Abstract
Liposarcoma is a type of soft tissue sarcoma that exhibits poor survival and a high recurrence rate. Treatment is generally limited to surgery and radiation, which emphasizes the need for better understanding of this disease. Because very few in vivo and in vitro models can reproducibly recapitulate the human disease, we generated several xenograft models from surgically resected human dedifferentiated liposarcoma. All xenografts recapitulated morphological and gene expression characteristics of the patient tumors after continuous in vivo passages. Importantly, xenograftability was directly correlated with disease-specific survival of liposarcoma patients. Thus, the ability for the tumor of a patient to engraft may help identify those patients who will benefit from more aggressive treatment regimens. Gene expression analyses highlighted the association between xenograftability and a unique gene expression signature, including down-regulated PTEN tumor-suppressor gene expression and a progenitor-like phenotype. When treated with the PI3K/AKT/mTOR pathway inhibitor rapamycin alone or in combination with the multikinase inhibitor sorafenib, all xenografts responded with increased lipid content and a more differentiated gene expression profile. These human xenograft models may facilitate liposarcoma research and accelerate the generation of readily translatable preclinical data that could ultimately influence patient care.
Liposarcoma (LPS), a neoplasm arising within adipose tissue that is thought to originate from mesenchymal stem cells,1–3 is a type of soft tissue sarcoma and is associated with poor patient outcomes.4–6 LPS is classified into three histological subtypes: well-differentiated/dedifferentiated (WD/DD), pleomorphic, and myxoid/round cell. Of these, WD/DD is the most prevalent subtype. The extent of differentiation, as reflected by histological grade, remains the most important determinant of patient outcome. The main treatment options for patients with LPS are surgery and radiation, because chemotherapy typically has little effect on the overall prognosis for patients with locally advanced or metastatic disease.7,8
The establishment of a clinically relevant disease model is vital in studying the molecular mechanisms underlying LPS malignant transformation and evaluating emerging targeted therapies. Certain characteristic mutations of LPS have been identified, such as chromosome 12q amplification in WD/DD LPS and a chromosomal translocation in myxoid LPS4; however, our knowledge of the cell-of-origin and major pathway alterations associated with LPS is still limited. In the present study, we developed a direct LPS xenograft method by implanting and serially passaging freshly resected LPS samples from patients into immunodeficient mice. We demonstrated that these xenograft models not only recapitulate the patient tumors, but carry a unique gene expression signature and can be used for testing targeted treatment.
Materials and Methods
Collection of Tumor Samples
Patients with suspected LPS who underwent surgery at the University of California, Los Angeles (UCLA), were enrolled on an Institutional Review Board–approved tissue procurement protocol after giving written informed consent. Each surgically resected tumor was sliced into sections with a sterile razor blade and was divided as follows: one portion was immediately frozen and stored at −80°C for genomic and gene expression analyses, another portion of the tumor was fixed in 10% formalin for histology, and the remainder was minced into small pieces for xenograft implantation and tissue culture studies. Histological review by a sarcoma pathologist (S.M.D.) confirmed diagnosis, subtype, and grade of tumors used in the present study.
Xenograft Implantation and Passage
Transplantation studies were performed using NOD.CB17-Prkdcscid/J (common name, NOD-SCID) or NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ (common name, NSG) immunodeficient mice acquired from colonies bred in-house at UCLA. For the initial xenograft, approximately 100 mg of the patient tumor was cut into fragments (approximately 3 mm3), using a sterile razor blade. Mice were anesthetized with isoflurane, and the fur on the hindquarters was shaved and prepared with an antiseptic agent. An incision was made, and one of the tumor fragments (approximately 3 mm3) was inserted into the subdermal space. The incision was closed with either 5-0 Prolene suture or a skin stapler; the closure was removed on postoperative day 10. Mice were maintained in barrier cages on standard chow diet with food and water available ad libitum; 2 mg/mL sulfamethoxazole and 0.4 mg/mL trimethoprim was added to drinking water to prevent infection. All studies were performed in accordance with the UCLA Animal Research Committee and Division of Laboratory Animal Medicine (protocol no. 96-126). All surgery was performed under isoflurane anesthesia, and all efforts were made to minimize suffering.
Implanted samples were allowed a 6-month incubation period to form a palpable tumor. When successful engraftment occurred, tumors were collected before reaching 1.5 cm in diameter. Harvested tumors were separated into three fragments: for histopathology, gene expression analysis, and serial transplantation for establishing stable xenograft models or cell culture models.
Tissue Dissociation for Xenografts
Single-cell suspensions from tumors were generated using either mechanical dissociation or cell straining. For mechanical dissociation, the gentleMACS dissociator (Miltenyi Biotec, Auburn, CA) was used. Approximately 2 mg of tumor was added to a gentleMACS C tube containing 5 mL of enzymatic solution: 0.85 mg/mL Gibco collagenase (Life Technologies–Invitrogen, Carlsbad, CA) in Dulbecco’s modified Eagle’s medium (DMEM), 10% fetal bovine serum, 1% penicillin/streptomycin. The C tube was placed upside down onto the sleeve of the dissociator, and gentleMACS programs C and D were run in succession. The tube was removed from the machine, and the material was applied to a 70-μm cell strainer, without force, into a 50-mL tube. The cell strainer was rinsed with 10 mL of Dulbecco’s phosphate buffered saline (DPBS), and the resulting solution was centrifuged at 300 × g for 10 minutes, aspirated, and resuspended in DPBS. Another dissociation method involved cutting the tumor into 2- to 4-mm pieces and, using the blunt end of a syringe plunger, manually straining the pieces through a 100-μm cell filter into 5 to 10 mL of DPBS. The cells were centrifuged at 300 × g for 10 minutes, aspirated, and resuspended in DPBS.
For cryopreservation, tissue was manually strained through a 100-μm cell filter with the blunt end of a syringe into 1 to 2 mL of freezing medium (10% dimethyl sulfoxide and 90% fetal bovine serum). Cells were then gradually frozen at −80°C and cryopreserved for at least 24 hours before being rapidly thawed in a 37°C water bath. After addition of 10 mL of DPBS, the cells were centrifuged, the supernatant was discarded, and cells were resuspended in DPBS. Cells were counted on a hemocytometer using Trypan Blue exclusion and concentration was adjusted to 5 × 106 cells/mL. Mice were anesthetized with isoflurane, fur on the hindquarters was shaved, and the skin was prepared with an antiseptic agent. Approximately 0.5 × 106 to 1.0 × 106 cells were injected subcutaneously with a 28-gauge needle on a 0.5-mL insulin syringe.
Tissue Culture
Tumor tissue samples (200 to 300 mg) were minced with a scalpel and then were dissociated by carefully passing them in complete medium (basal medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin) through a 40-μm cell strainer. Three basal media were tested: DMEM, 1:1 DMEM/Ham’s F12 medium, and RPMI-1640 medium. Cells were spun down at 150 × g for 5 minutes, resuspended in 10 mL of red cell lysis buffer (150 mmol/L ammonium chloride, 1 mmol/L potassium bicarbonate, 0.1 mmol/L EDTA), and incubated at room temperature for 10 minutes. After 5 minutes of spinning down at 150 × g, cells were resuspended in complete medium and plated onto six-well dishes (106 cells per well). Cell adhesion was observed 24 hours after plating, and confluent plates were expanded. Cells were maintained in a 37°C incubator with a 5% CO2–enriched atmosphere and were monitored for cell adhesion.
For inoculation in mice, 70% confluent cells were rinsed with DPBS and treated with 0.25% trypsin–EDTA (Life Technologies–Invitrogen) until the cells separated from the dish. Complete medium was added to quench the trypsin reaction, and the cells were centrifuged at 150 × g for 5 minutes. The supernatant was aspirated and cells were resuspended in DPBS, counted on a hemocytometer using Trypan Blue exclusion, and adjusted to a concentration of 5 × 106 cells/mL. Mice were anesthetized with isoflurane, fur in the injection region was shaved, the skin was prepared with an antiseptic, and 0.5 × 106 to 1.0 × 106 cells were injected subcutaneously with a 28-gauge needle on a 0.5-mL insulin syringe.
Histology and Immunohistochemistry
The sample was fixed in 10% phosphate-buffered formalin overnight and then was transferred into 50% to 70% ethanol. Shortly afterward, the tissue was embedded in paraffin using standard protocols, cut into sections (4 μm thick), stained with H&E at the UCLA Translational Pathology Core Laboratory, and reviewed by a sarcoma pathologist (S.M.D). Slides were digitally scanned at ×400 magnification using an Aperio XL system (Aperio, Vista, CA), and images were analyzed using ImageScope version 11.1.2.752 (Aperio) and Adobe Photoshop CS4 version 11.0.2 software.
