Scholar]
- 14.Jakab G.J., Bassett D.J. Influenza virus infection, ozone exposure, and fibrogenesis. Am Rev Respir Dis. 1990;141:1307–1315. doi: 10.1164/ajrccm/141.5_Pt_1.1307. [DOI] [PubMed] [Google Scholar]
- 15.Jakab G.J., Astry C.L., Warr G.A. Alveolitis induced by influenza virus. Am Rev Respir Dis. 1983;128:730–739. doi: 10.1164/arrd.1983.128.4.730. [DOI] [PubMed] [Google Scholar]
- 16.Monticelli L.A., Sonnenberg G.F., Artis D. Innate lymphoid cells: critical regulators of allergic inflammation and tissue repair in the lung. Curr Opin Immunol. 2012;24:284–289. doi: 10.1016/j.coi.2012.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Witte E., Witte K., Warszawska K., Sabat R., Wolk K. Interleukin-22: a cytokine produced by T, NK and NKT cell subsets, with importance in the innate immune defense and tissue protection. Cytokine Growth Factor Rev. 2010;21:365–379. doi: 10.1016/j.cytogfr.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 18.Monticelli L.A., Sonnenberg G.F., Abt M.C., Alenghat T., Ziegler C.G., Doering T.A., Angelosanto J.M., Laidlaw B.J., Yang C.Y., Sathaliyawala T., Kubota M., Turner D., Diamond J.M., Goldrath A.W., Farber D.L., Collman R.G., Wherry E.J., Artis D. Innate lymphoid cells promote lung-tissue homeostasis after infection with influenza virus. Nat Immunol. 2011;12:1045–1054. doi: 10.1031/ni.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aujla S.J., Chan Y.R., Zheng M., Fei M., Askew D.J., Pociask D.A., Reinhart T.A., McAllister F., Edeal J., Gaus K., Husain S., Kreindler J.L., Dubin P.J., Pilewski J.M., Myerburg M.M., Mason C.A., Iwakura Y., Kolls J.K. IL-22 mediates mucosal host defense against Gram-negative bacterial pneumonia. Nat Med. 2008;14:275–281. doi: 10.1038/nm1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang S.C., Tan X.Y., Luxenberg D.P., Karim R., Dunussi-Joannopoulos K., Collins M., Fouser L.A. Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J Exp Med. 2006;203:2271–2279. doi: 10.1084/jem.20061308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boniface K., Bernard F.X., Garcia M., Gurney A.L., Lecron J.C., Morel F. IL-22 inhibits epidermal differentiation and induces proinflammatory gene expression and migration of human keratinocytes. J Immunol. 2005;174:3695–3702. doi: 10.4049/jimmunol.174.6.3695. [DOI] [PubMed] [Google Scholar]
- 22.Boniface K., Guignouard E., Pedretti N., Garcia M., Delwail A., Bernard F.X., Nau F., Guillet G., Dagregorio G., Yssel H., Lecron J.C., Morel F. A role for T cell-derived interleukin 22 in psoriatic skin inflammation. Clin Exp Immunol. 2007;150:407–415. doi: 10.1111/j.1365-2249.2007.03511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wolk K., Haugen H.S., Xu W., Witte E., Waggie K., Anderson M., Vom Baur E., Witte K., Warszawska K., Philipp S., Johnson-Leger C., Volk H.D., Sterry W., Sabat R. IL-22 and IL-20 are key mediators of the epidermal alterations in psoriasis while IL-17 and IFN-gamma are not. J Mol Med (Berl) 2009;87:523–536. doi: 10.1007/s00109-009-0457-0. [DOI] [PubMed] [Google Scholar]
- 24.Hoegl S., Bachmann M., Scheiermann P., Goren I., Hofstetter C., Pfeilschifter J., Zwissler B., Muhl H. Protective properties of inhaled IL-22 in a model of ventilator-induced lung injury. Am J Respir Cell Mol Biol. 2011;44:369–376. doi: 10.1165/rcmb.2009-0440OC. [DOI] [PubMed] [Google Scholar]