For immunohistochemical analysis, formalin-fixed, paraffin-embedded sections were deparaffinized in xylene and rehydrated in graded alcohol. Endogenous peroxidase activity was blocked with 3% hydrogen peroxide. Antigen retrieval was performed by boiling the sections in 0.01 mol/L citric acid buffer (pH 6.0) for 15 minutes. Sections were first blocked with 5% normal donkey serum in DPBS, and then incubated with primary antibody against Ki-67 (1:500; Vector Laboratories, Burlingame, CA), PTEN (1:100), p-S6 (Ser240/244, 1:100), or p-ERK (Thr202/Tyr204, 1:300; all from Cell Signaling Technology, Danvers, MA) overnight at 4°C. Sections were then incubated with 1:500 biotinylated secondary antibody (Jackson ImmunoResearch Laboratories, West Grove, PA) for 1 hour at room temperature. Antibody binding was detected with a Vectastain Elite ABC kit (PK-6100; Vector Laboratories) and visualized with 3,3′-diaminobenzidine (DAB; Vector Laboratories). Sections were counterstained in hematoxylin.
For visualizing lipids, frozen sections were stained in a 0.3% Oil Red O/isopropyl alcohol solution for 7 to 10 minutes at room temperature. Excess stain was washed out in 60% isopropyl alcohol, followed by washes in 30% isopropyl alcohol and distilled water. Slides were counterstained with hematoxylin and then rinsed with two changes of tap water; coverslips were mounted with glycerol. Slides were subsequently stored at 4°C. Images were captured on a BX60 microscope equipped with a Microfire camera (both from Olympus America, Center Valley, PA), using PictureFrame software version 2.1 (Optronics, Goleta, CA), and were analyzed with Adobe Photoshop CS4 software.
Treatment Studies
Tissue from an existing xenograft was dissociated using a 100-μm mesh cell strainer into DPBS. Viable cells were counted with Trypan Blue exclusion, centrifuged, and resuspended in equal portions of DPBS and Matrigel (BD Biosciences, San Jose, CA), and 5 × 105 cells were injected subcutaneously. Tumors were measured with calipers. When tumors reached approximately 5 mm in diameter, mice were randomly assigned into placebo (vehicle) or treatment groups, with a minimum of five mice per group. Tumor volume was measured twice weekly and calculated as tumor volume V = 0.52 a2b, where a is the shorter and b is the longer diameter.
Rapamycin powder (LC Laboratories, Woburn, MA) was reconstituted in 100% ethanol to a stock solution of 10 mg/mL and was stored at −20°C. Fresh working solution was made each time by diluting the stock solution to 1 mg/mL with vehicle (5.68% Tween-80, 5.68% polyethylene glycol 400 in water). Rapamycin (4 mg/kg) in vehicle or an equal volume of vehicle alone was administered via intraperitoneal injection daily. Sorafenib pills were weighed, then crushed into powder and stored at −20°C. Powder was dissolved into freshly made vehicle (12.5% ethanol, 12.5% cremophor in water) to 20 mg/mL. Sorafenib (70 mg/kg) in vehicle or an equal volume of vehicle alone was administered via oral gavage daily. Mice were treated daily with one of the following combinations: rapamycin vehicle and sorafenib vehicle, rapamycin drug and sorafenib vehicle, sorafenib drug and rapamycin vehicle, or rapamycin drug and sorafenib drug. Body weight and tumor measurements were collected every 3 to 4 days. Tumors were collected, and the mouse was sacrificed if the tumor reached 1.5 cm in diameter, or if the mouse lost >10% of its body weight within 1 week, whichever occurred first.
Triglyceride Quantification
Triglycerides were quantified using a triglyceride colorimetric assay kit according to the manufacturer’s protocol (Cayman Chemical, Ann Arbor, MI). Briefly, frozen tumor samples from the treatment study were weighed and homogenized in the manufacturer’s 1× standard diluent assay reagent. After applying the manufacturer’s triglyceride enzyme mixture, the triglyceride standard curve and samples were read at 530 nm using a Benchmark Plus plate reader (Bio-Rad Laboratories, Hercules, CA). Triglyceride levels for each sample were normalized to the weight of tissue homogenized.
RNA Isolation and Gene Expression Microarray
Patient tumor samples and xenograft passages were prepared and RNA was isolated as described previously.9 For the xenograft treatment studies, frozen tissue from LPSX1 and LPSX3 vehicle-only and combination treatment groups was homogenized with a MagNA Lyser instrument and RNA was extracted with a MagNa Pure Compact nucleic isolation system (both from Roche Diagnostics, Indianapolis, IN). A low-input QuickAmp one-color labeling kit (Agilent Technologies, Santa Clara, CA) was used for first-strand, second-strand, and in vitro transcription reactions. Samples were analyzed on Agilent SurePrint Human GE 8x60k microarrays and scanned on an Agilent DNA microarray scanner (both from Agilent Technologies) at the UCLA Clinical Microarray Core.
CGH and Gene Expression Analysis
DNA isolation and comparative genomic hybridization (CGH) procedures for patient samples have been described in detail previously.9 CGH Analytics software version 4.0 (Agilent Technologies) was used for copy number analysis, using the ADM2 algorithm (threshold 5) with fuzzy zero and centralization corrections to minimize background noise. A minimum of three consecutive probes was required to define a region as amplified or deleted.
In gene expression analysis, a two-tailed t-test was used for comparing expression between groups. For a gene represented by multiple probes, its representative P value was the lowest P value among the probes. Gene ontology analysis was performed using pathway databases downloaded from Gene Ontology (http://www.geneontology.org/GO.downloads.shtml) and the Broad Institute Molecular Signature Database (http://www.broadinstitute.org/gsea/msigdb/index.jsp).10,11 Fisher’s exact test was used to identify the gene ontology or biological pathway significantly enriched by differentially expressed genes between groups (P < 0.05, two-tailed t-test, and fold change >1.5). Cluster 3.0 software (http://bonsai.hgc.jp/∼mdehoon/software/cluster), based on the original algorithm of Eisen et al,12 was used in hierarchical clustering analysis, in which the expression profile of each gene was first transformed to z-scores and then samples were clustered by using average linkage method based on uncentered correlation coefficients as the distance metric among samples.
The Rank-Rank Hypergeometric Overlap algorithm (http://systems.crump.ucla.edu/rankrank)13 was used to compare gene expression data from our samples versus gene expression of preadipocytes collected after 0 and 3 days of induction in differentiation medium (the series of Söhle et al14; http://www.ncbi.nlm.nih.gov/geo; GEO accession no. GSE28628).
Statistical Analysis
Ordinal logistic regression was performed on clinical data from all 22 patients using STATA/SE version 12.1 software (StataCorp, College Station, TX). Multiple predictors for engraftment and passage capability were tested, including patient age at diagnosis, sex, location of tumor, sample origin (primary, recurrent, or metastatic tumor), chemotherapy before tumor sample collection, radiation therapy before tumor sample collection, grade, size of tumor, presence of multifocal disease, patient history of other cancer type, presence and/or development of metastasis, and histological subtype. The odds ratio (OR), a measure of effect size, was calculated for each significant variable in the univariate ordinal regression analysis, with appropriate confidence intervals (CI). Given the small sample size (n = 22), a trending level of P < 0.1 was used to reveal possible influential variables; in the final model, a P < 0.05 was used to determine statistical significance. Other statistical analyses (Mantel–Cox test, one-way analysis of variance, and Friedman test) were performed using GraphPad Prism version 5.04 software (GraphPad Software, San Diego, CA), with P < 0.05 considered significant.
Results
Ability to Engraft and Passage LPS Tumor Correlates with Disease-Specific Survival
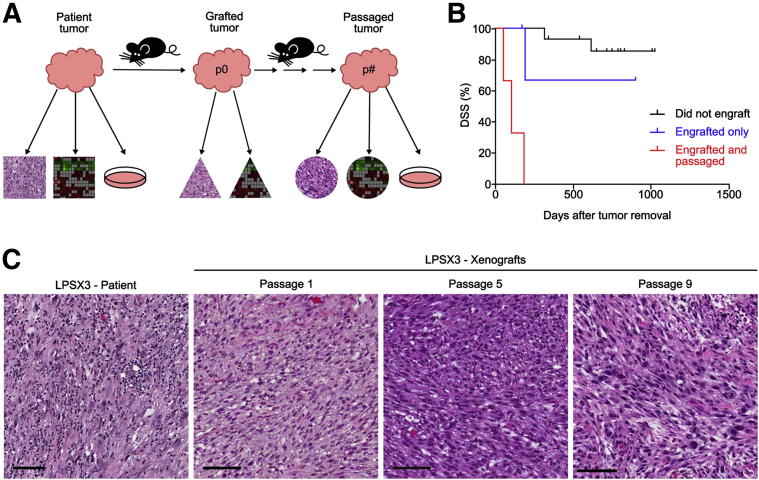
To generate clinically relevant LPS models for investigating molecular etiology and evaluating response to targeted therapies, we used a xenotransplantation approach by subcutaneously implanting fresh, surgically resected LPS samples into NOD/SCID or NOD/SCID/IL2γ−/− (NSG) mice (Figure 1A). Over a 2-year period, we implanted 22 surgically resected tumors (myxoid/round cell, n = 5; WD, n = 7; and DD, n = 10). These tumors varied in grade, site of origin, size, and treatment history (Table 1). Of the 10 DD samples, 8 came from tumors that had both WD and DD components, but all DD areas were pathologically diagnosed as high grade, and a sarcoma pathologist (S.M.D.) selected DD areas for processing. Within 6 months of implantation, seven patient tumors successfully grew in mice (2/5 myxoid/round cell, 0/7 WD, and 5/10 DD), resulting in a 32% engraftment rate (Table 1). All seven engrafted LPS tumors were high grade, and three of these xenografts, LPSX1, LPSX2, and LPSX3, could be serially passaged in mice (14% of all tumors, 43% of engrafted tumors) for 10 to 17 passages. In contrast, cells isolated directly from patient tumors failed to be serially passaged under the same in vitro culture conditions (data not shown).
Figure 1.
Xenograft strategy and correlation of DSS with tumor xenograftability. A: Schematic of study design. Surgically resected LPS and xenografted tumors were portioned for histopathological, gene expression, and in vitro and in vivo studies. B: Patients whose tumors could be serially passaged (n = 3) had significantly shortened median survival, compared with patients whose tumors engrafted but could not be passaged (n = 4; P = 0.01) and with those whose tumors did not engraft at all (n = 15; P < 0.0001, Mantel–Cox test). For surviving patients, tick marks indicate the latest follow-up. C: Histopathological features of the patient tumors are maintained in xenografts over serial passages. Shown is a representative passageable patient sample (LPSX3; H&E stain) and serial passages of the resulting xenografts. Scale bar = 100 μm.
Table 1.
Clinical and Pathological Data for Study Samples
| ID | Histology | Grade | Origin of sample | Site | Size (cm) | Treatment | Sex | Age | Patient status |
|---|---|---|---|---|---|---|---|---|---|
| Tumors that did not engraft | |||||||||
| 1 | Myxoid | Low | Primary | Thigh | 29 | XRT | M | 39 | NED |
| 2 | Myxoid/RC | Int. | Primary | Thigh | 16 | XRT | M | 28 | NED |
| 3 | Myxoid/RC | High | Primary | Thigh | 9 | XRT | M | 47 | NED |
| 4 | WD | Low | Primary | RP | 15 | F | 58 | NED | |
| 5 | Cellular WD | Low | Primary | RP | 41 | XRT | F | 51 | NED |
| 6 | WD | Low | Primary | RP | 32 | AC | M | 61 | AWD |
| 7 | WD | Low | Recurrent | RP | 15 | M | 72 | NED | |
| 8 | Cellular WD | Low | Recurrent | RP | 3 | XRT | M | 49 | NED |
| 9 | Cellular WD | Low | Recurrent | RP | 20 | XRT | M | 54 | AWD |
| 10 | WD | Low | Recurrent | RP | 22 | XRT | M | 43 | AWD |
| 11 | DD∗ | High | Primary | RP | 31 | XRT | F | 57 | NED |
| 12 | DD∗ | High | Primary | RP | 30 | M | 86 | DOD | |
| 13 | DD∗ | High | Primary | Abdomen | 13 | XRT | M | 66 | AWD |
| 14 | DD∗ | High | Recurrent | RP | 20 | XRT | F | 60 | NED |
| 15 | DD∗ | High | Recurrent | RP | 5 | XRT | F | 86 | DOD |
| Tumors that engrafted only | |||||||||
| 16 | Myxoid/RC | High | Primary | Gluteal | 16 | AC, XRT | F | 29 | NED |
| 17 | Myxoid/RC | High | Metastatic | Abdomen | 22 | AC | M | 66 | AWD |
| 18 | DD | High | Recurrent | RP | 5 | AC, XRT | F | 58 | NED |
| 19 | DD∗ | High | Metastatic | Flank | 4 | NC, XRT | F | 61 | DOD |
| Tumors that engrafted and could be passaged | |||||||||
| LPSX1 | DD | High | Recurrent | Abdomen | 12 | NC, XRT | F | 51 | DOD |
| LPSX2 | DD∗ | High | Recurrent | Thigh | 15 | XRT | M | 90 | DOD |
| LPSX3 | DD∗ | High | Primary† | RP | 27 | NC | M | 75 | DOD |
F, female; M, male; AC, adjuvant chemotherapy; AWD, alive with disease; DD, dedifferentiated; DOD, died of disease; Int., intermediate; NC, neoadjuvant chemotherapy; NED, no evidence of disease; RC, round cell; RP, retroperitoneum; WD, well-differentiated; XRT, radiation therapy.
Dedifferentiated arising from a well-differentiated tumor. All DD areas were pathologically diagnosed as high grade, and only DD areas were selected for processing.
The patient presented with metastatic lesions, but the sample came from the primary tumor.
To determine whether any clinicopathological factors influenced the ability to establish a xenograft, we evaluated multiple patient characteristics for each sample. Given the small sample size (n = 22), this was largely an exploratory analysis to determine possible clinical predictors of a tumor’s successful passage and/or engraftment. Univariate analysis of 12 clinical variables revealed that the size of tumor (P = 0.084; OR = 0.90; 95% CI = 0.80–1.01) and presence of multifocal disease (P = 0.058; OR = 15.35; 95% CI = 0.91–257.27) correlated with the ability of an LPS patient sample to be engrafted and/or passaged. On multivariate analysis, tumor size (P = 0.052; OR = 0.85, CI = 0.72–1.00) and presence of multifocal disease (P = 0.026; OR = 38.6, CI = 1.53–969.13) remained significant. Given the small sample size, however, and high variability in the data, these results should be interpreted with caution.
Next, we aimed to specifically evaluate the association of engraftment and passageability with disease-specific survival (DSS). As expected, the ability to engraft and be passaged significantly correlated with poor DSS on univariate analysis (P < 0.0001, Mantel–Cox test). Patients whose samples could be xenografted and serially passaged died of disease within 200 days of tumor removal (n = 3), in sharp contrast to those patients whose tumors did not grow (n = 15; P < 0.0001, Mantel–Cox test) (Figure 1B). Patients whose samples engrafted but could not be passaged also (n = 4) had a significantly better survival than those whose samples could be passaged (n = 3; P = 0.01, Mantel–Cox test) (Figure 1B). Such a correlation is consistent with grade-associated DSS studies from our research group on 46 independent LPS patients and another group's study of 61 patients showing that patients with high-grade LPS had significantly shortened survival, compared with patients with low grade LPS tumors.15,16 Taken together, these data suggest that the xenograftability of LPS can be used for studying key molecular and genetic alterations associated with the aggressive phenotype of LPS.
Xenografted Tumors Recapitulate Histopathological and Gene Expression Features of the Patient Tumors
To confirm that the xenografts recapitulate the patient disease, we compared surgically resected patient tumors with their derivatives (ie, xenograft tumors that had been passaged in vivo). Representative histological images from the patient tumor LPSX3 and its resulting xenografts passaged in vivo for 1 to 9 passages demonstrated that no major morphological changes occurred during serial passage (Figure 1C). We also compared in vitro cultured cells and different tissue dissociation and maintenance methods before transplantation into mice and found no resulting histopathological differences or proliferation rates, as measured by Ki-67 index (Supplemental Figure S1). These analyses demonstrated that in vivo passaged and culture-derived xenografts maintained the histological features of the patient tumors.