- 25.Sonnenberg G.F., Nair M.G., Kirn T.J., Zaph C., Fouser L.A., Artis D. Pathological versus protective functions of IL-22 in airway inflammation are regulated by IL-17A. J Exp Med. 2010;207:1293–1305. doi: 10.1084/jem.20092054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zenewicz L.A., Yancopoulos G.D., Valenzuela D.M., Murphy A.J., Stevens S., Flavell R.A. Innate and adaptive interleukin-22 protects mice from inflammatory bowel disease. Immunity. 2008;29:947–957. doi: 10.1016/j.immuni.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sugimoto K., Ogawa A., Mizoguchi E., Shimomura Y., Andoh A., Bhan A.K., Blumberg R.S., Xavier R.J., Mizoguchi A. IL-22 ameliorates intestinal inflammation in a mouse model of ulcerative colitis. J Clin Invest. 2008;118:534–544. doi: 10.1172/JCI33194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pickert G., Neufert C., Leppkes M., Zheng Y., Wittkopf N., Warntjen M., Lehr H.A., Hirth S., Weigmann B., Wirtz S., Ouyang W., Neurath M.F., Becker C. STAT3 links IL-22 signaling in intestinal epithelial cells to mucosal wound healing. J Exp Med. 2009;206:1465–1472. doi: 10.1084/jem.20082683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Braciale T.J. Immunologic recognition of influenza virus-infected cells. I. Generation of a virus-strain specific and a cross-reactive subpopulation of cytotoxic T cells in the response to type A influenza viruses of different subtypes. Cell Immunology. 1977;33:423–436. doi: 10.1016/0008-8749(77)90170-8. [DOI] [PubMed] [Google Scholar]
- 30.Crowe C.R., Chen K., Pociask D.A., Alcorn J.F., Krivich C., Enelow R.I., Ross T.M., Witztum J.L., Kolls J.K. Critical role of IL-17RA in immunopathology of influenza infection. J Immunol. 2009;183:5301–5310. doi: 10.4049/jimmunol.0900995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pociask D.A., Chen K., Choi S.M., Oury T.D., Steele C., Kolls J.K. gammadelta T cells attenuate bleomycin-induced fibrosis through the production of CXCL10. Am J Pathol. 2011;178:1167–1176. doi: 10.1016/j.ajpath.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patel S., Lyons-Weiler J. caGEDA: a web application for the integrated analysis of global gene expression patterns in cancer. Appl Bioinformatics. 2004;3:49–62. doi: 10.2165/00822942-200403010-00007. [DOI] [PubMed] [Google Scholar]
- 33.Draghici S., Khatri P., Tarca A.L., Amin K., Done A., Voichita C., Georgescu C., Romero R. A systems biology approach for pathway level analysis. Genome Res. 2007;17:1537–1545. doi: 10.1101/gr.6202607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tarca A.L., Draghici S., Khatri P., Hassan S.S., Mittal P., Kim J.S., Kim C.J., Kusanovic J.P., Romero R. A novel signaling pathway impact analysis. Bioinformatics. 2009;25:75–82. doi: 10.1093/bioinformatics/btn577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khatri P., Voichita C., Kattan K., Ansari N., Khatri A., Georgescu C., Tarca A.L., Draghici S. Onto-Tools: new additions and improvements in 2006. Nucleic Acids Res. 2007;35:W206–W211. doi: 10.1093/nar/gkm327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alcorn J.F., Ckless K., Brown A.L., Guala A.S., Kolls J.K., Poynter M.E., Irvin C.G., van der Vliet A., Janssen-Heininger Y.M. Strain-dependent activation of NF-kappaB in the airway epithelium and its role in allergic airway inflammation. Am J Physiol Lung Cell Mol Physiol. 2010;298:L57–L66. doi: 10.1152/ajplung.00037.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakagome K., Imamura M., Kawahata K., Harada H., Okunishi K., Matsumoto T., Sasaki O., Tanaka R., Kano M.R., Chang H., Hanawa H., Miyazaki J., Yamamoto K., Dohi M. High expression of IL-22 suppresses antigen-induced immune responses and eosinophilic airway inflammation via an IL-10-associated mechanism. J Immunol. 2011;187:5077–5089. doi: 10.4049/jimmunol.1001560. [DOI] [PubMed] [Google Scholar]