We also looked at specific genetic alterations associated with LPS, to confirm that the xenograft model accurately reflects the patient disease. Previous work by our research group9 and others17–21 identified characteristic amplicons on chromosome 12q in most WD and DD LPS tumors. Our CGH array analysis showed that passageable patient tumors indeed carry amplifications in this region (Supplemental Figure S2A). Amplicon-associated genes (eg, MDM2 and CDK4) showed similar up-regulation in both the patient tumors and their corresponding xenografts (Supplemental Figure S2B). These analyses indicated that the xenograft models recapitulate both histopathological and molecular features of the patient disease and therefore are suitable models for studying LPS.
Xenograftability Is Associated with Aggressive Proliferation and Invasion Signatures and with Up-Regulated PI3K/AKT/mTOR Pathway
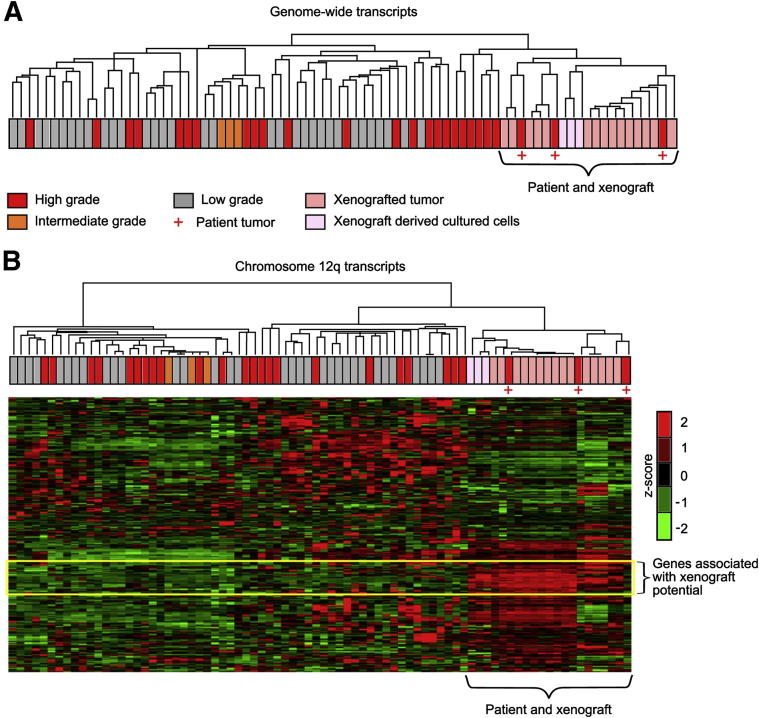
To understand the mechanisms underlying xenograftability and the associated decrease in DSS, we performed genome-wide gene expression analysis. Our unsupervised cluster analysis showed that the patient tumors cluster alongside their xenografts and xenograft-derived cultured cells (Figure 2A). The analysis also showed that gene expression of patient tumors and their xenograft derivatives was closely associated with that of other high-grade patient LPS tumors in our database, suggesting that mining the unique gene expression signature within this cluster may help identify the molecular mechanisms underlying the aggressive behavior of this subset of LPS. An example is our analysis of chromosome segment 12q13.3∼q23.1 (Figure 2B), which includes the commonly detected MDM2–CDK4 amplicon (Supplemental Figure S2A). We found that the patient tumors, xenografts, and xenograft-derived cultured cells maintain their own cluster and carry a unique gene expression signature (Figure 2B). Further global supervised transcriptome analyses identified approximately 1000 genes whose expressions are differentially altered between the passaged tumors (n = 3) and tumors that could be engrafted but not passaged (n = 3; tumor 16 was not included in gene expression analysis) (P < 0.05, two-tailed t-test, fold change >1.5). Importantly, several cell proliferation and invasion-related pathways were significantly up-regulated in the passaged xenograft samples (Supplemental Table S1).
Figure 2.
Gene expression analysis reveals unique features of xenograftable tumors. A: Unsupervised hierarchical cluster analysis based on 41,000 transcripts separates the xenograftable patient tumors and their xenografts from other LPS samples. B: Further analysis of transcripts of chromosome 12q amplicons (12q13.3∼q23.1) reveals a set of genes specifically associated with the xenograftable patient tumors and their xenografts (yellow box).
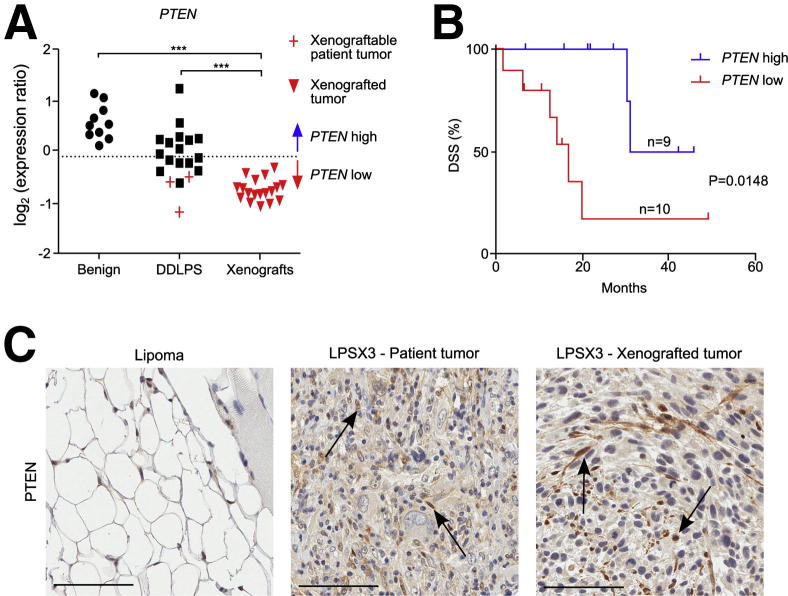
We focused on the alteration of the mTOR pathway, because we hypothesized that PTEN, a tumor suppressor upstream of mTOR, is responsible for the dys-regulated mTOR pathway. Although CGH analysis of the patient tumors from which the xenografts were derived showed no genetic alterations at the PTEN locus,9 expression analysis showed significantly decreased PTEN levels in DD LPS (n = 16), compared with benign lipoma samples (n = 10) (P < 0.001, one-way analysis of variance) (Figure 3A). The patient samples and their xenografts also showed low PTEN protein levels, except that stroma and blood vessels had higher PTEN intensity (Figure 3C; Supplemental Figure S3A). Furthermore, patient tumors and their derived xenografts had the lowest PTEN expression levels of all DD LPS (Figure 3, A and C), supporting the notion that decreased PTEN expression, most likely due to epigenetic mechanisms, may up-regulate the PI3K/AKT/mTOR pathway. Using the expression levels of the benign tumors as a baseline (below the 95% CI from the mean expression of benign samples; Figure 3A), we stratified the DD LPS into two groups based on PTEN expression levels (Figure S3B). Importantly, low PTEN expression was correlated with poor DSS (n = 19; P = 0.0148, Mantel–Cox test) (Figure 3B), which suggests that the PTEN-controlled PI3K/AKT/mTOR pathway is a potential prognostic signature and therapeutic target for DD LPS.
Figure 3.
PTEN gene and protein expression in xenograftable tumors. A: Gene expression of PTEN is lower in DD LPS, compared with benign lipomas, but patient tumors and their derived xenograft tumors show even lower gene expression. Mean expression of benign samples is indicated by a dotted line. B: DSS is significantly shortened in patients with low PTEN expression (P = 0.0148, Mantel–Cox test). The cutoff for low versus high expression was below the 95% CI from the mean expression of benign samples (shown in A). C: Immunohistochemical analysis of PTEN shows high positive staining in a benign lipoma, as well as of stromal cells (arrows) in LPSX3 patient tumor and derived xenograft. Scale bar = 100 μm.
LPS Xenografts with Low PTEN Expression Are Sensitive to anti-mTOR and Multikinase Inhibitor Treatment
To test whether DD LPS with down-regulated PTEN expression and up-regulated mTOR pathway activity is sensitive to targeted inhibition of the PI3K/AKT/mTOR pathway, we treated our xenograft models with rapamycin, a specific inhibitor of mTOR. Although rapamycin has been used clinically for soft tissue sarcoma, it has not been formally evaluated in LPS patients. Recent reports indicate that inhibition of mTOR kinase or the PI3K/AKT pathway may cause feedback-dependent activation of upstream AKT and receptor tyrosine kinase signaling,22–24 leading to therapeutic resistance. We therefore included the multitargeted tyrosine kinase inhibitor sorafenib in our study design. Sorafenib as a single agent has been used in phase 2 trials for advanced soft tissue sarcomas, including LPS, but failed to show significant clinical efficacy.25 We reasoned that combining sorafenib with rapamycin might block the rapamycin-induced feedback activation of upstream kinases and thus lead to sustainable therapeutic effect.