- 38.Ramsey C., Kumar A. H1N1: viral pneumonia as a cause of acute respiratory distress syndrome. Curr Opin Crit Care. 2011;17:64–71. doi: 10.1097/MCC.0b013e3283427259. [DOI] [PubMed] [Google Scholar]
- 39.Nakajima N., Sato Y., Katano H., Hasegawa H., Kumasaka T., Hata S., Tanaka S., Amano T., Kasai T., Chong J.M., Iizuka T., Nakazato I., Hino Y., Hamamatsu A., Horiguchi H., Tanaka T., Hasegawa A., Kanaya Y., Oku R., Oya T., Sata T. Histopathological and immunohistochemical findings of 20 autopsy cases with 2009 H1N1 virus infection. Mod Pathol. 2012;25:1–13. doi: 10.1038/modpathol.2011.125. [DOI] [PubMed] [Google Scholar]
- 40.Fukushi M., Ito T., Oka T., Kitazawa T., Miyoshi-Akiyama T., Kirikae T., Yamashita M., Kudo K. Serial histopathological examination of the lungs of mice infected with influenza A virus PR8 strain. PLoS One. 2011;6:e21207. doi: 10.1371/journal.pone.0021207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simonian P.L., Wehrmann F., Roark C.L., Born W.K., O’Brien R.L., Fontenot A.P. gammadelta T cells protect against lung fibrosis via IL-22. J Exp Med. 2010;207:2239–2253. doi: 10.1084/jem.20100061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taube C., Tertilt C., Gyulveszi G., Dehzad N., Kreymborg K., Schneeweiss K., Michel E., Reuter S., Renauld J.C., Arnold-Schild D., Schild H., Buhl R., Becher B. IL-22 is produced by innate lymphoid cells and limits inflammation in allergic airway disease. PLoS One. 2011;6:e21799. doi: 10.1371/journal.pone.0021799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo H., Topham D.J. Interleukin-22 (IL-22) production by pulmonary natural killer cells and the potential role of IL-22 during primary influenza virus infection. J Virol. 2010;84:7750–7759. doi: 10.1128/JVI.00187-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kumar P., Thakar M.S., Ouyang W., Malarkannan S. IL-22 from conventional NK cells is epithelial regenerative and inflammation protective during influenza infection. Mucosal Immunol. 2013;6(1):69–82. doi: 10.1038/mi.2012.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paget C., Ivanov S., Fontaine J., Renneson J., Blanc F., Pichavant M., Dumoutier L., Ryffel B., Renauld J.C., Gosset P., Si-Tahar M., Faveeuw C., Trottein F. Interleukin-22 is produced by invariant natural killer T lymphocytes during influenza A virus infection: potential role in protection against lung epithelial damages. J Biol Chem. 2012;287:8816–8829. doi: 10.1074/jbc.M111.304758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hamada H., Garcia-Hernandez Mde L., Reome J.B., Misra S.K., Strutt T.M., McKinstry K.K., Cooper A.M., Swain S.L., Dutton R.W. Tc17, a unique subset of CD8 T cells that can protect against lethal influenza challenge. J Immunol. 2009;182:3469–3481. doi: 10.4049/jimmunol.0801814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nagalakshmi M.L., Rascle A., Zurawski S., Menon S., de Waal Malefyt R. Interleukin-22 activates STAT3 and induces IL-10 by colon epithelial cells. Int Immunopharmacol. 2004;4:679–691. doi: 10.1016/j.intimp.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 48.Aggarwal S., Xie M.H., Maruoka M., Foster J., Gurney A.L. Acinar cells of the pancreas are a target of interleukin-22. J Interferon Cytokine Res. 2001;21:1047–1053. doi: 10.1089/107999001317205178. [DOI] [PubMed] [Google Scholar]
- 49.Radaeva S., Sun R., Pan H.N., Hong F., Gao B. Interleukin 22 (IL-22) plays a protective role in T cell-mediated murine hepatitis: iL-22 is a survival factor for hepatocytes via STAT3 activation. Hepatology. 2004;39:1332–1342. doi: 10.1002/hep.20184. [DOI] [PubMed] [Google Scholar]