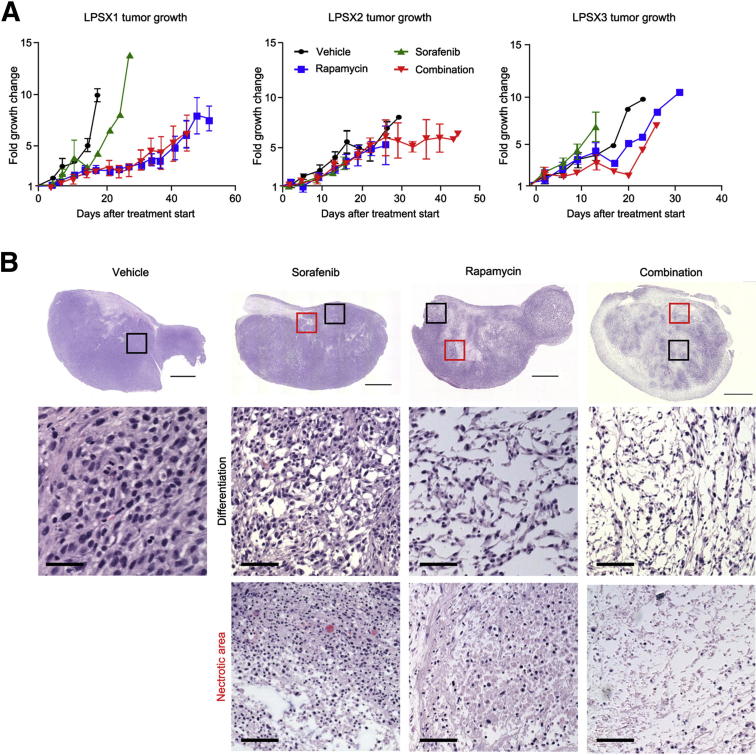
The three passageable LPS xenografts (LPSX1, LPSX2, and LPSX3) were treated daily with no drug (vehicle), rapamycin only, sorafenib only, or a combination of rapamycin and sorafenib. Tumor growth was monitored over the course of 3 to 7 weeks, or until tumors reached 1.5 cm in diameter. LPSX1 treated with rapamycin or combination therapy showed significantly reduced tumor growth, as measured by tumor volume, compared with either the vehicle-only or sorafenib-treated groups; however, LPSX2 and LPSX3 showed no reduction in tumor size, regardless of treatment (Figure 4A; Supplemental Figure S4).
Figure 4.
Tumor response to single and combination treatments. A: Tumor growth curves. LPSX1 showed slowed growth when treated with rapamycin or combination treatment, but LPSX2 and LPSX3 did not show slowed tumor growth in response to any of the treatments. B: Histological analysis shows evidence of treatment response, including necrosis and morphological changes suggestive of differentiation. Representative images of LPSX3 (H&E stain) at low (top row) and high magnification (middle and bottom rows) show differentiation and necrotic areas. Boxed areas correspond (although not to scale) to the high-power images of differentiation (black boxes) and necrosis (red boxes). Data are expressed as means ± SEM. n = 3 or 4 per treatment group. Scale bars: 2.5 mm (low-power images); 50 μm (high-power images).
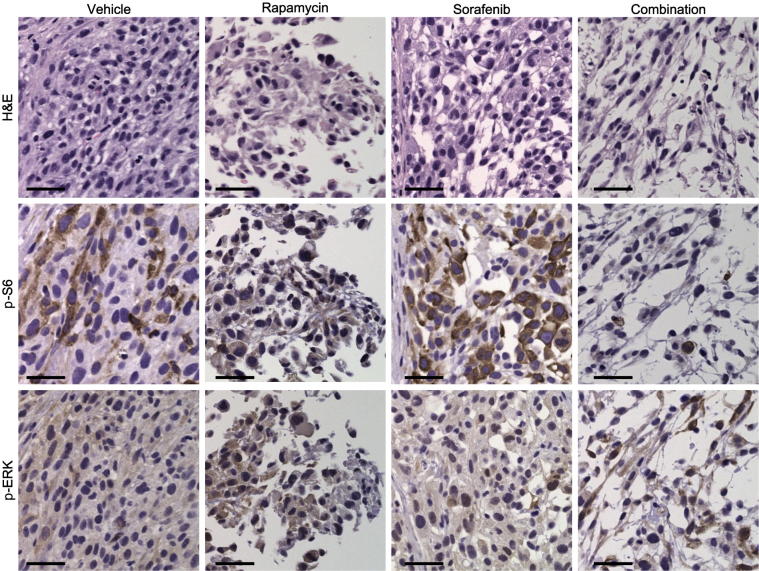
There are significant limitations to the current standard size-based RECIST criteria for monitoring response to therapy in soft tissue sarcomas,26 because tumors may undergo necrosis or differentiation without significant changes in tumor size. We therefore resected all xenograft tumors and examined the effects of various treatments at the histopathological level. In contrast to the size measurements, all three xenograft models responded to rapamycin and to combination treatment with significant necrosis and reduced cellularity, which was clearly identified in H&E-stained sections (Figure 4B; Supplemental Figure S5). Although the vehicle-only and sorafenib-treated tumors retained high cellular density, rapamycin-treated tumors were more differentiated with lower cellular density. In support of our hypothesis, combination treatment enhanced the effects of rapamycin, with more necrotic areas and further reduced cellularity (Figure 4B; Supplemental Figure S5). In addition to evaluating histopathological changes, we assessed the intended pathway targets of the drugs using immunohistochemistry. p-S6 was chosen as a surrogate marker of mTOR activity, and p-ERK was chosen as a downstream surrogate marker of sorafenib treatment. As expected, the levels of p-S6 decreased in response to rapamycin or combination treatment. p-ERK levels were relatively equal across treatment groups, with positive staining present in both the nucleus and the cytoplasm (Figure 5; Table 2). Although ERK is known to translocate to the nucleus when phosphorylated, this activated form of the protein has been observed in the cytoplasm in human melanoma cancer samples.27 These histopathological changes suggest that, even though tumor size did not differ among treatment groups of LPSX2 and LPSX3, the tumors responded to rapamycin or combination treatments similarly to LPSX1.
Figure 5.
Response of intracellular targets to single and combination treatment. Representative images from consecutive sections of LPSX3 show down-regulation of p-S6 and relatively stable p-ERK levels from immunohistochemical staining in response to rapamycin, sorafenib, and combination treatment or vehicle alone. Scale bar = 50 μm.
Table 2.
Quantification of Immunohistochemical Analysis in LPS Xenografts
| Marker and xenograft | Staining score (density/intensity)∗ |
|||
|---|---|---|---|---|
| Vehicle | Rapamycin | Sorafenib | Combination | |
| p-S6 | ||||
| LPSX1 | 2/3+ | 1/1+ | 2/2+ | 1/1+ |
| LPSX2 | 2/3+ | 1/1+ | 2/2+ | 1/1+ |
| LPSX3 | 2/3+ | 1/1+ | 2/2+ | 1/1+ |
| p-ERK | ||||
| LPSX1 | 2/2+ | 3/2+ | 3/2+ | 3/2+ |
| LPSX2 | 1/1+ | 1/1+ | 2/3+ | 2/1+ |
| LPSX3 | 2/1+ | 2/1+ | 2/2+ | 2/1+ |
Staining density is scored as <1, negative staining; 1, 1% to 25% of tumor cells staining; 2, 26% to 74% of tumor cells staining; 3, ≥75% of tumor cells staining. Staining intensity is scored as 1+, weak; 2+, moderate; 3+, strong.
Xenograftable DD LPS Are More Progenitor-Like, and Combination Treatment Induces Differentiation Resulting in Decreased Progenitor Expression Signature and Increased Lipid Content
Accumulated evidence suggests that different subtypes of LPS may represent developmental blockages of adipocyte differentiation from mesenchymal stem cells.2 Inducing differentiation could therefore be a potential therapeutic strategy for LPS.28,29 One of the morphological changes we observed after rapamycin or combination treatment was the presence of cells that more closely resembled adipocytes, suggesting potential differentiation. We therefore first wanted to determine whether the patient tumors that generated serially passaged xenografts are more progenitor-like and, second, whether the combination treatment can alleviate the differentiation blockage.
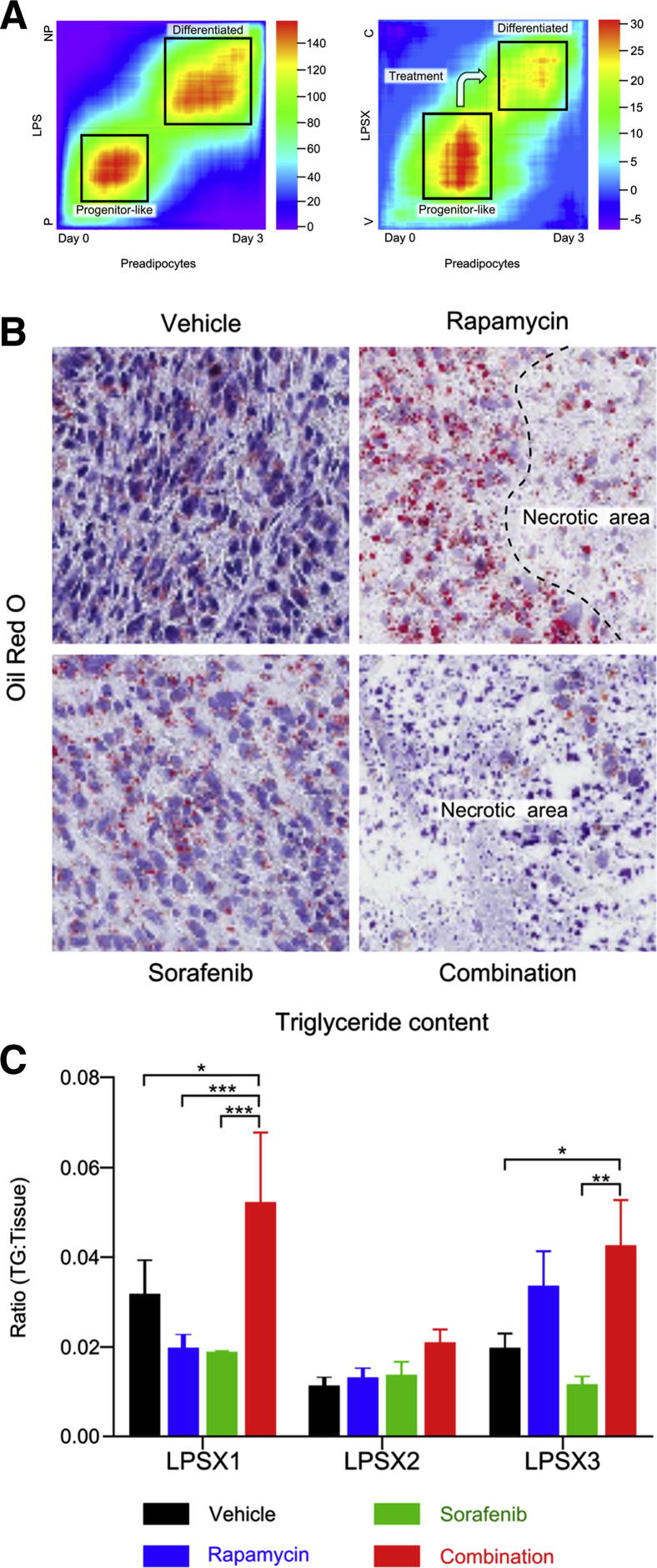
To compare the levels of differentiation of the patient samples, we first used the rank–rank hypergeometric overlap algorithm to compare the overlap of differentially expressed genes in passaged versus nonpassaged patient tumors with the differentially expressed genes of preadipocytes and differentiated adipocytes (0 days versus 3 days after differentiation; the series of Söhle et al14). The heat map generated from these comparisons (Figure 6A, LPS) showed greatest similarity in the gene expression profile between the passaged tumors and undifferentiated preadipocytes, whereas the nonpassaged tumors showed the greatest similarity to preadipocytes at day 3 after differentiation. We then performed the same analysis, comparing the gene expression of vehicle-treated and combination-treated samples with that of preadipocytes at 0 and 3 days after differentiation (the series of Söhle et al),14 and found that the combination treatment induced these DD LPS samples to initiate the differentiation process (Figure 6A, LPSX).
Figure 6.
Combination treatment induces dedifferentiated liposarcoma (DD LPS) differentiation. A: Heat maps compare the gene expression signatures of patient tumors that were passaged (P) versus nonpassaged (NP) (left) or vehicle (V) versus combination (C) treated tumors (right) to the signature of preadipocytes at day 0 versus day 3 after differentiation. The color indicates the log10 value of the hypergeometric enrichment P value between the two signature gene sets. The high-heat areas show high correlation between the genes up-regulated at day 0 (undifferentiated) preadipocytes and the passaged (P) and vehicle (V) treated tumors, whereas the nonpassaged (NP) and combination (C) tumors are most similar to preadipocytes at day 3, after differentiation. B: Oil Red O staining of LPSX3 reveals changes in lipid droplet size and intensity in response to treatments. C: Triglyceride quantification indicates increased lipid content in all three xenografts when treated with combination therapy. Data are expressed as means ± SEM. n = 4 to 8 per treatment group. *P < 0.05; **P < 0.01; and ***P < 0.001, one-way analysis of variance. Original magnification, ×400. TG, triglycerides.
To further confirm the differentiation phenotype, we analyzed the fat content of the tumors. We first looked for the presence of lipid droplets by staining with Oil Red O (Figure 6B). Rapamycin significantly increased both the number and the size of lipid droplets, especially near necrotic areas; combination-treated tumors had a similar extent of Oil Red O–positive cells (data not shown), but with massive necrosis (Figure 6B). Oil Red O staining also showed significant regional variations (data not shown). To provide more quantitative measurement, we extracted triglycerides from vehicle- and drug-treated tumor samples. Although the response to the individual drugs varied, there was a consistent increase in triglyceride levels in tumors treated with combination therapy (Figure 6C). These results suggest that the passaged tumors are less differentiated and more stem-like or progenitor-like, at least in part because of up-regulated PI3K pathway. Rapamycin and sorafenib combination treatment not only caused massive tumor necrosis, but also led to adipogenic differentiation of DD LPS tumors that have reduced PTEN levels or hyperactive PI3K/AKT/mTOR pathway.
Discussion
The xenograft system demonstrated in the present study successfully recapitulates the human disease and creates a flexible and useful model for studying the molecular mechanisms underlying LPS. This model also provides a foundation on which new targets can be developed and novel therapeutics tested.
This xenograft model system has the potential for prognostic and therapeutic value. To confirm such potential, however, a larger number of samples would need to be tested. Of the clinical variables we tested (including age, sex, location of the tumor, and history of prior treatment; Table 1), only size of tumor and presence of multifocal disease correlated with graftability. Moreover, the capability of the tumor to engraft and survive serial passaging correlated directly with poor DSS, which is similar to results from a recent study using a renal cell carcinoma xenograft model.30 Thus, the ability of a tumor sample to engraft may help identify those patients who will benefit from more aggressive treatment regimens. Additionally, because the median interval to disease recurrence is approximately 16 months for patients with high-grade LPS,31 xenografts generated from patient biopsies or resected tumors at the time of initial presentation can be molecularly profiled and interrogated with targeted therapies. This would allow physicians to define appropriate treatment paradigms and to institute personalized therapies that could be used to prevent or treat disease recurrence.
Our gene expression analysis identified alterations unique to the patient tumors that could be xenografted and serially passaged, which suggests that these tumors are more progenitor-like than the nonpassageable xenografts. One of the malignant signatures identified through the present study is up-regulated mTOR pathway activation and lower PTEN gene and protein expression, which suggests that these tumors may have increased sensitivity to the mTOR inhibitor rapamycin. Because rapamycin treatment alone often results in therapeutic resistance, we also included the multikinase inhibitor sorafenib. Histological evaluation of the tumors revealed a noticeable response in all three passageable xenografts to the combination therapy, with increased necrosis, decreased cell density, and changes in cell morphology. Further analysis of lipid content and gene expression revealed that the tumors treated with a combination of rapamycin and sorafenib became more differentiated.