- 50.Xiang X., Gui H., King N.J., Cole L., Wang H., Xie Q., Bao S. IL-22 and non-ELR-CXC chemokine expression in chronic hepatitis B virus-infected liver. Immunol Cell Biol. 2012;90:611–619. doi: 10.1038/icb.2011.79. [DOI] [PubMed] [Google Scholar]
- 51.Zhang W., Chen Y., Wei H., Zheng C., Sun R., Zhang J., Tian Z. Antiapoptotic activity of autocrine interleukin-22 and therapeutic effects of interleukin-22-small interfering RNA on human lung cancer xenografts. Clin Cancer Res. 2008;14:6432–6439. doi: 10.1158/1078-0432.CCR-07-4401. [DOI] [PubMed] [Google Scholar]
- 52.Foster R.G., Golden-Mason L., Rutebemberwa A., Rosen H.R. Interleukin (IL)-17/IL-22-producing T cells enriched within the liver of patients with chronic hepatitis C viral (HCV) infection. Dig Dis Sci. 2012;57:381–389. doi: 10.1007/s10620-011-1997-z. [DOI] [PubMed] [Google Scholar]
- 53.Narasaraju T., Ng H.H., Phoon M.C., Chow V.T. MCP-1 antibody treatment enhances damage and impedes repair of the alveolar epithelium in influenza pneumonitis. Am J Respir Cell Mol Biol. 2010;42:732–743. doi: 10.1165/rcmb.2008-0423OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Le Goffic R., Arshad M.I., Rauch M., L‘Helgoualc’h A., Delmas B., Piquet-Pellorce C., Samson M. Infection with influenza virus induces IL-33 in murine lungs. Am J Respir Cell Mol Biol. 2011;45:1125–1132. doi: 10.1165/rcmb.2010-0516OC. [DOI] [PubMed] [Google Scholar]
- 55.Kumar P.A., Hu Y., Yamamoto Y., Hoe N.B., Wei T.S., Mu D., Sun Y., Joo L.S., Dagher R., Zielonka E.M., Wang de Y., Lim B., Chow V.T., Crum C.P., Xian W., McKeon F. Distal airway stem cells yield alveoli in vitro and during lung regeneration following H1N1 influenza infection. Cell. 2011;147:525–538. doi: 10.1016/j.cell.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tohyama M., Yang L., Hanakawa Y., Dai X., Shirakata Y., Sayama K. IFN-alpha enhances IL-22 receptor expression in keratinocytes: a possible role in the development of psoriasis. J Invest Dermatol. 2012;132:1933–1935. doi: 10.1038/jid.2011.468. [DOI] [PubMed] [Google Scholar]
- 57.Brand S., Beigel F., Olszak T., Zitzmann K., Eichhorst S.T., Otte J.M., Diepolder H., Marquardt A., Jagla W., Popp A., Leclair S., Herrmann K., Seiderer J., Ochsenkuhn T., Goke B., Auernhammer C.J., Dambacher J. IL-22 is increased in active Crohn’s disease and promotes proinflammatory gene expression and intestinal epithelial cell migration. Am J Physiol Gastrointest Liver Physiol. 2006;290:G827–G838. doi: 10.1152/ajpgi.00513.2005. [DOI] [PubMed] [Google Scholar]
- 58.Zemke A.C., Snyder J.C., Brockway B.L., Drake J.A., Reynolds S.D., Kaminski N., Stripp B.R. Molecular staging of epithelial maturation using secretory cell-specific genes as markers. Am J Respir Cell Mol Biol. 2009;40:340–348. doi: 10.1165/rcmb.2007-0380OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ramirez M.I., Pollack L., Millien G., Cao Y.X., Hinds A., Williams M.C. The alpha-isoform of caveolin-1 is a marker of vasculogenesis in early lung development. J Histochem Cytochem. 2002;50:33–42. doi: 10.1177/002215540205000104. [DOI] [PubMed] [Google Scholar]
- 60.Pandit K.V., Corcoran D., Yousef H., Yarlagadda M., Tzouvelekis A., Gibson K.F., Konishi K., Yousem S.A., Singh M., Handley D., Richards T., Selman M., Watkins S.C., Pardo A., Ben-Yehudah A., Bouros D., Eickelberg O., Ray P., Benos P.V., Kaminski N. Inhibition and role of let-7d in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2010;182:220–229. doi: 10.1164/rccm.200911-1698OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Epithelial morphologic findings in naive C57BL/6 and IL-22−/− mice (n = 4). Large and small airways. Original magnification, ×600.