Xenograft models of renal cell carcinoma generated from patient samples have been shown to reflect clinical response to targeted therapy,30 demonstrating the utility of these models for preclinical evaluation of new therapies. The data presented here show that a combination of rapamycin and sorafenib has potential as a differentiation therapy for PTEN-low or PI3K/AKT/mTOR pathway–activated DD LPS. New mTOR inhibitors that have a greater ability to inhibit mTORC1 activity,32 or dual inhibitors of mTOR and PI3K that are able to prevent the AKT feedback loop,33 may further improve the efficacy of such combination therapies. Another option is to include PPARγ ligands as part of treatment. A study using a myxoid LPS mouse model demonstrated that addition of PPARγ agonists enhanced the tumor differentiation achieved by trabectedin treatment.29 In the present study, we have identified a malignant signature for stratification of patients who may be responsive to differentiation therapy, which offers a promising avenue to explore for future targeted and differentiation therapy of DD LPS.
Acknowledgments
We thank the patients and their families for participating in our research studies, Clara Magyar for assisting with the Oil Red O analysis, the UCLA Translational Pathology Core Laboratory staff for assisting in tissue procurement and performing histology services, the UCLA Clinical Microarray Core staff for performing mRNA array analysis, and the Division of Laboratory Animal Medicine staff for overseeing the daily care of the mice.
Footnotes
Supported in part by the University of California predoctoral Tumor Biology Program and Graduate Division Dissertation Year Fellowship (K.B.S.), postdoctoral Tumor Biology Program (L.M.T.), and seed grants from the Institute for Molecular Medicine, Jonsson Comprehensive Cancer Center, at UCLA; and NIH grants T32 CA009056 (K.B.S.), T32 CA009056 (L.M.T.), and P50 CA086306 (H.W. and F.C.E.).
Current address W.D.T.: Memorial Sloan-Kettering Cancer Center, New York, New York.
Supplemental Data
Methodology tested within the study. A: Various tissue dissociation methods tested (left) did not affect the proliferation of the xenografted tumors, as indicated by Ki-67 index (right). B: The tissue dissociation method does not affect tumor histology, as shown by LPSX3 passage 9. Scale bar = 100 μm. Data are expressed as box-and-whisker plots to indicate median, interquartile range, and maximum and minimum values. n = 5 per technique. P = 0.79, Friedman test.
Serial transplantation maintains genomic and gene expression profiles of patient tumors. A: CGH analysis shows areas of chromosome 12 amplified in xenograftable tumors. B: Genes amplified in patient tumors, such as CDK4 and MDM2, are expressed at similar levels in the patient tumors and corresponding xenografted tumors (fold change compared with patient sample).
PTEN expression. A: Immunohistochemical analysis of PTEN (arrows) from patient and xenograft tumors from LPSX1 and LPSX2. B: PTEN expression is significantly different between PTEN-high and PTEN-low DD LPS tumors. Data are expressed as box-and-whisker plots to indicate median, interquartile range, and maximum and minimum values. ∗∗∗P < 0.001, one-way analysis of variance. Scale bar: 100 μm.
Doubling times of treated xenografts. Only LPSX1 showed a significant difference in the tumor doubling time when treated with rapamycin or a combination of rapamycin and sorafenib. LPSX2 and LPSX3 showed a similar trend, but the difference did not reach statistical significance. For each treatment group, the means ± SEM is indicated along with individual data points. n = 4 to 8 per treatment group. ∗∗P < 0.01, ∗∗∗P < 0.001, one-way analysis of variance.
Histologic analysis shows evidence of response to combination treatment, including necrosis and morphologic changes suggestive of differentiation. Representative images of LPSX1 and LPSX2 at low (left column) and high (center and right columns) magnification show differentiation and necrotic areas. Regions boxed in black and red correspond (although not to scale) to the high-power images of differentiation and necrosis, respectively. Scale bars: 2.5 mm (low-power images); 100 μm (high-power images).
References
- 1.Mariani O., Brennetot C., Coindre J.M., Gruel N., Ganem C., Delattre O., Stern M.H., Aurias A. JUN oncogene amplification and overexpression block adipocytic differentiation in highly aggressive sarcomas. Cancer Cell. 2007;11:361–374. doi: 10.1016/j.ccr.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 2.Matushansky I., Hernando E., Socci N.D., Matos T., Mills J., Edgar M.A., Schwartz G.K., Singer S., Cordon-Cardo C., Maki R.G. A developmental model of sarcomagenesis defines a differentiation-based classification for liposarcomas. Am J Pathol. 2008;172:1069–1080. doi: 10.2353/ajpath.2008.070284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Snyder E.L., Sandstrom D.J., Law K., Fiore C., Sicinska E., Brito J., Bailey D., Fletcher J.A., Loda M., Rodig S.J., Dal Cin P., Fletcher C.D.M. Jun amplification and overexpression are oncogenic in liposarcoma but not always sufficient to inhibit the adipocytic differentiation programme. J Pathol. 2009;218:292–300. doi: 10.1002/path.2564. [DOI] [PubMed] [Google Scholar]
- 4.Dalal K.M., Antonescu C.R., Singer S. Diagnosis and management of lipomatous tumors. J Surg Oncol. 2008;97:298–313. doi: 10.1002/jso.20975. [DOI] [PubMed] [Google Scholar]
- 5.Skubitz K.M., D’Adamo D.R. Sarcoma. Mayo Clin Proc. 2007;82:1409–1432. doi: 10.4065/82.11.1409. [DOI] [PubMed] [Google Scholar]
- 6.Cormier J.N., Pollock R.E. Soft tissue sarcomas. CA Cancer J Clin. 2004;54:94–109. doi: 10.3322/canjclin.54.2.94. [DOI] [PubMed] [Google Scholar]
- 7.Kasper B., Gil T., Awada A. Treatment of patients with advanced soft tissue sarcoma: disappointment or challenge? Curr Opin Oncol. 2007;19:336–340. doi: 10.1097/CCO.0b013e32812143ef. [DOI] [PubMed] [Google Scholar]
- 8.Eilber F.C., Eilber F.R., Eckardt J., Rosen G., Riedel E., Maki R.G., Brennan M.F., Singer S. The impact of chemotherapy on the survival of patients with high-grade primary extremity liposarcoma. Ann Surg. 2004;240:686–695. doi: 10.1097/01.sla.0000141710.74073.0d. discussion 695–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tap W.D., Eilber F.C., Ginther C., Dry S.M., Reese N., Barzan-Smith K., Chen H.W., Wu H., Eilber F.R., Slamon D.J., Anderson L. Evaluation of well-differentiated/de-differentiated liposarcomas by high-resolution oligonucleotide array-based comparative genomic hybridization. Genes Chromosomes Cancer. 2011;50:95–112. doi: 10.1002/gcc.20835. [DOI] [PubMed] [Google Scholar]
- 10.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Harris M.A., Hill D.P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J.C., Richardson J.E., Ringwald M., Rubin G.M., Sherlock G., The Gene Ontology Consortium Gene ontology: tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eisen M.B., Spellman P.T., Brown P.O., Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Plaisier S.B., Taschereau R., Wong J.A., Graeber T.G. Rank-rank hypergeometric overlap: identification of statistically significant overlap between gene-expression signatures. Nucleic Acids Res. 2010;38:e169. doi: 10.1093/nar/gkq636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Söhle J., Machuy N., Smailbegovic E., Holtzmann U., Grönniger E., Wenck H., Stäb F., Winnefeld M. Identification of new genes involved in human adipogenesis and fat storage. PLoS One. 2012;7:e31193. doi: 10.1371/journal.pone.0031193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chopra S., Li Y., Gui D., Eilber F.C., Dry S.M. Primary retroperitoneal liposarcomas: mitoses in non-adipogenic zones correlate with survival. Mod Pathol. 2011;24(Suppl 1):11A. (abstract) [Google Scholar]
- 16.Evans H.L. Atypical lipomatous tumor, its variants, and its combined forms: a study of 61 cases, with a minimum follow-up of 10 years. Am J Surg Pathol. 2007;31:1–14. doi: 10.1097/01.pas.0000213406.95440.7a. [DOI] [PubMed] [Google Scholar]
- 17.Taylor B.S., Barretina J., Maki R.G., Antonescu C.R., Singer S., Ladanyi M. Advances in sarcoma genomics and new therapeutic targets. Nat Rev Cancer. 2011;11:541–557. doi: 10.1038/nrc3087. [Erratum appeared in Nat Rev Cancer 2011, 11:685] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singer S., Socci N.D., Ambrosini G., Sambol E., Decarolis P., Wu Y., O’Connor R., Maki R., Viale A., Sander C., Schwartz G.K., Antonescu C.R. Gene expression profiling of liposarcoma identifies distinct biological types/subtypes and potential therapeutic targets in well-differentiated and dedifferentiated liposarcoma. Cancer Res. 2007;67:6626–6636. doi: 10.1158/0008-5472.CAN-07-0584. [DOI] [PubMed] [Google Scholar]
- 19.Pedeutour F., Forus A., Coindre J.M., Berner J.M., Nicolo G., Michiels J.F., Terrier P., Ranchere-Vince D., Collin F., Myklebost O., Turc-Carel C. Structure of the supernumerary ring and giant rod chromosomes in adipose tissue tumors. Genes Chromosomes Cancer. 1999;24:30–41. [PubMed] [Google Scholar]
- 20.Willén H., Akerman M., Dal Cin P., De Wever I., Fletcher C.D., Mandahl N., Mertens F., Mitelman F., Rosai J., Rydholm A., Sciot R., Tallini G., Van den Berghe H., Vanni R. Comparison of chromosomal patterns with clinical features in 165 lipomas: a report of the CHAMP Study Group. Cancer Genet Cytogenet. 1998;102:46–49. doi: 10.1016/s0165-4608(97)00292-6. [DOI] [PubMed] [Google Scholar]
- 21.Fletcher C.D., Akerman M., Dal Cin P., de Wever I., Mandahl N., Mertens F., Mitelman F., Rosai J., Rydholm A., Sciot R., Tallini G., van den Berghe H., van de Ven W., Vanni R., Willén H. Correlation between clinicopathological features and karyotype in lipomatous tumors. A report of 178 cases from the Chromosomes and Morphology (CHAMP) Collaborative Study Group. Am J Pathol. 1996;148:623–630. [PMC free article] [PubMed] [Google Scholar]
- 22.Chandarlapaty S., Sawai A., Scaltriti M., Rodrik-Outmezguine V., Grbovic-Huezo O., Serra V., Majumder P.K., Baselga J., Rosen N. AKT inhibition relieves feedback suppression of receptor tyrosine kinase expression and activity. Cancer Cell. 2011;19:58–71. doi: 10.1016/j.ccr.2010.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodrik-Outmezguine V.S., Chandarlapaty S., Pagano N.C., Poulikakos P.I., Scaltriti M., Moskatel E., Baselga J., Guichard S., Rosen N. mTOR kinase inhibition causes feedback-dependent biphasic regulation of AKT signaling. Cancer Discov. 2011;1:248–259. doi: 10.1158/2159-8290.CD-11-0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun S.Y., Rosenberg L.M., Wang X., Zhou Z., Yue P., Fu H., Khuri F.R. Activation of Akt and eIF4E survival pathways by rapamycin-mediated mammalian target of rapamycin inhibition. Cancer Res. 2005;65:7052–7058. doi: 10.1158/0008-5472.CAN-05-0917. [DOI] [PubMed] [Google Scholar]
- 25.von Mehren M., Rankin C., Goldblum J.R., Demetri G.D., Bramwell V., Ryan C.W., Borden E. Phase 2 Southwest Oncology Group-directed intergroup trial (S0505) of sorafenib in advanced soft tissue sarcomas. Cancer. 2012;118:770–776. doi: 10.1002/cncr.26334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eisenhauer E.A., Therasse P., Bogaerts J., Schwartz L.H., Sargent D., Ford R., Dancey J., Arbuck S., Gwyther S., Mooney M., Rubinstein L., Shankar L., Dodd L., Kaplan R., Lacombe D., Verweij J. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 27.Houben R., Vetter-Kauczok C.S., Ortmann S., Rapp U.R., Broecker E.B., Becker J.C. Phospho-ERK staining is a poor indicator of the mutational status of BRAF and NRAS in human melanoma. J Invest Dermatol. 2008;128:2003–2012. doi: 10.1038/jid.2008.30. [DOI] [PubMed] [Google Scholar]
- 28.Tontonoz P., Singer S., Forman B.M., Sarraf P., Fletcher J.A., Fletcher C.D.M., Brun R.P., Mueller E., Altiok S., Oppenheim H., Evans R.M., Spiegelman B.M. Terminal differentiation of human liposarcoma cells induced by ligands for peroxisome proliferator-activated receptor gamma and the retinoid X receptor. Proc Natl Acad Sci USA. 1997;94:237–241. doi: 10.1073/pnas.94.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Charytonowicz E., Terry M., Coakley K., Telis L., Remotti F., Cordon-Cardo C., Taub R.N., Matushansky I. PPARgamma agonists enhance ET-743-induced adipogenic differentiation in a transgenic mouse model of myxoid round cell liposarcoma. J Clin Invest. 2012;122:886–898. doi: 10.1172/JCI60015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sivanand S., Peña-Llopis S., Zhao H., Kucejova B., Spence P., Pavia-Jimenez A., Yamasaki T., McBride D.J., Gillen J., Wolff N.C., Morlock L., Lotan Y., Raj G.V., Sagalowsky A., Margulis V., Cadeddu J.A., Ross M.T., Bentley D.R., Kabbani W., Xie X.J., Kapur P., Williams N.S., Brugarolas J. A validated tumorgraft model reveals activity of dovitinib against renal cell carcinoma. Sci Transl Med. 2012;4 doi: 10.1126/scitranslmed.3003643. 137ra175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eilber F.C., Brennan M.F., Riedel E., Alektiar K.M., Antonescu C.R., Singer S. Prognostic factors for survival in patients with locally recurrent extremity soft tissue sarcomas. Ann Surg Oncol. 2005;12:228–236. doi: 10.1245/ASO.2005.03.045. [DOI] [PubMed] [Google Scholar]
- 32.Peterson T.R., Sengupta S.S., Harris T.E., Carmack A.E., Kang S.A., Balderas E., Guertin D.A., Madden K.L., Carpenter A.E., Finck B.N., Sabatini D.M. mTOR Complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell. 2011;146:408–420. doi: 10.1016/j.cell.2011.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mazzoletti M., Bortolin F., Brunelli L., Pastorelli R., Di Giandomenico S., Erba E., Ubezio P., Broggini M. Combination of PI3K/mTOR Inhibitors: antitumor activity and molecular correlates. Cancer Res. 2011;71:4573–4584. doi: 10.1158/0008-5472.CAN-10-4322. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Methodology tested within the study. A: Various tissue dissociation methods tested (left) did not affect the proliferation of the xenografted tumors, as indicated by Ki-67 index (right). B: The tissue dissociation method does not affect tumor histology, as shown by LPSX3 passage 9. Scale bar = 100 μm. Data are expressed as box-and-whisker plots to indicate median, interquartile range, and maximum and minimum values. n = 5 per technique. P = 0.79, Friedman test.
Serial transplantation maintains genomic and gene expression profiles of patient tumors. A: CGH analysis shows areas of chromosome 12 amplified in xenograftable tumors. B: Genes amplified in patient tumors, such as CDK4 and MDM2, are expressed at similar levels in the patient tumors and corresponding xenografted tumors (fold change compared with patient sample).
PTEN expression. A: Immunohistochemical analysis of PTEN (arrows) from patient and xenograft tumors from LPSX1 and LPSX2. B: PTEN expression is significantly different between PTEN-high and PTEN-low DD LPS tumors. Data are expressed as box-and-whisker plots to indicate median, interquartile range, and maximum and minimum values. ∗∗∗P < 0.001, one-way analysis of variance. Scale bar: 100 μm.
Doubling times of treated xenografts. Only LPSX1 showed a significant difference in the tumor doubling time when treated with rapamycin or a combination of rapamycin and sorafenib. LPSX2 and LPSX3 showed a similar trend, but the difference did not reach statistical significance. For each treatment group, the means ± SEM is indicated along with individual data points. n = 4 to 8 per treatment group. ∗∗P < 0.01, ∗∗∗P < 0.001, one-way analysis of variance.
Histologic analysis shows evidence of response to combination treatment, including necrosis and morphologic changes suggestive of differentiation. Representative images of LPSX1 and LPSX2 at low (left column) and high (center and right columns) magnification show differentiation and necrotic areas. Regions boxed in black and red correspond (although not to scale) to the high-power images of differentiation and necrosis, respectively. Scale bars: 2.5 mm (low-power images); 100 μm (high-power images).